Abstract
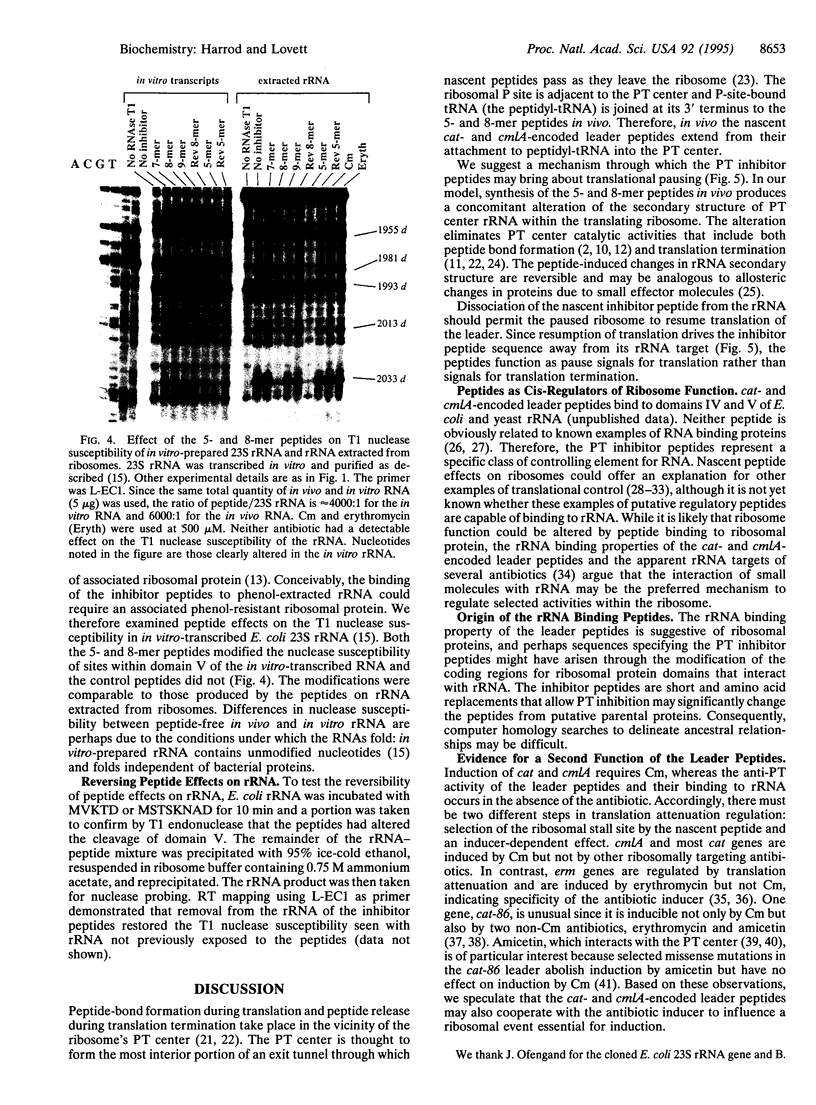
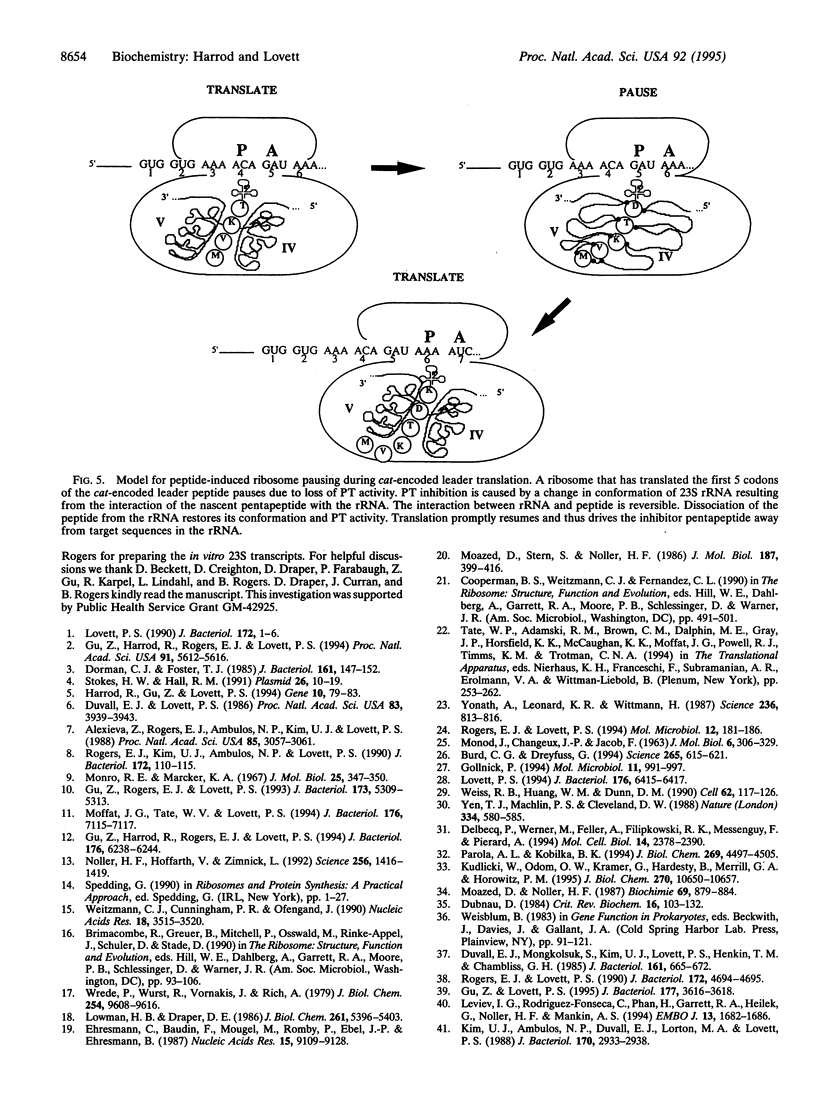
Peptides of 5 and 8 residues encoded by the leaders of attenuation regulated chloramphenicol-resistance genes inhibit the peptidyltransferase of microorganisms from the three kingdoms. Therefore, the ribosomal target for the peptides is likely to be a conserved structure and/or sequence. The inhibitor peptides "footprint" to nucleotides of domain V in large subunit rRNA when peptide-ribosome complexes are probed with dimethyl sulfate. Accordingly, rRNA was examined as a candidate for the site of peptide binding. Inhibitor peptides MVKTD and MSTSKNAD were mixed with rRNA phenol-extracted from Escherichia coli ribosomes. The conformation of the RNA was then probed by limited digestion with nucleases that cleave at single-stranded (T1 endonuclease) and double-stranded (V1 endonuclease) sites. Both peptides selectively altered the susceptibility of domains IV and V of 23S rRNA to digestion by T1 endonuclease. Peptide effects on cleavage by V1 nuclease were observed only in domain V. The T1 nuclease susceptibility of domain V of in vitro-transcribed 23S rRNA was also altered by the peptides, demonstrating that peptide binding to the rRNA is independent of ribosomal protein. We propose the peptides MVKTD and MSTSKNAD perturb peptidyltransferase center catalytic activities by altering the conformation of domains IV and V of 23S rRNA. These findings provide a general mechanism through which nascent peptides may cis-regulate the catalytic activities of translating ribosomes.
Full text
PDF
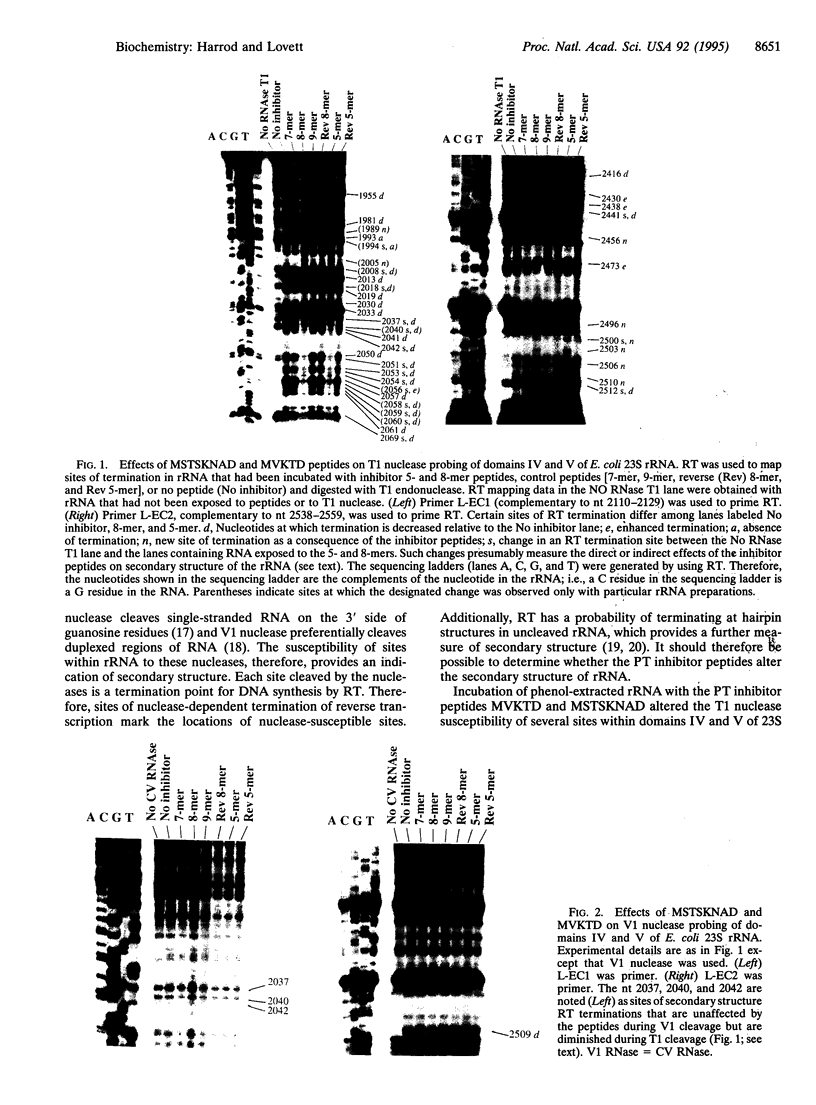
Images in this article
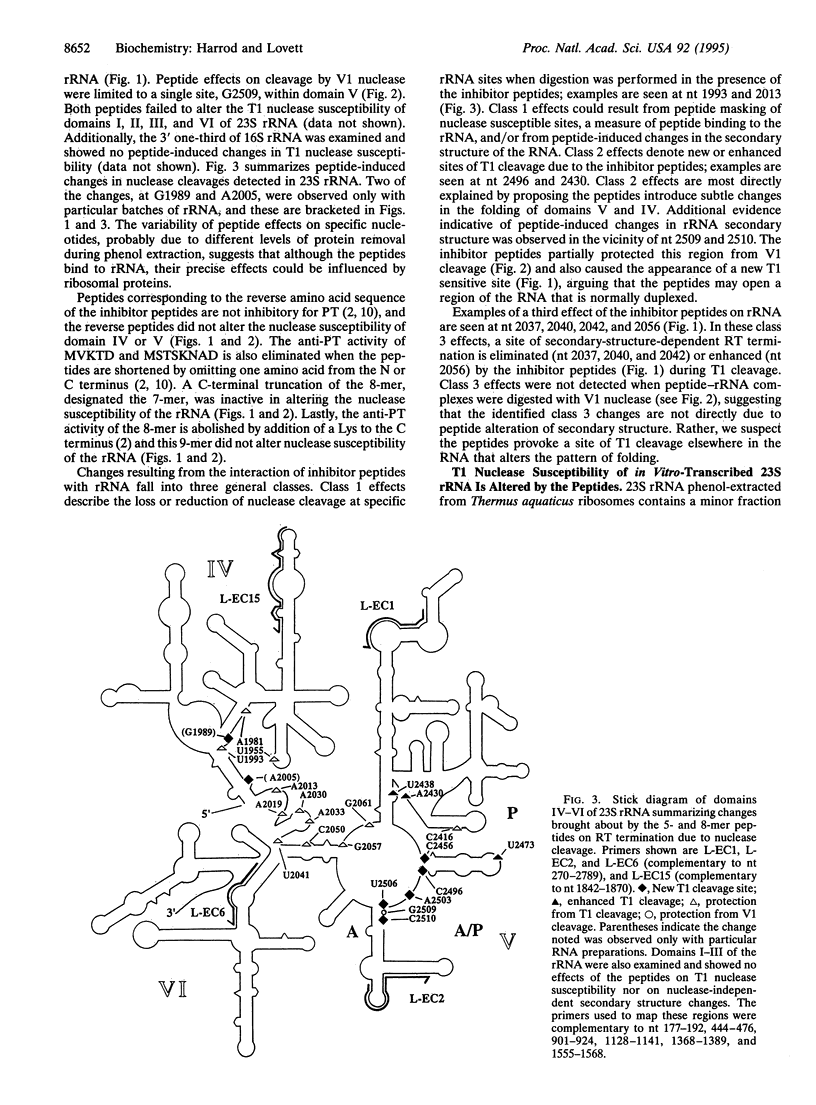
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alexieva Z., Duvall E. J., Ambulos N. P., Jr, Kim U. J., Lovett P. S. Chloramphenicol induction of cat-86 requires ribosome stalling at a specific site in the leader. Proc Natl Acad Sci U S A. 1988 May;85(9):3057–3061. doi: 10.1073/pnas.85.9.3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd C. G., Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994 Jul 29;265(5172):615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- Delbecq P., Werner M., Feller A., Filipkowski R. K., Messenguy F., Piérard A. A segment of mRNA encoding the leader peptide of the CPA1 gene confers repression by arginine on a heterologous yeast gene transcript. Mol Cell Biol. 1994 Apr;14(4):2378–2390. doi: 10.1128/mcb.14.4.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorman C. J., Foster T. J. Posttranscriptional regulation of the inducible nonenzymatic chloramphenicol resistance determinant of IncP plasmid R26. J Bacteriol. 1985 Jan;161(1):147–152. doi: 10.1128/jb.161.1.147-152.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau D. Translational attenuation: the regulation of bacterial resistance to the macrolide-lincosamide-streptogramin B antibiotics. CRC Crit Rev Biochem. 1984;16(2):103–132. doi: 10.3109/10409238409102300. [DOI] [PubMed] [Google Scholar]
- Duvall E. J., Lovett P. S. Chloramphenicol induces translation of the mRNA for a chloramphenicol-resistance gene in Bacillus subtilis. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3939–3943. doi: 10.1073/pnas.83.11.3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvall E. J., Mongkolsuk S., Kim U. J., Lovett P. S., Henkin T. M., Chambliss G. H. Induction of the chloramphenicol acetyltransferase gene cat-86 through the action of the ribosomal antibiotic amicetin: involvement of a Bacillus subtilis ribosomal component in cat induction. J Bacteriol. 1985 Feb;161(2):665–672. doi: 10.1128/jb.161.2.665-672.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehresmann C., Baudin F., Mougel M., Romby P., Ebel J. P., Ehresmann B. Probing the structure of RNAs in solution. Nucleic Acids Res. 1987 Nov 25;15(22):9109–9128. doi: 10.1093/nar/15.22.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollnick P. Regulation of the Bacillus subtilis trp operon by an RNA-binding protein. Mol Microbiol. 1994 Mar;11(6):991–997. doi: 10.1111/j.1365-2958.1994.tb00377.x. [DOI] [PubMed] [Google Scholar]
- Gu Z., Harrod R., Rogers E. J., Lovett P. S. Anti-peptidyl transferase leader peptides of attenuation-regulated chloramphenicol-resistance genes. Proc Natl Acad Sci U S A. 1994 Jun 7;91(12):5612–5616. doi: 10.1073/pnas.91.12.5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z., Harrod R., Rogers E. J., Lovett P. S. Properties of a pentapeptide inhibitor of peptidyltransferase that is essential for cat gene regulation by translation attenuation. J Bacteriol. 1994 Oct;176(20):6238–6244. doi: 10.1128/jb.176.20.6238-6244.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z., Lovett P. S. A gratuitous inducer of cat-86, amicetin, inhibits bacterial peptidyl transferase. J Bacteriol. 1995 Jun;177(12):3616–3618. doi: 10.1128/jb.177.12.3616-3618.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z., Rogers E. J., Lovett P. S. Peptidyl transferase inhibition by the nascent leader peptide of an inducible cat gene. J Bacteriol. 1993 Sep;175(17):5309–5313. doi: 10.1128/jb.175.17.5309-5313.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrod R., Gu Z., Lovett P. S. Analysis of the secondary structure that negatively regulates inducible cat translation by use of chemical probing and mutagenesis. Gene. 1994 Mar 11;140(1):79–83. doi: 10.1016/0378-1119(94)90734-x. [DOI] [PubMed] [Google Scholar]
- Kim U. J., Ambulos N. P., Jr, Duvall E. J., Lorton M. A., Lovett P. S. Site in the cat-86 regulatory leader that permits amicetin to induce expression of the gene. J Bacteriol. 1988 Jul;170(7):2933–2938. doi: 10.1128/jb.170.7.2933-2938.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudlicki W., Odom O. W., Kramer G., Hardesty B., Merrill G. A., Horowitz P. M. The importance of the N-terminal segment for DnaJ-mediated folding of rhodanese while bound to ribosomes as peptidyl-tRNA. J Biol Chem. 1995 May 5;270(18):10650–10657. doi: 10.1074/jbc.270.18.10650. [DOI] [PubMed] [Google Scholar]
- Leviev I. G., Rodriguez-Fonseca C., Phan H., Garrett R. A., Heilek G., Noller H. F., Mankin A. S. A conserved secondary structural motif in 23S rRNA defines the site of interaction of amicetin, a universal inhibitor of peptide bond formation. EMBO J. 1994 Apr 1;13(7):1682–1686. doi: 10.1002/j.1460-2075.1994.tb06432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett P. S. Nascent peptide regulation of translation. J Bacteriol. 1994 Nov;176(21):6415–6417. doi: 10.1128/jb.176.21.6415-6417.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett P. S. Translational attenuation as the regulator of inducible cat genes. J Bacteriol. 1990 Jan;172(1):1–6. doi: 10.1128/jb.172.1.1-6.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowman H. B., Draper D. E. On the recognition of helical RNA by cobra venom V1 nuclease. J Biol Chem. 1986 Apr 25;261(12):5396–5403. [PubMed] [Google Scholar]
- MONOD J., CHANGEUX J. P., JACOB F. Allosteric proteins and cellular control systems. J Mol Biol. 1963 Apr;6:306–329. doi: 10.1016/s0022-2836(63)80091-1. [DOI] [PubMed] [Google Scholar]
- Moazed D., Noller H. F. Chloramphenicol, erythromycin, carbomycin and vernamycin B protect overlapping sites in the peptidyl transferase region of 23S ribosomal RNA. Biochimie. 1987 Aug;69(8):879–884. doi: 10.1016/0300-9084(87)90215-x. [DOI] [PubMed] [Google Scholar]
- Moazed D., Stern S., Noller H. F. Rapid chemical probing of conformation in 16 S ribosomal RNA and 30 S ribosomal subunits using primer extension. J Mol Biol. 1986 Feb 5;187(3):399–416. doi: 10.1016/0022-2836(86)90441-9. [DOI] [PubMed] [Google Scholar]
- Moffat J. G., Tate W. P., Lovett P. S. The leader peptides of attenuation-regulated chloramphenicol resistance genes inhibit translational termination. J Bacteriol. 1994 Nov;176(22):7115–7117. doi: 10.1128/jb.176.22.7115-7117.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monro R. E., Marcker K. A. Ribosome-catalysed reaction of puromycin with a formylmethionine-containing oligonucleotide. J Mol Biol. 1967 Apr 28;25(2):347–350. doi: 10.1016/0022-2836(67)90146-5. [DOI] [PubMed] [Google Scholar]
- Noller H. F., Hoffarth V., Zimniak L. Unusual resistance of peptidyl transferase to protein extraction procedures. Science. 1992 Jun 5;256(5062):1416–1419. doi: 10.1126/science.1604315. [DOI] [PubMed] [Google Scholar]
- Parola A. L., Kobilka B. K. The peptide product of a 5' leader cistron in the beta 2 adrenergic receptor mRNA inhibits receptor synthesis. J Biol Chem. 1994 Feb 11;269(6):4497–4505. [PubMed] [Google Scholar]
- Rogers E. J., Kim U. J., Ambulos N. P., Jr, Lovett P. S. Four codons in the cat-86 leader define a chloramphenicol-sensitive ribosome stall sequence. J Bacteriol. 1990 Jan;172(1):110–115. doi: 10.1128/jb.172.1.110-115.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers E. J., Lovett P. S. Erythromycin induces expression of the chloramphenicol acetyltransferase gene cat-86. J Bacteriol. 1990 Aug;172(8):4694–4695. doi: 10.1128/jb.172.8.4694-4695.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers E. J., Lovett P. S. The cis-effect of a nascent peptide on its translating ribosome: influence of the cat-86 leader pentapeptide on translation termination at leader codon 6. Mol Microbiol. 1994 Apr;12(2):181–186. doi: 10.1111/j.1365-2958.1994.tb01007.x. [DOI] [PubMed] [Google Scholar]
- Stokes H. W., Hall R. M. Sequence analysis of the inducible chloramphenicol resistance determinant in the Tn1696 integron suggests regulation by translational attenuation. Plasmid. 1991 Jul;26(1):10–19. doi: 10.1016/0147-619x(91)90032-r. [DOI] [PubMed] [Google Scholar]
- Weiss R. B., Huang W. M., Dunn D. M. A nascent peptide is required for ribosomal bypass of the coding gap in bacteriophage T4 gene 60. Cell. 1990 Jul 13;62(1):117–126. doi: 10.1016/0092-8674(90)90245-A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitzmann C. J., Cunningham P. R., Ofengand J. Cloning, in vitro transcription, and biological activity of Escherichia coli 23S ribosomal RNA. Nucleic Acids Res. 1990 Jun 25;18(12):3515–3520. doi: 10.1093/nar/18.12.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrede P., Wurst R., Vournakis J., Rich A. Conformational changes of yeast tRNAPhe and E. coli tRNA2Glu as indicated by different nuclease digestion patterns. J Biol Chem. 1979 Oct 10;254(19):9608–9616. [PubMed] [Google Scholar]
- Yen T. J., Machlin P. S., Cleveland D. W. Autoregulated instability of beta-tubulin mRNAs by recognition of the nascent amino terminus of beta-tubulin. Nature. 1988 Aug 18;334(6183):580–585. doi: 10.1038/334580a0. [DOI] [PubMed] [Google Scholar]
- Yonath A., Leonard K. R., Wittmann H. G. A tunnel in the large ribosomal subunit revealed by three-dimensional image reconstruction. Science. 1987 May 15;236(4803):813–816. doi: 10.1126/science.3576200. [DOI] [PubMed] [Google Scholar]