Abstract
Background
Alpha-hemolysin (Hla) is a major virulence factor in the pathogenesis of Staphylococcus aureus infection, being active against a wide range of host cells. Although hla is ubiquitous in S. aureus, its genetic diversity and variation in expression in different genetic backgrounds is not known. We evaluated nucleotide sequence variation and gene expression profiles of hla among representatives of hospital (HA) and community-associated (CA) S. aureus clones.
Methods
51 methicillin-resistant S. aureus and 22 methicillin-susceptible S. aureus were characterized by PFGE, spa typing, MLST and SCCmec typing. The internal regions of hla and the hla promoter were sequenced and gene expression was assessed by RT-PCR.
Results
Alpha-hemolysin encoding- and promoter sequences were diverse, with 12 and 23 different alleles, respectively. Based on phylogenetic analysis, we suggest that hla may have evolved together with the S. aureus genetic background, except for ST22, ST121, ST59 and ST93. Conversely, the promoter region showed lack of co-evolution with the genetic backgrounds. Four non-synonymous amino acid changes were identified close to important regions of hla activity. Amino acid changes in the RNAIII binding site were not associated to hla expression. Although expression rates of hla were in general strain-specific, we observed CA clones showed significantly higher hla expression (p = 0.003) when compared with HA clones.
Conclusion
We propose that the hla gene has evolved together with the genetic background. Overall, CA genetic backgrounds showed higher levels of hla expression than HA, and a high strain-to-strain variation of gene expression was detected in closely related strains.
Introduction
Staphylococcus aureus is a human opportunistic pathogen responsible for a wide range of infections that can vary in its clinical presentation and severity. Methicillin-resistant S. aureus (MRSA) emerged in 1960 in the United Kingdom and has been a major problem in hospitals (HA-MRSA) worldwide during the last 40 years; however since the late 1990s, MRSA has been emerging as a leading cause of severe infection also in the community, in individuals without recent health-care contact (CA-MRSA) [1], [2].
CA-MRSA present distinct genetic backgrounds from their hospital counterparts, are more susceptible to antibiotics other than beta-lactams, carry the smallest staphylococcal cassette chromosome mec types (SCCmec IV or V), and have higher virulence capacity [1], [2], [3]. The underlying reasons behind the enhanced virulence of CA-MRSA appear to be multiple including a different capacity to overcome host cell response [4], different distribution of mobile genetic elements carrying virulence determinants [5], allelic variation in virulence determinants located in the core genome and in mobile genetic elements [6], and different levels of expression and protein production of virulence determinants (alpha-hemolysin, collagen adhesin, staphylokinase, coagulase, lipase, enterotoxins C3 and Q, V8 protease and cysteine protease) [7], [8], [9].
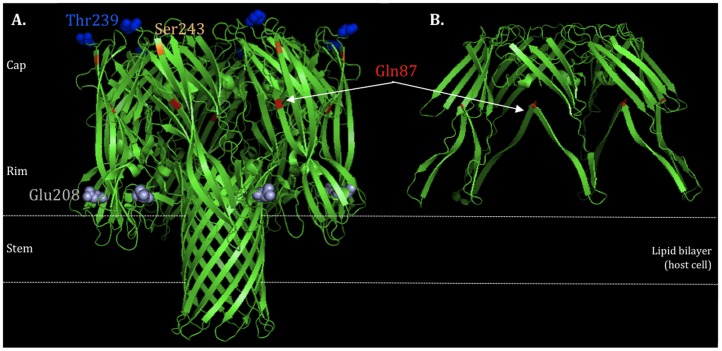
The alpha-hemolysin or α-toxin (Hla), is one of the major virulence determinants implicated in the pathogenesis of S. aureus, associated to severe skin and soft tissue infections (SSTI), necrotizing pneumonia and even sepsis [10]. Hla is the most prominent S. aureus cytotoxin that can act against a wide range of host cells including erythrocytes, epithelial cells, endothelial cells, T cells, monocytes and macrophages [10], [11], [12]. The gene encoding Hla is located in the core genome and is expressed as a water-soluble monomer (33.2 kD) that assembles to form a membrane-bound heptameric β-barrel pore (232.4 kD) on susceptible cells leading to cell death and lysis [11]. The overall structure is mushroom-like, divided into three domains: 1) Cap domain: largely hydrophobic, defining the entry of the pore; 2) Rim domain: underside of the Cap, in close proximity to membrane bilayer; 3) Stem domain: part of the transmembrane channel, forming the membrane-perforating β-barrel pore (Figure 1) [10], [11]. Hla expression is mainly controlled by the global toxin accessory gene regulator (agr), via the regulatory effector molecule RNAIII [13]. While agr provides the first and most important mechanism of up-regulation of hla, expression can also be modulated by other regulators, such as SaeR, SarZ, ArlS [14], [15], [16] (up-regulators) and Rot, SarT [17](down-regulators).
Figure 1. HLA protein structure.
A) wildtype (highlighted the non-synonymous mutations Gln87, Glu208, Thr239 and Ser243) and B) truncated protein due to a stop codon at Gln87. Structure generated by the program PyMOL v.1.6.
Although polymorphisms in the hla promoter region have been described [18], the range of genetic diversity and evolution of this toxin has never been assessed in a large representative S. aureus collection. Furthermore, although differences in hla expression have been described between community- and hospital-associated MRSA, these studies have been performed with a limited number of CA-MRSA epidemic clones [9], or almost exclusively with representatives of the USA300 clone [19], [20], [21]. To better understand the evolutionary history of hla and its importance as a virulence factor for CA-MRSA, in this study we compared the hla nucleotide sequence and expression among the major epidemic and minor CA and HA clones, including both MRSA and MSSA strains.
Materials and Methods
Ethics Statement
Isolates were obtained from routine diagnostic and were analyzed anonymously and only the isolates, not humans, were studied. All data was collected according to the European Parliament and Council decision for the epidemiological surveillance and control of communicable disease in the European community. Ethical approval and informed consent were for that reason not required.
Bacterial collection
A total of 73 S. aureus, including 51 MRSA and 22 MSSA were analyzed in this study. Strains were collected in 13 different countries (Belgium, Bulgaria, Czech Republic, Denmark, Greece, Netherlands, Portugal, Romania, Spain, Sweden, United Kingdom, USA and Brazil), between 1961 and 2009 from both community (n = 46) and hospital (n = 27). The strains comprised a total of 52 spa types and 23 sequence types (STs) (see Table S1).
Strains were defined as belonging to CA or HA clones if they contained the same or related genetic backgrounds as the reference CA-MRSA and HA-MRSA epidemic control strains, based on ST, spa type and SCCmec (in case of MRSA).
Media and bacterial growth conditions
Before RT-PCR analysis, strains were grown overnight at 37°C on tryptic soy agar plates (TSA). Bacterial growth experiments were performed by growing bacteria in tryptic Soy Broth (TSB) at 37°C with shake and measuring OD (600 nm) each hour in the follow up automatic incubator Bioscreen C (Oy Growth Curves AB, Helsinki, Finland). Plates of 100-well honeycomb (Oy Growth Curves AB, Helsinki, Finland) were filled with 300 µl/well of overnight culture diluted to OD600 = 0.05 in TSB growth medium. Three individual growth experiments (SetC, SetD and SetE) were performed for each strain and named accordingly e.g. HLZ6C, HLZ6D and HLZ6E (see Figure S2.I to III).
Nucleotide sequence of hla and promoter region
Chromosomal DNA was extracted from overnight cultures, using the boiling method (100°C for 10 min followed by centrifuged at 13.000 g for 5 minutes). Two sets of primers were designed to span the most polymorphic regions within the hla gene and hla promoter (considered as the region located −600 bp from hla starting codon), after alignment of sequences available on NCBI for S. aureus. One set of primers (Forward: hla-F_CGAAAGGTACCATTGCTGGT; Reverse: hla-R_CCAATCGATTTTATATCTTTC) amplified an internal fragment of the hla gene (nt 1170419–1170982, CP000730.1) and the other set (Forward: hlaPro-F_CACTATATTAAAAATACATAC; Reverse: hlaPro-R_GTTGTTACTGAGCTGAC) amplified an internal fragment of the hla promoter region (nt 1171289–1171773, CP000730.1) (Figure S3). PCR products were sequenced (Macrogen Europe, Amsterdam, The Netherlands) and sequences were analyzed using SeqMan (DNAstar, Lasergene v9, Madison, WI, USA). To each unique hla promoter (P) and gene sequence (hla) - allotype - a single Arabic number was attributed (e.g. P1, P2; hla1, hla2). Gene and promoter sequences were deposited in GenBank (accession numbers KM019547–KM019606; KM019607–KM019674).
Phylogenetic analysis
Phylogenetic relatedness was analyzed using the MEGA5 v5.05 software (http://www.megasoftware.net/) for gene, promoter region and concatenated sequences obtained from 1) gene with promoter region and 2) seven MLST alleles from the 23 representative STs within the collection. Phylogenetic trees were constructed using the Neighbor-Joining clustering method, and 1000 bootstrap replicates, which assigns confidence values for the groupings in the tree.
Moreover, nucleotide diversity (ND) between the two clusters was calculated based on the estimation of the average evolutionary divergence over sequence pairs within the two groups, where the number of base substitutions per site from averaging over all sequence pairs within each group are compared using the maximum composite likelihood model [22].
Detection of recombination
Alignments from the hla gene, hla promoter and internal fragments of each of the seven MLST gene were screened for the occurrence of putative recombination events using Recombination Detection Program version 4 (RDP4) (http://web.cbio.uct.ac.za/) with the default settings (with highest acceptable probability value of 0.05). Identification of recombinant sequences recombination breakpoints and major parent was determined using simultaneously nine recombination detection methods (RDP, BOOTSCAN, GENECONV, MAXCHI, CHIMAERA, SISCAN, PhylPro, LARD and 3SEQ. The “minor parent” is considered a sequence closely related to that from which sequences in the proposed recombinant region may have been derived (the presumed donor). The “major parent” was considered as a sequence closely related to that from which the greater part of the recombinant’s sequence may have been derived.
RT-PCR analysis
Culture growth was stopped at late exponential phase, when alpha-toxin is described to have maximal activity [23], corresponding to the time-points 1) 3 hours 30 min in one group (65 strains) and 2) 4 hours 30 min in another (8 strains). Total RNA was extracted from three biological replicates. Cells were mechanically disrupted with FastPrep-24 Instrument (MP Biomedicals, Solon, OH, USA) and RNA was protected using RNA Protect (Qiagen, Valencia, USA). RNA was extracted automatically using the QIAsymphony platforms (Qiagen, Valencia, USA) with QIAsymphony RNA kit (Qiagen, Valencia, USA).
The RT-PCR assay was performed on a 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA) using the following primers and TaqMan probes: Hla RT_F: TAATGAATCCTGTCGCTAATGCC; HlaRT_R: CACCTGTTTTTACTGTAGTATTGCTTCC; Hla RT Probe: 6FAM-AAACCGGTACTACAGATAT-MGBNFQ. The RT-PCR reaction was performed using the EZ RT-PCR Core Reagents (Applied Biosystems, Foster City, USA), in which RNA is reverse transcribed and amplified in a single reaction. The following PCR protocol was used: 50°C for 2 min, 60°C for 30 min, 95°C for 5 min, followed by 42 cycles of 95°C for 20 sec and 62°C for 1 min. The 16S gene was used as internal or reference control. The primers used for 16S RNA amplification were those previously described [24].
RT-PCR data analysis
The relative hla gene expression was calculated based on the Ct (RT-PCR output) of the gene of interest (Ct hla) as compared to the Ct of the internal control (Ct 16S) as follows: Delta Ct = Ct hla- Ct 16S. The lower the Delta Ct the higher is the amount of hla mRNA and the more the gene is expressed. The reproducibility of the assay was evaluated by the calculation of the arithmetic mean of the relative expression of the three biological replicates (Mean Delta Ct1–3 = Average (Delta Ct1; Delta Ct2; Delta Ct3). The reproducibility of RT-PCR reaction was evaluated by the calculation of the standard deviation (STDEV) of Delta Ct obtained for each biological replica (Delta Ct1; Delta Ct2; Delta Ct3). Values were considered valid when at least two Ct readings exist with STDEV<2.
Protein structure visualization (pyMOL)
The protein structure was modeled using PyMOL v.1.6 (http://www.pymol.org/) if a nucleotide mutation gave rise to a stop codon.
Statistical analysis
The statistical analysis was performed using the Graphpad Prism 6 (http://www.graphpad.com/scientific-software/prism/), with the two-tailed Student’s t-test to determine whether the differences of mean expression rates (MSSA versus MSSA; HA backgrounds versus CA backgrounds) were statistically significant (p≤0.05).
Regression tree analysis was used to explore which variables could be related with the hla expression [25]. Trees explain the variation of a single response variable (in this study the hla mRNA expression) by repeatedly splitting the data into more homogeneous groups, using combinations of explanatory variables (in our case, the ST, spa type, MRSA, MSSA and the type of SCCmec).
Results
Analysis of polymorphisms in the hla gene and hla promoter
The sequence analysis of the internal region of hla and the hla promoter region among the 73 strains identified a total of 12 hla and 23 promoter region different sequences (allotypes) (Table 1). We obtained no amplification products for hla and hla promoter region in one and 13 strains, respectively, which probably result from misparing of the primers used.
Table 1. Summary of molecular characterization, sequence variation and relative expression rates of S. aureus strains collection.

|
Isolate ID | SCCmec | spa type | MLST | Branch1 | Promotor Allotype | Gene Allotype | Nonsynonymous Mutation | Hla Expression (Mean Delta Ct)2 | Stddev Delta Ct 3 | Expression (High/Low) |
| 1 | HLZ6 | II | t002 | ST5 | L | P4 | hla1 | D208E | 8.69 | 2* | Low |
| 2 | BK2464 | II | t002 | ST5 | L | nt | hla1 | D208E | 5.37 | 1 | High |
| 3 | HBR73 | II | t067 | ST5 | L | P5 | hla1 | D208E | 8.75 | 1 | Low |
| 4 | C013 | VI | t002 | ST5 | L | P3 | hla1 | D208E | 6.84 | 1 | Low |
| 5 | HDES26 | VI | t062 | ST5 | L | P3 | hla1 | D208E | 8.01 | 1 | Low |
| 6 | HDE288 | VI | t311 | ST5 | L | P3 | hla1 | D208E | 6.67 | 1 | Low |
| 7 | HSA29 | – | t002 | ST5 | L | P3 | hla1 | D208E | Not Valid | – | Not Valid |
| 8 | HDE461 | IV | t022 | ST22 | H | P10 | hla12 | S239T; T243S | 6.60 | 1 | Low |
| 9 | HAR22 | IV | t022 | ST22 | H | P11 | hla13 | S239T; T243S | 6.43 | 1 | Low |
| 10 | HSMB280 | IV | t032 | ST22 | H | P10 | hla12 | S239T; T243S | 4.71 | 1 | High |
| 11 | LBM12 | IV | t747 | ST1806 | H | nt | hla12 | S239T; T243S | 9.28 | 1 | Low |
| 12 | HSMB184 | – | t5951 | ST1806 | H | P10 | hla12 | S239T; T243S | 6.74 | 1 | Low |
| 13 | HPH2 | II | t018 | ST36 | H | P7 | hla8 | D208E; S239T; stop codon | 8.02 | 2* | Low |
| 14 | HAR24 | II | t018 | ST36 | H | nt | hla8 | D208E; S239T; stop codon | 9.62 | 2* | Low |
| 15 | DEN4415 | II | t021 | ST36 | H | P7 | hla8 | D208E; S239T; stop codon | 8.95 | 2* | Low |
| 16 | C563 | IV | t015 | ST45 | H | nt | hla10 | S239T | 7.02 | 1 | Low |
| 17 | C036 | V | t015 | ST45 | H | nt | hla10 | S239T | 6.24 | 0 | Low |
| 18 | HAR38 | IV | t004 | ST45 | H | P7 | hla10 | S239T | 10.38 | 1 | Low |
| 19 | HFX77 | III | t037 | ST239 | L | P1 | hla4 | – | 8.74 | 2* | Low |
| 20 | HUC343 | IIIA | t037 | ST239 | L | P1 | hla4 | – | 8.27 | 0 | Low |
| 21 | HU25 | IIIA | t138 | ST239 | L | P1 | hla4 | – | 8.17 | 1 | Low |
| 22 | BK1953 | IA | t051 | ST247 | L | P1 | hla4 | – | 7.71 | 1 | Low |
| 23 | HPV107 | IA | t051 | ST247 | L | P1 | hla4 | – | 7.56 | 0 | Low |
| 24 | HSJ419 | IA | t725 | ST247 | L | P1 | hla4 | – | 8.23 | 1 | Low |
| 27 | E2125 | I | t051 | ST247 | L | P1 | hla4 | – | 7.29 | 0 | Low |
| 25 | 10395 | I | t008 | ST250 | L | P2 | hla4 | – | 8.15 | 1 | Low |
| 26 | COL | I | t008 | ST250 | L | P1 | hla4 | – | 8.01 | 1 | Low |
| 28 | HFX74 | IV | t008 | ST8 | L | P1 | hla4 | – | 6.46 | 1 | Low |
| 29 | USA300 | IV | t008 | ST8 | L | P1 | hla4 | – | 6.19 | 3* | Low |
| 30 | C438 | IV | t024 | ST8 | L | P1 | hla4 | – | 6.07 | 1 | Low |
| 31 | C574B | IV | t1257 | ST612 | L | P1 | hla4 | – | Not Valid | – | Not Valid |
| 32 | LBM27 | – | t024 | ST8 | L | P1 | hla4 | – | 8.12 | 0 | Low |
| 33 | LBM74 | – | t008 | ST8 | L | P1 | hla4 | – | 5.87 | 1 | Low |
| 34 | C270 | IV | t1381 | ST1 | L | P17 | hla2 | – | 8.81 | 1 | Low |
| 35 | USA400 | IV | t127 | ST1 | L | P17 | hla2 | – | 6.01 | 2* | Low |
| 36 | LBM36 | – | t127 | ST1 | L | P18 | hla2 | – | 11.09 | 1 | Low |
| 37 | C577 | IV | t216 | ST59 | L | P20 | hla5 | – | 5.35 | 0 | High |
| 38 | C583 | IV | t437 | ST59 | L | P19 | hla5 | – | 5.31 | 1 | High |
| 39 | C434 | V | t437 | ST59 | L | P19 | hla5 | – | 9.14 | 1 | Low |
| 40 | C018 | IV | t1819 | ST93 | L | nt | hla7 | – | 5.16 | 1 | High |
| 41 | C491 | IV | t202 | ST93 | L | P21 | hla7 | – | 5.45 | 0 | High |
| 42 | LBM54 | IV | t011 | ST398 | H | P12 | hla11 | – | 4.46 | 2* | High |
| 43 | C482 | IV | t011 | ST398 | H | P13 | hla11 | – | 3.25 | 1 | High |
| 44 | C496 | VII | t108 | ST398 | H | nt | hla11 | – | 2.85 | 1 | High |
| 45 | LBM40 | – | t034 | ST398 | H | P12 | hla11 | – | 5.37 | 1 | High |
| 46 | C017 | IV | t019 | ST30 | H | nt | hla9 | D208E; S239T | 4.53 | 0 | High |
| 47 | C385 | IV | t019 | ST30 | H | P7 | hla9 | D208E; S239T | 7.25 | 1 | Low |
| 48 | C479 | IV | t019 | ST30 | H | nt | hla9 | D208E; S239T | 8.10 | 1 | Low |
| 71 | HUC585 | – | t342 | ST30 | H | P7 | hla9 | D208E; S239T | 5.14 | 1 | High |
| 69 | HFF204 | – | t318 | ST30 | H | P9 | hla9 | D208E; S239T | 6.23 | 1 | Low |
| 70 | HFA30 | – | t012 | ST30 | H | P8 | hla8 | D208E; S239T; stop codon | 7.94 | 1 | Low |
| 49 | HSJO7 | IV | t148 | ST72 | L | P14 | hla1 | D208E | 6.56 | 1 | Low |
| 50 | USA700 | IV | t148 | ST72 | L | P14 | hla1 | D208E | 5.76 | 0 | Low |
| 51 | COO3 | IV | t791 | ST72 | L | P15 | hla1 | D208E | 6.28 | 1 | Low |
| 52 | SAMS1024 | IV | t1346 | ST1810 | L | P14 | hla1 | D208E | 4.78 | 1 | High |
| 53 | HUC594 | – | t148 | ST72 | L | P14 | hla1 | D208E | 8.36 | 1 | Low |
| 54 | HFA28 | – | t126 | ST72 | L | P14 | hla1 | D208E | 4.56 | 2* | High |
| 55 | C238 | – | t3682 | ST72 | L | P14 | hla1 | D208E | 4.64 | 1 | High |
| 56 | C168 | IV | t044 | ST80 | L | P16 | hla1 | D208E | 8.20 | 0 | Low |
| 57 | C485 | IV | t044 | ST80 | L | P16 | hla1 | D208E | 5.72 | 1 | High |
| 58 | C014 | IV | t131 | ST80 | L | P16 | hla1 | D208E | 4.87 | 0 | High |
| 59 | LBM25 | – | t1509 | ST15 | L | P2 | hla1 | D208E | 6.69 | 0 | Low |
| 60 | C157 | – | t084 | ST15 | L | P2 | hla1 | D208E | 4.86 | 1 | High |
| 61 | C230 | – | t346 | ST15 | L | P2 | hla1 | D208E | 9.03 | 2* | Low |
| 62 | HBA33 | – | t258 | ST25 | L | P6 | hla1 | D208E | 5.73 | 1 | High |
| 63 | C095 | – | t2909 | ST25 | L | P6 | hla1 | D208E | 4.16 | 1 | High |
| 64 | C141 | – | t081 | ST25 | L | P6 | hla1 | D208E | 4.50 | 2* | High |
| 65 | HBA34 | IV | t308 | ST121 | L | nt | hla6 | – | 5.62 | 1 | High |
| 66 | HUC574 | – | t435 | ST121 | L | P1 | hla6 | – | 5.19 | 1 | High |
| 67 | HUC587 | – | t159 | ST121 | L | P2 | hla6 | – | 5.09 | 1 | High |
| 68 | HUC578 | – | t284 | ST121 | L | P1 | hla6 | – | 7.10 | 2* | Low |
| 72 | LBM23 | – | t100 | ST9 | L | P22 | hla1 | D208E | 5.48 | 2* | High |
| 73 | HFX84 | – | t267 | ST97 | L | P23 | hla3 | – | 9.03 | 1 | Low |
H: High polymorphism; L: Low polymorphism;
Mean Delta Ct1–3 = Average (Delta Ct1; Delta Ct2; Delta Ct3), Delta Ct = Ct hla−Ct 16S; Not valid: only one Ct reading;
*low reproducibility between three CT values (Stddv≤2). nt: non typable; Stddv: standard deviation.
From the 12 hla (hla1–12), we observed that only a single hla-allotype was found among representatives of a specific ST, except for ST22 (hla12; hla13) and ST30 (hla8; hla9) where two different alleles were identified. On the other hand, the most frequent alleles, hla1 (33.3%, n = 24) and hla4 (20.8%, n = 15), were identified in more than one ST.
Regarding the nucleotide changes identified in the hla, some correspond to non-synonymous mutations (E208, T239 and S243) and, in one particular case, to a stop codon (Table 1 and 2). The substitutions observed did not correspond to any difference in the charge or polarity of the amino acid (aa). However, changes in molecular weight were observed: i) changes from aa D208 to aa E208 (D208E) and from aa S239 to T239 (S239T) gave rise to a higher molecular weight aa; and ii) change from aa T243 to S243 (T243S) resulted in a lower molecular weight aa; of note all changes occurred in the Rim domain of the protein. In a particular case, the aa change gave rise to a stop codon located in the CAP domain, in strains of ST36. Protein structure modeling showed that a protein of about one third of its real size is produced, truncated at the Gln87 (Figure 1, A and B). The truncation is in the outside part of the domain, suggesting that this will affect the capacity of the Hla to form cell wall pores, and ultimately to induce hemolysis.
Table 2. Strains data distribution based on promoter allotypes.
| Promotor allotype | Gene allotype | Non Synonymous Mutation | Isolates Molecular Characterization | Expression Category | ||
| CA backgrounds | ST398 | P13 | hla11 | – | ST398-IV, t011 | High expression |
| P12 | ST398, t034 | High expression | ||||
| NT | ST398-VII, t108 | High expression | ||||
| P12 | ST398-IV, t011 | High expression* | ||||
| ST25 | P6 | hla1 | D208E | ST25, t258 | High expression | |
| ST25, t081 | High expression* | |||||
| ST25, t2909 | High expression | |||||
| ST9 | P22 | hla1 | D208E | ST9, t100 | High expression* | |
| ST93 | P21 | hla7 | – | ST93-IV, t202 | High expression | |
| NT | ST93-IV, t1819 | High expression | ||||
| ST121 | P2 | hla6 | – | ST121, t159 | High expression | |
| P1 | ST121, t435 | High expression | ||||
| NT | ST121-IV, t308 | High expression | ||||
| P1 | ST121, t284 | Low expression* | ||||
| ST72 | P14 | hla1 | D208E | ST72-IV, t148 | High expression | |
| P14 | ST72, t3682 | High expression | ||||
| P14 | ST1810-IV, t1346 | High expression | ||||
| P14 | ST72, t126 | High expression* | ||||
| P15 | ST72-IV, t791 | Low expression | ||||
| P14 | ST72-IV, t148 | Low expression | ||||
| P14 | ST72, t148 | Low expression | ||||
| ST80 | P16 | hla1 | D208E | ST80-IcV, t131 | High expression | |
| ST80-IV, t044 | High expression | |||||
| ST80-IV, t044 | Low expression | |||||
| ST30 | P7 | hla9 | D208E; S239T | ST30, t342 | High expression | |
| NT | ST30-IV, t019 | High expression | ||||
| P7 | ST30-IV, t019 | Low expression | ||||
| P9 | ST30, t318 | Low expression | ||||
| NT | ST30-IV, t019 | Low expression | ||||
| P8 | hla8 | D208E; S239T; stop codon | ST30, t012 | Low expression | ||
| ST15 | P2 | hla1 | D208E | ST15, t084 | High expression | |
| ST15, t346 | Low expression* | |||||
| ST15, t1509 | Low expression | |||||
| ST59 | P20 | hla5 | – | ST59-IV, t216 | High expression | |
| P19 | ST59-IV, t437 | Low expression | ||||
| P19 | ST59-V, t437 | Low expression | ||||
| ST1 | P17 | hla2 | – | ST1-IV, t1381 | Low expression | |
| P17 | ST1-IV, t127 | Low expression* | ||||
| P18 | ST1, t127 | Low expression | ||||
| ST8 | P1 | hla4 | – | ST8-IV, t008 | Low expression | |
| ST8-IV, t024 | Low expression | |||||
| ST8-IV, t008 | Low expression* | |||||
| ST8, t008 | Low expression | |||||
| ST612-IV, t1257 | Not valid** | |||||
| ST8, t024 | Low expression | |||||
| ST97 | P23 | hla3 | – | ST97, t267 | Low expression | |
| HA backgrounds | ST22 | P10 | hla13 | S239T; T243S | ST22-IV, t032 | High expression |
| P10 | hla12 | ST22-IV, t022 | Low expression | |||
| P11 | ST22-IV, t022 | Low expression | ||||
| P10 | ST1806, t5951 | Low expression | ||||
| NT | ST1806-IV, t747 | Low expression | ||||
| ST5 | NT | hla1 | D208E | ST5-II, t002 | High expression | |
| P3 | ST5-VI, t002 | Low expression | ||||
| P3 | ST5-VI, t062 | Low expression | ||||
| P3 | ST5-VI, t311 | Low expression | ||||
| P4 | ST5-II, t002 | Low expression* | ||||
| P3 | ST5, t002, | Not valid** | ||||
| P5 | ST5-II, t067 | Low expression | ||||
| ST36 | P7 | hla8 | D208E; S239T; stop codon | ST36-II, t018 | Low expression* | |
| P7 | ST36-II, t021 | Low expression* | ||||
| NT | ST36-II, t01 | Low expression* | ||||
| ST45 | NT | hla10 | S239T | ST45-IV, t015 | Low expression | |
| NT | ST45-V, t015 | Low expression | ||||
| P7 | ST45-IV, t004 | Low expression | ||||
| ST239 | P1 | hla4 | – | ST239-IIIA, t037 | Low expression | |
| ST239-III, t037 | Low expression* | |||||
| – | ST239-IIIA, t138 | Low expression | ||||
| ST247 | P1 | hla4 | – | ST247-I, t051 | Low expression | |
| ST247-IA, 051 | Low expression | |||||
| ST247-IA, t051 | Low expression | |||||
| ST247-IA, t725 | Low expression | |||||
| ST250 | P1 | hla4 | – | ST250-I, t008 | Low expression | |
| P2 | ST250-I, t008 | Low expression |
(*)(**) relative expression values not valid (SDV≤2 or only one CT reading).
A high number of sequence variations were identified in the hla promoter region, (n = 23) (P1–23) (Table 1 and 2). Although we found that some STs were associated to a specific promoter allotype, and some promoters were identified in a single ST, we also identified cases where single STs were associated to different promoters (8 out of 23) and examples in which a single promoter allotype was associated to different STs (5 out of 23). This is the case of the most frequent promoter (P1) that was found in about one third of the strains analyzed (25.4%, n = 16), including several different STs.
A particular highly polymorphic region corresponding to nt −22 to −24 from the start codon, was found in the majority (16 out of 23) of the promoter allotypes (exceptions P1, P6, P13, P14, P15, P18 and P23). These polymorphisms are located in the vicinity of RNAIII binding site [26]; however, we could not find a direct correlation between a particular nucleotide sequence and a specific expression pattern (high or low expression). For example, the sequence TTT, observed in two strains belonging to ST398 that have a high level expression, was also observed in strains with low expression belonging to other genetic backgrounds (ST8, ST239, ST247, ST250, ST36, ST45 and ST22).
Alpha-hemolysin evolutionary history
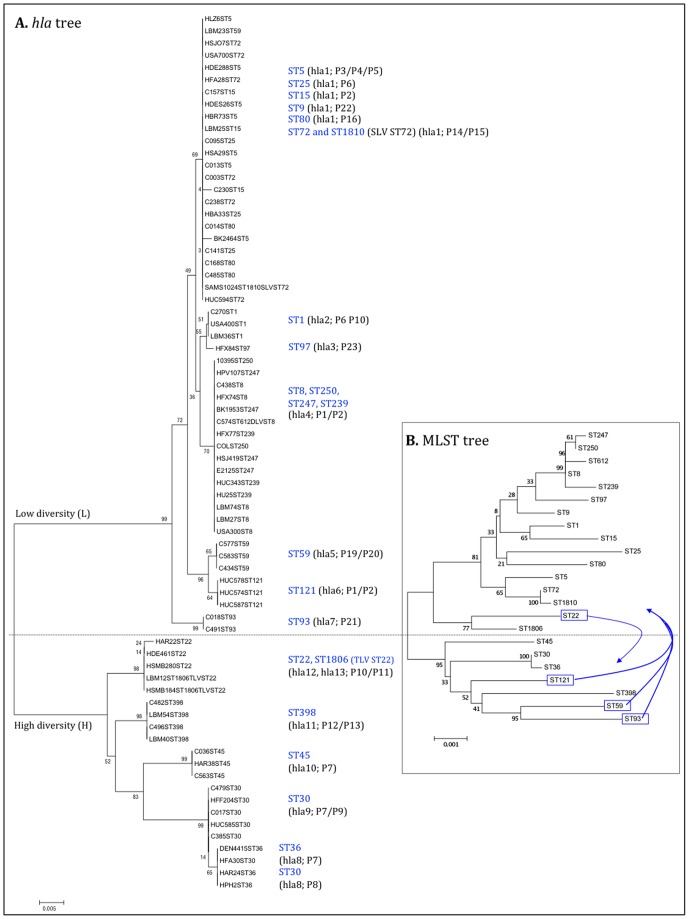
In order to better understand the evolution of hla gene within the S. aureus population, we constructed phylogenetic trees from the hla and hla promoter sequences, separately or concatenated (Figure 2, A) and compared it with the tree constructed from the concatenated sequences of the seven housekeeping genes used in MLST, including all the STs represented in the strain collection described here (Figure 2, B).
Figure 2. Phylogenetic trees of hla gene (A) and concatenated sequences of MLST alleles (B) from 23 STs representatives of the strains collection.
The tree was constructed using MEGA 5 with Neighbour-joining method and bootstrap values provided as percents over 1000 replications. Branch length values are indicated and the percentage of replicate trees (bootstrap test) are shown next to the branches. The dashed line indicates the separation of the two evolutionary branches.
The phylogenetic tree constructed for the hla gene showed two distinct major clusters with different evolutionary clocks that differed in their nucleotide diversity (ND, see Materials and Methods): cluster (L) with lower diversity (ND = 0.005), and cluster H with higher diversity (ND = 0.019). Cluster L included more than 70% of strains (71.2%, n = 52), and five sub-clusters; Cluster H contained about 29% of the strains (28.8%, n = 21), and comprised four minor sub-clusters including hla8–hla12 alleles, which were found in strains of ST30, ST36, ST45, ST398 and ST22.
As opposed to the phylogenetic tree constructed from hla gene, the one constructed from the promoter region did not show two distinct evolutionary branches (Figure S1). Moreover, dissimilar subgroup clustering was noticed in the tree constructed from the promoter gene sequence. For example, ST45, ST30 and ST36 backgrounds were clustered together in the promoter sequence-based tree whereas in the hla sequence-based tree ST45 was placed separately from ST30 and ST36 cluster (branch H). The same type of observations can be drawn for most of STs. Overall the promoter region showed to be more diverse than the hla gene sequence among the different backgrounds.
On the other hand, when we compared the phylogenetic tree constructed with the hla gene with that constructed from MLST concatenated genes, the same type of division into two distinct main clusters was observed (Figure 2). Moreover, the majority of STs were equally distributed between the two clusters in the two trees. The only exceptions were ST22, ST121, ST59 and ST93 that in the two trees have exchanged their positions from one cluster to the other (Figure 2, B-blue arrows).
Detection of recombination in hla gene, hla promoter and MLST genes
To understand if recombination could explain the incongruence found between the trees constructed from hla and MLST concatenated genes, we screened the hla gene, hla promoter and each MLST gene for recombination events using the RDP4 software.
The SiScan and 3Seq methods detected one recombination event in the hla gene. This event corresponded to a fragment ending in positions 385–410 of the hla alignment, however the beginning breakpoint was not possible to determine. In the collection analyzed this event was detected in five isolates belonging to ST22 or related STs (HSMB280, HDE461, HAR22 and LBM12 (TLV ST22) and HSMB184 (TLV ST22)) and four isolates of ST398 (LBM54, LBM40, C496, C482_ST398). The ST30 HFF204 strain was identified as the minor parent (97.8% identity with ST22 strains and −99.3% identity with ST398 strains) and ST121 strain HUC587 was identified as the major parent (with 100% identity to ST398 strains and 93.5–95.2% identity with ST22 strains) of the recombining fragment. A trace signal of recombination of this same event was also identified among ST45 isolates; however this signal was not statistically significant. Interestingly all the recombination events were detected in strains belonging to the high genetic diversity cluster in the tree constructed from hla gene. In the hla promoter region no recombination events were detected.
We have performed the same type of analysis using the internal sequences of each of the seven housekeeping used in MLST scheme, including the alleles present in all STs identified in this study, however no recombination events were detected in any of the genes.
Altogether the data gathered suggest that for the majority of strains hla gene evolved together with the genetic background. The different clustering of ST22 and ST121 strains, in the trees constructed from MLST concatenated genes and hla gene, may derive from recombination events occurring in the hla gene. Similarly these type of events might explain the genetic diversity observed in cluster H in the hla tree in strains belonging to ST22, ST398, ST45, ST30 and ST36 (H cluster of hla tree).
Expression of alpha-hemolysin
The expression of alpha-hemolysin in the 73 strains was assessed by RT-PCR, in three biological replicates. Fifteen of the 73 strains (20.5%) were excluded from the final analysis, either because a single valid determination for Delta Ct (N = 2) was obtained or because CT obtained from the different biological replicates were not reproducible (N = 13).
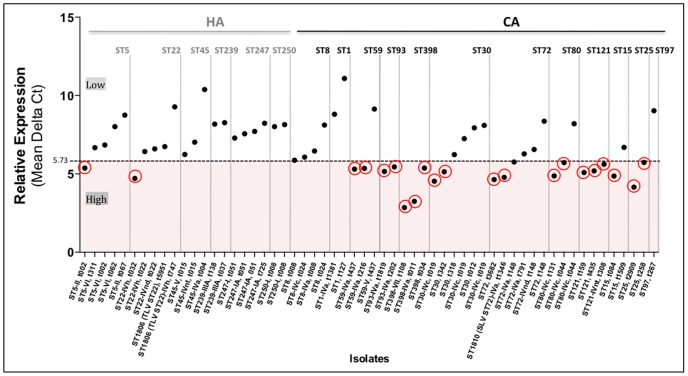
The analysis of the regression tree split the response variable into two distinct groups, according to the spa type of the strains. There was a group of strains with mean Delta Ct1–3≤5.73, that was classified as a high expression group and a second group with a mean Delta Ct1–3>5.73 classified as a low expression group (Table 1, Table 2 and Figure 3). Overall the regression tree explained 60% of the variance in the data. This is mostly because there were strains expressing a low or high mean Delta Ct that were classified in the same spa type; those were the cases of spa types t002, t019, t044 and t437.
Figure 3. HA and CA strains relative expression distribution.
Mean of expression rates from three biological replicates. Dashed line corresponding to the mean Ct value 5.73 results from the regression tree analysis which split strains in two distinct groups, at spa type level: a) high expression group - corresponding to strains with Mean Delta Ct≤5.73 and b) low expression group- corresponding to strains with Mean Delta Ct>5.73). Highlighted in red are the high expressing strains.
Furthermore, we explored in each of the spa types what other explanatory variables (ST, MRSA, MSSA and type of SCCmec) could differentiate the inclusion of some strains in the low or high expression group, but we found no associations with the variables we measured in the study.
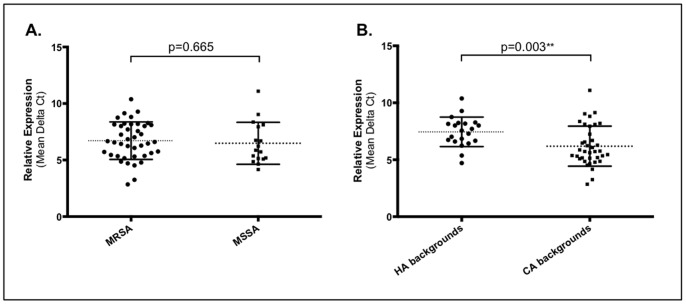
We observed that the hla expression level varied within strains of the same ST (Figure 3; Table 1 and 2). In fact, in some cases the same ST comprised strains with both high and low levels of expression (ST5, ST15, ST22, ST30, ST59, ST72 and ST80). Moreover, we found that the expression rates did not differ significantly (P = 0.665) between MRSA and MSSA strains. However, we did find a correlation between the hla expression and the origin of the genetic backgrounds. Actually, strains of CA genetic backgrounds showed, in general, higher mean expression rates than strains of HA backgrounds (p = 0.003) (Figure 4). Among the 21 strains (36.2%, 21 out of 58) with high expression level, only two (9.5%) belonged to HA backgrounds (ST22-IVh, t032 and ST5-II, t002) whereas the majority (90.5%, n = 19) were represented by CA backgrounds (Table 1 and Table 2). Moreover, two additional CA strains, ST72-IVa-t148 and ST8-MSSA-t008, showed expression rates near the cutoff value (5.73), with 5.76 and 5.87, respectively. These were considered as belonging to the low-level expression group.
Figure 4. Distribution of the relative hla expression.
Mean of relative expression of three independent readings. Expression comparison between a) MRSA and MSSA and b) HA and CA backgrounds using the Two-tailed Student’s t-test. Statistically significance (p≤0.05) (**).
The three strains with the highest expression rate were ST398-VII-t108 (2.85), ST398-IVa-t011 (3.25) and ST25-MSSA-t2909 (4.16) and strains with the lowest rate were ST1806 (TLV ST22)-IVh-t747 (9.28), ST45-IVa-t004 (10.38) and ST1-MSSA-t127 (11.09).
We observed that some promoters and gene alleles (P6, P12/P13, P21; and hla7, hla9, hla11) were exclusively associated to a high expression level profile, while others (P3/P4/P5, P7, P8/P9, P11, P15, P17/P18, P23; and hla4, hla8, hla10) were exclusively associated to a low expression level (Table 1 and 2). But we also found promoter and gene allotypes that were associated to both high and low expression levels.
Discussion
Although Hla is one of the most important S. aureus virulence factors [10], to the best of our knowledge, this is the first study in which the variation in hla nucleotide sequence and gene expression was assessed in such a large and representative collection.
We found that the nucleotide sequence of hla was highly diverse. The high degree of diversity found within hla is in accordance to results obtained for other exotoxins, which are generally highly polymorphic [27]. Four non-synonymous substitutions (Q87 stop codon, D208E, S239T and T243S) were identified, that are located in two structural protein domains which are essential for Hla oligomerization and pore formation (Rim and Cap) [11], [28], [29]. The impact of these amino acid (aa) changes on hla activity is uncertain. If by one hand, the aa changes described implicate differences in the molecular weight of the aa, that can have influence in the three dimensional structure stability and activity of the protein; on the other hand these aa changes did not match any of the aa previously described to be essential for Hla pore formation.
Furthermore, Walker and Bayley showed that multiple mutations in this same region (residues spanning Hla235–250) did not alter Hla activity in terms of binding, oligomerization or lysis. Thus, it would not be expected that S239T or T243S had significant biological impact in terms of toxin function. The unique mutation with an identified role in Hla function is the stop codon found in the ST36 and ST30 strains that was previously described by DeLeo and co-authors [30] to hinder toxin production and to originate a less virulent strain in a murine infection model. The true effect of the non-synonymous substitutions identified in our study in the activity of the protein would have to be tested by the construction of site directed mutagenesis mutants and by performing binding, oligomerization, hemolysis and in vivo models assays.
The construction of phylogenetic trees from the hla defined the existence of two clusters with different levels of genetic diversity suggesting that hla is evolving at different rates in different genetic backgrounds. Interestingly, the most diverse cluster included the clonal types which are presently more disseminated or that emerged recently (like ST398). This might be related to the fact that these clones still need to evolve to evade the human immune system and not enough time as elapsed for the most adapted allele to have been selected [31]. On the other hand the recombination events detected in the hla gene in this study were all in strains belonging to the high genetic diversity cluster, suggesting that this mechanism might have been important in the most recent hla evolution and diversification.
Interestingly, the phylogenetic tree constructed from the hla gene was similar to that constructed from MLST genes, in the sense that both trees distributed the different STs similarly in two main clusters. This observation suggests that hla gene has evolved together with the S. aureus genetic background. A similar type of correlation with the genetic background was previously described for adhesins, either located in the core genome (clfA, clfB, fnbA, map, sdrC, and spa) or accessory genome (ebpS, fnbB, sdrD, and sdrE) [32]. Although this was the case for the great majority of STs, we observed that four STs (ST22, ST121, ST59, ST93) were located in different clusters in the hla and MLST trees. Our results suggest that recombination occurring at the hla level, might explain the different clustering of strains belonging to ST22 and ST121. No recombination events were, however, detected in MLST genes or hla sequences of strains belonging to ST59 and ST93, suggesting that their displacement in the two trees could derive from different phenomena, like random mutation.
It was previously suggested that CA-MRSA expressed more hla than HA-MRSA [9]. Results from our study allowed us to extend this conclusion to virtually all epidemic CA, but also in two particular cases of HA genetic backgrounds. The CA strains belonging to ST398, ST25, ST121 and ST93 showed uniformly high relative expression rates and strains belonging to ST36, ST45, ST239, ST247 and ST250 showed uniformly low expression rates. To understand if in fact these patterns of expression are characteristic of these clones, more strains within each clone should be studied for hla expression. Nevertheless, we could not correlate the hla expression rate with any particular polymorphism within the promoter or any aa substitution in the hla gene. The results suggest that hla regulation is probably a result of combination of factors which are redundant, rather than associated to a single genetic event. In fact, it has been demonstrated by several authors that alpha-hemolysin is part of a complex regulatory network, that includes the main two-component systems (TCS) – Agr – that in turn is controlled by a diverse pool of regulatory networks that coordinately interact in response to external stimulus and cell signals, namely others TCS (SaeRS, ArlRS and SrrAB), alternative sigma factors (σB), and transcription factors (e.g. SarS, SarT, Rot, SarA, SarZ) [33], [34].
We showed that hla evolved together with the genetic background. Moreover, the most epidemic CA-MRSA genetic backgrounds express more hla than the most epidemic HA-MRSA genetic backgrounds. However, the finding of frequent strain-to-strain variation in the expression level of hla within strains of the same clonal types suggests that hla polymorphisms cannot be used as genetic markers of virulence and investigators should remain cautious when inferring conclusions for the entire MRSA population from studies performed with a limited number of strains.
Supporting Information
Phylogenetic trees of the hla gene, promoter gene and concatenated sequences of both. The tree was constructed using MEGA 5 with Neighbour-joining method and bootstrap values provided as percents over 1000 replications. Branch length values are indicated and the percentage of replicate trees (bootstrap test) are shown next to the branches. The dashed line indicates the separation of the two evolutionary branches (L and H).
(TIF)
I. Growth curves for triplicates of each S. aureus strain – Set C. II. Growth curves for triplicates of each S. aureus strain – Set D. III. Growth curves for triplicates of each S. aureus strain – Set E.
(TIFF)
Internal sequences of hla promoter (highlighted blue) and hla gene (highlighted orange) used for analysis in this study. Primers used are highlighted. The sequence shown corresponds to the promoter and hla regions of USA300 strain from our collection blasted against USA300_TCH1516.
(TIF)
Molecular characterization of the 73 MRSA and MSSA strains included in this study [35]–[50].
(DOC)
Funding Statement
This work was funded by project Ref. P-99911 from Fundação Calouste Gulbenkian (http://www.gulbenkian.pt/Institucional/pt/Homepage) and Project PTDC/BIA-MIC/3195/2012 from Fundação para a Ciência e Tecnologia (http://www.fct.pt/) awarded to HdL; Project PTDC/BIA-EVF/117507/2010 from Fundação para a Ciência e Tecnologia (http://www.fct.pt/) awarded to MM; and through grant Ref. Pest-OE/EQB/LAO004/2011 from Fundação para a Ciência e Tecnologia (FCT), Portugal. A. Tavares was supported by grant SFRH/BD/44220/2008 from FCT. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Deurenberg RH, Stobberingh EE (2009) The molecular evolution of hospital- and community-associated methicillin-resistant Staphylococcus aureus . Curr Mol Med 9: 100–115. [DOI] [PubMed] [Google Scholar]
- 2. David MZ, Daum RS (2010) Community-associated methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clin Microbiol Rev 23: 616–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Otto M (2013) Community-associated MRSA: What makes them special? Int J Med Microbiol. [DOI] [PMC free article] [PubMed]
- 4. Kobayashi SD, Voyich JM, Burlak C, DeLeo FR (2005) Neutrophils in the innate immune response. Arch Immunol Ther Exp (Warsz) 53: 505–517. [PubMed] [Google Scholar]
- 5. Baba T, Takeuchi F, Kuroda M, Yuzawa H, Aoki K, et al. (2002) Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359: 1819–1827. [DOI] [PubMed] [Google Scholar]
- 6. Diep BA, Gill SR, Chang RF, Phan TH, Chen JH, et al. (2006) Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus . Lancet 367: 731–739. [DOI] [PubMed] [Google Scholar]
- 7. Burlak C, Hammer CH, Robinson MA, Whitney AR, McGavin MJ, et al. (2007) Global analysis of community-associated methicillin-resistant Staphylococcus aureus exoproteins reveals molecules produced in vitro and during infection. Cell Microbiol 9: 1172–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Loughman JA, Fritz SA, Storch GA, Hunstad DA (2009) Virulence gene expression in human community-acquired Staphylococcus aureus infection. J Infect Dis 199: 294–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li M, Cheung GY, Hu J, Wang D, Joo HS, et al. (2010) Comparative analysis of virulence and toxin expression of global community-associated methicillin-resistant Staphylococcus aureus strains. J Infect Dis 202: 1866–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Berube BJ, Bubeck Wardenburg J (2013) Staphylococcus aureus alpha-Toxin: Nearly a Century of Intrigue. Toxins (Basel) 5: 1140–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Song L, Hobaugh MR, Shustak C, Cheley S, Bayley H, et al. (1996) Structure of staphylococcal alpha-hemolysin, a heptameric transmembrane pore. Science 274: 1859–1866. [DOI] [PubMed] [Google Scholar]
- 12. Valeva A, Palmer M, Bhakdi S (1997) Staphylococcal alpha-toxin: formation of the heptameric pore is partially cooperative and proceeds through multiple intermediate stages. Biochemistry 36: 13298–13304. [DOI] [PubMed] [Google Scholar]
- 13. Novick RP, Ross HF, Projan SJ, Kornblum J, Kreiswirth B, et al. (1993) Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J 12: 3967–3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ballal A, Ray B, Manna AC (2009) sarZ, a sarA family gene, is transcriptionally activated by MgrA and is involved in the regulation of genes encoding exoproteins in Staphylococcus aureus . J Bacteriol 191: 1656–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liang X, Yu C, Sun J, Liu H, Landwehr C, et al. (2006) Inactivation of a two-component signal transduction system, SaeRS, eliminates adherence and attenuates virulence of Staphylococcus aureus . Infect Immun 74: 4655–4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liang X, Zheng L, Landwehr C, Lunsford D, Holmes D, et al. (2005) Global regulation of gene expression by ArlRS, a two-component signal transduction regulatory system of Staphylococcus aureus . J Bacteriol 187: 5486–5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schmidt KA, Manna AC, Gill S, Cheung AL (2001) SarT, a repressor of alpha-hemolysin in Staphylococcus aureus . Infect Immun 69: 4749–4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liang X, Hall JW, Yang J, Yan M, Doll K, et al. (2011) Identification of single nucleotide polymorphisms associated with hyperproduction of alpha-toxin in Staphylococcus aureus . PLoS One 6: e18428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bubeck Wardenburg J, Bae T, Otto M, Deleo FR, Schneewind O (2007) Poring over pores: alpha-hemolysin and Panton-Valentine leukocidin in Staphylococcus aureus pneumonia. Nat Med 13: 1405–1406. [DOI] [PubMed] [Google Scholar]
- 20. Bubeck Wardenburg J, Patel RJ, Schneewind O (2007) Surface proteins and exotoxins are required for the pathogenesis of Staphylococcus aureus pneumonia. Infect Immun 75: 1040–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Inoshima I, Inoshima N, Wilke GA, Powers ME, Frank KM, et al. (2011) A Staphylococcus aureus pore-forming toxin subverts the activity of ADAM10 to cause lethal infection in mice. Nat Med 17: 1310–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tamura K, Nei M, Kumar S (2004) Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci U S A 101: 11030–11035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vandenesch F, Kornblum J, Novick RP (1991) A temporal signal, independent of agr, is required for hla but not spa transcription in Staphylococcus aureus . J Bacteriol 173: 6313–6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zielinska AK, Beenken KE, Joo HS, Mrak LN, Griffin LM, et al. (2011) Defining the strain-dependent impact of the Staphylococcal accessory regulator (sarA) on the alpha-toxin phenotype of Staphylococcus aureus . J Bacteriol 193: 2948–2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. De’ath G, Fabricius KE (2000) Classification and regression trees: a powerful yet simple thechnique for ecological data analysis. Ecology. Ecology 81: 3178–3192. [Google Scholar]
- 26. Morfeldt E, Taylor D, von Gabain A, Arvidson S (1995) Activation of alpha-toxin translation in Staphylococcus aureus by the trans-encoded antisense RNA, RNAIII. EMBO J 14: 4569–4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wilson GJ, Seo KS, Cartwright RA, Connelley T, Chuang-Smith ON, et al. (2011) A novel core genome-encoded superantigen contributes to lethality of community-associated MRSA necrotizing pneumonia. PLoS Pathog 7: e1002271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Montoya M, Gouaux E (2003) Beta-barrel membrane protein folding and structure viewed through the lens of alpha-hemolysin. Biochim Biophys Acta 1609: 19–27. [DOI] [PubMed] [Google Scholar]
- 29. Walker B, Bayley H (1995) Key residues for membrane binding, oligomerization, and pore forming activity of staphylococcal alpha-hemolysin identified by cysteine scanning mutagenesis and targeted chemical modification. J Biol Chem 270: 23065–23071. [DOI] [PubMed] [Google Scholar]
- 30. DeLeo FR, Kennedy AD, Chen L, Bubeck Wardenburg J, Kobayashi SD, et al. (2011) Molecular differentiation of historic phage-type 80/81 and contemporary epidemic Staphylococcus aureus . Proc Natl Acad Sci U S A 108: 18091–18096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Castillo-Ramirez S, Harris SR, Holden MT, He M, Parkhill J, et al. (2011) The impact of recombination on dN/dS within recently emerged bacterial clones. PLoS Pathog 7: e1002129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kuhn G, Francioli P, Blanc DS (2006) Evidence for clonal evolution among highly polymorphic genes in methicillin-resistant Staphylococcus aureus . J Bacteriol 188: 169–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Novick RP (2003) Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol Microbiol 48: 1429–1449. [DOI] [PubMed] [Google Scholar]
- 34. Thoendel M, Kavanaugh JS, Flack CE, Horswill AR (2011) Peptide signaling in the staphylococci. Chem Rev 111: 117–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tavares A, Miragaia M, Rolo J, Coelho C, de Lencastre H (2013) High prevalence of hospital-associated methicillin-resistant Staphylococcus aureus in the community in Portugal: evidence for the blurring of community-hospital boundaries. Eur J Clin Microbiol Infect Dis 32: 1269–1283. [DOI] [PubMed] [Google Scholar]
- 36. Oliveira DC, Tomasz A, de Lencastre H (2001) The evolution of pandemic clones of methicillin-resistant Staphylococcus aureus: identification of two ancestral genetic backgrounds and the associated mec elements. Microb Drug Resist 7: 349–361. [DOI] [PubMed] [Google Scholar]
- 37. Roberts RB, de Lencastre A, Eisner W, Severina EP, Shopsin B, et al. (1998) Molecular epidemiology of methicillin-resistant Staphylococcus aureus in 12 New York hospitals. MRSA Collaborative Study Group. J Infect Dis 178: 164–171. [DOI] [PubMed] [Google Scholar]
- 38. Aires-de-Sousa M, Correia B, de Lencastre H (2008) Changing patterns in frequency of recovery of five methicillin-resistant Staphylococcus aureus clones in Portuguese hospitals: surveillance over a 16-year period. J Clin Microbiol 46: 2912–2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rolo J, Miragaia M, Turlej-Rogacka A, Empel J, Bouchami O, et al. (2012) High Genetic Diversity among Community-Associated Staphylococcus aureus in Europe: Results from a Multicenter Study. PLoS One 7: e34768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Conceicao T, Tavares A, Miragaia M, Hyde K, Aires-de-Sousa M, et al. (2010) Prevalence and clonality of methicillin-resistant Staphylococcus aureus (MRSA) in the Atlantic Azores islands: predominance of SCCmec types IV, V and VI. Eur J Clin Microbiol Infect Dis 29: 543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sa-Leao R, Santos Sanches I, Dias D, Peres I, Barros RM, et al. (1999) Detection of an archaic clone of Staphylococcus aureus with low-level resistance to methicillin in a pediatric hospital in Portugal and in international samples: relics of a formerly widely disseminated strain? J Clin Microbiol 37: 1913–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Amorim ML, Aires de Sousa M, Sanches IS, Sa-Leao R, Cabeda JM, et al. (2002) Clonal and antibiotic resistance profiles of methicillin-resistant Staphylococcus aureus (MRSA) from a Portuguese hospital over time. Microb Drug Resist 8: 301–309. [DOI] [PubMed] [Google Scholar]
- 43. Milheirico C, Oliveira DC, de Lencastre H (2007) Multiplex PCR strategy for subtyping the staphylococcal cassette chromosome mec type IV in methicillin-resistant Staphylococcus aureus: ‘SCCmec IV multiplex’. J Antimicrob Chemother 60: 42–48. [DOI] [PubMed] [Google Scholar]
- 44. Richardson JF, Reith S (1993) Characterization of a strain of methicillin-resistant Staphylococcus aureus (EMRSA-15) by conventional and molecular methods. J Hosp Infect 25: 45–52. [DOI] [PubMed] [Google Scholar]
- 45. Oliveira DC, Milheirico C, Vinga S, de Lencastre H (2006) Assessment of allelic variation in the ccrAB locus in methicillin-resistant Staphylococcus aureus clones. J Antimicrob Chemother 58: 23–30. [DOI] [PubMed] [Google Scholar]
- 46. Faria NA, Oliveira DC, Westh H, Monnet DL, Larsen AR, et al. (2005) Epidemiology of emerging methicillin-resistant Staphylococcus aureus (MRSA) in Denmark: a nationwide study in a country with low prevalence of MRSA infection. J Clin Microbiol 43: 1836–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sanches IS, Ramirez M, Troni H, Abecassis M, Padua M, et al. (1995) Evidence for the geographic spread of a methicillin-resistant Staphylococcus aureus clone between Portugal and Spain. J Clin Microbiol 33: 1243–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. de Lencastre H, Chung M, Westh H (2000) Archaic strains of methicillin-resistant Staphylococcus aureus: molecular and microbiological properties of isolates from the 1960s in Denmark. Microb Drug Resist 6: 1–10. [DOI] [PubMed] [Google Scholar]
- 49. Crisostomo MI, Westh H, Tomasz A, Chung M, Oliveira DC, et al. (2001) The evolution of methicillin resistance in Staphylococcus aureus: similarity of genetic backgrounds in historically early methicillin-susceptible and -resistant isolates and contemporary epidemic clones. Proc Natl Acad Sci U S A 98: 9865–9870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. McDougal LK, Steward CD, Killgore GE, Chaitram JM, McAllister SK, et al. (2003) Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J Clin Microbiol 41: 5113–5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogenetic trees of the hla gene, promoter gene and concatenated sequences of both. The tree was constructed using MEGA 5 with Neighbour-joining method and bootstrap values provided as percents over 1000 replications. Branch length values are indicated and the percentage of replicate trees (bootstrap test) are shown next to the branches. The dashed line indicates the separation of the two evolutionary branches (L and H).
(TIF)
I. Growth curves for triplicates of each S. aureus strain – Set C. II. Growth curves for triplicates of each S. aureus strain – Set D. III. Growth curves for triplicates of each S. aureus strain – Set E.
(TIFF)
Internal sequences of hla promoter (highlighted blue) and hla gene (highlighted orange) used for analysis in this study. Primers used are highlighted. The sequence shown corresponds to the promoter and hla regions of USA300 strain from our collection blasted against USA300_TCH1516.
(TIF)
Molecular characterization of the 73 MRSA and MSSA strains included in this study [35]–[50].
(DOC)