Abstract
In zebrafish, ventricular myosin heavy chain (vmhc) gene is initially expressed at the anterior lateral mesoderm, thereafter its expression is restricted to the cardiac ventricle. The transcriptional control mechanisms in regulating chamber-specific expression of myosin heavy chains are not well defined. We isolated and analyzed zebrafish vmhc upstream region to examine the spatial and temporal regulation of vmhc using transgenic and transient expression techniques. Promoter deletion analyses defined a basal promoter region sufficient to drive vmhc expression in the ventricle and an upstream fragment necessary for repressing ectopic vmhc expression in the atrium. The transcriptional mechanism that prevents vmhc expression in the atrium is mediated through Nkx2.5 binding elements (NKE). We have further discovered that paired-related homeobox transcriptional factor 2 (Prx2/S8)-like binding elements are required for promoting vmhc expression, and Prrx1b, a Prx-related homeobox protein, participates in the regulation of vmhc expression with other transcriptional factors.
Introduction
In zebrafish, cardiac development begins during the early stages of embryogenesis (Stainier, 2001). At the late blastula stages, myocardial progenitors are located throughout the ventral and lateral region of the zebrafish embryo. Ventricular progenitors are positioned closer to the margin than atrial progenitors (Keegan et al., 2004). By the 15-somite stage, myocardial progenitor cells differentiate into cardiomyocytes and start to express cardiac myosin light chain-2 (cmlc2) (Yelon et al., 1999). These cells then segregate into ventricular and atrial myocytes, expressing ventricular myosin heavy chain (vmhc) and atrial myosin heavy chain (amhc), respectively (Yelon et al., 1999; Berdougo et al., 2003). After cardiac looping and chamber formation, vmhc expression is restricted in the ventricle, whereas amhc is expressed in the atrium. Both vmhc and amhc are thought to be the earliest myocardial markers for revealing ventricular and atrial cell lineages.
In mouse, two MHC isoforms, α-MHC and β-MHC, are expressed in the primitive heart tube (Lyons et al., 1990). As cardiac chambers form, β-MHC expression is restricted to the ventricle, and α-MHC levels increase in atrial cells. β-MHC is gradually replaced by α-MHC after birth (Lyons et al., 1990). In human, myosin heavy chain 7 (MYH7), an isoform of β-MHC, is predominantly expressed in the ventricle, whereas expression of myosin heavy chain 6 (MYH6/α-MHC) is restricted in the atrium (Everett, 1986). Mutations in MYH7/β-MHC are frequent causes of familial hypertrophic and dilated cardiomyopathies (Seidman and Seidman, 2001). MYH6/α-MHC mutations are commonly detected to be associated with atrial septal defects (Ching et al., 2005). Expression of α-MHC and β-MHC is independently controlled but coordinately regulated. GATA factors, Mef2 transcription factors, and steroid hormone receptors act synergistically to activate α-MHC expression (Molkentin et al., 1996; Lee et al., 1997). During embryogenesis, β-MHC is expressed as part of the cardiac myogenic program under the control of Nkx-2.5, Mef-2, and GATA transcription factors (Morimoto et al., 1999; Morkin, 2000; Meissner et al., 2007). Analysis of its promoter regions also reveals negative thyroid hormone response elements and a transcriptional enhancer factor-1 binding site (Flink et al., 1992; Edwards et al., 1994). However, little is known about cis-elements and regulatory factors in regulating chamber-specific expression of myosin heavy chains.
Transgenic zebrafish that express enhanced green fluorescent protein (EGFP) under the control of tissue-specific promoters are useful tools for following cell movements, visualizing dynamic gene expression patterns and dissecting regulatory transcription elements in live embryos (Long et al., 1997; Motoike et al., 2000; Zhang and Rodaway, 2007). In this study, we generated a stable line of vmhc:EGFP transgenic zebrafish using a 1952-bp upstream regulatory sequence. A minimal promoter containing Prx2/S8-like binding elements was defined to direct vmhc expression, and an upstream fragment (−1772/−1522) containing Nkx2.5 binding sites was identified to repress the ectopic vmhc expression in the atrium. We revealed NKE-mediated atrial repression for governing ventricle-specific expression, and discovered that zebrafish Prx-related homeobox factor Prrx1b participated in the regulation of vmhc expression.
Results
Generation of vmhc:EGFP transgenic zebrafish
To establish a stable transgenic zebrafish with EGFP expression in the ventricle of hearts, we cloned a 1952-bp vmhc upstream region containing its promoter from genomic DNA (GenBank GI:ID 163644330) using PCR. The vmhc upstream fragments were subcloned into pEGFP-1 vectors and resulted in a fusing construct (pvmhc-EGFP1). We microinjected linearized vmhc-EGFP1 constructs into one- to two-cell embryos, and examined transient EGFP expression at 48 hour post fertilization (hpf). 30–40% of injected embryos displayed EGFP expression in the ventricle, and four Go founders (line 1 to line 4) produced F1 embryos expressing EGFP in the ventricle. Line 1, 2 and 3 exhibited ectopic EGFP expression in the craniofacial region. About 50% of F2 offspring embryos carried ventricular EGFP expression by outcrossing F1 fish with wild-type fish, suggesting that the germline transmission rate of transgene complied with Mendelian inheritance law.
vmhc:EGFP expression patterns in transgenic embryos
In zebrafish, cardiac progenitor cells derive from the anterior lateral plate mesoderm at 12 hpf (Stainier, 2001). The ventricular sarcomere gene vmhc is first expressed in ventricular cardiomyocyte precursors at 16 hpf (Yelon et al., 1999). In Tg(vmhc:EGFP) embryos, EGFP expression was first observed as the bilateral strips in the lateral plate mesoderm at 18 hpf (Fig. 1A), 2 hours later than endogenous vmhc expression revealed by in situ hybridization (Fig. 1F). These EGFP-expressing cells gradually migrated and coalesced into a cardiac cone at 20 hpf (Fig. 1B). When the heart tube formed at 24 hpf, EGFP expression was observed in the outflow region, the presumptive ventricle in the heart tube (Fig. 1C). After cardiac looping and chamber demarcation, EGFP expression was restricted in the ventricle at 48 hpf, when compared with EGFP expression in both chambers, the ventricle and the atrium, in Tg(cmlc2:EGFP) embryos (Burns et al., 2005) (Fig. 1D, E). The observed EGFP expression was consistent with the temporal and spatial expression of endogenous vmhc expression in the lateral mesoderm (Fig. 1F), the heart cone (Fig. 1G), the heart tube (Fig. 1H), and the ventricle (Fig. 1I). However, in Tg(vmhc:EGFP) embryos, EGFP expression was only observed in the ventricle and not in somites, whereas endogenous vmhc transcripts were detected in both the ventricle and somites (Fig. 1H). These findings suggest that the vmhc upstream fragment (1952 bp) is sufficient to drive EGFP expression in the ventricle, but lacks cis-elements for directing EGFP expression in somites.
Figure 1. Spatial and temporal expression of vmhc:EGFP in zebrafish embryos.
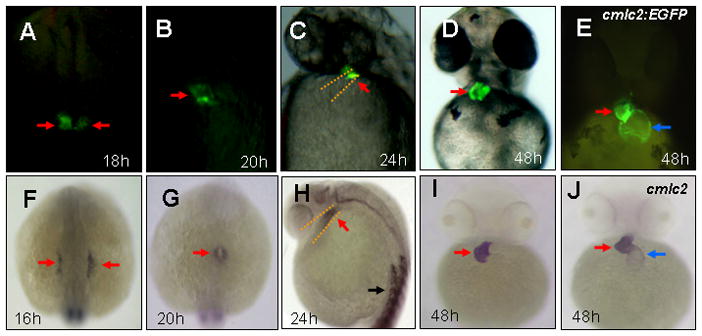
A–E: Fluorescent optics revealing expression of vmhc-EGFP in the lateral plate mesoderm (A), the cardiac cone (B), the ventricular portion of the heart tube (C) and the ventricle (D) in Tg[vmhc:EGFP] embryos, as well as cmlc2-EGFP expression in the ventricle and the atrium in Tg[cmlc2-EGFP] embryos (E). F–J: Whole-mount in situ analysis showing expression of vmhc in the lateral plate mesoderm (F), the cardiac cone (G), the ventricular portion of the heart tube (H) and the ventricle (I), as well as cmlc2 expression in both cardiac chambers (J). Dorsal views with anterior to the top (A,B,F,G). Lateral views (C,H). Ventral views (D,E,I, J). Red arrows: Ventricular myocytes: Blue arrows: Atrial myocytes. Black arrows: somites.
vmhc promoter analysis in live zebrafish embryos
The amino acid sequences of zebrafish Vmhc are 75% identical to mammalian β-MHC/MYH7 and 74% identical to mammalian α-MHC/MYH6. However, based on the similar ventricular-specific expression pattern of vmhc and β-MHC/MYH7 during heart development, vmhc is most likely the ortholog of β-MHC/MYH7. Thus, we compared the upstream regulatory region of zebrafish vmhc with mouse β-MHC and human MYH7, and identified several consensus cis-elements related to myocardial development, including binding sites of Nkx2.5, MEF2, GATA and Prx2/S8 transcription factors (Fig. 2).
Figure 2. Schematic representations of consensus binding elements within 1952-bp upstream region of zebrafish vmhc/mouse β-MHC/human MYH7, as well as a deletion series in the zebrafish vmhc promoter region.
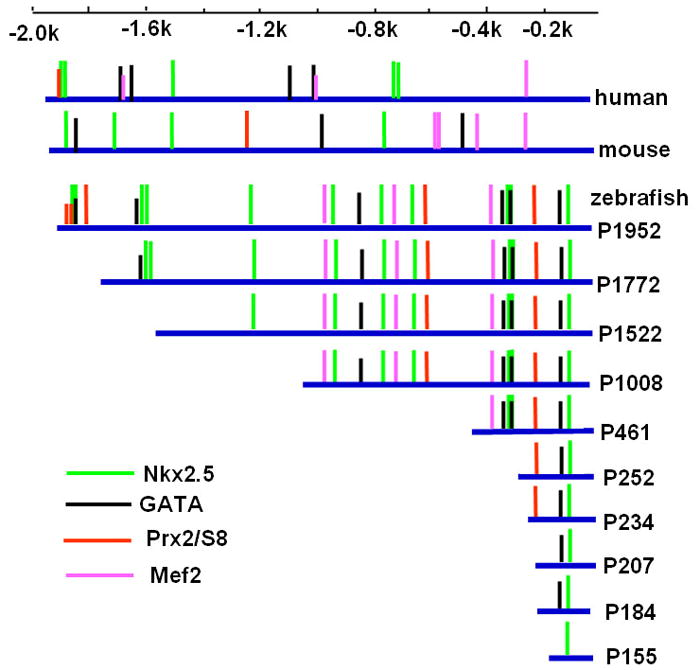
Vertical lines represent consensus binding sites for transcriptional factors of Nkx family, GATA family, Prx2/S8, and Mef2. All promoter deletions (blue lines) are linked to EGFP and SV40-polyA sequence (not shown).
To identify the minimal promoter that was sufficient for directing vmhc expression in the ventricle, a series of constructs containing progressive deletions from the 5′-end of 1952-bp fragment were generated by PCR using pvmhc-EGFP1 as a template. The resulting six deletion constructs (P1772, P1522, P1008, P461, P252 and P155) contained fragments of 1772 bp, 1522 bp, 1008 bp, 461 bp, 252 bp and 155 bp, respectively, which were linked to the EGFP gene and SV40 polyA signal (Fig. 2). The P1952 construct was generated as a non-deletion control. When P1952 was microinjected into one-cell embryos, approximately 39% of the injected embryos had EGFP expression in the ventricle and none exhibited EGFP expression in somites (Fig. 3E). This result was similar to those obtained with the original pvmhc-EGFP1 construct. Embryos microinjected with constructs P1772, P1522, P1008, P461 or P252 displayed gradual reduction in percentages of EGFP expression in ventricles (Fig. 3E). Notably, EGFP expression was not detected at all embryos microinjected with P155. To precisely map the minimal active promoter, three additional deletion constructs, P234, P207 and P184 were generated and microinjected into one-cell embryos. Only embryos injected with P234 had EGFP expression in the ventricle, and embryos injected with P207 or P184 did not show any EGFP expression at all (Fig. 3C,D;E). Thus, the 234-bp regulatory element contains the basal promoter that is sufficient to drive EGFP expression in the ventricle. This also suggests that 27 nucleotides spanning the region from −234 to −206 contain positive cis-elements required for vmhc expression.
Figure 3. Effects of vmhc promoter deletions on transgene EFFP expression in the ventricle.
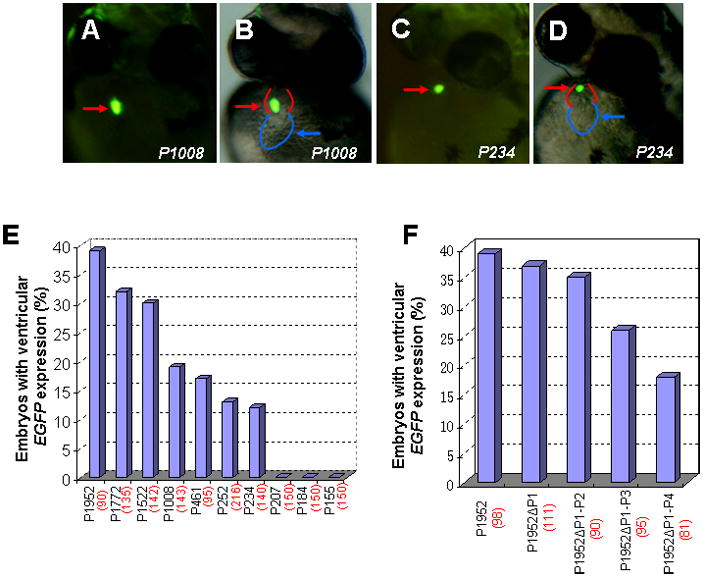
A–D: Embryos injected with a series of distal promoter deletion constructs at one or two cell stages were examined for transient EGFP expression at 48 hpf using fluorescent microscopy. Fluorescent optics showing ventricular EGFP expression in embryos injected with P1008 (A, B) and reduced EGFP expression in embryos with P234 injection (C, D). Reduced EGFP fluorescence typically correlates with smaller vmhc promoter. Red and blue lines sketch the ventricle and atrium, respectively. E: Bar graph showing percentages of embryos that express EGFP in the ventricle in microinjection of each deletion construct (P1952, P1772, P1522, P1008, P461, P252 and P234). F: Bar graph depicting percentages of ventricular EGFP-expressing embryos injected with Prx2/S8-deletion constructs (P1952 ΔP1, P1952 ΔP1-2, P1952 ΔP1-3, P1952Δ P1-4). The total number of embryos injected with each construct is shown in parenthesis. Ventral views (A–D).
Prx2/S8-like homeodomain elements are necessary for directing vmhc expression
The 27 nucleotides within the basal promoter (AACTAAATTAGCCCTCCGCT ATCAGAGAA) contained a homeobox transcription factor Prx2/S8-like binding site (AACTAAATTAGC). Prx2/S8 transcriptional factor, belonging to the family of paired-related homeobox gene family, are highly expressed in mesenchymal tissues throughout development, including the developing cardiovascular system (Leussink et al., 1995). The mouse Prx2/S8 homeodomain binds the 11-bp consensus element (ANC/TC/TAATTAA/GC) (de Jong et al., 1993). We hypothesized that the Prx2/S8-like element was necessary to mediate vmhc expression in the ventricle. To test this, five primers, each having two altered bases (cysteine) within the 27-bp core sequence of Prx2/S8-binding site were used to generate mutant forms of P234 constructs (Table 1). Embryos injected with constructs P234M1 or P234M2, each containing two base substitutions in the core sequence, showed completely absence of EGFP expression in the ventricle (Table 1). Mutation of two bases adjacent to the core sequence (P234M3 and P234M4) caused a marked decrease of EGFP expression in the ventricle (Table 1). In contrast, dinucleotide replacement (P234M5) that was 8-nucleotide apart from the conserved Prx2/S8-binding site resulted in the ventricular EGFP expression comparable to the one occurred in P234 injections (Table 1).
Table 1. Mutations in the Prx2/S8-binding site and their effects on EGFP expression in the ventricle.
| Construct | EGFP Expression | |
|---|---|---|
| P234: | 5′-CTAAATTAGCCCTCCGCTATCAGAGAA | 12% (n = 140) |
| P234M1: | 5′-CTAAATTCCCCCTCCGCTATCAGAGAA | 0% (n = 126) |
| P234M2: | 5′-CTAAACCAGCCCTCCGCTATCAGAGAA | 0% (n = 130) |
| P234M3: | 5′-CTAAATTAGGGCTCCGCTATCAGAGAA | 2% (n = 118) |
| P234M4: | 5′-CCCAATTAGCCCTCCGCTATCAGAGAA | 2% (n = 109) |
| P234M5: | 5′-CTAAATTAGCCCTCCGCCCTCAGAGAA | 10% (n = 105) |
Underlined: Prx2/S8 consensus binding site. n: total number of microinjections for each construct.
There were five conserved Prx2/S8-like binding sites within 1952 bp regulatory element. To test functional redundancy of these Prx2/S8 binding sites, we deleted these sites in P1952 constructs and established P1952 ΔP1, P1952 ΔP1-2, P1952 ΔP1-3, P1952 Δ P1-4 constructs with deletion of one-, two-, three- and five-Prx2/S8 binding sites, respectively. Embryos injected with P1952 ΔP1 or P1952 ΔP1-2 constructs did not significantly reduce EGFP expression percentages in ventricles compared to P1952-injected embryos (Fig. 3F). P1952 ΔP1-3 injection exhibited a marked reduction in EGFP expression, and embryos injected with P1952Δ P1-4 displayed a largest EGFP reduction (Fig. 3F). However, deletion of all Prx2/S8 elements did not completely eliminate vmhc expression in the P1952 constructs. Collectively, these data suggest that Prx2/S8-like homeodomain elements, together with other cis-elements, are necessary for directing vmhc expression.
vmhc promoter harbors atrial repression elements to govern ventricle-specific expression
Careful examination of embryos injected with deletion constructs revealed that P1522 injection resulted in approximately 20% embryos having EGFP expression in the atrium (Fig. 4A,B;E). The percentages of embryos with the atrial ectopic expression varied among deletion constructs, including P1772, P1008, P461, P252 and P234 (Fig. 4E). However, none of embryos injected with P1952, P207, P184 or P155 displayed ectopic expression in the atrium (Fig. 4E). Since nonspecific EGFP expression in tissues other than heart was typically observed less than 5% of embryos injected with these constructs, this suggests that the atrial ectopic expression is sequence specific, possibly due to regulatory cis-elements in the vmhc promoter. Microinjections of P1772 only caused a small percentage (3%) of embryos having atrial EGFP expression, a substantial reduction of ~ 17% when compared with the atrial ectopic expression in P1522 injections. However, the ventricular expression percentage of P1522 was comparable to P1772 (30% vs. 32%, Fig. 4E). These data suggest existence of negative cis-elements in the distal region (−1772/−1522) that prevent the atrium-specific expression. Compared to P1522, microinjections of P1008 and P461 progressively diminished embryos with the atrial expression to baseline levels, paralleling with those observed in ventricular expression, suggesting that positive elements exist in the proximal region (−1522/−461) to enable vmhc expression in both chambers.
Figure 4. vmhc promoter deletions caused ectopic EGFP expression in the atrium.

A–D: Fluorescent optics revealing the ectopic EGFP expression in the atrium and its normal ventricular expression in embryos with P1522 injection (A,B), as well as reduced EGFP expression in both chambers with P461 injection (C,D). Red and blue lines sketch the ventricle and atrium, respectively. E: Bar graph showing the percentage of embryos that express EGFP in the ventricle (blue) or the atrium (magenta) in embryos injected with each construct. The total number of embryos injected with each construct is shown in parenthesis. The ventricular expression data is the same as Fig.3.
Within the 251 bp region (−1772/−1522), there were two Nkx2.5 binding elements (NKE) and one GATA binding site (Fig. 2). Recent data indicate that Nkx2.5 and Nkx2.7 reduction causes a surplus of atrial cardiomyocyte number in zebrafish (Targoff et al., 2008), implicating that Nkx genes may have roles in atrial repression. In addition, Nkx2.5 and dHand act synergistically to modulate the expression of ventricular homeobox gene Irx4 in mouse (Bruneau et al., 2000). We thus deleted two Nkx2.5 binding elements in P1952 construct to generate P1952-ΔNKE constructs. Deletion of both Nkx2.5 elements caused ectopic EGFP expression in the atrium, which was comparable to the atrial expression percentage of P1522 injection (Fig. 4E). These data suggest that Nkx2.5 elements in the distal region (−1772/−1522) are required for repressing vmhc expression in the atrium.
Prrx1b, a Paired-related homeobox protein, participates in the regulation of vmhc expression
Our sequence analyses revealed several conserved transcriptional factor binding sites, including Mef2 elements, Nkx2.5 sites, Prx2/S8 elements and GATA binding sites, in the vmhc upstream regulatory region (Fig. 2). We previously reported that mef2a knockdown reduced endogenous vmhc expression (Wang et al., 2005). To determine whether mef2a regulated vmhc expression directly via its promoter region, we examined the effects of mef2a knockdown on the transgene EGFP expression. We injected transgenic embryos Tg(vmhc:EGFP) using mef2a antisense splicing morpholinos (mef2a-MO) (Wang et al., 2005), and found that mef2a reduction caused the decreased expression of transgene EGFP in the ventricle (Fig. 5A, B). We validated the EGFP reduction using real-time PCR analysis (Fig. 5E). Although mef2a knockdown caused the reduced vmhc expression, mef2a reduction failed to eliminate vmhc expression. These findings suggest that multiple transcriptional factors participate in the transcriptional regulation of vmhc, including mef2a.
Figure 5. mef2a and prrx1b knockdown reduced transgene EGFP expression.

A–D: Lateral view showing reduced EGFP expression in embryos injected with mef2a-MO and prrx1b-MO (B,D), when compared to embryos injected with control morpholinos (A,C). Red arrows: ventricle. Blue arrows: atrium. E–F: Bar chart depicting relative EGFP expression folds in mef2a and prrx1b morphants versus control embryos injected with mismatched morpholinos. Three independent experiments were conducted. Error bars indicate standard deviation, and asterisks indicate statistical significance between morphant and control embryos (p<0.01). G: Schematic graph depicting the prrx1b gene structure and the inhibitory splicing donor site targeted by prrx1b-MO. Red bar: prrx1b morpholinos. Red arrowhead: stop codon. Black arrow: sequence primers.
Searching the zebrafish genome database using mouse and human Prx1 or Prx2 revealed two zebrafish Prx isoforms, prrx1a and prrx1b. While prrx1a expression was detected in epidermis, musculature and pharyngeal arch, prrx1b expression was observed in the zebrafish heart (Marques et al., 2008)(Thisse et al., ZFIN direct submission). We therefore elected prrx1b to investigate its roles in regulating vmhc expression. We designed prrx1b antisense splicing morpholino oligonucleotides (prrx1b-MO) to target the donor site of intron 2 (Fig. 5G). Microinjection of prrx1b-MO into one- to two-cell transgenic embryos Tg(vmhc:EGFP) caused a reduction in EGFP expression in ventricles compared to controls (Fig. 5C, D). We validated a reduction of transgene EGFP in prrx1b morphant embryos using real-time PCR analyses (Fig. 5F). We next isolated total RNAs from prrx1b morphants, performed RT-PCR using primers indicated and sequenced PCR products (Fig. 5G). We found that prrx1b-MO injection caused retention of intron2 leading to generation of in-framed stop codon and Prrx1b protein truncation (Supplement data). Collectively, these data suggest that Prrx1b participates in the regulation of vmhc transcription, together with other transcriptional factors.
Discussion
In this study we established a transgenic zebrafish line that expresses EGFP in the ventricle under the control of a vmhc promoter, and described an important NKE-mediated atrial repression mechanism for governing ventricle-specific expression. We further discovered that Prx2/S8-like elements are necessary for directing vmhc expression in the ventricle and demonstrated paired-related homeobox protein Prrx1b participated in the regulation of vmhc expression.
Our studies indicated that an upstream regulatory region (1952 bp) of vmhc was sufficient to recapitulate the ventricular expression of vmhc during heart development. EGFP expression in Tg(vmhc:EGFP) embryo was first detected at the anterior lateral plate mesoderm, then restricted in the ventricular portion of the heart tube, and late in the formed cardiac ventricle. The spatial and temporal control of vmhc expression lies in its regulatory cis-elements within 1952 bp. By promoter dissection, we observed that deletion of a distal region (−1772/−1522) caused the ectopic expression of vmhc in the atrium. This suggests existence of an inhibitory mechanism that mediates atrial repression, which in turn ensures vmhc expression in the ventricle. There are several known cis-elements in the 251 bp region, including one GATA element and two Nkx2.5 binding sites. Notably, deletion of two Nkx2.5 sites in the context of −1952 upstream region relieves the atrial repression. This is consistent with the most recent reporter (Zhang and Xu, 2009). In addition to the distal region (−1772/−1522), Zhang et al also reported presence of the atrial repression elements in the proximal region (−100/+300) that contains first two exons and the first intron. Our P1952 construct does not contain the proximal region (−100/+300) and is sufficient to drive EGFP expression in the ventricle but not in the atrium, suggesting that Nkx2.5 binding sites in the distal region (−1772/−1522) play essential roles in repressing vmhc in the atrium. Studies in model systems suggest that Nkx2.5 can either promote ventricular gene expression or repress atrial gene expression in the ventricle. For example, Nkx2.5 regulates ventricular expression of Irx4 in mouse (Yamagishi et al., 2001), and can prevent expression of atrial natriuretic factor (anf) in the ventricle in Xenopus (Small and Krieg, 2003). In zebrafish, Nkx proteins limit atrial cell differentiation while promoting ventricular cell development (Targoff et al., 2008). Our findings thus describe an important Nkx2.5-mediated atrial repression mechanism for governing ventricle-specific expression, a novel extension of the roles of Nkx2.5 in regulating chamber-specific differentiation. It would be interesting to determine whether the ventricular expression of mammalian myosin heavy chain is also regulated by the NKE-mediated repression mechanism.
We revealed that the 234-bp basal promoter contained Prx2/S8-like homeodomain binding site (AACTAAATTAGC) and drived EGFP expression in the ventricle. Deletions or mutations of Prx2/S8-binding sites reduced or eliminated vmhc expression in the ventricle, suggesting that Prx2/S8 binding site was necessary for mediating the basal expression of vmhc. The Prx2/S8 binding site resembles the consensus sequence of Antennapedia (Antp)-type homeodomain proteins (de Jong et al., 1993). Like Antp family and paired-related homeobox genes, homeobox transcription factors are commonly recognized as important regulators of organogenesis and embryonic development. Functional analyses indicated a major role of murine Prx2/S8 in craniofacial and limb skeletogenesis as well as patterning and positioning of the aortic arch and the cardiac outflow tract (Kuratani et al., 1994; Leussink et al., 1995; Bergwerff et al., 2000). Zebrafish genome contains Prrx1a and Prrx1b (two Prx-related genes) and their functions during cardiac development have not been previously reported in zebrafish. In this study, we elected prrx1b due to its expression in the heart, as prrx1a is expressed in epidermis, musculature and pharyngeal arch (Marques et al., 2008)(Thisse et al., ZFIN direct submission). prrx1b knockdown caused the reduction of transgene EGFP and slightly small size of the ventricle. These cardiac defects are similar to mef2a morphants that displayed both the reduction of transgene EGFP and small ventricle size. In combination with the presence of Prx/S8 elements and Mef2 binding sites in the vmhc promoter, these data suggest that both prrx1b and mef2a participate in the regulation of vmhc transcription via possibly interacting with the transgene promoter. It would be next important step to investigate whether Prrx1b interacts with other cofactors (e.g., Mef2a and Nkx2.5) to activate vmhc, and whether and how Prrx1b binds to the vmhc promoter. Our analyses also indicated that Prx2/S8 elements exist in the upstream region of mouse β-MHC and human MYH7, which was not previously revealed. It will be important and interesting to determine whether Prx2/S8 homeodomain element is necessary for directing the expression of mammalian β-MHC/MYH7, and whether Prx1/Prx2 transcriptional factors act independently or collaboratively with other cofactors to regulate expression of ventricular myosin heavy chains in mammals.
Materials and methods
Zebrafish strains
Zebrafish (Danio rerio) embryos were obtained from natural spawning between wild-type AB lines (Oregon stock centre). Embryos were staged and maintained at 28.5°C as described (Westerfield, 1995).
Generation of vmhc:EGFP transgenic lines
The vmhc upstream regulatory region (1952 bp) was amplified and cloned from the genomic sequence (GeneBank ID 163644330) using primers (forward: 5′-ACTCCGC GGAGGCCATGTGTCCTAAATTCTG; reversed: 5′-ATCGGATCCGAACACCAA CCATGAGATCACT). The PCR products were digested with SacI/BamH I, gel-purified and subcloned into pEGFP-1 vector (pvmhc-EGFP1). The cloned vmhc promoter region was subjected to sequencing. The pvmhc-EGFP1 plasmid DNA was linearized with restriction enzyme Sac I and isolated following electrophoresis in 1% melting agarose gel. Approximately 200–300 pg of linearized DNA was injected into one- to two-cell stage embryos. Mosaic embryos displaying ventricle-specific EGFP expression were raised to adulthood. Pairs of male and female founders were in-crossed. If EGFP expression was found in offspring from the founder pairs, EGFP-positive embryos derived from founder pairs were bred to F1 adult fish.
Search for transcriptional factor binding sites
The transcription factor binding sites in the upstream region of zebrafish vmhc, mouse β-MHC and human MYH7 were identified using the following computation programs: MatInspector, V7.1 (http://www.genomatix.de) and TFsearch, V1.3 (http://www.cbrc.jp/research/db/TFSEARCH.html)
Generation of deletion constructs
To identify the minimal promoter, a series of 5′-deletion constructs within the 1952-bp fragment was generated by PCR using pvmhc-EGFP1 as template. A total of 10 primers for vmhc promoter sequences and a primer for pEGFP-1 vector sequences were used to produce a series of 5′-distal deletion constructs. All deletion constructs contained deleted vmhc promoter region, EGFP gene and SV40 polyA signal. These constructs were P1952, P1772, P1522, P1008, P461, P252, P234, P207, P184 and P155, in which the numbers refer to the distal nucleotide positions upstream of the vmhc transcriptional start site. These PCR products were gel-purified and used directly for microinjections. Approximately 200–300 pg of each construct was injected into one- to two-cell stage embryos. Primers are listed below:
Pm1952: 5′-AGGCCATGTGTCCTAAATTCTG;
Pm1772: 5′-GAATGTGAGCACAAGTCGGAGTG;
Pm1522: 5′-CATTAACGTCTGCCAATTGCACG;
Pm1008: 5′-GATTCACTCTGCAGGAATTTGAC;
Pm461: 5′-GTTGTCTGTTAAACCCTCACAGG;
Pm252; 5′-CTTCCCCCTGTCTCTGAACTAAAT;
Pm234: 5′-CTAAATTAGCCCTCCGCTATCAGAGAA;
Pm207: 5′-CCTAAAATGGAGAGCTAACAAAT;
Pm184: 5′-CTCTACAAAGATAAGACTTAACTCAACTA;
Pm155: 5′-GCAGGTTCTTGTTTCTCGCTAA;
PmEGFP: 5′-GAACAACACTCAACCCTATCTCG.
Generation of deletions and mutations of Prx2/S8 or Nkx2.5 binding site
All Prx2/S8-deletion constructs were made one by one with QuickChange® site-directed mutagenesis kit (Stratagene). The Prx2/S8-deletion construct p1952Δp1 (−234/−218) was made using pvmhc-EGFP1 as a template, and the p1952Δp1-2 (−234/−218; −616/−600) was made using p1952Δp1 as a template. The p1952Δp1-3 (−234/−218; −616/−600; −1808/−1792) and the p1952Δp1-4 (−234/−218; −616/−600; −1808/−1792; −1862/−1846; −1855/−1939) were made in the same manner. Primers are listed below.
-
L1
5′-CTCTTCCCCCTGTCTCTGAACCTCCGCTATCAGAGAACCT
-
R1
5′-AGGTTCTCTGATAGCGGAGGTTCAGAGACAGGGGGAAGAG
-
L2
5′-CCCTCCTTAAGCTCTCTGGCATGAGGCATGCAGTTC
-
R2
5′-GAACTGCATGCCTCATGCCAGAGAGCTTAAGGAGGG
-
L3
5′-CATTCATGGATAATGTGAGACCAGTGAGAACAAAAT
-
R3
5′-ATTTTGTTCTCACTGGTCTCACATTATCCATGAATG
-
L4
5′-TACAGTACAGGCCTTATCCAGCCAGCTGTTATGCTG
-
R4
5′-CAGCATAACAGCTGGCTGGATAAGGCCTGTACTGTA
P1952ΔNKE constructs was made with QuickChange® site-directed mutagenesis kit (Stratagene), and primers used for P1952ΔNKE were listed below.
-
L1
5′-CAGAACAGAACAGTCTTATCTCCATGTTTATCTGTTGA
-
R1
5′-TCAACAGATAAACATGGAGATAAGACTGTTCTGTTCTG
Mutant Constructs P234M1, P234M2, P234M3, P234M4 and P234M5 were generated by PCR using mutant primers and each containing 2-bp mutant bases (Table 2; underlined). These primers were used in conjunction with the downstream EGFP primer for PCR using pvmhc-EGFP1 as the template. These PCR products were gel-purified and used directly for microinjections.
Microinjection of antisense morpholino oligonucleotides
prrx1b (5′-TGAGGTGTGAAGTTTACCTGAACTC) splicing morpholinos were designed and synthesized (Gene tools, inc.). Morpholinos were dissolved in 1X Danieau buffer (58 mM NaCl, 0.7 mM KCl, 0.4 mM MgSO4, 0.6 mM CaNO3, 5mM HEPES, pH7.6) and were injected into one- to two-cell embryos (10ng/embryo). EGFP expression was observed under SZX-RFL3 fluorescent lens attached to Olympus SZX12 microscope and was photographed using a DP70 digital camera. Merged white and fluorescent images were processed using Olympus DP Controller software.
Real-time PCR analysis
For relative quantification, the reactions were performed in a total volume of 20 μL, containing 10 μL of SYBR® Premix Ex Taq™ Master Mix (TaKaLa, Japan), 10 ng of cDNA, and 4 pmol of each primer. Real-time PCR was performed in Rotor-Gene 3000 Instrument (Corbett research, Australia). The samples were analyzed in triplicate. β-actin was used as an internal control. Assay results were collected and analyzed using the Rotor-Gene Real-Time Analysis Software 6.0 (Corbett research, Australia). Relative quantification of target gene expression was evaluated using the comparative CT method (Schmittgen and Livak, 2008). The primer sequences used are listed.
β-actin: 5′-TGCTGTTTTCCCCTCCATTG
β-actin: 5′-TCTGTCCCATGCCAACCAT
EGFP: 5′-AGCAAAGACCCCAACGAGAA
EGFP: 5′-GCGGCGGTCACGAACTC
Supplementary Material
Acknowledgments
We are grateful to members of our laboratories for comments on the manuscript and helpful discussions. We acknowledge Dong Liu, Yuexiang Wang and John Guan for invaluable assistance in promoter isolation, microinjection and fish cares. This research is supported in part by grants from NIH (TPZ) and American Heart Association (TPZ).
Funded by:
The National Institute of Health; Grant Number: HL073348
American Heart Association; Grant Number: 0455439B
Footnotes
Supplemental Information: Total RNAs were isolated from prrx1b morphant embryos, and subjected to RT-PCR using primers indicated (Fig. 5G). These PCR products were directly sequenced and in-framed stop codon in intron regions were identified and underlined. Base pairs in intron regions are indicated using small letters, and base pairs in the exon are labeled using large letters.
References
- Berdougo E, Coleman H, Lee DH, Stainier DY, Yelon D. Mutation of weak atrium/atrial myosin heavy chain disrupts atrial function and influences ventricular morphogenesis in zebrafish. Development. 2003;130:6121–6129. doi: 10.1242/dev.00838. [DOI] [PubMed] [Google Scholar]
- Bergwerff M, Gittenberger-de Groot AC, Wisse LJ, DeRuiter MC, Wessels A, Martin JF, Olson EN, Kern MJ. Loss of function of the Prx1 and Prx2 homeobox genes alters architecture of the great elastic arteries and ductus arteriosus. Virchows Arch. 2000;436:12–19. doi: 10.1007/pl00008193. [DOI] [PubMed] [Google Scholar]
- Bruneau BG, Bao ZZ, Tanaka M, Schott JJ, Izumo S, Cepko CL, Seidman JG, Seidman CE. Cardiac expression of the ventricle-specific homeobox gene Irx4 is modulated by Nkx2–5 and dHand. Dev Biol. 2000;217:266–277. doi: 10.1006/dbio.1999.9548. [DOI] [PubMed] [Google Scholar]
- Burns CG, Milan DJ, Grande EJ, Rottbauer W, MacRae CA, Fishman MC. High-throughput assay for small molecules that modulate zebrafish embryonic heart rate. Nat Chem Biol. 2005;1:263–264. doi: 10.1038/nchembio732. [DOI] [PubMed] [Google Scholar]
- Ching YH, Ghosh TK, Cross SJ, Packham EA, Honeyman L, Loughna S, Robinson TE, Dearlove AM, Ribas G, Bonser AJ, Thomas NR, Scotter AJ, Caves LS, Tyrrell GP, Newbury-Ecob RA, Munnich A, Bonnet D, Brook JD. Mutation in myosin heavy chain 6 causes atrial septal defect. Nat Genet. 2005;37:423–428. doi: 10.1038/ng1526. [DOI] [PubMed] [Google Scholar]
- de Jong R, van der Heijden J, Meijlink F. DNA-binding specificity of the S8 homeodomain. Nucleic Acids Res. 1993;21:4711–4720. doi: 10.1093/nar/21.20.4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards JG, Bahl JJ, Flink IL, Cheng SY, Morkin E. Thyroid hormone influences beta myosin heavy chain (beta MHC) expression. Biochem Biophys Res Commun. 1994;199:1482–1488. doi: 10.1006/bbrc.1994.1398. [DOI] [PubMed] [Google Scholar]
- Everett AW. Isomyosin expression in human heart in early pre- and post-natal life. J Mol Cell Cardiol. 1986;18:607–615. doi: 10.1016/s0022-2828(86)80968-3. [DOI] [PubMed] [Google Scholar]
- Flink IL, Edwards JG, Bahl JJ, Liew CC, Sole M, Morkin E. Characterization of a strong positive cis-acting element of the human beta-myosin heavy chain gene in fetal rat heart cells. J Biol Chem. 1992;267:9917–9924. [PubMed] [Google Scholar]
- Keegan BR, Meyer D, Yelon D. Organization of cardiac chamber progenitors in the zebrafish blastula. Development. 2004;131:3081–3091. doi: 10.1242/dev.01185. [DOI] [PubMed] [Google Scholar]
- Kuratani S, Martin JF, Wawersik S, Lilly B, Eichele G, Olson EN. The expression pattern of the chick homeobox gene gMHox suggests a role in patterning of the limbs and face and in compartmentalization of somites. Dev Biol. 1994;161:357–369. doi: 10.1006/dbio.1994.1037. [DOI] [PubMed] [Google Scholar]
- Lee Y, Nadal-Ginard B, Mahdavi V, Izumo S. Myocyte-specific enhancer factor 2 and thyroid hormone receptor associate and synergistically activate the alpha-cardiac myosin heavy-chain gene. Mol Cell Biol. 1997;17:2745–2755. doi: 10.1128/mcb.17.5.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leussink B, Brouwer A, el Khattabi M, Poelmann RE, Gittenberger-de Groot AC, Meijlink F. Expression patterns of the paired-related homeobox genes MHox/Prx1 and S8/Prx2 suggest roles in development of the heart and the forebrain. Mech Dev. 1995;52:51–64. doi: 10.1016/0925-4773(95)00389-i. [DOI] [PubMed] [Google Scholar]
- Long Q, Meng A, Wang H, Jessen JR, Farrell MJ, Lin S. GATA-1 expression pattern can be recapitulated in living transgenic zebrafish using GFP reporter gene. Development. 1997;124:4105–4111. doi: 10.1242/dev.124.20.4105. [DOI] [PubMed] [Google Scholar]
- Lyons GE, Schiaffino S, Sassoon D, Barton P, Buckingham M. Developmental regulation of myosin gene expression in mouse cardiac muscle. J Cell Biol. 1990;111:2427–2436. doi: 10.1083/jcb.111.6.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques IJ, Leito JT, Spaink HP, Testerink J, Jaspers RT, Witte F, van den Berg S, Bagowski CP. Transcriptome analysis of the response to chronic constant hypoxia in zebrafish hearts. J Comp Physiol [B] 2008;178:77–92. doi: 10.1007/s00360-007-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner JD, Umeda PK, Chang KC, Gros G, Scheibe RJ. Activation of the beta myosin heavy chain promoter by MEF-2D, MyoD, p300, and the calcineurin/NFATc1 pathway. J Cell Physiol. 2007;211:138–148. doi: 10.1002/jcp.20916. [DOI] [PubMed] [Google Scholar]
- Molkentin JD, Jobe SM, Markham BE. Alpha-myosin heavy chain gene regulation: delineation and characterization of the cardiac muscle-specific enhancer and muscle-specific promoter. J Mol Cell Cardiol. 1996;28:1211–1225. doi: 10.1006/jmcc.1996.0112. [DOI] [PubMed] [Google Scholar]
- Morimoto T, Hasegawa K, Kaburagi S, Kakita T, Masutani H, Kitsis RN, Matsumori A, Sasayama S. GATA-5 is involved in leukemia inhibitory factor-responsive transcription of the beta-myosin heavy chain gene in cardiac myocytes. J Biol Chem. 1999;274:12811–12818. doi: 10.1074/jbc.274.18.12811. [DOI] [PubMed] [Google Scholar]
- Morkin E. Control of cardiac myosin heavy chain gene expression. Microsc Res Tech. 2000;50:522–531. doi: 10.1002/1097-0029(20000915)50:6<522::AID-JEMT9>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Motoike T, Loughna S, Perens E, Roman BL, Liao W, Chau TC, Richardson CD, Kawate T, Kuno J, Weinstein BM, Stainier DY, Sato TN. Universal GFP reporter for the study of vascular development. Genesis. 2000;28:75–81. doi: 10.1002/1526-968x(200010)28:2<75::aid-gene50>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Seidman JG, Seidman C. The genetic basis for cardiomyopathy: from mutation identification to mechanistic paradigms. Cell. 2001;104:557–567. doi: 10.1016/s0092-8674(01)00242-2. [DOI] [PubMed] [Google Scholar]
- Small EM, Krieg PA. Transgenic analysis of the atrialnatriuretic factor (ANF) promoter: Nkx2-5 and GATA-4 binding sites are required for atrial specific expression of ANF. Dev Biol. 2003;261:116–131. doi: 10.1016/s0012-1606(03)00306-3. [DOI] [PubMed] [Google Scholar]
- Stainier DY. Zebrafish genetics and vertebrate heart formation. Nat Rev Genet. 2001;2:39–48. doi: 10.1038/35047564. [DOI] [PubMed] [Google Scholar]
- Targoff KL, Schell T, Yelon D. Nkx genes regulate heart tube extension and exert differential effects on ventricular and atrial cell number. Dev Biol. 2008;322:314–321. doi: 10.1016/j.ydbio.2008.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YX, Qian LX, Yu Z, Jiang Q, Dong YX, Liu XF, Yang XY, Zhong TP, Song HY. Requirements of myocyte-specific enhancer factor 2A in zebrafish cardiac contractility. FEBS Lett. 2005;579:4843–4850. doi: 10.1016/j.febslet.2005.07.068. [DOI] [PubMed] [Google Scholar]
- Westerfield M. The Zebrafish Book. Oregon: University of Oregon Press; 1995. [Google Scholar]
- Yamagishi H, Yamagishi C, Nakagawa O, Harvey RP, Olson EN, Srivastava D. The combinatorial activities of Nkx2.5 and dHAND are essential for cardiac ventricle formation. Dev Biol. 2001;239:190–203. doi: 10.1006/dbio.2001.0417. [DOI] [PubMed] [Google Scholar]
- Yelon D, Horne SA, Stainier DY. Restricted expression of cardiac myosin genes reveals regulated aspects of heart tube assembly in zebrafish. Dev Biol. 1999;214:23–37. doi: 10.1006/dbio.1999.9406. [DOI] [PubMed] [Google Scholar]
- Zhang R, Xu X. Transient and transgenic analysis of the zebrafish ventricular myosin heavy chain (vmhc) promoter: An inhibitory mechanism of ventricle-specific gene expression. Dev Dyn. 2009 doi: 10.1002/dvdy.21929. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XY, Rodaway AR. SCL-GFP transgenic zebrafish: in vivo imaging of blood and endothelial development and identification of the initial site of definitive hematopoiesis. Dev Biol. 2007;307:179–194. doi: 10.1016/j.ydbio.2007.04.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


