Abstract
Non-small cell lung cancer (NSCLC) is notorious for its paltry responses to first-line therapeutic regimens. In contrast to acquired chemoresistance, little is known about the molecular underpinnings of the intrinsic resistance of chemo-naïve NSCLC. Here we report that intrinsic resistance to paclitaxel in NSCLC occurs at a cell-autonomous level due to the uncoupling of mitotic defects from apoptosis. To identify components that permit escape from mitotic stress-induced death, we employed a genome-wide RNAi-based strategy, which combines a high-throughput toxicity screen with a live-cell imaging platform to measure mitotic fate. This strategy revealed that prolonging mitotic arrest with a small molecule inhibitor of the APC/Cyclosome could sensitize otherwise paclitaxel-resistant NSCLC. We also defined novel roles for CASC1 and TRIM69 in supporting resistance to spindle poisons. CASC1, which is frequently co-amplified with KRAS in lung tumors, is essential for microtubule polymerization and satisfaction of the spindle assembly checkpoint. TRIM69, which associates with spindle poles and promotes centrosomal clustering, is essential for formation of a bipolar spindle. Notably, RNAi-mediated attenuation of CASC1 or TRIM69 was sufficient to inhibit tumor growth in vivo. On the basis of our results, we hypothesize that tumor evolution selects for a permissive mitotic checkpoint, which may promote survival despite chromosome segregation errors. Attacking this adaptation may restore the apoptotic consequences of mitotic damage to permit the therapeutic eradication of drug-resistant cancer cells.
Keywords: mitotic slippage, pan-genomic RNAi screen, paclitaxel, CASC1, TRIM69
Introduction
Paclitaxel is a first-line chemotherapeutic agent that inhibits the dynamic instability of microtubules, thus preventing bi-orientation of chromosomes during mitosis (1). While the reduction of breast and ovarian tumor burden following taxane-based therapies demonstrates efficacy in these settings, responses in NSCLC are rarely curative, as only 30% of patients exhibit a partial response at best, indicating a widespread intrinsic resistance to anti-mitotic agents (2, 3). Thus, given the promise, yet limitation, of current anti-mitotic therapies, the identification of mechanisms supporting intrinsic resistance to paclitaxel in NSCLC is essential. Our goal here was to identify the cell autonomous components that permit escape from mitotic stress-induced cell death in a paclitaxel resistant NSCLC setting.
Efficacy of paclitaxel and other anti-mitotic agents hinges on the coupling of mitotic defects to cell death. By inhibiting the dynamic instability of microtubules, paclitaxel disrupts chromosome alignment, thereby preventing satisfaction of the spindle assembly checkpoint (SAC). The SAC is composed of sentinel proteins including MAD2 and BUBR1, which, in the absence of proper microtubule-kinetochore attachments, inhibit the activity of the Anaphase Promoting Complex/Cyclosome (APC/C) (4, 5). Nearly all tumor cells are sensitive to paclitaxel-induced mitotic defects and engage the SAC. However, live-cell imaging studies have revealed that the length and outcome of this mitotic arrest is variable within and among tumor cell lines. For example, apoptosis may be activated directly from mitotic arrest. Otherwise, arrested cells undergo mitotic slippage, defined as an aberrant exit in the presence of misaligned chromosomes thereby forming micronucleated cells that can either die, arrest or reenter a subsequent division cycle (6–8). This slippage from an SAC-mediated mitotic arrest has been implicated as a survival mechanism, as delaying mitotic exit, either by inhibiting an activator of the APC/C, CDC20, or overexpressing cyclin B1, can increase mitotic dwell time and cell death during or following mitosis (6, 9). The prolonged mitotic arrest may allow for accumulation of death signals that trigger apoptosis (10, 11). However, the molecular components that promote mitotic slippage, particularly in chemoresistant tumor cell lines are unclear.
Previously, we established a pan-genomic, siRNA-based screening platform to identify gene products that modulate sensitivity of NSCLC cells to paclitaxel (12). Here, we employ this platform to investigate the molecular components supporting resistance to paclitaxel in a NSCLC system that exhibits no loss of viability following exposure to doses as high as 1 μM. At single cell resolution, we find that resistant cells undergo mitotic slippage and survive as micronucleated cells, indicative that resistance is due to a regulatory setting that permits the deflection of mitotic stress-induced cell death. In agreement with this notion, a global screen discovered components, which when depleted, sensitize these chemorefractory cells to paclitaxel. Suppression of a cohort of these chemosensitizers induced a protracted mitotic arrest, which we find is essential for post-mitotic cell death. Functional elaboration of these chemosensitizers reveals that prolonging mitotic arrest can be accomplished either by direct inhibition of the APC/Cyclosome (APC/C), or through collateral spindle damage, due to the depletion of CASC1 or TRIM69, which we reveal here are novel regulators of mitotic spindle assembly in tumor cells. Thus, prolonging mitotic arrest, which can be achieved through multiple avenues, is a dominant mechanism to sensitize otherwise paclitaxel-resistant NSCLC cells.
Materials and Methods
Cells and Reagents
HBEC, NSCLC, 293 and HeLA cell were a gift from John Minna and Michael White. HeLa, H1299, HCC4017, HCC1171, H1299, H1155 and HBEC lines were validated by Short Tandem Repeat analysis. NSCLC cell lines were maintained in RPMI medium (Gibco) with 5% fetal bovine serum (FBS). HBEC cell lines were maintained in keratinocyte medium plus supplements (Gibco). HeLa and 293 cells were maintained in DMEM with 10% FBS. Paclitaxel (Sigma or Tocris), ProTAME (Boston Biochem) and Nocodazole (Sigma) were dissolved in Dimethyl Sulfoxide (DMSO). Cell-Titer Glo® (CTG) and APO-ONE® were obtained from Promega. Dharmafect transfection reagents were obtained from Thermo-Fisher. Control siRNA transfections were performed with either non-targeting siRNA pool (Dharmacon) or a pool targeting DLNB14(12).
Paclitaxel Dose Curves
Cell lines were plated in 96-well plates to reach 50% confluence at 48 hours, when they were exposed to paclitaxel for 48 hours. Cell viability assessed using the Cell-Titer Glo® assay.
Immunoblotting
Cells were lysed in boiling 2X Laemmli sample buffer and resolved by SDS-PAGE. Antibodies used as follows: Actin, ANAPC5, CASC1, GAPDH, MCL-1, PDE3B, Myc-A14, Myc-9E10, GST (Santa Cruz); Cleaved Caspase-3 (Epitomics); MAD2L1 (Covance); TRIM69, β-tubulin (Sigma); Pericentrin (Abcam); phospho-histone-H3 (Millipore).
Immunofluorescence
Cells were grown on glass coverslips, fixed in 3.7% formaldehyde, permeabilized with 0.5% Triton X-100 and blocked with PBTA (PBS, 0.1% Tween-20, 10 mg/ml bovine serum album). Slips were incubated with primary antibodies in PBTA and, following PBTA washes, incubated with AlexaFluor-conjugated secondary antibodies (Invitrogen). For BUBR1 staining, cells were fixed in cold methanol. For microtubule preservation, cells were extracted with BRB80 (80 mM PIPES pH6.8, 1 mM MgCl2, 5 mM EGTA) and 0.5% Triton X-100 for 30 seconds followed by a 0.5% glutaraldehyde fix and 0.1% NaBH4 quench. Slides were imaged on an Axioimager upright microscope (Zeiss) equipped with a charge-coupled device (CCD) camera or a Leica CTR5500 upright scope with a monochrome digital camera (Leica DFC340 FX). Quantitation of fluorescence was performed with ImageJ software and normalized to cytoplasmic fluorescence.
Stable cell line production
Cell lines stably expressing green fluorescent protein-histone H2B (GFP-H2B) or myc-TRIM69A were generated by retroviral transduction. Retrovirus was produced by transfection of 293GP cells with vesicular stomatitis virus G protein (VSV-G) and either pCLNCX-GFP-H2B (gift from Gray Pearson) or pLPCX-myc-TRIM69A. Virus was harvested 48 hours post-transfection and transduced cells were selected with Geneticin® (pCLNCX-GFP-H2B) or Puromycin (pLPCX-myc-TRIM69A).
High-content live–cell imaging
Cell lines stably expressing GFP-H2B were transfected in a 96-well plate and exposed to vehicle or paclitaxel 48 hours post transfection and, unless otherwise indicated, imaged immediately following the addition of paclitaxel. Imaging was performed on a BD Pathway™ 855 Bioimager using a 40x or a 20x high-numerical-aperture objective. Images were taken every 20 minutes for 48–72 hours. Image sequences were generated using ImageJ and manually quantified.
siRNA screen
A two-condition, triplicate analysis screen was performed in a 96-well plate format using a Thermo-Fisher library that targets 21,127 unique genes (12). HCC366 cells were transfected with 40 nM siRNAs for 48 hours, followed by exposure to vehicle or 10 nM paclitaxel for an additional 48 hours. Subsequently, CTG was added and values were read on an EnVision® Plate Reader. The screening protocol was identical to that previously described with the following exceptions: HCC366 cells were seeded at 1×104, transfected with Dharmafect 2 complexed siRNAs in RPMI medium and cells were treated at 48 hours post transfection with vehicle or paclitaxel and a final FBS concentration of 10% (12). High interest chemosensitizers were ranked by z-score (Supplemental methods).
siRNA Transfections
siRNA transfections were performed using Thermo-Fisher of Sigma siRNA pools. Pools were used at a final concentration of 50 nM for indicated times.
Quantitative Reverse Transcription PCR
Total RNA was collected using the GenElute Mammalian Total RNA Miniprep Kit (Sigma). 2μg total RNA was used in subsequent reverse transcription using the High-Capacity cDNA reverse transcription kit (Applied Biosystems). Quantitative reverse transcription PCR (qRT-PCR) was performed with TaqMan gene expression assays and ribosomal protein L27 (RPL27) was the endogenous control.
Colony formation assays
HCC366 cells were transfected in a 24-well format and exposed to 0 and 10 nM paclitaxel 48 hours post-transfection. 96 hours post transfection 2×103 viable cells were replated in a 6-well format. Cells were fed biweekly for up to 3 weeks prior to formaldehyde fixation and Geimsa staining. Colonies were counted manually.
Microtubule regrowth assay
Cells were exposed to 11 μM nocodazole for 1 hour, 72 hours post-transfection. Cells were then rinsed with PHEM buffer (60 mM PIPES, 25 mM HEPES, 10 mM EGTA, 2 mM MgCl2, 1 μM Paclitaxel) and placed in fresh media to recover for the time indicated. Cells were then permeabilized with 0.2% Triton X-100 for one minute then fixed and immunostained.
In vivo polymerized tubulin assay
72 hours post-transfection, cells were lysed in microtubule stabilizing buffer (100 mM PIPES, 2 M glycerol, 0.1 M MgCl2, 2 mM EGTA, 0.5% TritonX-100, 5 μM Paclitaxel). Subsequently, whole cell lysates samples were collected and centrifuged for 30 minutes at 4°C and 16,000g. The supernatant (monomeric tubulin) was collected and pellet (polymerized tubulin) resuspended in microtubule stabilizing buffer. Fractions were resolved by SDS-PAGE.
cDNA expression and plasmids
Cells were transfected with cDNA expression vectors using Lipofectamine 2000 (Invitrogen). Plasmids used: Tomato-H2B, pCMV-myc (Clontech), mCMV-myc-TRIM69A, pCMV-myc-TRIM69B and pCMV-myc-TRIM69A(C50S/C53S).
Tumor Xenografts
pLKO.1 vectors expressing short hairpin RNA (shRNA) were used to generate lentivirus to infect HCC366 cells. Target knockdown was assessed 72 hours post-infection. Cells were harvested 96 hours post-infection and 2 ×106 cells were injected into the flank of female NSG (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ JAX®) mice. 3 ×106 cells were used for taxol studies. All mice were maintained in sterile conditions according to an approved IACUC protocol and abiding by all UNC Animal Welfare guidelines. Tumor growth was monitored by caliper measurement at indicated time points. In paclitaxel experiments, mice were treated with 20 mg/kg of paclitaxel starting 10 days post injection, at a frequency of two days per week for 4 weeks.
Hematoxylin and eosin staining (H&E)
Tumors from xenograft mouse studies were excised and formalin fixed and paraffin embedded. Tissue was processed by routine microtomy into 5–6 micron sections for automated staining. At least 7 fields were manually quantitated for micronucleated cells.
TUNEL staining
Terminal deoxynucleotidyltransferase-mediated UTP end label (TUNEL) staining for apoptotic cells in tissue sections was performed using the DeadEnd Fluorometric TUNEL System (Promega) standard protocol. Apoptotic cells were labeled with fluorescein, and the sections were counterstained with propidium iodide. At least 6 fields were counted per sample.
Results
Pan-genomic screening reveals components that liberate tumor cells from mitotic stress induced death
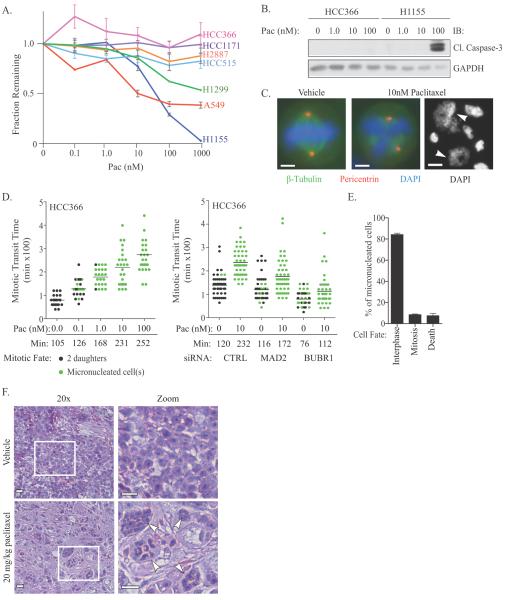
As a discovery platform for identifying mechanisms that promote resistance to paclitaxel-induced cell death, we selected HCC366 cells, a member of a cohort of NSCLC cell lines, which undergo limited viability loss and no activation of caspase-3 following exposure to escalating doses of paclitaxel (Figure 1A and B). While cell lines in this cohort exhibit minimal viability loss following paclitaxel exposure, a 10 nM dose is sufficient to induce multipolar spindles, misaligned chromosomes, and generate micronucleated progeny (Figure 1C, Supplemental 1A). Indicating that these cells have an intact SAC, live-cell imaging studies demonstrated that paclitaxel induced a MAD2 and BUBR1 dependent mitotic arrest that uniformly led to mitotic slippage and the generation of micronucleated progeny (Figure 1D). Single cell lineage tracing of HCC366 micronucleates, beginning 72 hours post paclitaxel exposure, revealed that the majority (92%) of these cells continued to survive for 40 hours and 8 % successfully underwent a subsequent division cycle (Figure 1E, Supplemental Figure 1B, Supplemental Video 1). Replating assays revealed that 40–50 % of treated cells were capable of colony formation, indicating that these cells, despite mitotic damage, maintain the capacity to proliferate (Supplemental Figure 1C). In contrast, a representative sensitive cell line, H1155, undergoes a dramatic mitotic arrest resolved by either cell death or micronucleation at 100 nM (Supplemental Figure 1D). The HCC366 mitotic slippage response was also recapitulated in vivo, as tumors grown subcutaneously in immune deficient mice exposed to paclitaxel were composed of micronucleated cells (Figure 1F). This finding suggests that HCC366 cells harbor a regulatory context in which mitotic stress is uncoupled from tumor cell death, leading to paclitaxel resistance and continued tumor cell proliferation.
Figure 1.
(A) Paclitaxel dose curves in indicated NSCLC cell lines. Each point is the mean ± Standard Error of the Mean (SEM) for 3 independent experiments. (B) Whole cell lysates (WCLs) from HCC366 and H1155 exposed to paclitaxel for 48 hours were immunoblotted as indicated. (C) Immunostaining of HCC366 cells treated for 24 hours. Arrowheads indicate micronucleated cells. Scale = 5 μm or 15 μm (right panel). (D) Single-cell lineage tracing of HCC366 cells stably expressing GFP-H2B (HCC366-GFP-H2B) for 48 hours post-paclitaxel exposure. Each circle represents a single cell. Bar is mean mitotic transit time, which is also indicated. Right panel: Cells were transfected with indicated siRNAs for 48 hours prior to drug exposure. (E) Quantitation of single-cell lineage tracing of HCC366-GFP-H2B micronucleates post exposure to 10 nM paclitaxel. Bars represent mean ± range for 100 cells over 2 experiments. (F) H&E staining of HCC366 subcutaneous tumor xenografts treated as indicated. Arrowheads specify micronucleated cells. Scale = 40 μm. Zoomed images are of white boxed area.
To comprehensively investigate the molecular basis for deflection of paclitaxel-induced death, we performed a pan-genomic, siRNA-based loss of function screen in the presence and absence of 10 nM paclitaxel in HCC366 cells (Supplemental Figure 1E and F). This analysis returned 49 candidate siRNA pools with a z-score of < −2.5 that correspond to annotated genes in the human GENE database (see Supplementary Methods, Supplementary Table 1 and Supplementary Table 2).
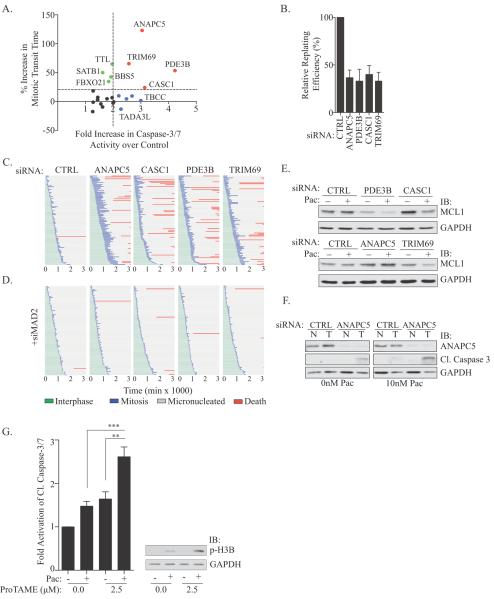
We further stratified these siRNAs using a secondary screening strategy to measure caspase-3/7 activity and mitotic transit time for 23 candidates selected from diverse functional categories (Figure 2A, Supplemental Figure 2A&B, Supplementary Table 1). Target mRNA knockdown was also evaluated (Supplemental Figure 2C). This counter screen returned nine targets whose inhibition activated apoptotic signaling, four of which increased mitotic transit time at least 20%. Colony formation assays revealed a 50% decrease in replating efficiency, indicating that the cell death observed was sufficient to reduce survival of the population (Figure 2B). This finding indicated that even in slippage prone NSCLC, a number of molecular mechanisms exist to enhance SAC robustness. Thus, we focused on this cohort of ANAPC5, PDE3B, CASC1 and TRIM69 for further mechanistic elaboration. All four of these siRNA pools met our off-target validation criteria of two independent siRNAs or non-redundant pools reproducing the apoptotic phenotype (Supplementary Figure 2D).
Figure 2.
(A) Graph of relative mitotic transit time (y) vs. relative caspase-3/7 activity (APO-ONE®) in HCC366 cells exposed to paclitaxel. A subset of siRNA gene targets is indicated. (B) Colony formation assay in HCC366s exposed to 10 nM paclitaxel for 48 hours. Bars represent mean ± SEM from n=3 independent experiments. (C) HCC366-GFP-H2B cells were transfected with indicated siRNAs for 48 hours followed by exposure to 10 nM paclitaxel. Live cell imaging commenced with paclitaxel treatment and single-cell lineage traces of 50 cells from 2 independent experiments were measured. (D) As in C, but in the presence of siMAD2. (E) WCLs from HCC366 cells transfected for 48 hours followed by exposure to vehicle or 10 nM paclitaxel for 48 hours were immunoblotted with indicated antibodies. (F) WCLs from non-tumorigenic HBEC3KT (N) and HCC366 (T) cells treated as in 2E were immunoblotted with indicated antibodies. (G) Left panel: Relative cleaved caspase-3/7 activity (APO-ONE®) in HCC366s treated with vehicle, 10 nM paclitaxel or 2.5 μM proTAME for 24 hours. Bars represent mean ± SEM for n=9 independent experiments. ** P <0.01 *** P < 0.0001 (unpaired two-tailed student's t-test). Right panel: Immunoblot of WCLs of HCC366s treated as in left panel.
Single-cell lineage tracing revealed that the prolonged mitotic arrest in target depleted cells resolved as mitotic slippage to generate micronucleated progeny; 40–50% of which subsequently died as measured by cell blebbing and a loss of movement (Figure 2C). Indicative of a functional conservation of these genes across multiple genetic backgrounds, their inhibition enhanced paclitaxel-induced mitotic arrest in HCC1171, HCC515 and H2887, cell lines that also exhibit a deflection of mitotic stress induced death (Supplemental Figure 2E). Thus, prolonging mitotic arrest induced a post-mitotic apoptotic response.
Restoring paclitaxel sensitivity requires a protracted mitotic arrest
Of these validated genes, ANAPC5 is the best characterized as encoding a scaffold in the multi-protein APC/Cyclosome complex, whose inhibition attenuates the proteolytic cascades required for anaphase onset (13). While roles for CASC1, TRIM69 and PDE3B in mitosis have not been described, their identification in a cohort with ANAPC5 suggests that they may contribute to APC/C inhibition. Indeed, co-depletion of CASC1, TRIM69 or PDE3B with MAD2 significantly reduced mitotic transit time in the presence of paclitaxel (Supplemental Figure 3A). Co-depletion also rescued micronucleated cell death as assessed by live-cell imaging and cleaved-caspase-3 protein levels (Figure 2D, Supplemental Figure 3B).
The pro-survival protein, MCL-1, is degraded during a prolonged mitosis and has been implicated as a regulator of apoptosis following exposure to anti-mitotic therapeutics (10, 11). Depletion of CASC1 or TRIM69 leads to a reduction in MCL-1 levels in a paclitaxel dependent manner while PDE3B inhibition led to a reduction in MCL-1 even in the absence of paclitaxel. Consistent with previous reports indicating that MCL-1 is degraded by residual APC/C activity during mitosis, siANAPC5 did not decrease MCL-1 levels (10) (Figure 2E). Cell death despite MCL-1 stabilization in ANAPC5 depleted cells indicates existence of MCL-1 independent apoptotic mechanisms. Taken together, these findings suggest that enhancing mitotic arrest by 20–50% in the presence of paclitaxel can be sufficient to engage a programmed cell death response in chemoresistant NSCLC.
Inhibition of APC/C is synthetic lethal with paclitaxel in chemorefractory NSCLC
Findings thus far indicate that, in principle, prolonging mitotic arrest by inhibiting the APC/C may be a therapeutic strategy to restore paclitaxel sensitivity in NSCLC. Importantly, suppression of ANAPC5 in non-tumorigenic cells (HBEC3KT) did not enhance the impact of paclitaxel, suggesting that tumor cells may have an enhanced sensitivity to APC/C inhibition (Figure 2F). The difference in response is unlikely due to cell cycle kinetics as the HBEC3KT and HCC366 cells proliferate at approximately the same rate (Supplemental Figure 3C). Together these data suggest that this combinatorial approach may exhibit a therapeutic window. Indeed, combining a sub-lethal dose of the APC/C inhibitor, proTAME, with paclitaxel in HCC366 cells enhanced the accumulation of mitotic cells and induced cleaved-caspase 3/7 above levels detected for either agent alone (Figure 2G, Supplemental Figure 3D) (14).
To further examine molecular mechanisms that contribute to the duration of mitotic arrest, we focused on CASC1 and TRIM69, whose inhibition displayed minimal, if any, single agent activity, but which collaborated with paclitaxel to both increase mitotic transit time and reduce MCL-1 levels.
CASC1 regulates microtubule function
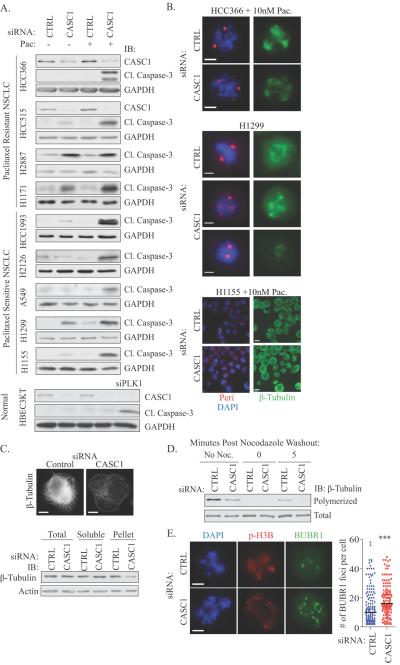
While limited information is available on the function of human CASC1, a polymorphism in the murine Casc1 gene at codon 60 (AGT (Ser) -> AAT (Asp)) is associated with susceptibility to urethane-induced lung carcinogenesis in mouse models (15, 16). The significance of these polymorphisms has not been realized in human cancer as none of the five human CASC1 isoforms harbor Asn or Ser at residue 60, and other polymorphisms in CASC1 have not been linked to NSCLC susceptibility or progression (17). However, TCGA analysis revealed an amplification of CASC1 in 15%, 11%, and 7% of lung (adenoma & squamous), ovarian and breast tumors, respectively (Supplemental Figure 4A)(18). This amplification typically co-occurs with KRAS, which is adjacent to CASC1 on chromosome 12. While the function of human CASC1 has not been documented, returning this gene as a top paclitaxel sensitizer in a genome-wide screen implies it may have essential roles in supporting cell division. We assessed the consequences of CASC1 depletion alone and in the presence of an otherwise innocuous dose of paclitaxel in a panel of NSCLC lines with differing paclitaxel sensitivity. In resistant cell lines, where we detected mitotic arrest phenotypes, we found a dependence on CASC1 for viability alone or in the presence of paclitaxel (Figure 3A). Additionally, siCASC1 is monogenic lethal or synthetic lethal with paclitaxel in 5 additional NSCLC cell lines, a defect we did not observe in an immortalized human bronchial epithelial (HBEC3KT) cell line (Figure 3A). Thus, CASC1 appears to be essential in both resistant and sensitive tumor settings, indicating that tumor cells develop a dependence on this protein for mitotic progression. Examination of individual cells indicated that CASC1 is essential for mitotic spindle integrity as inhibition led to: 1) exacerbation of the frequency of spindles with >2 pericentrin positive foci in HCC366 cells, 2) generation of multipolar or miniature spindles in H1299 cells and 3) induction of aberrant mitotic spindles in the presence of an otherwise innocuous dose of paclitaxel in H1155 cells (Figure 3B and Supplemental Figure 4B). Live-cell imaging of H1155 cells indicated that depletion of CASC1 prolonged mitotic arrest, which resolved as either aberrant mitotic exit or cell death in the presence of paclitaxel (Supplemental Figure 4C).
Figure 3.
(A) WCLs of NSCLC cells immunoblotted with indicated antibodies following siRNA transfection for 48 hours and subsequent treatment as indicated for 48 hours. siPLK1 was used as a positive control for cleaved caspase-3 detection in HBEC3KT cells. (B) As in A, except paclitaxel treatment was 24 hours prior to immunostaining. Scale = 5 μm. (C) Top panel: H1299 cells were transfected for 72 hours with indicated siRNAs followed by glutaraldehyde fix and immunostained. Scale =15 μm. Bottom panel: Immunoblots of H1299 cells following an in vivo polymerized tubulin assay (D) In vivo polymerized tubulin assay in H1299 cells transfected for 72 hours. Lysates were immunoblotted with indicated antibodies. (E) HCC366 cells treated as in B, were fixed in cold methanol and immunostained as indicated. Scale = 5 μm. BUBR1 foci were counted by manual inspection in 60 cells per experiment in 3 independent experiments. Bar indicates mean. *** P < 0.001, Mann-Whitney t-test.
Previous reports indicate that murine and chlamydomonas CASC1 (IC97) orthologues can bind microtubules (15, 19). Given that human and mouse CASC1 proteins are 66.7% identical and 92% identical in the microtubule binding region, we reasoned that the mitotic defects observed following CASC1 depletion could be due to a microtubule defect. We examined the microtubule network in H1299 cells, where siCASC1 alone had a potent impact on mitotic spindle formation and cell viability. siCASC1 diminished the polymerized tubulin network in interphase cells and attenuated microtubule regrowth after nocodazole washout (Figure 3C and 3D). Recently, SAC signaling has been correlated with levels of unattached kinetochores. Thus, increasing the number of unattached kinetochores only slightly could prolong mitotic arrest (20–22). An increase of BUBR1 positive kinetochores in HCC366 cells exposed to paclitaxel and depleted of CASC1 suggest that CASC1 may be supporting mitotic slippage by promoting microtubule-kinetochore attachments and reducing SAC strength (Figure 3E).
We further examined the impact of siCASC1 on microtubules in HBEC3KT alone, or following stable depletion of p53 in the absence (HBEC3KT-p53) and presence of oncogenic K-RAS (HBEC3KT-p53RASV12) (23). Here, we found that CASC1 was essential for microtubule polymerization in all settings, but only essential for viability following depletion of p53, thus suggesting the tumor-specific dependency on CASC1 for viability can be driven by a loss of tumor suppressor activity (Supplemental Figure 4D & 4E). Taken together, these findings suggest that combinatorial regimens that incorporate multiple anti-tubulin agents may enhance or restore sensitivity to NSCLC.
TRIM69 is a centrosomal and microtubule associated protein essential for mitotic spindle formation
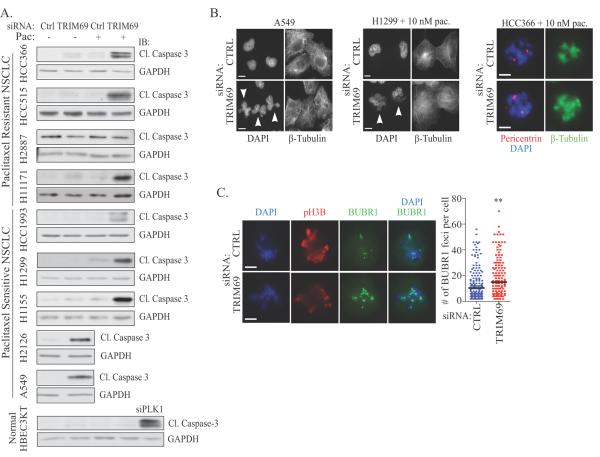
TRIM69 is a member of the TRIpartite Motif Containing (TRIM) protein family, which contains a RING E3 ligase domain, B box and coiled-coil domains (24). Two isoforms exist, A and B, with only the TRIM69A isoform containing a RING domain (Supplemental Figure 5A). Indicative that TRIM69A is an E3 ligase, we detected autoubiquitination of TRIM69A in an in vitro ubiquitination assay (Supplemental Figure 5B). We found that TRIM69 depletion, similar to CASC1, activated cell death signaling alone or in the presence of paclitaxel in multiple sensitive and resistant NSCLC backgrounds, with the exception of H2887. This defect was not observed in normal HBEC3KT cells (Figure 4A), thus implicating a dependency on TRIM69 for tumor cell survival. At the single cell level, we observed a number of mitotic defects including: 1) micronucleation (A549), 2) paclitaxel-enhanced micronucleation (H1299) and 3) multipolar spindles in the presence of paclitaxel (HCC366 & H1299) (Figure 4B and Supplemental Figure 5C). Depletion of TRIM69 also increased BUBR1 occupancy at kinetochores in paclitaxel-treated HCC366 cells (Figure 4C). Thus, we conclude that TRIM69 is essential for mitotic fidelity and may be essential for the proper attachment of microtubules to kinetochores.
Figure 4.
(A&B) As in 3A&B. Scale bars = 10 μm for A549 and H1299, 5 μm for HCC366. (C) As in 3E. ** P <0.05.
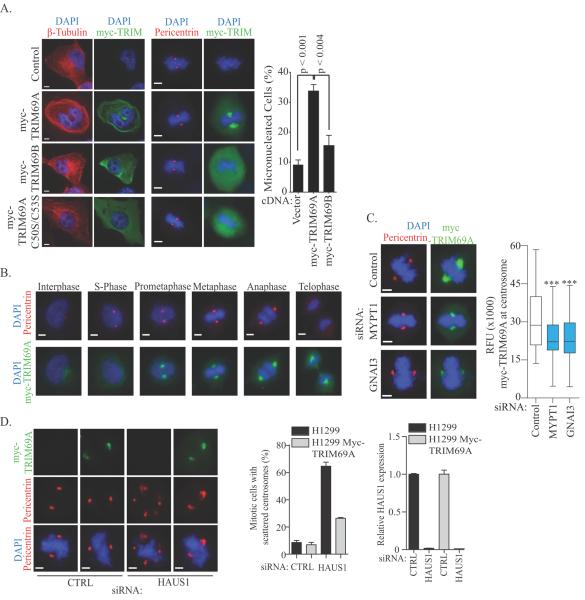
To further elucidate the mitotic function of TRIM69, we examined the localization of both isoforms of TRIM69 during the cell cycle. Examination of cells overexpressing myc-TRIM69A revealed localization to microtubules during interphase and centrosomes during mitosis (Figure 5A). Overexpressed myc-TRIM69A also induced microtubule bundling, rendering microtubules resistant to nocodazole and leading to the generation of micronucleated cells (Figure 5A and Supplemental Figure 5D). The localization of myc-TRIM69A to microtubules and centrosomes was dependent upon an intact RING domain as mutation of canonical cysteines in the zinc finger of TRIM69A abrogated microtubule and centrosomal localization of the overexpressed construct, indicating that the localization of TRIM69A is dependent upon E3-ligase activity and/or interaction partners (Figure 5A). Consistent with this finding, myc-TRIM69B, which lacks the RING domain, was dispersed throughout the cytoplasm during the cell cycle and did not display microtubule localization or induce alterations in microtubule stability (Figure 5A, Supplemental Figure 5D). These data suggest that TRIM69 may play a critical role in supporting mitotic fidelity.
Figure 5.
(A) HeLa cells transfected with indicated cDNAs for 48 hours were fixed and immunostained. Scale = 5μm. Right panel: Quantitation of cells from (A). Bars represent mean ± range for n=2 independent experiments. (B) Immunostaining of H1299-myc-TRIM69A representing each stage in the cell cycle based on nuclear morphology. Scale bar = 5 μm. (C) H1299 cells stably expressing myc-TRIM69A were transfected with indicated siRNA for 72 hours and immunostained with indicated antibodies. Plots represent relative fluorescence of myc-TRIM69A at centrosomes in > 150 cells across 3 independent experiments. *** P <0.0001 by two-tailed Mann Whitney test. Scale = 5 μm. (D) Left panel: H1299 cells were immunostained with indicated antibodies following siRNA transfection for 72 hours. Scale = 5 μm. Middle panel: Quantitation of cells from (D). Bars represent mean ± range from 2 independent experiments. Right panel: Bars represent mean mRNA expression levels of HAUS1 72 hours post-transfection.
We next studied the dynamics of myc-TRIM69A localization during the cell cycle, using H1299 cells with stable expression of myc-TRIM69A. In this setting, we found that myc-TRIM69A is recruited to centrosomes during prometaphase where it remains through telophase (Figure 5B and Supplemental 5E). myc-TRIM69A also associated with endogenous pericentrin in cells arrested in mitosis (Supplemental Figure 5F). The centrosomal localization of TRIM69A was further bolstered by proteomics data sets that indicate interactions between TRIM69 and a number of mitotic proteins, including centrosomal and cytoskeletal components (25–36) (Supplemental 5G). Indeed depletion of two centrosomal proteins that putatively interact with TRIM69, MYPT1 and GNAI3, attenuate centrosomal localization of myc-TRIM69A during mitosis (Figure 5C).
Tumor cells frequently amplify centrosomes, which can be induced by tumor suppressors and/or oncogenes. As multiple centrosomes can lead to multipolar mitosis, mitotic defects and cell death, tumor cells depend upon mechanisms that promote centrosomal clustering for cell survival (37). Proteomic studies suggest that TRIM69 interacts with the 8-member Augmin complex (also known as HAUS), which is essential for centrosomal clustering (31, 37). To evaluate a functional interaction between TRIM69 and the HAUS complex, we depleted HAUS1 in H1299 cells in the presence and absence of myc-TRIM69A. We find that spindle multipolarity induced by inhibition of HAUS1 can be reversed by overexpression of myc-TRIM69A, indicating that TRIM69A can promote centrosomal clustering (Figure 5D). Thus, inhibition of TRIM69A may enhance the vulnerability of tumor cells to paclitaxel by attenuating centrosome clustering, which may prolong mitotic arrest and allow for the reengagement of cell death signaling.
CASC1 and TRIM69 are required for in vivo tumor growth
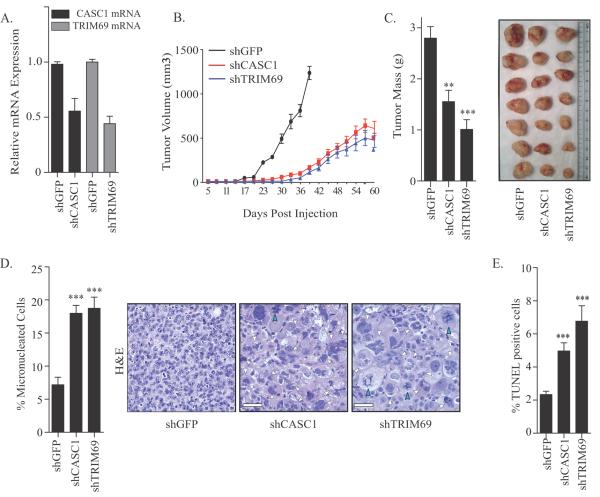
We next determined whether CASC1 and TRIM69 were essential for growth of tumors in vivo. Specifically, to achieve stable depletion of CASC1 and TRIM69, we introduced shRNAs targeting GFP, CASC1 or TRIM69 into HCC366 cells (Figure 6A). These cells were subcutaneously injected into immunodeficient mice, and we observed attenuation in growth of tumors expressing shTRIM69 and shCASC1 (Figure 6B and C). Indicating that the depletion of CASC1 or TRIM69 in vivo was sufficient to reduce mitotic fidelity, H&E staining on excised tumors revealed extensive micronucleation and aberrant mitotic figures (Figure 6D). Furthermore, TUNEL staining on tissue sections revealed enhanced cell death (Figure 6E). Taken together, these findings indicate that CASC1 and TRIM69 suppression is sufficient to impact mitotic fidelity and viability in vivo.
Figure 6.
(A) Bars represent mean mRNA expression levels ± range of CASC1 and TRIM69 in HCC366 cells expressing indicated hairpins from 2 independent infections. (B) Tumor growth curves for mice harboring HCC366s expressing indicated shRNAs. shGFP n=9, shCASC1 n=9 and shTRIM69 n=7. Mean ± standard deviation (SD). (C) Tumors expressing indicated short hairpins were excised 96 days following injection. Left panel: Bars represent mean mass of tumors ± SEM; shGFP n=7, shCASC1 n=10, shTRIM69 n=8. Right: Representative images of excised tumors. (D) Tumors from (B) were sectioned and H&E stained. Left: Quantitation of micronucleated cells in tumor sections. 6 independent tumors were examined for each indicated shRNA. Right: Representative images of H&E stained tumor sections. Scale = 50 μm. White arrowheads indicate micronucleated cells. Blue arrowheads indicate aberrant mitotic cells. (E) Tumors from (B) were sectioned and TUNEL stained. Bars indicate mean TUNEL positive cells ± SEM from 6 tumors for each shRNA condition. For all panels, ** P <0.01. *** P < 0.0001 (unpaired two-tailed student's t-test).
Discussion
While genomic studies have begun to isolate cohorts of patients that may be selectively sensitive to precision therapeutics, the majority of NSCLC patients are administered standard chemotherapeutic regimens that are comprised of DNA damage agents and anti-mitotics. These regimens are non-curative and in 70% of cases fail to reduce tumor burden (2, 3). The underlying basis for the intrinsic resistance of NSCLC to standard chemotherapy is poorly understood. Here, we use a genome-wide RNAi approach to define the molecular components that deflect cell death in the presence of mitotic damage. This work resulted in a number of new principles regarding chemoresistance and mitosis in tumor cells. First, a common feature to uncouple tumor cell death from an insurmountable spindle defect is mitotic slippage. Second, inhibition of specific proteins can increase mitotic transit time >20%, which is sufficient to activate cell death. Third, CASC1 is a novel regulator of microtubule dynamics that is essential for tumor, but not normal cell survival. Fourth, TRIM69 is a novel mitotic centrosomal resident protein that supports centrosomal clustering. In contrast to a previous screen in H1155 sensitive cells, we did not return multiple components of the γTURC or kinetochore complexes, suggesting that perturbing spindle assembly in general is not sufficient to enhance sensitivity in NSCLC cells resistant to paclitaxel(12). These findings suggest a new therapeutic strategy for chemoresistant NSCLC: combining anti-mitotic agents, such as paclitaxel, with agents, such as small molecule inhibitors of the APC/Cyclosome may enhance the potency of these first-line agents with limited impacts on normal tissues.
Many tumor cells exhibit a brief SAC dependent arrest and mitotic escape following exposure to spindle poisons in vitro and in vivo (6, 38). The frequency of this behavior suggests that tumor evolution may generally select for variants that are capable of bypassing prolonged mitotic arrest. Oncogenic changes such as p53 loss, EGFR activation or K-RAS mutation can induce mitotic spindle and centrosomal alterations, which could conceivably disrupt mitotic progression and lead to cell death in pre-tumorigenic lesions (39–41). Thus, those pre-neoplastic variants that bypass mitotic arrest could have a selective advantage, if they retain fecundity. A collateral consequence of this relationship would be an intrinsic resistance to anti-mitotic chemotherapeutics. Our work supports the hypothesis that reengaging a prolonged mitotic arrest can enhance sensitivity to anti-mitotics (6, 9). We find that elevation of mitotic transit time by 20–50% is sufficient to switch a paclitaxel resistant setting to a sensitive one. However, frank inhibition of the APC/C, through depletion of CDC20, is highly lethal in tumor cells and likely normal cells, given the requirement for CDC20 to activate the APC/C (9). Thus, single agent targeting of the APC/C could have a narrow or non-existent therapeutic window. Our finding that ANAPC5 depletion can exhibit a synthetic lethal interaction with paclitaxel, suggests that tumor cells may have an enhanced sensitivity to APC/C inhibition that may be exploited by combining sub-lethal inhibition of APC/C with low dose paclitaxel to produce a synthetic phenotype.
We also define a mitotic and microtubule role for the human CASC1 protein in tumor cells. While this gene has been linked with murine lung tumorigenesis, any functional role in human tumorigenesis has not been demonstrated. We find that tumor cells require CASC1 for microtubule polymerization and inhibition of CASC1 potentially impacts microtubule availability for the alignment of chromosomes, thereby increasing the number of unattached kinetochores and prolonging the SAC. The heightened dependency on tubulin regulation in tumor cells could be driven by oncogenic stress that may increase centrosomal clustering defects and attachment errors, thus creating an environment in which mild disruptions to the microtubule network can wreak havoc on chromosome segregation in tumor cells. As a recent reports suggest that mitosis is compromised in K-RAS backgrounds, the genetic link between CASC1 and K-RAS may allow for mitigation of transformation induced mitotic defects by increasing the expression of a microtubule regulatory protein in concert with an oncogene (40). Importantly, these finding suggests that one mechanism for prolonging the SAC in tumor cells may be to compromise microtubule function in parallel, but independent, manners by combining anti-tubulin agents.
TRIM69A is an E3 ligase with only limited characterized function, which we implicate as critical for mitotic fidelity. Specifically, TRIM69A appears to promote centrosomal clustering, which is an essential process to prevent multipolar spindles and decreased mitotic fidelity (37, 42, 43). Thus, interfering with centrosomal clustering may be a route to sensitize resistance cells to paclitaxel. Furthermore, the dependency of TRIM69 on MYPT1 and GNAI3 for centrosomal localization implicates TRIM69 in the regulation of phosphatase and receptor-independent G(ialpha) signaling during mitosis. MYPT1, which is localized to the spindle, kinetochores and centrosomes, is thought to regulate SAC checkpoint silencing though dephosphorylation of Aurora B and BUB1 (44–46). GNAI3 and other G(ialpha) subunits have been localized to the centrosome and depletion of GNAI3 induces cytokinesis defects, while overexpression increases mitotic transit time (47). Thus, TRIM69A activity may be essential for coordinating multiple molecular events that promote bipolar spindle formation and allow for rapid checkpoint silencing. Investigation into the role of TRIM69A regulation may reveal additional mechanisms governing spindle alignment and centrosome clustering.
Supplementary Material
Acknowledgements
We thank Charlene Ross for support with animal studies, Stephen Rogers for assistance with microtubule assays and Noah Sciaky for technical support with live-cell imaging.
Financial Support: This work was supported by Public Health Service grants CA154699 and CA128926 to A.W.W. from the National Cancer Institute and a research grant from Uniting Against Lung Cancer. R.S. and PT were supported by general medicine training grant T32GM007040-37. KMC was supported by the National Cancer Institute training grant CA071341-14. P.T. was supported by general NCI training grant NIH-CA071341-14.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to disclose
References
- 1.Jordan MA, Kamath K. How do microtubule-targeted drugs work? An overview. Curr Cancer Drug Targets. 2007;7(8):730–42. doi: 10.2174/156800907783220417. [DOI] [PubMed] [Google Scholar]
- 2.Schiller JH, Harrington D, Belani CP, Langer C, Sandler A, Krook J, et al. Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N Engl J Med. 2002;346(2):92–8. doi: 10.1056/NEJMoa011954. [DOI] [PubMed] [Google Scholar]
- 3.Ichiki M, Kawasaki M, Takayama K, Ninomiya K, Kuba M, Iwami F, et al. A multicenter phase II study of carboplatin and paclitaxel with a biweekly schedule in patients with advanced non-small-cell lung cancer: Kyushu thoracic oncology group trial. Cancer Chemother Pharmacol. 2006;58(3):368–73. doi: 10.1007/s00280-005-0168-3. [DOI] [PubMed] [Google Scholar]
- 4.Peters JM. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nature reviews Molecular cell biology. 2006;7(9):644–56. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- 5.Yu H. Regulation of APC-Cdc20 by the spindle checkpoint. Curr Opin Cell Biol. 2002;14(6):706–14. doi: 10.1016/s0955-0674(02)00382-4. [DOI] [PubMed] [Google Scholar]
- 6.Gascoigne KE, Taylor SS. Cancer cells display profound intra- and interline variation following prolonged exposure to antimitotic drugs. Cancer Cell. 2008;14(2):111–22. doi: 10.1016/j.ccr.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 7.Hornick JE, Bader JR, Tribble EK, Trimble K, Breunig JS, Halpin ES, et al. Live-cell analysis of mitotic spindle formation in taxol-treated cells. Cell Motil Cytoskeleton. 2008;65(8):595–613. doi: 10.1002/cm.20283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rieder CL, Maiato H. Stuck in division or passing through: what happens when cells cannot satisfy the spindle assembly checkpoint. Dev Cell. 2004;7(5):637–51. doi: 10.1016/j.devcel.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 9.Huang HC, Shi J, Orth JD, Mitchison TJ. Evidence that mitotic exit is a better cancer therapeutic target than spindle assembly. Cancer Cell. 2009;16(4):347–58. doi: 10.1016/j.ccr.2009.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harley ME, Allan LA, Sanderson HS, Clarke PR. Phosphorylation of Mcl-1 by CDK1-cyclin B1 initiates its Cdc20-dependent destruction during mitotic arrest. The EMBO journal. 2010;29(14):2407–20. doi: 10.1038/emboj.2010.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wertz IE, Kusam S, Lam C, Okamoto T, Sandoval W, Anderson DJ, et al. Sensitivity to antitubulin chemotherapeutics is regulated by MCL1 and FBW7. Nature. 2011;471(7336):110–4. doi: 10.1038/nature09779. [DOI] [PubMed] [Google Scholar]
- 12.Whitehurst AW, Bodemann BO, Cardenas J, Ferguson D, Girard L, Peyton M, et al. Synthetic lethal screen identification of chemosensitizer loci in cancer cells. Nature. 2007;446(7137):815–9. doi: 10.1038/nature05697. [DOI] [PubMed] [Google Scholar]
- 13.Schreiber A, Stengel F, Zhang Z, Enchev RI, Kong EH, Morris EP, et al. Structural basis for the subunit assembly of the anaphase-promoting complex. Nature. 2011;470(7333):227–32. doi: 10.1038/nature09756. [DOI] [PubMed] [Google Scholar]
- 14.Zeng X, Sigoillot F, Gaur S, Choi S, Pfaff KL, Oh DC, et al. Pharmacologic inhibition of the anaphase-promoting complex induces a spindle checkpoint-dependent mitotic arrest in the absence of spindle damage. Cancer Cell. 2010;18(4):382–95. doi: 10.1016/j.ccr.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu P, Wang Y, Vikis H, Maciag A, Wang D, Lu Y, et al. Candidate lung tumor susceptibility genes identified through whole-genome association analyses in inbred mice. Nat Genet. 2006;38(8):888–95. doi: 10.1038/ng1849. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Z, Futamura M, Vikis HG, Wang M, Li J, Wang Y, et al. Positional cloning of the major quantitative trait locus underlying lung tumor susceptibility in mice. Proc Natl Acad Sci U S A. 2003;100(22):12642–7. doi: 10.1073/pnas.2133947100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galbiati F, Pettinicchio A, Dragani TA, Manenti G. Allelic effects of mouse Pas1 candidate genes in human lung cancer cell lines. Cancer letters. 2006;244(2):176–81. doi: 10.1016/j.canlet.2005.12.015. [DOI] [PubMed] [Google Scholar]
- 18.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wirschell M, Yang C, Yang P, Fox L, Yanagisawa HA, Kamiya R, et al. IC97 is a novel intermediate chain of I1 dynein that interacts with tubulin and regulates interdoublet sliding. Molecular biology of the cell. 2009;20(13):3044–54. doi: 10.1091/mbc.E09-04-0276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Collin P, Nashchekina O, Walker R, Pines J. The spindle assembly checkpoint works like a rheostat rather than a toggle switch. Nature cell biology. 2013;15(11):1378–85. doi: 10.1038/ncb2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dick AE, Gerlich DW. Kinetic framework of spindle assembly checkpoint signalling. Nature cell biology. 2013;15(11):1370–7. doi: 10.1038/ncb2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heinrich S, Geissen EM, Kamenz J, Trautmann S, Widmer C, Drewe P, et al. Determinants of robustness in spindle assembly checkpoint signalling. Nature cell biology. 2013;15(11):1328–39. doi: 10.1038/ncb2864. [DOI] [PubMed] [Google Scholar]
- 23.Sato M, Vaughan MB, Girard L, Peyton M, Lee W, Shames DS, et al. Multiple oncogenic changes (K-RAS(V12), p53 knockdown, mutant EGFRs, p16 bypass, telomerase) are not sufficient to confer a full malignant phenotype on human bronchial epithelial cells. Cancer Res. 2006;66(4):2116–28. doi: 10.1158/0008-5472.CAN-05-2521. [DOI] [PubMed] [Google Scholar]
- 24.Hatakeyama S. TRIM proteins and cancer. Nature reviews Cancer. 2011;11(11):792–804. doi: 10.1038/nrc3139. [DOI] [PubMed] [Google Scholar]
- 25.Zhu M, Settele F, Kotak S, Sanchez-Pulido L, Ehret L, Ponting CP, et al. MISP is a novel Plk1 substrate required for proper spindle orientation and mitotic progression. The Journal of cell biology. 2013;200(6):773–87. doi: 10.1083/jcb.201207050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamashiro S, Yamakita Y, Totsukawa G, Goto H, Kaibuchi K, Ito M, et al. Myosin phosphatase-targeting subunit 1 regulates mitosis by antagonizing polo-like kinase 1. Dev Cell. 2008;14(5):787–97. doi: 10.1016/j.devcel.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang W, Zhang W, Han Y, Chen J, Wang Y, Zhang Z, et al. NELIN, a new F-actin associated protein, stimulates HeLa cell migration and adhesion. Biochemical and biophysical research communications. 2005;330(4):1127–31. doi: 10.1016/j.bbrc.2005.03.082. [DOI] [PubMed] [Google Scholar]
- 28.Snyers L, Thines-Sempoux D, Prohaska R. Colocalization of stomatin (band 7.2b) and actin microfilaments in UAC epithelial cells. European journal of cell biology. 1997;73(3):281–5. [PubMed] [Google Scholar]
- 29.Mulder J, Ariaens A, van den Boomen D, Moolenaar WH. p116Rip targets myosin phosphatase to the actin cytoskeleton and is essential for RhoA/ROCK-regulated neuritogenesis. Molecular biology of the cell. 2004;15(12):5516–27. doi: 10.1091/mbc.E04-04-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levi M, Maro B, Shalgi R. The conformation and activation of Fyn kinase in the oocyte determine its localisation to the spindle poles and cleavage furrow. Reproduction, fertility, and development. 2011;23(7):846–57. doi: 10.1071/RD11033. [DOI] [PubMed] [Google Scholar]
- 31.Lawo S, Bashkurov M, Mullin M, Ferreria MG, Kittler R, Habermann B, et al. HAUS, the 8-subunit human Augmin complex, regulates centrosome and spindle integrity. Current biology : CB. 2009;19(10):816–26. doi: 10.1016/j.cub.2009.04.033. [DOI] [PubMed] [Google Scholar]
- 32.Kao SC, Chen CY, Wang SL, Yang JJ, Hung WC, Chen YC, et al. Identification of phostensin, a PP1 F-actin cytoskeleton targeting subunit. Biochemical and biophysical research communications. 2007;356(3):594–8. doi: 10.1016/j.bbrc.2007.03.026. [DOI] [PubMed] [Google Scholar]
- 33.Hauge H, Patzke S, Aasheim HC. Characterization of the FAM110 gene family. Genomics. 2007;90(1):14–27. doi: 10.1016/j.ygeno.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 34.Hasegawa H, Hyodo T, Asano E, Ito S, Maeda M, Kuribayashi H, et al. The role of PLK1-phosphorylated SVIL in myosin II activation and cytokinetic furrowing. J Cell Sci. 2013;126(Pt 16):3627–37. doi: 10.1242/jcs.124818. [DOI] [PubMed] [Google Scholar]
- 35.Grimm-Gunter EM, Milbrandt M, Merkl B, Paulsson M, Plomann M. PACSIN proteins bind tubulin and promote microtubule assembly. Experimental cell research. 2008;314(10):1991–2003. doi: 10.1016/j.yexcr.2008.03.015. [DOI] [PubMed] [Google Scholar]
- 36.Fischer RS, Yarmola EG, Weber KL, Speicher KD, Speicher DW, Bubb MR, et al. Tropomodulin 3 binds to actin monomers. The Journal of biological chemistry. 2006;281(47):36454–65. doi: 10.1074/jbc.M606315200. [DOI] [PubMed] [Google Scholar]
- 37.Leber B, Maier B, Fuchs F, Chi J, Riffel P, Anderhub S, et al. Proteins required for centrosome clustering in cancer cells. Science translational medicine. 2010;2(33):33ra8. doi: 10.1126/scitranslmed.3000915. [DOI] [PubMed] [Google Scholar]
- 38.Orth JD, Kohler RH, Foijer F, Sorger PK, Weissleder R, Mitchison TJ. Analysis of mitosis and antimitotic drug responses in tumors by in vivo microscopy and single-cell pharmacodynamics. Cancer Res. 2011;71(13):4608–16. doi: 10.1158/0008-5472.CAN-11-0412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fukasawa K, Choi T, Kuriyama R, Rulong S, Vande Woude GF. Abnormal centrosome amplification in the absence of p53. Science. 1996;271(5256):1744–7. doi: 10.1126/science.271.5256.1744. [DOI] [PubMed] [Google Scholar]
- 40.Luo J, Emanuele MJ, Li D, Creighton CJ, Schlabach MR, Westbrook TF, et al. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell. 2009;137(5):835–48. doi: 10.1016/j.cell.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mardin BR, Isokane M, Cosenza MR, Kramer A, Ellenberg J, Fry AM, et al. EGF-induced centrosome separation promotes mitotic progression and cell survival. Dev Cell. 2013;25(3):229–40. doi: 10.1016/j.devcel.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Quintyne NJ, Reing JE, Hoffelder DR, Gollin SM, Saunders WS. Spindle multipolarity is prevented by centrosomal clustering. Science. 2005;307(5706):127–9. doi: 10.1126/science.1104905. [DOI] [PubMed] [Google Scholar]
- 43.Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460(7252):278–82. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.London N, Ceto S, Ranish JA, Biggins S. Phosphoregulation of Spc105 by Mps1 and PP1 regulates Bub1 localization to kinetochores. Current biology : CB. 2012;22(10):900–6. doi: 10.1016/j.cub.2012.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meadows JC, Shepperd LA, Vanoosthuyse V, Lancaster TC, Sochaj AM, Buttrick GJ, et al. Spindle checkpoint silencing requires association of PP1 to both Spc7 and kinesin-8 motors. Dev Cell. 2011;20(6):739–50. doi: 10.1016/j.devcel.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Posch M, Khoudoli GA, Swift S, King EM, Deluca JG, Swedlow JR. Sds22 regulates aurora B activity and microtubule-kinetochore interactions at mitosis. The Journal of cell biology. 2010;191(1):61–74. doi: 10.1083/jcb.200912046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cho H, Kehrl JH. Localization of Gi alpha proteins in the centrosomes and at the midbody: implication for their role in cell division. The Journal of cell biology. 2007;178(2):245–55. doi: 10.1083/jcb.200604114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.