Abstract
Purpose
Lymphocytic infiltration of tumors predicts improved survival in breast cancer patients. Previous studies have suggested that this survival benefit is confined predominantly to the basal-like subtype. Immune infiltration in ovarian tumors is also associated with improved prognosis. Currently, it is unclear what aspects of the immune response mediate this improved outcome.
Experimental Design
Using The Cancer Genome Atlas (TCGA) mRNA-seq data and a large microarray data set, we evaluated adaptive immune gene expression by genomic subtype in breast and ovarian cancer. To investigate B-cells observed to be prognostic within specific subtypes, we developed methods to analyze B-cell population diversity and degree of somatic hypermutation (SHM) from B-cell receptor (BCR) sequences in mRNA-seq data.
Results
Improved metastasis-free/progression-free survival was correlated with B-cell gene expression signatures, which were restricted mainly to the basal-like and HER2-enriched breast cancer subtypes and the immunoreactive ovarian cancer subtype. Consistent with a restricted epitope-driven response, a subset of basal-like and HER2-enriched breast tumors and immunoreactive ovarian tumors showed high expression of a low-diversity population of BCR gene segments. More BCR segments showed improved prognosis with increased expression in basal-like breast tumors and immunoreactive ovarian tumors compared with other subtypes. Basal-like and HER2-enriched tumors exhibited more BCR sequence variants in regions consistent with somatic hypermutation.
Conclusion
Taken together, these data suggest the presence of a productive and potentially restricted anti-tumor B-cell response in basal-like breast and immunoreactive ovarian cancers. Immunomodulatory therapies that support B-cell responses may be a promising therapeutic approach to targeting these B-cell infiltrated tumors.
Introduction
The role of tumor-infiltrating lymphocytes (TILs) in breast cancer is not fully understood, although multiple studies have shown an association between the presence of TILs and an improved prognosis (1–5). TILs in breast tumors are predominantly cytotoxic (CD8+) T-cells (6, 7), and the proportion of CD8+ T-cells may be prognostic (4, 5, 8). In contrast, TILs of the regulatory T-cell phenotype (CD4+CD25+FoxP3+ Tregs) are associated with poorer outcomes in breast cancer (9, 10). The role of B-cell TILs in human breast cancer is not as clear as that of T-cell TILs. Using gene expression profiling, our group and others have showed that gene signatures representing B-cells, plasmablasts, plasma cells and immunoglobulin predicted favorable clinical outcome in ER+ and ER− breast tumors(11–15). In this manuscript, these are referred to as B-cell signatures; while plasmablasts and plasma cells are known to infiltrate some breast tumors, we use the term “B-cell TIL” here to refer to any TIL in the B-cell lineage. The presence of B-cell TILs as assessed by immunohistochemistry (IHC) has also been shown to be an independent prognostic feature in breast cancer (16). Studies of small numbers of breast tumors have shown the B-cell response in these tumors to be clonally expanded, with evidence of having undergone class switching and somatic hypermutation (17–22). This strongly suggested that in some breast tumors there may be a clonally restricted, antigen-directed B-cell anti-tumor response. Several studies have identified auto-antibodies in breast cancer patients, including antibodies against improperly processed β-actin in some medullary breast cancers, although the association between such auto-antibodies and patient survival is unclear (18, 21, 23). Together, these findings provide evidence that B-cell TILs may be important in affecting breast cancer biology and progression.
Human breast cancer is a heterogeneous disease, with individual tumors varying according to morphology, natural history, and response to therapy. Gene expression analyses have identified at least five distinct genomic subtypes of breast cancer: luminal A, luminal B, HER2-enriched, basal-like, and claudin-low, as well as a normal-like group (24–28). The prognostic value of both T and B-cell TILs may be restricted to a subset of highly immune-infiltrated breast tumors (14). Basal-like breast tumors, in particular, appear to have beneficial TILs (5, 15). Multiple groups have identified signatures of lymphocyte-related gene expression that are overrepresented in basal-like breast tumors and predict better survival(14, 15); in contrast, luminal A breast tumors show low levels of lymphocytic infiltrate(5).
Comprehensive genomic profiling of multiple tumor types in TCGA has shown there is a strong similarity between basal-like breast cancer and serous ovarian cancer (24). These two tumor types exhibit a similar mutational spectrum and share many of the same driver events (i.e. TP53 loss, RB1 loss, c-MYC gain, etc.). Like basal-like breast cancer, many ovarian tumors are rich in TILs. Analysis of TCGA serous ovarian cancer gene expression identified four genomic subtypes: mesenchymal, proliferative, differentiated, and immunoreactive(29). The immunoreactive subtype, in particular, showed high expression of T-cell chemokine ligands and lymphocyte-related genes. Furthermore, a number of studies have shown the presence of T and B-cell TILs is a positive prognostic feature in ovarian cancer (30–33). As in breast cancer, the precise role of B-cell TILs is less understood than that of T-cell TILs. These data suggest that, like basal-like breast cancer, serous ovarian cancer may be a likely candidate for identifying a productive anti-tumor T-cell and/or B-cell TIL response.
If there is an effective, subtype-specific antitumor response mediated by B-cell TILs, this presents the possibility of subtype-specific immunogenic epitopes that could promote development of a subtype-specific antibody response. While some studies have identified antigen-directed TIL clones in breast tumors (18–22), currently the degree to which TILs are antigen-directed is unknown. The development of a mature B-cell response following antigen stimulation depends on a number of processes that occur during the germinal center reaction, including clonal expansion and antibody class switching (34, 35). While in the germinal center, B-cells undergo somatic hypermutation (SHM), whereby mutations at BCR loci are introduced to enhance B-cell receptor (BCR) affinity. Mutations occur preferentially in “hot spot” nucleotide positions, particularly within the antigen-binding complementarity-determining regions (CDRs), and they favor replacements and transitions(36). In a tumor-antigen driven response, the TIL population is expected to be enriched for one or more “dominant clones” exhibiting BCR characteristics consistent with somatic hypermutation. Here, using a novel approach to characterize B-cell responses from short read mRNA-seq data, we demonstrate subtype-specific enrichment of B-cell gene segments in basal-like and HER2-enriched breast cancer and in immunoreactive ovarian cancer. We show evidence of clonal restriction of the B-cell response in these three tumor types, and mutation patterns consistent with SHM in basal-like and HER2-enriched breast cancer. These findings suggest an important role for the endogenous B-cell response specifically in these tumor subtypes.
Methods
Data Sets
The breast cancer data set used for all analyses except the survival analysis of gene expression signatures was the TCGA data set of 819 mRNA-seq samples, comprising 728 breast tumors and 91 normal breast samples (see TCGA Data Portal at https://tcga-data.nci.nih.gov/tcga/, CGHub at https://cghub.ucsc.edu/). This 728 sample set is an extension of the 480 tumors previously profiled by microarray(24), but these 480 plus 350 new samples have all been assayed by mRNA-seq using Illumina 2×50bp sequencing as described by the TCGA in an evaluation of Lung Squamous samples(37). Gene expression values were represented as RSEM (RNA-seq by Expectancy-Maximization) data normalized within-sample to the upper quartile of total reads as previously described (37). These data and further details about data processing are available at the TCGA Data portal under the V2_MapSpliceRSEM workflow (https://tcga-data.nci.nih.gov/tcgafiles/ftp_auth/distro_ftpusers/anonymous/tumor/brca/cgcc/unc.edu/illuminahiseq_rnaseqv2/rnaseqv2/unc.edu_BRCA.IlluminaHiSeq_RNASeqV2.mage-tab.1.6.0/DESCRIPTION.txt). Genomic subtype was assigned within the set of 728 mRNA-seq samples using the PAM50 assay (38). The training set of breast samples used in the PAM50 assay is 50% clinically ER+, therefore the mRNA-seq data were normalized to reflect the training set (https://genome.unc.edu/pubsup/breastGEO/Guide%20to%20Intrinsic%20Subtyping%209-6-10.pdf). Based on clinical data taken from the TCGA Data portal on September, 2012 (https://tcga-data.nci.nih.gov/tcga/), of the 728 samples, 157 were ER−, 535 were ER+, 2 were ER-indeterminate, 29 did not have ER status assays performed, and 5 did not have available data, indicating that 77% of the mRNA-seq samples were ER+. To normalize the data similar to the PAM50 training dataset, in which 50% of samples are ER+, all 157 ER− samples were selected, as well as 157 randomly selected ER+ samples. The median gene expression for the PAM50 intrinsic gene list was calculated based on this subset of samples. To perform platform correction for mRNA-seq, these median values were then subtracted from all 728 samples prior to running the PAM50 assay as previously described(38) (https://genome.unc.edu/pubsup/breastGEO/PAM50.zip). Due to the short median follow-up time (17 months) of the TCGA data set, survival analysis of gene expression signatures were carried out using a microarray-based gene expression data set of 855 breast tumors with published intrinsic subtype calls (140 basal-like, 90 claudin-low, 144 HER2-enriched, 243 Luminal A, 162 Luminal B, 76 Normal-like) and clinical data (combined data from the following data sets: GSE2034, GSE12276, GSE2603, and the NKI295 (microarray-pubs.stanford.edu/wound_NKI/Clinical_Data_Supplement.xls)) (39). Survival analyses of BCR segment expression, however, used the TCGA RNA-seq data set. For all analyses of ovarian cancer, we used the TCGA serous ovarian cancer mRNA-seq data set, which, like the breast cancer data set, represents new mRNA-seq data, again using Illumina 2 × 50bp sequencing, on a subset of the 500 cases from the TCGA Ovarian project (29). This mRNA-seq data set consists of 266 tumors with follow-up data (https://tcga-data.nci.nih.gov/tcga/, https://cghub.ucsc.edu/). The TCGA barcodes and genomic subtypes of all breast and ovarian mRNA-seq samples are included as Supplementary Data 1. Unique sample IDs for downloading TCGA mRNA-seq data from CGHub are included as Supplementary Data 3.
Gene Expression Signatures and Survival Analyses
Immune gene expression signatures were established using unsupervised hierarchical clustering of mRNA-seq expression data for 728 breast tumor samples (see supplementary data). Gene dendrogram nodes corresponding to genes characteristically expressed in specific immune cell types were identified and validated through DAVID functional annotation clustering and IPA (Ingenuity®Systems, www.ingenuity.com)(40, 41). Gene lists for all five signatures are included in Supplementary Data 2. Additional lymphocyte gene signatures were obtained from published studies: IGG_Cluster(11), B_Cell(42), and B_Cell_60gene(12) are B-cell signatures, and T_Cell(42), CD8(42), LCK(43), and TNBC_T-Cell(15) are T-cell signatures, with the CD8 signature specifically representing CD8+ T-cells.
Survival analyses were performed by Kaplan-Meier analysis and log-rank testing, and hazard ratios were derived from the Cox proportional hazards model. For analysis of the prognostic value of BCR segment expression, samples were divided into high and low expression groups of equal number for Kaplan-Meier analysis and log-rank testing. To evaluate the prognostic value of gene expression signatures, the Cox proportional hazards model was used with each signature tested as a continuous variable. Multivariate survival analysis was performed by ANOVA comparing individual Cox proportional hazards models.
To provide a control for the number of prognostic BCR gene segments, 353 (the number of BCR gene segments tested) random genes were selected and p values were calculated for the association of each of these genes with overall survival (breast) or progression-free survival (ovarian) as in the BCR gene segments. The number of significant (p < 0.05) p values was calculated from this set and 95% confidence intervals were calculated through bootstrap resampling.
B-Cell Receptor Diversity
The method for estimating sequence diversity of a BCR gene segment for an individual sample/tumor using paired-end mRNA-seq data is outlined in Supplementary Figure 3. Read pairs mapping to the EntrezGene genomic location (www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=gene)(44) of a given BCR gene segment were identified (mapped by MapSplice(45)). The sequence of these read pairs was compared to the hg19 reference genome to identify non-reference bases. The genomic position and nucleotide identity of all non-reference bases was identified for each read pair. Each observed pattern of non-reference bases was then assigned a score representing the number of read-pairs containing exactly that pattern of non-reference bases. This set of observed patterns and their corresponding count was used to calculate the effective number of species, which is a diversity function isomorphic to Shannon entropy, as described by Jost et al.(46).
De novo Assembly and Somatic Hypermutation Analysis
De novo assembly of BCR variable (V) gene segments from paired-end mRNA-seq reads was performed using the Assembly-Based Re-Aligner (ABRA) algorithm (Mose et al., manuscript in preparation). To generate ABRA contigs, unmapped reads and reads mapping to a BCR variable region of interest were first split into overlapping k-mers where k=31. K-mers that were comprised exclusively of non-ambiguous bases with quality score > 20 were assembled into a de Bruijn graph. K-mers with fewer than 100 observations were then pruned from the graph. The graph was then traversed to produce all possible contigs. This set of contigs was used for somatic hypermutation analyses.
To determine if the sequence of a BCR variable gene segment was consistent with somatic hypermutation, the reference sequence for that gene segment was first established by Smith-Waterman alignment to each IMGT® (IMGT®, the international ImMunoGeneTics information system® http://www.imgt.org (founder and director: Marie-Paule Lefranc, Montpellier, France))(47–53) reference allele sharing the same gene segment family and selection of the closest match. The segment sequence was compared to the reference sequence, and mutated and non-mutated bases were counted within SHM hotspots (WRCY, RGYW, WA, and TW sequences) and non-hotspot regions(54). Mutated and non-mutated (i.e., reference and non-reference) bases were counted again within CDRs. Chi-square testing was used to determine if the distribution of mutated bases was consistent with the mutation pattern expected in SHM; chi-square testing was conducted separately for the whole V segment sequence and for CDRs.
The method for producing an overall estimate of the degree of somatic hypermutation for a BCR variable segment in mRNA-seq data is outlined in Supplementary Figure 4. ABRA contigs were first constructed for that V gene segment and quantitated by BWA paired-end alignment(55) of all unmapped reads and reads mapping to that segment to the set of ABRA contigs. Multiple mappings were allowed, as long as both read pairs mapped to the same contig. For each contig, the number of SHM hotspot mutations, SHM hotspot nonmutated bases, non-hotspot mutations, and non-hotspot non-mutated bases were counted for the whole segment and within CDRs. This information was weighted by the BWA alignment score for that contig. These weighted values were summed across all contigs and chi-squared testing was used to determine if the mutation pattern across the whole segment or within CDRs indicated the presence of SHM. For this analysis, only segments with a number of mapped reads greater than or equal to 0.04 × the length in bp of the segment (approximating even coverage of depth 2) were considered for sequence analysis in a given sample.
Results
B-cell gene expression signatures are prognostic in breast and ovarian cancer
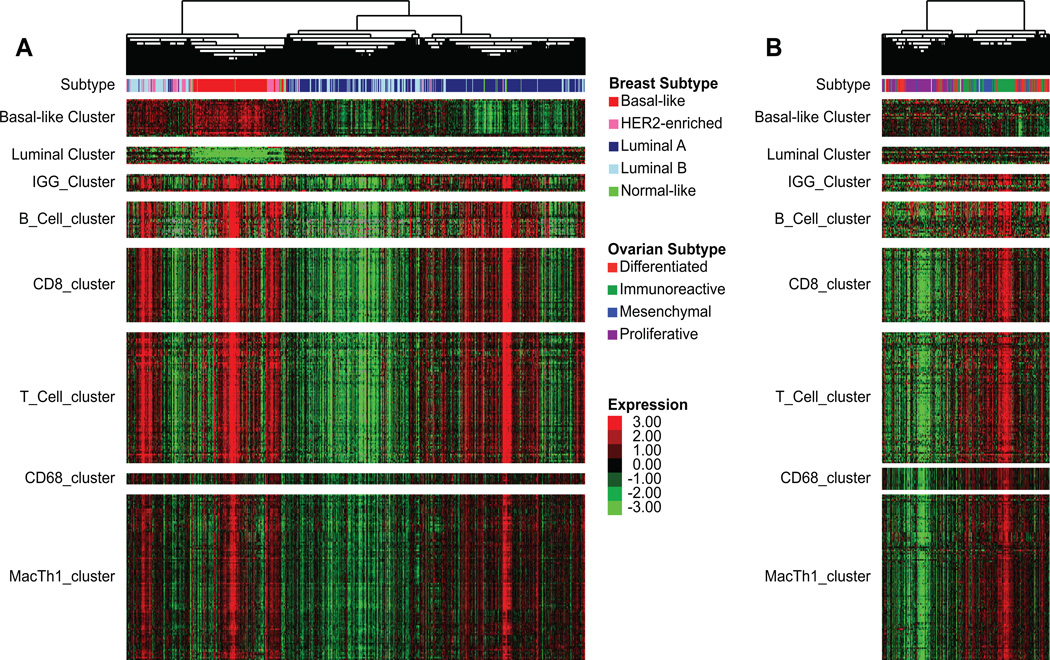
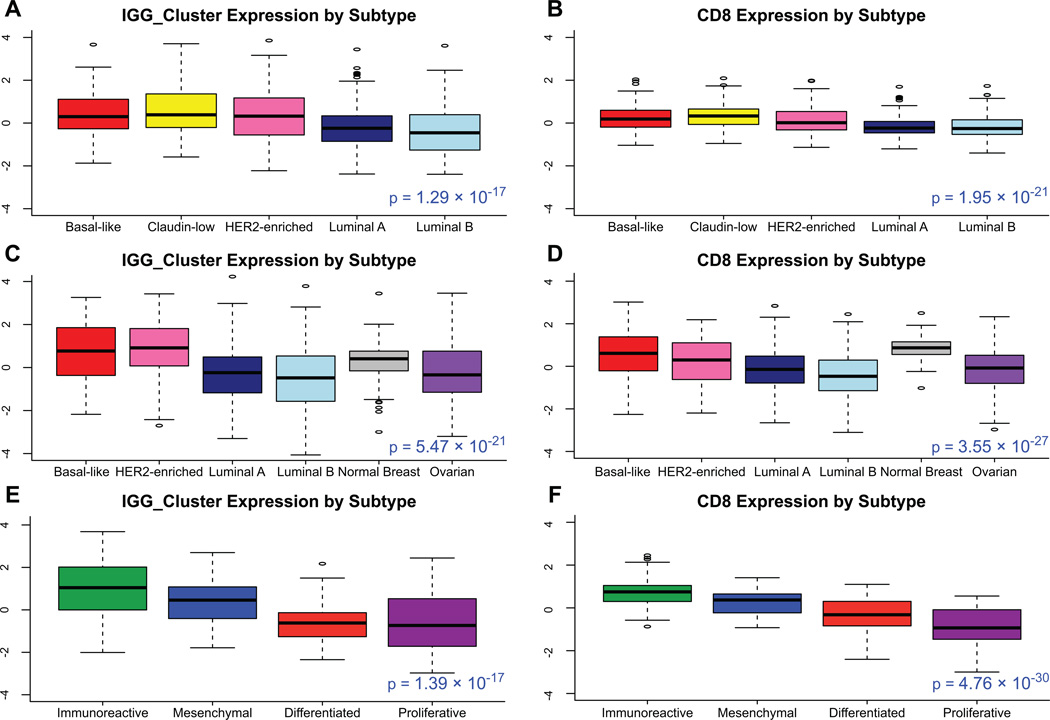
Increased expression of B-cell gene signatures has been shown to be favorably prognostic in breast cancer (11–13). To explore the role of B-cells and other lymphocyte cell types in the different intrinsic subtypes of breast cancer, immune cell associated genomic signatures were newly derived from unsupervised clustering of mRNA-seq data from 728 TCGA breast cancer samples (Figure 1A). Gene dendrogram nodes containing characteristic lymphocyte genes were selected as potential gene signatures. The identities of these signatures were confirmed through functional annotation analysis and gene pathway-based analysis (40, 41). These and other previously published lymphocyte gene signatures were used to confirm the prognostic value of TILs on a genomic level, and to assess whether this benefit is isolated to one or more intrinsic subtypes. We first evaluated a gene expression microarray data set of 855 breast tumors, using a univariate Cox proportional hazards model, for prognostic value by subtype of lymphocyte gene signatures(39). For the newly derived B_Cell_cluster signature derived from unsupervised clustering here, the IGG_cluster previously developed by our group(11), and three B-cell gene signatures generated by others(12, 14, 15), overall expression in breast tumors was greater in the basal-like and HER2-enriched subtypes (Figure 2 A,C). Similar to previous work (12, 16, 56), high expression was associated with better metastasis-free survival in basal-like and HER2-enriched tumors with greatest difference in hazards in the basal-like subtype (Table 1).
Figure 1.
A Concordant expression of genes from different immune cell types in a distinct subset of primarily basal-like, HER2-enriched, and normal-like tumors. Unsupervised hierarchical clustering of breast cancer mRNA-seq data (n=728); signatures B_Cell_cluster, CD8_cluster, T_Cell_cluster, CD68_cluster, and MacTh1_cluster are derived from this hierarchical clustering. B Expression of breast cancer-derived gene expression signatures in an ovarian cancer mRNA-seq data set (n=266).
Figure 2.
Box plots of expression of IGG_Cluster and CD8 gene expression signatures by subtype, with one-way ANOVA p value, in A, B microarray data from 855 breast tumors C, D mRNA-seq data from 819 breast samples (728 tumor, 91 normal breast) and 266 ovarian tumors E, F mRNA-seq data from 266 ovarian tumors. Expression of immune genes is high in basal-like, claudin-low, and HER2-enriched breast cancer subtypes and the immunoreactive ovarian cancer subtype.
Table 1.
Univariate Cox proportional hazards ratios and p values for immune gene signature expression versus metastasis-free survival (breast, n=855) or progression-free survival (ovarian, n=266). NS indicates non-significant p values (p ≥ 0.05).
| IGG_Cluster1 | TNBC_B-Cell2 | B_Cell1 | B_Cell_60gene3 | B_Cell_cluster4 | T_Cell1 | TNBC_T-Cell2 | LCK1 | CD81 | T_Cell_cluster4 | CD8_cluster4 | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | Hazard Ratio |
p Value | Hazard Ratio |
p Value | Hazard Ratio |
p Value | Hazard Ratio |
p Value | Hazard Ratio |
p Value | Hazard Ratio |
p Value | Hazard Ratio |
p Value | Hazard Ratio |
p Value | Hazard Ratio |
p Value | Hazard Ratio |
p Value | Hazard Ratio |
p Value | ||
| Breast | All Breast | 855 | 0.847 | 6.61E-04 | 0.901 | 1.56E-03 | 0.585 | 1.52E-02 | 0.888 | 4.79E-04 | 0.782 | 9.38E-04 | 0.562 | 5.31E-03 | 0.901 | 3.83E-02 | 0.888 | 3.37E-02 | NS | 0.858 | 2.27E-02 | 0.865 | 1.81E-02 | |
| Luminal A | 243 | NS | NS | NS | NS | NS | NS | NS | NS | NS | NS | NS | ||||||||||||
| Luminal B | 162 | NS | NS | NS | NS | NS | 0.304 | 1.59E-02 | NS | NS | NS | NS | NS | |||||||||||
| HER2-enriched | 144 | 0.755 | 4.67E-03 | 0.827 | 1.12E-02 | 0.323 | 9.16E-03 | 0.791 | 2.43E-03 | 0.641 | 4.34E-03 | 0.328 | 6.01E-03 | 0.745 | 4.74E-03 | 0.725 | 6.39E-03 | 0.658 | 3.29E-02 | 0.687 | 5.03E-03 | 0.705 | 4.15E-03 | |
| Basal-like | 140 | 0.599 | 1.20E-04 | 0.686 | 4.24E-05 | 0.17 | 2.39E-03 | 0.682 | 6.05E-05 | 0.491 | 3.56E-04 | 0.126 | 2.44E-04 | 0.644 | 1.00E-03 | 0.619 | 1.34E-03 | 0.472 | 1.53E-03 | 0.496 | 1.29E-04 | 0.548 | 2.35E-04 | |
| Claudin-low | 90 | NS | NS | NS | NS | NS | NS | NS | NS | NS | NS | NS | ||||||||||||
| Ovarian | All Ovarian | 266 | 0.845 | 7.40E-03 | NS | NS | 0.857 | 4.09E-03 | 0.856 | 5.63E-03 | NS | NS | NS | NS | NS | NS | ||||||||
| Immunoreactive | 77 | 0.675 | 7.43E-03 | NS | NS | 0.714 | 1.68E-02 | 0.725 | 1.22E-02 | 0.177 | 2.25E-02 | NS | NS | NS | 0.686 | 2.99E-02 | 0.707 | 4.28E-02 | ||||||
| Mesenchymal | 42 | 0.673 | 3.32E-02 | NS | NS | 0.729 | 4.31E-02 | NS | NS | NS | NS | NS | NS | NS | ||||||||||
| Differentiated | 65 | NS | NS | NS | NS | NS | NS | NS | NS | NS | NS | NS | ||||||||||||
| Proliferative | 78 | NS | NS | NS | NS | NS | NS | NS | NS | NS | NS | NS | ||||||||||||
Superscript indicates origin of gene signature (1=Fan, et al. 2011; 2=Rody, et al. 2011; 3=Schmidt, et al. 2008; 4=hierarchical clustering of breast cancer mRNA-seq data (n=728)).
We next performed a similar analysis on TCGA ovarian cancer data. Overall B-cell gene signature expression was increased in immunoreactive ovarian tumors (Figure 2E–F). Several B-cell gene signatures were prognostic for progression-free survival in the immunoreactive ovarian tumor subtype, which was not true for the other subtypes (Table 1). T-cell signatures (14, 15, 43) were also evaluated, and showed a similar pattern of expression and prognostic value (Fig 2B, D). In multivariate survival analysis of individual immune signatures with other clinical and genomic features in breast cancer, most B-cell and T-cell signatures remained significantly prognostic (Supplementary Table 1).
For both breast and ovarian tumors, B-cell gene signature expression strongly correlated with other lymphocyte gene signatures, including those representing T-cells and macrophages (Supplementary Figure 1). Likelihood ratio testing was performed to assess the independence of immune gene signatures as predictors of survival. Conditioning on clinical variables (node status, age, and hormone receptor status for breast; stage, grade, and age for ovarian) and adding either B-cell or T-cell gene signatures to the model, only one gene signature was needed to significantly improve the predictive power of the model (data not shown). In accordance with the high degree of correlation between immune cell signatures, adding further signatures for the same cell type did not improve the model. Each ordering of immune signatures was tested to ensure this finding was not specific to specific signatures. Including both B-cell and T-cell signatures in the model, in breast cancer only one signature significantly improved the model; in ovarian, one B-cell signature and one T-cell signature each significantly improved the model. Together, these analyses indicate an improved outcome for patients with specific subtypes of breast and ovarian cancer. This correlated with the presence of B-cells, plasmablasts and/or plasma cells in the tumor microenvironment, which suggests a productive endogenous B/plasma cell response may be present in the tumor microenvironment.
Specific B-cell receptor gene segment expression is prognostic in basal-like breast cancer
Next, we wished to determine if the B-cell gene signature found in patients with basal-like breast cancer was consistent with an antigen-specific response. Other groups have shown clonal expansion and somatic hypermutation in breast B-cell TILs, suggesting an antigendirected response in those samples (18, 20, 22). Actively responding antigen-specific B-cell populations are characterized by clonal expansion; thus, we expect B-cell clonal expansion in patients where an effective, antigen-directed anti-tumor response is occurring. Because the clonal diversity of a B-cell population can be inferred by the diversity of the BCRs they express, there should be a prognostic benefit in samples with increased expression of specific BCR gene segments (i.e. immunoglobulin heavy chain and light chain variable, joining, diversity, and constant region segments). It has been shown that the BCR protein from breast cancer TILs is mainly produced by plasma cells, not B-cells (13). Here, we will continue to use the term “B-cell” to refer to the heterogeneous group of BCR-producing cells in the B-cell lineage.
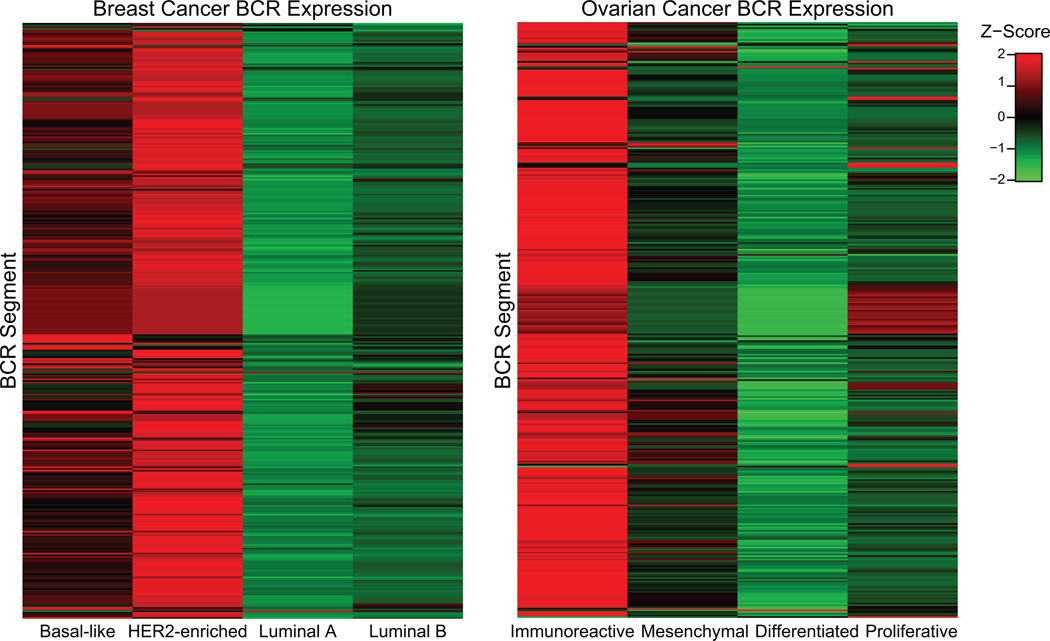
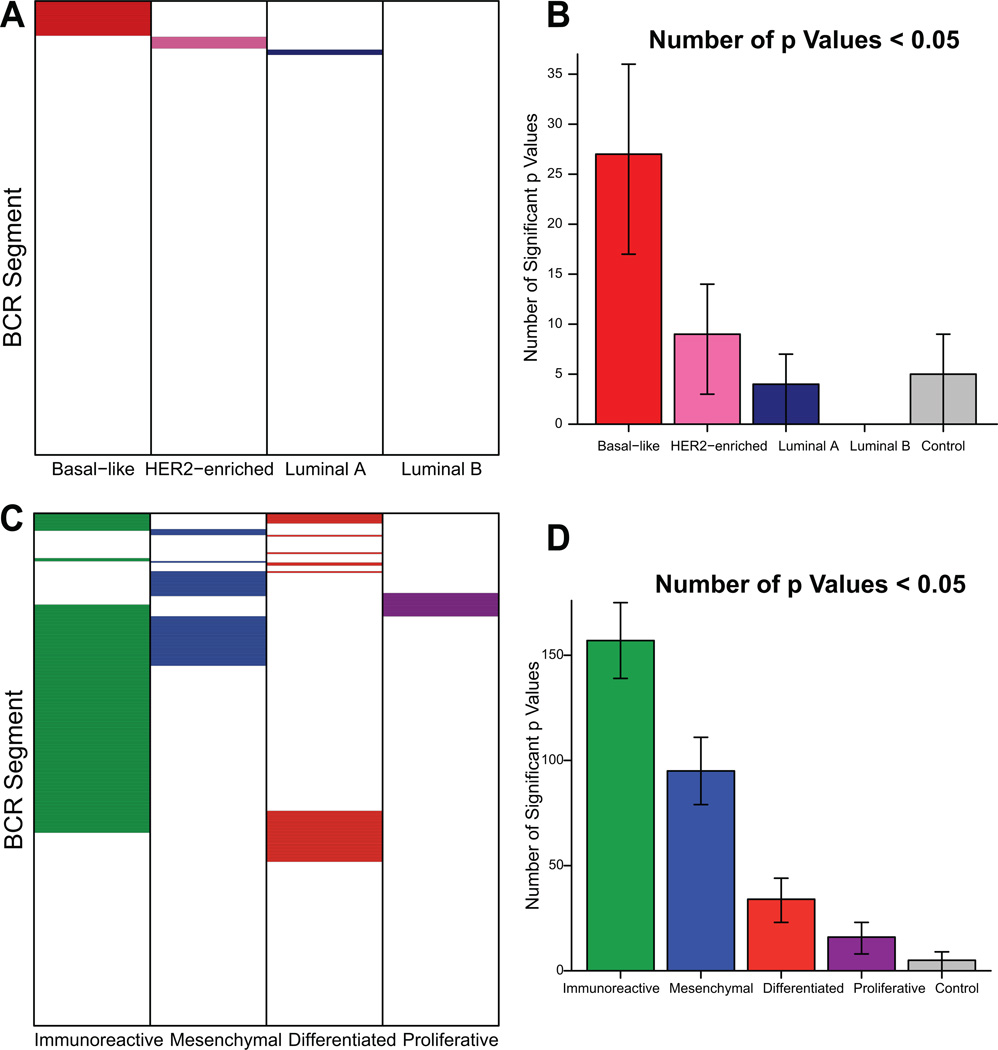
We first calculated the expression levels of all 353 BCR gene segments available in the IMGT® database across the breast and ovarian tumor data. Breast HER2-enriched and basal-like tumors, as well as ovarian immunoreactive and mesenchymal tumors, showed high expression widely across BCR segments (Figure 3). We analyzed prognosis by expression of each individual BCR segment, and then compared this to an identical number of randomly selected genes using a bootstrap procedure to assess the significance of this finding. In basal-like breast tumor subtypes, we identified a significantly greater number of BCR segments predictive of overall survival than expected by random sampling (Figure 4A–B). No other breast cancer subtype demonstrated a greater number of prognostic BCR segments than expected by chance (Figure 4A). Similarly, in patients with ovarian cancer significantly more BCR segments were predictive of progression-free survival in immunoreactive tumors than in the other subtypes (Figure 4C–D). The mesenchymal and differentiated ovarian subtypes also showed significantly more prognostic BCR segments than expected by chance. In breast tumors, this finding cannot be explained solely by increased overall expression of BCR gene segments in the basal-like subtype, as the highest expression of BCR segments was found in the HER2-enriched subtype (Figure 3). Prognostic segments were discovered in multiple gene families including IgHV, IgHJ, IgHC, IgKV, IgKJ, IgKC, IgLV, IgLJ, and IgLC. IgKC, which has been previously identified as prognostic in several solid tumor types including breast cancer (13), predicted progression-free survival in ovarian cancer but did not attain significance in the breast data set. Individual representative plots of overall or progression-free survival by high vs. low expression of representative prognostic segments are shown in Supplementary Figure 2.
Figure 3.
Expression level of all BCR segments across breast cancer subtypes and ovarian cancer. Expression is highest in basal-like and HER2-enriched breast cancer and immunoreactive ovarian cancer subtypes and highly correlated within groups.
Figure 4.
Expression level of BCR segments is preferentially predictive of improved overall survival in basal-like breast cancer and progression-free survival in immunoreactive ovarian cancer. A, C Grid of prognostic value of all BCR segments (colored cells represent positively prognostic segments). B, D Prognostic BCR segment distribution across subtypes, with bootstrap confidence intervals (95%). Control value represents 353 random non-BCR genes.
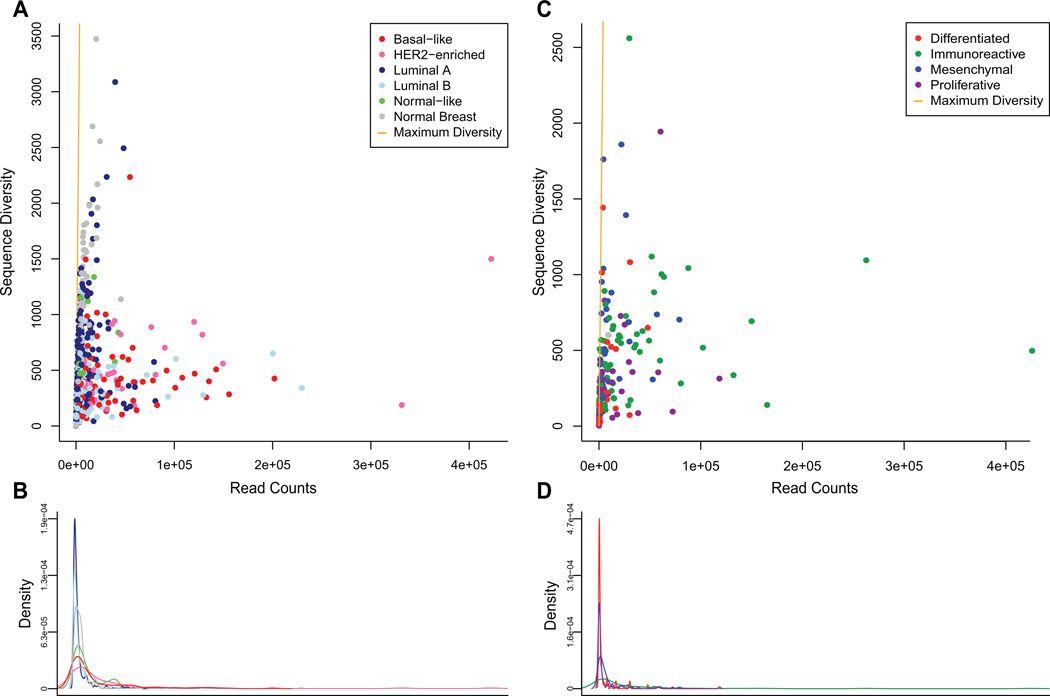
Since B-cells undergo somatic hypermutation following antigen stimulation in the germinal center reaction, reads mapping to each germline BCR gene segment are expected to contain many corresponding single nucleotide variations. Each group of mapped reads corresponding to a BCR gene segment would then exist as a population, the diversity of which should inversely relate to the degree of clonal expansion in the tumor infiltrate. We used the Shannon entropy index normalized as the effective number of species as a measure of diversity(46). For this analysis, we calculated the diversity per patient of each BCR gene segment; a description of this procedure is shown as Supplementary Figure 3. Basal-like, HER2-enriched, and luminal B breast tumors, and immunoreactive ovarian tumors, include a subset of tumors with high expression of low-diversity segments (Figure 5). This finding is consistent with the presence of a clonally-expanded B-cell population within those tumor subtypes that is absent in other subtypes.
Figure 5.
A Basal-like, HER2-enriched, and luminal B tumors show high expression of low-diversity BCR V segment pools (data averaged over top ten highest expressed V segments). B Density of V segment expression in breast cancer. C A subset of predominantly immunoreactive ovarian tumors show high expression of low-diversity BCR V segment pools (data averaged over top ten highest expressed V segments). D Density of V segment expression in ovarian cancer.
Analysis of somatic hypermutation patterns in mRNA-seq data
Somatic hypermutation in BCR gene segments is characterized by mutations that favor defined local sequence regional “hotspots” and CDRs, due to bias in the enzymatic activity that facilitates the mutation process(36). In order to evaluate the degree of somatic hypermutation represented in our data, we made use of the novel de novo assembly algorithm ABRA to assemble unique contigs from reads that map to each BCR variable (V) segment locus, followed by analysis of the contigs for the presence or absence of SHM. These contigs allowed us to analyze SHM mutation patterns across a V segment or its CDRs, rather than interrogating each mRNA-seq read pair separately. An overview of this method is given as Supplementary Figure 4.
We applied our method of analyzing somatic hypermutation in mRNA-seq data to the TCGA breast and ovarian data sets. For the top 10 most highly expressed BCR V gene segments in breast or ovarian tumors in our data sets, the basal-like and HER2-enriched breast subtypes were enriched for tumors with V gene segments consistent with SHM (Supplementary Figure 5). Immunoreactive ovarian tumors showed a high proportion of segments with mutation patterns suggestive of SHM, but it was not significantly higher than the proportion observed in other ovarian subtypes. The presence of SHM sequence characteristics from TILs is suggestive of the presence of antigen-experienced B-cells, potentially against tumor antigens, in the tumor microenvironment.
Discussion
We define here four characteristics of an active, antigen-driven, anti-tumor B-cell response that can be identified from mRNA-seq data, namely: 1) increased expression and prognostic value of B-cell gene signatures, 2) increased expression and prognostic value of BCR gene segments, 3) decreased diversity of highly expressed BCR gene segments, and 4) mutation patterns consistent with BCR somatic hypermutation. All four conditions were found in basal-like breast cancers, and three of these conditions were found for immunoreactive ovarian tumors and HER2-enriched breast tumors. These findings support the hypothesis that a productive B-cell-driven endogenous anti-tumor response may be generated in many basal-like breast and immunoreactive ovarian carcinomas. To our knowledge this represents the first inference of BCR repertoire characteristics from mRNA-seq data.
Investigations into the anti-cancer adaptive immune response have largely been focused on T-cells. Accordingly, current cancer immunotherapy is directed at modifying the T-cell immune response through modulating targets like CTLA4 and PD-L1. In this work, we show that the presence of tumor-infiltrating B-cells correlated with overall and progression-free survival suggesting that B-cells play an important role in anti-tumor immunity. We do show that the expression of B-cell genes was highly correlated with the expression of T-cell genes. By further demonstrating that in specific breast and ovarian cancer subtypes B-cell TILs are clonally expanded and enriched for somatic hypermutation, we provide evidence that B-cell TILs are not merely a surrogate marker for an anti-tumor T-cell response. While it is technically possible that previously expanded B-cell clones may be trafficked to the tumor independent of their antigen binding capability, previous studies showing clonal evolution within breast tumors make this unlikely(19–22), as does the association with specific tumor subtypes. Tumor antigen-directed B-cell responses, which we suggest are present in many basal-like breast and immunoreactive ovarian tumors, may provide a novel way to clinically target these tumor types. Interestingly PD-1 (PDCD1), which is expressed on activated T and B-cells, is currently a very promising target for immunotherapy. Previous work has shown that blocking the interaction of PD-1 with PDL1 and PDL-2 (PDCD1LG2) enhances the activation, proliferation, and cytokine production of human B-cells in the presence of TLR stimulation(57). As immunotherapy advances in breast cancer, it will be important to evaluate B-cell TILs to investigate if and how anti-cancer immunotherapies may modulate the B-cell compartment.
As more immunomodulatory treatments become available for cancer therapy, one critical issue is the identification of the specific cancer patients who may benefit from such therapy. This work highlights the subtype association of clonally restricted B-cell responses. Previous studies in ovarian cancer have been mixed as to the importance of B-cell TILs, perhaps because of the heterogeneity of the B-cell response across the subtypes of ovarian cancer. Milne, et al. highlighted the high-grade serous histologic subtype as being selectively associated with TILs predictive of disease-specific survival(32). Here, we further identify the immunoreactive genomic subtype of serous ovarian cancer as containing prognostic TILs. Among basal-like breast tumors, it is interesting to note that patients with high B-cell infiltration as assessed by gene signatures were also significantly younger than other patients with basal-like breast cancer (data not shown), corroborating previous work highlighting this group(58).
Other investigations of B-cell TILs in breast cancer have found the survival benefit associated with B-cell gene expression to be dependent on proliferation (12, 13, 56). However, we do not see this association in our data. While the basal-like and HER2-enriched subtypes are both highly proliferative, we observed no survival benefit for B-cell TILs in luminal B tumors, which are also characterized by high proliferation. Furthermore, likelihood ratio testing conditioning on clinical variables and genomic subtype (data not shown) demonstrated that proliferation did not significantly increase the predictive ability of the model in breast cancer.
This work again underscores the similarity between basal-like breast and ovarian tumors. Previous genomic studies have established that serous ovarian tumors resemble basal-like breast cancer in terms of their mutational profiles and DNA copy number changes(24). Here we show that this similarity extends to the immune component of the tumor microenvironment. In terms of the immune response, basal-like breast cancer bears more similarity to ovarian immunoreactive cancer than to the luminal A breast cancer subtype. This adds further weight to the notion that the therapeutic approach to basal-like breast and ovarian cancer could be similar.
The claudin-low subtype of breast cancer is also known to have abundant TILs(26), which we confirm by expression of TIL gene signatures (Figure 2A–B). However, unlike the basal-like and HER2-enriched subtypes, TIL abundance was not associated with a survival benefit within claudin-low tumors (Table 1). This could potentially be due to different immunosuppressive mechanisms within the tumor microenvironment, or it is possible that claudin-low tumors elicit a nonspecific inflammatory response in contrast with other high-immune breast tumors. We were unable to assess the BCR sequence diversity of claudin-low TILs as very few (fewer than ten) claudin-low tumors have been identified within the TCGA data set. If TILs within claudin-low tumors are not productive or antigen-directed, misclassification of these tumors may limit the effectiveness of immunomodulatory treatments within triple-negative breast cancers.
There are several standard approaches to analyzing the adaptive immune response present in the tumor microenvironment and tumor-draining lymph node. Immunohistochemistry and immunofluorescence can be performed, although specific antibodies often require frozen tissue; similarly, flow cytometry can be performed on frozen tissue. One of the benefits of the approach described here is the potential to perform the analysis on formalin-fixed paraffin embedded tissue as methods for mRNA-seq from these samples have been demonstrated(59), and will continue to improve and become standardized. As this is available on a substantially greater number of patients compared to frozen tissue, this approach could allow for a much larger group of patients to be analyzed for the presence of adaptive immune signatures. Indeed, there are a large and growing number of human tissue samples with available mRNA-seq data; through the methods described here, these samples may be analyzed for antigen-directed B-cell responses.
Given our data that the presence of B-cells in the tumor microenvironment in patients with basal-like and HER2-enriched breast cancers and immunoreactive ovarian cancer is predictive of outcome, the role that endogenous B-cells play at these sites of tumor growth is a critical question. Plasma cells could generate anti-tumor antibodies that could be important in the early control of the growth of breast cancer cells, but which may ultimately become lost during tumor progression. Alternatively, B-cells may function as antigen-presenting cells to activate tumor-specific T-cells, which in turn may be inhibited via immunosuppressive mechanisms such as the PD-1/PD-L1 interaction. Future work is needed to determine the mechanism by which B-cells affect tumor growth in these different molecular subtypes of cancer, and if and how this could be harnessed to improve endogenous anti-tumor immune responses.
The most difficult breast tumors to treat clinically are often of the “triple-negative” class, defined as such by the lack of cell-surface expression of estrogen receptor, progesterone receptor, and HER2(60). The majority (60–80%) of triple-negative tumors are basal-like (61), and thus the basal-like subtype represents a critical target for the development of novel therapeutics. The presence of BCR characteristics associated with overall survival and consistent with a productive anti-tumor endogenous B-cell response suggests that methods to enhance or induce anti-tumor B-cells in patients with basal-like breast cancer may be clinically efficacious.
Supplementary Material
Statement of Translational Relevance.
Immunomodulatory therapies such as those targeting CTLA4 and PD-L1 (CD274) have been shown to be effective in a number of tumor types. These treatments primarily target the T-cell component of the adaptive immune system. In this work, we provide further evidence that tumor-infiltrating B-cells are also important in anti-tumor immunity in basal-like breast and immunoreactive ovarian cancer, suggesting that immunomodulatory therapy may be effective in these tumor types. By identifying basal-like breast cancer as an important setting for immunomodulatory treatment, we provide a rationale for the use of targeted therapies in many clinically triple-negative breast cancers, where no targeted therapies currently exist.
Acknowledgements
We thank the TCGA Network for the mRNA-seq data for the Breast and Ovarian samples. We were supported by grants from the NCI Breast SPORE program grant P50-CA58223-09A1, The Cancer Genome Atlas (U24-CA143848-05), RO1-CA-138255, F31-CA177188-01, the Breast Cancer Research Foundation, and the University Cancer Research Fund.
Footnotes
Disclosure of Potential Conflicts of Interest
C.M.P is an equity stock holder, and Board of Director Member, of University Genomics/BioClassifier LLC. C.M.P and J.S.P are also listed as inventors on a patent application on the PAM50 assay.
References
- 1.Lee AH, Gillett CE, Ryder K, Fentiman IS, Miles DW, Millis RR. Different patterns of inflammation and prognosis in invasive carcinoma of the breast. Histopathology. 2006;48:692–701. doi: 10.1111/j.1365-2559.2006.02410.x. [DOI] [PubMed] [Google Scholar]
- 2.Yakirevich E, Izhak OB, Rennert G, Kovacs ZG, Resnick MB. Cytotoxic phenotype of tumor infiltrating lymphocytes in medullary carcinoma of the breast. Mod Pathol. 1999;12:1050–1056. [PubMed] [Google Scholar]
- 3.Rakha EA, Aleskandarany M, El-Sayed ME, Blamey RW, Elston CW, Ellis IO, et al. The prognostic significance of inflammation and medullary histological type in invasive carcinoma of the breast. Eur J Cancer. 2009;45:1780–1787. doi: 10.1016/j.ejca.2009.02.014. [DOI] [PubMed] [Google Scholar]
- 4.Mahmoud SM, Paish EC, Powe DG, Macmillan RD, Grainge MJ, Lee AH, et al. Tumor-infiltrating CD8+ lymphocytes predict clinical outcome in breast cancer. J Clin Oncol. 2011;29:1949–1955. doi: 10.1200/JCO.2010.30.5037. [DOI] [PubMed] [Google Scholar]
- 5.Liu S, Lachapelle J, Leung S, Gao D, Foulkes WD, Nielsen TO. CD8+ lymphocyte infiltration is an independent favorable prognostic indicator in basal-like breast cancer. Breast Cancer Res. 2012;14:R48. doi: 10.1186/bcr3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ben-Hur H, Cohen O, Schneider D, Gurevich P, Halperin R, Bala U, et al. The role of lymphocytes and macrophages in human breast tumorigenesis: an immunohistochemical and morphometric study. Anticancer Res. 2002;22:1231–1238. [PubMed] [Google Scholar]
- 7.Leong PP, Mohammad R, Ibrahim N, Ithnin H, Abdullah M, Davis WC, et al. Phenotyping of lymphocytes expressing regulatory and effector markers in infiltrating ductal carcinoma of the breast. Immunol Lett. 2006;102:229–236. doi: 10.1016/j.imlet.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 8.Dobrzanski MJ, Reome JB, Hylind JC, Rewers-Felkins KA. CD8-mediated type 1 antitumor responses selectively modulate endogenous differentiated and nondifferentiated T cell localization, activation, and function in progressive breast cancer. J Immunol. 2006;177:8191–8201. doi: 10.4049/jimmunol.177.11.8191. [DOI] [PubMed] [Google Scholar]
- 9.Gobert M, Treilleux I, Bendriss-Vermare N, Bachelot T, Goddard-Leon S, Arfi V, et al. Regulatory T cells recruited through CCL22/CCR4 are selectively activated in lymphoid infiltrates surrounding primary breast tumors and lead to an adverse clinical outcome. Cancer Res. 2009;69:2000–2009. doi: 10.1158/0008-5472.CAN-08-2360. [DOI] [PubMed] [Google Scholar]
- 10.Xu L, Zhou Y, Xiao DM, Qin M, Luo JM, Tang XY. The change of CD4+ CD25high CCR6+ regulatory T cells in breast cancer patients. Sichuan Da Xue Xue Bao Yi Xue Ban. 2010;41:415–419. [PubMed] [Google Scholar]
- 11.Fan C, Prat A, Parker JS, Liu Y, Carey LA, Troester MA, et al. Building prognostic models for breast cancer patients using clinical variables and hundreds of gene expression signatures. BMC Med Genomics. 2011;4:3. doi: 10.1186/1755-8794-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmidt M, Bohm D, von Torne C, Steiner E, Puhl A, Pilch H, et al. The humoral immune system has a key prognostic impact in node-negative breast cancer. Cancer research. 2008;68:5405–5413. doi: 10.1158/0008-5472.CAN-07-5206. [DOI] [PubMed] [Google Scholar]
- 13.Schmidt M, Hellwig B, Hammad S, Othman A, Lohr M, Chen Z, et al. A comprehensive analysis of human gene expression profiles identifies stromal immunoglobulin kappa C as a compatible prognostic marker in human solid tumors. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18:2695–2703. doi: 10.1158/1078-0432.CCR-11-2210. [DOI] [PubMed] [Google Scholar]
- 14.Teschendorff AE, Miremadi A, Pinder SE, Ellis IO, Caldas C. An immune response gene expression module identifies a good prognosis subtype in estrogen receptor negative breast cancer. Genome Biol. 2007;8:R157. doi: 10.1186/gb-2007-8-8-r157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rody A, Karn T, Liedtke C, Pusztai L, Ruckhaeberle E, Hanker L, et al. A clinically relevant gene signature in triple negative and basal-like breast cancer. Breast Cancer Res. 2011;13:R97. doi: 10.1186/bcr3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mahmoud SM, Lee AH, Paish EC, Macmillan RD, Ellis IO, Green AR. The prognostic significance of B lymphocytes in invasive carcinoma of the breast. Breast Cancer Res Treat. 2012;132:545–553. doi: 10.1007/s10549-011-1620-1. [DOI] [PubMed] [Google Scholar]
- 17.Coronella JA, Telleman P, Truong TD, Ylera F, Junghans RP. Amplification of IgG VH and VL (Fab) from single human plasma cells and B cells. Nucleic acids research. 2000;28:E85. doi: 10.1093/nar/28.20.e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hansen MH, Nielsen H, Ditzel HJ. The tumor-infiltrating B cell response in medullary breast cancer is oligoclonal and directed against the autoantigen actin exposed on the surface of apoptotic cancer cells. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:12659–12664. doi: 10.1073/pnas.171460798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coronella JA, Telleman P, Kingsbury GA, Truong TD, Hays S, Junghans RP. Evidence for an antigen-driven humoral immune response in medullary ductal breast cancer. Cancer research. 2001;61:7889–7899. [PubMed] [Google Scholar]
- 20.Coronella JA, Spier C, Welch M, Trevor KT, Stopeck AT, Villar H, et al. Antigen-driven oligoclonal expansion of tumor-infiltrating B cells in infiltrating ductal carcinoma of the breast. Journal of immunology. 2002;169:1829–1836. doi: 10.4049/jimmunol.169.4.1829. [DOI] [PubMed] [Google Scholar]
- 21.Hansen MH, Nielsen HV, Ditzel HJ. Translocation of an intracellular antigen to the surface of medullary breast cancer cells early in apoptosis allows for an antigen-driven antibody response elicited by tumor-infiltrating B cells. Journal of immunology. 2002;169:2701–2711. doi: 10.4049/jimmunol.169.5.2701. [DOI] [PubMed] [Google Scholar]
- 22.Nzula S, Going JJ, Stott DI. Antigen-driven clonal proliferation, somatic hypermutation, and selection of B lymphocytes infiltrating human ductal breast carcinomas. Cancer research. 2003;63:3275–3280. [PubMed] [Google Scholar]
- 23.Coronella-Wood JA, Hersh EM. Naturally occurring B-cell responses to breast cancer. Cancer immunology, immunotherapy: CII. 2003;52:715–738. doi: 10.1007/s00262-003-0409-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prat A, Perou CM. Deconstructing the molecular portraits of breast cancer. Mol Oncol. 2011;5:5–23. doi: 10.1016/j.molonc.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12:R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herschkowitz JI, Simin K, Weigman VJ, Mikaelian I, Usary J, Hu Z, et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007;8:R76. doi: 10.1186/gb-2007-8-5-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cancer Genome Atlas Research N. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gooden MJ, de Bock GH, Leffers N, Daemen T, Nijman HW. The prognostic influence of tumour-infiltrating lymphocytes in cancer: a systematic review with meta-analysis. British journal of cancer. 2011;105:93–103. doi: 10.1038/bjc.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hwang WT, Adams SF, Tahirovic E, Hagemann IS, Coukos G. Prognostic significance of tumor-infiltrating T cells in ovarian cancer: a meta-analysis. Gynecol Oncol. 2012;124:192–198. doi: 10.1016/j.ygyno.2011.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Milne K, Kobel M, Kalloger SE, Barnes RO, Gao D, Gilks CB, et al. Systematic analysis of immune infiltrates in high-grade serous ovarian cancer reveals CD20, FoxP3 and TIA-1 as positive prognostic factors. PLoS One. 2009;4:e6412. doi: 10.1371/journal.pone.0006412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nielsen JS, Sahota RA, Milne K, Kost SE, Nesslinger NJ, Watson PH, et al. CD20+ tumor-infiltrating lymphocytes have an atypical CD27- memory phenotype and together with CD8+ T cells promote favorable prognosis in ovarian cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18:3281–3292. doi: 10.1158/1078-0432.CCR-12-0234. [DOI] [PubMed] [Google Scholar]
- 34.Maul RW, Gearhart PJ. Controlling somatic hypermutation in immunoglobulin variable and switch regions. Immunologic research. 2010;47:113–122. doi: 10.1007/s12026-009-8142-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vinuesa CG, Linterman MA, Goodnow CC, Randall KL. T cells and follicular dendritic cells in germinal center B-cell formation and selection. Immunological reviews. 2010;237:72–89. doi: 10.1111/j.1600-065X.2010.00937.x. [DOI] [PubMed] [Google Scholar]
- 36.Odegard VH, Schatz DG. Targeting of somatic hypermutation. Nature reviews Immunology. 2006;6:573–583. doi: 10.1038/nri1896. [DOI] [PubMed] [Google Scholar]
- 37.Cancer Genome Atlas Research N. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harrell JC, Prat A, Parker JS, Fan C, He X, Carey L, et al. Genomic analysis identifies unique signatures predictive of brain, lung, and liver relapse. Breast Cancer Res Treat. 2012;132:523–535. doi: 10.1007/s10549-011-1619-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 41.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic acids research. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Palmer C, Diehn M, Alizadeh AA, Brown PO. Cell-type specific gene expression profiles of leukocytes in human peripheral blood. BMC Genomics. 2006;7:115. doi: 10.1186/1471-2164-7-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rody A, Holtrich U, Pusztai L, Liedtke C, Gaetje R, Ruckhaeberle E, et al. T-cell metagene predicts a favorable prognosis in estrogen receptor-negative and HER2-positive breast cancers. Breast Cancer Res. 2009;11:R15. doi: 10.1186/bcr2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic acids research. 2005;33:D54–D58. doi: 10.1093/nar/gki031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang K, Singh D, Zeng Z, Coleman SJ, Huang Y, Savich GL, et al. MapSplice: accurate mapping of RNA-seq reads for splice junction discovery. Nucleic acids research. 2010;38:e178. doi: 10.1093/nar/gkq622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jost L. Entropy and diversity. Oikos. 2006;113:363–375. [Google Scholar]
- 47.Lefranc MP, Giudicelli V, Ginestoux C, Bodmer J, Muller W, Bontrop R, et al. IMGT, the international ImMunoGeneTics database. Nucleic acids research. 1999;27:209–212. doi: 10.1093/nar/27.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ruiz M, Giudicelli V, Ginestoux C, Stoehr P, Robinson J, Bodmer J, et al. IMGT, the international ImMunoGeneTics database. Nucleic acids research. 2000;28:219–221. doi: 10.1093/nar/28.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lefranc MP. IMGT, the international ImMunoGeneTics database. Nucleic acids research. 2001;29:207–209. doi: 10.1093/nar/29.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lefranc MP. IMGT, the international ImMunoGeneTics database. Nucleic acids research. 2003;31:307–310. doi: 10.1093/nar/gkg085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lefranc MP, Giudicelli V, Kaas Q, Duprat E, Jabado-Michaloud J, Scaviner D, et al. IMGT, the international ImMunoGeneTics information system. Nucleic acids research. 2005;33:D593–D597. doi: 10.1093/nar/gki065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lefranc MP, Clement O, Kaas Q, Duprat E, Chastellan P, Coelho I, et al. IMGT-Choreography for immunogenetics and immunoinformatics. In silico biology. 2005;5:45–60. [PubMed] [Google Scholar]
- 53.Lefranc MP, Giudicelli V, Ginestoux C, Jabado-Michaloud J, Folch G, Bellahcene F, et al. IMGT, the international ImMunoGeneTics information system. Nucleic acids research. 2009;37:D1006–D1012. doi: 10.1093/nar/gkn838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rogozin IB, Pavlov YI, Bebenek K, Matsuda T, Kunkel TA. Somatic mutation hotspots correlate with DNA polymerase eta error spectrum. Nat Immunol. 2001;2:530–536. doi: 10.1038/88732. [DOI] [PubMed] [Google Scholar]
- 55.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nagalla S, Chou JW, Willingham MC, Ruiz J, Vaughn JP, Dubey P, et al. Interactions between immunity, proliferation and molecular subtype in breast cancer prognosis. Genome Biol. 2013;14:R34. doi: 10.1186/gb-2013-14-4-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thibult ML, Mamessier E, Gertner-Dardenne J, Pastor S, Just-Landi S, Xerri L, et al. PD-1 is a novel regulator of human B-cell activation. International Immunology. 2013;25:129. doi: 10.1093/intimm/dxs098. [DOI] [PubMed] [Google Scholar]
- 58.Schmidt M, Hengstler JG, von Torne C, Koelbl H, Gehrmann MC. Coordinates in the universe of node-negative breast cancer revisited. Cancer research. 2009;69:2695–2698. doi: 10.1158/0008-5472.CAN-08-4013. [DOI] [PubMed] [Google Scholar]
- 59.Adiconis X, Borges-Rivera D, Satija R, DeLuca DS, Busby MA, Berlin AM, et al. Comparative analysis of RNA sequencing methods for degraded or low-input samples. Nature methods. 2013;10:623–629. doi: 10.1038/nmeth.2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Carey LA, Dees EC, Sawyer L, Gatti L, Moore DT, Collichio F, et al. The triple negative paradox: primary tumor chemosensitivity of breast cancer subtypes. Clin Cancer Res. 2007;13:2329–2334. doi: 10.1158/1078-0432.CCR-06-1109. [DOI] [PubMed] [Google Scholar]
- 61.Rakha EA, Ellis IO. Triple-negative/basal-like breast cancer: review. Pathology. 2009;41:40–47. doi: 10.1080/00313020802563510. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.