Abstract
The p97 AAA (ATPase associated with diverse cellular activities), also called VCP (valosin-containing protein), is an important therapeutic target for cancer and neurodegenerative diseases. p97 forms a hexamer composed of two AAA domains (D1 and D2) that form two stacked rings, and an N-terminal domain that binds numerous cofactor proteins. The interplay between the three domains in p97 is complex, and a deeper biochemical understanding is needed in order to design selective p97 inhibitors as therapeutic agents. It is clear that the D2 ATPase domain hydrolyzes ATP in vitro, but whether D1 contributes to ATPase activity is controversial. Here, we use Walker A and B mutants to demonstrate that D1 is capable of hydrolyzing ATP, and show for the first time that nucleotide binding in the D2 domain increases the catalytic efficiency (kcat/Km) of D1 ATP hydrolysis 280-fold, by increasing kcat 7-fold and decreasing Km about 40-fold. We further show that an ND1 construct lacking D2 but including the linker between D1 and D2 is catalytically active, resolving a conflict in the literature. Applying enzymatic observations to small-molecule inhibitors, we show that four p97 inhibitors (DBeQ, ML240, ML241, and NMS-873) have differential responses to Walker A and B mutations, to disease-causing IBMPFD mutations, and to the presence of the N-domain binding cofactor protein p47. These differential effects provide the first evidence that p97 cofactors and disease mutations can alter p97 inhibitor potency and suggest the possibility of developing context-dependent inhibitors of p97.
Keywords: p97/VCP AAA ATPase, IBMPFD/ALS, steady-state kinetics, SPR, p97 inhibitor
Introduction
The highly conserved and abundant p97 protein belongs to the type II AAA ATPase enzyme family and converts chemical energy from ATP hydrolysis into mechanical energy to unfold proteins or disassemble protein complexes. p97 plays a critical role in cellular processes such as Golgi membrane reassembly (1), membrane transport (2,3), degradation of proteins by the ubiquitin–proteasome system (UPS) (4,5), regulation of myofibril assembly (6), cell division (7), and protein aggregation (8–10). Because p97 functions in many protein homeostatic regulatory processes, it is a potential therapeutic target for cancer and neurodegenerative diseases (11–13).
Each monomer of p97 ATPase is composed of three domains (the N, D1 ATPase, and D2 ATPase domains) joined by linker regions. D1 and D2 each form a hexameric ring, which stack to form a cylindrical structure, as revealed by X-ray crystallography (14,15). Although it is clear that the D2 domain hydrolyzes ATP in vitro, the level of D1-specific ATPase activity varies among different reports (16–18). Furthermore, genetic studies in yeast reporting D1 ATPase activity as an essential function are controversial. For example, some mutants that inhibit ATP hydrolysis in the D1 domain affect growth (17), whereas others do not (19). However, a consistent observation is that the nucleotide-binding state of D1 is tightly controlled (15,18). Thus, it is important to clarify the role of the D1 domain in vitro, providing hypotheses that can then be tested in cells.
Adjacent to the D1 domain is the N domain, which functions as a protein-protein interaction site that binds p97 cofactors (20,21). By recruiting cofactor proteins, the N domain links the mechanochemical ATPase activity of p97 to many substrate proteins, accounting in part for the broad range of cellular p97 functions. For example, cofactor p47 is required for p97-mediated membrane fusion (22), by mediating binding of p97 to syntaxin 5 during the reassembly of Golgi stacks at the end of mitosis (23). Binding of p47 to the N-terminus alters the structure and function of p97 by significantly reducing the diameter of the p97 ring (22) and inhibiting ATPase activity (24). Hence, p47 supports communication between the N, D1 and D2 domains of p97.
There are several additional lines of evidence for conformational crosstalk among the three domains of p97. Conformational changes occur during the ATPase cycle, including rotation of the D1 and D2 rings with respect to each other, changes in the hexameric ring diameter, and positioning of the N-domain (25–29). Additionally, pathogenic mutations in the N and D1 domains alter nucleotide binding to the D1 domain and ATPase activity in the D2 domain (29–31). Defining interactions between the D1 and D2 domains could provide opportunities for developing small-molecule inhibitors with diverse mechanisms of inhibition and perhaps diverse cellular effects.
In an effort to better understand the mechanistic and cellular functions of p97, a few potent and specific small molecule inhibitors of p97 ATPase activity have been identified via high-throughput screening, followed by chemical optimization (32–35). Published reversible p97 ATPase inhibitors fall into two classes – ATP-competitive (DBeQ, ML240, ML241) and noncompetitive (NMS-873) inhibitors. NMS-873 is thought to inhibit p97 by binding at the D1–D2 interdomain linker and stabilizing the D2-ADP-bound form, thus breaking the catalytic cycle. Both competitive and noncompetitive inhibitors affect multiple p97-dependent processes in cells, including ubiquitin fusion degradation (UFD), endoplasmic reticulum associated degradation (ERAD), autophagy, and cancer cell growth. However, investigations have not yet shown whether competitive compounds inhibit ATPase activity arising from only D2 or from both D1 and D2, and whether inhibitors are influenced by nucleotide binding states, disease mutations, or cofactor binding.
In this study, we enzymatically characterized a set of p97 constructs and mutations to elucidate the effect of small-molecule p97 inhibitors on D1 and D2 domain ATPase activities. We first clarified the in vitro activity of the D1 domain. We found that an ND1-linker truncation is a robust, catalytically competent ATPase whose activity is similar to that of the D1 domain in full-length p97. Interestingly, D1 showed ATPase activity in the full-length protein only when the D2 domain was capable of binding nucleotides. Using our enzymatically characterized set of p97 proteins, we addressed the domain specificity of four potent p97 inhibitors on wild type (WT) p97. We then evaluated whether inhibition was sensitive to pathogenic mutations in the ND1 domain or to the presence of bound p47, a p97 binding protein. DBeQ and NMS-873 inhibited both ATPase domains, whereas ML240 and ML241 were specific for D2. In addition, inhibition of D2 by ML240 and ML241 was altered by a pathogenic mutation in ND1 and upon p47 binding, indicating domain communication within p97. Together, our results provide the framework for developing domain, cofactor-complex, and pathway specific inhibitors (32), with the ultimate goal of validating p97 as a potential therapeutic target.
Results
The Human p97 D1 Domain is a Competent ATPase
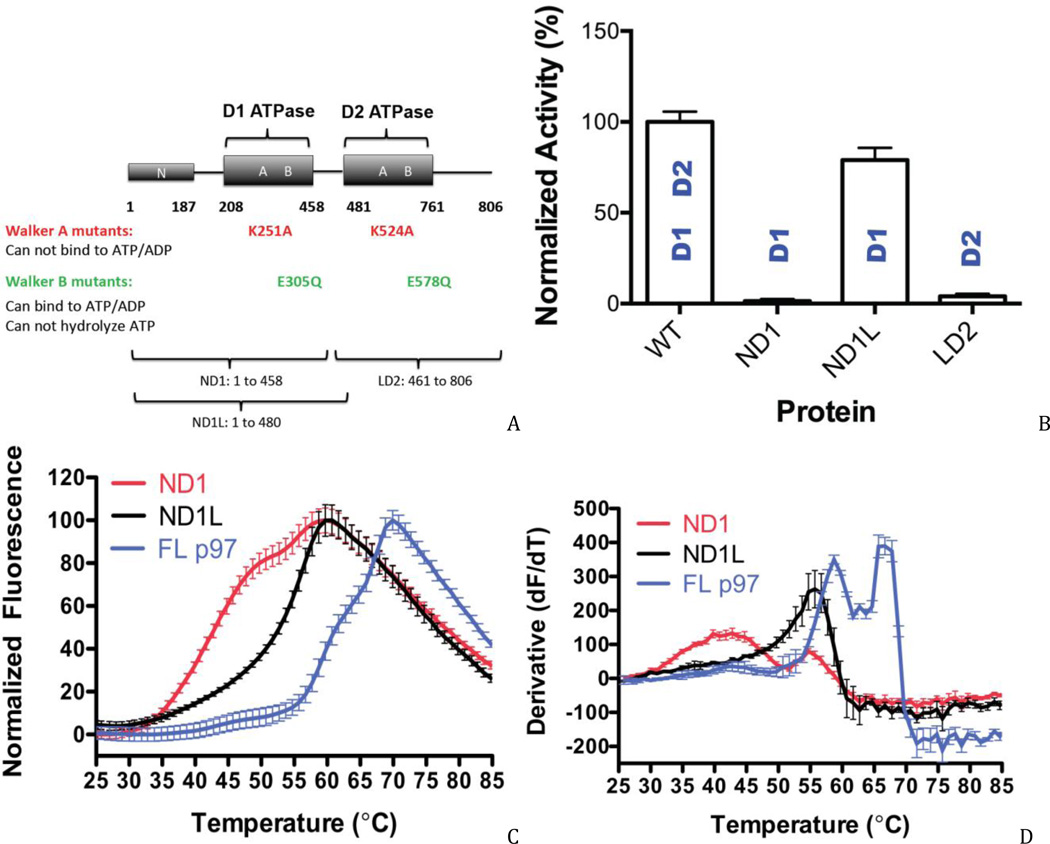
To resolve the controversy over whether the isolated D1 domain can hydrolyze ATP in vitro, we prepared two truncated constructs: ND1 (residues 1–458) without the D1–D2 linker region, and ND1L (1–480), which included the linker region (Fig. 1A). ND1 was used previously to conclude that D1 was inactive (16), whereas ND1L has been used in crystallography studies (31) and was recently reported to be catalytically active (29). Specific activities with 600 µM ATP at room temperature for human and mouse p97 were 5.2±0.3 and 5.1±0.2 nmol Pi/min/nmol p97, respectively. We found that the ATPase activity of ND1 was 1.4% of the WT level, whereas ND1L had 79% of the WT activity (at 600 uM ATP; Fig. 1B). Thus, ND1L was 56-fold more active than ND1. As a comparison to ND1L, we also prepared a truncation containing the D1–D2 linker region plus the D2 domain (LD2; amino acids 461–806). LD2 existed in dynamic multimer-monomer equilibrium (Figs. S1), and thus it was not surprising that its ATPase activity was only 4% of WT values (Fig. 1B).
Figure 1. Comparison of WT and isolated domain variants.
(A) Human p97 mutants and variants generated in this study. (B) ATPase activities of WT p97, ND1, ND1L, and LD2 domains with 0.01% Triton X-100 at 600 µM ATP. (C, D) The linker region (a.a. 459–480; SNPSALRETVVEVPQVTWEDIG) stabilized the D1 domain. Differential scanning fluorimetry revealed enhanced stability of the ND1 domain with the linker peptide (i.e., a 13 °C higher melting temperature). Melting temperatures: ND1 (Tm 41.9 °C, 55.7 °C), ND1L (Tm 55.5 °C) and full-length p97 (Tm 59.7 °C, 66.0 °C).
To address the difference in activities between ND1 and ND1L, we evaluated their stabilities by differential scanning fluorimetry (DSF; Figs. 1C and D) and nucleotide binding by surface plasmon resonance (SPR; Figs. 2A and B). Thermal stability studies showed that addition of the linker strongly stabilized ND1 by 13 °C, to a temperature that approaches the first melting transition of full-length p97. Additionally, ND1 showed multiple melting transitions, whereas ND1L gave a single, highly cooperative melting curve. The increased thermal stability and single-state melting curve suggested that ND1L formed a more stable protein than ND1. To determine whether ND1 bound weakly to nucleotides, leading to low catalytic activity, we monitored binding of nucleotides to ND1 and ND1L by SPR at 20 °C, well below the melting temperature for ND1. Both ND1 and ND1L bound to ATP, ATPγS, or ADP with similar binding affinities and kinetics (within 2-fold) (Fig. S2). Hence, the linker region had only a small effect on nucleotide-binding affinity, which did not account for the 56-fold increase in ATPase activity. Rather, we concluded that the linker region rendered the D1 domain a competent ATP hydrolase mainly by substantially increasing its thermal stability. We cannot rule out that the linker also favorably altered the conformational dynamics of the protein.
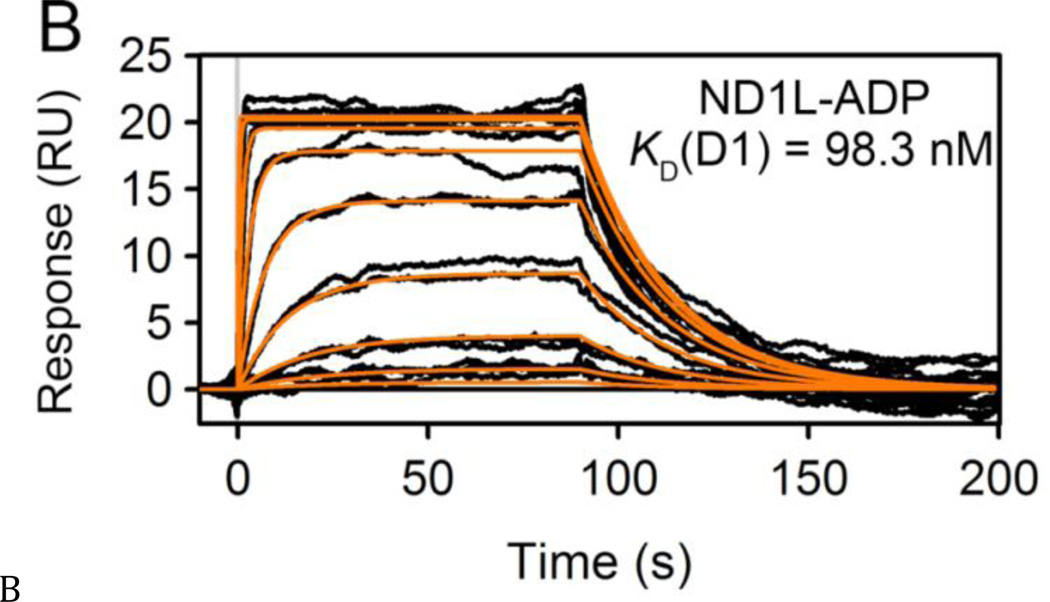
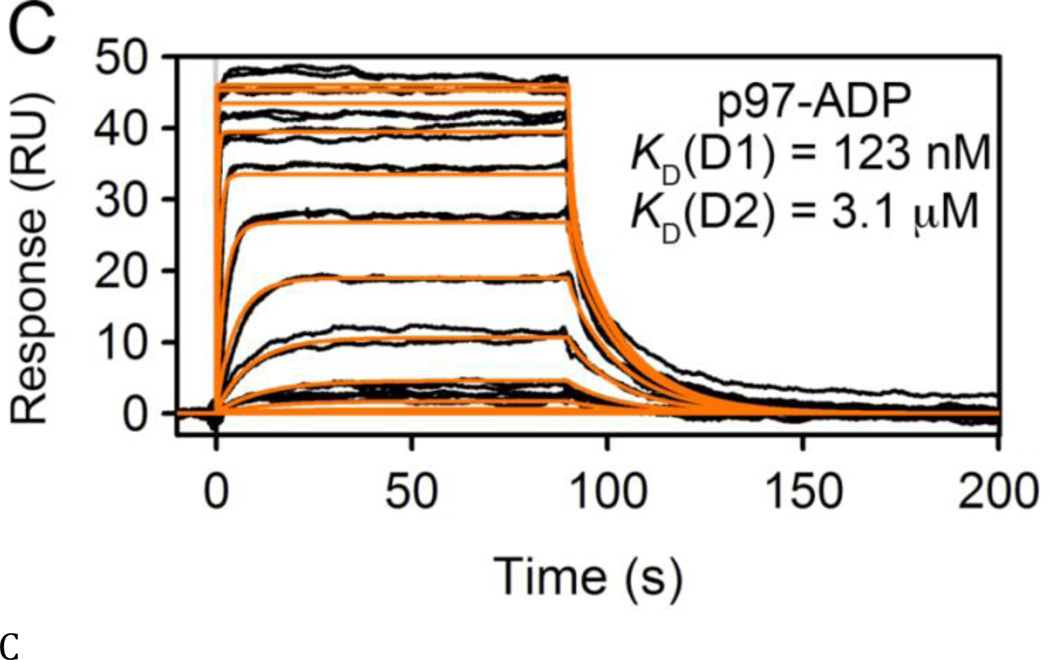
Figure 2. Analysis of nucleotide binding to p97.
(A, B) SPR sensorgrams for ADP (0–16.67 µM, 3-fold dilutions) binding to the ND1 (A) and ND1L (B) domains of p97. Response data were fit to a one-site kinetic model (orange lines) to obtain kinetic binding parameters. (C) SPR sensorgrams for ADP (0–150 µM, 3-fold dilutions), binding to full-length p97. Data were fit to a two-site kinetic binding model to obtain KD values for the two ATPase domains in p97.
By directly comparing the two constructs that were used in the previous studies, we identified the source of the conflicting results on D1 activity. We demonstrated that ND1L can hydrolyze ATP, which is in a good agreement with the recent report by Tang and coworkers (29). In contrast, ND1 cannot hydrolyze ATP, which is consistent with the earlier report by Song and coworkers (16).
Nucleotide Binding and Enzyme Kinetic Analysis of Walker A and B Mutants
We next directly compared the nucleotide binding affinities and activity of ND1L to full-length WT p97 and active-site mutants. Walker A mutations (D1-K251A and D2-K524A) disrupt ATP and ADP binding, whereas Walker B mutations (D1-E305Q and D2-E578Q) disrupt activation of hydrolysis but allow ATP and ADP binding (16,17,36) (Fig. 1A). For nucleotide binding analysis, full-length WT p97 and Walker A mutants were assayed by SPR to compare the binding affinities of the D1 domain in the context of the full-length p97 protein versus isolated ND1 domains (Figs. 2C, S2, and Table S2). Nucleotides bound to WT p97 with two distinct KD values. The tight binding affinity (132, 131, and 36.7 nM for ADP, ATP, ATPgS, respectively) corresponded to the KD values obtained for ND1, ND1L, and the D2-K524A mutant (which is unable to bind nucleotides in the D2 domain). Thus, the D1 site bound to nucleotides with KD values in the 100 nM range. Based on these results and the binding to the D1-K251A mutant (which is unable to bind nucleotides in the D1 domain), we ascribed the second, weaker binding affinity to the D2 domain (3.9, 5.5, and 1.1 µM for ADP, ATP, ATPgS, respectively). It is noteworthy that analysis of nucleotide binding by SPR resulted in KD values similar to those obtained by fluorescence polarization (see below) and tryptophan fluorescence (18,31), but gave ~10-fold tighter binding affinities compared to values from ITC (18,31). Overall, our SPR results supported the previous studies showing that D1 bound nucleotides more tightly than did D2 (18,35).
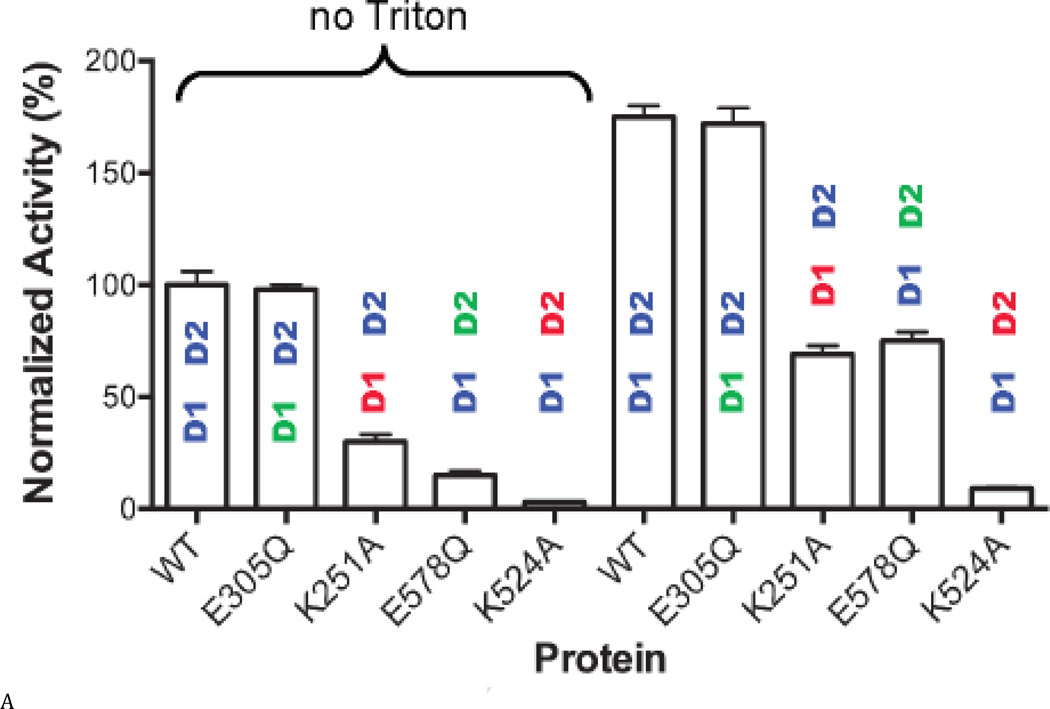
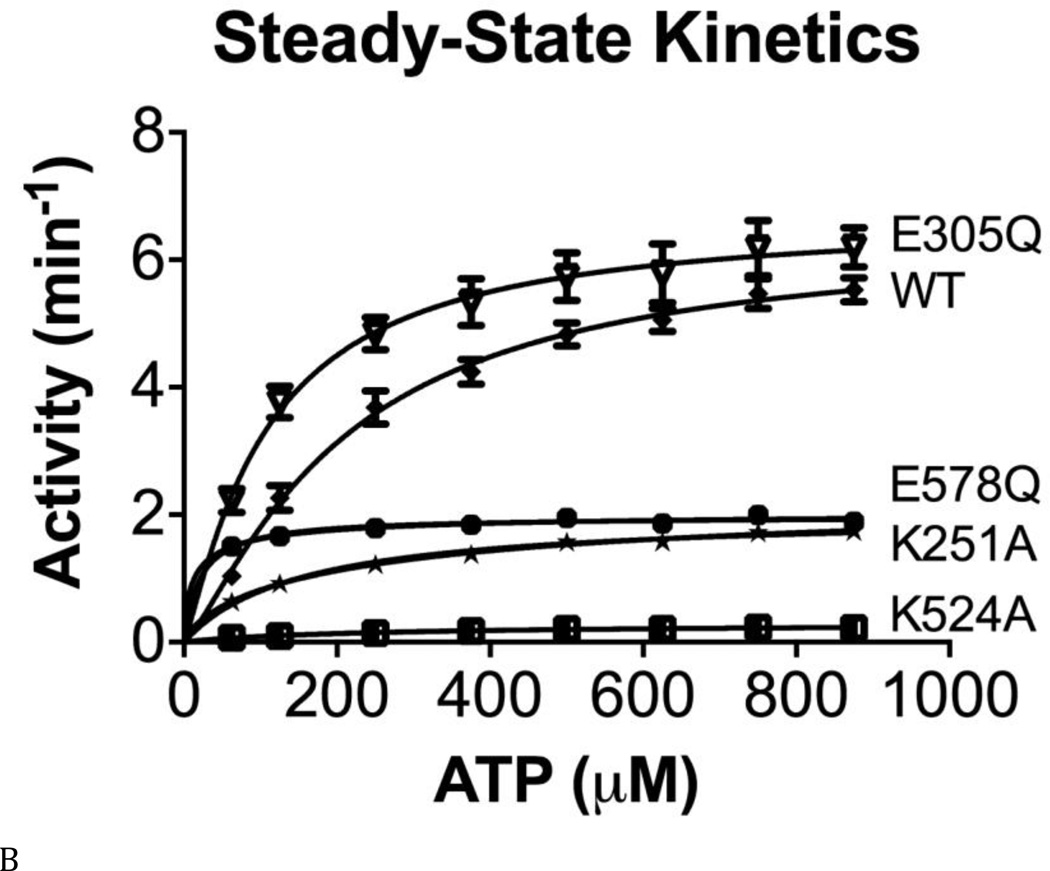
We next measured the activity of the Walker A and B mutations, to evaluate how nucleotide binding or ATP hydrolysis in one domain may alter the ATPase activity of the other domain. We found that Walker A mutations lowered ATPase activity more than did the Walker B mutations, supporting the importance of nucleotide binding in one domain for ATPase activity of the other domain (Fig. 3A, black font indicates the active domain in each protein). We included 0.01% Triton X-100 in our standard reaction buffer and observed a 1.7 fold increase in ATPase activity for WT p97 upon addition of Triton X-100 (Fig. 3A). In general, the increase in ATPase activity by Triton X-100 was greater for the D1-active Walker B mutant (D2-E578Q) compared to the D2-active Walker B mutant (D1-E305Q) (Fig. 3A).
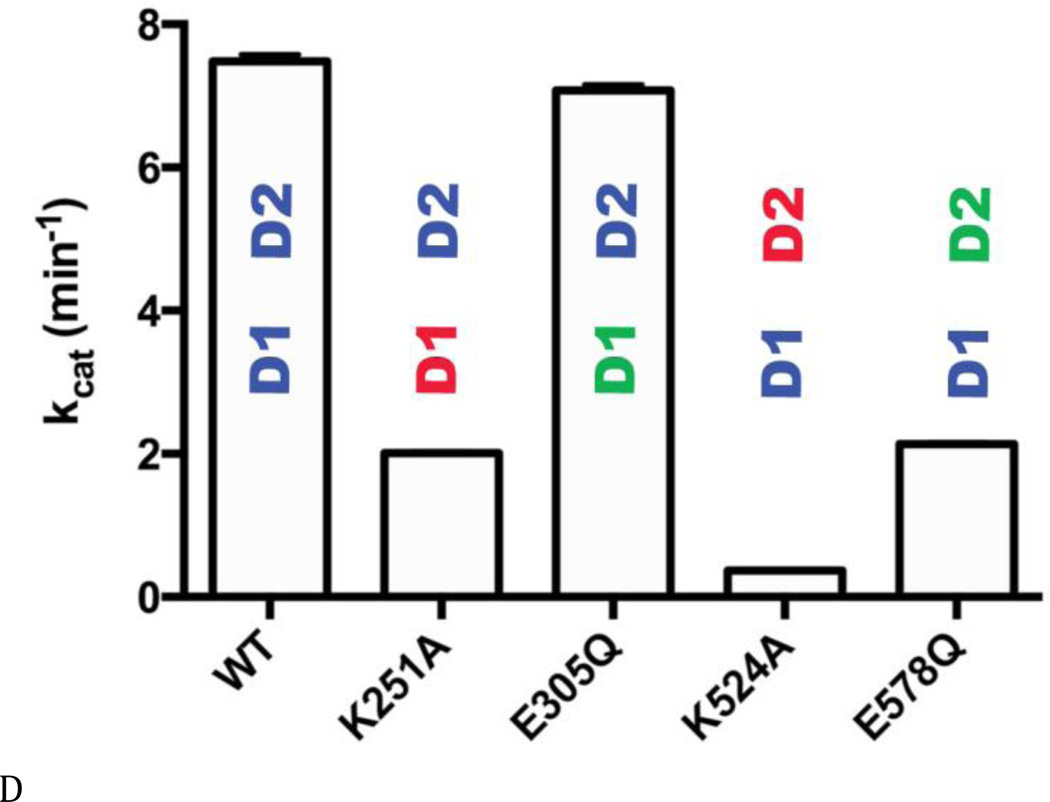
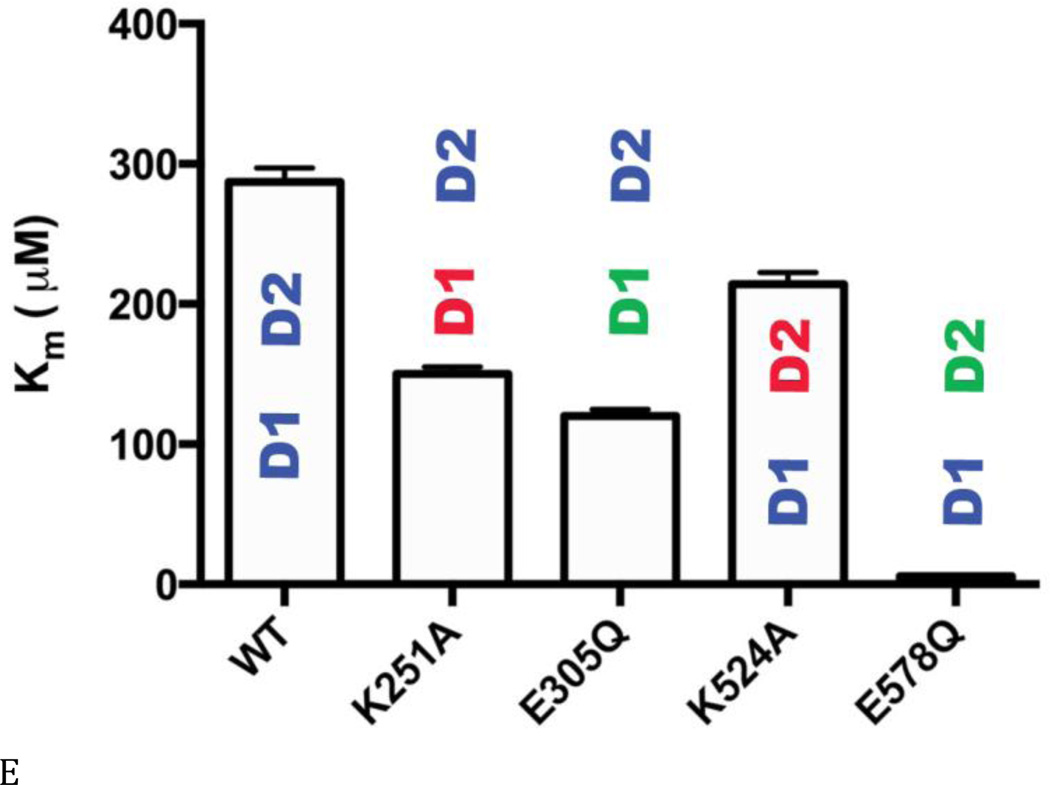
Figure 3. Steady state kinetic analyses of human p97 ATPase activity.
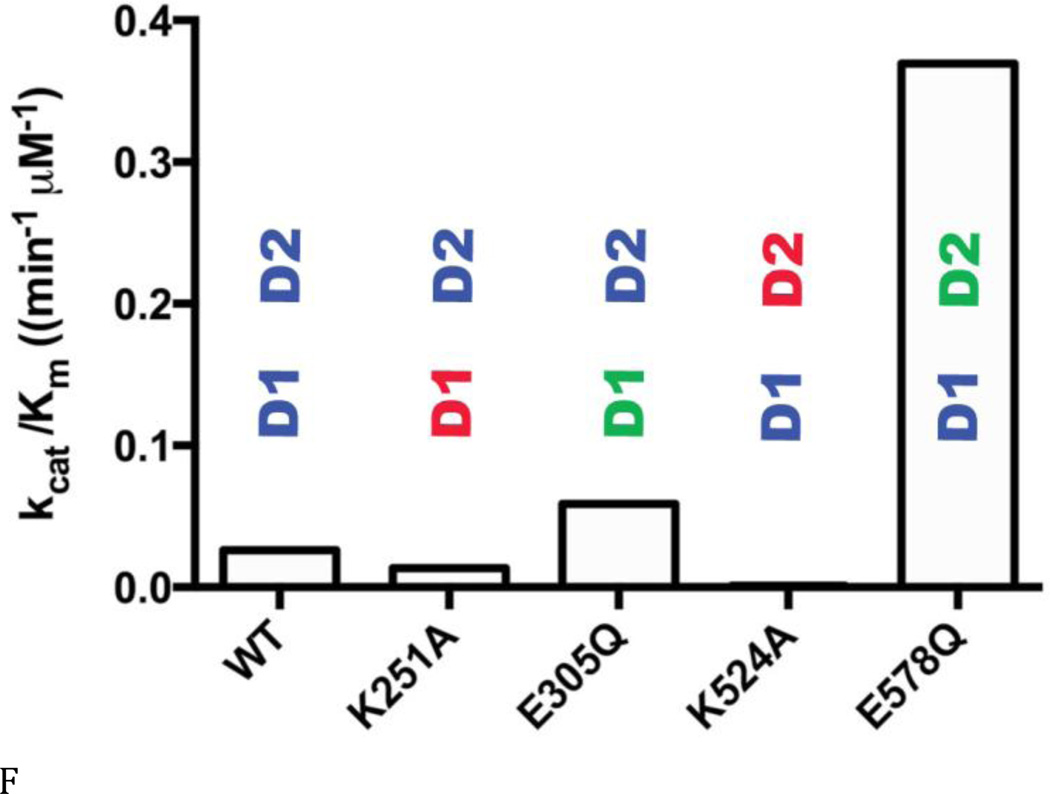
(A) ATPase activities of WT and Walker A and B mutants for D1 (D1-K251A; D1-E305Q) and D2 (D2-K524A; D2-E578Q), with or without 0.01% Triton X-100 at 200 µM ATP. Blue lettering indicates the ATPase active domain in each protein. Red lettering indicates the Walker A mutant. Green lettering indicates the Walker B mutant. (B) and (C) Michaelis-Menten plots of ATP hydrolysis for WT and mutant p97. From (D) to (F), black font indicates the ATPase active domain in each protein. (D) WT p97 hydrolyzes 7.48 ± 0.01 ATP molecules per minute per monomer (turnover number, kcat, min−1). The Walker A mutation of D2 (D2-K524A) decreases kcat 22-fold. (E) The apparent Michaelis-Menten constant, Km, of WT p97 is 287 ± 10 µM. The Walker B mutation of D2 (D2-E578Q) decreases Km 50-fold. (F) The catalytic efficiency (kcat/Km) for WT p97 is 0.026 min−1 µM−1. A 2-fold decrease for the Walker A mutation of D1 (D1-K251A), a 3-fold increase for the Walker B mutation of D1 (D1-E305Q), a 15-fold decrease for the Walker A mutation of D2 (D2-K524A), and a 14-fold increase for the Walker B mutation of D2 (D2-E578Q) together suggest that D1 is a catalytically competent ATPase, when D2 is able to bind to nucleotides.
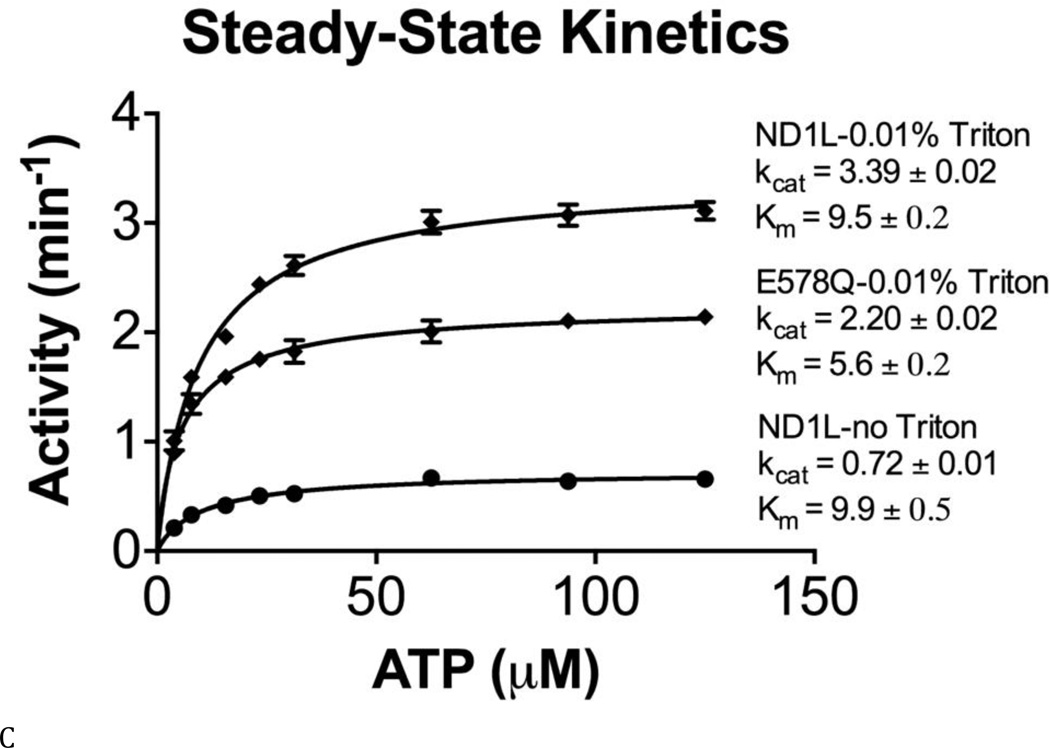
These activity data prompted us to measure steady-state kinetic constants (kcat, Km, and kcat/Km), in order to understand the enzymology of D1 ATPase activity in the context of full-length p97 and to evaluate the crosstalk between the D1 and D2 domains (Table S3 and Figs. 3B and C). Published kinetic studies have focused on only full-length WT p97 (16) and the full-length D1-E305Q mutant (37). Our data for these constructs were consistent with the published values (16,38). The most striking deleterious effects of Walker mutations were the 22-fold reduction in kcat (from 7.5 to 0.29 min−1) (Fig. 3D) and the10-fold reduction in catalytic efficiency (kcat/Km; from 0.026 to 0.0013 min−1uM−1) (Fig. 3F) for the Walker A mutation in the D2 domain (D2-K524A), compared to WT. Thus, the strongest effect on catalysis came from blocking binding of nucleotides to the D2 domain. Simply blocking catalysis of D2 without blocking nucleotide binding (D2-E578Q) gave a modest 3-fold effect on kcat and actually increased catalytic efficiency through a reduction in Km (kcat/Km from 0.026 to 0.39 min−1uM−1). D1-E305Q also showed a slightly higher kcat/Km than WT. Taken with D2-E578Q, the data suggested a cross-inhibitory effect on the activity of the D1 and D2 domains. These observations were consistent with the negative cooperativity between the D1 and D2 rings shown for mouse p97 (28) and with the recent demonstration that the presence of D2 inhibited D1 activity (29). Finally, it is noteworthy that ND1L gave the same kinetic constants as D2-E578Q (Table S3 and Fig. 3C). Thus, ND1L can serve as an accurate kinetic model of the D1 domain in the full-length p97.
It is noteworthy that adding detergent (Triton X-100) to the assay buffer was critical for observing robust ND1L activity (Fig. 3C). Triton X-100 increased kcat 4.7-fold, and had no effect on Km. This result suggested that detergent facilitated catalysis, perhaps by aiding the release of prebound ADP from the hexamer (18,29). It was also likely that detergent prevented protein aggregation and reduced protein binding to the surface of the plastic assay plates. We have included 0.01% Triton X-100 in our standard reaction buffer (32,34) and detergents like Triton X-100 and Tween-20 are frequently added when testing smallmolecule inhibitors to prevent compounds from forming colloids that can cause non-specific enzyme inhibition (39).
ATP-Competitive p97 Inhibitors, ML240 and ML241 are Selective for the D2 domain
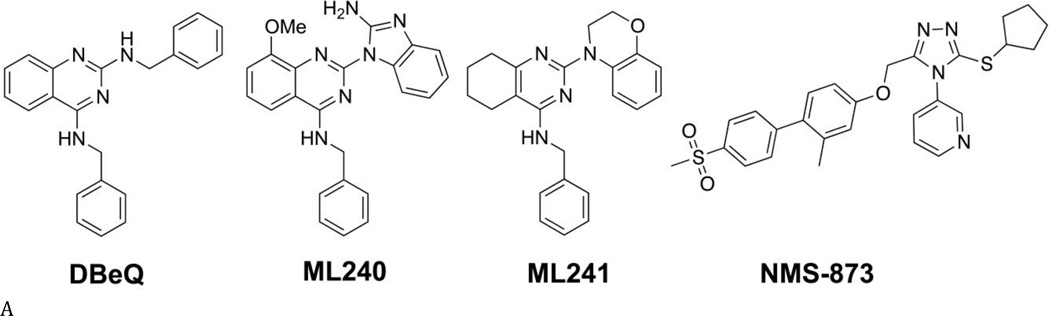
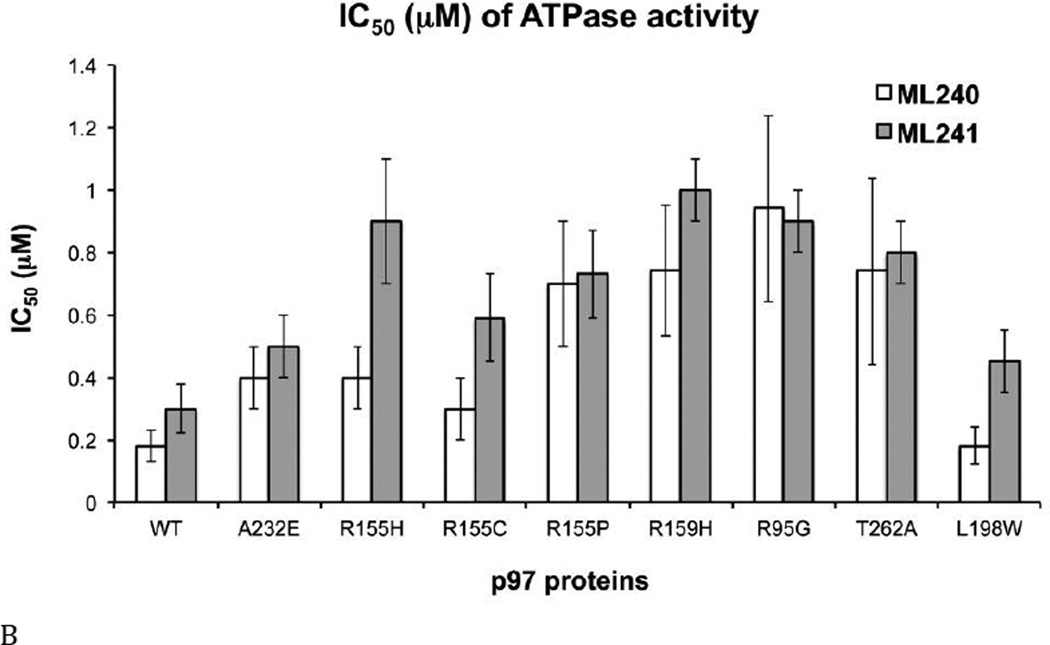
We previously identified three potent, ATP-competitive p97 inhibitors (DBeQ, ML240, and ML241; Fig. 4A) (32,34). Nerviano Medical Sciences and Genentech, Inc., identified NMS-873 as a potent non-ATP-competitive p97 inhibitor, with potencies in the low nanomolar range (Table S4) (33,35). To investigate whether these four p97 compounds inhibit the ATPase activity of D1, D2, or both domains, IC50 values were determined for WT p97 and five variants in the presence of 200 µM or 875 µM ATP. Lower potencies were observed for inhibition of WT p97 (roughly 2 to 5-fold increases in IC50; Fig. 4 and Tables S4 and S5).
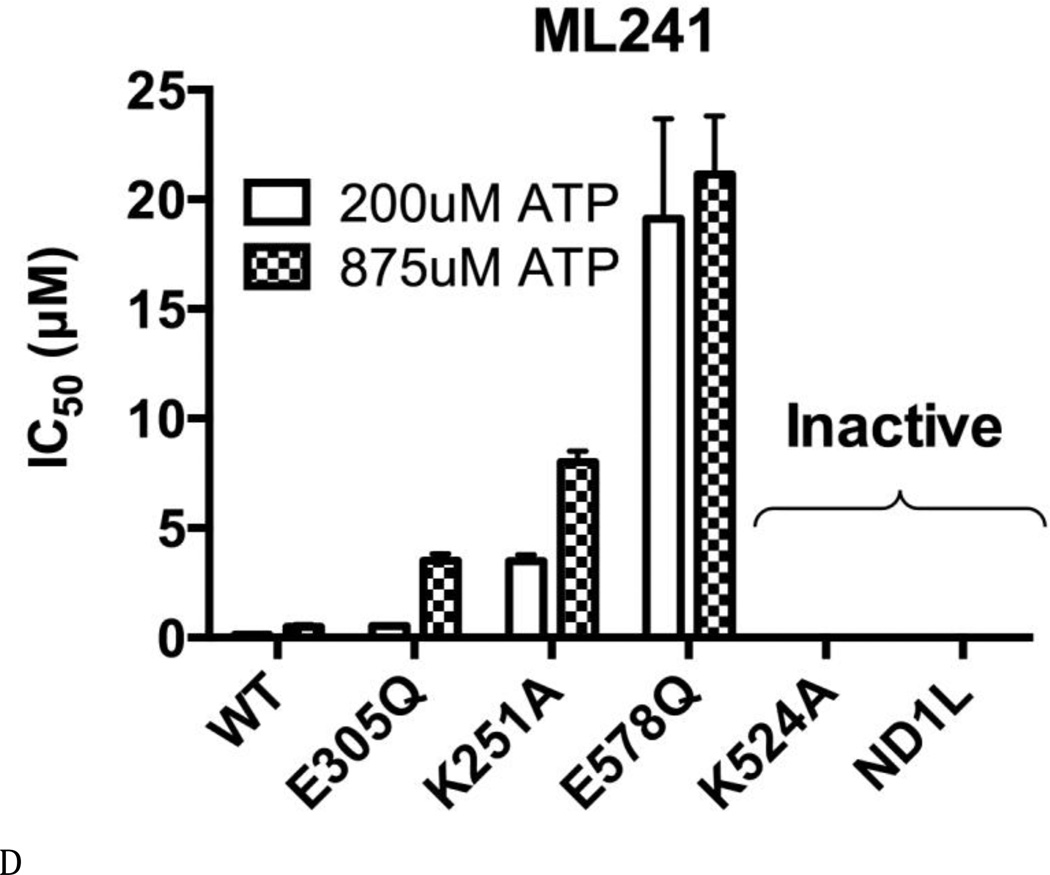
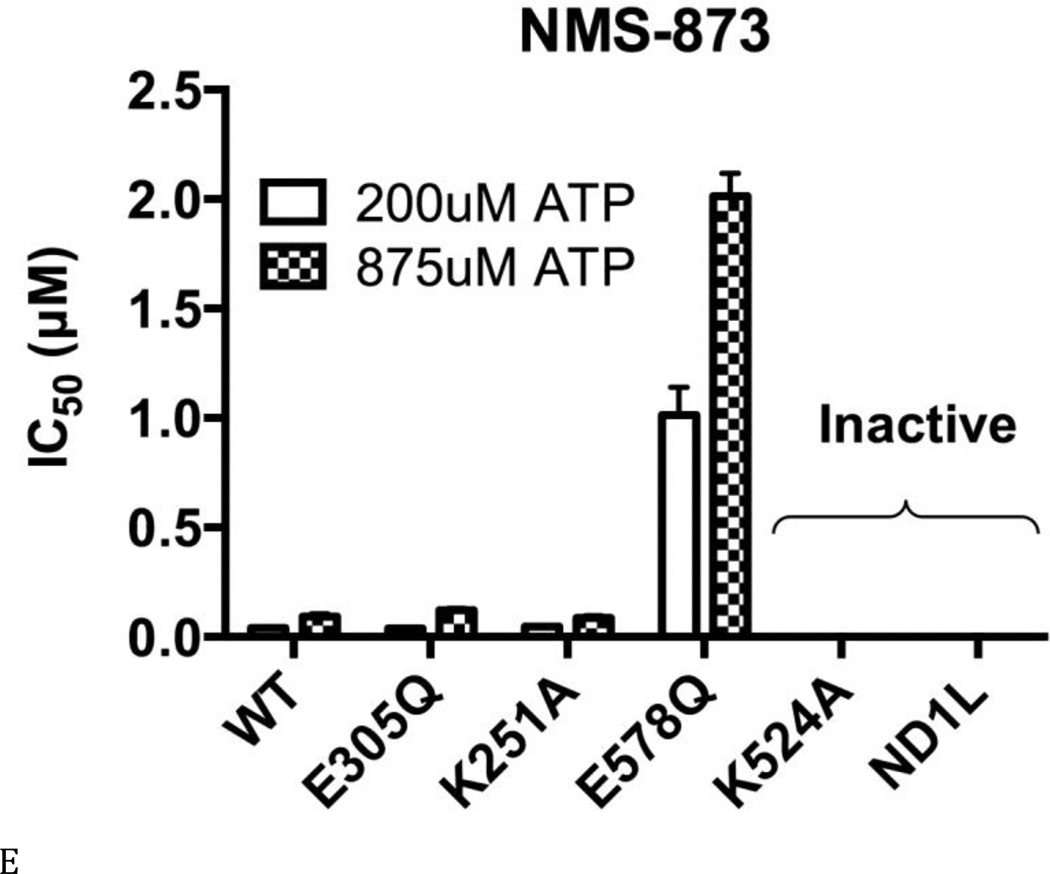
Figure 4. ML240 and ML241 selectively inhibit the D2 domain.
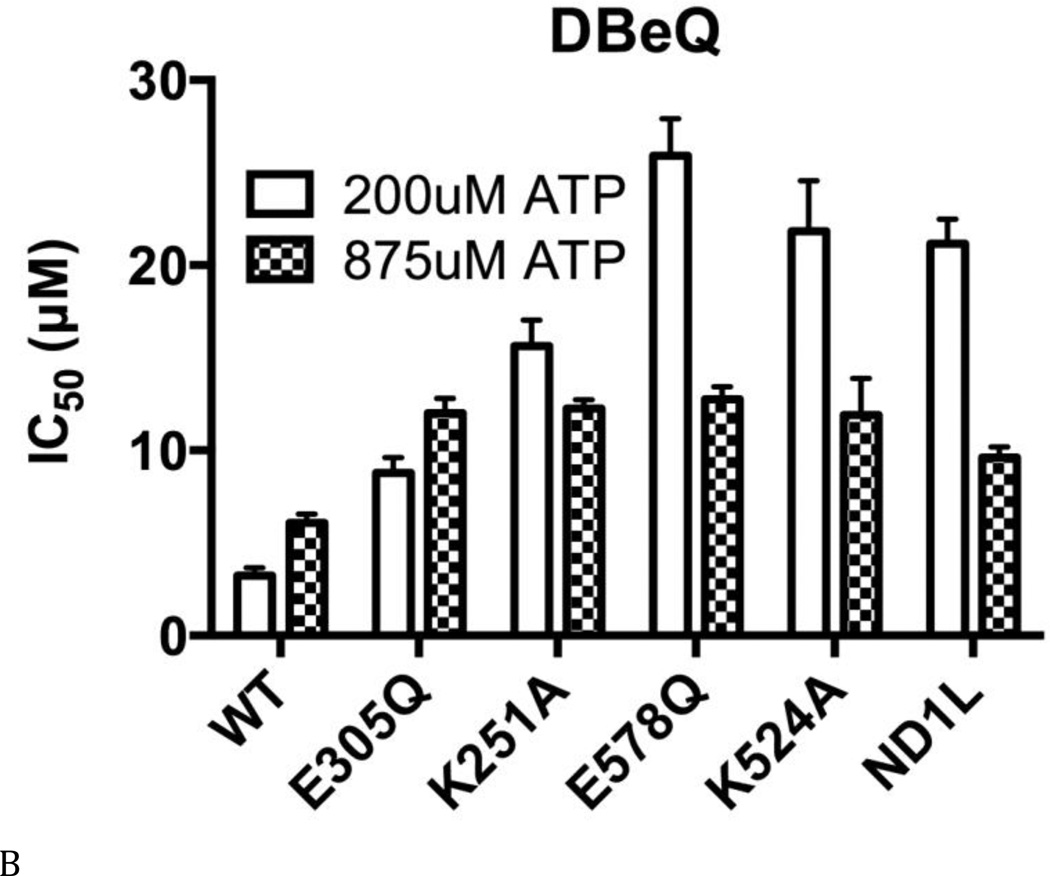
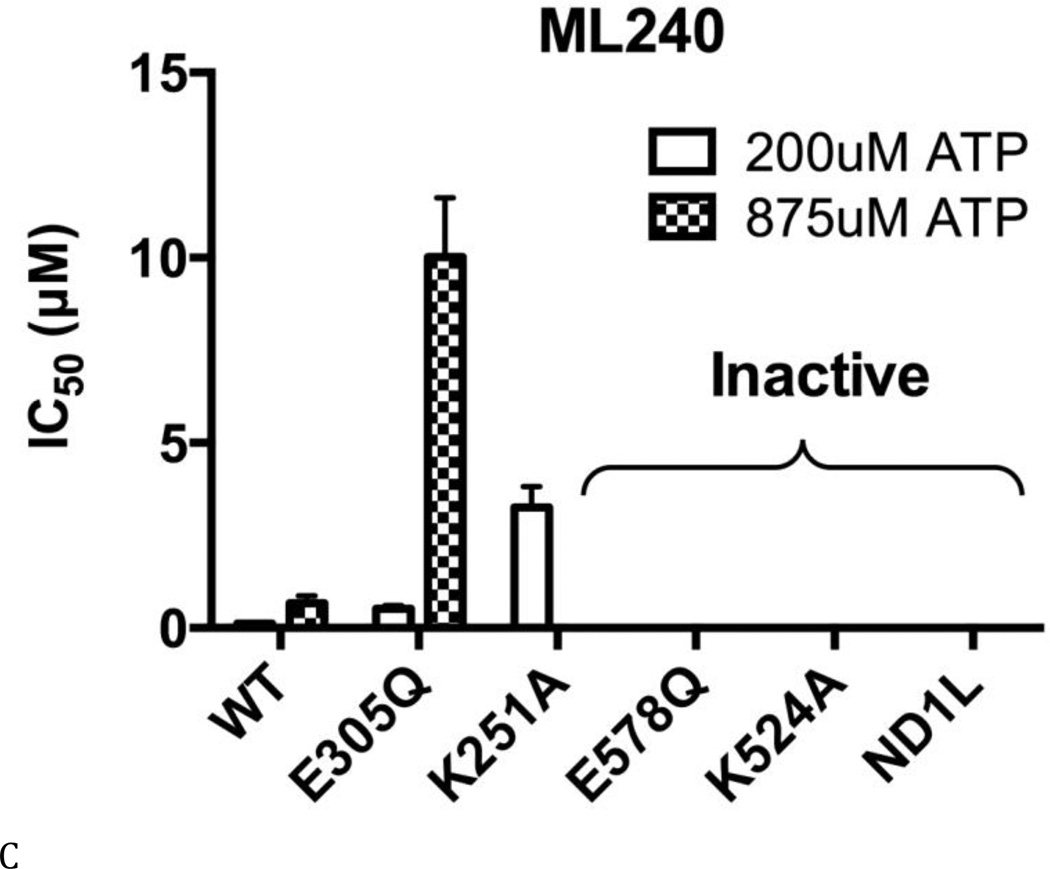
(A) Structures of DBeQ, ML240, ML241, and NMS-873. (B) to (E) IC50 (µM) of WT, D1-E305Q, D1-K251A, D2-E578Q, D2-K524A, and ND1L for (B) DBeQ, (C) ML240, (D) ML241, and (E) NMS-873 with 200 µM ATP or 875 µM ATP.
The ATP competitive inhibitors were sensitive to Walker A and B mutations in both domains. There was a 2.7- to 10-fold increase in IC50 for D1-E305Q (Walker B mutant in the D1 domain) for DBeQ, ML240, and ML241 (Figs. 4 B–D). These increases in IC50 can be partly explained by the 2.4-fold decrease in Km for D1-E305Q, because ATP competitive inhibitors are sensitive to the levels of ATP binding (Fig. 3E). In contrast to the smaller effects found for the D1 D1-E305Q mutant, ML240 was inactive toward D2 mutants D2-E578Q and D2-K524A, and toward ND1L. ML241 was also inactive towards D2-K524A and ND1L, and showed 147-fold weaker inhibition for D2-E578Q. Thus, both ML240 and ML241 were D2 selective inhibitors (Figs. 4 C and D). DBeQ was less selective, showing 8 to 10-fold weaker inhibition of D2-E578Q, D2-K524A, and ND1L (Fig. 4B). The “D2 specific” inhibitors were nevertheless sensitive to changes in D1. For example, the D1 D1-K251A mutation increased the IC50 for ML240 and ML241 by 27-fold with 200 µM ATP and ML240 can not inhibit D1 D1-K251A with 875 µM ATP, suggesting that the architecture of the D2 active site was changed significantly due to the mutation and/or due to the absence of nucleotide binding to D1. This result was consistent with our kinetic analysis of D1-K251A (Fig. 3B), which also showed strong effects of this mutation on the activity of the D2 domain.
The allosteric inhibitor NMS-873 sensed mutations in the D2 domain, but was insensitive to D1 mutations (Tables S4 and S5, Fig. 4E). In agreement with the published data for Walker A mutants, NMS-873 inhibited D1-K251A and WT p97 similarly, but could not bind and inhibit D2-K524A (35). Unexpectedly, NMS-873 inhibited D2-E578Q (IC50 1.0 µM, or 25-fold higher than for WT p97, Fig. 4E), suggesting that maintaining D2 in the ATP-bound state allowed NMS-873 to bind and prevent D1 from hydrolyzing ATP. In addition to the proposed mechanism, whereby NMS-873 prevented ADP release from D2, our D2-E578Q data suggested that NMS-873 locked p97 into a conformation that may have also prevented ADP release from D1. The ATP-binding sites in D1 and D2 are similar for DBeQ binding, because the IC50 values for D1-E305Q, D1-K251A, D2-E578Q, D2-K524A, and the isolated ND1L are almost identical at higher concentrations of ATP (Table S5). Therefore, DBeQ inhibits both D1 and D2 ATPase activities similarly. However, ML240 and ML241 behave quite differently. They are much more sensitive to Walker A and B mutations and cannot inhibit D2-E578Q, D2-K524A, and ND1L as potently as seen for D1-E305Q. Thus, they are more selective for inhibiting D2 ATPase activity. Even though NMS-873 does not bind to ATP binding sties, its ability to bind to p97 and inhibit D1 ATPase is diminished for the D2-K524A mutant (35). However, when D1 and D2 can both bind nucleotides (as in D2-E578Q), NMS-873 inhibits D1 ATP hydrolysis, but 25-fold less potent as compared to D1-E305Q p97. In summary, the four inhibitors fall into three classes: NMS-873, which binds at the D1-D2 interface, seems to inhibit ATPase activity from both D1 and D2 domains; the ATP-competitive inhibitors DBeQ can inhibit both D1 and D2 ATPase activity; and ML240 and ML241 are selective against the D2 domain.
Taken together, the D1 ATPase activity is more difficult to block by p97 inhibitors (Figs. 4 B to E, D2-E578Q). This is consistent with results of our steady state kinetic analysis of D2-E578Q, the mutant with the lowest Km for ATP (Figs. 3B, C, and E) and tighter nucleotide binding to D1 (Table S2 and Figs. 2 and 5A). Furthermore, all ATP-competitive inhibitors (DBeQ, ML240, and ML241) displayed lower inhibitory potencies (higher IC50 values) toward all ATPase-defective mutants (Walker A/B mutants), compared to the WT. On the other hand, the non-ATP-competitive inhibitor NMS-873 showed similar potencies toward the D1 mutants (D1-E305Q and D1-K251A), compared to the WT, but much lower potencies toward D2 mutants (D2-E578Q and D2-K524A). In other words, the mutants in general are more resistant to inhibitors, especially the D2 Walker A mutant (D2-K524A). Such resistance seems expected, due to the significant impairment of their steady state behavior toward ATP.
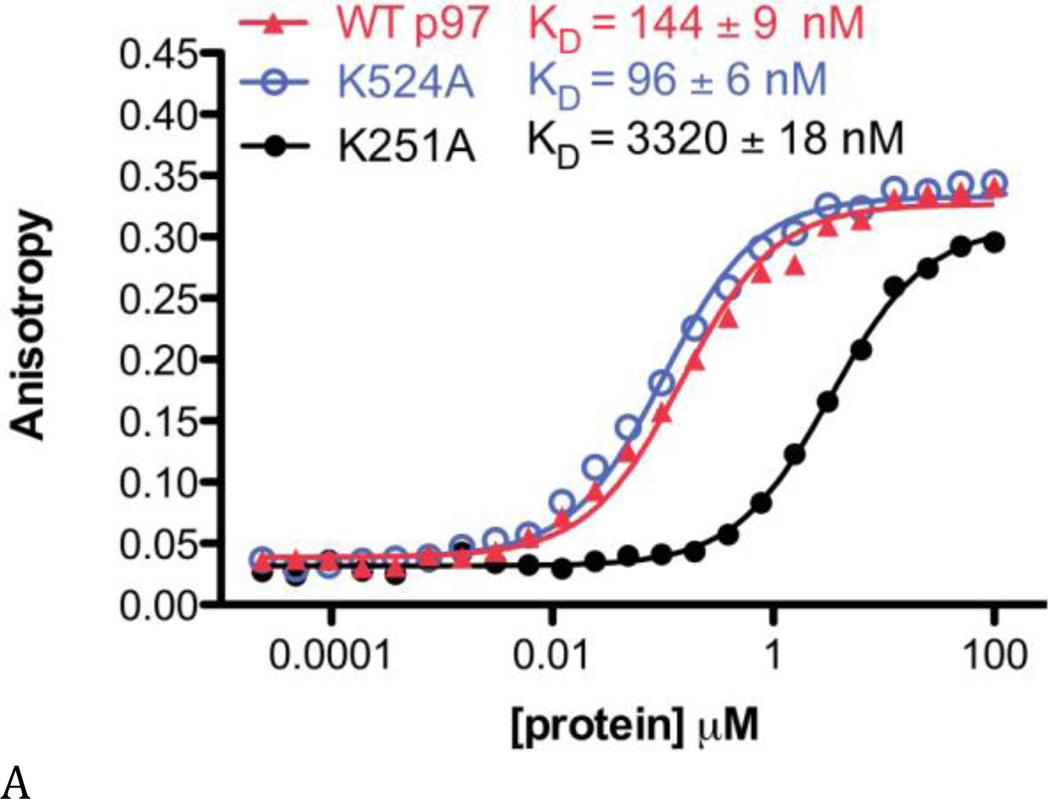
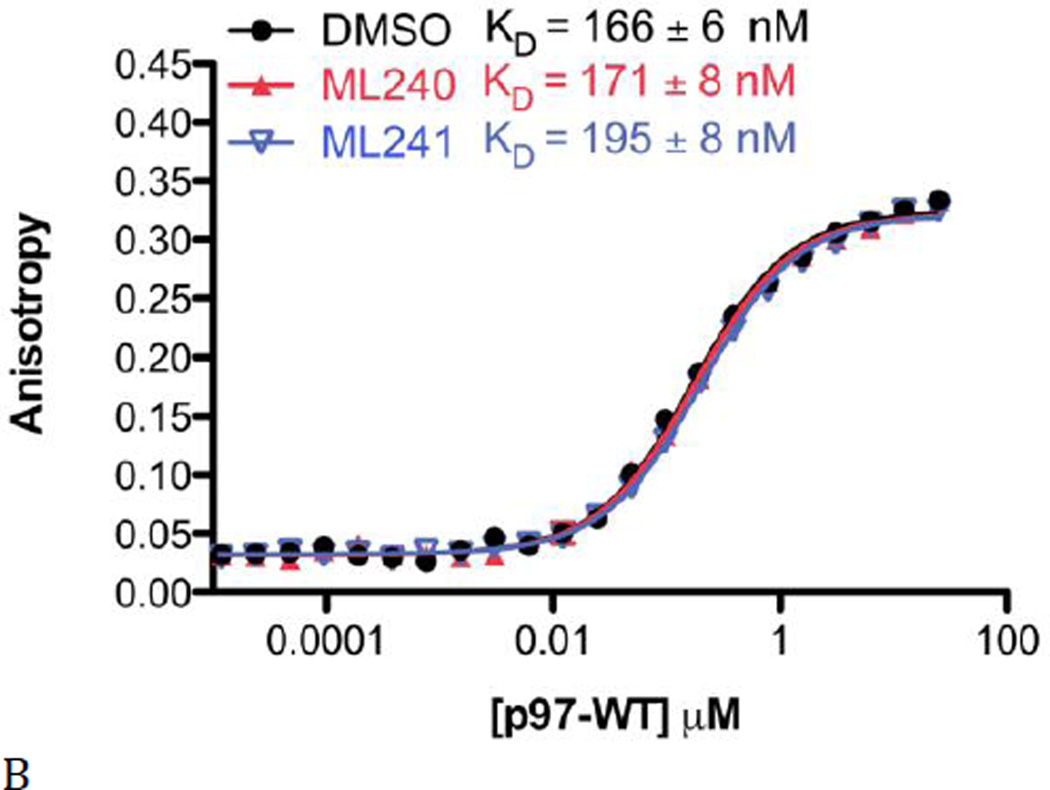
Figure 5. Binding of D2 selective inhibitors does not alter ATP binding to the D1 domain.
(A) Fluorescence anisotropy measurements for BODIPY-FL-ATP binding to WT p97 and Walker A mutants. (B) Binding of the BODIPY-FL-ATP probe in the presence of the D2 specific inhibitors, ML240 and ML241. Data were determined in triplicate, and errors in binding affinities shown are standard errors of the mean of the fit.
To determine if D2 domain-selective inhibitors affect ATP binding to the D1 domain, we performed experiments with fluorescent BODIPY-FL-ATP. Fluorescence anisotropy was detected when BODIPY-FL-ATP bound to WT p97 (Fig. 5A, red line). The KD values for BODIPY-FL-ATP binding were similar for WT p97 and the Walker A mutation in the D2 domain (D2-K524A, Fig. 5A, blue line), indicating that nucleotide binding monitored in WT p97 was predominantly to D1. As anticipated by SPR data, BODIPY-FL-ATP does bind to D2 (D1-K251A, Figure 5A, black line) with a ~23-fold weaker affinity than for D1. These results are also consistent with the binding of other fluorescent probes to WT p97 and Walker A mutants (18,35). We then carried out binding experiments in the presence of 50 µM ML240 and ML241 and demonstrated that inhibiting D2 with ML240 and ML241 did not affect BODIPY-FL-ATP binding to D1 (Fig. 5B).
IBMPFD/ALS Mutations and p47 Cofactor Binding Affect Potency of the D2-selective p97 Inhibitors
Mutations at the ND1 interface of p97 cause IBMPFD/ALS (40,41), a devastating degenerative disorder that has no known treatment. Therefore, p97 inhibitors that can correct the increased ATPase activity of mutants to WT levels might be useful for exploration in animal models, as a possible starting point for research on treatment approaches.
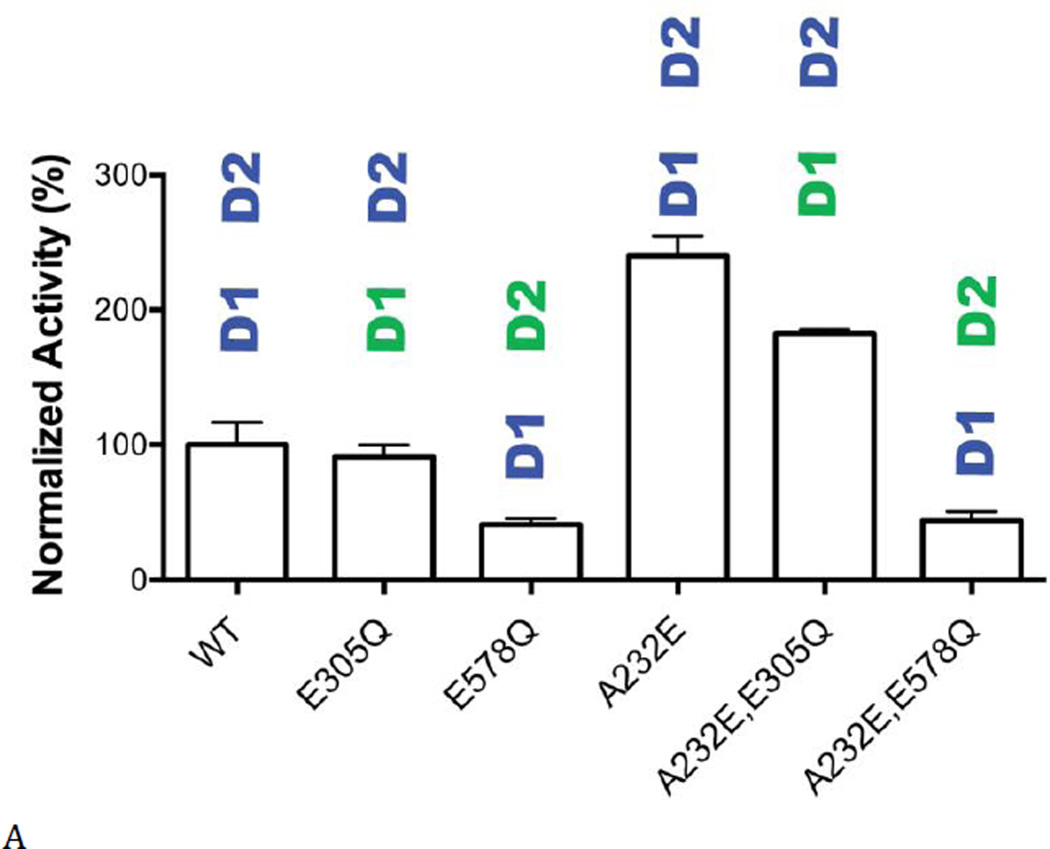
To begin to investigate this opportunity, we prepared a series of IBMPFD p97 mutants and tested the activity of ML240 and ML241. We first confirmed a prior report (38) that the elevated ATPase activity from mutants such as A232E arose from turnover in D2 (Fig. 6A). As expected, when D1 activity was knocked out by D1-E305Q, D2 activity was increased by A232E, whereas A232E had no affect on D1-mediated ATPase activity (D2-E578Q). We then determined IC50 values for the D2-selective inhibitors, ML240 and ML241, against eight mutants that cause disease (Fig. 6B). Interestingly, some mutants were less sensitive to the inhibitors. For example, the IC50 value for ML240 was 5-fold higher for the R95G mutant, compared to WT p97. This result demonstrated that the conformation at the ND1 interface altered binding of inhibitors to the D2 ATP site. Our data suggest that it may be possible to develop a mutant selective inhibitor. Selective inhibition of the mutant subunit may reduce the pool of active p97 mutants (or hexamers containing mutants) within cells, so that cells use predominantly WT hexamers. However, additional work will be required to develop mutant-selective inhibitors.
Figure 6. IBMPFD/ALS p97 mutations affect potency of ML240 and ML241.
(A) ATPase activities of WT p97 and the A232E disease mutant (with Walker B mutations) indicated that the elevated ATPase activity originated from D2. (B) Inhibition of ATPase activity of WT p97 and eight disease mutants by ML240 and ML241.
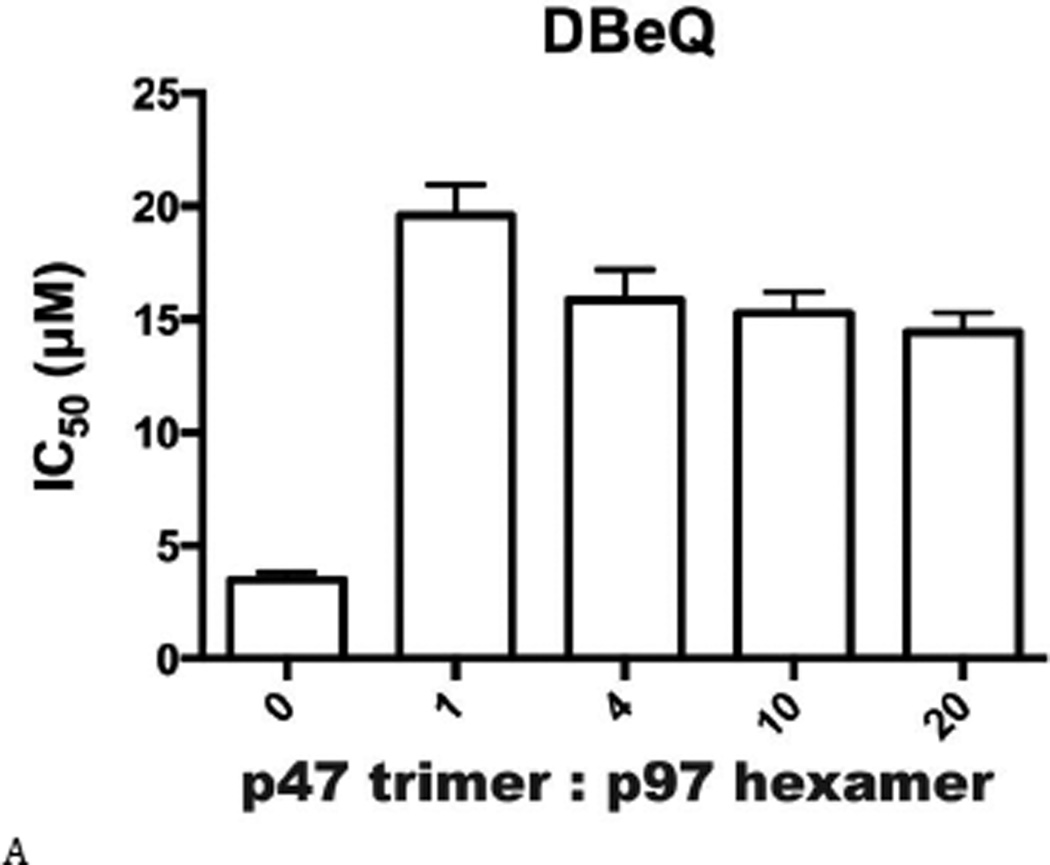
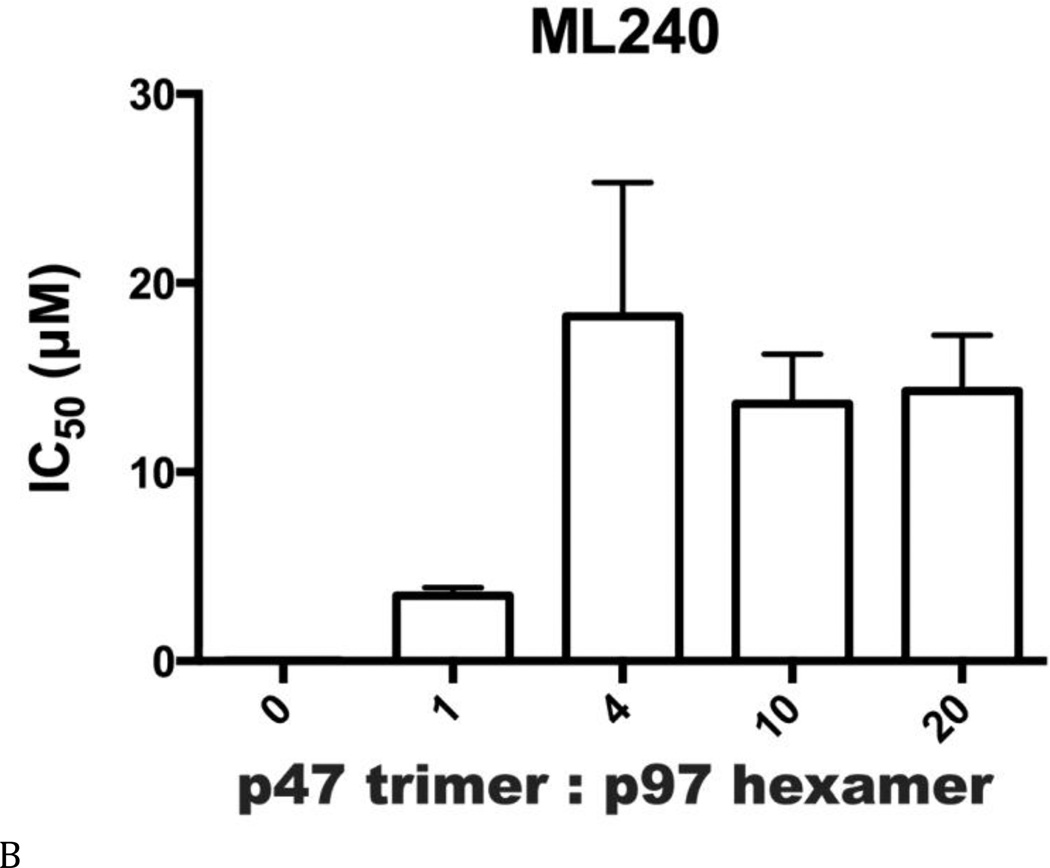
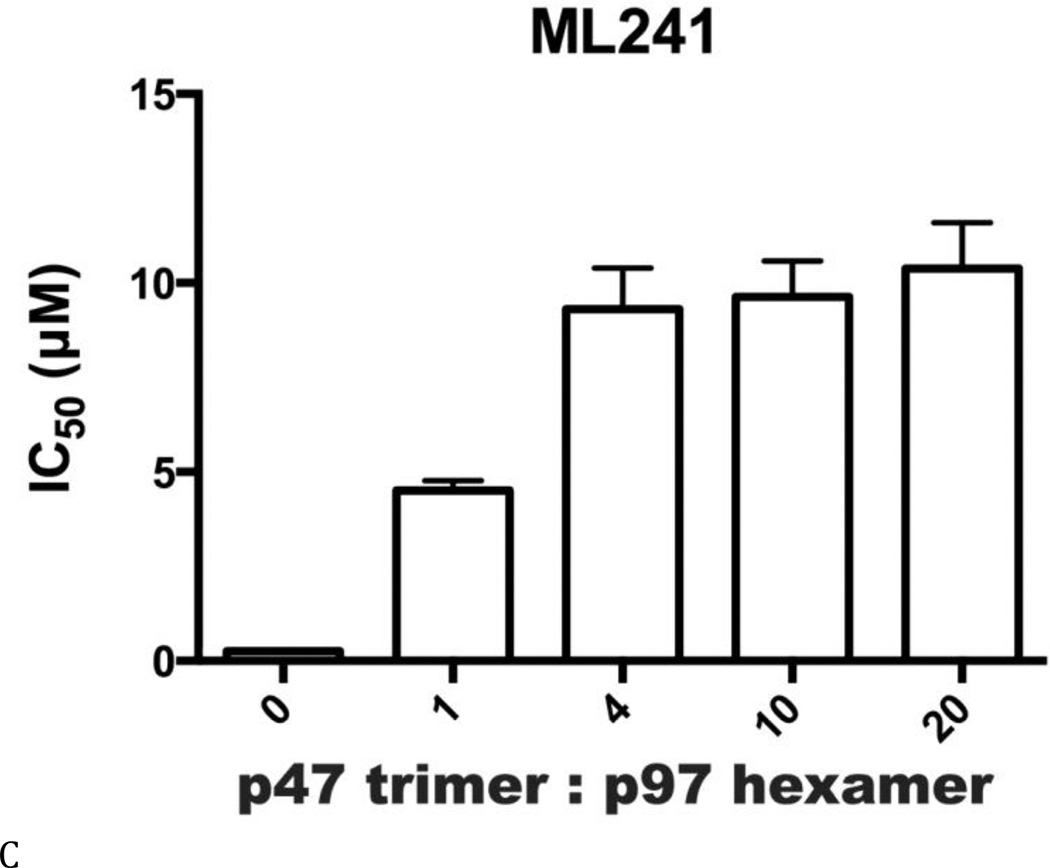
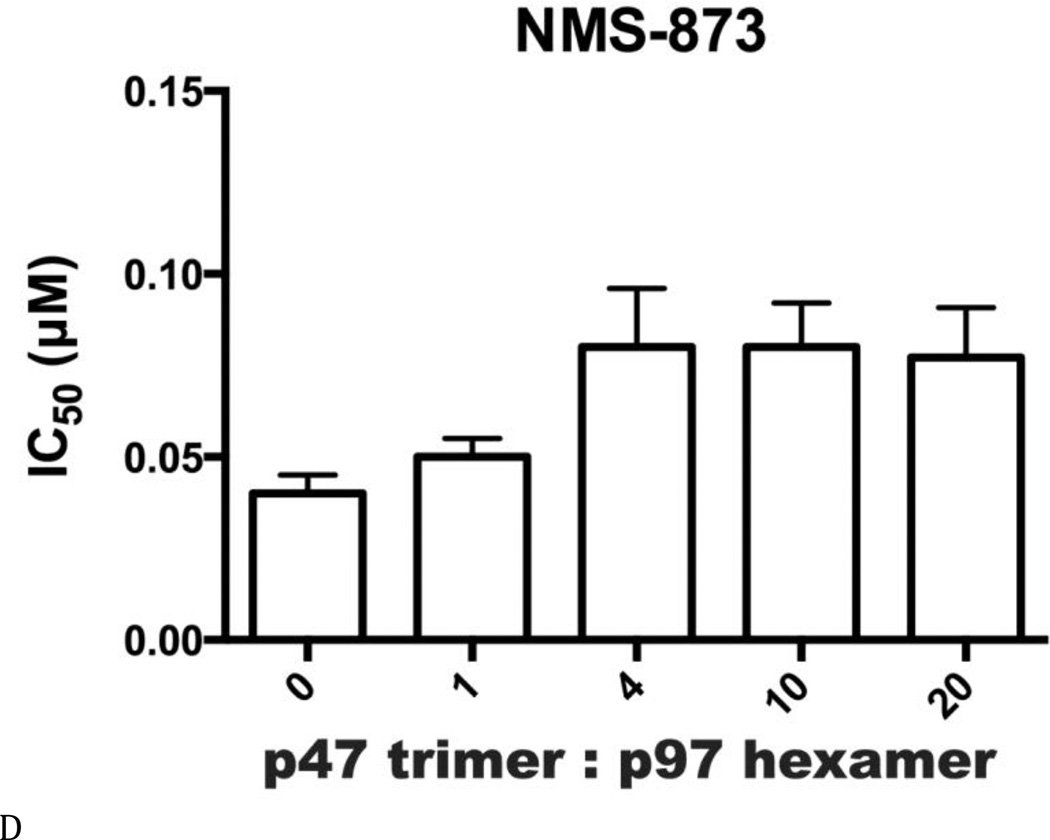
Inhibitor potency was clearly affected by IBMPFD/ALS mutations and by the nucleotide binding state of the D1 domain of p97, even though these mutations are distal to the small-molecule binding site in the D2 domain. By extension, we hypothesized that cofactor binding to p97 might also be critical for p97 inhibitor potency. Therefore, we evaluated inhibitor potency for p97–p47 complexes at four concentrations of p47 (Table S6 and Fig. 7). All concentrations of p47 increased the IC50 of DBeQ 4- to 6-fold (Fig. 7A). More dramatically, p47 trimer concentrations greater than 16.7 nM increased IC50 values of ML240 and ML241 more than 49-fold and 37-fold, respectively (Figs. 7B and C). In contrast, only a minor 2-fold effect was detected for the allosteric inhibitor NMS-873 (Fig. 7D). This result provided the first demonstration that a major p97 cofactor, p47, can influence the potency of ATP-competitive p97 inhibitors.
Figure 7. A Major p97 cofactor, p47, decreased potency of ML240 and ML241.
IC50 values (µM) for p97 inhibitors against p97-p47 mixtures for (A) DBeQ, (B) ML240, (C) ML241, and (D) NMS-873.
Discussion
Our interest is to understand the function of the D1 and D2 p97 ATPase domains, through kinetic analysis of mutants, and use of small molecule inhibitors. The impetus to evaluate effects of p97 inhibitors came from work on the D1 inhibitor, Eeyarestatin I (27), and the N-domain inhibitor, Xanthohumo1 (28). Neither of these compounds blocks full-length p97 ATPase activity, but they inhibit some p97-dependent cellular activities. The results suggest that modifying activity of either the N or D1 domain by small molecules may alter different p97 functions in cells. Moreover, the human disorder known as IBMPFD/ALS is associated with dominantly inherited, missense mutations in the N, ND1 linker, and D1 domains (that cause high p97 ATPase activity), but not by mutations in D2 (29,30).
The AAA ATPase p97 is a dynamic molecular machine that consists of two ATPase domains, D1 and D2, which function to drive the molecular remodeling of protein substrates. We first did a systematic comparison of D1 and D2 ATPase activity to resolve the controversy on whether D1 is an active ATPase. With the active ND1L mutant and Walker A and B mutants for both D1 and D2, we carried out steady-state kinetic analyses and dose-dependent titrations with 4 p97 inhibitors against these mutants.
Our steady-state kinetic analyses provide molecular insights into the roles of both ATPase domains, and also indicate that nucleotide binding and p97 inhibitors modulate each domain interdependently. We show that the ND1L domain is a catalytically competent ATPase, resolving a debate in the literature. Tang and coworkers (29) demonstrated ATPase activity in an ND1L construct (Fig. 1A), whereas two other labs published that ND1 was inactive (16,18). Moreover, our results provide a plausible explanation for the conflicting reports on whether D1 is an active ATPase. We show that the D1–D2 linker region (residues 459–480) dramatically improves thermal stability (Fig. 1D). However, both ND1 and ND1L are capable of binding nucleotides when the temperature is below the melting temperature (Figs. 2A and B). These data are consistent with X-ray crystallography studies that reveal identical active-site structural arrangements for ND1, ND1L, and the ND1 domain in the full-length enzyme (14,15,31,42). One possible explanation for the 56-fold increase in activity in the ND1L construct is that the linker region positions a water molecule into a catalytically competent orientation to hydrolyze ATP.
In this study, we provide quantitative analysis of kinetic constants for the D1 and D2 ATPase domains of human p97, by using point mutations at the conserved Walker A and B positions of D1 and D2 (16,17,36) and an isolated ND1L domain. Additionally, we examine the binding kinetics and dissociation constants for Walker A mutations and isolated ND1 domains. Our estimated Km of 287 µM for ATP at room temperature for human p97 is within the range of published values. Specifically, estimated Km values for p97 isolated from rat liver are 550 µM or 620 µM (24,43) for ATP at 37 °C. Three studies found Km values of 330 µM (16), 360 µM (38), or 49 µM (44) for recombinant mouse p97 purified from E. coli. The Km for the C. elegans p97 homolog is 390 µM (37). Some variation among different reports may be due to differences in assay methods.
Our results showed that ATP hydrolysis within the D1 domain of a full-length p97 protein containing a Walker B mutation in D2 (D2-E578Q) exhibited Michaelis-Menten behavior almost identical to that for ND1L (Fig. 3C, Table S3). Consistent with the enzymology, isolated ND1L was found to bind nucleotides with similar affinities as the D1 domain in full-length p97 (Figs. 2 and S2). Furthermore, the Km of D1 (D2-E578Q) for ATP was ~20-fold less than the D2 value (D1-E305Q) (Fig. 3E), consistent with the 10-fold tighter nucleotide binding to D1 than to D2 (Table S2) (18). Although the D1 maximal velocity was about 3.5 times lower than for WT p97 (Figs. 3C and D), the kcat/Km for D1 is actually 14-fold higher than the WT value, indicating that D1 is a catalytically competent ATPase within full-length p97.
Previous studies hinted that intra- and inter-domain communication within the p97 hexamer is important for p97 activity (17,27–29,37). We found that preventing nucleotide binding in the D2 domain using a Walker A mutant (D2-K524A) drastically impaired both the D1 Km for ATP (~40-fold weaker) and catalytic activity (7-fold lower kcat), as compared to a D2-E578Q mutant (Fig. 3A). This striking finding indicates that nucleotide binding to D2 is required for D1 activity in full-length p97. Hence, domain communication within the p97 hexamer critically controls the capacity of both domains to hydrolyze ATP. Moreover, the tighter Km for D1 may imply that D1 ATPase activity is more important under stress conditions, when cellular ATP concentration is depleted (45–48).
In contrast to our experiments with ND1L, a truncation strategy was not suitable for investigating the catalytic efficiency of the D2 ATPase, because isolated D2 cannot form stable hexamers (Fig. S1) (49), Instead, we focused on D2 ATP hydrolysis within the full-length p97 protein. The Walker B mutation at D1 (D1-E305Q) resulted in a 2.4-fold decrease in Km for the D2 domain, which increased its catalytic efficiency (kcat/Km) by ~ 3-fold (Table S3). Conversely, the Walker A mutation in D1 (D1-K251A) prevented D2 from reaching maximal WT activity, no matter how much ATP was present. Because the Km values are similar for D1-D1-K251A and D1-E305Q (Figs. 3A and D), we conclude that nucleotide binding to D1 is essential to achieve maximal kcat values for D2 (18,37).
Together, our results support the presence of domain communication within the full-length WT p97 ATPase, in which nucleotide binding to one ATPase domain is essential for activity of the other domain. Further work is needed to decipher the precise roles and the molecular mechanisms of this communication. In particular, the importance of D1 ATPase activity in cells is controversial (17,19,29), and our enzymatic studies with mutants pave the way to investigate the cellular role of D1 activity, cooperativity between D1 and D2, and the extent to which p97 cofactors are involved in regulating such communication.
Our results also have implications for the design of p97 inhibitors for use in cancer and neurodegeneration. We have generated a set of tools to develop D1- and D2-domain specific inhibitors, and clearly demonstrated the existence of two types of ATP-competitive inhibitors. DBeQ inhibits both D1 and D2, whereas ML240 and ML241 only inhibit the D2 ATPase domain (Figs. 4 A–C and Tables S4 and S5). Moreover, inhibition by ML240 and ML241 is sensitive to IBMPFD/ALS mutations in p97 (Fig. 6B), suggesting that IBMPFD/ALS-selective inhibitors might be developed via either structure-based drug design or high-throughput screens.
Another significant finding in this study is the roughly 50-fold decrease in ML240 potency in the presence of p47, providing the first evidence that p97 cofactors can affect the potency of ATP-competitive inhibitors (Figs. 7A–C, and Table S6). These results favor an interpretation that the p97–p47 complex exists in a conformation that cannot be inhibited by ML240 and ML241 as effectively as by the allosteric inhibitor NMS-873. Indirect evidence suggests that p97 ATP binding sites exist in different conformations in the presence of p47, because p47 dramatically changes the global p97 conformation (21,22,50) and lowers the maximal velocity of ATPase activity by 5-fold (24). Thus, it may be possible to develop complex-specific p97 inhibitors for p47 and other p97 cofactor proteins (51). It is enticing to consider whether ML240 and ML241 may inhibit a subset of p97 complexes in cells, whereas NMS-873 may be a broad-spectrum inhibitor. Future in vivo studies are needed to compare anti-cancer potencies and general toxicities of distinct classes of p97 inhibitors. Taken together, our results provide a new way to consider domain communication in p97 and the tools to develop D1- or D2-domain selective, complex-specific, and disease-mutant-selective p97 inhibitors.
Materials and Methods
Materials
Plasmids used are listed in Supplemental Table S1.
BIOMOL Green ATPase Assay
Purified p97 (25 µL of 50 µM; final concentration in the reaction was 25 nM) was diluted in 40 mL of assay buffer [10 mL of 5× assay buffer A (1× = 50 mM Tris pH 7.4, 20 mM MgCl2, 1 mM EDTA,) mixed with 30 mL water and 50 µL 0.5M TCEP, 50 µL 10% Triton] to make the enzyme solution. Some experiments represented in Figures 3A and 3C were performed without 0.01% Triton X-100. In these experiments, 50 µL 10% Triton was not added to the assay buffer. For all other experiments, 50 µL 10% Triton was included. 40 µL of enzyme solution was dispensed into each well of a 96 well plate. The WT p97 ATPase assay was carried out by adding 10 µL of 1000 µM or 4375 µM ATP (Roche, pH 7.5) to each well and incubating the reaction at room temperature for 35 or 15 min, respectively Reactions were stopped by adding 50 µL of BIOMOL Green reagent (Enzo Life Sciences). Absorbance at 635 nm was measured after 4 min on the Synergy Neo Microplate Reader (BioTek). To determine the steady-state kinetic constant, eight final ATP concentrations were used (62.5, 125, 250, 375, 500, 625, 750, 875 µM) or (3.9, 7.8, 15.6, 23.4, 31.25, 62.5, 93.8, 125 µM). For mutants, reaction times were adjusted according to specific activities, to give acceptable absorbance readings. Michaelis-Menten constants were calculated from eight replicates by fitting data to Equation 1 using GraphPad Prism 6.0.
| (1) |
where ν is the initial velocity, kcat is the turnover number, [E]t is the total enzyme concentration, [S] is the substrate concentration, and Km is the concentration of substrate to reach half maximal velocity.
ATPase Assay for Determining IC50 Values of Inhibitors
Purified p97 (25 µL of a 50 µM stock solution) was diluted in 30 mL of assay buffer [10 mL of 5×assay buffer A (1×= 50 mM Tris pH 7.4, 20 mM MgCl2, 1 mM EDTA,) mixed with 20 mL of water, 50 µL of 0.5M TCEP, and 50 µL of 10% Triton X-100] to make the enzyme solution. 30 µL of enzyme solution was dispensed into each well of a 96 well plate. The test compound (10 µL) or DMSO (12%, 10 µL) was then added to each well, and the plate was incubated at room temperature for 10 min. The reaction was initiated by adding 10 µL of 1 mM ATP (pH 7.5) to each well at room temperature. After 30 min, the reaction was stopped by adding 50 µL of BIOMOL Green reagent. Absorbance at 635 nm was measured after 4 min. DBeQ was assayed at a range of concentrations (0, 0.04, 0.12, 0.37, 1.1, 3.3, 10, 30 µM). The other compounds were assayed at (0, 0.013, 0.04, 0.12, 0.37, 1.1, 3.3, and 10 µM). IC50 values were calculated from four replicates by non-linear regression analysis with Equation 2 using GraphPad Prism 6.0.
| (2) |
where X is the compound concentration.
Surface Plasmon Resonance (SPR)
The kinetic parameters for the direct binding of nucleotides to p97 were measured on a Biacore 4000 instrument. Neutravidin-coated sensor chips were prepared on CM5 chips, as follows: the surface was activated by injecting a 1:1 mixture of 60 mM N-hydroxysuccinimide and 240 mM 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide for 7 min, followed by a 7 min injection of 0.25 mg/mL NeutrAvidin (Thermo Scientific) in 10 mM acetic acid pH 4.5. The surface was then blocked by a 2 min injection of 1 M ethanolamine, pH 8.3.
p97 proteins containing site-specific biotinylation (AviTag; Avidity) were expressed and purified from E. coli (see Supplemental materials). Purified protein was immobilized in 10 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM TCEP, 0.05% Tween 20. Protein was immobilized to 4000–6000 resonance units (RUs) by injecting 100–500 µg/mL of protein for 1 min at 25 °C. Protein immobilization was followed by a 2 min injection of 0.2 mg/mL amino-PEG-biotin (Thermo Scientific) to block any remaining biotin binding sites. A reference surface was created using the same protocol with omission of the protein injection step.
Binding of ADP (Sigma A2754), ATP (promega V703A) and ATP' S (Sigma A1388) were measured in duplicate in SPR running buffer (25 mM Tris pH 7.5, 150 mM NaCl, 10 mM MgCl2, 0.5 mM TCEP, 0.05% Tween 20) at 20 °C. Nucleotides were injected at a flow rate of 30 µL/min for 90–180 seconds, with a dissociation time of 180–600 seconds.
SPR experiments were carried out in the presence of 0.05% Tween-20, whereas the BIOMOL Green ATPase assays were carried out in the presence of 0.01% Triton X-100. Because Tween-20 and the BIOMOL Green reagent are incompatible, to compare the ATPase activities of WT p97 in the presence of Triton X-100 and Tween-20, we used the ADP-Glo assay. Similar to the BIOMOL green assay, we found the addition of 0.01% Triton X-100 or Tween-20 modestly increased WT p97 activity by 1.79- and 1.97-fold, respectively. The addition of 0.05% Tween-20 further increased the activity by 2.8-fold over no detergent conditions.
Differential Scanning Fluorimetry (DSF)
The thermal stabilities of p97 ND1 and ND1L domains were measured on a Mx3005P q-PCR (Stratagene) instrument in a 24-well Thermo-Fast PCR plate (Thermo Scientific). Four replicates of 20 µL of ND1, ND1L or full-length p97 (2.0 µM) were mixed with 5 µL of a 1:200 dilution of SYPRO Orange Protein Gel Stain (Sigma) in 25 mM Tris, pH 7.5, 150 mM NaCl, 10 mM MgCl2, 0.5 mM TCEP. Samples were heated from 25 °C to 100 °C at 1 °C/min, and fluorescence (Ex = 492 nM, Em = 610 nM) was measured every minute. Curves were normalized, and the melting temperature was calculated from the first derivative using the FIT for DSF website (skuld.bmsc.washington.edu/FIT/).
Fluorescence Anisotropy
Fluorescence anisotropy experiments were completed in black, low volume, non-binding 384-well plates (Corning) in 25 mM Tris, pH 7.5, with 150 mM NaCl, 10 mM MgCl2, and 0.5 mM TCEP, by mixing 10 µL of protein (200 µM – 48 pM; 2-fold dilutions) with 10 µL of BODIPY-FL-ATP ligand (Life Technologies; 20 nM). After a 30-minute incubation, fluorescence polarization was measured on an Analyst HT plate reader (Molecular Devices) using 485 nm and 530 nm excitation and emission filters, respectively. No time-dependent change in anisotropy was observed over 30 minutes (e.g., due to catalytic turnover of the BODIPY-FL-ATP probe). Background fluorescence (protein only) was subtracted from the parallel and perpendicular intensities before calculating anisotropy. The KD of BODIPY-FL-ATP was determined with GraphPad Prism 5.0, by Equations 3 and 4 to account for ligand depletion:
| (3) |
| (4) |
where [RL] is the concentration of the p97-BODIPY-FL-ATP complex, [L] is the concentration of BIODPYFL- ADP, [R] is the concentration of p97, A is anisotropy, Af is the anisotropy of BODIPY-FL ATP, and Ab is the anisotropy of the p97-BODIPY-FL-ATP complex.
To determine if DBeQ, ML240, or ML241 altered nucleotide binding to the D1 domain of p97, 5 µL of WT p97 (100 µM – 24 pM; 2-fold dilutions) was mixed with 5 µL of 5% DMSO or compound (200 µM in 5% DMSO buffer) and 10 µL of 20 nM BODIPY-FL-ATP.
Materials and additional methods, including procedures for protein expression, are available in supplemental data.
Supplementary Material
Highlights.
Potent p97 inhibitors are developed, but whether they inhibit D1 activity is unknown.
Inhibitors can differentially target D1, D2, and p97-cofactor complexes.
This represents a step toward domain- and complex-selective p97inhibitors.
ML240 and ML241 are D2-selective inhibitors and p47 influences their potency.
Acknowledgment
We thank Michelina Iacovino for critical reading and suggestions on this manuscript. We thank David Myszka for assistance in fitting SPR data in CLAMP. The project was in part supported by the National Center for Advancing Translational Sciences through UCLA CTSI Grant UL1TR000124 and the LA BioMed Seed Grant program (20826-01). This project has been funded in part with federal funds from the National Cancer Institute, National Institutes of Health, under Chemical Biology Consortium Contract No. HHSN261200800001E and by the National Human Genome Research Institute of the National Institutes of Health under Award Number U54HG005031 (Jeffrey Aubé, PI). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. government. RJD is an Investigator of the Howard Hughes Medical Institute, and TFC is a member of UCLA Jonsson Comprehensive Cancer Center.
Abbreviations
- ERAD
endoplasmic reticulum associated degradation
- SPR
surface plasmon resonance
- UFD
ubiquitin fusion degradation
- UPS
ubiquitin-proteasome system
- WT
wild type
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rabouille C, Levine TP, Peters JM, Warren G. An NSF-like ATPase, p97, and NSF mediate cisternal regrowth from mitotic Golgi fragments. Cell. 1995;82:905–914. doi: 10.1016/0092-8674(95)90270-8. [DOI] [PubMed] [Google Scholar]
- 2.Ye Y, Meyer HH, Rapoport TA. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature. 2001;414:652–656. doi: 10.1038/414652a. [DOI] [PubMed] [Google Scholar]
- 3.Ye Y, Shibata Y, Yun C, Ron D, Rapoport TA. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature. 2004;429:841–847. doi: 10.1038/nature02656. [DOI] [PubMed] [Google Scholar]
- 4.Golbik R, Lupas AN, Koretke KK, Baumeister W, Peters J. The Janus face of the archaeal Cdc48/p97 homologue VAT: protein folding versus unfolding. Biol. Chem. 1999;380:1049–1062. doi: 10.1515/BC.1999.131. [DOI] [PubMed] [Google Scholar]
- 5.Richly H, Rape M, Braun S, Rumpf S, Hoege C, Jentsch S. A series of ubiquitin binding factors connects CDC48/p97 to substrate multiubiquitylation and proteasomal targeting. Cell. 2005;120:73–84. doi: 10.1016/j.cell.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 6.Janiesch PC, Kim J, Mouysset J, Barikbin R, Lochmuller H, Cassata G, Krause S, Hoppe T. The ubiquitin-selective chaperone CDC-48/p97 links myosin assembly to human myopathy. Nat. Cell Biol. 2007;9:379–390. doi: 10.1038/ncb1554. [DOI] [PubMed] [Google Scholar]
- 7.Cao K, Nakajima R, Meyer HH, Zheng Y. The AAA-ATPase Cdc48/p97 regulates spindle disassembly at the end of mitosis. Cell. 2003;115:355–367. doi: 10.1016/s0092-8674(03)00815-8. [DOI] [PubMed] [Google Scholar]
- 8.Boyault C, Gilquin B, Zhang Y, Rybin V, Garman E, Meyer-Klaucke W, Matthias P, Muller CW, Khochbin S. HDAC6-p97/VCP controlled polyubiquitin chain turnover. EMBO J. 2006;25:3357–3366. doi: 10.1038/sj.emboj.7601210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyault C, Zhang Y, Fritah S, Caron C, Gilquin B, Kwon SH, Garrido C, Yao TP, Vourc'h C, Matthias P, Khochbin S. HDAC6 controls major cell response pathways to cytotoxic accumulation of protein aggregates. Genes Dev. 2007;21:2172–2181. doi: 10.1101/gad.436407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ju JS, Miller SE, Hanson PI, Weihl CC. Impaired protein aggregate handling and clearance underlie the pathogenesis of p97/VCP-associated disease. J. Biol. Chem. 2008;283:30289–33099. doi: 10.1074/jbc.M805517200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fessart D, Marza E, Taouji S, Delom F, Chevet E. P97/CDC-48: proteostasis control in tumor cell biology. Cancer letters. 2013;337:26–34. doi: 10.1016/j.canlet.2013.05.030. [DOI] [PubMed] [Google Scholar]
- 12.Stolz A, Hilt W, Buchberger A, Wolf DH. Cdc48: a power machine in protein degradation. Trends Biochem. Sci. 2011;36:515–523. doi: 10.1016/j.tibs.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 13.Kakizuka A. Roles of VCP in human neurodegenerative disorders. Biochem. Soc. Trans. 2008;36:105–108. doi: 10.1042/BST0360105. [DOI] [PubMed] [Google Scholar]
- 14.Huyton T, Pye VE, Briggs LC, Flynn TC, Beuron F, Kondo H, Ma J, Zhang X, Freemont PS. The crystal structure of murine p97/VCP at 3.6A. J. Struct. Biol. 2003;144:337–348. doi: 10.1016/j.jsb.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 15.DeLaBarre B, Brunger AT. Complete structure of p97/valosin-containing protein reveals communication between nucleotide domains. Nat. Struct. Biol. 2003;10:856–863. doi: 10.1038/nsb972. [DOI] [PubMed] [Google Scholar]
- 16.Song C, Wang Q, Li CC. ATPase activity of p97-valosin-containing protein (VCP). D2 mediates the major enzyme activity, and D1 contributes to the heat-induced activity. J. Biol. Chem. 2003;278:3648–3655. doi: 10.1074/jbc.M208422200. [DOI] [PubMed] [Google Scholar]
- 17.Ye Y, Meyer HH, Rapoport TA. Function of the p97-Ufd1-Npl4 complex in retrotranslocation from the ER to the cytosol: dual recognition of nonubiquitinated polypeptide segments and polyubiquitin chains. J. Cell Biol. 2003;162:71–84. doi: 10.1083/jcb.200302169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Briggs LC, Baldwin GS, Miyata N, Kondo H, Zhang X, Freemont PS. Analysis of nucleotide binding to p97 reveals the properties of a tandem AAA hexameric ATPase. J. Biol. Chem. 2008;283:13745–11352. doi: 10.1074/jbc.M709632200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Esaki M, Ogura T. ATP-bound form of the D1 AAA domain inhibits an essential function of Cdc48p/p97. Biochem. Cell Biol. 2010;88:109–117. doi: 10.1139/o09-116. [DOI] [PubMed] [Google Scholar]
- 20.Dreveny I, Kondo H, Uchiyama K, Shaw A, Zhang X, Freemont PS. Structural basis of the interaction between the AAA ATPase p97/VCP and its adaptor protein p47. EMBO J. 2004;23:1030–1039. doi: 10.1038/sj.emboj.7600139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uchiyama K, Jokitalo E, Kano F, Murata M, Zhang X, Canas B, Newman R, Rabouille C, Pappin D, Freemont P, Kondo H. VCIP135, a novel essential factor for p97/p47-mediated membrane fusion, is required for Golgi and ER assembly in vivo. J. Cell. Biol. 2002;159:855–866. doi: 10.1083/jcb.200208112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kondo H, Rabouille C, Newman R, Levine TP, Pappin D, Freemont P, Warren G. p47 is a cofactor for p97-mediated membrane fusion. Nature. 1997;388:75–78. doi: 10.1038/40411. [DOI] [PubMed] [Google Scholar]
- 23.Rabouille C, Kondo H, Newman R, Hui N, Freemont P, Warren G. Syntaxin-5 is a common component of the NSF- and p97-mediated reassembly pathways of Golgi cisternae from mitotic Golgi fragments in vitro. Cell. 1998;92:603–610. doi: 10.1016/s0092-8674(00)81128-9. [DOI] [PubMed] [Google Scholar]
- 24.Meyer HH, Kondo H, Warren G. The p47 co-factor regulates the ATPase activity of the membrane fusion protein, p97. FEBS Lett. 1998;437:255–257. doi: 10.1016/s0014-5793(98)01232-0. [DOI] [PubMed] [Google Scholar]
- 25.Rouiller I, DeLaBarre B, May AP, Weis WI, Brunger AT, Milligan RA, Wilson-Kubalek EM. Conformational changes of the multifunction p97 AAA ATPase during its ATPase cycle. Nat. Struct. Biol. 2002;9:950–957. doi: 10.1038/nsb872. [DOI] [PubMed] [Google Scholar]
- 26.Noi K, Yamamoto D, Nishikori S, Arita-Morioka K, Kato T, Ando T, Ogura T. High-speed atomic force microscopic observation of ATP-dependent rotation of the AAA+ chaperone p97. Structure. 2013;21:1992–2002. doi: 10.1016/j.str.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 27.Davies JM, Tsuruta H, May AP, Weis WI. Conformational changes of p97 during nucleotide hydrolysis determined by small-angle X-Ray scattering. Structure. 2005;13:183–195. doi: 10.1016/j.str.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 28.Beuron F, Flynn TC, Ma J, Kondo H, Zhang X, Freemont PS. Motions and negative cooperativity between p97 domains revealed by cryo-electron microscopy and quantised elastic deformational model. J. Mol. Biol. 2003;327:619–629. doi: 10.1016/s0022-2836(03)00178-5. [DOI] [PubMed] [Google Scholar]
- 29.Tang WK, Xia D. Altered Intersubunit Communication Is the Molecular Basis for Functional Defects of Pathogenic p97 Mutants. J. Biol. Chem. 2013;288:36624–33635. doi: 10.1074/jbc.M113.488924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Halawani D, LeBlanc AC, Rouiller I, Michnick SW, Servant MJ, Latterich M. Hereditary inclusion body myopathy-linked p97/VCP mutations in the NH2 domain and the D1 ring modulate p97/VCP ATPase activity and D2 ring conformation. Mol. Cell Biol. 2009;29:4484–4494. doi: 10.1128/MCB.00252-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang WK, Li D, Li CC, Esser L, Dai R, Guo L, Xia D. A novel ATP-dependent conformation in p97 N-D1 fragment revealed by crystal structures of disease-related mutants. EMBO J. 2010;29:2217–2229. doi: 10.1038/emboj.2010.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chou T-F, Brown SJ, Minond D, Nordin BE, Li K, Jones AC, Chase P, Porubsky PR, Stoltz BM, Schoenen FJ, Patricelli MP, Hodder P, Rosen H, Deshaies RJ. Reversible inhibitor of p97, DBeQ, impairs both ubiquitin-dependent and autophagic protein clearance pathways. Proc. Natl. Acad. Sci. USA. 2011;108:4834–4839. doi: 10.1073/pnas.1015312108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Polucci Pea, D'Alessio R. Alkylsulfanyl-1,2,4-triazoles, a new class of allosteric valosine containing protein inhibitors. Synthesis and structure-activity relationships. J. Med. Chem. 2013;56:437–450. doi: 10.1021/jm3013213. [DOI] [PubMed] [Google Scholar]
- 34.Chou TF, Li K, Frankowski KJ, Schoenen FJ, Deshaies RJ. Structure-activity relationship study reveals ML240 and ML241 as potent and selective inhibitors of p97 ATPase. Chem Med Chem. 2013;8:297–312. doi: 10.1002/cmdc.201200520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Magnaghi P, Isacchi A, et al. Covalent and allosteric inhibitors of the ATPase VCP/p97 induce cancer cell death. Nat. Chem. Biol. 2013;9:548–556. doi: 10.1038/nchembio.1313. [DOI] [PubMed] [Google Scholar]
- 36.Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nishikori S, Esaki M, Yamanaka K, Sugimoto S, Ogura T. Positive cooperativity of the p97 AAA ATPase is critical for essential functions. J. Biol. Chem. 2011;286:15815–15820. doi: 10.1074/jbc.M110.201400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Niwa H, Ewens CA, Tsang C, Yeung HO, Zhang X, Freemont PS. The role of the N-domain in the ATPase activity of the mammalian AAA ATPase p97/VCP. J. Biol. Chem. 2012;287:8561–8570. doi: 10.1074/jbc.M111.302778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McGovern SL, Caselli E, Grigorieff N, Shoichet BK. A common mechanism underlying promiscuous inhibitors from virtual and high-throughput screening. J. Med. Chem. 2002;45:1712–1722. doi: 10.1021/jm010533y. [DOI] [PubMed] [Google Scholar]
- 40.Watts GD, Wymer J, Kovach MJ, Mehta SG, Mumm S, Darvish D, Pestronk A, Whyte MP, Kimonis VE. Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nat. Genet. 2004;36:377–381. doi: 10.1038/ng1332. [DOI] [PubMed] [Google Scholar]
- 41.Weihl CC, Miller SE, Hanson PI, Pestronk A. Transgenic expression of inclusion body myopathy associated mutant p97/VCP causes weakness and ubiquitinated protein inclusions in mice. Hum. Mol. Genet. 2007;16:919–928. doi: 10.1093/hmg/ddm037. [DOI] [PubMed] [Google Scholar]
- 42.Zhang X, Freemont PS, et al. Structure of the AAA ATPase p97. Mol. Cell. 2000;6:1473–1484. doi: 10.1016/s1097-2765(00)00143-x. [DOI] [PubMed] [Google Scholar]
- 43.Zhang L, Ashendel CL, Becker GW, Morre DJ. Isolation and characterization of the principal ATPase associated with transitional endoplasmic reticulum of rat liver. J. Cell Biol. 1994;127:1871–1883. doi: 10.1083/jcb.127.6.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.DeLaBarre B, Christianson JC, Kopito RR, Brunger AT. Central pore residues mediate the p97/VCP activity required for ERAD. Mol. Cell. 2006;22:451–462. doi: 10.1016/j.molcel.2006.03.036. [DOI] [PubMed] [Google Scholar]
- 45.Imamura H, Nhat KP, Togawa H, Saito K, Iino R, Kato-Yamada Y, Nagai T, Noji H. Visualization of ATP levels inside single living cells with fluorescence resonance energy transfer-based genetically encoded indicators. Proc. Natl. Acad. Sci. USA. 2009;106:15651–15656. doi: 10.1073/pnas.0904764106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leppanen O, Bjornheden T, Evaldsson M, Boren J, Wiklund O, Levin M. ATP depletion in macrophages in the core of advanced rabbit atherosclerotic plaques in vivo. Atherosclerosis. 2006;188:323–330. doi: 10.1016/j.atherosclerosis.2005.11.017. [DOI] [PubMed] [Google Scholar]
- 47.St-Pierre J, Brand MD, Boutilier RG. Mitochondria as ATP consumers: cellular treason in anoxia. Proc. Natl. Acad. Sci. USA. 2000;97:8670–8674. doi: 10.1073/pnas.140093597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chandel NS, Budinger GR. The cellular basis for diverse responses to oxygen. Free Radic Biol Med. 2007;42:165–174. doi: 10.1016/j.freeradbiomed.2006.10.048. [DOI] [PubMed] [Google Scholar]
- 49.Wang Q, Song C, Li CC. Hexamerization of p97-VCP is promoted by ATP binding to the D1 domain and required for ATPase and biological activities. Biochem. Biophys. Res. Commun. 2003;300:253–260. doi: 10.1016/s0006-291x(02)02840-1. [DOI] [PubMed] [Google Scholar]
- 50.Rouiller I, Butel VM, Latterich M, Milligan RA, Wilson-Kubalek EM. A major conformational change in p97 AAA ATPase upon ATP binding. Mol Cell. 2000;6:1485–1490. doi: 10.1016/s1097-2765(00)00144-1. [DOI] [PubMed] [Google Scholar]
- 51.Chou TF, Deshaies RJ. Development of p97 AAA ATPase inhibitors. Autophagy. 2011;7:1091–1092. doi: 10.4161/auto.7.9.16489. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.