Abstract
The effects of UVR on the skin include tanning, carcinogenesis, immunomodulation, and synthesis of vitamin D, among others. Melanocortin 1 receptor polymorphisms correlate with skin pigmentation, UV sensitivity, and skin cancer risk. This article reviews pathways through which UVR induces cutaneous stress and the pigmentation response. Modulators of the UV tanning pathway include sunscreen agents, MC1R activators, adenylate cyclase activators, phosphodiesterase 4D3 inhibitors, T oligos, and MITF regulators such as histone deacetylase (HDAC)-inhibitors. UVR, as one of the most ubiquitous carcinogens, represents both a challenge and enormous opportunity in skin cancer prevention.
Keywords: ultraviolet radiation, melanocyte, melanoma
INTRODUCTION
The incidence of melanoma and non-melanoma skin cancers has continued to rise over the past few decades. The etiology is multifactorial with discrete genetic pathways and environmental factors. Although genetic factors may contribute significantly, environmental factors can be modified to potentially decrease the risk of developing deadly diseases such as melanoma. Exposure to ultraviolet radiation (UVR) from sunlight is well established as a significant risk factor for melanoma development. However, indoor tanning is a source of preventable UVR exposure that represents a growing, multi-billion dollar industry (Levine et al, 2005). UVR is a major environmental risk factor that contributes to carcinogenesis through DNA damage and immune modulation via inflammatory and immunosuppressive pathways (Tran et al, 2008; Liu and Fisher 2010; D'Orazio et al, 2013; Weinstock 2013). It has long been appreciated that tanning, through increasing epidermal melanin content, is the skin's major photoprotective response against acute and chronic UV damage. DNA damage from UVR induces signaling cascades that ultimately lead to activation of pigmentation machinery to produce the tanning effect. This process can be synthetically perturbed at different points along the pathway to upregulate driver signals or to suppress inhibitory feedback, thereby promoting a UVR-independent protective tanning response. These strategies range from broad, such as transcriptional activators, to narrow, such as molecular analogues. Because the UV tanning pathway is essential for both melanogenesis and protection from skin cancers, we summarize here the consequences of UV signaling pathway deficiencies and strategies to regulate the UV signaling pathway.
FEATURES OF UVR AND UV-INDUCED MUTAGENESIS
UVR, spanning the 200 to 400 nm wavelengths of the electromagnetic spectrum, is a high energy component of solar radiation. UVR is divided into three categories based on wavelength: UVA (400–320 nm), UVB (320–290 nm), and UVC (290–200 nm). Over 95% of UVA and 1–10% of UVB radiation reaches the earth’s surface, while almost 100% of solar UVC is absorbed by the atmosphere and the ozone layer. Thus, most of the research on the effects of UVR has focused on UVA and UVB. A history of sunburn in childhood and continued unprotected exposure to UVR through adolescence and adulthood contribute to skin cancer risk. However, many adolescents and adults continue to seek a tan, either from direct sun exposure or from tanning beds.
UVR directly targets macromolecules in the skin such as proteins, lipids, and nucleic acids, with the latter resulting in signature mutations characteristically found in melanomas and other skin cancers. When these mutations occur within genes regulating apoptosis, cell cycle progression, and genetic repair machinery, they may initiate oncogenic transformation (Fisher and James 2010; Schulman and Fisher 2009). UVR photoexcitation of the direct chromophore DNA produces excited electron states and toxic by-products, leading to direct and indirect DNA damage. This often produces signature mutations dependent on the insult and mechanism of damage. We will focus on mutations resulting from UVA and UVB specifically.
UVA radiation, upon exciting endogenous chromophores, can generate reactive oxygen species (ROS) capable of causing oxidative DNA damage. Through generation of singlet oxygen (1O2) or type-1 photosensitization reactions, UVA is able to cause oxidative base modifications, predominately at guanine bases. This process leads to generation of 7,8-dihydro-8-oxoguanine (8-oxoG) lesions, which have been shown to induce specific DNA mutations if not repaired. (Garibyan and Fisher 2010). The major UVA-induced mutations are G→T transversions and G→A transitions. Like UVB, UVA may also trigger DNA damage through cyclobutane pyrimidine dimer (CPD) formation.
UVB contact with DNA activates a photochemical reaction that usually occurs between adjacent pyrimidine nucleotides and leads to formation of photoproducts known as CPDs and pyrimidine 6-4 pyrimidones. After the formation of CPDs and pyrimidine 6-4 pyrimidone photoproducts, either spontaneous reversion may occur (for CPDs), or DNA repair enzymes participate in the correction of the damage. Incorrect repair of these damaged DNA lesions leads to mutations in epidermal cells that may initiate oncogenesis. When UVB induced CPDs and pyrimidine 6-4 pyrimidones are incorrectly resolved, certain signature mutations may form, including C→T and CC→TT transition mutations (Garibyan and Fisher 2010; Tran et al, 2008).
These characteristic mutations are not exclusively induced by UVR from sunlight. DeMarini and colleagues compared the mutagenic effects of radiation from three common sources using Salmonella assays and determined that mutagenic ability was most potent in radiation from tanning salon beds, followed by sunlight. White fluorescent light represented the least mutagenic source of radiation. The most common mutations were G:C→A:T transitions. The CC→TT transitions characteristic of UVB exposure represented 83% of mutations induced by tanning bed radiation exposure, demonstrating that both solar and non-solar sources of UV radiation are capable of inflicting signature UV mutations (DeMarini et al, 1995; Besaratinia and Pfeifer 2008).
While UVB mutations have comprised the majority of the traditional UVR-associated mutations, little overlap exists between these mutations and those observed in codon V600 of the BRAF gene, the most common location of the well established BRAF mutations in melanoma. BRAF V600 variants can be attributed to G→A transitions and T→A, T→G, and G→T transversions (Thomas et al, 2006; Besaratinia and Pfeifer 2008). In contrast, traditional UVB-induced mutations from exposure to sunlight are characterized by single or tandem C→T transitions at dipyrimidine nucleotides. Damage from UVA radiation has been characterized more recently, with one mechanism being the generation of DNA cross-links and lesions through oxidative damage from UVA-induced photosensitization reactions. Certain of these UVA-induced DNA lesions resemble mutations in BRAF V600 variants from sun-exposed melanomas, suggesting a greater role for UVA in melanomagenesis than traditionally thought (Garibyan and Fisher 2010). Importantly, BRAF V600 mutations may also occur in non-sun-exposed malignancies, such as colon, lung, and thyroid, potentially consistent with oxidative damage as a common carcinogenic mechanism (in those cases independent of UVA). Other important melanoma-associated genes such as INK4A, PTEN, FGFR2, and N-RAS may also possess mutations attributable to UVR (Mar et al, 2013).
UV SIGNALING PATHWAYS FOR TANNING
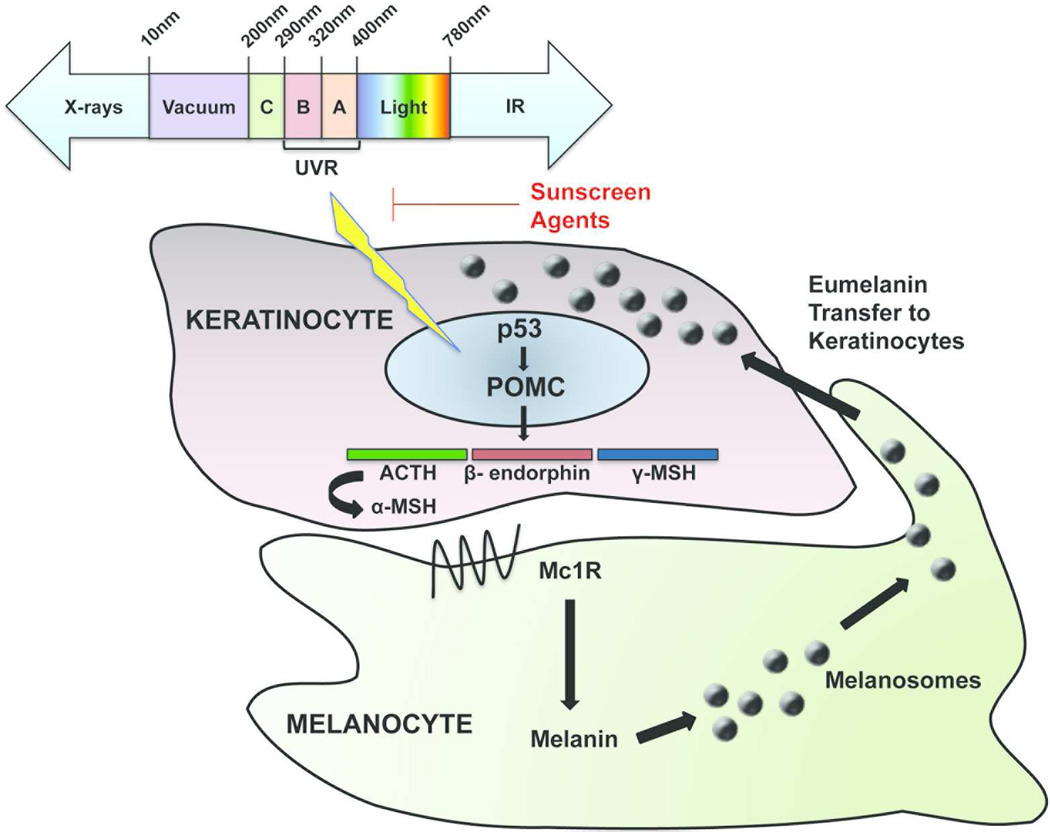
The core component of the skin response to sunlight is the epidermal melanin unit (EMU), comprised of the melanocyte and its associated keratinocytes. UV exposure induces DNA damage in keratinocytes and results in stabilization of the p53 tumor suppressor protein. This promotes p53 transcriptional activation of proopiomelanocortin (POMC), which is enzymatically cleaved to produce α -melanocyte stimulating hormone (α -MSH). α -MSH is released by keratinocytes and binds the MC1R on melanocytes. MC1R activation by α-MSH triggers an increase in cAMP levels within the melanocytes, which increase transcription of microphthalmia-associated transcription factor (MITF) via CREB/ATF1. Binding of MITF to the E-box sequences in promoter regions triggers transcription of numerous pigmentation genes (Tran et al, 2008; Hearing 2011, 2011). These genes act to synthesize, mature, and traffic melanin, the most common types of which are brown-black eumelanin and yellow-red pheomelanin. The melanin is packaged in melanosomes which are exported to keratinocytes, where they localize over the nucleus and may protect the genomic material from further UVR-induced damage (Figure 1 and Figure 2).
Figure 1. The epidermal melanin unit and tanning response to UV radiation.
UV radiation induces DNA damage, which leads to activation of p53. In turn, p53 stimulates transcriptional upregulation of the proopiomelanocortin (POMC) gene, which is post-translationally processed to adrenocorticotrophic hormone (ACTH), α-melanocyte-stimulating hormone (MSH), and β-endorphin. Secreted α-MSH binds to the melanocortin 1 receptor (MC1R) on melanocytes, leading to production of melanin. The melanin is packaged within melanosomes and transported back to keratinocytes, where they localize over the nucleus as part of the protective tanning response to UV radiation.
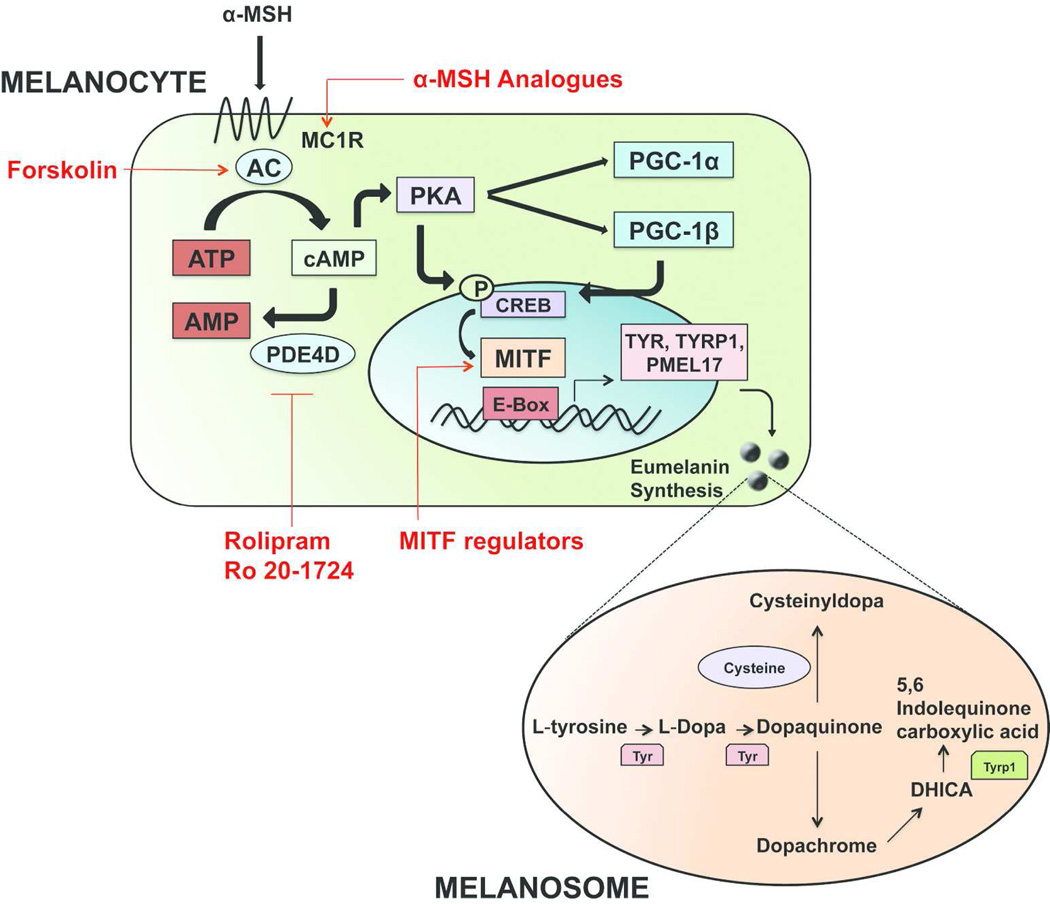
Figure 2. Melanin synthesis and strategies to regulate the tanning response.
Secreted α-MSH from keratinocytes binds MC1R on melanocytes, leading to upregulation of cAMP, which stimulates expression of MITF. MITF then transcriptionally activates expression of enzymatic machinery including tyrosinase and tyrosinase-related protein 1 (Tyrp1), which are critical in the synthesis of melanin within melanosomes. Tyrosinase catalyzes the initial conversion of tyrosine to DOPA and dopaquinone. Dopaquinone may then combine with cysteine to form the pheomelanin precursor cysteinyldopa, or it may enter a separate pathway catalyzed in part by Tyrp1 to produce the eumelanin precursor. The matured melanin is then transported in vesicles called melanosomes to the overlying epidermal keratinocytes. Strategies such as MC1R activators, adenylate cyclase activators, Phosphodiesterase 4D3 inhibitors, and MITF regulators are shown to regulate the UV tanning response by targeting different components of this pathway.
CONSEQUENCES OF UV SIGNALING PATHWAY DEFICIENCY
The loss of p53
As an important regulator of the genotoxic response, p53 is a key tumor suppressor gene that is mutated frequently in human cancer, including skin cancers. The p53 protein regulates multiple signaling pathways in response to stimuli such as DNA damage, oxidative stress, hypoxia, heat shock, membrane compromise, and other stresses (Cui et al, 2007). p53 is thought to participate in DNA repair via multiple mechanisms, including control of cell cycle checkpoint activity as well as regulation of the DNA repair machinery. Lesions with mutant p53 are readily found in UV-exposed hairless mouse skin and sun-exposed healthy human skin. These mutations tend to be localized to dipyrimidine sequences and consist of C→T or CC→TT transitions (Beaumont et al, 2008; Weinstock 2013).
MC1R mutations in skin cancers
The MC1R gene is highly polymorphic in humans, with over 80 variants identified. Certain variants are closely associated with red hair color (RHC) phenotype, which is accompanied by fair skin, poor tanning ability, high sunburn risk, and the highest risk of melanoma for any skin pigmentation type. Other MC1R polymorphic variants with weaker melanoma associations are known as “non-red hair color” (NRHC) variants. Three RHC variants of MC1R that are associated with fair skin and poor tanning are Arg151Cys, Arg160Trp, and Asp294His (Han et al, 2006). The 151Cys variant was associated with increased risks of the three types of skin cancer after controlling for hair color, skin color, and other skin cancer risk factors. Women with medium or olive skin color carrying one non-red hair color allele and one red hair color allele had the highest risk of melanoma (Han et al, 2006; Fargnoli et al, 2010).
One mechanism by which MC1R polymorphisms affect melanoma risk may be through repair of DNA damage (Kadekaro et al, 2005). Human melanocyte cultures exposed to varying levels of UVR were found to have CPD levels that correlated with MC1R genotype and function. In the melanocytes with non-functional MC1R, treatment with forskolin to directly activate adenylate cyclase appeared to enhance CPD repair (Hauser et al, 2006).
Eumelanin and pheomelanin synthesis contributes to melanomagenesis
In addition to its direct effects on DNA damage repair, MC1R may also affect oncogenic drivers through regulation of pigmentation. MC1R signaling and cysteine availability govern the balance in production of eumelanin and pheomelanin. The amino acid cysteine is required for pheomelanin synthesis but not eumelanin synthesis. When MC1R signaling is strong, cysteine stores are insufficient to keep pace with the rate of generation of pigment precursors, and eumelanin production is favored. When the MC1R signal is weak as in redhead melanocytes, cysteine stores keep pace with the slower formation of pigment precursors, leading to formation of cysteine-containing pheomelanin. In our 2012 study using redhead mice with inactivating MC1R mutations, UVR was not necessary for increased melanoma development in these mice when compared to black mice expressing an activating BRAF mutation in their melanocytes. This study supports carcinogenic potential of the pheomelanin synthetic pathway through an UVR-independent mechanism.
Oxidative stress appeared to play a role in pheomelanin-mediated melanomagenesis (Mitra et al, 2012). We hypothesize two possible mechanistic pathways to explain the observed pheomelanin-dependent oxidative DNA damage that drives melanomagenesis. First, pheomelanin might generate ROS that directly or indirectly cause oxidative DNA damage. Second, pheomelanin synthesis might consume cellular antioxidant stores and make the cell more vulnerable to other endogenous ROS (Morgan et al, 2013).
STRATEGIES TO REGULATE THE PIGMENTATION SIGNALING PATHWAY
The UV signaling pathway can be synthetically perturbed at different points to regulate the activity of MC1R, adenylate cyclase, cAMP, and MITF. Such strategies could induce a UV-independent tanning response, potentially conferring a photoprotective effect against UVR-mediated melanomagenesis. Here we will discuss targetable processes at each level in detail.
MC1R activators (analogues of α-MSH)
In addition to the use of sunscreen agents, one strategy for melanoma prevention is based on analogs of α-MSH that function as MC1R agonists (Marwaha et al, 2005). These include products such as melanotan I, melanotan II, afamelanotide, Ac-His-D-Phe-Arg-Trp-NH2, and n-Pentadecanoyl- and 4-Phenylbutyryl-His-D-Phe-Arg-Trp-NH2. Those analogs were more potent than α-MSH itself in stimulating melanogenesis, as well as reducing apoptosis, decreasing release of hydrogen peroxide, and enhancing repair of DNA photoproducts in melanocytes exposed to UVR. The photoprotective and other biological effects of α-MSH analogues await full determination (Hadley et al, 1998; Langan et al, 2010; Schulze et al, 2013; Miller and Tsao 2010).
Some pathologic processes can alter levels of α-MSH and indirectly affect melanogenesis. α-MSH, like ACTH and thyroid-stimulating hormone (TSH), is secreted by the anterior pituitary gland. In Addison’s disease (chronic adrenal insufficiency), lack of negative feedback from cortisol induces the anterior pituitary to produce greater levels of ACTH. As a by-product, more MSH is also produced, leading to hyperpigmented lesions in these patients. The classical hypothalamic-pituitary-adrenal (HPA) axis is a negative feedback neuroendocrine pathway that is essential for the systemic response to external or internal stress. Emerging evidence has indicated that a fully functional cutaneous equivalent participates in the response of skin to local stress as well as other homeostatic contexts (Slominski and Wortsman 2000; Zbytek et al, 2006; Slominski et al, 2007, 2007, 2012). This local system can modulate the function of skin and follicular melanin units following UVR exposure and maintain or restore immune privilege in hair follicles. In the tanning pathway, the EMU comprised of the keratinocyte and melanocyte can be recognized as a functional equivalent of the HPA axis in the skin.
Adenylate cyclase activation
Another strategy to promote the tanning response is through direct stimulation of adenylate cyclase activity downstream of MC1R. UVR-induced tanning is defective in numerous fair-skinned individuals, some of whom possess functional disruption of the MC1R. While UVR is capable of inducing α-MSH production in keratinocytes, loss of MC1R function in red-haired mouse models results in inability to produce a tanning response upon UV exposure. However, pigmentation can be rescued by topical application of the cAMP agonist forskolin. This process can occur without UVR, demonstrating that the pigmentation machinery is available despite the absence of functional MC1R (D'Orazio et al, 2006).
Alternative strategies
Cyclic AMP is an ATP-derived second messenger that functions in signal transduction for a variety of intracellular pathways. Levels of cAMP are controlled by its production, catalyzed by adenylate cyclase, and its hydrolysis, catalyzed by the phosphodiesterase class of enzymes. Phosphodiesterase 4D3 (PDE4D3) was identified as a direct target of the MSH/cAMP/MITF pathway (Khaled et al, 2010). Its activation creates a negative feedback loop that induces refractoriness to chronic stimulation of the cAMP pathway in melanocytes. This highlights a potent mechanism controlling melanocyte differentiation that may be amenable to pharmacologic manipulation (Khaled et al, 2010). Telomere-related oligonucleotides (T-oligos) also have shown promise in augmenting the tanning pathway while bypassing UV-stimulation to confer a protective effect on skin (Arad et al, 2006). This strategy was born from an understanding of telomeric derived oligonucleotides as inducers of DNA repair responses in melanocytes, as well as concomitant inducers of melanogenesis (Atoyan et al, 2007; Gilchrest et al, 2009).
Regulation of MITF through direct targeting and modification of post transcriptional processes
Finally, strategies to regulate the tanning response may focus on MITF, which is required for melanocyte development and is an amplified oncogene in a fraction of human melanomas. In addition to its control of critical pigmentation genes, MITF also regulates target genes essential to cell cycle progression, apoptosis, and differentiation (Levy et al, 2006). Therefore, pharmacologic suppression of MITF is of potential interest in a variety of clinical settings. However, MITF is not known to contain intrinsic catalytic activity amenable to direct small molecule inhibition (Flaherty et al, 2012). An alternative drug-targeting strategy is to identify and interfere with lineage-restricted mechanisms required for MITF expression. Multiple histone deacetylase (HDAC)-inhibitor drugs potently suppress MITF expression in melanocytes, melanoma, and clear cell sarcoma cells (which are sometimes pigmented). Although HDAC inhibitors may affect numerous cellular targets, they have been shown to suppress skin pigmentation upon topical application in mice (Yokoyama et al, 2008). High throughput screens to identify additional small molecules capable of modulating MITF activity are currently being conducted in the authors’ lab, and candidate leads are under development.
A germline missense substitution in MITF (Mi-E318K) was found to occur in families with high incidences of melanoma in Australia, United States, Great Britain, and France (Bertolotto et al, 2011; Yokoyama et al, 2011). Codon 318 is located in a small-ubiquitin-like modifier (SUMO) consensus site (PsiKXE) (Miller et al, 2005) and Mi-E318K ablated that SUMOylation event on MITF. The Mi-E318K mutation measurably increases MITF’s transcriptional activity. An additional key post-translational modification on MITF is its phosphorylation by MAPK (Hemesath et al, 1998), which subsequently targets MITF for ubiquitination and proteolysis (Wu et al, 2000). More recently, it was shown that MITF is targeted by the de-ubiquitinase USP13, a theoretically drug-able protease whose suppression results in strong downregulation of MITF protein levels (Zhao et al, 2011).
Other MITF gene regulators and MITF gene co-factors
In addition to directly targeting MITF, potential strategies to regulate the tanning response can target factors upstream of MITF or genes that serve as co-factors for MITF. The peroxisome proliferator-activated receptor gamma coactivator proteins PGC-1α and PGC-1β are key mediators of α-MSH activation of MITF. PGC-1α and PGC-1β are stabilized through α-MSH signaling via phosphorylation by protein kinase A (PKA). The PGC-1 proteins subsequently activate MITF transcription, and inhibition of the proteins blocks expression of MITF and its target genes in the tanning pathway.
Recent studies in humans revealed polymorphisms in PGC-1β that associated with ability to tan and protection against melanoma (Shoag et al, 2013). YY1, which functions as both a transcriptional repressor and activator, also cooperates with M-MITF to regulate the expression of the piebaldism gene KIT and multiple additional pigmentation genes (Li et al, 2012).
CONCLUSIONS
Tanning represents increased melanization of the epidermis following UV exposure. The UV tanning pathway is a DNA damage-related stress and injury response. Targeting components of the UV tanning pathway through small molecules such as α-MSH analogues may be one strategy to modulate skin pigmentation. α-MSH analogues would likely be less potent on the MC1R loss-of-function variants that are most frequently found in melanoma patients, but they might still function. The strategies targeting components downstream of MC1R show potential in rescuing deficiencies of the UV tanning pathway. These include adenylate cyclase activators, PDE4D3 inhibitors, and T-oligos. Additional interventions which may suppress key melanoma survival factors include MITF regulators such as HDAC-inhibitors and candidates from ongoing high throughput screens for MITF regulators. Strategies may also target MITF post-transcriptional modification processes such as SUMO modification, dimerization, and ubiquitination/deubiquitination. Future mechanism-based studies of UVR are needed to help completely elucidate molecular pathways responsible for the carcinogenic effects of UVR on the melanocyte lineage. We hope to develop better strategies to regulate pigmentation and in doing so, identify further opportunities for prevention, early detection, and treatment of melanoma.
ACKNOWLEDGMENTS
We thank members of the Fisher lab, particularly E. Roydon Price, for invaluable discussions and advice. We apologize to the authors of many papers that have significantly contributed toward the current state of the melanoma and pigmentation fields but could not be acknowledged here due to space constraints. This work was supported by grants to DEF from NIH (1P01 CA163222-01; R01 AR043369-17; R01CA150226; R21CA175907) and The Dr. Miriam and Sheldon G. Adelson Medical Research Foundation.
Abbreviations
- a-MSH
alpha melanocyte-stimulating hormone
- BCC
basal cell carcinoma
- SCC
squamous cell carcinoma
- NMSC
nonmelanoma skin cancer
- ROS
reactive oxygen species
- UVR
ultraviolet radiation
- UVA
ultraviolet A light
- UVB
ultraviolet B light
- UVC
ultraviolet C light
- MC1R
melanocortin 1 receptor
- HDAC
histone deacetylase
- PDE4D3
Phosphodiesterase 4D3
- MITF
microphthalmia-associated transcription factor
- POMC
proopiomelanocortin
- ACTH
adrenocorticotropic hormone
- RHC
red hair color
- TSH
thyroid-stimulating hormone
- NRHC
non-red hair color
- HPA
hypothalamic-pituitary-adrenal
- cAMP
cyclic adenosine monophosphate
- ERK
extracellular signal-regulated kinase
- T-oligos
telomere-derived oligonucleotides
- SUMO
small-ubiquitin-like modifier
- PKA
protein kinase A
- PGC-1α
peroxisome proliferator-activated receptor-γ coactivator-1α
- Dopa
dihydroxyphenylalanine
- Tyrp1
tyrosinase-related protein 1
Footnotes
CONFLICT OF INTEREST
The authors state no conflict interests.
REFERENCES
- Arad S, Konnikov N, Goukassian DA, et al. T-oligos augment UV-induced protective responses in human skin. FASEB J. 2006;20:1895–1897. doi: 10.1096/fj.06-5964fje. [DOI] [PubMed] [Google Scholar]
- Atoyan RY, Sharov AA, Eller MS, et al. Oligonucleotide treatment increases eumelanogenesis, hair pigmentation andmelanocortin-1 receptor expression in the hair follicle. Exp Dermatol. 2007;16:671–677. doi: 10.1111/j.1600-0625.2007.00582.x. [DOI] [PubMed] [Google Scholar]
- Beaumont KA, Shekar SN, Cook AL, et al. Red hair is the null phenotype of MC1R. Hum Mutat. 2008;29:E88–E94. doi: 10.1002/humu.20788. [DOI] [PubMed] [Google Scholar]
- Bertolotto C, Lesueur F, Giuliano S, et al. A SUMOylation-defective MITF germline mutation predisposes to melanoma and renal carcinoma. Nature. 2011;480:94–98. doi: 10.1038/nature10539. [DOI] [PubMed] [Google Scholar]
- Besaratinia A, Pfeifer GP. Sunlight ultraviolet irradiation and BRAF V600 mutagenesis in human melanoma. Hum Mutat. 2008;29:983–991. doi: 10.1002/humu.20802. [DOI] [PubMed] [Google Scholar]
- Cui R, Widlund HR, Feige E, et al. Central role of p53 in the suntan response and pathologic hyperpigmentation. Cell. 2007;128:853–864. doi: 10.1016/j.cell.2006.12.045. [DOI] [PubMed] [Google Scholar]
- D'Orazio J, Jarrett S, Amaro-Ortiz A, et al. UV Radiation and the Skin. Int J Mol Sci. 2013;14:12222–12248. doi: 10.3390/ijms140612222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Orazio JA, Nobuhisa T, Cui R, et al. Topical drug rescue strategy and skin protection based on the role of Mc1r inUV-induced tanning. Nature. 2006;443:340–344. doi: 10.1038/nature05098. [DOI] [PubMed] [Google Scholar]
- DeMarini DM, Shelton ML, Stankowski LF., Jr Mutation spectra in Salmonella of sunlight, white fluorescent light, and lightfrom tanning salon beds: induction of tandem mutations and role of DNA repair. Mutat Res. 1995;327:131–149. doi: 10.1016/0027-5107(94)00179-9. [DOI] [PubMed] [Google Scholar]
- Fargnoli MC, Gandini S, Peris K, et al. MC1R variants increase melanoma risk in families with CDKN2A mutations: ameta-analysis. Eur J Cancer. 2010;46:1413–1420. doi: 10.1016/j.ejca.2010.01.027. [DOI] [PubMed] [Google Scholar]
- Fisher DE, James WD. Indoor tanning--science, behavior, and policy. N Engl J Med. 2010;363:901–903. doi: 10.1056/NEJMp1005999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty KT, Hodi FS, Fisher DE. From genes to drugs: targeted strategies for melanoma. Nat Rev Cancer. 2012;12:349–361. doi: 10.1038/nrc3218. [DOI] [PubMed] [Google Scholar]
- Garibyan L, Fisher DE. How sunlight causes melanoma. Curr Oncol Rep. 2010;12:319–326. doi: 10.1007/s11912-010-0119-y. [DOI] [PubMed] [Google Scholar]
- Gilchrest BA, Eller MS, Yaar M. Telomere-mediated effects on melanogenesis and skin aging. J Investig Dermatol Symp Proc. 2009;14:25–31. doi: 10.1038/jidsymp.2009.9. [DOI] [PubMed] [Google Scholar]
- Hadley ME, Hruby VJ, Blanchard J, et al. Discovery and development of novel melanogenic drugs. Melanotan-I and -II. Pharm Biotechnol. 1998;11:575–595. doi: 10.1007/0-306-47384-4_25. [DOI] [PubMed] [Google Scholar]
- Han J, Kraft P, Colditz GA, et al. Melanocortin 1 receptor variants and skin cancer risk. Int J Cancer. 2006;119:1976–1984. doi: 10.1002/ijc.22074. [DOI] [PubMed] [Google Scholar]
- Hauser JE, Kadekaro AL, Kavanagh R, et al. Melanin content and MC1R function independently affect UVR-induced DNA damage in cultured human melanocytes. Pigment Cell Melanoma Res. 2006;19:303–314. doi: 10.1111/j.1600-0749.2006.00315.x. [DOI] [PubMed] [Google Scholar]
- Hearing VJ. Determination of melanin synthetic pathways. J Invest Dermatol. 2011;131:E8–8E11. doi: 10.1038/skinbio.2011.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hearing VJ. Milestones in melanocytes/melanogenesis. J Invest Dermatol. 2011;131:E1. doi: 10.1038/skinbio.2011.1. [DOI] [PubMed] [Google Scholar]
- Hemesath TJ, Price ER, Takemoto C, et al. MAP kinase links the transcription factor Microphthalmia to c-Kit signaling in melanocytes. Nature. 1998;391:298–301. doi: 10.1038/34681. [DOI] [PubMed] [Google Scholar]
- Kadekaro AL, Kavanagh R, Kanto H, et al. α-Melanocortin and endothelin-1 activate anti-apoptotic pathways and reduce DNA damage in human melanocytes. Cancer Res. 2005;65:4292–4299. doi: 10.1158/0008-5472.CAN-04-4535. [DOI] [PubMed] [Google Scholar]
- Khaled M, Levy C, Fisher DE. Control of melanocyte differentiation by a MITF-PDE4D3 homeostatic circuit. Genes Dev. 2010;24:2276–2281. doi: 10.1101/gad.1937710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langan EA, Nie Z, Rhodes LE. Melanotropic peptides: more than just 'Barbie drugs' and 'sun-tan jabs'. Br J Dermatol. 2010;163:451–455. doi: 10.1111/j.1365-2133.2010.09891.x. [DOI] [PubMed] [Google Scholar]
- Levine JA, Sorace M, Spencer J, Siegel DM. The indoor UV tanning industry: a review of skin cancer risk, health benefit claims, and regulation. J Am Acad Dermatol. 2005;53:1038–1044. doi: 10.1016/j.jaad.2005.07.066. [DOI] [PubMed] [Google Scholar]
- Levy C, Khaled M, Fisher DE. MITF: master regulator of melanocyte development and melanoma oncogene. Trends Mol Med. 2006;12:406–414. doi: 10.1016/j.molmed.2006.07.008. [DOI] [PubMed] [Google Scholar]
- Li J, Song JS, Bell RJ, et al. YY1 regulates melanocyte development and function by cooperating with MITF. PLoS Genet. 2012;8:e1002688. doi: 10.1371/journal.pgen.1002688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JJ, Fisher DE. Lighting a path to pigmentation: mechanisms of MITF induction by UV. Pigment Cell Melanoma Res. 2010;23:741–745. doi: 10.1111/j.1755-148X.2010.00775.x. [DOI] [PubMed] [Google Scholar]
- Mar VJ, Wong SQ, Li J, et al. BRAF/NRAS wild-type melanomas have a high mutation load correlating with histologic and molecular signatures of UV damage. Clin Cancer Res. 2013;19:4589–4598. doi: 10.1158/1078-0432.CCR-13-0398. [DOI] [PubMed] [Google Scholar]
- Marwaha V, Chen YH, Helms E, et al. T-oligo treatment decreases constitutive and UVB-induced COX-2 levels throughp53- and NFkappaB-dependent repression of the COX-2 promoter. J Biol Chem. 2005;280:32379–32388. doi: 10.1074/jbc.M503245200. [DOI] [PubMed] [Google Scholar]
- Miller AJ, Tsao H. New insights into pigmentary pathways and skin cancer. Br J Dermatol. 2010;162:22–28. doi: 10.1111/j.1365-2133.2009.09565.x. [DOI] [PubMed] [Google Scholar]
- Mitra D, Luo X, Morgan A, et al. An ultraviolet-radiation-independent pathway to melanoma carcinogenesis in thered hair/fair skin background. Nature. 2012;491:449–453. doi: 10.1038/nature11624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan AM, Lo J, Fisher DE. How does pheomelanin synthesis contribute to melanomagenesis?: Two distinctmechanisms could explain the carcinogenicity of pheomelanin synthesis. Bioessays. 2013;35:672–676. doi: 10.1002/bies.201300020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman JM, Fisher DE. Indoor ultraviolet tanning and skin cancer: health risks and opportunities. Curr Opin Oncol. 2009;21:144–149. doi: 10.1097/CCO.0b013e3283252fc5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze F, Erdmann H, Hardkop LH, et al. Eruptive naevi and darkening of pre-existing naevi 24 h after a single mono-dose injection of melanotan II. Eur J Dermatol. 2013 doi: 10.1684/ejd.2013.2227. [DOI] [PubMed] [Google Scholar]
- Shoag J, Haq R, Zhang M, et al. PGC-1 coactivators regulate MITF and the tanning response. Mol Cell. 2013;49:145–157. doi: 10.1016/j.molcel.2012.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slominski A, Wortsman J. Neuroendocrinology of the skin. Endocr Rev. 2000;21:457–487. doi: 10.1210/edrv.21.5.0410. [DOI] [PubMed] [Google Scholar]
- Slominski A, Wortsman J, Tuckey RC, et al. Differential expression of HPA axis homolog in the skin. Mol Cell Endocrinol. 2007;265–266:143–149. doi: 10.1016/j.mce.2006.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slominski AT, Zmijewski MA, Skobowiat C, et al. Sensing the environment: regulation of local and global homeostasis by the skin'sneuroendocrine system. Adv Anat Embryol Cell Biol. 2012;212:v–vii. 1–115. doi: 10.1007/978-3-642-19683-6_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas NE, Berwick M, Cordeiro-Stone M. Could BRAF mutations in melanocytic lesions arise from DNA damage induced byultraviolet radiation. J Invest Dermatol. 2006;126:1693–1696. doi: 10.1038/sj.jid.5700458. [DOI] [PubMed] [Google Scholar]
- Tran TT, Schulman J, Fisher DE. UV and pigmentation: molecular mechanisms and social controversies. Pigment Cell Melanoma Res. 2008;21:509–516. doi: 10.1111/j.1755-148X.2008.00498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstock MA. Epidemiology and UV exposure. J Invest Dermatol. 2013;133:E11–E12. doi: 10.1038/skinbio.2013.178. [DOI] [PubMed] [Google Scholar]
- Wu M, Hemesath TJ, Takemoto CM, et al. c-Kit triggers dual phosphorylations, which couple activation and degradation of the essential melanocyte factor Mi. Genes Dev. 2000;14:301–312. [PMC free article] [PubMed] [Google Scholar]
- Yokoyama S, Feige E, Poling LL, et al. Pharmacologic suppression of MITF expression via HDAC inhibitors in themelanocyte lineage. Pigment Cell Melanoma Res. 2008;21:457–463. doi: 10.1111/j.1755-148X.2008.00480.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama S, Woods SL, Boyle GM, et al. A novel recurrent mutation in MITF predisposes to familial and sporadic melanoma. Nature. 2011;480:99–103. doi: 10.1038/nature10630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zbytek B, Wortsman J, Slominski A. Characterization of a ultraviolet B-induced corticotropin-releasinghormone-proopiomelanocortin system in human melanocytes. Mol Endocrinol. 2006;20:2539–2547. doi: 10.1210/me.2006-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Fiske B, Kawakami A, et al. Regulation of MITF stability by the USP13 deubiquitinase. Nat Commun. 2011;2:414. doi: 10.1038/ncomms1421. [DOI] [PubMed] [Google Scholar]