Abstract
Pore-forming toxins (PFTs) are a class of pathogen-secreted molecules that oligomerize to form transmembrane channels in cellular membranes. Determining the mechanism for how PFTs bind membranes is key to understanding their role in disease and possible ways to block their action. Vibrio vulnificus, an aquatic pathogen responsible for severe food poisoning and septicemia in humans, secretes a PFT called Vibrio vulnificus hemolysin (VVH), which contains a single C-terminal targeting domain predicted to resemble a β-trefoil lectin fold. In order to understand the selectivity of the lectin for glycan motifs, we expressed the isolated VVH β-trefoil domain and used glycan-chip screening to identify that VVH displays a preference for terminal galactosyl groups including N-acetyl-D-galactosamine (GalNAc) and N-acetyl-D-lactosamine (LacNAc). The X-ray crystal structure of the VVH lectin domain solved to 2.0 Å resolution reveals a heptameric ring arrangement similar to the oligomeric form of the related, but inactive, lectin from Vibrio cholerae cytolysin. Structures bound to glycerol, GalNAc, and LacNAc outline a common and versatile mode of recognition allowing VVH to target a wide variety of cell-surface ligands. Sequence analysis in light of our structural and functional data suggests that VVH may represent an earlier step in the evolution of Vibrio PFTs.
Keywords: bacterial pathogenesis, carbohydrate, crystallography, cytolysin, pore-forming toxin
Introduction
Vibrio vulnificus is an emerging human pathogen that causes severe food-poisoning and opportunistic infections with a mortality rate exceeding 50% 1. In contrast to the primarily gut-localized pathogenesis of the historically significant Vibrio cholerae bacterium, V. vulnificus escapes the gastrointestinal tract to cause primary septicemia and septic shock, particularly in individuals with liver disease or who are immuno-compromised 1. Furthermore, V. vulnificus will enter skin lesions exposed to seawater leading to cellulitis and necrotic skin infections, albeit with a lower mortality rate 2. In support of its role as a human pathogen, V. vulnificus produces a number of virulence factors, including the cytolytic pore-forming toxin (PFT) Vibrio vulnificus hemolysin/cytolysin (VVH), a product of the vvhA gene. Recent evidence from mouse models suggests that VVH may work in conjunction with the V. vulnificus multifunctional autoprocessing RTX (MARTXVv) toxin to facilitate rapid growth, inflammation, and epithelial necrosis in the intestine 3. Most strikingly, removal of these two factors alone rendered a clinical strain of V. vulnificus unable to cause infection in a mouse disease model 3.
Although its three-dimensional structure has not been determined, VVH belongs to a larger family of toxins found in both gram-positive and gram-negative pathogens that likely share a similar three-dimensional structure first identified in Staphylococcus aureus α-hemolysin 4. Typically secreted as water-soluble monomers, these toxins bind to target membranes, oligomerize into a pre-pore intermediate 5, and then undergo a structural rearrangement that forms transmembrane channels in the cell membrane. PFTs across this family have been shown to lyse a broad array of target cells, including intestinal cells, neutrophils, and erythrocytes. Alternatively, some toxins may play a non-lytic role by triggering inflammation 6 or activating membrane metalloproteases to break down focal adhesions allowing bacteria to penetrate epithelial barriers 7.
To facilitate binding to cell membranes, PFTs may contain binding sites or additional domains that recognize specific motifs found on target host cells. Protein receptors have been identified for several VVH-homologous Staphylococcal toxins including ADAM 10 as a receptor for α-hemolysin 7, CCR5 as a receptor for leukotoxin ED 8, and C5a receptors as targets of Panton-Valentine Leukocidin 9. Protein receptors have not yet been identified for VVH, but many toxins within the Vibrio family contain one or two domains attached to their carboxyl-termini with sequence and structural similarity to carbohydrate-binding lectins 10,11. Like many PFTs, VVH may also utilize cholesterol in the membrane to recognize eukaryotic cells 12.
Sequence analysis of VVH suggests that it has a single C-terminal domain that resembles an R-type lectin 13. R-type lectins are common carbohydrate-binding motifs exemplified by the B-chain of the plant toxin ricin from Ricinus communis 14, but also found in bacteria, fungi, and animals 15. Other examples of R-type lectins include the hemagglutinin HA1 from Clostridium botulinum 16, the mushroom-derived LSL toxin 17, the human mannose receptor 18,19, bacterial glycosyltransferases 20, and bacterial glycosidases 21. These domains possess a β-trefoil fold with a pseudo three-fold axis of symmetry relating three potential carbohydrate-binding sites with characteristic QxW motifs and otherwise highly divergent sequences 13. The number of active binding sites varies, with demonstrated examples containing one, two, or three functional sites that typically bind to terminal sugar motifs found on cell-surface glycans. Many R-type lectins, including the ricin B-chain, prefer to bind D-galactoside moieties (D-galactose and N-acetyl-D-galactosamine) of varying complexity 22. This versatile fold has been adopted to other specificities, including 4-O-sulfated N-acetyl-D-galactosamine (GalNAc) termini for mannose receptors 23 and polymeric xylans for the Streptomyces lividans xylanase CBM13 21. Lectins with β-trefoil folds typically bind monosaccharide sugars in solution with micro- to high-millimolar affinity, but will bind to cells with nano- to low-micromolar affinity resulting from multivalent binding to multi-saccharide motifs on surface glycans (particularly Galβ1-4GlcNAc-R) 15. VVH has been shown to interact with methyl-β-cyclodextran 24, providing evidence that the lectin domain may possess carbohydrate-binding activity. Several mutations in and around the lectin domain also appear to inactivate the toxin 25,26.
A number of Vibrio hemolysins/cytolysins related to VVH possess a second lectin domain with a characteristic β-prism fold following the β-trefoil domain. The best structurally-characterized example of this addition is in Vibrio cholerae cytolysin (VCC), which contains a β-prism lectin (Figure 1) similar to the plant lectins jacalin 27 and artocarpin 28. Interestingly, while the VCC β-prism domain recognizes complex N-glycans with roughly 100 nM affinity, its β-trefoil domain appears to be inactive 29.
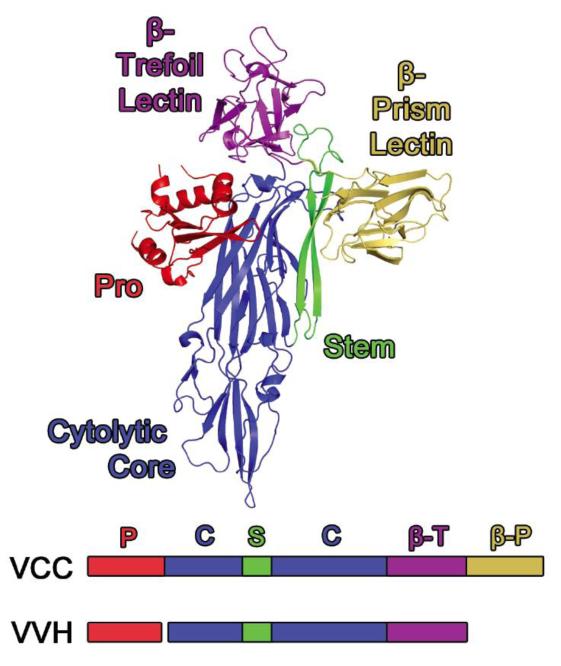
Figure 1. Comparison between Vibrio cholerae Cytolysin and Vibrio vulnificus Hemolysin.
The crystal structure of VCC (PDB 1XEZ) serves as a model for VVH with two main exceptions. The VVH toxin does not contain a β-prism lectin domain (gold) and the chaperone-like pro-domain is expressed as a separate gene product (vvhB). The β-trefoil domain (magenta) is attached to the C-terminus of the cytolytic core (blue). The stem region (green) is an amphiphilic loop that inserts into the membrane following oligomerization of the toxin (heptamer in VCC).
Could VVH represent an evolutionary predecessor to toxins with two lectin domains? To test this possibility, we subjected the isolated VVH β-trefoil domain to glycan-chip screening and identified that it indeed binds galactosyl ligands with micromolar affinity. We characterized binding to several of these ligands and solved the high-resolution crystal structure of the isolated VVH β-trefoil domain bound to two of these sugars. Our results indicate a scenario whereby a β-trefoil lectin domain with micromolar affinity is supplanted by a β-prism lectin with nanomolar affinity. This may have led to an improvement in the ability of the toxin to target cells at the low concentrations typically found at the site of infection, or a change in the glycan markers recognized by the toxin. These results show how a toxin scaffold may be fine-tuned to affect activity across a family of homologous proteins and provide a better understanding of how Vibrio PFTs target cells during infection.
Results
Glycan Screen Reveals Glycan Specificity
To investigate the glycan specificity of the VVH lectin, we expressed the isolated C-terminal lectin domain fused to GFPUV, which allowed for the isolation of soluble protein. Initial attempts to express and purify full-length VVH or the isolated lectin alone were unsuccessful. We used a structural alignment based on the VCC crystal structure to identify the ideal place to truncate the domain and chose a construct containing residues A338 to L471. Following removal of the fusion partner, the purified VVH lectin was labeled with AlexaFluor 488 and submitted to the Consortium for Functional Glycomics (www.functionalglycomics.org) for screening against a combinatorial array of 610 glycans commonly found on mammalian membranes. Even though the possible number of glycans found in the human body is estimated be quite large (100,000-500,000) due to variations that occur on their termini, smaller glycan arrays may capture much of the information encoded in the human glycome 30. Glycan screening is particularly powerful in that it allows one to not only identify which glycans are bound by a particular lectin, but also which glycans are not bound 31. Since the array contains entire glycans along with fragments of these often branched molecules, the specific carbohydrate footprint of the lectin on the larger glycans can be identified. Lectins often display one of two behaviors; specific binding to a small subset of glycans on the chip or broad binding to a large range of glycans with a common shared motif 31.
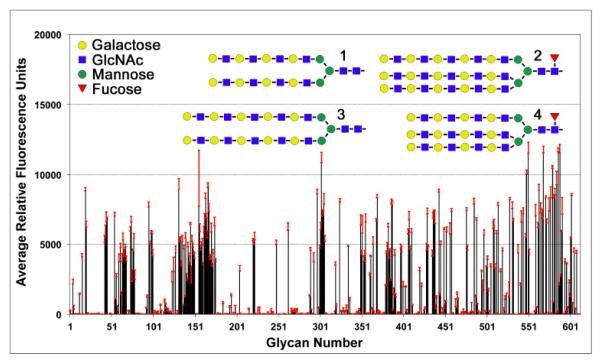
Glycan screening of VVH at several concentrations indicated a broad recognition pattern (Figure 2 and Table 1), with approximately 200 glycans scoring over 1,200 RFUs (relative fluorescence units; 10% of the maximum signal) for the 50 μg/ml VVH sample. As a point of reference, earlier glycan screening (using a nearly identical mammalian array) of the VCC β-prism domain resulted in only 20 glycans scoring greater than 10% of the maximum signal 29. For VVH, a few select terminal groups dominated the glycan results, with 63 of the top 100 glycans terminating in a Galβ1-4GlcNAc-(N-acetyl-D-lactosamine or LacNAc) moiety (Table 1). The remaining glycans contained a Galβ1-4-(8 of 100), a GalNAc-(9 of 100) or a Galβ1-3-(20 of 100) terminal moiety. Approximately one quarter of the glycans were additionally modified, with sulfate, fucose, and/or sialic acid groups attached to the terminal or penultimate sugars in the C2 or C6 positions. The top four hits consisted of bi- and triantennary N-glycans containing a high density of LacNAc units (4-6) on each branch (Figure 2). Glycans ranked 100-200 (representing 1359-5165 RFUs) also display a preference for LacNAc and Galβ1-3GlcNAc-R groups, with further fucose and neuraminic acid substitutions found on the terminal galactose ring for some glycans.
Figure 2. Glycan array data.
VVH displays a broad glycan recognition profile with a preference for terminal LacNAc groups. Labeled VVH lectin (50 μg/ml) was screened against a panel of 610 mammalian-derived carbohydrate ligands by the Consortium for Functional Glycomics with six replicates. Data are presented as the average relative fluorescence units and errors bars (in red) denote the standard deviation from four replicates (after dropping the highest and lowest of six measurements). Schematic representations of the top four glycans identified in the screen are shown above the chart data. Additional analysis of these results is provided in Table 1.
Table 1.
Summary of glycan-chip data
| Terminal Group | Number (out of top 100) |
|---|---|
|
| |
| Galβ1-4GlcNAc- | 63 |
| Galβ1-3GlcNAc- | 14 |
| Galβ1-4Glc- | 8 |
| Galβ1-3GalNAc- | 6 |
| GalNAcβ1-4GlcNAc- | 6 |
| GalNAcβ1-3Gal- | 2 |
| GalNAcβ1-3GlcNAc- | 1 |
|
| |
| Total | 100 |
| Terminal Modifications | Number |
|
| |
| 6-sulfo- | 11 |
| Fucα1-2- | 10 |
| NeuAcα2-6- | 5 |
| GlcNAcβ1-6- | 1 |
|
| |
| Total | 27 (24 glycans) |
Quantification of Sugar Binding by Isothermal Titration Calorimetry
Because glycan screening does not provide absolute numbers regarding the strength of glycan interactions, we conducted binding studies using isothermal titration calorimetry (ITC) to quantify the binding of putative VVH ligands. Our results (Table 2 and Supplemental Figure 1) confirmed that the VVH β-trefoil domain binds LacNAc with a measured dissociation constant (Kd) of 152 μM. Our data best fit to a stoichiometry of one LacNAc molecule per VVH molecule, suggesting that only one of the three potential binding sites in the β-trefoil is active. To further understand the specificity of the domain, we looked at binding of the monosaccharide D-galactose, a fragment of LacNAc, which exhibited a similar Kd of 173 μM. Binding to D-lactose and GalNAc gave slightly tighter dissociation constants of 121 μM and 131 μM, respectively. Together, these results suggest that the lectin binds primarily to the galactose moiety and that acetylation of either the galactose or glucose moieties had little effect on binding.
Table 2.
Isothermal Titration Calorimetry data for VVH p-trefoil Lectin Domain
| Compound | Ka (M−1 × 103) |
Kd (μM) | ΔH (kcal/mol) |
TΔS (kcal/mol) |
ΔG (kcal/mol) |
Reduced X2 (× 10−2) |
|---|---|---|---|---|---|---|
| D-Galactose | 5.79 (4.20,6.81) |
173 (147,238) |
−8.66 (−7.80,−13.10) |
−3.53 (−2.57,−8.16) |
−5.13 (−4.94,−5.23) |
5.78 |
| N –Acetyl-D-Galactosamine | 7.66 (5.99,9.50) |
131 (105,167) |
−6.87 (−5.53,−9.53) |
−1.57 (−0.11,−4.38) |
−5.30 (−5.15,−5.42) |
1.95 |
| D-Galactose 6-Sulfate | 1.63 (0.80,2.13) |
612 (469,1147) |
−12.99 (−9.95,NF) |
−8.61 (−5.41,NF) |
−4.38 (−3.96,−4.54) |
2.99 |
| D-Lactose | 8.24 (7.23,9.34) |
121 (107,139) |
−13.97 (−12.29,−16.37) |
−8.63 (−6.88,−11.11) |
−5.34 (−5.26,−5.41) |
2.16 |
| N-Acetyl-D-Lactosamine | 6.58 (5.61,7.61) |
152 (131,178) |
−10.49 (−8.93,−12.99) |
−5.28 (−3.64,−7.88) |
−5.21 (−5.11,−5.29) |
0.86 |
Binding was not detected with N-acetylglucosamine or N-acetylneuraminic acid. T=298K.
Parentheses denote 95% confidence limits as calculated by SEDPHAT. NF: Not found. See also Figure S1.
To determine if the lectin domain is only specific for galactosyl sugars, we tested binding to N-acetylglucosamine (GlcNAc) by ITC and were unable to detect any binding. We also received negative results with N-acetylneuraminic acid, which suggests that while 6-position sialylation of the terminal galactosyl sugar is tolerated by the lectin, there is no affinity for neuraminic acid by itself. As seen in our glycan data, the VVH lectin will bind LacNAc molecules with 6-sulfate substitutions; a motif found in keratan sulfate and associated with some tumors 32. We observed that binding to free D-galactose 6-sulfate was only slightly weaker than D-galactose, with a measured Kd of 612 μM.
Crystallization of the VVH β-Trefoil Lectin Domain
Our glycan-binding data indicate that the VVH β-trefoil domain represents a functional lectin able to bind glycans that terminate in galactosyl groups. In order to understand the basis of this preference and compare it to the non-functional VCC β-trefoil domain, we solved the crystal structure of the VVH β-trefoil alone and bound to two ligands identified in our screen. Purified VVH β-trefoil protein was subjected to sparse matrix crystallization screens and crystals were obtained in a condition containing 100 mM TRIS pH 8.25 and 6% PEG 8000. Crystals of different morphologies were obtained under the same condition when the ligands GalNAc and LacNAc were included in the crystallization trial. X-ray data collected on the apo, GalNAc and LacNAc crystals indicated three different space groups with different unit cell dimensions (Table 3).
Table 3.
X-ray and Refinement Statistics
| Crystal | Apo (Glycerol) | GalNAc | LacNAc |
|---|---|---|---|
| Resolution Limits (Å) | 48.5 - 2.0 (2.07- 2.0)* |
49.6 - 2.0 (2.07- 2.0) |
48.0 – 2.1 (2.18- 2.1) |
| Space Group | P212121 | P21 | I2 |
| Cell dimensions a, b, c (Å); α,β,γ(°) |
75.8, 118.2, 149.3; 90, 90, 90 |
55.4, 79.8, 148.7; 90, 89.5, 90 |
55.6, 79.8, 326.1; 90, 94.1, 90 |
| Total Reflections (N) | 756,224 | 491,799 | 375,224 |
| Unique Reflections (N) | 91,312 (8,999) | 83,325 (5,960) | 83,123 (8,250) |
| Redundancy (%) | 8.3 (8.1) | 5.9 (4.0) | 4.5 (4.3) |
| Completeness (%) | 100.0 (100.0) | 95.0 (68.5) | 99.8 (99.9) |
| Rsym (%) | 17.5 (69.0) | 16.1 (98.9) | 11.3 (69.3) |
| Rpim (%) | 6.6 (25.6) | 7.1 (46.8) | 5.8 (34.9) |
| I/σI (Mn(I/sd)) | 17.2 (2.2) | 18.5 (1.9) | 24.0 (3.0) |
| Refinement Statistics | |||
| Atoms (non-hydrogen) | 8,022 | 8,067 | 7,717 |
| Waters | 915 | 845 | 529 |
| R-work (%) | 16.6 | 15.8 | 16.2 |
| R-free (%) | 20.0 | 19.1 | 18.8 |
| R.m.s. dev. bonds (Å) | 0.010 | 0.011 | 0.013 |
| R.m.s. dev. angles (°) | 1.18 | 1.23 | 1.28 |
| Average B-factor | 36.5 | 36.2 | 52.1 |
| Ramachandran statistics | |||
| Favored (%) | 95.1 | 96.9 | 95.9 |
| Allowed (%) | 4.9 | 3.1 | 4.1 |
| Outliers (%) | 0.0 | 0.0 | 0.0 |
Numbers in parentheses denote highest resolution shell.
Self-rotation plots of the diffraction data revealed a 7-fold non-crystallographic symmetry (NCS) axis consistent with the heptameric symmetry seen in the oligomeric structure of the full-length VCC toxin 33. Molecular replacement trials using a heptameric VCC β-trefoil ring extracted from the VCC structure (PDB 3O44) were successful in giving a solution with good crystal packing interactions and favorable R-factors. In order to reduce possible phase bias from the VCC search model, low-resolution phases from the molecular replacement solution were extended with 7-fold averaging and solvent density modification (see Materials and Methods). Final high-resolution data were obtained to 2.0 Å resolution for the apo and GalNAc crystals and to 2.1 Å resolution for the LacNAc crystals. All three structures were refined to final Rfree values less than 20% (Table 3).
VVH β-Trefoil Lectin Domain Structure
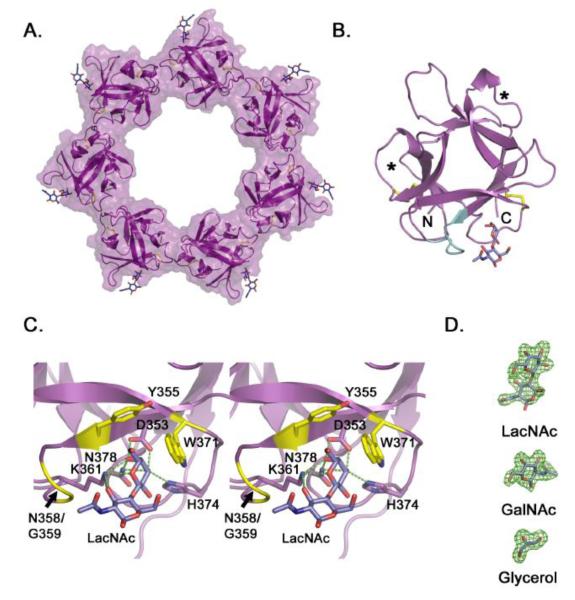
The VVH β-trefoil domain formed a heptameric ring in all three crystal forms with approximate inner and outer diameters of 50 Å and 100 Å, respectively (Figure 3A). Individual monomers in the ring exhibited an expected R-lectin β-trefoil fold with two disulfide bridges within the first and second QxW repeats. The interface between subunits in the ring buried on average 396.5 Å2 of surface area according to the PISA method 34 as implemented in Coot 35. Consistent with our ITC data, ligand electron density was only observed in one of the three potential sugar-binding pockets (Figure 3B,C). Crystal packing interactions between VVH rings in the GalNAc and LacNAc structures led to a variation in overall temperature factors in the structures, with some subunits exhibiting better overall electron density than others (Supplemental Figure 2). The analysis of ligand interactions was performed on the domain with the lowest overall B-factors and best ligand electron density. Inspection of the ligand-binding pocket in the crystals grown without added ligand indicated clear electron density for bound glycerol, which was used as a cryoprotectant in all three crystal forms (Figure 3D).
Figure 3. Crystal structure of the VVH β-trefoil lectin.
A. Cartoon representation of the isolated VVH lectin domain crystallized in the presence of LacNAc. Density for LacNAc was clearly interpretable for six of seven subunits, with the remaining LacNAc molecule modeled in to produce this figure. B. Crystal structure of one isolated β-trefoil domain from the heptamer. The two internal disulfide bonds (yellow) are shown in stick representation and the extended loop (Y355-T362) that contacts the ligand is colored in cyan. Asterisks identify the approximate location of carbohydrate binding pockets in β-trefoil domains with multiple binding sites. C. Close-up view of the binding pocket displayed in stereo. Predicted hydrogen bonds are shown as green dotted lines and non-bonded interactions are colored yellow. Predictions were made using the LigPlot+ software 51. D. Electron omit density (green) contoured at 3 σ for representative ligands from the three structures.
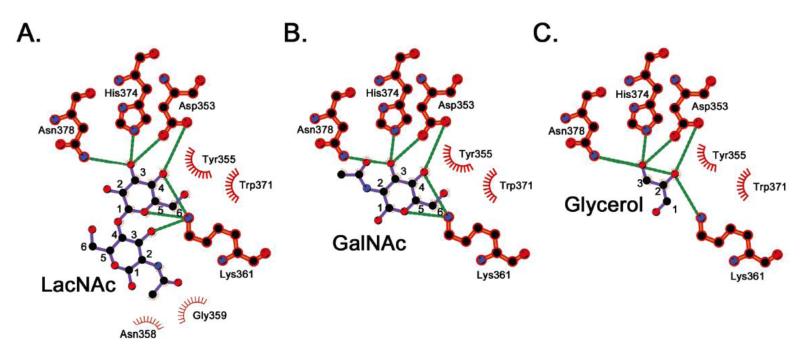
Crystals grown in the presence of LacNAc showed clear density for LacNAc molecules in six of seven subunits in the ring. In the seventh subunit, crystal packing interactions leave too little space for LacNAc binding and glycerol appears to have filled in the remaining space. Close inspection of the LacNAc binding site indicates a number of polar and non-polar interactions involved in recognition of the disaccharide ligand (Figure 3C and 4A) involving approximately 250 Å2 of buried surface area. Similar to interactions seen in other R-type lectins 13, a key tryptophan (W371) stacks against one side of the galactose ring accompanied by a second aromatic residue (Y355) perpendicular to the ring. A second feature common to previously characterized lectins is an aspartate residue (D353) that forms hydrogen bonds with the O3 and O4 hydroxyl groups on the galactose ring. Flanking both sides of the ring and forming additional stabilizing hydrogen bonds are K361 and H374 as well as the amine group of N378. Most of the specific interactions between the protein and the dissaccharide involve the galactose group, with the exception of a single hydrogen bond involving K361 and non-polar interactions with N358 and G359. The loop containing these residues seemingly wraps around the GlcNAc ring interacting with the C2 acetyl group (Figures 3B,C and 4A).
Figure 4. Specific ligand interactions.
Schematic representations of interactions between residues in the binding site and the three ligands are shown in (A), (B), and (C) for LacNAc, GalNAc, and glycerol, respectively. Hydrogen bonds are shown as green dotted lines and nonbonded interactions are denoted by spoked arcs. Carbon atoms are numbered for comparison. Images were generated using LigPlot+ v.1.4.5 51.
Structures of the VVH β-trefoil lectin bound to GalNAc and glycerol indicate nearly identical interactions between the protein and ligand, as compared to the LacNAc structure (Figure 4B,C). The GalNAc ligand buries approximately 180 Å2 of surface area with the C2 acetyl group stacking against the H374 imidazole ring. Glycerol mimics the C3/C4/C5 backbone of the galactose ring seen in LacNAc and GalNAc burying approximately 120 Å2 of surface area. Together, these results are consistent with the ITC data, and help explain the pattern of glycan recognition observed in the glycan-chip data. Because the bulk of the interactions between the VVH lectin and the glycan are with the terminal galactose moiety, the most important driver of selectivity is the stereochemistry of the terminal sugar. This is further supported by our ITC data, which shows that GlcNAc and N-acetylneuraminic acid do not bind to VVH. The identity of the penultimate sugar is not as important, with glucosyl and galactosyl groups both represented in the glycan-chip data with and without C2 acetyl groups. The structure also indicates that the terminal galactosyl O2 and O6 hydroxyl groups do not participate in hydrogen-bonding interactions and are sterically unoccluded. This explains why modification of these positions with sulfate, fucose, neuraminic acid, and GlcNAc groups is generally tolerated.
Relationship to VCC and Other β-Trefoil Lectins
To investigate how the VVH domain relates to other β-trefoil lectins, we conducted BLAST 36 and DALI 37 searches to look for sequence and structural homologs and created a sequence alignment using MEGA 38. For our alignment (Figure 5A), we selected representative β-trefoil domains from species within the larger family of related pore-forming toxins. For comparison, we also included the ricin β-trefoil domain, which has a single functional binding site 14, and the Streptomyces lividans CBM13 lectin 21, which has three functional carbohydrate-binding sites and scored near the top of our DALI search (1.9 Å RMSD and Z-factor of 17.3). In all, the DALI search returned over 800 domains in the protein data bank (PDB) with RMSD values ranging roughly between 1.7 and 3.0 Å.
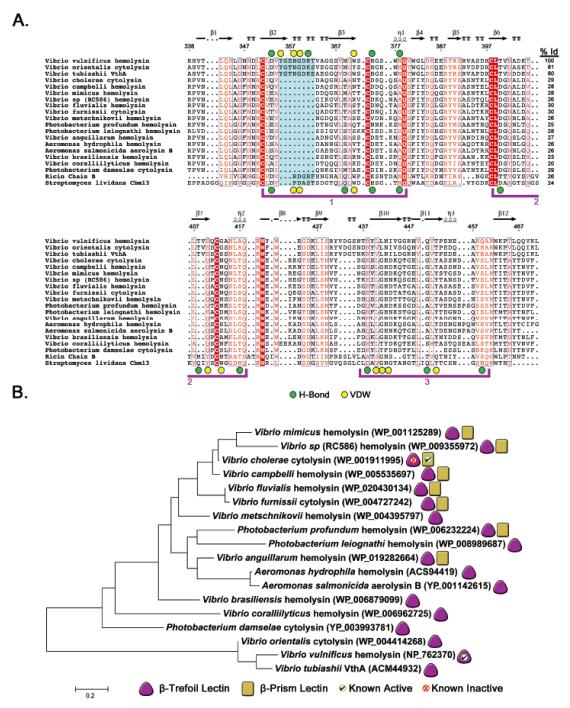
Figure 5. Sequence alignment of VVH and related β-trefoil domains.
A. A BLAST 36 search using the VVH β-trefoil protein sequence was conducted and representative sequences from related toxins aligned using the Muscle 52 algorithm as implemented in MEGA v. 5.2.2 38 and prepared using ESPript 3.0 (http://espript.ibcp.fr) 53. Sequences from β-trefoil domains from ricin B-chain and S. lividans CBM13 were included at the bottom of the alignment for reference. Three potential binding sites are bracketed in purple. The ricin domain has an active sugar-binding site in bracket 1 and CBM13 has three active binding sites. VVH residues involved in hydrogen bonding or van der Waals interactions are denoted as green or yellow circles, respectively, above the alignment. Residues involved in similar interactions in CBM13 are marked similarly below the alignment. The extended loop that contacts the penultimate carbohydrate unit in VVH is shaded in cyan and the degree of sequence identity between VVH and each toxin is listed in the right column. The numbers above the alignment are based on the complete VVH sequence and secondary structure is represented as calculated by ESPript. B. Evolutionary history as inferred using the maximum likelihood method based on the JTT matrix-based model as implemented in MEGA v. 5.2.2. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site (111 total positions). Toxins with a β-trefoil domain are denoted with a purple triangle and the subset with β-prism domains are marked with a yellow rectangle. Domains for which we have obtained positive binding results using glycan-chip screening are marked with a green checkmark and domains that did not exhibit activity are marked with a red ‘X’.
Our previous results suggest that the β-trefoil domain of VCC is inactive 29. The β-trefoil lectin domains of VCC and VVH share 29% sequence identity and superimpose with a RMSD of 1.8 Å (Figure 5A and Supplemental Figure 3A). Examination of a sequence alignment between both domains helps shed light on the reason VCC is inactive while VVH binds carbohydrates. The important residues that coordinate the sugar ring in VVH including D353 and W371 are replaced by dissimilar amino acids in VCC (a glutamine and an asparagine, respectively). Most importantly, the entire eight-residue loop in VVH that contacts the GlcNAc sugar ring and contains Y355 and K361 is missing in VCC (Figure 3B, 5A and Supplemental Figure 3A). With the exception of the closely related toxins from Vibrio orientalis and Vibrio tubiashii (with 81% and 80% sequence identity to VVH, respectively), all of the remaining Vibrio toxins we identified in our BLAST search are also missing this eight-amino acid stretch and contain substitutions at the other positions that coordinate carbohydrate ligands in VVH. Conversely, the ricin and S. lividans lectins contain this loop (although three residues shorter) and maintain the important aspartate and tryptophan residues that coordinate glycans in VVH.
If the first QxW domain is inactive in most of the Vibrio PFT β-trefoil lectins, what about the second and third domains? Comparison with the S. lividans CBM13 lectin, (which has three functional domains) suggests that both of these QxW domains are inactive in all of the toxins examined. While most of the toxins do contain an aspartate residue in the coordinating position found in the CBM13 second QxW domain, they do not maintain any of the other residues that form a functional binding site and are missing a coordinating aspartate in the third QxW domain. In light of these results, we predict that the β-trefoil lectin domain is inactive in all of the toxins we examined, with the exception of the two toxins closely related to VVH (V. orientalis and V. tubiashii).
Analytical Ultracentrifugation
In order to determine whether the heptamer observed in the crystal structure also exists in solution, we conducted a sedimentation velocity analytical ultracentrifugation experiment on the purified VVH lectin. Analysis of sedimentation velocity data using the ls-g(s*) method 39 shows a primary peak close to 2 Svedbergs (S) that slightly increases with increasing protein concentration (Supplemental Figure 4). The molecular weight deduced from discrete analysis of the lower concentration samples is consistent with a monomer and the shift suggests weak-self association. To further characterize this interaction, a sedimentation equilibrium experiment was conducted at three protein concentrations at 44,000 RPM. A single ideal species model gave a fit molecular weight of 15,511 Da, close to the sequence-predicted weight of 15,370 Da. Global fitting of the three concentrations to a monomer-heptamer model using the predicted monomer molecular weight gave Ka = 6.98 (5.28, 8.96) × 1022 M−6, with values in parenthesis signifying 95% confidence intervals (RMS deviation 0.01862). This value is consistent with an effective Kd of approximately 150 μM and suggests that the heptamer should be the predominant species at the concentrations used for crystallization (325 μM). However, the Kd also suggests that the monomer is likely to dominate at the concentration used for glycan screening (50 μM) reducing the possibility that avidity effects were represented in the chip data.
Discussion
Pore-forming toxins utilize a variety of strategies for targeting the membrane of host cells. In toxins structurally related to VVH, carbohydrate-binding lectin domains appear to be a common mechanism for attaching to glycosylated proteins on the cell surface. Structurally similar toxins from Staphyloccocus aureus including α- and γ-hemolysins do not contain any C-terminal lectin domains, but rather utilize lipid and protein receptors for cell targeting. Looking more broadly at this family of homologous toxins, it appears that the two carbohydrate-binding lectin domains were obtained at different times as the toxin genes evolved to their current state. The fact that the VVH β-trefoil domain forms a heptameric ring by itself is an interesting result, since this is not an arrangement commonly seen in structures of β-trefoil domains throughout the PDB. This finding suggests that the lectin domain is not merely tethered to the toxin core, but has incorporated the symmetry of the assembled toxin into its own inter-domain interfaces.
To further investigate the relationship between the VVH and VCC toxins, we compared the structure of the VVH ring with the β-trefoil ring removed from the heptameric VCC crystal structure (Supplemental Figure 3B). Our analysis indicates similar interfaces between subunits, with 421.1 Å2 and 396.5 Å2 of buried surface area, respectively. The RMSD between alpha carbons for the two rings is 2.4 Å2, with VVH containing five predicted hydrogen bonds and one salt bridge between interfaces and VCC containing 10 hydrogen bonds. Both proteins have several hydrophobic residues in the interface including a conserved leucine at position 416 in VVH and position 528 in VCC. Considering the similarity of the two ring structures, we conclude that the VVH ring seen in our crystal structure is likely physiologically relevant and that the full-length VVH toxin is likely to assemble into a heptameric oligomeric state similar to VCC, although we are unaware of any direct evidence for this arrangement. Since the overall sequence identity between VVH and VCC is close to 30% with key structural residues conserved in the cytolysin domain 11, the VCC oligomeric structure is likely an acceptable model for understanding the overall structure of the VVH oligomer.
Our results reveal that the β-trefoil domain of VVH has micromolar affinity for galactosyl-terminating groups including LacNAc, which is a common motif found on a large number of cell-surface glycans, sometimes with large numbers of repeats 40. As has been suggested for other pore-forming toxins, binding to cell surface receptors helps concentrate the toxin on the membrane surface, thus facilitating oligomerization and pore formation 41. Whether the toxin needs to disengage from the glycan receptors to assemble into the final pore state is not known, but the flexibility of glycan chains may allow for continued binding through the assembly process. On the other hand, based on the VCC oligomer structure the final position of the VVH binding pocket is likely to sit approximately 80 Å above the cell surface and longer glycans or taller glycosylated proteins may be necessary to bridge the gap. The entire VCC pro-toxin is a monomer in solution {Olson:2005hl} and likely to bind to the membrane initially as a monomer. If we assume a similar monomeric state for the VVH secreted toxin, then the heptameric ring seen in the VVH crystal structure is unlikely to form until the toxin has oligomerized on the membrane and should be considered as part of a larger ring containing the cytolysin domains. This means that possible avidity effects are unlikely to be significant until oligomers have already formed on the cell surface.
In contrast to VCC, which targets a more limited array of branched N-glycans with nanomolar affinity 29, VVH exhibits a broader affinity for terminal galactosyl groups. These specificities overlap however, since complex N-glycans often contain LacNAc groups on their branched arms (Figure 2). Our data also indicate that the common 6C-position addition of neuraminic acid to terminal LacNAc groups, as well as additional sulfate or fucose modifications, are unlikely to prevent binding by the VVH β-trefoil lectin.
How does VVH relate to the larger family of homologous PFTs? A phylogenetic tree constructed from the alignment of β-trefoil domains (Figure 5B) from homologous toxins indicates that VVH falls on a separate branch that shares 30% sequence identity with the rest of the toxin family. Our glycan-binding data and sequence analysis suggests that the three genes falling on this branch may represent the only functional β-trefoil domains in the entire tree. If we assume that the first toxin to acquire a β-trefoil domain had carbohydrate-binding activity, then VVH may represent one of the few toxins that never lost this activity over time. If this is true, we suggest two possible evolutionary pathways that might have lead to the current distribution of lectin domains. Both pathways involve the acquisition of an active β-trefoil domain by some evolutionary predecessor. In one scenario, acquisition of the second lectin, the β-prism domain (which possesses nanomolar over micromolar affinity), rendered the β-trefoil domain less useful, resulting in its loss of activity. Because the β-trefoil domain had incorporated in to the assembly mechanism of the toxin, it was impossible to lose. Later, some bacteria lost their β-prism domain, resulting in a number of toxins with a single non-functional β-trefoil domain. In the second scenario, the β-trefoil carbohydrate-binding activity was lost first, leading to the non-functional domains seen in the current tree. Subsequent acquisition of the β-prism domain later restored glycan-binding activity to a subset of toxins, which would then contain both a non-functional β-trefoil domain as well as a functional β-prism domain.
The driving force for the acquisition or loss of these domains may have to do with maximizing the affinity of these toxins for target membranes or a specialization of targeting towards specific cell types to fill a particular environmental niche. As we have seen represented in the binding data, switching from an active β-trefoil domain to an active β-prism domain results in tighter binding, yet less broad recognition. The characterization of more toxins within this family will be necessary to determine the true extent of lectin activity across these related toxins, as well as in determining whether the gain or loss of glycan-binding activity was accompanied by a compensatory gain or loss of binding to a different type of receptor. The acquisition of other toxins or virulence factors may also have an effect on the relevance of these toxins in different disease pathologies. The bacteria represented in the tree range from demonstrated human pathogens like V. cholerae and V. vulnificus, to fish and invertebrate pathogens like A. salmonicida and V. corallilyticus. Further understanding the targets and targeting mechanisms of these toxins will be instrumental in understanding and combating these important bacterial pathogens.
Material and Methods
Protein Expression and Purification
The gene fragment encoding the VVH lectin domain (residues A338 to L471) was cloned from Vibrio vulnificus genomic DNA (strain CMCP6) by PCR amplification and inserted into the plasmid pNGFP-BC (courtesy of Dr. Eric Gouaux). The resulting construct creates a thrombin-cleavable N-terminal fusion with a His-tagged green fluorescent protein (GFPUV) mutant optimized for bacterial expression 42. For protein production, the resulting construct was transformed into SHuffle T7 Express Competent E. coli (New England Biolabs Inc.) grown overnight at 37 °C in Luria Broth (LB) with constant shaking. The next day, the overnight culture was diluted 30-fold into fresh LB and grown to an optical density at 600 nm (OD600) of approximately 0.6. Following induction with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG), the protein was expressed for eight hours at 30 °C. Cells were pelleted for 10 minutes at 17,500 × g and resuspended in 20 ml per 1 L of culture of TBS buffer (20 mM TRIS pH 7.5, 150 mM NaCl) containing 10 mM imidazole and protease inhibitor cocktail (Sigma). Cells were lysed by passing 3x through an EmusiFlex-C5 homogenizer at ~17,000 psi and cellular debris removed by spinning for 20 minutes in a Sorvall SS-34 rotor at 48,000 × g. The cleared supernatant was loaded onto a pre-equilibrated 5 ml Ni-NTA column (GE Healthcare, Piscataway, NJ), the column was washed with 10 volumes of TBS buffer + 40 mM imidazole, and the protein was eluted in 15 ml of TBS buffer + 250 mM imidazole. The fusion protein was cleaved by treating with a 1:1000 wt/wt ratio of human α-thrombin (Haematologic Technologies Inc., Essex Junction, VT) and separated from the free VVH lectin by passage over a Superose 200 16/60 gel filtration column (GE Healthcare).
Glycan Screening
Purified VVH lectin was labeled with fluorescent dye using the Molecular Probes Alexa Fluor 488 Labeling Kit (Life Technologies Corp., Carlsbad, CA) following the manufacturer’s instructions. After labeling, the protein was separated from unbound dye by running over a Superose 200 10/300 gel filtration column. Labeling efficiency was determined to be approximately 10% by absorption spectroscopy and by visualizing labeled protein run on an SDS-PAGE gel with a Typhoon FLA 9400 imager (GE Healthcare). Labeled protein was sent to the Consortium for Functional Glycomics (www.functionalglycomics.org), who screened the VVH lectin against Version 5.1 of their printed array containing 610 mammalian glycans. Data is reported in average relative fluorescence units (RFU) of four of six measurements after dropping the highest and lowest point. The full results obtained from the screen are available on the CFG website. Labeled VVH was screened at concentrations of 2, 10, 50, and 200 μg/ml. Data for the 50 μg/ml concentration exhibited lower overall standard deviations than the 200 μg/ml concentration and were used in the final analysis.
Isothermal Titration Calorimetry (ITC)
Purified VVH lectin-GFPUV fusion protein was exchanged into 20 mM TRIS pH 7.5, 150 mM NaCl using a Bio-Gel P-6 desalting column (Bio-Rad Laboratories, Inc., Hercules, CA) and concentrated to 130 μM using an Amicon Ultra-4 30,000 Da-cutoff centrifugal concentrator. ITC was performed using a using a MicroCal VP-ITC Isothermal Titration Calorimeter (GE Healthcare). To the sample cell, degassed protein (1.44 ml) was added and titrated with thirty 5-μl injections at 25 °C of the following carbohydrates: D-galactose (5 mM), N-acetyl-D-galactosamine (5 mM), D-lactose (5 mM), N-acetyl-D-lactosamine (5 mM), N-acetyl-D-neuraminic acid (5 mM), and D-galactose 6-sulfate (5 mM). As a control, buffer alone was titrated with each sugar to calculate the heat of dilution. ITC data were processed using the program NITPIC v. 1.0.3 43 and fit to a single-site binding model using the ITC module of SEDPHAT v. 10.54d 44.
Analytical Ultracentrifugation
Sedimentation velocity analysis was conducted at 20°C and 50,000 RPM using interference optics with a Beckman-Coulter XL-I analytical ultracentrifuge. Double sector synthetic boundary cells equipped with sapphire windows were used to match the sample and reference menisci. The rotor was equilibrated under vacuum at 20°C and after a period of ~1 hr, the rotor was accelerated to 50,000 RPM. Interference scans were acquired at 60-second intervals for 6 hours. Samples were prepared at 0.26, 0.634, 1.25 and 2.03 mg/ml in 20mM TRIS, 150 mM NaCl, pH 7.47. The data were analyzed using SEDFIT to produce ls-g(s*) distributions39. The data were also fit to a single discrete species model using SEDFIT. Sedimentation equilibrium analysis was performed at 20°C and 44,000 RPM using interference optics with a Beckman-Coulter XL-I analytical ultracentrifuge. Samples at 0.076, 0.157 and 0.313 mg/ml were loaded into external-loading, six-channel centerpieces and water blanks were subtracted. Data were fit to a monomer-heptamer equilibrium model using HeteroAnalysis45.
Crystallization and X-ray Data Collection
Purified VVH lectin in TBS buffer (10 mM TRIS pH 7.5, 100 mM NaCl, 1 mM EDTA) was concentrated to 5 mg/ml and subjected to crystallization screens using the hanging drop method. Crystals were obtained in a condition containing 100 mM TRIS pH 8.25 and 6% PEG 8000 in the presence of 2 mM N-acetyl-D-lactosamine, 2 mM N-acetyl-D-galactosamine, or without any added sugar. The crystal morphology and space group was dependent on which carbohydrate ligands were included in the crystallization drop. Crystals were soaked briefly in mother liquor containing 20% glycerol before being flash-frozen in liquid nitrogen. Initial X-ray diffraction data on apo and LacNAc crystals were collected using Oxford Xcalibur Nova X-ray Generator with an Onyx CCD detector at 100 K and processed using CryAlisPro software (Oxford Diffraction) and Scala46. Final high-resolution data for all three crystal forms were collected at beamline X25 of the National Synchrotron Light Source at Brookhaven National Laboratory and processed using HKL2000 47.
Structure Determination and Refinement
Self-rotation functions calculated by the program polarrfn 46indicated at least one 7-fold non-crystallographic symmetry (NCS) axis in all three crystal forms. A search model containing a heptameric arrangement of the Vibrio cholerae cytolysin β-trefoil lectin domain (residues 459-579, PDB code 3O44) was used as a molecular replacement search model in the orthorhombic crystal data using the program Phaser 48. Successive searches gave a unique solution with good packing also consistent with the self-rotation data. Maps calculated using phases from the molecular replacement solution were poor. To improve the map quality while removing possible model bias, we utilized an approach to extend low-resolution phases from the molecular replacement solution to higher resolution taking advantage of 7-fold symmetry constraints and solvent flattening. Low resolution phases to 5 Å were improved through 100 cycles of solvent flattening, histogram matching, and 7-fold NCS averaging using the program DM 46. Phases were extended by the same method from 5.0-2.8 Å in 1000 steps resulting in a final mean figure of merit of 0.779. The resulting maps were readily interpretable and automatic building of >90% of the structure accomplished by ARP/wARP 49 resulting in an initial R-work of 29.5%. Manual rebuilding and refinement was carried out iteratively using Coot 35 and phenix.refine 50. The resulting structure was used as a molecular replacement search model to phase all three high-resolution data sets, which were further refined to obtain the final statistics listed in Table 3.
Supplementary Material
Highlights.
V. vulnificus hemolysin has a single lectin domain targeting glycans on cells
Glycan-chip screening reveals a preference for galactosyl-terminating glycans
Isothermal titration calorimetry indicates ~150 μM affinity for sugars
The lectin forms a heptameric ring in the X-ray crystal structure
The lectin contains an extended loop missing in related toxins with inactive domains
Acknowledgements
Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award number R15 AI101977. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We thank Dr. Karla Satchell for providing us with the V. vulnificus genomic DNA. Glycan screening was conducted by the Protein-Glycan Interaction Resource of the Consortium for Functional Glycomics, which is supported by NIH grant R24 GM098791. Data for this study were measured at beamline X25 of the National Synchrotron Light Source. Financial support comes principally from the Offices of Biological and Environmental Research and of Basic Energy Sciences of the US Department of Energy, and from the National Center for Research Resources (P41RR012408) and the National Institute of General Medical Sciences (P41GM103473) of the National Institutes of Health. Thanks to Dr. Annie Heroux for assistance in X-ray data collection and processing and to Dr. Bing Hao for allowing us access to her X-ray home source.
Abbreviations
- CFG
Consortium for Functional Glycomics
- GalNAc
N-acetyl-D-galactosamine
- GFPUV
green-fluorescent protein (UV)
- GlcNAc
N-acetyl-D-glucosamine
- ITC
isothermal titration calorimetry
- LacNAc
N-acetyl-D-lactosamine
- PDB
protein data bank
- PFT
pore-forming toxin
- VCC
Vibrio cholerae cytolysin
- VVH
Vibrio vulnificus hemolysin
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers:
Coordinates and structure factors have been deposited into the PDB (www.rcsb.org) under the accession numbers 4OWJ, 4OWK, and 4OWL.
References
- 1.Jones MK, Oliver JD. Vibrio vulnificus: disease and pathogenesis. Infect. Immun. 2009;77:1723–1733. doi: 10.1128/IAI.01046-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oliver JD. Wound infections caused by Vibrio vulnificus and other marine bacteria. Epidemiol. Infect. 2005;133:383–391. doi: 10.1017/s0950268805003894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jeong H-G, Satchell KJF. Additive function of Vibrio vulnificus MARTX(Vv) and VvhA cytolysins promotes rapid growth and epithelial tissue necrosis during intestinal infection. PLoS Pathog. 2012;8:e1002581. doi: 10.1371/journal.ppat.1002581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Song L, Hobaugh MR, Shustak C, Cheley S, Bayley H, Gouaux JE. Structure of staphylococcal α-hemolysin, a heptameric transmembrane pore. Science. 1996;274:1859–1866. doi: 10.1126/science.274.5294.1859. [DOI] [PubMed] [Google Scholar]
- 5.Walker B, Braha O, Cheley S, Bayley H. An intermediate in the assembly of a pore-forming protein trapped with a genetically-engineered switch. Chem Biol. 1995;2:99–105. doi: 10.1016/1074-5521(95)90282-1. [DOI] [PubMed] [Google Scholar]
- 6.Perret M, Badiou C, Lina G, Burbaud S, Benito Y, Bes M, Cottin V, Couzon F, Juruj C, Dauwalder O, Goutagny N, Diep BA, Vandenesch F, Henry T. Cross-talk between Staphylococcus aureus leukocidins-intoxicated macrophages and lung epithelial cells triggers chemokine secretion in an inflammasome-dependent manner. Cell. Microbiol. 2012;14:1019–1036. doi: 10.1111/j.1462-5822.2012.01772.x. [DOI] [PubMed] [Google Scholar]
- 7.Wilke GA, Wardenburg JB. Role of a disintegrin and metalloprotease 10 in Staphylococcus aureus alpha-hemolysin-mediated cellular injury. Proc. Natl. Acad. Sci. U.S.A. 2010;107:13473–13478. doi: 10.1073/pnas.1001815107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alonzo F, Kozhaya L, Rawlings SA, Reyes-Robles T, DuMont AL, Myszka DG, Landau NR, Unutmaz D, Torres VJ. CCR5 is a receptor for Staphylococcus aureus leukotoxin ED. Nature. 2013;493:51–55. doi: 10.1038/nature11724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spaan AN, Henry T, van Rooijen WJM, Perret M, Badiou C, Aerts PC, Kemmink J, de Haas CJC, van Kessel KPM, Vandenesch F, Lina G, van Strijp JAG. The staphylococcal toxin panton-valentine leukocidin targets human c5a receptors. Cell Host Microbe. 2013;13:584–594. doi: 10.1016/j.chom.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 10.Olson R, Gouaux E. Vibrio cholerae cytolysin is composed of an α-hemolysin-like core. Protein Sci. 2003;12:379–383. doi: 10.1110/ps.0231703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olson R, Gouaux E. Crystal Structure of the Vibrio cholerae cytolysin (VCC) pro-toxin and its assembly into a heptameric transmembrane pore. J. Mol. Biol. 2005;350:997–1016. doi: 10.1016/j.jmb.2005.05.045. [DOI] [PubMed] [Google Scholar]
- 12.Yu H-N, Lee Y-R, Park K-H, Rah S-Y, Noh E-M, Song E-K, Han M-K, Kim B-S, Lee S-H, Kim J-S. Membrane cholesterol is required for activity of Vibrio vulnificus cytolysin. Arch. Microbiol. 2007;187:467–473. doi: 10.1007/s00203-007-0214-0. [DOI] [PubMed] [Google Scholar]
- 13.Hazes B. The (QxW)3 domain: a flexible lectin scaffold. Protein Sci. 1996;5:1490–1501. doi: 10.1002/pro.5560050805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rutenber E, Ready M, Robertus JD. Structure and evolution of ricin B chain. Nature. 1987;326:624–626. doi: 10.1038/326624a0. [DOI] [PubMed] [Google Scholar]
- 15.Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME. R-type Lectins. Cold Spring Harbor Laboratory Press; Cold Spring Harbor (NY): 2009. [PubMed] [Google Scholar]
- 16.Inoue K, Sobhany M, Transue TR, Oguma K, Pedersen LC, Negishi M. Structural analysis by X-ray crystallography and calorimetry of a haemagglutinin component (HA1) of the progenitor toxin from Clostridium botulinum. Microbiology (Reading, Engl.) 2003;149:3361–3370. doi: 10.1099/mic.0.26586-0. [DOI] [PubMed] [Google Scholar]
- 17.Mancheño JM, Tateno H, Goldstein IJ, Martínez-Ripoll M, Hermoso JA. Structural analysis of the Laetiporus sulphureus hemolytic pore-forming lectin in complex with sugars. J. Biol. Chem. 2005;280:17251–17259. doi: 10.1074/jbc.M413933200. [DOI] [PubMed] [Google Scholar]
- 18.Liu Y, Chirino AJ, Misulovin Z, Leteux C, Feizi T, Nussenzweig MC, Bjorkman PJ. Crystal structure of the cysteine-rich domain of mannose receptor complexed with a sulfated carbohydrate ligand. J. Exp. Med. 2000;191:1105–1116. doi: 10.1084/jem.191.7.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.East L, Isacke CM. The mannose receptor family. Biochim. Biophys. Acta. 2002;1572:364–386. doi: 10.1016/s0304-4165(02)00319-7. [DOI] [PubMed] [Google Scholar]
- 20.Hagen, Ten KG, Fritz TA, Tabak LA. All in the family: the UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferases. Glycobiology. 2003;13:1R–16R. doi: 10.1093/glycob/cwg007. [DOI] [PubMed] [Google Scholar]
- 21.Notenboom V, Boraston AB, Williams SJ, Kilburn DG, Rose DR. High-resolution crystal structures of the lectin-like xylan binding domain from Streptomyces lividans xylanase 10A with bound substrates reveal a novel mode of xylan binding. Biochemistry. 2002;41:4246–4254. doi: 10.1021/bi015865j. [DOI] [PubMed] [Google Scholar]
- 22.Nicolson GL, Blaustein J. The interaction of Ricinus communis agglutinin with normal and tumor cell surfaces. Biochim. Biophys. Acta. 1972;266:543–547. doi: 10.1016/0005-2736(72)90109-5. [DOI] [PubMed] [Google Scholar]
- 23.Fiete DJ, Beranek MC, Baenziger JU. A cysteine-rich domain of the “mannose” receptor mediates GalNAc-4-SO4 binding. Proc. Natl. Acad. Sci. U.S.A. 1998;95:2089–2093. doi: 10.1073/pnas.95.5.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sugiyama H, Kashimoto T, Ueno S, Susa N. Inhibition of binding of Vibrio vulnificus hemolysin (VVH) by MβCD. J. Vet. Med. Sci. 2013;75:649–652. doi: 10.1292/jvms.12-0387. [DOI] [PubMed] [Google Scholar]
- 25.Kashimoto T, Ueno S, Koga T, Fukudome S, Ehara H, Komai M, Sugiyama H, Susa N. The aromatic ring of phenylalanine 334 is essential for oligomerization of Vibrio vulnificus hemolysin. J Bacteriol. 2010;192:568–574. doi: 10.1128/JB.01049-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miyoshi S-I, Abe Y, Senoh M, Mizuno T, Maehara Y, Nakao H. Inactivation of Vibrio vulnificus hemolysin through mutation of the N- or C-terminus of the lectin-like domain. Toxicon. 2011;57:904–908. doi: 10.1016/j.toxicon.2011.03.013. [DOI] [PubMed] [Google Scholar]
- 27.Sankaranarayanan R, Sekar K, Banerjee R, Sharma V, Surolia A, Vijayan M. A novel mode of carbohydrate recognition in jacalin, a Moraceae plant lectin with a beta-prism fold. Nat. Struct. Biol. 1996;3:596–603. doi: 10.1038/nsb0796-596. [DOI] [PubMed] [Google Scholar]
- 28.Jeyaprakash AA, Srivastav A, Surolia A, Vijayan M. Structural basis for the carbohydrate specificities of artocarpin: variation in the length of a loop as a strategy for generating ligand specificity. J. Mol. Biol. 2004;338:757–770. doi: 10.1016/j.jmb.2004.03.040. [DOI] [PubMed] [Google Scholar]
- 29.Levan S, De S, Olson R. Vibrio cholerae Cytolysin Recognizes the Heptasaccharide Core of Complex N-Glycans with Nanomolar Affinity. J. Mol. Biol. 2013;425:944–957. doi: 10.1016/j.jmb.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rillahan CD, Paulson JC. Glycan Microarrays for Decoding the Glycome. Annu. Rev. Biochem. 2011;80:797–823. doi: 10.1146/annurev-biochem-061809-152236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Drickamer K, Taylor ME. Glycan arrays for functional glycomics. Genome Biol. 2002;3:REVIEWS1034. doi: 10.1186/gb-2002-3-12-reviews1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Honke K, Taniguchi N. Sulfotransferases and sulfated oligosaccharides. Med Res Rev. 2002;22:637–654. doi: 10.1002/med.10020. [DOI] [PubMed] [Google Scholar]
- 33.De S, Olson R. Crystal structure of the Vibrio cholerae cytolysin heptamer reveals common features among disparate pore-forming toxins. Proc. Natl. Acad. Sci. U.S.A. 2011;108:7385–7390. doi: 10.1073/pnas.1017442108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 35.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 36.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 37.Holm L, Rosenström P. Dali server: conservation mapping in 3D. Nucleic Acids Research. 2010;38:W545–9. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schuck P, Rossmanith P. Determination of the sedimentation coefficient distribution by least-squares boundary modeling. Biopolymers. 2000;54:328–341. doi: 10.1002/1097-0282(20001015)54:5<328::AID-BIP40>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 40.Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME, Sasisekharan R. Glycomics. Cold Spring Harbor Laboratory Press; Cold Spring Harbor (NY): 2009. [PubMed] [Google Scholar]
- 41.Abrami L, van der Goot FG. Plasma membrane microdomains act as concentration platforms to facilitate intoxication by aerolysin. J Cell Biol. 1999;147:175–184. doi: 10.1083/jcb.147.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kawate T, Gouaux E. Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure. 2006;14:673–681. doi: 10.1016/j.str.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 43.Keller S, Vargas C, Zhao H, Piszczek G, Brautigam CA, Schuck P. High-precision isothermal titration calorimetry with automated peak-shape analysis. Anal. Chem. 2012;84:5066–5073. doi: 10.1021/ac3007522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Houtman JCD, Brown PH, Bowden B, Yamaguchi H, Appella E, Samelson LE, Schuck P. Studying multisite binary and ternary protein interactions by global analysis of isothermal titration calorimetry data in SEDPHAT: application to adaptor protein complexes in cell signaling. Protein Sci. 2007;16:30–42. doi: 10.1110/ps.062558507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cole JL. Analysis of heterogeneous interactions. Meth. Enzymol. 2004;384:212–232. doi: 10.1016/S0076-6879(04)84013-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Collaborative Computational Project, Number 4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 47.Otwinowski Z, Minor W. Processing of X-ray diffraction data. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 48.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat Protoc. 2008;3:1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Laskowski RA, Swindells MB. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J Chem Inf Model. 2011;51:2778–2786. doi: 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
- 52.Edgar R. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gouet P, Robert X, Courcelle E. ESPript/ENDscript: Extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Research. 2003;31:3320–3323. doi: 10.1093/nar/gkg556. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.