Mammalian germ cells must reprogram their epigenomes to establish germ cell potency. However, little is known about how DNA methylation patterns are erased and reset during development. The piRNA pathway protects germ cell genomes against mobile genetic elements. Molaro et al. show that DNA methylation can be separated into two waves: one occurs by default, and the other is dependent on the piRNA pathway. Two mechanistically and temporally distinct waves thus play essential roles in resetting germ cell methylation marks at each generation.
Keywords: DNA methylation, piRNA, retrotransposon, germ cells, LINE, LTR
Abstract
During development, mammalian germ cells reprogram their epigenomes via a genome-wide erasure and de novo rewriting of DNA methylation marks. We know little of how methylation patterns are specifically determined. The piRNA pathway is thought to target the bulk of retrotransposon methylation. Here we show that most retrotransposon sequences are modified by default de novo methylation. However, potentially active retrotransposon copies evade this initial wave, likely mimicking features of protein-coding genes. These elements remain transcriptionally active and become targets of piRNA-mediated methylation. Thus, we posit that these two waves play essential roles in resetting germ cell epigenomes at each generation.
The bulk of mammalian genomes is composed of a diversity of mobile genetic elements (Levin and Moran 2011) whose potentially deleterious transposition is controlled in part by DNA methylation (Walsh et al. 1998; Bourc’his and Bestor 2004). Mammalian germ cells are derived from somatic precursors (Ginsburg et al. 1990), and genome-wide epigenetic patterns must be erased and reset to establish germ cell potency (Surani et al. 2007). This occurs, at least in part, via a nearly complete erasure of DNA methylation in primordial germ cells (PGCs) (Monk et al. 1987; Reik et al. 2001; Hajkova et al. 2002; Lees-Murdock et al. 2003; Guibert et al. 2012). This has the potential to expose germ cells to a burst of transposon activity.
Although the mechanisms leading to erasure of DNA methylation have come under scrutiny (Hackett et al. 2012), little is known about the targeting of de novo methylation in germ cells. The piRNA pathway is a conserved, small RNA-based immune system that protects germ cell genomes against transposons. In mammals, the piRNA pathway has been implicated in directing transposon methylation in PGCs during germ cell development. PGCs express two PIWI proteins: MILI (or PIWIL2), which initiates expression at approximately embryonic day 12.5 (E12.5), and MIWI2 (or PIWIL4), which is evident from approximately E14.5 when male PGCs cell cycle-arrest until they re-enter cycle at approximately postnatal day 3 (P3). Whereas MILI is exclusively cytoplasmic, MIWI2 localizes to PGC nuclei in a MILI-dependent fashion at approximately E15.5 (Kuramochi-Miyagawa et al. 2001, 2004; Aravin et al. 2007, 2008; Carmell et al. 2007; Aravin and Bourc’his 2008).
piRNAs exhibit substantial diversity, comprising the full spectrum of transposon sequences that make up roughly half of the mouse genome. Thus, piRNAs could be responsible for directing the deposition of DNA methylation over a large portion of the mouse genome. We previously observed that thousands of repeat copies evade de novo methylation in primate sperm despite highly similar copies being successfully targeted (Molaro et al. 2011). Thus, targeting for de novo methylation may be a more complex process, which takes into account more than simply the expression of a similar piRNA. We therefore took an unbiased approach toward understanding the role of piRNAs in shaping the epigenetic landscape of germ cell genomes.
Results and Discussion
Features of Mili mutant spermatocyte methylomes
To investigate the impact of piRNA-directed DNA methylation on germ cell reprograming, we generated genome-wide maps of DNA methylation in male E13.5 PGCs and of fully reprogrammed wild-type and Mili−/− spermatocytes. We chose these cell types because they represent two possible endpoints of the reprogramming process, with a nadir of methylation in E13.5 PGCs having risen to roughly somatic levels by the spermatocyte stage. Mili−/− spermatocyte methylomes were produced from two independent replicates and sequenced to average levels of 5× and 10×, respectively; these were well correlated but were combined to increase confidence in methylation calls (Supplemental Table S1). Wild-type methylomes were independently validated by comparison with publicly available data sets (Supplemental Table S1; Kobayashi et al. 2012; Seisenberger et al. 2012).
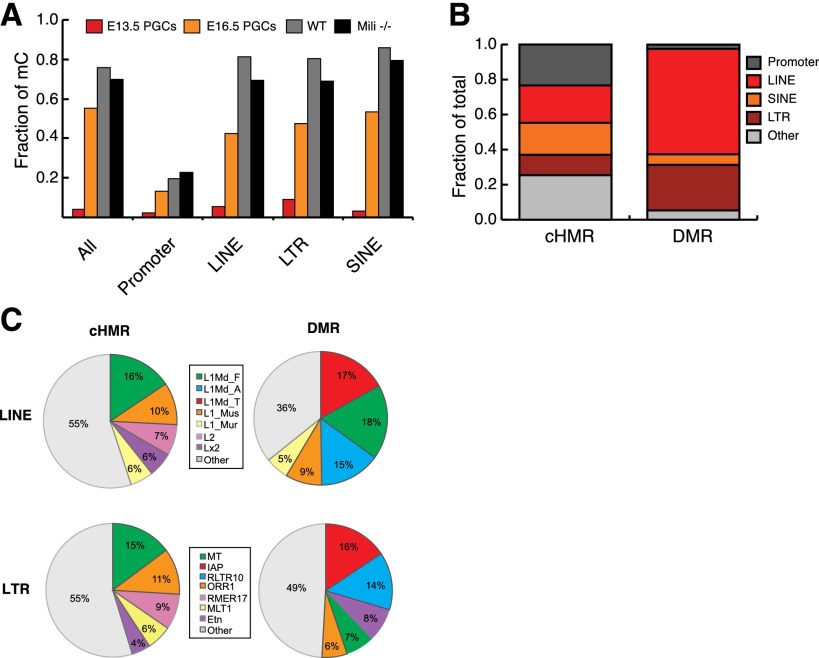
As expected, E13.5 PGCs had few methylated CpG sites, whereas spermatocyte genomes were highly methylated. The nearly complete loss of CpG methylation at E13.5 was apparent across all genomic annotations (Fig. 1A). Nevertheless, PGCs displayed a substantial fraction of retrotransposons that resist reprogramming (Lane et al. 2003; Seisenberger et al. 2012). Spermatocytes showed methylation levels similar to those seen in somatic cell types (Kaaij et al. 2013). Contrary to our expectations, wild-type and Mili−/− spermatocytes displayed very similar overall methylation levels (76% and 70%, respectively). This was also true if a focus was placed on retrotransposon sequences (Fig. 1A), suggesting that most repeat methylation is re-established through a nonselective wave of “default” de novo methylation rather than being piRNA-dependent.
Figure 1.
Impact of RNA-directed DNA methylation during germ cell development. (A) Fraction of methylated CpG dinucleotides in wild-type (WT; gray) or Mili−/− (black) spermatocytes and male E13.5 PGCs (red). E16.5 PGCs (orange) from Seisenberger et al. (2012) are also shown for comparison. (B) Proportion of cHMRs or DMRs annotated as promoter, LINE, SINE, LTR, or other. (C) Relative abundance of LINE and LTR subfamilies among cHMRs and DMRs (only the top five are colored for display).
piRNA-dependent and piRNA-independent de novo methylation
To characterize the impact of the piRNA pathway on the epigenome, we identified contiguous domains of low methylation, termed hypomethylated regions (HMRs) (Hodges et al. 2011; Molaro et al. 2011), in wild-type and mutant animals. Overall, most HMRs were found at the same genomic location in both genotypes (defined as constitutive HMRs [cHMRs], ∼60,000 regions); however, clear differences were seen over repeat loci, consistent with the proposed role of piRNAs in targeting transposons for methylation (Supplemental Table S2). HMRs in Mili−/− spermatocytes that were fully methylated in wild-type animals we refer to as differentially methylated regions or mutant DMRs (for differentially methylated regions). We found a total of ∼17,800 high-confidence mutant DMRs. These tended to overlap retrotransposons (∼90%) and were greatly enriched for the LINE (long interspersed nuclear element) and LTR (long terminal repeat) classes (Fig. 1B). These regions overlap 34,191 individual LINE and LTR copies (∼2.5% of all genomic insertions) (Fig. 1C; Supplemental Table S3); we refer to these as piRNA-dependent transposons, with the implication that they require a functional piRNA pathway to acquire de novo methylation. The distribution of piRNA-dependent sites is different from what is observed for cHMRs, where repeats occupy only ∼45% of all regions, essentially as expected by chance (Fig. 1B). These covered a total of 57,168 LINE and LTR copies (∼3% of all genomic insertions). We refer to these as constitutively hypomethylated transposons (Fig. 1B,C; Supplemental Table S3). The vast majority of LINE and LTR elements (∼1.4 million copies; ∼95% of all genomic insertions) do not belong to either group and are considered to be de novo methylated independently of the piRNA pathway or by “default” (for an illustration of these groups of retrotransposons, see Supplemental Fig. S1). It should be noted that these include both insertions that are nearly full length, although they might be diverged from the consensus, and fragmentary insertions. Thus, it might be useful to consider that these sites are not all “elements,” per se, but instead represent simply genomic transposon content.
A comparison of the transposons corresponding to DMRs and cHMRs revealed that copies belonging to active subfamilies of LINE and LTR elements comprised a large proportion of DMRs (Fig. 1C; Goodier and Kazazian 2008; Sookdeo et al. 2013). For example, copies of young L1 subfamilies L1Md_T/A make up to 32% of all LINE-associated DMRs, whereas cHMRs proportionately overlap more copies belonging to older LINE subfamilies (i.e., Lx2 and L2) (Fig. 1C). Interestingly, the “middle-aged” subfamilies (for example, L1Md_F) showed an equal representation in DMRs and cHMRs. This was also true if the enrichment of transposon subfamilies was measured against all HMRs in wild-type or Mili−/− spermatocyte methylomes (Supplemental Table S4). For example, more copies of IAPs and other active mouse endogenous retroviruses (ERVs) appear hypomethylated in mutant germ cells when compared with other less active subfamilies (Supplemental Table S4). This implies that distinct mechanisms direct methylation over different classes of elements during germ cell development and that piRNA-directed de novo methylation is biased toward retrotransposon subfamilies that have been recently expanding in the mouse genome.
Two distinct waves of de novo methylation operate during germ cell development
An examination of methylation levels over repeat subfamilies in wild-type PGCs at the intermediate time point E16.5 (Seisenberger et al. 2012) supported the hypothesis that de novo methylation operates in successive “waves” (Fig. 1A; Supplemental Table S5). At E16.5, de novo methylation over most sites had already occurred (Walsh et al. 1998; Lees-Murdock et al. 2003; Kato et al. 2007; Aravin et al. 2008; Kuramochi-Miyagawa et al. 2008). The bulk of L1 and ERVs displayed intermediate to high average methylation, similar to levels observed genome-wide (Fig. 1A; Supplemental Table S5). However, subfamilies of repeats that showed dependence on the piRNA pathway, those that were enriched in mutant DMRs in spermatocytes, appeared to resist this first wave and showed methylation levels closer to those of gene promoters. Roughly 70% of such regions showed an average methylation <0.4 (or 40%) (Supplemental Table S5). For example, a close examination of the promoters of L1 and ETn copies that were associated with mutant DMRs verified that these remain unmethylated at E16.5 and have evaded the initial default wave (Supplemental Fig. S3)
This apparent delay in de novo methylation of piRNA-dependent elements mirrors another delay that occurs during the erasure phase of reprogramming. Many mutant-specific DMRs show incomplete demethylation at E11.5 (Seisenberger et al. 2012). Roughly 50% retain average methylation levels of >0.3, as compared with ∼20% genome-wide and <10% for other HMRs (Supplemental Table S5). However, these regions achieve complete erasure by E13.5. Thus, on average, piRNA-targeted retrotransposon copies are more subject to the active phase of erasure, remaining methylated for a longer period of time in developing PGCs (Hajkova et al. 2002; Guibert et al. 2012; Hackett et al. 2012).
piRNAs selectively impact DNA methylation over regulatory regions
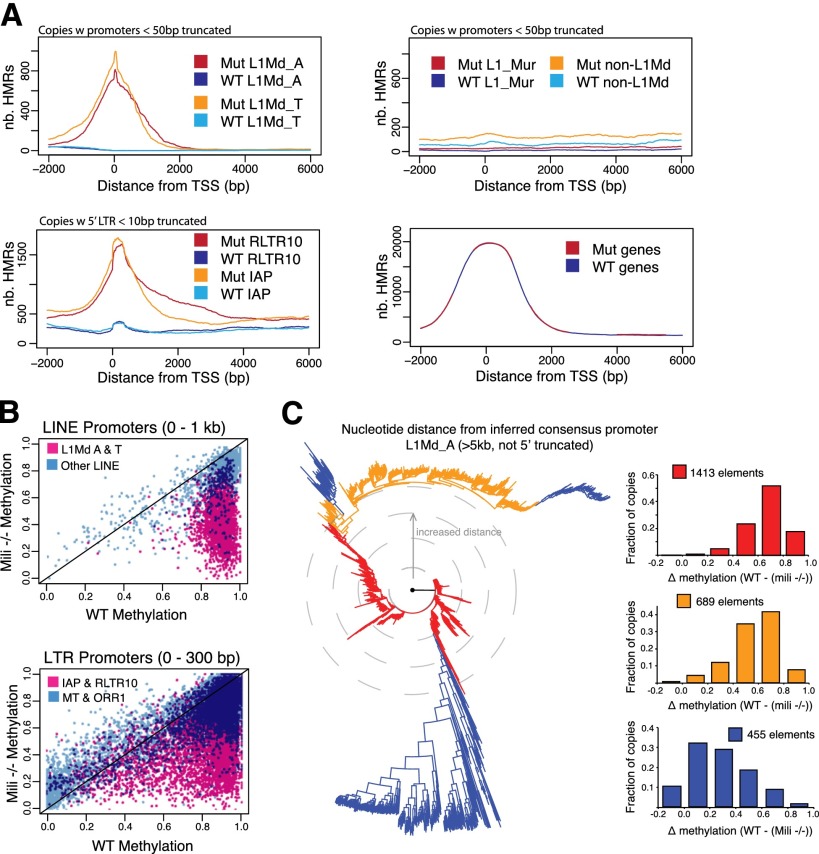
Successful propagation of retrotransposons relies on enhancers and other regulatory elements driving transcription of genomic RNAs in germ cells. Related retrotransposon subfamilies often vary in their 5′ regulatory regions, reflecting their exploitation of a diversity of host–factor interactions (Levin and Moran 2011; Feschotte and Gilbert 2012). piRNAs map over both the regulatory regions and the bodies of retrotransposons, suggesting that methylation across the entire element might be affected in Mili mutants (Aravin et al. 2008; Kuramochi-Miyagawa et al. 2008). An examination of the location of HMRs within mobile elements indicated that their promoter regions were most prone to change upon loss of the piRNA pathway (Fig. 2A). Notably, constitutively hypomethylated transposon insertions and gene promoters were also preferentially hypomethylated over their transcriptional start sites (TSSs), suggesting some commonality in the mechanisms used to evade default methylation (Fig. 2A; Supplemental Fig. S1). Previous studies have shown that hypomethylated domains are strongly enriched for transcription factor-binding sites and that transcription factor binding can be sufficient to prevent de novo DNA methylation (Hodges et al. 2011; Lienert et al. 2011; Molaro et al. 2011). This strongly suggests that the regulatory sequences of piRNA-targeted transposons present features resembling genomic regions that naturally evade the nonselective default wave of de novo methylation in germ cells, such as the promoters of protein-coding genes.
Figure 2.
Regulatory features associated with piRNA dependence. (A) Distribution of HMRs along copies of LINE (top) and LTR (bottom left) subfamilies. Wild-type (WT) and mutant (Mut) traces are shown in distinct colors. (Bottom right) A similar analysis along 1000 randomly selected genes is also shown. (B) Average promoter methylation in Mili−/− (Y-axis) and wild-type (X-axis) spermatocytes. Each dot represents a single transposon insertion. (C, left panel) Neighbor-joining tree of all L1Md_A promoters aligned using ClustalW2. Increased distance from the root signifies increased variation from consensus. For each colored group, the right panels display the frequency of copies with distinct methylation differences between wild-type and Mili−/− spermatocytes.
piRNA dependence is a property of individual transposon insertions
Within each subfamily, individual retrotransposon copies did not show uniform average methylation levels between the wild type and mutant (Fig. 2B). The promoters of some insertions were strong DMRs, whereas others displayed intermediate to low degrees of differential methylation. This prompted us to investigate the relationship between promoter sequence divergence from the consensus and dependency on the piRNA pathway. Examining individual full-length (>5 kb, not 5′ truncated) L1Md_A genomic insertions, we found that promoters displaying the least divergence from the inferred consensus showed the greatest degree of differential methylation (Fig. 2C). Groups of insertions that have drifted to the greatest extent from the consensus promoter showed the opposite trend. Overall, differentially methylated L1 promoters also tended to preserve higher CpG densities, perhaps indicating protection from long-term deamination (Supplemental Fig. S2A).
Because of their transposition mechanism, LINEs tend to be truncated upon insertion into a new genomic location. We examined the degree to which piRNA-dependent retrotransposon copies were truncated or intact. This revealed that piRNA dependency of both L1 and LTR elements correlated with elements being full length (Supplemental Fig. S2B). Interestingly, IAP elements were an exception to this rule, as constitutively hypomethylated copies appeared longer, on average, than their piRNA-dependent counterparts. Considered together, these results suggest that the piRNA pathway is responsible for controlling mainly those copies with features of active elements, including active promoters and intact, full-length sequences.
Transcription of piRNA-dependent retrotransposon copies during development
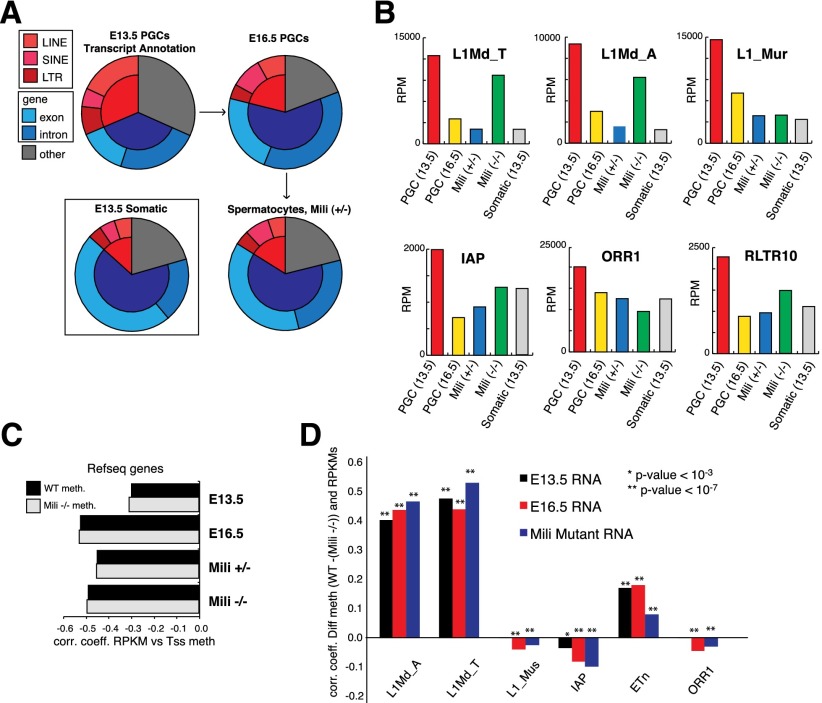
The youngest mobile elements are generally those closest to the consensus, and our data indicated that these behaved similarly to genes in the resistance of their regulatory regions to default methylation. We therefore examined the transcription profiles of transposon families before, during, and after de novo methylation has occurred (Fig. 3A). E13.5 PGCs showed substantial retrotransposon expression (Fig. 3A). This seemed to result from pervasive expression of a broad range of retrotransposon subfamilies, as previously noted in PGCs (Singh et al. 2013). The fraction of retrotransposon-derived reads rapidly decreased as PGCs initiated the first wave of de novo methylation, dropping from ∼30% of all mapped reads in E13.5 PGCs to ∼20% in E16.5 PGCs and to <15% in spermatocytes. In contrast, cosorted neighboring somatic cells never showed >10% of retrotransposon reads, suggesting that the transient burst in element transcription is PGC-specific (Fig. 3A; Supplemental Table S6). We validated this observation by quantitative RT–PCR (qRT–PCR) using an independently purified cohort of E13.5 cells, comparing results from various L1 primer sets with results from PGCs and somatic cells (Supplemental Fig. S4A).
Figure 3.
Transient reactivation of retrotransposon transcription in developing germ cells. (A) Annotation of mapped reads from RNA-seq libraries. (B) Read abundance, by element type (in reads per million mapped reads [RPM]), in E13.5 PGCs (red), E16.5 PGCs (yellow), Mili+/− spermatocytes (blue), Mili−/− spermatocytes (green), and E13.5 somatic tissues (gray). (C) Spearman correlation coefficients between TSS methylation in wild-type (WT) or Mili−/− spermatocytes and RNA expression (in reads per kilobase per million mapped reads [RPKM]) for all RefSeq genes. (D) Spearman correlation coefficient between differential methylation (wild type − [Mili−/−]) and RPKM values for all individual copies within LINE and LTR subfamilies.
Although all of our results are compatible with substantial levels of transposon expression occurring following genome-wide erasure, it is formally possible that transposon expression does not substantially change, while overall expression of protein-coding genes decreases. Some prior studies failed to report increases in transposon expression similar to those that we observed here (Seisenberger et al. 2012). All such studies measured only mature, polyA-selected transcripts. In our case, the use of random primers might capture a greater diversity of RNA species.
Whereas E13.5 PGCs displayed a bias toward LINE and LTR annotated reads as compared with SINEs (short interspersed nuclear elements) (∼25% vs. ∼5%), the latter constituted the highest fraction from E16.5 onward (∼10%) (Fig. 3A). When we investigated the distribution of RNA sequencing (RNA-seq) reads over LINE and LTR subfamilies (in reads per million [RPM]) (see the Supplemental Material), all showed high RPM values in E13.5 PGCs, and these were reduced to levels seen in somatic cells by the spermatocyte stage. Our data suggest that the high fraction of transposon-derived reads at E13.5 results from the combined expression of many subfamilies within the LINE and LTR class (Fig. 3B; Supplemental Table S7). However, only a few subfamilies displayed increased RPMs in Mili−/−− spermatocytes as compared with heterozygous siblings. This is consistent with piRNAs largely affecting the youngest and most active transposon families by transcriptional silencing (i.e., L1Md_T compared with L1_mur) (Fig. 3B).
Individual genes with HMRs over their promoter regions or covering upstream regulatory regions showed stronger expression levels than genes without associated HMRs (Fig. 3B). At every stage of germ cell maturation, RNA abundance (in reads per kilobase per million [RPKM]) (see the Supplemental Material) showed a strong negative correlation with average methylation at gene TSSs. The correlation was lowest at E13.5 and reaches high levels as early as E16.5 (0.31–0.53) (Fig. 3C), consistent with transcriptional programs moving from generally permissive to germ cell maturation-restricted (Molaro et al. 2011; Seisenberger et al. 2012).
Individual piRNA-dependent retrotransposon copies are expressed in developing PGCs
To determine which transposon copies, according to subfamily, contribute most to RNA levels, we ranked the expression of individual retrotransposons copies (in RPKM) (see the Supplemental Material) and correlated these with differential promoter methylation in Mili−/− spermatocytes (Fig. 3D). Some subfamilies showed a strong positive correlation between RNA levels and the presence of Mili-dependent DMRs, while for other families, the correlation was lower or even negative (Fig. 3D). piRNA-dependent L1 copies showed a strong and specific correlation between methylation and read abundance at E16.5 and in mutant spermatocytes (i.e., L1Md_T) (also see Supplemental Table S8) despite an overall reduction in LINE transcription at this stage (Fig. 3A).
Our results are consistent with a picture in which LINE expression at E13.5 seems to originate from many individual copies belonging to diverse subfamilies, whereas E16.5 LINE expression is probably dominated by the transcription of fewer elements within specific subfamilies whose silencing is piRNA-dependent. Such elements may show relatively greater expression at E16.5 than they do at E13.5, perhaps in part because pervasive element expression has been suppressed by default methylation (Supplemental Fig. S4B).
LTR transcript levels in wild-type or mutant spermatocytes did not vary as dramatically as did L1, instead remaining at relatively constant levels across development (Fig. 3B; Supplemental Fig. S4B). This may be attributable to a high constitutive rate of LTR transcription, which would mask any changes in the expression of individual copies. IAP elements, despite contributing substantially to piRNAs, displayed weaker correlations with differential promoter methylation than L1 copies, indicating that slightly different mechanisms might ultimately lead to L1 and IAP silencing (De Fazio et al. 2011). It is also formally possible that similarity among IAP copies negates our ability to detect copy-specific changes using current sequencing data sets.
piRNA biogenesis begins in E13.5 PGCs
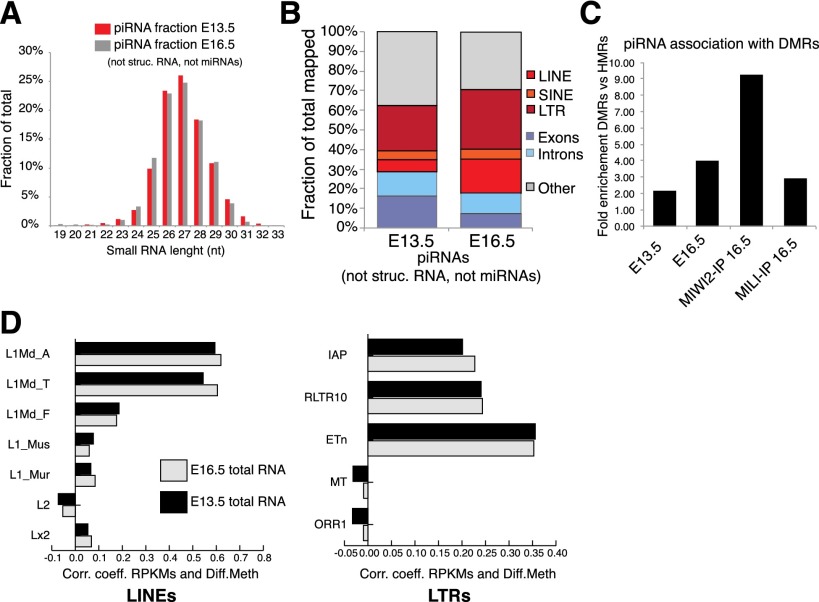
To investigate the nature of the piRNAs targeting differentially methylated transposons, we sequenced 24- to 33-nucleotide (nt) small RNAs from male genital ridges at E13.5 and compared these with libraries representing either total RNA or MILI or MIWI2 immunoprecipitates from E16.5 genital ridges (Aravin et al. 2008). E13.5 genital ridges contained an abundant fraction of reads resembling piRNAs, consistent with MILI being expressed at this stage (Supplemental Table S9; Aravin et al. 2008). These piRNAs displayed a strong 5′ U bias (80% of all reads) (Supplemental Table S10) and a size range typical of this small RNA class (24–30 nt) (Fig. 4A). These piRNAs are likely to represent the most primary population produced in PGCs. E13.5 piRNAs were less strongly enriched for transposon-derived reads than those from E16.5 (∼30% vs. ∼50%) (Fig. 4B). This contrasts with relative RNA abundance at E16.5 (Fig. 3A), indicating that LINE and LTR piRNAs may be secondarily amplified, most probably via ping-pong (Aravin et al. 2008; Kuramochi-Miyagawa et al. 2008). Thus, it remains possible that in addition to their directing perdurant transcriptional gene silencing via DNA methylation, piRNAs could also impact transposon activity via PTGS, protecting germ cells against transposition even during intervals when methylation levels are low.
Figure 4.
Secondary amplification of retrotransposon piRNAs is correlated with MILI-dependent DNA methylation. (A) Length distribution of non-microRNA and nonstructural RNA reads in total small RNA libraries cloned from E13.5 genital ridges (red). E16.5 small RNAs (Aravin et al. 2008) are shown for comparison. (B) Comparison of the piRNA fraction annotation in total small RNA libraries at E13.5 and E16.5. (C) Fold enrichment of piRNA over DMRs as compared with cHMRs in E13.5 and E16.3 total RNA libraries and in MIWI2 and MILI immunoprecipitated RNAs (IP) (Aravin et al. 2008). (D) Correlation coefficient between TSS differential methylation (see Fig. 3C) and piRNA abundance for all individual copies of distinct LINE and LTR subfamilies.
A distinct piRNA population targets retrotransposon copies for de novo methylation
piRNAs had a strong tendency to be enriched over DMRs as compared with cHMRs (Fig. 4C). This enrichment rose between E13.5 and E16.5 and peaked for MIWI2-associated piRNAs, in which piRNAs mapping to DMRs were enriched ∼10-fold (Fig. 4C). These enrichments were also very strong when comparing DMRs to default methylated regions (Supplemental Table S11). These observations suggest that a piRNA population at E16.5 and in MIWI2 complexes drives piRNA-dependent de novo methylation but that constitutively hypomethylated transposons somehow evade targeting by both this and the default de novo methylation pathways via unknown mechanisms. Finally, when focusing on individual repeat copies, a strong correlation was detected between differential TSS methylation and piRNA abundance (Fig. 4D; Supplemental Table S12). Similar to what we reported with transcript quantification (Fig. 3D), retrotransposon copies showing the most differential methylation in Mili mutants contributed the most to piRNA populations (i.e., L1Md_T vs. L1_mus).
Concluding remarks
Considered as a whole, the data presented here suggest that de novo methylation occurs in two hierarchically distinct waves, leading to the unique epigenetic signature of germ cells. Reminiscent of what has been observed in plants (Slotkin et al. 2009; Calarco et al. 2012), upon epigenetic reprogramming of PGCs, active repeats are transiently reactivated and converted into a primary pool of piRNAs. When a first wave of nonselective default de novo methylation is engaged past E13.5, the vast majority of the genome progressively regains methylation, and the relative abundance of repeats is reduced. Nevertheless, a fraction of retrotransposon copies evades this first wave and, by mirroring the behavior of protein-coding genes, remains transcriptionally active. These transcripts are available to engage in secondary piRNA amplification. This adaptively programs MIWI2 complexes and ultimately contributes specificity to the active secondary wave of de novo methylation (Supplemental Fig. S5). This is clearly dependent on the piRNA pathway, indicating that the marks that ultimately direct this methylation must be set during the period from E16.5 to about P3, when MIWI2 vanishes from germ cell nuclei. The nature of this primary signal and the precise mechanism and timing of methylation of piRNA-dependent sites have yet to be determined.
Materials and methods
Mouse strains
All strains used in this study were maintained on a C57BL/6 background. For determination of wild-type spermatocyte methylomes, mice were purchased from Charles River Laboratories. The Mili knockout strain was obtained from Haifan Lin (Yale University) (described in Kuramochi-Miyagawa et al. 2004). For PGC isolation, Oct4-EGFP mice described in Lengner et al. (2007) were purchased from the Jackson Laboratory.
Cell sorting
Spermatocytes were FACS-sorted (Aria II, BD Bioscience) from wild-type and Mili mutant animals based on DNA content using Hoechst staining (described in Bastos et al. 2005) and positive staining for the Alexa 647-conjugated Ep-Cam antibody (CD326, clone G8.8 from Biolegend). E13.5 and E16.5 PGCs were sorted using Oct4-EGFP-positive cells. GFP-negative cells (“somatic”) were also collected.
Shotgun bisulfite library preparation and sequencing
Shotgun bisulfite sequencing was performed as described in Molaro et al. (2011). Briefly, purified genomic DNA was sheared to an average size of 200–300 bp, end-repaired, and A-tailed. Illumina paired-end adaptors were ligated for 30 min at 25°C. The ligated products were bisulfite-converted and amplified (15 cycles) by PCR. Amplicons were paired-end-sequenced on the Illumina GAII or HiSeq platform (76PE and 100PE).
Small RNA cloning
Small RNA cloning from total RNA was performed as described in Aravin et al. (2008).
RNA-seq
RNA from sorted PGCs and adult spermatocytes was extracted using Trizol (Invitrogen). Following DNase treatment, each sample was subjected to reverse transcription and linear amplification using the Ovation RNA-seq system according to the manufacturer’s protocol (Nugen). Both oligo-dT and random priming were used during this procedure. Finally, double-stranded cDNAs were subjected to a standard Illumina paired-end genomic library preparation and sequenced to a size of 76 base pairs.
Also see the Supplemental Material
Acknowledgments
We acknowledge members of the Hannon and Smith laboratories for advice on experimental design, and especially Assaf Gordon, the Cold Spring Harbor Laboratory Bioinformatics Shared Resource, Emily Lee, and Maria Mosquera for technical help. We acknowledge the Cold Spring Harbor Laboratory animal facility for help with mouse handling. This research was supported through a National Institutes of Health (NIH) stimulus grant (no. 5RC2HD064459-01) and R37GM062534 to G.J.H., and NIH R01 grant HG006015 to A.D.S. Raw sequencing data were deposited for download on Sequence Read Archive under accession numbers SRP037785, SRP037807, and SRP037987.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.244350.114.
References
- Aravin AA, Bourc’his D 2008. Small RNA guides for de novo DNA methylation in mammalian germ cells. Genes Dev 22: 970–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ 2007. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science 316: 744–747 [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Bourc’his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ 2008. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell 31: 785–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastos H, Lassalle B, Chicheportiche A, Riou L, Testart J, Allemand I, Fouchet P 2005. Flow cytometric characterization of viable meiotic and postmeiotic cells by Hoechst 33342 in mouse spermatogenesis. Cytometry A 65: 40–49 [DOI] [PubMed] [Google Scholar]
- Bourc’his D, Bestor TH 2004. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature 431: 96–99 [DOI] [PubMed] [Google Scholar]
- Calarco JP, Borges F, Donoghue MT, Van Ex F, Jullien PE, Lopes T, Gardner R, Berger F, Feijo JA, Becker JD, et al. 2012. Reprogramming of DNA methylation in pollen guides epigenetic inheritance via small RNA. Cell 151: 194–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmell MA, Girard A, van de Kant HJ, Bourc’his D, Bestor TH, de Rooij DG, Hannon GJ 2007. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell 12: 503–514 [DOI] [PubMed] [Google Scholar]
- De Fazio S, Bartonicek N, Di Giacomo M, Abreu-Goodger C, Sankar A, Funaya C, Antony C, Moreira PN, Enright AJ, O’Carroll D 2011. The endonuclease activity of Mili fuels piRNA amplification that silences LINE1 elements. Nature 480: 259–263 [DOI] [PubMed] [Google Scholar]
- Feschotte C, Gilbert C 2012. Endogenous viruses: insights into viral evolution and impact on host biology. Nat Rev Genet 13: 283–296 [DOI] [PubMed] [Google Scholar]
- Ginsburg M, Snow MH, McLaren A 1990. Primordial germ cells in the mouse embryo during gastrulation. Development 110: 521–528 [DOI] [PubMed] [Google Scholar]
- Goodier JL, Kazazian HH Jr 2008. Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell 135: 23–35 [DOI] [PubMed] [Google Scholar]
- Guibert S, Forne T, Weber M 2012. Global profiling of DNA methylation erasure in mouse primordial germ cells. Genome Res 22: 633–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett JA, Sengupta R, Zylicz JJ, Murakami K, Lee C, Down TA, Surani MA 2012. Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science 339: 448–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajkova P, Erhardt S, Lane N, Haaf T, El-Maarri O, Reik W, Walter J, Surani MA 2002. Epigenetic reprogramming in mouse primordial germ cells. Mech Dev 117: 15–23 [DOI] [PubMed] [Google Scholar]
- Hodges E, Molaro A, Dos Santos CO, Thekkat P, Song Q, Uren PJ, Park J, Butler J, Rafii S, McCombie WR, et al. 2011. Directional DNA methylation changes and complex intermediate states accompany lineage specificity in the adult hematopoietic compartment. Mol Cell 44: 17–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaaij LT, van de Wetering M, Fang F, Decato B, Molaro A, van de Werken HJ, van Es JH, Schuijers J, de Wit E, de Laat W, et al. 2013. DNA methylation dynamics during intestinal stem cell differentiation reveals enhancers driving gene expression in the villus. Genome Biol 14: R50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y, Kaneda M, Hata K, Kumaki K, Hisano M, Kohara Y, Okano M, Li E, Nozaki M, Sasaki H 2007. Role of the Dnmt3 family in de novo methylation of imprinted and repetitive sequences during male germ cell development in the mouse. Hum Mol Genet 16: 2272–2280 [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Sakurai T, Imai M, Takahashi N, Fukuda A, Yayoi O, Sato S, Nakabayashi K, Hata K, Sotomaru Y, et al. 2012. Contribution of intragenic DNA methylation in mouse gametic DNA methylomes to establish oocyte-specific heritable marks. PLoS Genet 8: e1002440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Kimura T, Yomogida K, Kuroiwa A, Tadokoro Y, Fujita Y, Sato M, Matsuda Y, Nakano T 2001. Two mouse piwi-related genes: miwi and mili. Mech Dev 108: 121–133 [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Kimura T, Ijiri TW, Isobe T, Asada N, Fujita Y, Ikawa M, Iwai N, Okabe M, Deng W, et al. 2004. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development 131: 839–849 [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri TW, et al. 2008. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev 22: 908–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane N, Dean W, Erhardt S, Hajkova P, Surani A, Walter J, Reik W 2003. Resistance of IAPs to methylation reprogramming may provide a mechanism for epigenetic inheritance in the mouse. Genesis 35: 88–93 [DOI] [PubMed] [Google Scholar]
- Lees-Murdock DJ, De Felici M, Walsh CP 2003. Methylation dynamics of repetitive DNA elements in the mouse germ cell lineage. Genomics 82: 230–237 [DOI] [PubMed] [Google Scholar]
- Lengner CJ, Camargo FD, Hochedlinger K, Welstead GG, Zaidi S, Gokhale S, Scholer HR, Tomilin A, Jaenisch R 2007. Oct4 expression is not required for mouse somatic stem cell self-renewal. Cell Stem Cell 1: 403–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin HL, Moran JV 2011. Dynamic interactions between transposable elements and their hosts. Nat Rev Genet 12: 615–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lienert F, Wirbelauer C, Som I, Dean A, Mohn F, Schubeler D 2011. Identification of genetic elements that autonomously determine DNA methylation states. Nat Genet 43: 1091–1097 [DOI] [PubMed] [Google Scholar]
- Molaro A, Hodges E, Fang F, Song Q, McCombie WR, Hannon GJ, Smith AD 2011. Sperm methylation profiles reveal features of epigenetic inheritance and evolution in primates. Cell 146: 1029–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk M, Boubelik M, Lehnert S 1987. Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development. Development 99: 371–382 [DOI] [PubMed] [Google Scholar]
- Reik W, Dean W, Walter J 2001. Epigenetic reprogramming in mammalian development. Science 293: 1089–1093 [DOI] [PubMed] [Google Scholar]
- Seisenberger S, Andrews S, Krueger F, Arand J, Walter J, Santos F, Popp C, Thienpont B, Dean W, Reik W 2012. The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol Cell 48: 849–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P, Li AX, Tran DA, Oates N, Kang ER, Wu X, Szabo PE 2013. De novo DNA methylation in the male germ line occurs by default but is excluded at sites of H3K4 methylation. Cell Reports 4: 205–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin RK, Vaughn M, Borges F, Tanurdzic M, Becker JD, Feijo JA, Martienssen RA 2009. Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136: 461–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AD, Chung WY, Hodges E, Kendall J, Hannon G, Hicks J, Xuan Z, Zhang MQ 2009. Updates to the RMAP short-read mapping software. Bioinformatics 25: 2841–2842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sookdeo A, Hepp CM, McClure MA, Boissinot S 2013. Revisiting the evolution of mouse LINE-1 in the genomic era. Mob DNA 4: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surani MA, Hayashi K, Hajkova P 2007. Genetic and epigenetic regulators of pluripotency. Cell 128: 747–762 [DOI] [PubMed] [Google Scholar]
- Walsh CP, Chaillet JR, Bestor TH 1998. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat Genet 20: 116–117 [DOI] [PubMed] [Google Scholar]