A key function of the DNA damage response is to facilitate the bypass of replication fork-stalling DNA lesions. Template switch reactions allow such a bypass and involve the formation of DNA joint molecules (JMs) between sister chromatids. However, the regulation of JM resolution is poorly understood. Pfander and colleagues elucidate a mechanism in yeast that critically controls JM resolution by the Mus81–Mms4 endonuclease. The data show that the conserved complex comprising scaffold proteins Dbp11 and Slx4 integrates cellular inputs and regulates the activation of the JM-resolving nuclease Mus81.
Keywords: DNA damage response, cell cycle, post-replicative repair, homologous recombination, joint molecule resolution
Abstract
A key function of the cellular DNA damage response is to facilitate the bypass of replication fork-stalling DNA lesions. Template switch reactions allow such a bypass and involve the formation of DNA joint molecules (JMs) between sister chromatids. These JMs need to be resolved before cell division; however, the regulation of this process is only poorly understood. Here, we identify a regulatory mechanism in yeast that critically controls JM resolution by the Mus81–Mms4 endonuclease. Central to this regulation is a conserved complex comprising the scaffold proteins Dpb11 and Slx4 that is under stringent control. Cell cycle-dependent phosphorylation of Slx4 by Cdk1 promotes the Dpb11–Slx4 interaction, while in mitosis, phosphorylation of Mms4 by Polo-like kinase Cdc5 promotes the additional association of Mus81–Mms4 with the complex, thereby promoting JM resolution. Finally, the DNA damage checkpoint counteracts Mus81–Mms4 binding to the Dpb11–Slx4 complex. Thus, Dpb11–Slx4 integrates several cellular inputs and participates in the temporal program for activation of the JM-resolving nuclease Mus81.
Intrinsically and extrinsically induced DNA lesions can compromise the integrity of the genetic information and threaten cell viability. DNA lesions are particularly dangerous during S phase, when faithful DNA replication relies on two intact DNA strands. DNA lesions hamper the progression of replication forks and thereby the complete duplication of chromosomes. Moreover, replication forks that are stalled at DNA lesion sites can collapse and cause chromosome breaks and genome instability (Branzei and Foiani 2010).
Eukaryotes possess two fundamentally different mechanisms to bypass DNA lesions that affect one of the parental DNA strands: translesion synthesis (TLS) and template switching. TLS employs specialized polymerases (translesion polymerases) that in many cases are able to replicate the damaged strand but with a reduced fidelity (Prakash et al. 2005). On the other hand, during template switching, the genetic information is copied from the newly synthesized, undamaged sister chromatid. This mechanism is therefore error-free in principle, yet its precise mechanism remains poorly understood. Template switching is a complex process that can be initiated by different recombination-based mechanisms (homologous recombination [HR] and error-free post-replicative repair [PRR]) (Branzei et al. 2008). The choice between the different bypass mechanisms is regulated by ubiquitin and SUMO modifications of the replication protein PCNA at sites of stalled replication forks (Pfander et al. 2005).
Template switch mechanisms involve the formation of DNA joint molecules (JMs; also referred to as sister chromatid junctions [SCJs] or X molecules) as repair intermediates (Branzei et al. 2008). In order to allow completion of DNA replication and faithful chromosome segregation, these X-shaped DNA structures need to be disentangled before sister chromatids are separated during mitosis. To date, three enzymatic activities—the topoisomerase-containing Sgs1–Top3–Rmi1 complex (STR) as well as the Mus81–Mms4 and Yen1 structure-specific endonucleases—were shown to process JMs in budding yeast (Liberi et al. 2005; Blanco et al. 2010; Mankouri et al. 2011; Szakal and Branzei 2013). These three activities can be distinguished by their mechanism (termed dissolution for STR and resolution for Mus81–Mms4 and Yen1) (Gaillard et al. 2003; Ip et al. 2008; Cejka et al. 2010) but show a partial functional overlap. Moreover, they are differentially regulated during the cell cycle: Whereas the STR activity appears to be cell cycle-independent, the activity of Mus81–Mms4 is stimulated by CDK-mediated and Cdc5 (budding yeast Polo-like kinase)-mediated phosphorylation and peaks in mitosis (Matos et al. 2011, 2013; Gallo-Fernández et al. 2012; Szakal and Branzei 2013). Accordingly, the Mus81 regulation is assumed to create a hierarchy, with STR acting as a primary resolution pathway and Mus81–Mms4 acting as a salvage pathway. How Mus81–Mms4 phosphorylation by cell cycle kinases facilitates this temporal regulation of JM resolution pathways remains hardly understood.
The bypass of DNA lesions during replication is additionally regulated by the DNA damage checkpoint, the main cellular signaling pathway in response to DNA damage (Harrison and Haber 2006). As the primary purpose of the checkpoint is the stabilization of stalled replication forks (Branzei and Foiani 2010), its activation is a fundamental requirement for all fork repair and reactivation reactions. Notably, the checkpoint has been suggested to be involved in the choice of the JM resolution pathway, since precocious activation of the Mus81–Mms4 endonuclease is observed in checkpoint-deficient mutants (Szakal and Branzei 2013). However, it remains to be clarified how this second layer of regulation of JM resolution is achieved on a molecular level and how it is linked to cell cycle regulation.
Here, we identify an evolutionarily conserved protein complex comprising two scaffold proteins, Slx4 and Dpb11/TopBP1, as an important regulator of JM resolution by Mus81–Mms4. We show that the formation of the Slx4–Dpb11 complex is regulated by the cell cycle stage. An slx4 mutant, compromised specifically in Dpb11 binding, exhibits hypersensitivity to the replication fork-stalling drug MMS, a delay in the resolution of X-shaped DNA JMs, and a reduced propensity to form crossovers (COs). The function of the Slx4–Dpb11 scaffold in JM resolution correlates with the finding that Dpb11 binds to the Mus81–Mms4 endonuclease. This association is restricted to mitosis, since it is dependent on the mitotic kinase Cdc5. Moreover, the checkpoint acts antagonistically to the regulation of JM resolution by Slx4 and Dpb11, as we found that partial inactivation of the DNA damage checkpoint can compensate for defects in formation of the Slx4–Dpb11 scaffold complex.
Results
An evolutionarily conserved and phosphorylation-dependent interaction between Slx4 and Dpb11/TopBP1
Dpb11 and its human homolog, TopBP1, are critical regulators of the cellular DNA damage response and interact with several DNA replication, repair, and checkpoint proteins (Garcia et al. 2005; Germann et al. 2011). In these protein complexes, Dpb11/TopBP1 specifically binds to phosphorylated proteins via its tandem BRCT domains (Yu 2003; Garcia et al. 2005). A key role of Dpb11/TopBP1 is to function as a scaffold, bringing together specific sets of proteins via several interaction surfaces. In budding yeast, two Dpb11 complexes have been described in detail, which regulate replication initiation (with Sld3 and Sld2) (Tanaka et al. 2007; Zegerman and Diffley 2007) and the DNA damage checkpoint (with Rad9, the 9-1-1 complex, and Mec1–Ddc2) (Mordes et al. 2008; Navadgi-Patil and Burgers 2008; Puddu et al. 2008; Pfander and Diffley 2011), respectively (Fig. 1A). Recently, a third Dpb11 complex with Slx4 and Rtt107 was identified (Ohouo et al. 2010, 2012). In this latter complex, Slx4 appears to inhibit the formation of the Dpb11 DNA damage checkpoint complex (Ohouo et al. 2012).
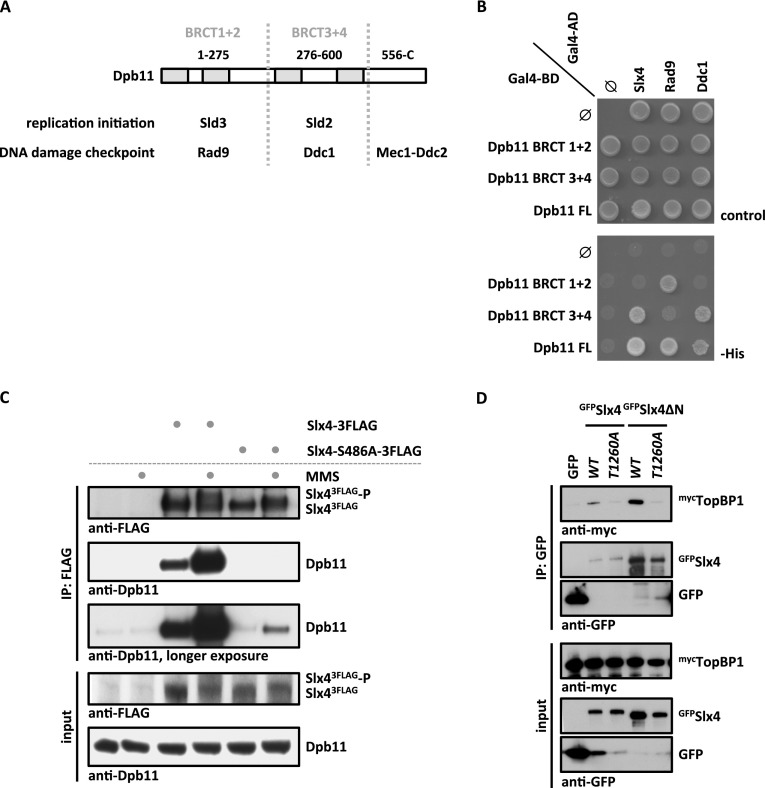
Figure 1.
An evolutionarily conserved, phosphorylation-dependent interaction between Slx4 and Dpb11/TopBP1. (A) Schematic diagram of Dpb11 domain structure depicted with its interaction partners in replication initiation and DNA damage checkpoint. (B) Slx4 binds to the BRCT3+4 domain of Dpb11. Two-hybrid analysis of GAL4-BD fused to full-length Dpb11 or to BRCT1+2 and BRCT3+4 fragments and of GAL4-AD fusions with Slx4, Rad9, and Ddc1. (C) The Slx4–Dpb11 interaction is reduced by mutation of Slx4 Ser486 and is regulated by DNA damage. Coimmunoprecipitation of endogenous Dpb11 with Slx43Flag or phosphorylation-deficient Slx4-S486A3Flag from undamaged cells or cells treated for 30 min with 0.033% MMS. (D) Human TopBP1 and Slx4 interact dependent on Thr1260 of Slx4. Coimmunoprecipitation of human mycTopBP1 with GFPSlx4 or N-terminally truncated GFPSlx4ΔN after transient overexpression in HEK293T cells. Slx4 or Slx4ΔN was expressed either as wild type (WT) or a T1260A phosphorylation-deficient variant.
In the course of our studies of Dpb11 function, we identified an interaction between a Dpb11 fragment that includes the tandem BRCT repeats 3 and 4 (BRCT3+4) and Slx4 using a two-hybrid screen. To confirm this finding, we tested the binding of different Dpb11 constructs to Slx4 and known Dpb11 binders. As observed before (Puddu et al. 2008; Pfander and Diffley 2011), we found that Rad9 binds to BRCT1+2 of Dpb11, whereas Ddc1 binds to BRCT3+4 (Fig. 1B). For Slx4, we found an interaction with full-length Dpb11 and the BRCT3+4 fragment but not with the BRCT1+2 domain (Fig. 1B). When we tested binding of Slx4 from cell extracts to recombinant, purified fragments of Dpb11, Slx4 also bound to BRCT3+4, albeit weaker than to the full-length protein (Supplemental Fig. S1A). Moreover, ablation of Dpb11 Thr451, which is predicted to be part of the BRCT3+4 phospho-protein-binding surface (Rappas et al. 2011), partially inhibited the Slx4–Dpb11 interaction (Supplemental Fig. S1B). A recent report suggested that the Dpb11 BRCT1+2 domain is involved in Slx4 binding (Ohouo et al. 2012). However, although our data do not rule out a contribution of BRCT1+2 in overall binding, our two independent lines of evidence clearly demonstrate that BRCT3+4 of Dpb11 significantly contributes to Slx4 binding.
Next, we mapped the Dpb11-binding site on Slx4 starting from a fragment (amino acids 461–738) that was common to all Slx4 clones identified in our initial Dpb11 two-hybrid screen. Truncated variants that begin at amino acid 490 failed to interact with Dpb11 (Supplemental Fig. S1C), indicating that the region between amino acid 461 and amino acid 490 is important for Dpb11 interaction. As several examples indicate that Dpb11 binds phosphorylated S/TP motifs, we tested all S/TP motifs within the C-terminal part of Slx4 for their ability to mediate Dpb11 binding. Indeed, we found that alteration of Ser486 in Slx4 into a nonphosphorylatable alanine residue (slx4-S486A mutant) reduced Dpb11 binding in a two-hybrid system (Supplemental Fig. S1D). Moreover, whereas immunoprecipitation of wild-type Slx4 efficiently copurified endogenous Dpb11 from cell extracts, in particular following MMS treatment, the Slx4–Dpb11 interaction was strongly decreased in extracts from cells expressing the slx4-S486A mutant, even after induction of DNA damage (Fig. 1C; see also Ohouo et al. 2012). Furthermore, the phospho-S486-containing peptide was specifically enriched (17-fold), when Dpb11 immunoprecipitations were analyzed by quantitative mass spectrometry (MS) (Supplemental Fig. S4A). We therefore conclude that the Slx4–Dpb11 interaction involves the BRCT3+4 region of Dpb11 and a region of Slx4 harboring the phosphorylated residue S486.
We further tested whether also the human homologs TopBP1 and Slx4 are binding partners. Indeed, we detected a specific interaction of TopBP1 and Slx4 or an N-terminally truncated version of Slx4 after transient transfection in human embryonic kidney (HEK) 293T cells (Fig. 1D). In contrast to the yeast proteins, we did not observe a stimulation of TopBP1 binding to Slx4 by DNA damage (Supplemental Fig. S1E). Human Slx4 is substantially larger than yeast Slx4, with an overall sequence conservation of only 17.9%. Nonetheless, we identified a conserved short linear motif present in Slx4 proteins from different eukaryotes that comprises Ser486 in budding yeast and Thr1260 in humans (Supplemental Fig. S2). Mutation of Thr1260 to a nonphosphorylatable alanine (T1260A) in human Slx4 reduced the interaction with TopBP1 (Fig. 1D), suggesting that this residue may function analogously to Ser486 in budding yeast. These data suggest the presence of a novel, evolutionarily conserved motif in Slx4 that functions in Dpb11/TopBP1 binding.
Cdk1-dependent phosphorylation of Slx4 regulates binding to Dpb11
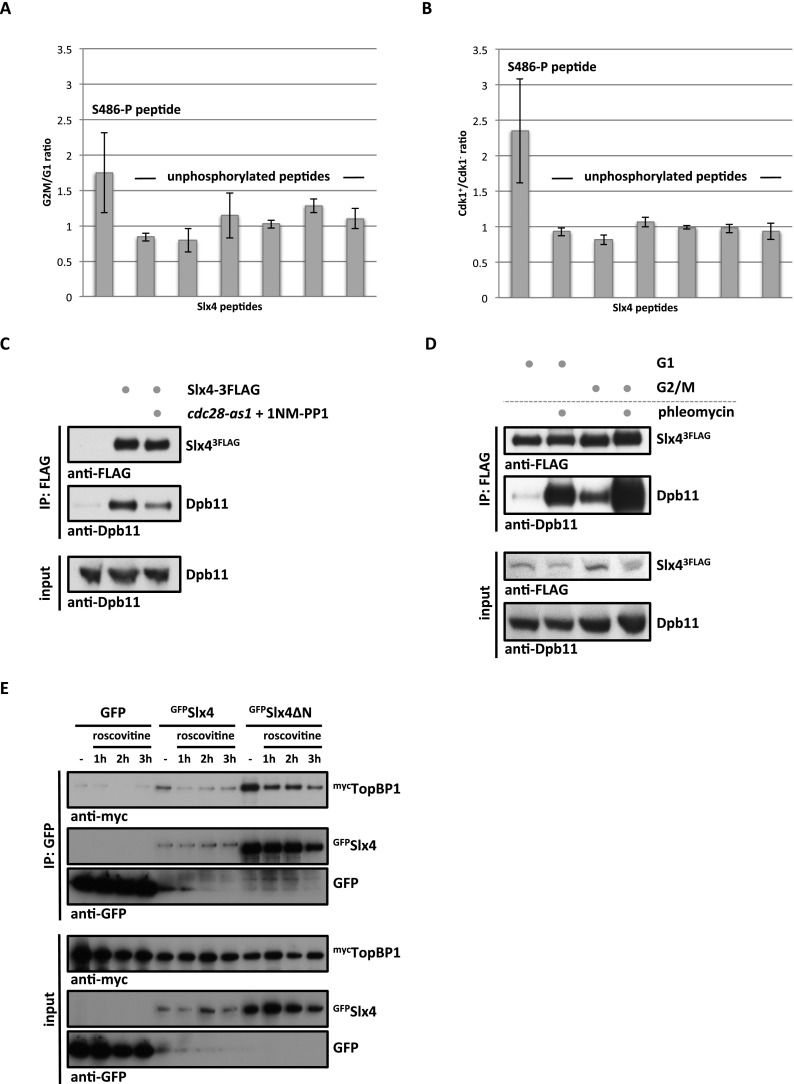
In order to unravel the regulation of the Slx4–Dpb11-binding surface, we quantified the relative amount of Ser486 phosphorylation under different cellular conditions using SILAC-based quantitative MS. We observed a specific increase of Ser486 phosphorylation in G2/M-arrested cells compared with G1-arrested cells, indicating that the analyzed Slx4 phosphorylation is cell cycle-regulated (Fig. 2A). In agreement with Ser486 matching the consensus target sequence for phosphorylation by cyclin-dependent kinase Cdk1 (S/TPxK) (Holt et al. 2009), we observed a marked reduction of Ser486 phosphorylation in G2/M-arrested cells when Cdk1 activity was abrogated using the cdc28-as1 allele (Bishop et al. 2000) in combination with 1NM-PP1 inhibitor treatment (Fig. 2B). Notably, we also detected reduced Slx4 binding to Dpb11 when Cdk1 was inhibited (Fig. 2C).
Figure 2.
The Slx4–Dpb11/TopBP1-binding interface is cell cycle-regulated by Cdk1 phosphorylation of Slx4. (A) Ser486 phosphorylation is cell cycle-regulated. Relative abundance of the Slx4 480–489 phospho-peptide and six unmodified Slx4 peptides was measured by SILAC-based quantitative MS using 15N213C6 lysine (Lys8) and compared between Slx4 isolated from G1- and G2/M-arrested cells. H/L ratios for individual peptides were normalized to total Slx4 ratios. Error bars represent standard deviations from two independent experiments, including label switch. (B) S486 phosphorylation depends on Cdk1. Analysis as in A but comparing Slx4 from G2/M-arrested cells with normal Cdk1 activity with cells in which Cdk1 has been inactivated using the cdc28-as1 allele and 500 nM 1NM-PP1. (C) The Slx4–Dpb11 interaction is regulated by CDK. Coimmunoprecipitation of Dpb11 and Slx43Flag from G2/M-arrested cells or G2/M-arrested cells in which Cdk1 has been inactivated as in B. (D) The Slx4–Dpb11 interaction is regulated by cell cycle phase and DNA damage. Experiment as in C but with G1- and G2/M-arrested cells, which were either damaged by 50 μg/mL phleomycin or left untreated. (E) Binding of human Slx4 and TopBP1 is regulated by CDK phosphorylation. Coimmunoprecipitation of mycTopBP1 with GFPSlx4 and GFPSlx4ΔN after transient overexpression in HEK293T cells. Cells were left untreated or treated with 10 μg/mL roscovitine for the indicated times to inhibit CDK activity.
In addition to cell cycle-dependent regulation, we also observed a stimulation of Slx4–Dpb11 binding by DNA damage (Figs. 1C, 2D, Supplemental Fig. S1F). When Slx4 binding to recombinant GST-Dpb11 was tested, the DNA damage-dependent stimulation was less pronounced (Supplemental Fig. S1A), substantiating the notion that the Slx4–Dpb11 interaction may be additionally regulated by a damage-induced post-translational modification of Dpb11. On the other hand, Slx4 harbors several sites that can be targeted by kinases of the DNA damage checkpoint pathway. Mutation of seven sites in Slx4 partially inhibits its binding to Dpb11 (Ohouo et al. 2010), and the corresponding mutant shows phenotypes similar to those of slx4-S486A (Supplemental Fig. S3). As we cannot fully exclude pleiotropic defects for this mutant, we focused our analysis on slx4-S486A.
Taken together, our findings suggest that the Slx4–Dpb11 complex integrates at least two cellular signals: (1) cell cycle state through Cdk1 phosphorylation of Slx4 at Ser486 and (2) the presence of DNA damage through checkpoint kinase phosphorylation of several sites on Slx4 and perhaps on Dpb11.
Interestingly, the CDK regulation of this interaction is conserved between yeast and humans, since addition of the CDK inhibitor roscovitine reduced binding of Slx4 and TopBP1 (Fig. 2E).
The Slx4–Dpb11 complex is required for the response to replication fork stalling
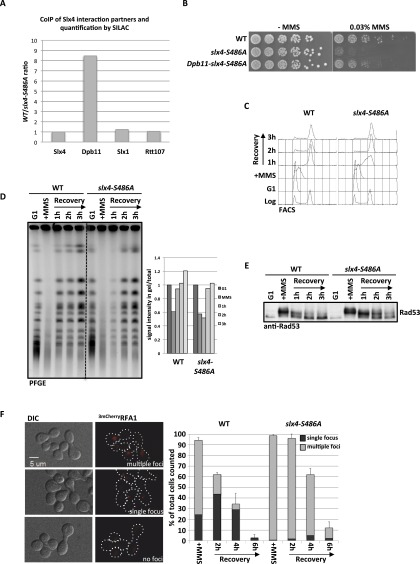
Budding yeast Slx4 is known to bind to several DNA repair proteins (Slx1, Rtt107, and Rad1–Rad10) (Mullen et al. 2001; Roberts et al. 2006; Flott et al. 2007; Ohouo et al. 2010). However, whether these interaction partners are part of only one or several distinct complexes is unknown. While Slx4 has several independent DNA repair functions in budding yeast (Flott et al. 2007), until now, a detailed phenotypic characterization has only been conducted for slx4Δ deletion mutants. To test the specificity of the Dpb11-binding-deficient slx4-S486A phosphorylation site mutant, we compared its binding partners with those of wild-type Slx4 using quantitative proteomics. Indeed, we found that the mutant protein (Slx4-S486A3Flag) displayed eightfold reduced binding to Dpb11 (Fig. 3A). This variant still bound Slx1 and Rtt107 as efficiently as wild-type Slx4, indicating that Ser486 phosphorylation is specifically relevant for the Dpb11 interaction (Fig. 3A; see Supplemental Fig. S4A for specific Slx4 interactors). We thus took advantage of the slx4-S486A separation-of-function mutant to reveal a specific role of the Slx4–Dpb11 complex.
Figure 3.
Mutation of slx4-S486A results in a specific defect in binding to Dpb11 and the response to stalled replication forks. (A) The slx4-S486A mutant leads to a specific defect in binding to Dpb11. Relative enrichment of Slx4 interactors (see Supplemental Fig. S4A) found in purifications of wild-type (WT) Slx43Flag versus Slx4-S486A3Flag as determined by SILAC-based quantitative MS. Values >1 indicate a reduced binding to the Slx4-S486A relative to wild-type Slx4. (B) The slx4-S486A mutant, but not a Dpb11–slx4-S486A-fusion, is hypersensitive to MMS. Wild type or strains expressing slx4-S486A or the Dpb11–slx4-S486A-fusion from the SLX4 promoter as only a copy of SLX4 were spotted in fivefold serial dilutions on MMS-containing medium and assayed for growth after 2 d. (C,D) Replication fork stalling is prolonged in the slx4-S486A mutant. Cells were treated with a pulse of MMS during S phase, and recovery was analyzed by FACS (C; to measure cellular DNA content) and pulsed-field gel electrophoresis (D; to measure intact, fully replicated chromosomes). (D) For quantification, the fluorescence signal of chromosomes that migrated into the gel was divided by the total signal, including the pocket, and all signals were normalized to the G1 sample from each strain. (E) The DNA damage checkpoint is inactivated with reduced kinetics in the slx4-S486A mutant. Cells were treated as in C, and checkpoint activity was determined by anti-Rad53 Western blot. (F) The slx4-S486A mutant shows increased DNA damage foci and delayed recovery after transient MMS treatment in S phase. DNA damage sites were visualized by the ssDNA-binding RFA13mCherry after transient MMS treatment during S phase. Cells were sorted into three categories: multiple, dispersed RFA1 foci; one RFA1 focus; and no RFA1 foci. Values are from two independent experiments, counting 100–150 cells per strain and time point. Error bars represent standard deviations.
Using different DNA-damaging agents, we observed that the slx4-S486A mutant is particularly sensitive to MMS and, to a lesser extent, 4-NQO (Fig. 3B; Supplemental Fig. S4B), two reagents that create toxicity through replication fork stalling. Notably, the mutant was not sensitive to reagents that generate DNA strand breaks or interstrand cross-links, consistent with a recombination rate that was similar to wild type (Supplemental Fig. S4B,C). Remarkably, expression of a fusion protein of the phospho-site mutant variant of Slx4 with Dpb11 (Dpb11–Slx4-S486A) rescued the MMS hypersensitivity phenotype almost to wild-type levels (Fig. 3B), suggesting that binding of Slx4 to Dpb11 is crucial for tolerance of replication fork-stalling lesions.
Next, we tested whether the response to stalled replication forks is aberrant in the slx4-S486A mutant. To this end, we treated synchronized cells with a pulse of MMS in early S phase. Under these conditions, the slx4-S486A mutant completed DNA replication with slightly slower kinetics compared with wild-type cells (Fig. 3C, 1-h time point). Also, the appearance of fully replicated and repaired chromosomes, as visualized by pulsed-field gel electrophoresis, was delayed (Fig. 3D, 1-h time point). This finding indicates that stalled replication fork structures or repair intermediates persist longer in the absence of the Slx4–Dpb11 complex. Additionally, the DNA damage checkpoint activation was prolonged in slx4-S486A cells (Fig. 3E), as determined by the phosphorylation status of the checkpoint kinase Rad53. This effect was specific for MMS treatment and could not be observed in cells in which double-strand breaks were induced by zeocin or phleomycin inside or outside of S phase (Supplemental Fig. S4D).
Defects in a checkpoint-antagonistic pathway (checkpoint “dampening”) (Ohouo et al. 2012) in slx4 mutants could, in principle, lead to prolonged checkpoint activation and could thereby indirectly lead to slow S-phase kinetics and DNA damage hypersensitivity. Alternatively, persistence of unrepaired DNA lesions or DNA repair intermediates could lead to very similar phenotypes. In order to discriminate between the two possibilities, we examined the DNA damage levels during recovery from an MMS pulse in wild-type and slx4-S486A cells. To this end, we investigated the appearance and disappearance of nuclear foci formed by the ssDNA-binding protein RPA after MMS treatment in S phase. Indeed, slx4-S486A cells contained more RPA foci, which persisted longer than in wild-type cells (Fig. 3F). Therefore, we conclude that unrepaired DNA lesions or DNA repair intermediates that contain ssDNA persist in slx4-S486A mutants. This finding does not necessarily exclude a role of Slx4 as a regulator of the DNA damage checkpoint yet strongly suggests an additional direct function of the Slx4–Dpb11 complex in the repair of replication fork structures.
The Slx4–Dpb11 complex promotes Mus81–Mms4-dependent JM resolution
As our findings pointed to a function of the Slx4–Dpb11 complex in the response and repair of MMS-induced lesions, we next investigated whether the complex is involved in the DNA damage bypass. Therefore, we tested possible functions in HR and error-prone or error-free PRR. From several lines of genetic evidence, we conclude that the Slx4–Dpb11 complex is not exclusively involved in either PRR or HR (Supplemental Fig. S5). First, the slx4-S486A mutation enhanced the MMS hypersensitivity of mutants defective in error-free PRR (double mutant with either mms2Δ, rad5-KT538,539AA, or rad5-C914S), error-prone PRR (double mutant with either rev1Δ, rev3Δ, or rad30Δ), or HR (double mutant with rad51Δ) (Supplemental Fig. S5A). Second, spontaneous mutagenesis, a hallmark of error-prone PRR, was not significantly altered in slx4-S486A mutants (Supplemental Fig. S5B). Third, recombination rates, as determined by a direct repeat recombination assay, were similar between wild-type and slx4-S486A strains (Supplemental Fig. S4C). Fourth, siz1Δ or srs2ΔC mutations, which cause an up-regulation of HR at stalled replication forks (Pfander et al. 2005), did not alleviate the MMS hypersensitivity of slx4-S486A mutants (Supplemental Fig. S5C).
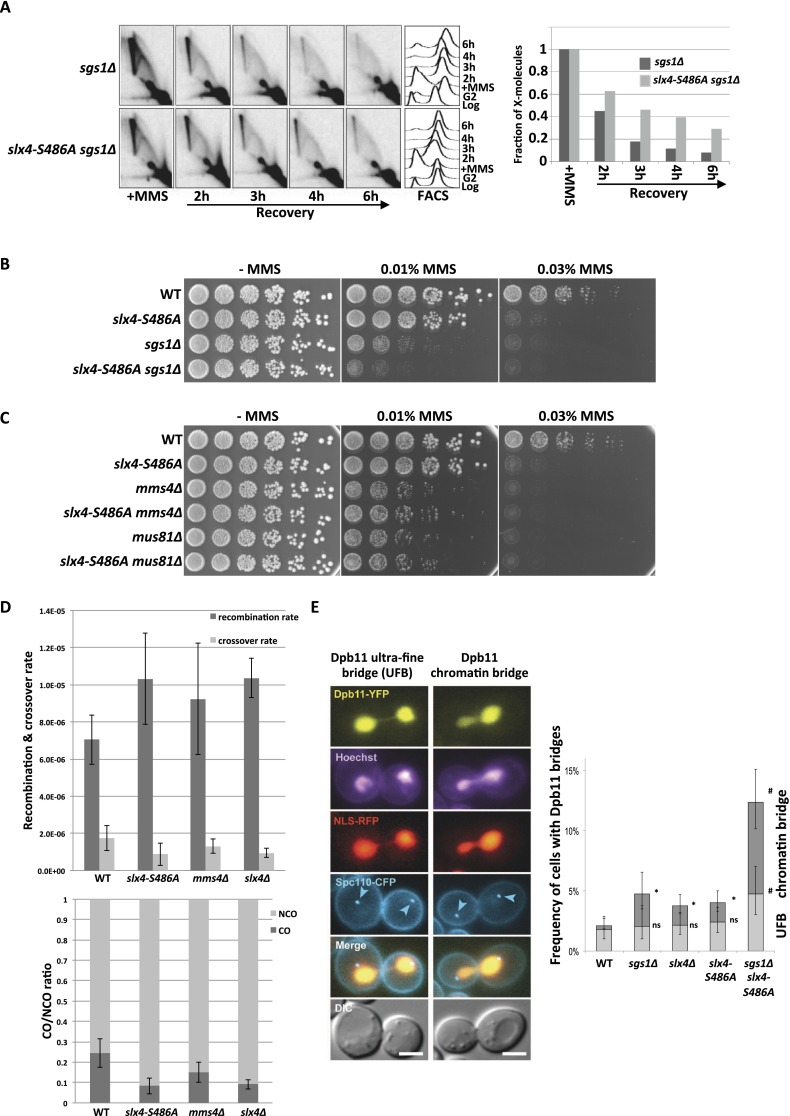
The nonepistatic relationship of the slx4-S486A mutant to PRR or HR pathways could be explained if Slx4 and Dpb11 participated in a step common to both error-free PRR and HR because, in such a scenario, both pathways would be affected by the slx4-S486A mutation. Both HR and error-free PRR operate via template switching in order to bypass the replication fork-stalling lesion by copying the undamaged information from the sister chromatid. A critical step in template switching is the final removal of X-shaped DNA intermediates (JMs) that link the two sister chromatids (Mankouri et al. 2013). JM removal pathways act, in principle, independently of the pathway by which JMs have been created (Branzei et al. 2008; for mus81Δ phenotypes, see Interthal and Heyer 2000; Li and Brill 2005). To test whether the Slx4–Dpb11 complex is involved in this late step, we visualized these DNA intermediates in an sgs1Δ mutant (deficient in JM dissolution) by two-dimensional (2D) gel electrophoresis (Liberi et al. 2005; Mankouri et al. 2011). In this mutant, MMS treatment in S phase leads to enhanced levels of JMs, which subsequently disappear during late S, G2, and M phase (Szakal and Branzei 2013). The additional mutation of slx4-S486A in the sgs1Δ background does not alter the formation of JMs, indicating that the Slx4–Dpb11 complex is not required at early steps (Supplemental Fig. S6A). Interestingly, however, during the recovery from the MMS treatment, JMs are more slowly resolved in the sgs1Δ slx4-S486A double mutant compared with the sgs1Δ single mutant (Fig. 4A). A similar effect can be observed using an slx4Δ mutant and conditionally inactivated SGS1 in the same experimental setup (Supplemental Fig. S6B). Consistently, we observed an enhanced MMS sensitivity for the sgs1Δ slx4-S486A double mutant compared with the respective single mutants (Fig. 4B). From these experiments, we conclude that the Slx4–Dpb11 complex is involved in the resolution of JMs that are supposedly intermediates arising from a template switch reaction and that this complex functions in a pathway parallel to dissolution by the STR complex.
Figure 4.
The Dpb11 binding-deficient slx4-S486A mutant causes defects in the Mus81–Mms4-dependent JM resolution. (A) JM structures are resolved slower in sgs1Δ slx4-S486A cells. X-shaped JMs were visualized as spike signal in 2D gels in sgs1Δ and sgs1Δ slx4-S486A cells that have been treated with a pulse of MMS in S phase. (B) MMS sensitivity is enhanced in the sgs1Δ slx4-S486A double mutant compared with each single mutant. Analysis of the MMS hypersensitivity phenotype as in Figure 3B. (C) The MMS hypersensitivity of mms4Δ and mus81Δ mutants is not further enhanced by an additional slx4-S486A mutation. Experiment as in B. (D) The slx4-S486A mutation leads to a reduced CO formation. COs and NCOs from an interchromosomal recombination assay using arg4 heteroalleles on chromosome V and VIII (Robert et al. 2006) were determined using a PCR-based strategy. (Top panel) Recombination and CO rates were determined by fluctuation analysis using a maximum likelihood approach. (Bottom panel) CO ratio is quotient of CO rate and overall recombination rate. Error bars represent standard deviations of two to 11 independent experiments. (E) Dpb11 anaphase bridge structures occur more frequently when JM dissolution and the Dpb11–Slx4 interaction are defective. (Right panel) Quantification of Dpb11 ultrafine bridges (UFBs) or chromatin bridges in wild-type (WT), sgs1Δ, slx4Δ, slx4-S486A, and slx4-S486A sgs1Δ strains. Cells express Dpb11-YFP, NLS-RFP as a marker of the nucleoplasm and Spc110-CFP as a marker of the spindle pole body. DNA is stained with Hoechst. (Left panel) Images of representative anaphase cells are shown. Bar, 3 μm. Error bars indicate 95% confidence intervals. Significance is as follows: (*) P < 0.01 (compared with wild type); (#) P < 0.01 (compared with the single mutants); (ns) not significantly different from wild type.
To elucidate a potential role of the Slx4–Dpb11 complex in a resolution mechanism, we investigated the genetic interaction with Mus81–Mms4. Indeed, the MMS sensitivities of slx4-S486A mms4Δ or slx4-S486A mus81Δ double mutants were identical to those of mms4Δ or mus81Δ single mutants (Fig. 4C). This suggests that the Slx4–Dpb11 complex acts in the Mus81–Mms4 pathway. The same epistatic relationship was seen between mms4Δ and slx4-S486A when we investigated JM resolution by 2D gel electrophoresis when the STR complex was inactivated using the Tc-sgs1 allele (Supplemental Fig. S6C). We note that the MMS hypersensitivity and the JM resolution defect of the slx4-S486A mutant are less pronounced compared with the deletion mutants that fully abolish Mus81 function (Fig. 4C; Supplemental Fig. S6C), suggesting that not all functions of the Mus81–Mms4 endonuclease depend on the Slx4–Dpb11 complex.
We also tested the involvement of other structure-specific endonucleases (Slx1, Rad1–Rad10, and Yen1) (Tomkinson et al. 1993; Fricke and Brill 2003; Coulon 2006; Ip et al. 2008), specifically of Slx1, as it associates with the Slx4–Dpb11 complex (Supplemental Fig. S4A). We found that rad1Δ showed an additive phenotype with slx4-S486A, while slx1Δ and yen1Δ mutants were not hypersensitive to MMS (Supplemental Fig. S6D; Fricke and Brill 2003; Coulon 2006; Blanco et al. 2010). We therefore conclude that these factors either are not involved in the resolution of template switch intermediates by Mus81 and the Slx4–Dpb11 complex or (in case of Slx1 and Yen1) have a function that can be taken over by a redundant pathway in the respective deletion mutant. Interestingly, the yen1Δ mutation caused an increase of MMS sensitivity specifically of the sgs1Δ slx4-S486A double mutant (Supplemental Fig. S6E), suggesting that Yen1 function becomes specifically important if the STR complex is inactive and function of the Slx4–Dpb11 complex is reduced.
The balance between STR-dependent JM dissolution and Mus81-dependent JM resolution is reflected in the ratio of CO to non-CO (NCO) products (Ira et al. 2003; Ho et al. 2010; Mankouri et al. 2013), since STR-mediated dissolution will not yield COs, while Mus81-mediated resolution can generate CO products. We therefore analyzed the rates of CO formation in the slx4-S486A mutant with a recombination assay using interchromosomal arg4 heteroalleles (Robert et al. 2006; Szakal and Branzei 2013). Despite a slight increase in overall recombination rates, we measured a reduction in CO rates in the slx4-S486A mutant compared with wild-type cells (Fig. 4D). We therefore conclude that the Slx4–Dpb11 complex is an important regulator of JM removal pathways and that it acts by stimulating JM resolution, inhibiting JM dissolution, or both.
Persistent JMs interfere with the separation of sister chromatids in mitosis. Under circumstances in which JMs are not resolved before anaphase, these repair intermediates are thought to give rise to anaphase bridges between the dividing DNA masses (Chan et al. 2007; Mankouri et al. 2013). Consistent with a role in the resolution of JMs, Dpb11 localizes to DNA bridges between the separated chromosome masses in anaphase (Germann et al. 2014). Dpb11-containing anaphase bridges can be observed with a low frequency in undamaged cells (<5%) and are induced upon MMS treatment, suggesting that they arise from replication fork stalling (Germann et al. 2014). Interestingly, the occurrence of Dpb11 bridges is increased in sgs1Δ cells (Germann et al. 2014), indicating that the localization of Dpb11 to chromatin bridges reflects its action in a resolution mechanism. We observed a pronounced increase of cells containing Dpb11 bridges when the sgs1Δ and slx4-S486A mutants were combined (Fig. 4E). The genetic requirements for Dpb11 bridges are therefore highly similar to those for persistent JMs (Fig. 4A), supporting a role for Dpb11 and Slx4 in JM resolution. In line with this model, we observed frequent colocalization of either Slx4YFP or Mus81YFP with Dpb11CFP-positive bridges that is further enhanced in sgs1Δ cells (Supplemental Fig. S7A). We also noticed a colocalization of Slx4, Mus81, and Dpb11 in DNA damage foci yet to a lesser extent (Supplemental Fig. S7B). Overall, the data in Figure 4 provide strong support for an involvement of the Slx4–Dpb11 complex in JM resolution by Mus81–Mms4.
Mus81–Mms4 interacts with the Slx4–Dpb11 complex during mitosis in a Cdc5-dependent fashion
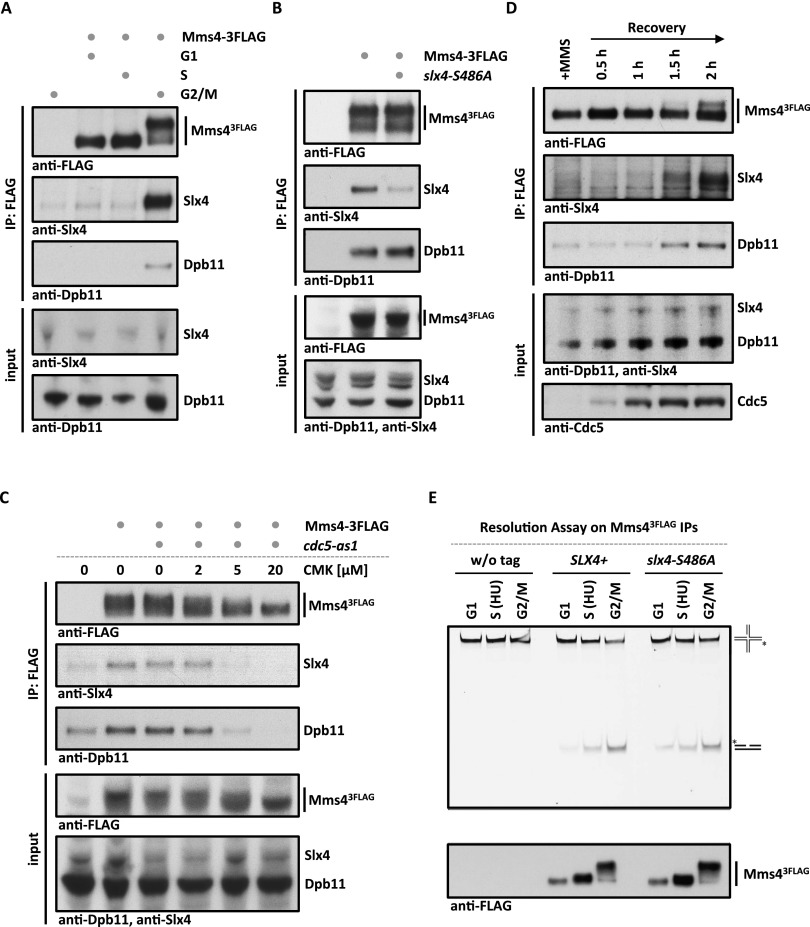
To elucidate how the Slx4–Dpb11 complex regulates Mus81 function, we investigated a possible physical interaction. In previous studies using asynchronously growing yeast cells, no binding of Slx4 to Mus81–Mms4 was detectable (Schwartz et al. 2012). However, we detected Mms4 as a cell cycle-specific interactor if Slx43Flag immunoprecipitations were investigated by SILAC MS (such as in Fig. 2A). Moreover, when we arrested cells in G2/M by nocodazole treatment, immunopurification of Mms43Flag copurified Dpb11 and Slx4 (Fig. 5A). Notably, this interaction is highly cell cycle-specific, as it could not be observed in G1- or S-phase cells (Fig. 5A). We determined, using an unbiased SILAC MS approach, that Dpb11, Slx4, and Rtt107 are among the best interactors of Mus81–Mms4 in G2/M-arrested cells (Supplemental Fig. S8A).
Figure 5.
Slx4, Dpb11, and Mus81–Mms4 form a Cdc5-dependent complex at the G2/M cell cycle stage. (A) Mms4 binds to Dpb11 and Slx4 specifically in G2/M. Coimmunoprecipitation samples of Mms43Flag from G1, S, or G2/M cells were tested for binding to Dpb11 and Slx4. (B) Slx4-S486A is partially lost from the Slx4–Dpb11–Mms4–Mus81 complex, suggesting that Dpb11 bridges the interaction between Mms4 and Slx4. Mms43Flag coimmunoprecipitation were carried out as in A but from G2/M-arrested wild-type (WT) or slx4-S486A mutant cells. (C) The Dpb11–Mms4 interaction is dependent on the Polo-like kinase Cdc5. cdc5-as1 was inhibited by 2, 5, and 20 μM CMK in G2/M-arrested cells. Mms43Flag coimmunoprecipitation was performed as in A. (D) Cdc5 hyperphosphorylated Mus81–Mms4 binds to Slx4 and Dpb11 during recovery from MMS damage. Cells were treated with a 30-min pulse of 0.03% MMS. Mms43Flag coimmunoprecipitations were performed from samples after 0, 30, 60, 90, and 120 min of recovery in nocodazole-containing medium. (E) Cell cycle regulation of Mus81–Mms4 nuclease activity remains intact in the slx4-S486A mutant. Mms43Flag and control immunoprecipitations (see the bottom panel for immunoprecipitation samples) from cells arrested in their cell cycle by α factor, HU, or nocodazole were incubated with a fluorescence-labeled nicked Holliday junction substrate.
Next, we tested whether Dpb11, Slx4, and Mus81–Mms4 form a single protein complex. Indeed, the three proteins comigrated at a size of ∼33 S (Supplemental Fig. S8B, fractions 18–20, apparent molecular weight 1.1–1.2 MDa) when the eluate of an Mms43Flag purification from G2/M cells was subjected to a glycerol gradient centrifugation. When we analyzed the complex architecture by a two-hybrid approach, we detected a direct interaction of Dpb11 and Mms4 that is independent of Slx4 (Supplemental Fig. S8C). Moreover, when we immunoprecipitated Mms43Flag in the slx4-S486A background, we observed a reduction of Slx4, but not Dpb11, binding to Mms43Flag (Fig. 5B). These findings thus suggest that Dpb11, Slx4, and Mus81–Mms4 are part of a multiprotein complex in which Dpb11 acts as a bridge between Slx4 and Mus81–Mms4.
We observed that Dpb11 and Slx4 could be partially eluted from Mms4-containing beads using λ-phosphatase treatment (Supplemental Fig. S8D), suggesting that the binding is at least in part dependent on protein phosphorylation. Previous work has established that Mus81 activity is decisively up-regulated in mitosis in response to a sequential phosphorylation of Mms4 by CDK and the Polo-like kinase Cdc5 (Matos et al. 2011; Gallo-Fernández et al. 2012; Saugar et al. 2013; Szakal and Branzei 2013). We therefore used two systems to interfere with Cdc5 activity: the cdc5-as1 analog-sensitive allele, which we inhibited using chloromethylketone (CMK) (Snead et al. 2007), and transcriptional shutoff of pGAL-CDC5 using glucose repression. Both types of Cdc5 inactivation resulted in a loss of the slower-migrating species of Mms4 in gels and at the same time diminished the binding of Dpb11 and Slx4 to Mms43Flag (Fig. 5C; Supplemental Fig. S9A). In order to rule out indirect effects, we tested whether Cdk1 activity was uninfluenced under conditions of Cdc5 inhibition/shutoff and saw that neither the interaction between Slx4 and Dpb11 nor phosphorylation of a CDK target site on Rad9 (T474) (Pfander and Diffley 2011) was influenced by Cdc5 inactivation (Supplemental Fig. S9B,C). Together with our results on the architecture of the Slx4–Dpb11–Mms4–Mus81 complex, these experiments suggest that binding of Mms4 to Dpb11 is regulated by Cdc5 phosphorylation.
We also tested whether the formation of the Slx4–Dpb11–Mms4–Mus81 was regulated upon DNA damage. We found that Mms43Flag bound similar amounts of Dpb11 and Slx4 after phleomycin or mock treatment of G2/M-arrested cells (Supplemental Fig. S9D). Moreover, we could also observe formation of the Slx4–Dpb11–Mms4–Mus81 complex during recovery from MMS pulse treatment during S phase (Fig. 5D). However, this binding occurred only once Cdc5 became active, as visualized by the slower-migrating form of Mms4, indicating that even after DNA damage, the Dpb11–Mms4 interaction is dependent on Cdc5 (Fig. 5D).
Given that the cell cycle regulation of Mus81 activity and the cell cycle regulation of the Slx4–Dpb11–Mms4–Mus81 complex formation have the same requirements, we tested whether the up-regulation of Mus81 nuclease activity requires Slx4 and Dpb11. We analyzed in vitro resolution of nicked Holliday junctions, Holliday junctions, and model replication fork structures on immunopurified Mus81–Mms4 and found that the enhanced activity of mitotic Mus81 is similar, independently of whether Mus81 was purified from wild-type or slx4-S486A cells (Fig. 5E; Supplemental Fig. S9E). Therefore, we conclude that cell cycle kinases regulate Mus81 by at least two mechanisms: direct up-regulation of the catalytic activity, which can be reconstituted in vitro, and an up-regulation through formation of an Slx4–Dpb11–Mms4–Mus81 complex, which could be seen in vivo.
The DNA damage checkpoint regulates the Slx4–Dpb11-dependent Mus81 function
The DNA damage checkpoint prevents collapse of stalled replication forks and thereby is fundamentally required for all aspects of the response to stalled replication forks (Branzei and Foiani 2010). Moreover, the checkpoint was also suggested to counteract Cdc5-dependent Mus81 activation, since premature Mms4 phosphorylation by Cdc5 was observed after MMS treatment of checkpoint-deficient cells (Szakal and Branzei 2013). Possible explanations for this phenomenon are a faster S-phase progression in the checkpoint mutants or a direct inhibition of Cdc5 activity by the checkpoint (Zhang et al. 2009).
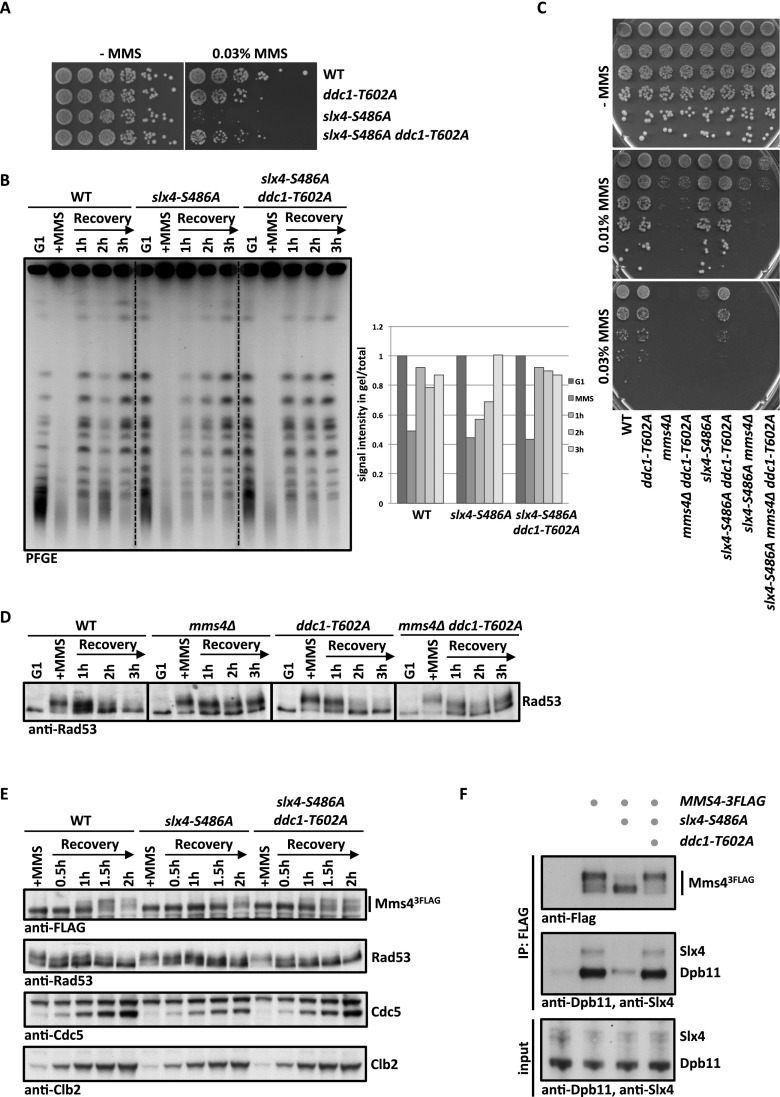
To address these possibilities, we investigated the influence of the DNA damage checkpoint on Slx4–Dpb11–Mms4–Mus81 complex function. Interestingly, we found that a partial defect in DNA damage checkpoint signaling alleviated the phenotypes of the slx4-S486A mutant (Fig. 6A,B; Supplemental Fig. S10A,B; see also Ohouo et al. 2012). In these experiments, we used three distinct mutants, which were partially impaired in checkpoint signaling: ddc1-T602A (defective in Dpb11-dependent Rad9 recruitment (Puddu et al. 2008), dot1Δ (defective in chromatin-dependent Rad9 recruitment) (Giannattasio et al. 2005), and rad53-3HA (a hypomorphic Rad53 allele) (Cordon-Preciado et al. 2006). All three mutants partially suppressed the hypersensitivity of slx4-S486A to chronic exposure of MMS (Fig. 6A; Supplemental Fig. S10A). Furthermore, the recovery from MMS treatment as judged by the reappearance of fully replicated chromosomes in PFGE and reappearance of unphosphorylated Rad53 was enhanced in slx4-S486A ddc1-T602A cells compared with slx4-S486A cells (Fig. 6B; Supplemental Fig. S10B).
Figure 6.
Partial inactivation of the DNA damage checkpoint rescues slx4-S486A phenotypes in an MMS4-dependent manner. (A) The DNA damage repsonse defect of slx4-S486A is suppressed by partial inactivation of the DNA damage checkpoint. Wild type (WT), slx4-S486A, the partial checkpoint mutant ddc1-T602A, and the slx4-S486A ddc1-T602A double mutant were spotted in fivefold serial dilutions on MMS-containing plates. (B) The prolonged replication fork stalling of the slx4-S486A mutant is rescued by the ddc1-T602A mutation. Cells were cell cycle-synchronized and treated with a 30-min pulse of 0.033% MMS in S phase. Recovery of fully replicated chromosomes was analyzed by pulsed-field gel electrophoresis. Quantification as in Figure 3D. (C) A complete defect in Mus81 activity (mms4Δ) cannot be rescued by checkpoint inactivation. The MMS hypersensitivity phenotypes of slx4-S486A, mms4Δ, and ddc1-T602A mutants and double and triple mutant combinations were analyzed as in A. (D) The checkpoint recovery defect of mms4Δ mutants is not rescued by a partial checkpoint mutant. Cells were treated as in B, and checkpoint activity was measured by Rad53 phosphorylation. (E,F) Cdc5-dependent hyperphosphorylation of Mms4 and concomitant binding to Dpb11 and Slx4 occur earlier during recovery from replication fork stalling in slx4-S486A ddc1-T602A double mutants compared with slx4-S486A mutants. (E) Cells were treated with a 40-min pulse of 0.033% MMS in S phase. The Cdc5-dependent Mms43Flag phosphorylation shift was measured by anti-Flag Western blot, checkpoint activity was measured by Rad53 phosphorylation, and cell cycle progression was followed by anti-Clb2 and anti-Cdc5 Westerns. (F) Wild-type, slx4-S486A, and slx4-S486A ddc1-T602A cells that contain MMS43Flag were harvested during the recovery phase (2.5 h after MMS removal) and subjected to anti-Flag immunoprecipitation. Coimmunoprecipitation samples were tested for binding to Dpb11 and Slx4.
A plausible interpretation of these results is that a partial inactivation of the checkpoint may compensate for a reduced or delayed formation of the Slx4–Dpb11–Mms4–Mus81 complex. Such compensation may occur by either a direct up-regulation of the Slx4–Dpb11–Mms4–Mus81 complex or hyperactivation of a Mus81-independent salvage pathway. We therefore tested whether the observed rescue would depend on Mms4. Consistent with a direct influence of the checkpoint on the Slx4–Dpb11–Mms4–Mus81 complex, a partial inactivation of the checkpoint did not rescue the MMS hypersensitivity of the mms4Δ or mms4Δ slx4-S486A mutants (Fig. 6C). In contrast, the sgs1Δ slx4-S486A or yen1Δ slx4-S486A double mutants could be rescued by additional mutation of ddc1-T602A (Supplemental Fig. S10C), suggesting that neither STR nor Yen1 activity is required for the rescue. Furthermore, mms4Δ ddc1-T602A mutants show a slow checkpoint recovery after a pulse of MMS in S phase that is similar to mms4Δ cells (Fig. 6D). These results suggest that the rescue of slx4-S486A mutants upon partial checkpoint inactivation is due to the action of Mms4–Mus81.
Furthermore, when we transiently exposed cells to MMS during S phase and released them into a G2/M arrest, we observed that the Cdc5-dependent phosphorylation shift of Mms4, which in this experiment serves as a marker for the interaction with Slx4–Dpb11, was slightly delayed in slx4-S486A cells compared with wild-type cells (Fig. 6E), probably because of a slower S-phase progression (see Fig. 3C). Importantly, the additional partial inactivation of the checkpoint (slx4-S486A ddc1-T602A) (Fig. 6E,F) allowed Cdc5-dependent Mms4 phosphorylation to occur earlier. Concomitantly, the binding of Mms4 to Dpb11 and Slx4 was rescued by partial checkpoint inactivation when immunoprecipitations were performed during the recovery phase (Fig. 6F). The occurrence of Mms4 phosphorylation in the two mutants inversely correlated with DNA damage checkpoint activation (Rad53 phosphorylation) (Fig. 6E). It needs to be emphasized that Slx4–Dpb11 interaction is reduced, but not abolished, in the slx4-S486A mutant (Figs. 1B, 3A). The results in Figure 6, E and F, therefore suggest that the Slx4–Dpb11–Mms4–Mus81 complex can form earlier and potentially to a larger extent in the slx4-S486A ddc1-T602A mutant compared with the slx4-S486A single mutant. This offers a straightforward explanation for the rescue of the slx4-S486A mutant phenotypes by partial inactivation of the DNA damage checkpoint.
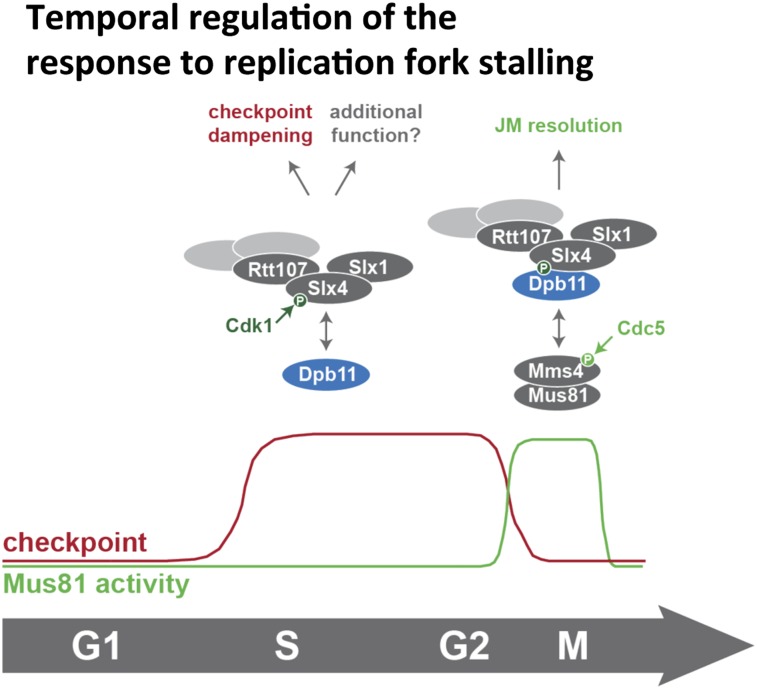
Taken together, we therefore identified an intricate regulatory mechanism of the Mus81 endonuclease, which critically depends on the formation of an Slx4–Dpb11–Mms4–Mus81 complex. The formation of this complex is activated by cell cycle stage-specific signaling and antagonized by the DNA damage checkpoint. Remarkably, complex formation and the direct control of Mus81 catalytic activity occur with similar timing, at the G2/M transition (Fig. 7).
Figure 7.
Model of the temporal response to replication fork stalling and its regulation by Slx4–Dpb11 complexes.
Discussion
In this study, we describe a new facet of JM resolution following the bypass of DNA damage via template switch recombination. We describe a multiprotein complex containing Slx4, Dpb11, and Mus81–Mms4. This complex is cell cycle-controlled by at least two mechanisms: Cdk1-dependent phosphorylation of Slx4 and Cdc5-dependent phosphorylation of Mms4, and Dpb11 acts as a reader of both modifications. The conservation of the Slx4–Dpb11/TopBP1 interaction and its cell cycle regulation suggests that a similar complex may be involved in JM resolution in human cells. Importantly, the inhibition of Slx4 binding to Dpb11 causes phenotypes that are indicative of JM resolution defects, and we therefore infer that the association with Slx4 and Dpb11 promotes Mus81 function.
Slx4–Dpb11 multiprotein complex formation correlates with DNA JM resolution
The starting point of our analysis was a multiprotein complex containing Slx4, Dpb11, Slx1, and Rtt107 (Ohouo et al. 2010, 2012). In order to characterize a putative function of this complex in DNA repair, we tested whether the Slx4–Dpb11 complex would transiently interact with DNA repair enzymes and found an interaction with the Mus81–Mms4 structure-specific endonuclease specifically in mitotic cells. Based on the findings that the slx4-S486A mutant impairs complex formation and results in JM resolution defects, we propose that the Slx4–Dpb11 complex regulates Mus81–Mms4 activity. Our binding studies furthermore indicate a direct Dpb11–Mms4 interaction. Given the nature of Dpb11 as a scaffold protein, it appears likely that Dpb11 operates by tethering Mus81 to other activities that collaborate in the resolution reaction or targeting Mus81 to JM structures.
An intricate feature of the Slx4–Dpb11 complex is its complexity, as it involves four scaffold proteins: Dpb11, Slx4, Rtt107, and Mms4. An obvious advantage of such a multiscaffold complex is that its formation depends on several interaction surfaces, which offer numerous possibilities for regulation. The assembly of the complex therefore allows the integration of different cellular signals (for example, cell cycle and DNA damage), or one specific signal may control complex assembly by several mechanisms. Such a setup includes features of multisite phosphorylation systems, which have the ability to create switch-like transitions (Nash et al. 2001). Moreover, a multiscaffold complex may allow the assembly and coordination of different enzymatic activities (see below).
Our work has identified Mus81 as one catalytically active component of the Slx4–Dpb11 complex; a second one could potentially be Slx1. Recently, the Mus81 and Slx1 endonucleases from humans and mice have been shown to cooperate in the resolution of Holliday junctions in an Slx4-dependent manner (Wyatt et al. 2013). While our results suggest that also in budding yeast, Mus81 and Slx1 may be part of the same complex, we did not observe any specific defects in the response to MMS-induced replication fork stalling for slx1Δ cells (Supplemental Fig. S6D). Therefore, we conclude that either Slx1 is not involved in Mus81-dependent JM resolution in budding yeast or a functionally redundant nuclease compensates for the defects of the slx1Δ mutant.
Cell cycle regulation of the response to replication fork stalling and JM resolution
The cellular response to replication fork-stalling DNA lesions is intimately linked to the progression of the cell cycle. First, the primary problem, fork stalling, arises specifically in S phase. Moreover, the cells are required to finish the repair/bypass process at the latest in mitosis, when sister chromatids need to be accurately separated, and any remaining links between the chromatids have to be resolved.
In this study, we characterized two Dpb11 interactors: Slx4 and Mms4. Both proteins are phosphorylation targets of CDKs; however, Mms4 is additionally phosphorylated by the Polo-like kinase Cdc5 (Matos et al. 2011; Gallo-Fernández et al. 2012; Szakal and Branzei 2013). Interestingly, the Slx4–Dpb11 and Mms4–Dpb11 interactions display distinct cell cycle specificities: We observed a strong Slx4–Dpb11 interaction in asynchronous cultures as well as in S-phase and mitotic cells (Figs. 1C, 2C), while the Mms4–Dpb11 interaction was highly specific for mitosis (Fig. 5A). Accordingly, we found that the Mms4–Dpb11 interaction requires Cdc5, suggesting that Dpb11 can act as a reader of phosphorylation events that are initiated by different cell cycle kinases.
The cell cycle regulation of the Mus81–Mms4 association with the Slx4–Dpb11 complex correlates exactly with the known activity profile of Mus81 (Matos et al. 2011). Notably, the multiprotein complex is not the only mechanism of cell cycle regulation: Our in vitro resolution assays suggest that Cdc5 phosphorylation of Mus81–Mms4 directly stimulates Mus81 independently of complex formation. Therefore, we conclude that at least two parallel pathways of cell cycle regulation exist that promote appropriate Mus81 function in mitosis.
To date, it remains uncertain why cells restrict the activity of Mus81 until mitosis. The temporal regulation of Mus81 channels a large proportion of JMs into the Sgs1–Top3–Rmi1 dissolution pathway (Matos et al. 2011; Szakal and Branzei 2013). It has therefore been speculated that Sgs1-dependent dissolution, which leads to a NCO outcome (Ira et al. 2003), may be beneficial for cells dividing by a mitotic cell cycle. A second reason for restricting Mus81 activity may be the necessity to achieve temporal separation of the lesion bypass reaction and the JM resolution reaction (Saugar et al. 2013). Mus81 could impede the bypass reaction, given its relatively broad substrate specificity to a range of DNA structures (e.g., replication forks, D-loop structures, and Holliday junctions).
Intriguingly, the differences in the temporal regulation of the Slx4–Dpb11 and Mms4–Dpb11 interactions suggest that the composition of the Slx4–Dpb11 complex changes from the replication-associated template switch to the resolution reaction. Supporting the idea of several distinct Slx4–Dpb11 complexes is the fact that not all features of the Slx4–Dpb11 interaction (for example, enhanced binding after DNA damage) are seen in the Slx4–Dpb11–Mms4–Mus81 complex. It therefore appears plausible that Slx4–Dpb11 may associate with stalled replication forks already in S phase, while Mus81–Mms4 joins the complex in mitosis. It is tempting to speculate that the Slx4–Dpb11 complex may chaperone the DNA lesion site/repair intermediates until resolution (Fig. 7).
Regulation of JM resolution by the DNA damage checkpoint
The DNA damage checkpoint antagonizes JM resolution by Mus81 (Fig. 6; Szakal and Branzei 2013), and it has been suggested that Slx4 may act as negative regulator (“dampener”) of the checkpoint by competing with binding of the checkpoint mediator Rad9 to Dpb11 (Ohouo et al. 2012). The JM resolution phenotypes of the slx4-S486A mutant could therefore, in principle, be explained by an indirect effect arising from checkpoint hyperactivation. Given the extensive ties between checkpoint and DNA repair pathways, the presented in vivo experiments cannot rule out a contribution of checkpoint misregulation to the observed JM resolution phenotypes.
We favor, however, a more direct role of Slx4 and Dpb11 in JM resolution for two reasons. First, the formation of the Slx4–Dpb11–Mms4–Mus81 complex and its regulation correlate with the temporal activation of Mus81. Second, the “dampener” model cannot account for all observed phenotypes. For example, the rescue of the MMS hypersensitivity of the slx4-S486A mutant by a covalent fusion with Dpb11 cannot be explained by competition, since in the fusion mutant, cells express two copies of full-length Dpb11 (one endogenous, one fused to Slx4), and therefore even more Dpb11 molecules (not less) are able to engage in checkpoint signaling complexes. Moreover, our analysis of RPA foci suggests that DNA lesions or repair intermediates persist and accumulate in the absence of a functional Slx4–Dpb11 complex, indicative of a role for Slx4 and Dpb11 in DNA repair.
Importantly, we found that the checkpoint regulates the formation of the Slx4–Dpb11–Mms4–Mus81 complex: Partial inhibition of the checkpoint enables Cdc5-dependent hyperphosphorylation of Mms4, which allows Dpb11 binding to occur earlier during the recovery from an MMS pulse and thereby reverses the effect of the slx4 mutant. These findings suggest that at least in the slx4-S486A mutant background, the DNA damage checkpoint antagonizes the Slx4–Dpb11–Mms4–Mus81 complex. Partial inactivation of the checkpoint may therefore extend the temporal window during which Mus81 is active, which we propose to be beneficial in mutants with reduced JM resolution activity such as slx4-S486A. Whether this inhibitory mechanism takes place on the level of Cdc5 regulation in general (Zhang et al. 2009; Matos et al. 2013) or by specifically regulating Mms4 phosphorylation by Cdc5 remains to be determined. The important implication of this finding is that the activation of Mus81 is temporally restricted by two pathways: activation by cell cycle kinases and inhibition by the DNA damage checkpoint.
The Slx4–Dpb11 complex is conserved among eukaryotes
In addition to the mechanistic studies of the budding yeast Slx4–Dpb11 complex, we also provide the first evidence that at least parts of this complex may be found in human cells as well, since Slx4 and TopBP1 interact in a manner that depends on CDK phosphorylation of Slx4. It is worth noting that not all aspects of the protein network that controls resolution of JMs are conserved through evolution: While in human cells, Slx4 binds directly to Mus81–Eme1, this interaction appears to be absent in budding yeast (Fekairi et al. 2009; Muñoz et al. 2009; Svendsen et al. 2009; Schwartz et al. 2012). Given that both Slx4 and Mms4 bind to Dpb11, our data suggest that Dpb11 may serve as a bridge between the two proteins. Although a direct interaction between Slx4 and Mus81–Mms4 cannot be definitively excluded, it appears as if the bridging interaction with Dpb11 in yeast may replace the direct interaction of Slx4 and Mus81 in human cells. Importantly, similar to our results in yeast, a stimulation of Slx4 binding to Mus81–Eme1 after phosphorylation by CDK and Polo-like kinase was observed in mitotic human cells as well (Wyatt et al. 2013). At this point, it seems therefore very likely that in both systems, JM resolution is promoted by a cell cycle-regulated complex containing several scaffold proteins.
Materials and methods
Yeast strains
All yeast strains are based on W303. Genotypes are listed in Supplemental Material.
Interaction assays
Coimmunoprecipitations of yeast extracts were performed using anti-Flag agarose resin (Sigma). Bound proteins were eluted with 3× Flag-peptide (Sigma).
For GST pull-downs, GST-Dpb11 or GST-tagged protein fragments were recombinantly expressed and purified as described (Pfander and Diffley 2011). Pull-downs were performed with ammonium sulphate-precipitated (57%) yeast extracts on glutathione sepharose 4B (GE Healthcare).
For coimmunoprecipitations from HEK293T, GFP-tagged proteins were transiently overexpressed and precipitated using GFP-Trap magnetic beads (Chromotek).
Nuclease activity assays
Nuclease assays on Mms43Flag immunoprecipitations were done as described (Matos et al. 2011).
DNA gel electrophoresis
PFGE and 2D gel analysis of DNA intermediates were performed as previously described (Karras and Jentsch 2010; Szakal and Branzei 2013).
Mutation and recombination assays
Mutation rates were determined using a CAN1 forward mutation assay. Interchromosomal recombination rates were determined using a direct repeat system using leu2 heteroalleles (Aguilera and Klein 1988). CO rates were determined using a system harboring two arg4 alleles on chromosome V and VIII (Robert et al. 2006; Szakal and Branzei 2013). In all, rates were determined by fluctuation analysis using a maximum likelihood approximation (Pfander et al. 2005).
Microscopy and immunofluorescence
Microscopy experiments were carried out as described (Germann et al. 2014).
A detailed methods description is provided in the Supplemental Material.
Acknowledgments
We thank U. Kagerer for excellent technical assistance, and C. Biertümpfel, J. Diffley, and H. Klein for providing plasmids, strains, and antibodies. We are grateful to Z. Storchova for help with microscopy analysis of RPA foci and thank S. Jentsch, M. Räschle, Z. Storchova, and members of the Jentsch and Pfander laboratories for stimulating discussion and critical reading of the manuscript. We thank the Max-Planck Institute of Biochemistry core facility for MS analysis. This work was supported by the Max-Planck Society and the German Research Council (Deutsche Forschungsgemeinschaft) to B.P.; the European Research Council, the Italian Association for International Cancer Research, and The Italian Foundation for Cancer Research (FIRC) to D.B.; and the Danish Agency for Science, Technology, and Innovation, the Villum Kann Rasmussen Foundation, the Lundbeck Foundation, and the European Research Council to M.L. S.C.S.B. is supported by a fellowship from the German Chemical Industry Association (VCI). B.S and D.B. performed the 2D gel experiments of Figure 4A and Supplemental Figure S6A–C and analyzed the data. M.L. performed the imaging experiments of Figure 4E and Supplemental Figure S7 and analyzed the data. J.M. performed the in vitro nuclease assays of Figure 5E and Supplemental Figure S9E and analyzed the data. B.H.H. did the bioinformatics analysis of Supplemental Figure S2. All other experiments were performed and analyzed by D.G., L.N.P., S.C.S.B, L.W., S.S., and B.P. B.P. wrote the paper, and all authors commented on the manuscript.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.240515.114.
References
- Aguilera A, Klein HL 1988. Genetic control of intrachromosomal recombination in Saccharomyces cerevisiae. I. Isolation and genetic characterization of hyper-recombination mutations. Genetics 119: 779–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop AC, Ubersax JA, Petsch DT, Matheos DP, Gray NS, Blethrow J, Shimizu E, Tsien JZ, Schultz PG, Rose MD, et al. 2000. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature 407: 395–401 [DOI] [PubMed] [Google Scholar]
- Blanco MG, Matos J, Rass U, Ip SCY, West SC 2010. Functional overlap between the structure-specific nucleases Yen1 and Mus81–Mms4 for DNA-damage repair in S. cerevisiae. DNA Repair (Amst) 9: 394–402 [DOI] [PubMed] [Google Scholar]
- Branzei D, Foiani M 2010. Maintaining genome stability at the replication fork. Nat Rev Mol Cell Biol 11: 208–219 [DOI] [PubMed] [Google Scholar]
- Branzei D, Vanoli F, Foiani M 2008. SUMOylation regulates Rad18-mediated template switch. Nature 456: 915–920 [DOI] [PubMed] [Google Scholar]
- Cejka P, Plank JL, Bachrati CZ, Hickson ID, Kowalczykowski SC 2010. Rmi1 stimulates decatenation of double Holliday junctions during dissolution by Sgs1–Top3. Nat Struct Mol Biol 17: 1377–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K-L, North PS, Hickson ID 2007. BLM is required for faithful chromosome segregation and its localization defines a class of ultrafine anaphase bridges. EMBO J 26: 3397–3409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordon-Preciado V, Ufano S, Bueno A 2006. Limiting amounts of budding yeast Rad53 S-phase checkpoint activity results in increased resistance to DNA alkylation damage. Nucleic Acids Res 34: 5852–5862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulon S 2006. Rad22Rad52-dependent repair of ribosomal DNA repeats cleaved by Slx1–Slx4 endonuclease. Mol Biol Cell 17: 2081–2090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekairi S, Scaglione S, Chahwan C, Taylor ER, Tissier A, Coulon S, Dong M-Q, Ruse C, Yates JR, Russell P, et al. 2009. Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases. Cell 138: 78–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flott S, Alabert C, Toh GW, Toth R, Sugawara N, Campbell DG, Haber JE, Pasero P, Rouse J 2007. Phosphorylation of Slx4 by Mec1 and Tel1 regulates the single-strand annealing mode of DNA repair in budding yeast. Mol Cell Biol 27: 6433–6445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricke WM, Brill SJ 2003. Slx1–Slx4 is a second structure-specific endonuclease functionally redundant with Sgs1–Top3. Genes Dev 17: 1768–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard P-HL, Noguchi E, Shanahan P, Russell P 2003. The endogenous Mus81–Eme1 complex resolves Holliday junctions by a nick and counternick mechanism. Mol Cell 12: 747–759 [DOI] [PubMed] [Google Scholar]
- Gallo-Fernández M, Saugar I, Ortiz-Bazán MÁ, Vázquez MV, Tercero JA 2012. Cell cycle-dependent regulation of the nuclease activity of Mus81–Eme1/Mms4. Nucleic Acids Res 40: 8325–8335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia V, Furuya K, Carr AM 2005. Identification and functional analysis of TopBP1 and its homologs. DNA Repair (Amst) 4: 1227–1239 [DOI] [PubMed] [Google Scholar]
- Germann SM, Oestergaard VH, Haas C, Salis P, Motegi A, Lisby M 2011. Dpb11/TopBP1 plays distinct roles in DNA replication, checkpoint response and homologous recombination. DNA Repair (Amst) 10: 210–224 [DOI] [PubMed] [Google Scholar]
- Germann SM, Schramke V, Pedersen RT, Gallina I, Eckert-Boulet N, Oestergaard VH, Lisby M 2014. TopBP1/Dpb11 binds DNA anaphase bridges to prevent genome instability. J Cell Biol 204: 45–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannattasio M, Lazzaro F, Plevani P, Muzi-Falconi M 2005. The DNA damage checkpoint response requires histone H2B ubiquitination by Rad6–Bre1 and H3 methylation by Dot1. J Biol Chem 280: 9879–9886 [DOI] [PubMed] [Google Scholar]
- Harrison JC, Haber JE 2006. Surviving the breakup: the DNA damage checkpoint. Annu Rev Genet 40: 209–235 [DOI] [PubMed] [Google Scholar]
- Ho CK, Mazón G, Lam AF, Symington LS 2010. Mus81 and Yen1 promote reciprocal exchange during mitotic recombination to maintain genome integrity in budding yeast. Mol Cell 40: 988–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt LJ, Tuch BB, Villén J, Johnson AD, Gygi SP, Morgan DO 2009. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science 325: 1682–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Interthal H, Heyer WD 2000. MUS81 encodes a novel helix–hairpin–helix protein involved in the response to UV- and methylation-induced DNA damage in Saccharomyces cerevisiae. Mol Gen Genet 263: 812–827 [DOI] [PubMed] [Google Scholar]
- Ip SCY, Rass U, Blanco MG, Flynn HR, Skehel JM, West SC 2008. Identification of Holliday junction resolvases from humans and yeast. Nature 456: 357–361 [DOI] [PubMed] [Google Scholar]
- Ira G, Malkova A, Liberi G, Foiani M, Haber JE 2003. Srs2 and Sgs1–Top3 suppress crossovers during double-strand break repair in yeast. Cell 115: 401–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karras GI, Jentsch S 2010. The RAD6 DNA damage tolerance pathway operates uncoupled from the replication fork and is functional beyond S phase. Cell 141: 255–267 [DOI] [PubMed] [Google Scholar]
- Li M, Brill SJ 2005. Roles of SGS1, MUS81, and RAD51 in the repair of lagging-strand replication defects in Saccharomyces cerevisiae. Curr Genet 48: 213–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberi G, Maffioletti G, Lucca C, Chiolo I, Baryshnikova A, Cotta-Ramusino C, Lopes M, Pellicioli A, Haber JE, Foiani M 2005. Rad51-dependent DNA structures accumulate at damaged replication forks in sgs1 mutants defective in the yeast ortholog of BLM RecQ helicase. Genes Dev 19: 339–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankouri HW, Ashton TM, Hickson ID 2011. Holliday junction-containing DNA structures persist in cells lacking Sgs1 or Top3 following exposure to DNA damage. Proc Natl Acad Sci 108: 4944–4949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankouri HW, Huttner D, Hickson ID 2013. How unfinished business from S-phase affects mitosis and beyond. EMBO J 32: 2661–2671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos J, Blanco MG, Maslen S, Skehel JM, West SC 2011. Regulatory control of the resolution of DNA recombination intermediates during meiosis and mitosis. Cell 147: 158–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos J, Blanco MG, West SC 2013. Cell-cycle kinases coordinate the resolution of recombination intermediates with chromosome segregation. Cell Rep 1–11 [DOI] [PubMed] [Google Scholar]
- Mordes DA, Nam EA, Cortez D 2008. Dpb11 activates the Mec1–Ddc2 complex. Proc Natl Acad Sci 105: 18730–18734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen JR, Kaliraman V, Ibrahim SS, Brill SJ 2001. Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics 157: 103–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz IM, Hain K, Déclais A-C, Gardiner M, Toh GW, Sanchez-Pulido L, Heuckmann JM, Toth R, Macartney T, Eppink B, et al. 2009. Coordination of structure-specific nucleases by human SLX4/BTBD12 is required for DNA repair. Mol Cell 35: 116–127 [DOI] [PubMed] [Google Scholar]
- Nash P, Tang X, Orlicky S, Chen Q, Gertler FB, Mendenhall MD, Sicheri F, Pawson T, Tyers M 2001. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature 414: 514–521 [DOI] [PubMed] [Google Scholar]
- Navadgi-Patil VM, Burgers PM 2008. Yeast DNA replication protein Dpb11 activates the Mec1/ATR checkpoint kinase. J Biol Chem 283: 35853–35859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohouo PY, Bastos de Oliveira FM, Almeida BS, Smolka MB 2010. DNA damage signaling recruits the Rtt107–Slx4 scaffolds via Dpb11 to mediate replication stress response. Mol Cell 39: 300–306 [DOI] [PubMed] [Google Scholar]
- Ohouo PY, de Oliveira FMB, Liu Y, Ma CJ, Smolka MB 2012. DNA-repair scaffolds dampen checkpoint signalling by counteracting the adaptor Rad9. Nature 493: 120–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfander B, Diffley JFX 2011. Dpb11 coordinates Mec1 kinase activation with cell cycle-regulated Rad9 recruitment. EMBO J 30: 4897–4907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfander B, Moldovan GL, Sacher M, Hoege C, Jentsch S 2005. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature 436: 428–433 [DOI] [PubMed] [Google Scholar]
- Prakash S, Johnson RE, Prakash L 2005. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu Rev Biochem 74: 317–353 [DOI] [PubMed] [Google Scholar]
- Puddu F, Granata M, Di Nola L, Balestrini A, Piergiovanni G, Lazzaro F, Giannattasio M, Plevani P, Muzi-Falconi M 2008. Phosphorylation of the budding yeast 9-1-1 complex is required for Dpb11 function in the full activation of the UV-induced DNA damage checkpoint. Mol Cell Biol 28: 4782–4793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappas M, Oliver AW, Pearl LH 2011. Structure and function of the Rad9-binding region of the DNA-damage checkpoint adaptor TopBP1. Nucleic Acids Res 39: 313–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert T, Dervins D, Fabre F, Gangloff S 2006. Mrc1 and Srs2 are major actors in the regulation of spontaneous crossover. EMBO J 25: 2837–2846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts TM, Kobor MS, Bastin-Shanower SA, Ii M, Horte SA, Gin JW, Emili A, Rine J, Brill SJ, Brown GW 2006. Slx4 regulates DNA damage checkpoint-dependent phosphorylation of the BRCT domain protein Rtt107/Esc4. Mol Biol Cell 17: 539–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saugar I, Vazquez MV, Gallo-Fernandez M, Ortiz-Bazan MA, Segurado M, Calzada A, Tercero JA 2013. Temporal regulation of the Mus81–Mms4 endonuclease ensures cell survival under conditions of DNA damage. Nucleic Acids Res 41: 8943–8958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz EK, Wright WD, Ehmsen KT, Evans JE, Stahlberg H, Heyer WD 2012. Mus81–Mms4 functions as a single heterodimer to cleave nicked intermediates in recombinational DNA repair. Mol Cell Biol 32: 3065–3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snead JL, Sullivan M, Lowery DM, Cohen MS, Zhang C, Randle DH, Taunton J, Yaffe MB, Morgan DO, Shokat KM 2007. A coupled chemical-genetic and bioinformatic approach to Polo-like kinase pathway exploration. Chem Biol 14: 1261–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svendsen JM, Smogorzewska A, Sowa ME, O’Connell BC, Gygi SP, Elledge SJ, Harper JW 2009. Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell 138: 63–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szakal B, Branzei D 2013. Premature Cdk1/Cdc5/Mus81 pathway activation induces aberrant replication and deleterious crossover. EMBO J 32: 1155–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Umemori T, Hirai K, Muramatsu S, Kamimura Y, Araki H 2007. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature 445: 328–332 [DOI] [PubMed] [Google Scholar]
- Tomkinson AE, Bardwell AJ, Bardwell L, Tappe NJ, Friedberg EC 1993. Yeast DNA repair and recombination proteins Rad1 and Rad10 constitute a single-stranded-DNA endonuclease. Nature 362: 860–862 [DOI] [PubMed] [Google Scholar]
- Wyatt HDM, Sarbajna S, Matos J, West SC 2013. Coordinated actions of SLX1–SLX4 and MUS81–EME1 for Holliday junction resolution in human cells. Mol Cell 52: 234–247 [DOI] [PubMed] [Google Scholar]
- Yu X 2003. The BRCT domain is a phospho-protein binding domain. Science 302: 639–642 [DOI] [PubMed] [Google Scholar]
- Zegerman P, Diffley JFX 2007. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature 445: 281–285 [DOI] [PubMed] [Google Scholar]
- Zhang T, Nirantar S, Lim HH, Sinha I, Surana U 2009. DNA damage checkpoint maintains Cdh1 in an active state to inhibit anaphase progression. Dev Cell 17: 541–551 [DOI] [PubMed] [Google Scholar]