Abstract
Inositol requiring enzyme-1 (IRE1) is highly conserved from yeasts to humans. Upon endoplasmic reticulum (ER) stress, IRE1 activates X-box-binding protein 1 (XBP1) by unconventional splicing of XBP1 mRNA, which activates unfolded protein response (UPR) to restore ER homeostasis. In mice, IRE1α plays an essential role in extraembryonic tissues. However, its precise action during the early stage of development is unknown. In this study, the gain and loss-of-function analyses were used to investigate the function of Xenopus IRE1α (xIRE1α). The effects of xIRE1α during embryo development were detected with RT-PCR and whole mount in situ hybridization. ER stress was induced by tunicamycin. The apoptotic cells were measured by TUNNEL assays. Although both gain and loss of xIRE1α function had no significant effect on Xenopus embryogenesis, knockdown of xIRE1α could rescue tunicamycin-induced developmental defects and apoptosis. The finding indicates that xIRE1α is not required for embryogenesis but is required for tunicamycin-induced developmental defects and apoptosis in Xenopus laevis.
Keywords: IRE1α, Xenopus laevis, tunicamycin, developmental defects
INTRODUCTION
The endoplasmic reticulum (ER) plays an important role in the synthesis and modification of secretary and membrane proteins in all eukaryotic cells. However, the function of the ER is disrupted when the inflow of unfolded polypeptide chains exceeds the folding or processing capacity of the ER, which is called ER stress. A series of adaptive responses, called the unfolded protein response (UPR), can up-regulate the transcription of various genes to increase the activity of protein-folding and protein-degradation for maintaining ER homeostasis. However, the persistent activation of UPR will trigger the cell death pathway because of prolonged ER dysfunction.
The UPR is transduced through three forms of ER-resident transmembrane sensors: IRE1, PKR-like ER Kinase (PERK), and activating transcription factor 6 (ATF6). The IRE1-dependent branch is highly evolutionarily conserved from yeasts to humans[1]-[4]. IRE1 is an ER-located type I transmembrane protein with a kinase domain and RNase domain in the cytosolic region. It plays a central role in the ER stress response. Upon ER stress, IRE1 is activated and the signal is transduced to the cytosol by the sequential dimerization/multimerization, trans-autophosphorylation, and activation of its endoribonuclease[5]-[7]. The specific activity of the endoribonuclease is responsible for the unconventional cytosolic splicing of HAC1 in yeast or excision of the 26-nucleotide intron of the X-box-binding protein 1 (XBP-1) transcription factor in metazoan organisms. The removal of the intron causes a frame shift and results in production of a spliced XBP1 mRNA, and encodes the active transcription factor XBP1s from the unspliced XBP1 mRNA (XBP1U)[8],[9]. In Xenopus, there is another form of frame shift generated by the complete removal of exon 4[10]. The active form of XBP1 up-regulates chaperones to enhance protein folding and genes that mediate ER-associated degradation (ERAD) to target degradation of misfolded proteins in ER stress response[4]. Therefore, the splicing of XBP1 mRNA is a major event to mediate the UPR. Tunicamycin (TM) is an inducer of UPR and it was reported that TM induced severe ER stress, apparently with increased XBP1 splicing, and caused serious developmental defects in Xenopus[11].
In mammals, two IRE1 paralogues (IRE1α and IRE1β) have been reported. IRE1α is expressed ubiquitously and IRE1β primarily in intestinal epithelial cells. The two isoforms appear to have the same in vitro activities; subcellular localizations, and downstream target (XBP1 mRNA)[5]-[7]. However, they play a different role in embryo development. IRE1β deletion does not lead to significant developmental defects in mammals[12],[13]. IRE1α is confirmed to be expressed ubiquitously in fetal and adult mice and to be essential for mammalian developmental processes[5]. Knockout of IRE1α caused widespread developmental defects, leading to embryonic death after 12.5 days of gestation in mice[14], which suggested an important role of IRE1α during development. To date, it has been reported that, during development, IRE1α is required for B-cell differentiation[14], exocrine tissues[15] and placental development and embryonic viability[16]. However, the data could not explain why IRE1α-/- embryos died at early stage.
Recently, two IRE1 homologues in Xenopus (xIRE1α and xIRE1β) were identified, and differentially expressed during embryogenesis. It was reported that xIRE1β is not only required for cytoplasmic splicing of xXBP1 pre-mRNA but also for mesoderm formation[17]. Although the expression pattern of IRE1α during embryogenesis was reported, the function of IRE1α during embryogenesis was unknown until now. In this study, knockdown and over-expression of IRE1α analyses were used to study the role of IRE1α during Xenopus embryogenesis. The results showed that knockdown of IRE1α had no significant effects on germ layer formation but it rescued TM induced developmental defects in Xenopus (X.) laevis.
MATERIALS AND METHODS
Ethics statement
The care of X. laevis (Nasco), the in vitro fertilization procedure, and embryo study were performed according to protocols approved by the Ethics Committee of Nanjing Medical University (permit number: 20080205).
Embryo manipulation and TM treatment
X. laevis eggs were obtained via in vitro fertilization, dejellied with 2% cysteine hydrochloride (pH 7.8–8.0) and cultured in 0.1×MBSH (8.8 mmol/L NaCl, 0.24 mmol/L NaHCO3, 0.1 mmol/L KCl, 0.082 mmol/L MgSO4, 0.041 mmol/L CaCl2, 0. 033 mmol/L Ca(NO3)2, and 1 mmol/L HEPES, pH 7.4). Embryonic stages were determined according to Nieuwkoop and Faber[18]. TM (Sigma, St. Louis, MO, USA) was dissolved in dimethylsulfoxide (DMSO) to obtain a stock solution (5 mg/mL). Four-cell stage embryos were cultured in 0.1×MBSH containing TM (2 μg/μL). Embryos were grown in 0.1×MBSH without TM were used as control. When control sibling embryos reached mid-blastula stage, treated embryos were transferred to fresh culture media without TM.
In vitro transcription of RNA, antisense morpholino oligonucleotides and microinjection
Plasmids pCS2+-xIRE1α were linearized with NotI. Capped mRNA for microinjection was synthesized with SP6 mMessage mMachineTM kit (Ambion, USA) and cleaned-up with RNeasy kit (Qiagen, Germany). The antisense morpholino oligonucleotide (Gene Tools, USA) used for xIRE1α functional knockdown (IRE1α MO) was 5′-AAGAGAACCGCCAGAGGCGCCATGT-3′; an antisense morpholino oligonucleotide XBP(C)MO designed to inhibit the cytoplasmic splicing of xXBP1 was 5′-GACATCTGGGCCTGCTCCTGCTGCA-3′; a standard control mopholino oligonucleotide, CoMO) was 5′-CCTCTTACCTCAGTTACAATTTATA-3′. One nanogram of xIRE1α and 50 ng of IRE1α MO were injected into four blastomeres at 4-cell stage for scoring the phenotype and marker gene analysis.
Gene expression analysis
Total RNA from embryos was extracted, digested by DNase I and purified by RNeasy kit (Qiagen, Hilden,Germany). First strand cDNA was synthesized with RevertAidTM first strand cDNA synthesis kit (Fermentas, Barlington, ON, Canada). Semi-quantitative RT-PCR was performed and primers for xXBP1 splicing and Xbra, Xsox17a, α-actin, XMyoD, and Xnot were described previously[10]. In parallel, ODC was amplified to confirm equal amounts and integrity of different RNA preparations. Real time RT-PCR was performed and primers for chop were as described previously[19].
Whole mount in situ hybridization
Whole mount in situ hybridization was performed according to standard procedures[20].
TUNNEL assay
TUNNEL assay on whole embryos was done using the protocol previously described[21].
Statistical analysis
Statistical analyses were performed with SPSS 15.0 software (SPSS Inc, Chicago, IL, USA). Student's t-test was used to compare the differences among groups. A P value of less than 0.05 was considered to be statistically significant.
RESULTS
IRE1α is not required for early stage development of X. laevis
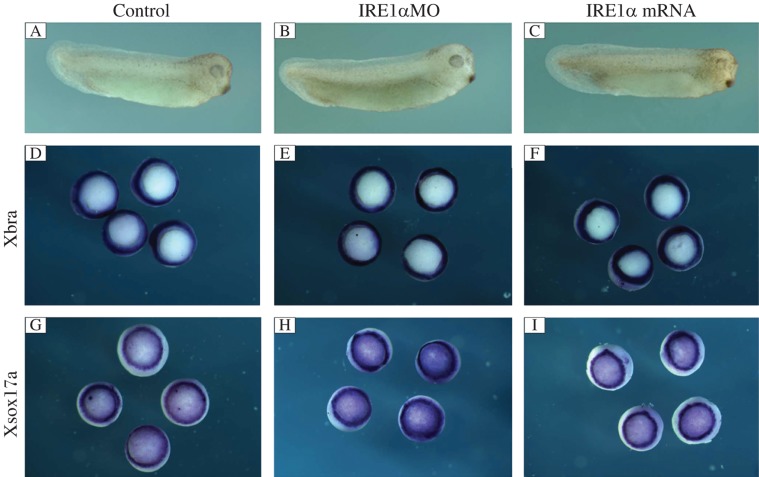
The xIRE1α expression pattern during early stage of development was reported[17], but the role of xIRE1α is still unknown. To elucidate the function of xIRE1α during early embryonic development, the role of xIRE1α was focused on germ layer formation by gain of function (GOF) and loss of function (LOF) experiments. Different doses (from 0.5 to 1.5 ng/embryo) of xIRE1α mRNA were injected into all blastomeres at 4-cell stage, respectively. All the injected embryos at the tailbud stage were nearly normal (Fig. 1C). Whole mount in situ hybridization of embryos injected with 1.5 ng xIRE1α revealed that the expression of the pan-mesodermal marker gene Xbra and the endodermal gene Xsox17a at stage 10.5 was not significantly changed, compared with control MO injected embryos (Fig. 1F and I).
Fig. 1. xIRE1α does not affect mesoderm and endoderm formation.
A: control MO injected embryo at tailbud stage. B and C: embryo injected with 50 ng xIRE1α MO or 1.5 ng xIRE1α mRNA. Overexpression or knockdown of xIRE1α did not change the expression of Xbra (D–F) and Xsox17a (G–I) at stage 10.5.
Next, a LOF analysis was performed by using an antisense morpholino oligonucleotide (IRE1α MO) directed against xIRE1α[22]. To examine the embryonic phenotype in response to xIRE1α knockdown, 50 ng IRE1α MO was injected into 4 blastomeres of 4-cell stage embryos. Interestingly, the injected embryos showed completely different phenotype from that of the xIRE1β knockdown embryos. Even until the tailbud stage, the xIRE1α knockdown embryos still looked normal (Fig. 1B). Further detection of the germ layer marker expression with whole mount in situ hybridization also showed no change (Fig. 1E and H), compared with control MO injected embryos. Therefore, GOF and LOF of IRE1α could not cause an apparent change of phenotype. These results show that xIRE1α is not required for development of X. laevis at early stage.
Knockdown of xIRE1α rescued the developmental defects induced by TM
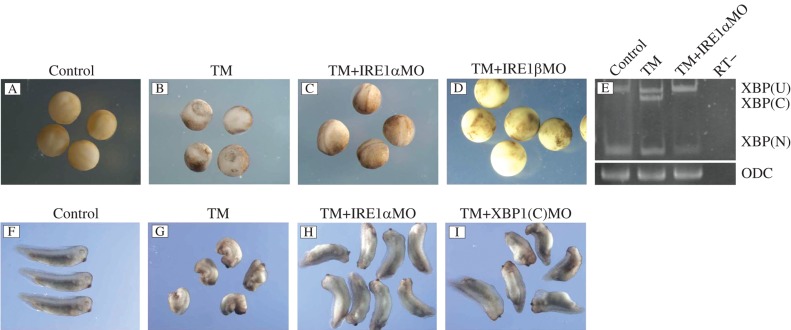
TM is the inducer of UPR, and it was reported that TM induced severe ER stress with an apparent increase in XBP1 splicing and caused serious developmental defects in Xenopus[19]. We speculated that, if XBP1 splicing is repressed, the developmental defects could be rescued. We injected 50 ng XBP1(C) MO into the 4 blastomeres at 4 cell stage embryos treated with 2 μg/mL TM. As shown in Fig. 2, when control MO injected embryos reached the tailbud stage (Fig. 2F), TM-treated embryos still exhibited open neural folds and developed severe inhibition of anterior-posterior axis elongation and loss of head structure (Fig. 2G), while the developmental abnormality caused by TM was rescued in XBP1(C) MO injected embyos. The rescued embryos only showed a shorter axis, compared with the control sibling embryos (Fig. 2I).
Fig. 2. Developmental defects caused by tunicamycin (TM) were rescued with the injection of IRE1αMO and xXBP1(C) MO.
A: control MO injected embryos at stage 18. B: embryos treated with 2 μg/mL TM at stage 18. C: embryos treated with 2 μg/mL TM and injected with 50 ng IRE1αMO at stage 18. D: embryos treated with 2 μg/mL TM and injected with 50 ng IRE1βMO at stage 18. E: rescue effect on XBP1 splicing. TM treatment led to an increase of xXBP1(C) while TM treatment together with xIRE1α MO injection rescued the change in embryos at stage 11. F: control MO injected embryos at stage 32. G: embryos treated with 2 μg/mL TM at stage 32. H: embryos treated with 2 μg/mL TM and injected with 50 ng IRE1αMO at stage 32. I: embryos treated with 2 μg/mL TM and injected with 50 ng XBP1(C) MO at stage 32. RT-: no-reverse transcriptase control.
Since both IRE1α and IRE1β have the same effect on XBP1 splicing, we detected if the rescue effect is dependent on the xIRE1α/XBP1 or xIRE1β/XBP1 pathway. At the neurula stage, untreated embryos developed normal blastopore closure and neural fold formation (Fig. 2A), while TM-treated embryos showed no blastopore closure and no clearly visible neural fold formation (Fig. 2B). We tried to do the rescue with injection of 50 ng of IRE1β MO into the embryos with the same treatment, but the rescue failed and the embryos died before or during gastrulation (Fig. 2D). Then, 50 ng of IRE1α MO was injected, as described above, and successful rescue was observed. The embryos looked nearly normal at the neurula stage (Fig. 2C) and showed a slightly shorter axis at the tailbud stage (Fig. 2H).
We also detected the rescue effect on XBP1 splicing after IRE1α MO injection with RT-PCR. It was shown that at stage 11, in control MO injected embryos, the cytoplasmic splicing was undetectable while the cytoplasmic splice form of xXBP1 (xXBP1(C)) was present in TM-treated embryos (Fig. 2E). After IRE1α MO injection, xXBP1(C) was undetectable.
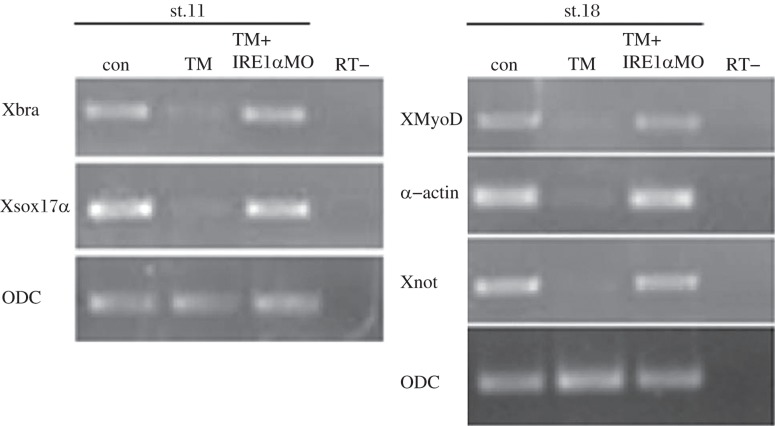
Since TM treatment destroyed germ layer formation, we tested the rescue effect of IRE1α MO on the expression of germ layer markers. As shown in Fig. 3, endodermal gene sox17a, mesodermal gene xbra, XMyoD, and α-actin and neuroectodermal marker Xnot were significantly inhibited in TM-treated embryos, while after knockdown of xIRE1α, the expression of these genes was recovered to nearly normal. These results suggested that knockdown of IRE1α could rescue the developmental defects through the xIRE1α /XBP1 pathway.
Fig. 3. Rescue effects of xIRE1α knockdown on gene expression.
Control, TM treated and rescued embryos were collected at stage 11 or 18 and subjected to RT-PCR. Expression of mesodermal, endodermal, and neuroectodermal genes were inhibited in TM-treated embryos and rescued by xIRE1α knockdown. RT-: no-reverse transcriptase control.
Knockdown of xIRE1α rescued apoptosis induced by TM
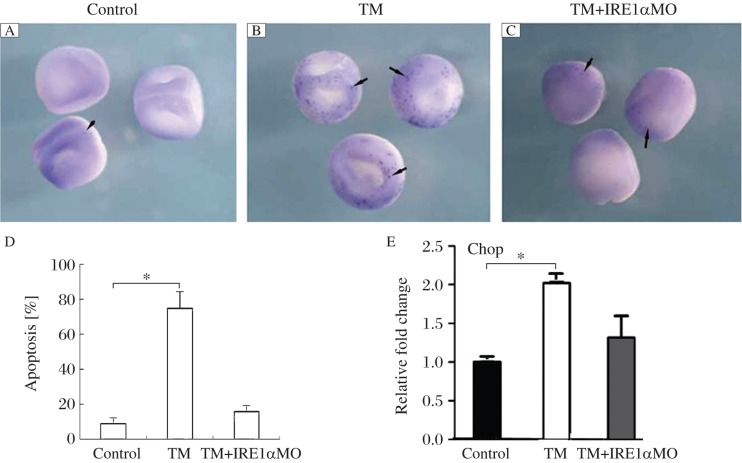
Increased ER stress triggers the apoptosis pathway, which was confirmed in TM treated embryos. The apoptotic cells were detected in most of the treated embryos (75%, n = 40, Fig. 4B), which was one of the possibilities that caused the developmental defects and even death of treated embryos. Then, we detected the rescue effects of xIRE1α knockdown on apoptosis with TUNNEL assay. As shown in Fig. 4C, compared with TM-treated embryos, embryos injected with IRE1α MO and TM treatment showed dramatically decreased apoptotic signal.
Fig. 4. xIRE1α knockdown rescued the apoptosis in embryos of stage 18 with TM treatment by TUNNEL.
A: The control embryos without TM treatment showed few staining indicating apoptosis (arrow). B: The TM-treated embryos showed significant signal for apoptosis (arrows). C: The embryos were injected with 50 ng of IRE1αMO at 4 cell stage, then treated with TM, and the apoptotic signal was decreased (arrows). D: A quantitative presentation is given. E: Expression of CHOP is inhibited in TM-treated embryos and rescued by xIRE1α knockdown. *P < 0.05 compared with control embryos.
CHOP codes for a bZIP transcription factor involved in the apoptosis pathway mediated by ER stress or DNA damage[23]. The higher increase in CHOP expression was observed in embryos treated with TM, while in xIRE1α knockdown embryos, the expression of CHOP decreased nearly to the normal level.These results clearly showed that knockdown of xIRE1α protected embryos from ER stress-induced cell death in X. laevis.
DISCUSSION
It has been established that ER stress is associated with various diseases[24]-[27] and ER stress-related molecules play an important role during embryo development[28]-[30]. IRE1α is the most evolutionarily conserved branch of the UPR and, upon activation, initiates the unconventional splicing of the mRNA encoding the transcriptional factor XBP1 to attenuate ER stress by mediating UPR. In the mammal, IRE1α and IRE1β have the same effect in mediating UPR, but they play different role in embryo development[11],[12]. Recently, both IRE1α and IRE1β were identified in X. lavies. Unexpectedly, besides mediating XBP1splicing, IRE1β played an essential role in germ layer formation[17], which has not been previously reported in mammals. The findings from the mammal model show that the function of IRE1α appears to be more important than that of IRE1β. Thus, we focused our study on the function of IRE1α in germ layer formation during X. lavies embryo development. However, we found that IRE1α is not required for germ layer formation during the early stage development of X. lavies.
Although knockdown of xIRE1α and xIRE1β can inhibit the splicing of xXBP1[17],[21], their phenotype at the early stage of embryo development is different. These data also suggested that XBP1 may be not the only downstream molecule and other pathways mediate the IRE1 function. On the other hand, XBP1 may have functions independent of xIRE1α and xIRE1β. In the studies of XBP1 or IRE1α knockout mice, their different phenotypes also support the possibility.[15],[31]
Here we showed that xIRE1α knockdown did not show effects as strong as the effects caused by knockdown of xIRE1β, which suggests that xIRE1α and xIRE1β play different roles during the development. Furthermore, xIRE1β is essential in germ layer formation, while xIRE1α could be important in organogenesis. In the mammal, IRE1α plays an essential function in extraembryonic tissues and IRE1a disruption causes histological abnormality of the pancreatic acinar and exhibits mild hypoinsulinemia, hyperglycemia, and a low-weight trend[15]. The previous study reported an expression pattern which showed that xIRE1α is expressed in the pancreas[17]. This suggested that xIRE1α could play a role in pancreas formation. The function of hIRE1α and the correlation between pancreas and hIRE1α will be investigated in future study.
Since the endogenous xIRE1α showed no effect on germ layer formation, we then detected the role of xIRE1α activated by ER stress. In a previous report[21], we found that embryos treated with TM showed significant abnormality during development and germ layer formation was obviously inhibited. In those embryos, xXBP1 splicing was enhanced, which suggested that the xIRE1/xXBP1 pathway was involved. We wonder that, if the xIRE1/xXBP1 pathway is inhibited, whether the developmental defects can be rescued. There are two ways to inhibit the xIRE1/xXBP1 pathway, one is to block the splicing of XBP1 to decrease the active form of XBP1; the other is to block the upstream gene IRE1. We tried to knockdown XBP1splicing and xIRE1α expression, and observed that both xXBP1 MO and xIRE1α MO had the rescue effects. However, when we tried to knockdown xIRE1β in TM treated embryos, the embryos died at early stage. These data indicated that it was not the xIRE1β/xXBP1 pathway, but the xIRE1α/xXBP1 pathway that was essential in TM caused developmental defects.
The splicing of XBP1 is increased in the case of overexpression of xXBP1 and TM treatment, but the following phenotype is different, which indicates that XBP1 could induce different kinds of downstream target genes and be multifunctional. In addition to the induction of UPR-related genes by ER stress, recent studies demonstrated that XBP1 also induces genes in cell type- and condition-specific manner[32]. In TM treated embryos xXBP1 showed UPR-related function mainly. Thus, xIRE1α knockdown can get rescue effect in TM treated embryos. Moreover, knockdown of xIRE1α showed better rescue effect than that showed in XBP1 knockdown embryos indicated that xIRE1α has still other functions independent of XBP1.
In this study, the function of xIRE1α was compared during development, with TM treated embryos. The results showed that the phenotypes in TM treated embryos were significantly different from those during development, and indicated that IRE1α, in normal and ER stressed embryos, play different roles during embryogenesis. In summary, we showed that xIRE1α was not required for germ layer formation but was required for XBP1 splicing during embryogenesis, and knockdown of xIRE1α could rescue TM induced developmental defects and apoptosis in X. laevis.
References
- 1.Anelli T, Sitia R. Protein quality control in the early secretory pathway. EMBO J. 2008;27:315–27. doi: 10.1038/sj.emboj.7601974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schröder M. Endoplasmic reticulum stress responses. Cell Mol Life Sci. 2008;65:862–94. doi: 10.1007/s00018-007-7383-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schröder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem. 2005;74:739–89. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- 4.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–29. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 5.Tirasophon W, Welihinda AA, Kaufman RJ. A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev. 1998;12:1812–24. doi: 10.1101/gad.12.12.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang XZ, Harding HP, Zhang Y, Jolicoeur EM, Kuroda M, Ron D. Cloning of mammalian Ire1 reveals diversity in the ER stress responses. EMBO J. 1998;17:5708–17. doi: 10.1093/emboj/17.19.5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iwawaki T, Hosoda A, Okuda T, Kamigori Y, Nomura-Furuwatari C, Kimata Y, et al. Translational control by the ER transmembrane kinase/ ribonuclease IRE1 under ER stress. Nat Cell Biol. 2001;3:158–64. doi: 10.1038/35055065. [DOI] [PubMed] [Google Scholar]
- 8.Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–6. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 9.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–91. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 10.Cao Y, Knöchel S, Oswald F, Donow C, Zhao H, Knöchel W. XBP1 forms a regulatory loop with BMP-4 and suppresses mesodermal and neural differentiation in Xenopus embryos. Mech Dev. 2006;123:84–96. doi: 10.1016/j.mod.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 11.Lipson KL, Fonseca SG, Urano F. Endoplasmic reticulum stress-induced apoptosis and auto-immunity in diabetes. Curr Mol Med. 2006;6:71–7. doi: 10.2174/156652406775574613. [DOI] [PubMed] [Google Scholar]
- 12.Bertolotti A, Wang X, Novoa I, Jungreis R, Schlessinger K, Cho JH, et al. Increased sensitivity to dextran sodium sulfate colitis in IRE1beta-deficient mice. J Clin Invest. 2001;107:585–93. doi: 10.1172/JCI11476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iqbal J, Dai K, Seimon T, Jungreis R, Oyadomari M, Kuriakose G, et al. IRE1beta inhibits chylomicron production by selectively degrading MTP mRNA. Cell Metab. 2008;7:445–55. doi: 10.1016/j.cmet.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang K, Wong HN, Song B, Miller CN, Scheuner D, Kaufman RJ. The unfolded protein response sensor IRE1alpha is required at 2 distinct steps in B cell lymphopoiesis. J Clin Invest. 2005;115:268–81. doi: 10.1172/JCI21848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iwawaki T, Akai R, Kohno K. IRE1a disruption causes histological abnormality of exocrine tissues, increase of blood glucose level, and decrease of serum immunoglobulin level. PLoS ONE. 2010;5:e13052. doi: 10.1371/journal.pone.0013052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iwawaki T, Akai R, Yamanaka S, Kohno K. Function of IRE1 alpha in the placenta is essential for placental development and embryonic viability. Proc Natl Acad Sci U S A. 2009;106:16657–62. doi: 10.1073/pnas.0903775106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yuan L, Cao Y, Oswald F, Knöchel W. IRE1beta is required for mesoderm formation in Xenopus embryos. Mech Dev. 2008;125:207–22. doi: 10.1016/j.mod.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 18.Nieuwkoop PD, Faber J. Normal Table of Xenopus laevis (Daudin), 2nd ed. Elsevier/North Holland; Amsterdam: 1967. pp. 163–88. [Google Scholar]
- 19.Yuan L, Cao Y, Knöchel W. Endoplasmic reticulum stress induced by tunicamycin disables germ layer formation in Xenopus laevis embryos. Dev Dyn. 2007;236:2844–51. doi: 10.1002/dvdy.21299. [DOI] [PubMed] [Google Scholar]
- 20.Harland RM. In situ hybridization: an improved wholemount method for Xenopus embryos. Methods Cell Biol. 1991;36:685–95. doi: 10.1016/s0091-679x(08)60307-6. [DOI] [PubMed] [Google Scholar]
- 21.Hensey C, Gautier J. Programmedcell death during Xenopus development: a spatio-temporal analysis. Dev Biol. 1998;203:36–48. doi: 10.1006/dbio.1998.9028. [DOI] [PubMed] [Google Scholar]
- 22.Yuan Li, Li Xinxin, Feng Jiaojiao, Yin Chenyang, Yuan Fang, Wang Xinru. IRE1α is essential for Xenopus pancreas development. J Biomed Res. 2014;28:123–31. doi: 10.7555/JBR.28.20130076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marciniak SJ, Yun CY, Oyadomari S, Novoa I, Zhang Y, Jungreis R, et al. doi: 10.1101/gad.1250704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matus S, Lisbona F, Torres M, Leon C, Thielen P, Hetz C. The stress rheostat: an interplay between the unfolded protein response (UPR) and autophagy in neurodegeneration. Curr Mol Med. 2008;8:157–72. doi: 10.2174/156652408784221324. [DOI] [PubMed] [Google Scholar]
- 25.Moenner M, Pluquet O, Bouchecareilh M, Chevet E. Integrated endoplasmic reticulum stress responses in cancer. Cancer Res. 2007;67:10631–4. doi: 10.1158/0008-5472.CAN-07-1705. [DOI] [PubMed] [Google Scholar]
- 26.Todd DJ, Lee AH, Glimcher LH. The endoplasmic reticulum stress response in immunity and autoimmunity. Nat Rev Immunol. 2008;8:663–74. doi: 10.1038/nri2359. [DOI] [PubMed] [Google Scholar]
- 27.Wu J, Rutkowski DT, Dubois M, Swathirajan J, Saunders T, Wang J. ATF6 alpha optimizes long-term endoplasmic reticulum function to protect cells from chronic stress. Dev Cell. 2007;13:351–64. doi: 10.1016/j.devcel.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 28.Yamamoto K, Sato T, Matsui T, Sato M, Okada T, Yoshida H. Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6α and XBP1. Dev Cell. 2007;3:365–76. doi: 10.1016/j.devcel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 29.Harding HP, Zeng H, Zhang Y, Jungries R, Chung P, Plesken H, et al. Diabetes mellitus and exocrine pancreatic dysfunction in Perk-/- mice reveals a role for translational control in secretory cell survival. Mol Cell. 2001;7:1153–63. doi: 10.1016/s1097-2765(01)00264-7. [DOI] [PubMed] [Google Scholar]
- 30.Hetz C, Glimcher LH. Fine-tuning of the unfolded protein response: Assembling the IRE1alpha interactome. Mol Cell. 2009;35:551–61. doi: 10.1016/j.molcel.2009.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee AH, Chu GC, Iwakoshi NN, Glimcher LH. XBP-1 is required for biogenesis of cellular secretory machinery of exocrine glands. EMBO J. 2005;24:4368–80. doi: 10.1038/sj.emboj.7600903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Acosta-Alvear D, Zhou Y, Blais A, Tsikitis M, Lents NH, Arias C, et al. XBP1 controls diverse cell type- and condition-specific transcriptional regulatory networks. Mol Cell. 2007;27:53–66. doi: 10.1016/j.molcel.2007.06.011. [DOI] [PubMed] [Google Scholar]