Abstract
Long-range PCR remains a flexible, fast, efficient and cost-effective choice for sequencing candidate genomic regions in a small number of samples, especially when combined with next-generation sequencing (NGS) platforms. Several long-range DNA polymerases are advertised as being able to amplify up to 15 kb or longer genomic DNA. However, their real-world performance characteristics and their suitability for NGS remain unclear. We evaluated six long-range DNA polymerases (Invitrogen SequalPrep, Invitrogen AccuPrime, TaKaRa PrimeSTAR GXL, TaKaRa LA Taq Hot Start, KAPA Long Range HotStart and QIAGEN LongRange PCR Polymerase) to amplify three amplicons, with sizes of 12.9 kb, 9.7 kb, and 5.8 kb, respectively. Subsequently, we used the PrimeSTAR enzyme to amplify entire BRCA1 (83.2 kb) and BRCA2 (84.2 kb) genes from nine subjects and sequenced them on an Illumina MiSeq sequencer. We found that the TaKaRa PrimeSTAR GXL DNA polymerase can amplify almost all amplicons with different sizes and Tm values under identical PCR conditions. Other enzymes require alteration of PCR conditions to obtain optimal performance. From the MiSeq run, we identified multiple intronic and exonic single-nucleotide variations (SNVs), including one mutation (c.5946delT in BRCA2) in a positive control. Our study provided useful results for sequencing research focused on large genomic regions.
Since its inception, the polymerase chain reaction (PCR) has become one of the most indispensible tools in molecular biology to clone small DNA fragments1,2. However, traditionally PCR reactions were limited by the maximum size of amplified fragments. In 1992, Barnes3 developed new PCR conditions to allow for amplification of up to 5 kb. Long-range PCR increased the size of amplicons from 3–5 kb to over 30 kb by modifying the polymerases. These technical advances have brought the speed and simplicity of PCR to genomic mapping and sequencing, and have facilitated studies in molecular genetics4,5. When combined with sequencing, long-range PCR can achieve higher sensitivity and provide a faster and more cost effective tool for detecting genetic variations6,7.
Multiple long-range DNA polymerases are commercially available to amplify long genomic fragments. Some of them are advertised as being able to amplify up to 15 kb or longer genomic DNA and can work well for specific genomic regions under highly optimized conditions. However, little is known in literature (except manufacturer's flyer) on the advantage and disadvantages of each enzyme, and we are not sure about their real-world performance on randomly chosen amplicons. Since many next generation sequencing (NGS) experiments can benefit from long-range PCR, knowing the different characteristics of enzymes will have a significant impact on selecting enzymes and optimizing experimental conditions. Therefore, we compared six long-range DNA polymerases and attempted to amplify three amplicons with various sizes, to identify enzymes that have good performance with minimal requirements for condition optimization. Subsequently, we chose one enzyme to amplify the entire BRCA1 and BRCA2 genes (including introns and exons) for sequencing to further evaluate its performance for NGS.
A new generation of personal genome sequencers, such as the Illumina MiSeq and Ion Torrent PGM, are becoming popular in research and clinical settings. These sequencers have lower throughput and higher per-base-cost than Illumina HiSeq or Ion Proton, but their versatility and flexibility made them ideal for small labs where investigators prefer fast turn-around time. For example, the MiSeq sequencer allows assembly of small genomes or detection of variants in candidate regions with high accuracy, and the latest model can now generate 2 × 300 paired-end reads and up to 15 Gb of data in a single run. A previous study has successfully used long-range PCR to sequence BRCA1 and BRCA2 by Illumina Genome Analyzer II, and they have only tested one enzyme, the Invitrogen's SequalPrep8. In the current study, we selected Illumina MiSeq sequencer to determine if the combined use of long-range PCR and MiSeq can work well to identify exonic and intronic mutations in two important genes known to confer susceptibility to breast cancer.
Methods
Enzymes and Amplicons
We evaluated six commercially available long-range enzymes including SequalPrep polymerase (Invitrogen, Carlsbad, CA), AccuPrime Taq DNA Polymerase (Invitrogen, Carlsbad, CA), PrimeSTAR GXL polymerase (TaKaRa Bio, Shiga, Japan), LA Taq Hot Start Version Polymerase (TaKaRa Bio, Osaka, Japan), KAPA long Range HotStart DNA polymerase (KAPA Biosystems, Wobum, MA) and QIAGEN LongRange PCR Polymerase (Hilden, Germany). These enzymes were selected based on our knowledge at the time of the experiments, and based on Internet search. However, this is not a comprehensive list, and we acknowledge that other similar enzymes are also commercially available, such as New England Biolabs Phusion HF Polymerase and LongAmp Taq DNA Polymerase, Roche Expand Long Range DNA Polymerase, etc. Readers should not assume that the six enzymes used in the current study to be superior than those not included here.
Three amplicons were selected as the targets for comparing six long-range PCR enzymes, due to their variable lengths and variable Tm values for primers. The PCR primers of Brca1.1, 1.6 and 2.8 were synthesized by Integrated DNA technologies (Coralville, IA). The three PCR amplicons have sizes of 12.9 kb, 9.7 kb and 5.8 kb, and Tm values are 54°C, 63.3°C and 54.5°C, respectively (Table 1).
Table 1. Primer Sequences for the BRCA1 and BRCA2 Genomic Region.
| Primer ID | Genomic sequence (GRCh37/hg19) | Primer Sequence | Tm (°C) | Length of Amplicon (bp) |
|---|---|---|---|---|
| Brca1.1# | Chr17: 41194339-41207207 | 5′-ACCCCAACATTGATTCCTTTC-3′ | 53.2 | 12896 |
| 5′-CACAGGGAGAAAGTCTGCAAG-3′ | 55.7 | |||
| Brca1.2 | Chr17: 41207139-41215625 | 5′-GGGAGCTGAGAAAGCAGCCAGC-3′ | 62.9 | 8487 |
| 5′-TCGGCAGGAATCCATGTGCAGC-3′ | 62.4 | |||
| Brca1.3 | Chr17: 41215424-41229078 | 5′-AGCAGAAGAACGTGCTCTTTTCACGG-3′ | 61.2 | 13655 |
| 5′-ACAGTCTTCAATGTGGAGGCAGTAGGG-3′ | 61.8 | |||
| Brca1.4 | Chr17: 41227538-41239085 | 5′-CTGGATTGAAGATGGGTGAGA-3′ | 54.1 | 11548 |
| 5′-TTTCCTGTACCTTGCCAACAC-3′ | 55.5 | |||
| Brca1.5 | Chr17: 41238861-41251840 | 5′-GGCAATCCTGAAGAAGTGGA-3′ | 54.8 | 12980 |
| 5′-ACAAAGCAGCGGATACAACC-3′ | 55.8 | |||
| Brca1.6# | Chr17: 41246132-41255853 | 5′-GGGGAGGCTTGCCTTCTTCCG-3′ | 62.9 | 9722 |
| 5′-CTGTGCCCGGCCGGTAAAACC-3′ | 63.6 | |||
| Brca1.7 | Chr17: 41255072-41264003 | 5′-GCCATGGCACCCAGCTGAAGTA-3′ | 62.1 | 8932 |
| 5′-CTGGGAGCGATACCCCCATGCT-3′ | 63.4 | |||
| Brca1.8 | Chr17: 41258624-41271477 | 5′-GCCATGAAAAGATAATCTCACAACTGC-3′ | 56.6 | 12854 |
| 5′-GGTGGCTCTGCTTATATACACAACTGG-3′ | 59.0 | |||
| Brca1.9 | Chr17: 41270116-41281112 | 5′-GAAAGGTTTCACTGAGGTGAGACTA-3′ | 56.2 | 10997 |
| 5′-ACAAGTTAGCTTTTCCTCCACATC-3′ | 55.3 | |||
| Brca2.1 | Chr13: 32888055-32900396 | 5′-CTCCCCCACAAAAAGGGGACAAAGC-3′ | 62.3 | 12342 |
| 5′-ACAAACTCCCACATACCACTGGGGG-3′ | 62.6 | |||
| Brca2.2 | Chr13: 32900267-32911248 | 5′-CACCACAAAGAGATAAGTCAGGTAT-3′ | 54.3 | 10982 |
| 5′-TCGTTTACACAAGTCAAGTCTG-3′ | 52.8 | |||
| Brca2.3 | Chr13:32910302-32923088 | 5′-GCCACTGTGCCCAAACACTACC-3′ | 60.9 | 12787 |
| 5′-TGTGCCTGGCCTCAATTCACCA-3′ | 61.6 | |||
| Brca2.4 | Chr13:32922508-32935813 | 5′-TGACCCACAGTAAGGCACATC-3′ | 57.0 | 13306 |
| 5′-GCCCTCTTCTACCATTTGTGC-3′ | 56.1 | |||
| Brca2.5 | Chr13:32933399-32945178 | 5′-GGCCTTATGGTAGATTCCTCCCCCG-3′ | 62.5 | 11780 |
| 5′-TGGGCCTCCACATATTTTGCTGCTT-3′ | 61.3 | |||
| Brca2.6 | Chr13:32945170-32958766 | 5′-GGAGGCCCAACAAAAGAGAC-3′ | 56.1 | 13597 |
| 5′-GTGGTTTAGCCGGACTCCTC-3′ | 57.6 | |||
| Brca2.7 | Chr13: 32958373-32969763 | 5′-GGCAACTGTACAGGCAGACA-3′ | 57.5 | 11391 |
| 5′-GTCTGGGTTCTGCATTCGAT-3′ | 55.0 | |||
| Brca2.8# | Chr13: 32969199-32974976 | 5′-TTAATTGCCCATGAACCTCAG-3′ | 53.0 | 5778 |
| 5′-TATTGACTTGTATTGTGTTCGCTGT-3′ | 54.9 |
#The three amplicons selected for comparing six long-range PCR enzymes.
After comparing these six long-range PCR enzymes, we used PrimeSTAR to amplify all amplicons for the entire BRCA1/2 genes. Seventeen pairs of primers were synthesized by Integrated DNA technologies (Coralville, IA), where nine covered BRCA1 and eight covered BRCA2, with sizes ranging from 5.8 kb to 13.6 kb (Table 1). Most of the primers were taken from Ozcelik et al8, and three pairs of primers were designed by Primer39.
Reaction mixture and PCR conditions
To evaluate the performance of different enzymes, we tested each enzyme to amplify DNA samples from de-identified human subjects. The study was reviewed and approved by the Institutional Review Board of the University of Southern California (#HS-14-00425). Each of six long-range PCR enzymes was used to amplify three amplicons using the same genomic DNA sample as the template. Because the amplification protocols of long-range PCR enzymes were different for different enzymes, all experiments were designed according to the reaction mixture and cycling conditions on the manual of the corresponding enzymes, and we also optimized PCR conditions according to the preliminary results for each enzyme. Reactions were performed using the Eppendorf Master Cycler (Hamburg, Germany). To measure the success of a long-range PCR amplification, the final PCR product was run on 0.8% agarose gel and visualized by staining with GelGreen Nucleic Acid Stain (Biotium, Hayward, CA). These amplicons were generated using the reaction mixture and PCR conditions listed in Table 2.
Table 2. The protocol of the long-range PCR reactions for different enzymes.
| Enzymes | The composition of reaction mixture | PCR conditions |
|---|---|---|
| SequalPrep | 70 ng template DNA, 2 μL of a 2.5 μmol/L primers, 2 μL 10× Reaction Buffer, 1 μL PCR enhancer, 0.4 μL DMSO, 0.36 μL Polymerase and sterilized distilled water up to 20 μL | 94°C 2 minutes |
| 10 cycles | ||
| 94° 10 seconds | ||
| Tm-5° (55, 60 or 65°C) 30 seconds | ||
| 68°C 10/13 minutes | ||
| 25 cycles | ||
| 94°C 10 seconds | ||
| Tm-5° (55, 60 or 65°C) 30 seconds | ||
| 68°C 10/13 minutes plus 20 seconds | ||
| 72°C 5 minutes | ||
| hold at 4°C | ||
| AccuPrime | 70 ng template DNA, 3.2 μL of 2.5 μmol/L primers, 2 μL 10× Buffer II, 0.1 μL polymerase, and distilled water to 20 μL. | 94°C 30 seconds |
| 30 cycles | ||
| 94°C 30 seconds | ||
| 60°C 20 seconds | ||
| 68°C 13 minutes | ||
| Hold at 4°C | ||
| PrimeSTAR | 35 ng template DNA, 3.2 μL of 2.5 μmol/L primers, 4 μL 5× buffer, 1.6 μL dNTP, 0.4 μL Polymerase, and distilled water to 20 μL. | 30 cycles |
| 98°C 10 seconds | ||
| 68°C 10 minutes | ||
| Hold at 4°C | ||
| LA Taq | 35 ng template DNA, 4 μL of 2.5 μmol/L primers, 2 μL 10× Buffer II, 3.2 μL dNTP, 0.2 μL polymerase, and distilled water to 20 μL. | 94°C 1 minute |
| 30 cycles | ||
| 98°C 10 seconds | ||
| 68°C 12 minutes | ||
| 72°C 10 minutes | ||
| Hold at 4°C | ||
| KAPA 5–18 kb | 35 ng template DNA, 8 μL of 2.5 μmol/L primers, 2 μL 5× Buffer, 0.6 μL dNTP, 1.4 μL 25 mM Mgcl2, 0.2 μL polymerase(2.5 U/ μL), and distilled water to 20 μL | 94°C 30 seconds |
| 35 cycles | ||
| 94°C 20 seconds | ||
| 60°C 15 seconds | ||
| 68°C 12 minutes | ||
| 72°C 10 minutes | ||
| Hold at 4°C | ||
| QIAGEN | ||
| (0.1–10 kb) | 35 ng template DNA, 6.4 μL of 2.5 μmol/L primers, 2 μL Buffer, 1 μL dNTP, 0.2 μL enzyme, and distilled water to 20 μL. | 94°C 3 minutes |
| 35 cycles | ||
| 93°C 15 seconds | ||
| 62°C 30 seconds | ||
| 68°C 10 minutes | ||
| Hold at 4°C | ||
| (>10 kb) | 70 ng template DNA, 6.4 μL of 2.5 μmol/L primers, 2 μL Buffer, 1 μL dNTP, 0.2 μL enzyme, and distilled water to 20 μL. | 94°C 3 minutes |
| 10 cycles | ||
| 93°C 15 seconds | ||
| 62°C 30 seconds | ||
| 68°C 13 minutes | ||
| 28 cycles | ||
| 93°C 15 seconds | ||
| 62°C 30 seconds | ||
| 68°C 13 minutes plus 20 seconds | ||
| Hold at 4°C |
We further used the 2-step PCR condition of PrimeSTAR to amplify all amplicons in the BRCA1/2 genes. We found that the Brca 1.9 amplicon was difficult to amplify after re-designing multiple pairs of primers, possibly due to the presence of secondary structures during PCR amplification. After we added 0.4 μL dimethyl sulfoxide (DMSO) to 20 μL mixture reaction to interfere with the self-complementarity, the amplicon can be successfully amplified multiple times. All the other primers can be amplified using the standard 2-step protocol of PrimeSTAR.
Library preparation and NGS for PCR amplicons
We purified all the amplicons using the Agencourt AMPure XP PCR Purification systems (Beckman Coulter, Pasadena, CA) and quantified the starting DNA library using the Qubit dsDNA BR Assay system (Invitrogen, Carlsbad, CA). The sequencing library construction was performed according to the Nextera XT sample preparation guide (Illumina, San Diego, CA) that uses transposome to fragment and simultaneously adds adapter and barcoding sequences.
The pooled and barcoded libraries were subsequently sequenced using the MiSeq sequencer with v2 kits, which generated 250-base paired-end sequence reads.
Sequencing data analysis
The sequencing data analysis including quality control, mapping and variant calling was streamlined by SeqMule10, which consists of popular third party tools and then we used the wANNOVAR web server11 to annotate all the detected mutations.
First of all, sequencing data was evaluated with FastQC12. Short reads were aligned to reference genome (hg19) by BWA-MEM (version 0.7.4-r385)13 algorithm with default settings. Then we followed the GATK (Genome Analysis ToolKit) best practice to identify variants. GATK (version 2.8-1-g932cd3a)14 was used to realign reads and recalibrate base quality scores. Pre-processed BAM files were subjected to HaplotypeCaller of GATK for variant calling. The resulting SNPs were filtered by a set of filters including QD (quality by depth) <2.0, FS (Fisher strand bias test score) >60.0, MQ (root mean square of the mapping quality) <40.0, MappingQualityRankSum (mapping quality rank sum test score) <-12.5, ReadPosRankSum (read position rank sum test score) <−8.0. Indels were filtered by QD < 2.0, ReadPosRankSum < −20 and FS > 200. VQSR (Variant Quality Score Recalibration) method of GATK was not applicable due to limited number of variants. Then the wANNOVAR server was used to identify and annotate exonic and intronic variants, determine if the variants had been observed in public databases, and give predictions on whether non-synonymous variants were predicted to be deleterious based on multiple scoring systems.
Results
PCR results of six long-range PCR enzymes
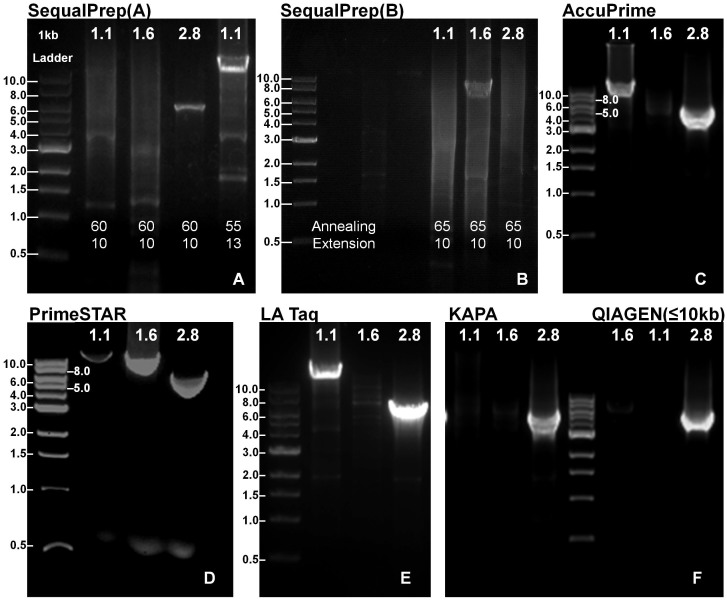
We selected six long-range PCR enzymes for examination, each of which was advertised to be able to generate amplicons up to 15 kb or more (Table 3). We evaluated them on three amplicons with sizes of 12.9 kb, 9.7 kb, and 5.8 kb and Tm values of 54°C, 63.3°C and 54.5°C, respectively (Table 1). In summary, we found that both PrimeSTAR and SequalPrep Polymerases can amplify all three targets. AccuPrime and LA Taq can only amplify the 12.9 kb and 5.8 kb targets. KAPA and QIAGEN long Range polymerase can amplify the 5.8 kb target but not the two other larger ones (Figure 1).
Table 3. A list of enzymes for long-range PCR reaction.
| Enzyme ID | Product | Company | Amplicon size (kb) | high-fidelity | Hot-start | For GC-rich templates | Proofreading activity | Reaction time (13 kb) | Price per 20 μL reaction |
|---|---|---|---|---|---|---|---|---|---|
| SequalPrep | SequalPrep Long PCR Polymerase | Invitrogen | 15 | Yes | Yes | - | - | ~7 h | $1.6 |
| AccuPrime | AccuPrime Taq DNA Polymerase | Invitrogen | 20 | Yes | Yes | - | Yes | ~7 h | $0.6 |
| PrimeSTAR | PrimeSTAR GXL DNA Polymerase | TaKaRa | >30 | Yes | Yes | Yes | - | ~5 h | $0.4 |
| LA Taq | LA Taq Hot Start Version | TaKaRa | >15 | - | Yes | - | - | ~6 h | $1.4 |
| KAPA | long Range HotStart DNA polymerase | KAPA | 20 | Yes | Yes | - | Yes | ~7 h | $0.5 |
| QIAGEN | LongRange PCR polymerase | QIAGEN | 40 | Yes | - | Yes | Yes | ~8.5 h | $1.0 |
| - | ACCUZYME DNA Polymerase | BIOLINE | 5 | Yes | - | - | Yes | - | - |
| - | KOD Hot Start DNA Polymerase | Novagen | 12 | Yes | Yes | Yes | Yes | - | - |
Figure 1. Gel electrophoresis of PCR products from the long-range PCR amplification by six enzymes.
(A) and (B): SequalPrep (Three amplicons were amplified using amplicon-specific annealing temperature and extension time). (C): AccuPrime (Amplicons for Brca1.1 and 2.8 with similar Tm values were amplified). (D): PrimeSTAR (Three amplicons were amplified using a unified two-step PCR condition.). (E): LA Taq (Amplicons for Brca1.1 and 2.8 were amplified). (F): KAPA and QIAGEN (Only the amplicon for Brca2.8 was amplified).
The six enzymes require different PCR conditions to work properly (Table 2). The PrimeSTAR enzyme can use a unified two-step PCR condition to amplify all three targets, making experimental design and implementation for PCR much easier in real-world settings, as one single thermocycler can be used to amplify all targets simultaneously. However, the SequalPrep needs to use amplicon-specific annealing temperature and extension time, which for the three amplicons were 55°C and 13 minutes, 60°C and 10minutes, 65°C and 10 minutes, respectively. Both the LA Taq and AccuPrime can amplify 12.9 kb and 5.8 kb amplicons with similar Tm values using one PCR condition. The KAPA enzyme can amplify the 5.8 kb target, only after using the annealing temperature of 55°C with 13 minutes extension time, which represents the “longer targets cycle conditions” in the user manual. When we used the “very long range” reaction mixture and cycle conditions for the QIAGEN enzyme, none of the three amplicons can be amplified; however, after using the “long range” PCR conditions, the shortest target (5.8 kb) can be amplified (Table 4). All experiments were repeated at least twice to confirm these findings.
Table 4. Primer-specific PCR conditions and results.
| SequalPrep | QIAGEN | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | AccuPrime | PrimeSTAR | LA Taq | KAPA | 0.1-10 kb | >10 kb | ||||||||||||||||||
| Sample | 1 | 2 | 3 | 1 | 1 | 2 | 3 | 1 | 2 | 3 | 1 | 2 | 3 | 1 | 2 | 3 | 1 | 2 | 3 | 2 | 1 | 3 | 1 | 2 | 3 |
| Annealing(°C) | 60 | 60 | 60 | 55 | 65 | 60 | - | - | 60 | 62 | 62 | ||||||||||||||
| Extension(min) | 10 | 10 | 10 | 13 | 10 | 13 | 2-step | 12 | 12 | 10 | 13 | ||||||||||||||
| Results | - | - | + | + | - | + | - | + | - | + | + | + | + | + | - | + | - | - | + | - | - | + | - | - | - |
In addition to reaction time and tolerance to variation of cycling conditions, we were also interested in cost per reaction for these enzymes, for practical purposes of large-scale applications. Comparing the reaction time and price among these six enzymes, the PrimeSTAR polymerase stands out with 5 hours of PCR time and a cost of $0.4 per 20 μL reactions. Therefore, we chose the PrimeSTAR enzyme for long-range PCR for our NGS experiments below.
Coverage and cost comparison of long-range PCR and custom capture arrays
To compare the target coverage of long-range PCR versus capture arrays, we used the Agilent SureDesign15 and NimbleGen SeqCap EZ Designs16 to design capture solutions for BRCA1/2. For Agilent solutions, we evaluated both SureSelect and HaloPlex. The designable coverage for exons is over 98% using all three designs. However, for exons and introns together, only HaloPlex design can achieve 96.6% coverage, yet SureSelect and SeqCap achieved coverage of 73.8% and 85.1%, respectively, suggesting reduced ability to cover intronic regions for these platforms. In comparison, the real-world performance of our long-range PCR method showed that it can get up to 100.00% coverage, even in a multiplex sequencing scenario where uneven sequencing depth exists across samples (Supplementary Table 1).
As with cost, both the SureSelect and HaloPlex might be four times as expensive as the long-range PCR method for library preparation, according to the quotes for capture probes and other reagents. However, this does not take into account labour costs or equipment costs, and some methods are more labour-intensive and error-prone than others. Furthermore, long-range PCR method may have higher specificity and uniformity than conventional capture method, and therefore requires lower sequencing coverage to obtain high-quality data17.
Targeted Amplification of BRCA1 and BRCA2 Genomic Regions
To evaluate the PrimeSTAR polymerase in NGS settings, we amplified the entire genomic regions of BRCA1 (chr17:41196312-41279500, GRCh37/hg19) and BRCA2 (chr13:32889617-32973809, GRCh37/hg19). Initially we followed the primer sets reported by Ozcelik et al8, but a few amplicons cannot be amplified, despite multiple attempts to alter cycling conditions. Therefore, we re-designed some primer pairs, with the updated primers listed in Table 1.
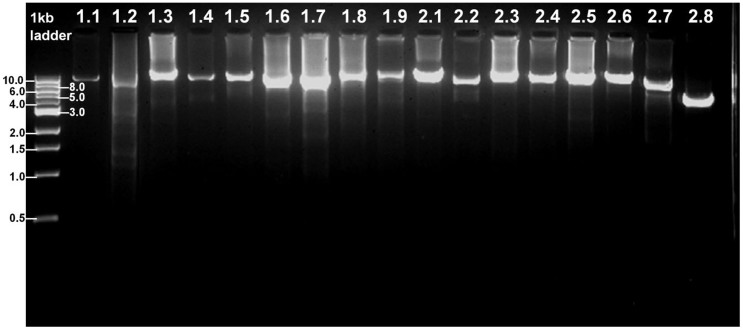
Nine DNA samples from peripheral blood of eight control subjects and one patient with hereditary breast cancer were used in our NGS experiments. Our goal is to evaluate if the experimental procedure can work consistently well among a group of samples and if a positive causal mutation can be identified reliably. For all samples, we were able to generate all the BRCA1/2 amplicons successfully, all of which display a single band with the expected size, without non-specific bands or smear (Figure 2).
Figure 2. Successful results of 17 long-range PCR amplifications spanning the complete genomic region of BRCA1/2and their flanking sequences.
Amplicons 1.1–1.9 cover BRCA1 and 2.1–2.8 cover BRCA2.
Sequencing the amplicons on MiSeq
We purified all amplicons, prepared sequencing libraries, and quantified the libraries using Qubit dsDNA BR Assay system (Invitrogen, Carlsbad, CA). Nine normalized libraries were pooled and sequenced together in one run on the Illumina MiSeq platform. Subsequently, we used BWA-MEM13 to align the sequencing reads, GATK software tool to call variants14, and the wANNOVAR web server11,18 to annotate variants detected from the sequencing data. On average, each sample had 4.6 million (range: 2.9–6.9) QC-passed reads, and 99.41% (range: 97.55% to 99.82%) of them can be properly aligned and paired. For each sample, 70.99% (range: 45.61% to 85.64%) of the reads can be mapped to the designed target region. The average coverage on the target regions was 2261X (range: 1285X to 3583X), and 93.75% (range:81.55% to 100.00%) of the target region had coverage of over 10 and 98% (range: 92.53% to 100%) of the target region was covered at least once (Supplementary Table 1).
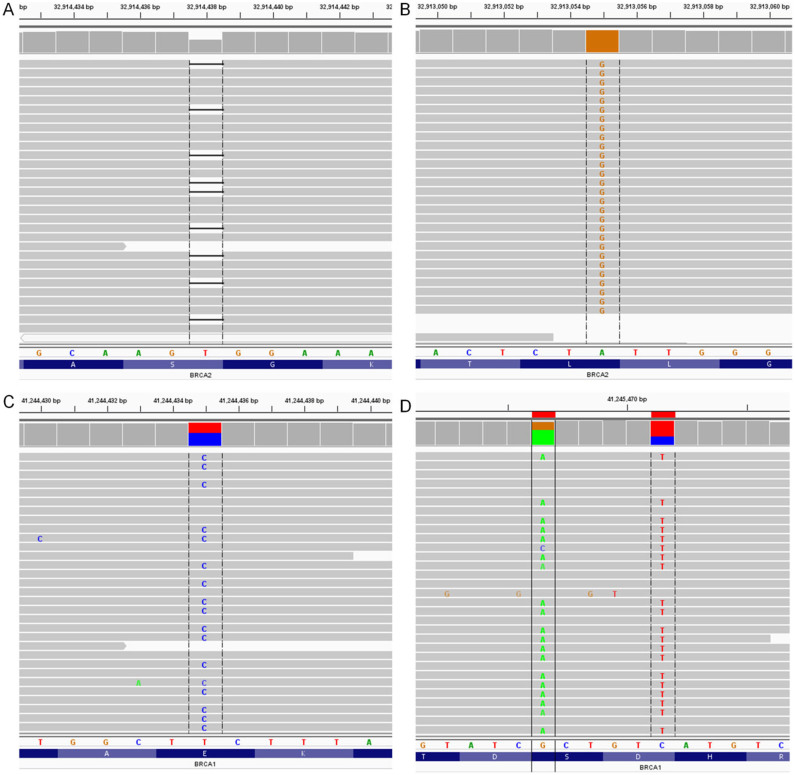
We examined the variant calls generated on these nine samples. On average, we identified 234 SNVs per sample, with the vast majority being non-coding variants. Based on variant annotation from the wANNOVAR web server, these nine samples carried 4, 8, 3, 7, 4, 2, 7, 7 and 6 non-synonymous SNVs, respectively. Additionally, we identified a nonframeshift deletion from one control subject and a frameshift deletion from the subject with hereditary breast cancer (Supplementary Table 2). This is a known disease causal mutation (c.5946delT in BRCA2) in the sample with hereditary breast cancer, and this mutation was verified by visualizing alignment on Integrative Genomics Viewer19 (Figure 3A). There are several mutations of unknown significance in other samples (Figure 3B–D). All non-synonymous SNVs found in our samples are listed in Supplementary Table 3.
Figure 3. Visual validation of mutations.
(A): A mutation (c.5946delT:p.S1982fs in BRCA2) from the sample with hereditary breast cancer; (B): A mutation (c.4563A > G in BRCA2, synonymous SNV) from all samples; (C): A mutation (c.2972A > G:p.E991G) from all samples; (D): Two mutations (c.1941C > T:p.S647S and c.1936G > A:p.D646N) in BRCA1 from sample 2, 7 and 8.
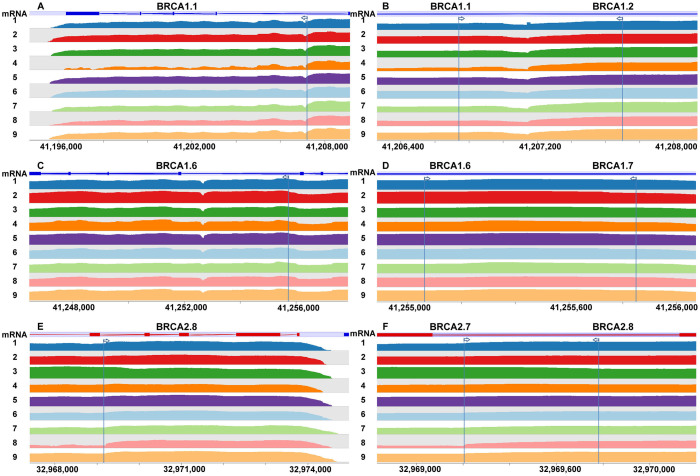
One potential advantage of long-range PCR-based NGS might be that the sequence coverage is more likely to be even, given that the same amount of starting DNA material is available for all fragments from the same amplicon. We used Wiggle plot in SeqMonk20 to view sequencing read depth of the three amplicons in BRCA1 and BRCA2 that were used in the comparative analysis of six enzymes (Figure 4). The coverage plot demonstrated that significant variations of read depth may still exist even for regions in the same amplicon from long-range PCR. Different amplicons (for example, BRCA1.1 and BRCA1.2) may also have different coverage, which may be improved by better sample normalization during library preparation. Additionally, we found that the relative sequencing depth for the same region tends to correlate across samples, suggesting that coverage correlates with certain sequence features such as GC content, repetitive sequence and Nextera restriction enzyme sites. At the rims of amplicons, coverage tends to be lower than the neighbouring region (e.g. BRCA1.1-BRCA1.2 junction). This loss of coverage can be recovered by larger overlapping between two amplicons (Figure 4D, 4F). Based on our observation, 1 kb overlapping of two amplicons seems to be sufficient (Figure 4F). In summary, long-range PCR is not immune to uneven sequence coverage typically observed in NGS experiments for capture arrays.
Figure 4. Visualization of sequencing read depth in SeqMonk for three amplicons previously used for comparing enzymes.
Left column shows amplicons BRCA1.1, BRCA1.6 and BRCA2.8 and their boundaries. Right column shows junction regions. Arrows indicate boundaries. Coverage is log2-transformed. The coverage plot demonstrates that significant variations of read depth may still exist within and between amplicons from long-range PCR. Loss of coverage may occur at junction of two amplicons (B), but can be recovered by larger overlapping of two amplicons (D, F).
Discussion
Long-range PCR has been commonly used to prepare specific high-molecular-weight DNA fragments for a variety of applications, including cloning, genome mapping and sequencing, and contig construction21. Generally speaking, to successfully amplify all amplicons in an experiment, one needs to change the annealing temperature and extension time, which are specific to each amplicon because the primers may have very different Tm values. In our experiment, we found that the TaKaRa PrimeSTAR GXL DNA Polymerase can amplify all amplicons of BRCA1/2 without altering experimental conditions, which we believe is an key advantage of using this enzyme when resources such as thermocycler is a limiting factor in research and clinical settings.
In addition to long-range PCR, a variety of other methods, such as solution-based capture, microarray-based capture, molecular inversion probes (MIPs) and multiplex PCR, have been used in target enrichment applications. Target enrichment is a highly effective way of reducing costs and saving time when only specific genomic regions (such as all exons in a gene, or a genomic region spanning a few GWAS loci) are of interest. Approaches based on capture, such as solution-based capture and microarray-based capture, achieve high-performance and have advantages for medium to large target regions (10–50 Mb)22. However, the microarray-based methods, such as Agilent SureSelect and HaloPlex, require large amounts of input DNA to be successful as well as expensive hardware working with microarray slides17,23; solution-based capture, such as NimbleGen SeqCap, is less extensively used because of performance issues24 but the solution-based capture techniques are constantly improving. Generally speaking, GC-rich segments were not well-represented in capture samples. This may be attributed to sequencing bias, as well as difficulty in capture for high GC template23. This is less a concern for the long-range PCR that “capture” large regions at once, especially for specific enzymes (such as PrimeSTAR GXL and QIAGEN LongRange PCR polymerase) that were optimized for amplifying GC-rich segments. MIPs are generally believed to be superior in terms of specificity, but far less amenable to multiple sample co-processing in a single reaction. Moreover, its design has to consider the uniqueness of each target region fragment and the most suitable hybridization conditions22. Long-range PCR has its unique niche, in that it does not require customized design by commercial vendors, and can be afforded by small laboratories when a small number of samples and continuous regions (such as full gene region including introns) are of interest.
Although mutations in the coding regions of BRCA1/2 have been heavily studied in previous genetic studies, potentially deleterious alterations may also reside in the less studied non-coding intronic sequences. For example, an insertion/deletion mutation in intron 24 (3′ UTR) of BRCA1 gene was found in one of the families with five breast cancer patients25. Additionally, a novel intronic mutation (IVS7 + 34_47delTTCTTTTCTTTTTT) and two unclassified intronic variations (IVS7 + 34_47delAAGAAAAGAAAAAA in the antisense strand and IVS7 + 50_63delTTCTTTTTTTTTTT in the sense strand) in BRCA1 were identified in a Thai family with a history of breast cancer26. Olgaet et al27 reported that an intronic mutation (c.6937 + 594T > G) can activate a cryptic exon in BRCA2 that disrupts the coding sequence in breast cancer families. For these regions, to gain a more comprehensive understanding of the genotype-phenotype relationships on BRCA1/2, it is necessary to examine both intronic and exonic regions.
In this study, we compared 6 long-range DNA polymerases for amplification of three amplicons, with sizes of 12.9 kb, 9.7 kb, and 5.8 kb, respectively, and found that the TaKaRa PrimeSTAR GXL DNA polymerase can amplify almost all amplicons with different sizes and Tm values under identical PCR conditions. We demonstrated that real-world performance for enzymes vary greatly between manufacturers, despite advertised performance characteristics, and how to couple long-range PCR with MiSeq sequencer which results in much faster turnaround time than previously possible. Overall, this report provides a practical guide on how to use long-range PCR to perform NGS on large genomic regions, especially when the entire gene regions including introns are of interest.
Author Contributions
H.J. performed the experiments and analyzed the data. Y.G. offered technical support for data analysis and SeqMule. W.Z. provided materials and advised on the design and implementation of the study. K.W. conceived and supervised the study.
Supplementary Material
Supplementary Dataset 123
Acknowledgments
We thank members of the Wang lab for helpful comments.
Footnotes
Yes, there is potential competing interest. WZ is an employee of Kingmed Diagnostics, a clinical laboratory that provides comprehensive lab testing services. KW is a board member and share holder of Tute Genomics, a bioinformatics consulting company for next-generation sequencing data. The study described in this manuscript did not involve Kingmed Diagnostics or Tute Genomics.
References
- Saiki R. et al. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science 230, 1350–1354 (1985). [DOI] [PubMed] [Google Scholar]
- Saiki R. et al. Diagnosis of sickle cell anemia and beta-thalassemia with enzymatically amplified DNA and nonradioactive allele-specific oligonucleotide probes. N. Engl. J. Med. 19, 537–541 (1988). [DOI] [PubMed] [Google Scholar]
- Barnes W. The fidelity of Taq polymerase catalyzing PCR is improved by an N-terminal deletion. Gene 112, 29–35 (1992). [DOI] [PubMed] [Google Scholar]
- Cheng S., Fockler C., Barnes W. & Higuchi R. Effective amplification of long targets from cloned inserts and human genomic DNA. Proc Natl Acad Sci USA. 91, 5695–5699 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias M. d. S. et al. Detection of novel mutations that cause autosomal dominant retinitis pigmentosa in candidate genes by long-range PCR amplification and next -generation sequencing. Mol. Vis. 19, 654–664 (2013). [PMC free article] [PubMed] [Google Scholar]
- Tan Y. et al. A novel long-range PCR sequencing method for genetic analysis of the entire PKD1 gene. J. Mol. Diagn. 14, 305–313 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knierim E., Lucke B., Schwarz J. M., Schuelke M. & Seelow D. Systematic comparison of three methods for fragmentation of long-range PCR products for next generation sequencing. PLoS ONE 6, e28240 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcelik H. et al. Long-range PCR and next-generation sequencing of BRCA1 and BRCA2 in breast cancer. J. Mol. Diagn. 14, 467–475 (2012). [DOI] [PubMed] [Google Scholar]
- Rozen S. & Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132, 365–386 (2000). [DOI] [PubMed] [Google Scholar]
- Guo Y., Lyon G. J. & Wang K. SeqMule <http://seqmule.usc.edu/> (2013) (Date of Access:01/12/2013). [Google Scholar]
- Chang X. & Wang K. wANNOVAR: annotating genetic variants for personal genomes via the web. J. Med. Genet. 49, 433–436 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S. FastQC <http://www.bioinformatics.babraham.ac.uk/projects/fastqc/> (2010) (Date of Access:01/12/2013). [Google Scholar]
- Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 1303, 1–3 (2013). [Google Scholar]
- McKenna A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agilent Technologies I. SureDesign <https://earray.chem.agilent.com/suredesign/> (2013) (Date of Access:01/12/2013). [Google Scholar]
- Roche NimbleGen I. NimbleGen SeqCap EZ design <http://www.nimblegen.com/products/seqcap/ez/designs/index.html> (2013) (Date of Access:01/12/2013). [Google Scholar]
- Mamanova L. et al. Target-enrichment strategies for next-generation sequencing. Nat. Methods 7, 111–118 (2010). [DOI] [PubMed] [Google Scholar]
- Wang K., Li M. & Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J. T. et al. Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S. SeqMonk <http://www.bioinformatics.babraham.ac.uk/projects/seqmonk/> (2007) (Date of Access:14/06/2014). [Google Scholar]
- Hogrefe H. H. & Borns M. C. Long-range PCR with a DNA polymerase fusion. Methods Mol. Biol. 687, 17–23 (2011). [DOI] [PubMed] [Google Scholar]
- Mertes F. et al. Targeted enrichment of genomic DNA regions for next-generation sequencing. Briefings in Functional Genomics 10, 374–386 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodi K. et al. Comparison of commercially available target enrichment methods for next-generation sequencing. J. Biomol. Tech. 24, 73–86 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bainbridge M. N. et al. Whole exome capture in solution with 3 Gbp of data. Genome Biol. 11, R62 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietschmann A. et al. Mutation analysis of BRCA1 and BRCA2 genes in Iranian high risk breast cancer families. J. Cancer Res. Clin. Oncol. 131, 552-558 (2005). [DOI] [PubMed] [Google Scholar]
- Ratanaphan A., Panomwan P., Canyuk B. & Maipang T. Identification of novel intronic BRCA1 variants of uncertain significance in a Thai hereditary breast cancer family. J Genet 90, 327–331 (2011). [DOI] [PubMed] [Google Scholar]
- Anczukow O. et al. BRCA2 deep intronic mutation causing activation of a cryptic exon: opening toward a new preventive therapeutic strategy. Clin. Cancer Res. 18, 4903–4909 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Dataset 123