Abstract
Myristoylated alanine-rich C-kinase substrate (MARCKS) is an actin binding protein substrate of protein kinase C (PKC) and critical for mouse and Xenopus development. Herein two MARCKS paralogs, marcksa and marcksb, are identified in zebrafish and the role of these genes in zebrafish development is evaluated. Morpholino-based targeting of either MARCKS protein resulted in increased mortality and a range of gross phenotypic abnormalities. Phenotypic abnormalities were classified as mild, moderate or severe, which is characterized by a slight curve of a full-length tail, a severe curve or twist of a full-length tail and a truncated tail, respectively. All three phenotypes displayed abnormal neural architecture. Histopathology of Marcks targeted embryos revealed abnormalities in retinal layering, gill formation and skeletal muscle morphology. These results demonstrate that Marcksa and Marcksb are required for normal zebrafish development and suggest that zebrafish are a suitable model to further study MARCKS function.
Introduction
Myristoylated Alanine-Rich C-kinase Substrate (MARCKS) is a ubiquitously expressed, acidic, rod shaped, actin binding protein. MARCKS is conserved among vertebrate species possessing three conserved domains; an amino-terminal myristoylation domain, a MH-2 domain and the phospho-site domain (PSD), which serves as the phosphorylation site for protein kinase C (PKC) (Aderem, 1992, Blackshear, 1993, Ali et al., 1997). Phosphorylation of MARCKS by PKC results in dissociation of MARCKS from the plasma membrane to the cytosol (Seykora et al., 1996, Tzlil et al., 2008), where MARCKS is dephosphorylated and is subsequently able to bind filamentous actin (F-actin) (Hartwig et al., 1992, Yarmola et al., 2001). F-actin bound MARCKS then returns to cell membranes (Myat et al., 1997), resulting in rearrangement of the actin cytoskeleton. Given its association with the actin cytoskeleton, previous experiments have elicited a role for MARCKS in cell spreading (Disatnik et al., 2002, Disatnik et al., 2004), phagocytosis (Fallman et al., 1992, Allen and Aderem, 1995), and endo- and exo-cytosis (Song et al., 1999, Li et al., 2001, Singer et al., 2004).
One of the first characterized roles for MARCKS was in development. Blackshear and colleagues generated a homozygous MARCKS deficient mouse strain by targeted disruption of the Marcks gene (also termed Macs), resulting in perinatal lethality and severe abnormalities. Abnormalities observed in Marcks deficient mice included decreased body size and severe muscular and neural abnormalities. The neural abnormalities observed included exencephaly, agenesis of the corpus callosum and defects in retinal layering (Stumpo et al., 1995), demonstrating a critical role for MARCKS in neural development (Blackshear et al., 1996, Scarlett and Blackshear, 2003, Weimer et al., 2009). Marcks mRNA expression is observed in embryonic mice as early as day 7.5 and expression is maximal by day 8.5–9.5 (Blackshear et al., 1996), correlating with the neural abnormalities observed in MARCKS deficient mice (Stumpo et al., 1995, Blackshear et al., 1996). Interestingly, adult mice and rats express lower levels of Marcks mRNA than developing animals (Patel and Kligman, 1987, Blackshear et al., 1996), further supporting a functional role for MARCKS during embryonic development. The cause of developmental defects in the absence of MARCKS can be attributed to the role of MARCKS in gastrulation. Blockade of MARCKS protein expression in Xenopus laevis embryogenesis resulted in defective morphogenetic movements during gastrulation, specifically during convergent extension (Iioka et al., 2004). MARCKS function is also required during myogenesis (Kim et al., 2000, Kim et al., 2002).
Herein, two zebrafish MARCKS genes, marcksa and marcksb, are identified and evaluated for their role in embryonic development. The marcksa and marcksb genes appear to be paralogs derived from a progenitor MARCKS gene. Expression of marcksa and marcksb transcripts are detected throughout embryonic development and morpholino-based targeting of either gene resulted in developmental defects reminiscent of those observed in MARCKS deficient mouse and Xenopus models. These results demonstrate that zebrafish is a viable model for the study of MARCKS in developmental biology and potentially other biological processes.
Materials and Methods
Rapid amplification of cDNA ends (RACE)
Invitrogen’s Gene Racer Kit (Invitrogen, Carlsbad, CA) was utilized following manufacturer’s recommendations for RACE. 5’ RACE was performed with cDNA generated from 5 µg of total RNA from the spleen, kidney and intestines of adult zebrafish per reaction. The primers utilized for RACE are as follows: Marcksa 5’ CTC CAA TAT TTC ACA CCC GGC ACC; Marcksa nested 5’ CGC TCC ACT GTA CAA CAG GGA AC; Marcksb 5’ TCC ATG TTT GTG CTC CTC GGA CTT; Marcksb nested 5’ AAA ACG GGC CTT TCA CCT CCT CC.
Bioinformatics
Peptide sequences were aligned by Clustal W (Larkin et al., 2007). For phylogenetic analysis, MARCKS and MARCKS-like protein sequences were aligned by Clustal W and neighbor joining trees were constructed from pairwise Poisson correlation distances with 2000 bootstrap replications with MEGA 4 software (Kumar et al., 2008). Accession numbers for both the protein alignment and phylogenetic tree are as follows: zebrafish Marcksa, GU563328; zebrafish Marcksb, GU563329; human MARCKS, NP_002347; mouse MARCKS, NP_032564; Xenopus laevis MARCKS, NP_001080075; chicken MARCKS, NP990811; salmon MARCKS, ACN11034; human MARCKS-like, NP_075385; mouse MARCKS-like, NP_034937; zebrafish Marcks-like, NP_998298; chicken MARCKS-like, NP_001074187.
For synteny analysis, genes flanking marcksa and marcksb were identified using the Entrez Gene database on the National Center for Biotechnology Information (NCBI) website. Homologous genes in human and mice were determined using the Homologene database on the NCBI website. The chromosomal location of the homologous genes in humans and mice were then determined by the Entrez Gene database.
Zebrafish maintenance and husbandry
Experiments involving live zebrafish were approved by the North Carolina State University Institutional Animal Care and Use Committee (IACUC). Wild type, EKK adult zebrafish (EKKwill Waterlife Resources, Ruskin, FL) were maintained in a recirculating aquarium facility (Aquatic Habitats, Apopka, FL) at 28° and were fed a commercial grade zebrafish diet. Zebrafish were mated and embryos were collected in egg water (0.005% Methylene blue and 60 µg/mL aquarium salt mixture).
mRNA isolation and RT-PCR
Twenty EKK embryos were collected at the indicated time and euthanized with 0.17% Tricaine methanesulfonate (Finquel MS-222, Argent Chemical Laboratories, Redmond, WA) and transferred to 1 mL RNA Later (Qiagen, Valencia, CA) and stored at −80°C. Samples were thawed and transferred into RLT buffer (Qiagen) with β-mercaptoethanol added and homogenized with a pellet pestle (Kimble Kontes, Vineland, NJ) and passed through QIAshredder™ columns (Qiagen). Qiagen’s RNeasy Protect mini kit was used to isolate mRNA following manufacturer’s suggestions with an on column DNase digestion step (RNase-free DNase Set, Qiagen). Complementary DNA (cDNA) synthesis was performed as previously described (Neuder et al., 2009).
The following primer (Integrated DNA Technologies, Coralville, IA) sequences and annealing temperatures were used for RT-PCR amplification of zebrafish marcksa, marcksb and β-actin (NM_131031): marcksa (annealing temperature 57.5°C) forward primer 5’ CAC AAA AAC AGC TGG AAA AG, reverse primer 5’ ATC GCT TCT GTG TTT CCA TC; marcksb (annealing temperature 65.8°C) forward primer 5’TCC AAA AAC GGA GCA AAA GAC GAG, reverse primer 5’ TTC GCT GGA AGC TTC GGG CTT; β-actin (annealing temperature 57.5°C) forward primer 5’ GGA GAA GAT CTG GCA TCA CAC CTT CTA C, reverse primer 5’ TGG TCT CGT GGA TAC CGC AAG ATT CCA T. The optimal annealing temperature was determined for each primer pair by the temperature gradient aspect of the MyIQ thermocycler (Bio-Rad, Hercules, CA). Amplitaq Gold PCR master mix (Applied Biosystems, Foster City, CA) was used for both annealing temperature gradient PCR and RT-PCR and manufacturer’s instructions were followed for a 40µL total PCR reaction. The thermal cycling conditions for RT-PCR are as follows: 94°C for 10 minutes, 35 cycles of [94°C for 15 sec, the appropriate annealing temperature for 60 sec, 72°C for 2 min], and 72°C for 10 min.
Morpholino injections and phenotypic observations
The following MOs were purchased from GeneTools (Philomath, OR): Marcksa translation blocking (MAT) 5’ CTG TTT TTG TGA ATT GCG CTC CCA T; Marcksa upstream translation blocking (MATU) 5’ AGC TCA ACA GAT CCC AAT ACA GAA C; Marcksb translation blocking (MBT) 5’ TTT TGG AGA TTT GTG CTC CCA TGC T; Marcksb splice blocking (MBS) 5’ ATA ATT AAA GTT ATT ACC TGC CCG T. Standard stocks of control and p53 MOs were purchased from GeneTools. MOs were suspended to 1 mM with sterile water and these stock solutions were stored at room temperature. Prior to loading MOs into calibrated needles, MOs were diluted in 0.5% phenol red to aid in visualization of injections. Injected embryos were transferred to a petri dish containing egg water after injection and incubated at 28°C with controlled light/dark cycles.
Embryos were dechorionated at 24 hpf using very fine forceps and phenotyped. For photomicroscopy, embryos were anesthetized in 0.017% Tricaine methanesulfonate and images were captured with a Nikon AZ100 microscope (Melville, NY) under bright field light.
Histopathology
Embryos were euthanized at 72 hpf with a 0.17% tricaine solution. Whole (dechorionated) embryos were then fixed in 10% neutral buffered formalin solution, routinely processed, embedded in paraffin, sectioned at 5µm, stained with hematoxylin and eosin (H & E) and examined via light microscopy.
Statistical Analysis
Statistical analysis was performed using Sigma Stat Software and a one-way ANOVA test. When a significant interaction was detected, a pairwise multiple comparison procedure (Fisher LSD method) was used to identify the source of the interaction. Significance was set at P < 0.05.
Results
Zebrafish express two MARCKS paralogs
It has been predicted that, subsequent to its divergence from mammals, the bony fish lineage experienced a whole genome duplication event hundreds of millions of years ago. The remnants of this genome duplication event are detected by the presence of paralogs of approximately half of the genes in the zebrafish genome (Taylor et al., 2001, Taylor et al., 2003). MARCKS appears to be one of such genes in zebrafish, as BLASTx searches using the human MARCKS protein as a query identified two zebrafish marcks cDNA sequences in GenBank. Both sequences were deposited as a result of a large scale cDNA sequencing effort (Strausberg et al., 2002). One sequence, which had been termed zebrafish marcks on both GenBank (NM_001015060) and the ZFIN database (ZDB-GENE-030131-1921), is located on chromosome 17. The other sequence, which has been termed zgc: 109978 on the GenBank (NM_001024404) and ZFIN database (ZDB-GENE-050522-145), is located on chromosome 20. Based on our sequence comparisons and phylogenetic analyses (below), we demonstrate that these genes are likely paralogs and have renamed zgc:109978 and marcks to be marcksa and marcksb, respectively. We PCR amplified, cloned and sequenced full-length cDNA sequences for both genes by rapid amplification of cDNA ends (RACE). We observed 11 nucleotide and seven amino acid differences between our marcksa cDNA sequence and its reference sequence (NM_001024404). We observed seven nucleotide and three amino acid differences between our marcksb cDNA sequence and its reference sequence (NM_001015060).
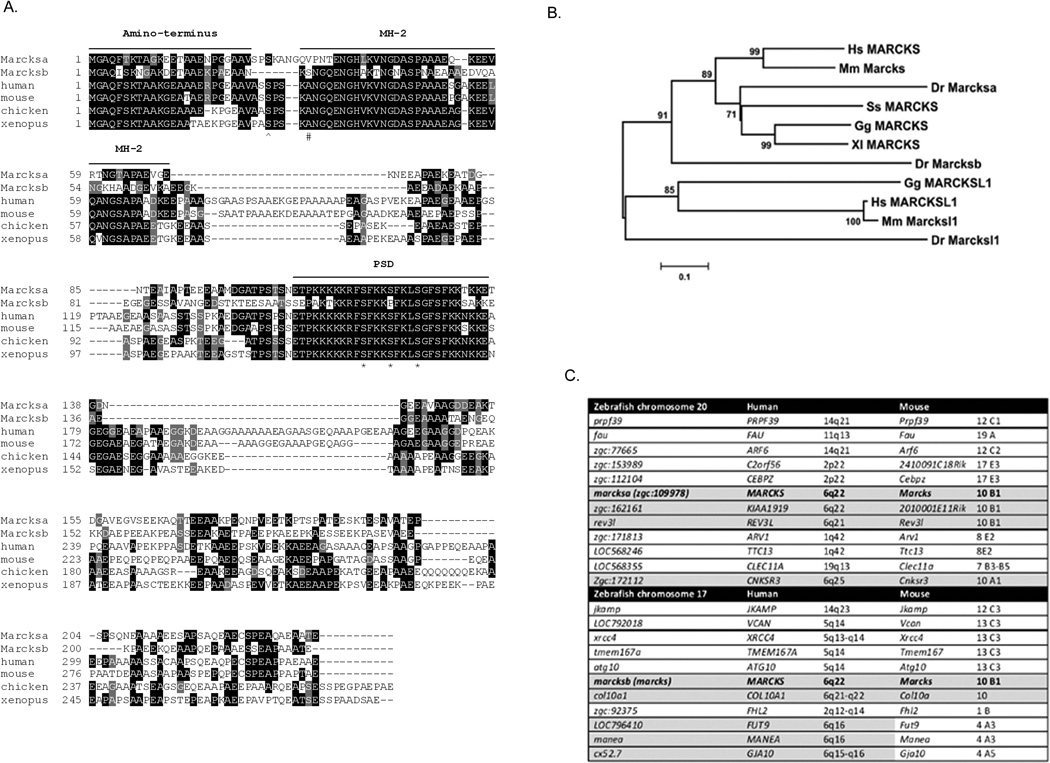
Zebrafish Marcksa and Marcksb are 40.6% identical. Marcksa is 48.8% identical to human MARCKS and 43.7% identical to mouse MARCKS. Marcksb is 41.9% identical to human MARCKS and 42.1% identical to mouse MARCKS. Protein sequence alignments reveal that the three conserved domains characteristic of MARCKS (Blackshear, 1993) (the myristoylated amino-terminus, the MH2 domain and PSD), are conserved in both zebrafish Marcks proteins (Fig. 1A). PKC phosphorylates MARCKS at Ser152, Ser 156 and Ser163 in rodents (Heemskerk et al., 1993) and it appears that zebrafish Marcksa could be phosphorylated at equivalent serine residues. However, zebrafish Marcksb appears to be capable of serine phosphorylation at only two of these sites. Instead of having a serine at amino acid 119 (analogous to Ser156 in murine MARCKS) zebrafish Marcksb encodes a proline at this position. Phosphorylation of MARCKS on Ser159 in humans (Ser152 in mice) occurs via the RhoA/ROCK pathways (Tanabe et al., 2006) and both zebrafish Marcksa and Marcksb encode serines at this position, presumably also capable of being phosphorylated. Phosphorylation of MARCKS occurs in chick neurons on Ser25 (Zolessi et al., 2004) and zebrafish Marcksa has a serine at that corresponding residue but zebrafish Marcksb does not. However, Marcksb does have a serine residue at position 26 that may be a candidate serine residue capable of phosphorylation in a similar manner as Ser25 in other species. Taken together, these observations suggest that zebrafish Marcks proteins are homologous to MARCKS proteins expressed by other vertebrate species. The evidence also suggests non-redudant roles for Marcksa and Marcksb in zebrafish.
Figure 1.
Zebrafish Marcksa and Marcksb are similar to other vertebrate MARCKS sequences. (A) Zebrafish Marcksa and Marcksb protein sequences are aligned with MARCKS sequences from human, mouse, chicken and Xenopus. Black shading indicates identical amino acid alignment; gray shading indicates functionally similar amino acids. The three conserved domains of MARCKS are indicated: the myristoylated amino-terminus, the MH-2 domain and the phospho-site domain (PSD). The three serine residues phosphorylated by PKC are identified by an asterisk (*) and Ser25, which is phosphorylated in neural tissue, is denoted by a caret (^). The pound sign (#) denotes a candidate serine in Marcksb that may also be phosphorylated given its proximity to Ser25 in MARCKS expressed by other species. (B) Phylogenetic comparison of MARCKS and MARCKS-like protein sequences from vertebrate species. Branch lengths are measured in terms of amino acid substitutions with the scale indicated below the tree. Hs = human, Homo sapiens; Mm = mouse, Mus musculus; Dr = zebrafish, Danio rerio; Ss = salmon, Salmo salar; Gg = chicken, Gallus gallus; Xl = frog, Xenopus laevis. (C) Zebrafish marcksa and marcksb loci both share conserved synteny with the MARCKS locus in human and mice. Genes flanking marcksa on chromosome 20 and marcksb on chromosome 17 were determined and the chromosomal location of orthologous genes in humans and mice were determined. Gray shaded areas indicate conserved synteny.
Phylogenetic analysis of vertebrate MARCKS and MARCKS-like protein (MLP) demonstrate that zebrafish Marcksa and Marcksb are more similar to MARCKS than to MLP (Fig. 1B). MLP is a member of the MARCKS family of proteins, sharing approximately 50% amino acid identity with MARCKS (Arbuzova et al., 2002, Sundaram et al., 2004). Further, Marcksa is more closely related to other vertebrate MARCKS proteins than Marcksb (Fig. 1B).
A comparison of genes flanking both zebrafish marcksa and marcksb demonstrate conserved synteny with the MARCKS locus on human chromosome 6q and mouse chromosome 10 (Fig. 1C). On zebrafish chromosome 20 marcksa is flanked by zgc:162161, rev3l and zgc:172112 which are orthologous to KIAA1919, REV3L and CNSKR3 on human chromosome 6q and 2010001E11Rik, Rev3l and Cnksr3 on mouse chromosome 10. On zebrafish chromosome 17 marcksb is flanked by col10a1, LOC796410, manea and cx52.7 which are orthologous to COL10A1, FUT9, MANEA and GJA10 on human chromosome 6q: of these genes, only Col10a is present on mouse chromosome 10. This data suggests that marcksa and marcksb are indeed paralogs.
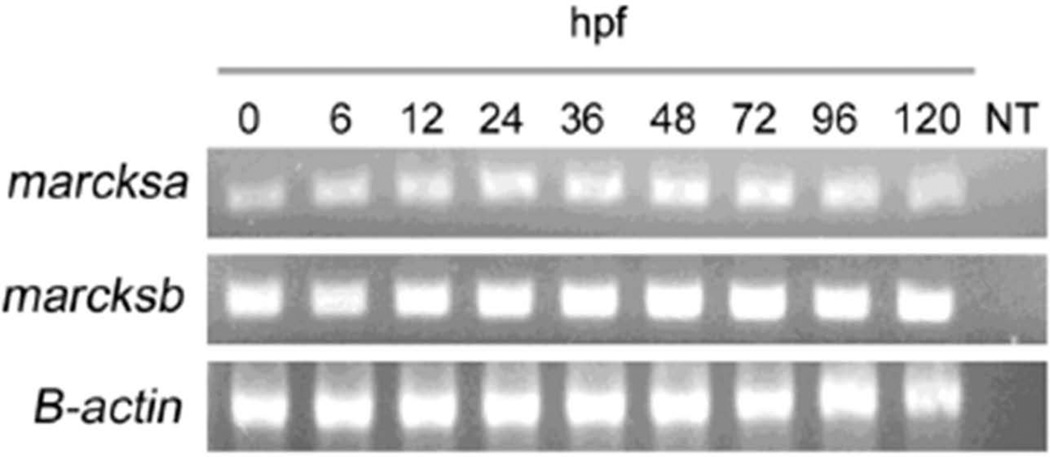
A large scale RNA in situ hybridization project revealed that marcksa and marcksb are expressed in developing zebrafish embryos, with uniform expression from the single cell stage to 60 hours post fertilization (hpf). Both marcksa and marcksb were described in a non-spatially restricted pattern and high expression was evident in the brain and notochord (Thisse et al., 2004). We validated these results by determining the mRNA expression of marcksa and marcksb during zebrafish development by reverse transcriptase PCR (RT-PCR). Maternal transcripts of marcksa and marcksb are detected in the fertilized egg (0 hpf) and somatic expression maintains consistent mRNA levels through 120 hpf (Fig. 2). Given the lack of a commercially available zebrafish specific anti-MARCKS antibody and differences in the amino acid sequences between mouse, human and zebrafish MARCKS proteins; we were unable to detect zebrafish Marcks protein expression by western blot (unpublished observation).
Figure 2.
Zebrafish marcksa and marcksb are expressed throughout early development. Reverse transcriptase PCR was performed to detect zebrafish marcksa, marcksb and β-actin transcripts from embryo derived cDNAs. Age of embryos is indicated above each lane in hours post fertilization (hpf). Data is representative of 3 separate experiments and NT indicates no template control.
Morpholino-based targeting of zebrafish Marcksa and Marcksb results in increased mortality and abnormal phenotypes
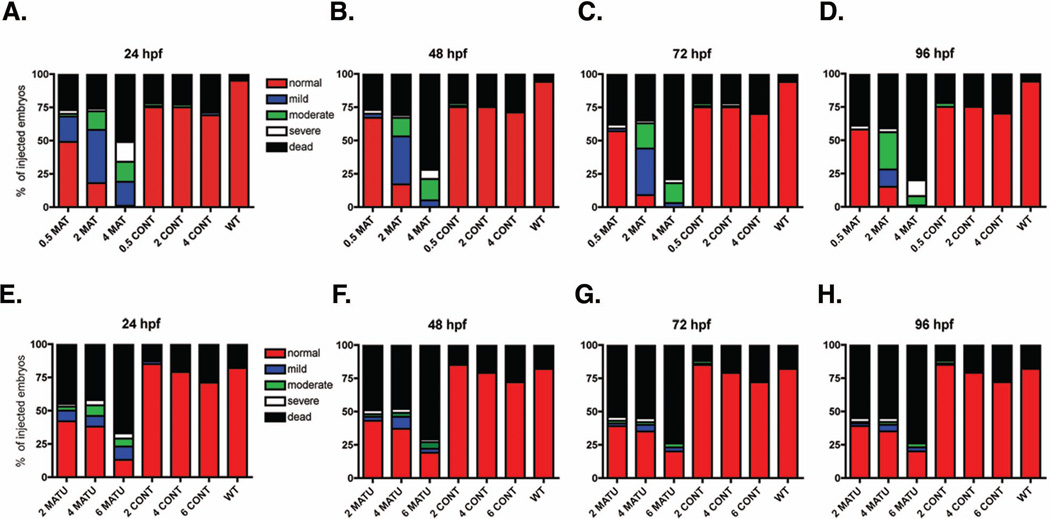
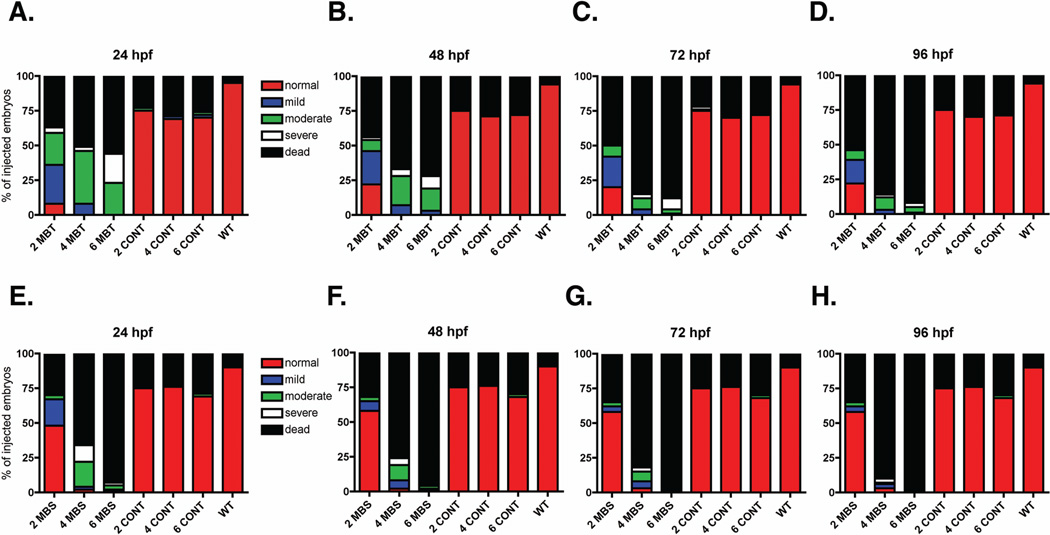
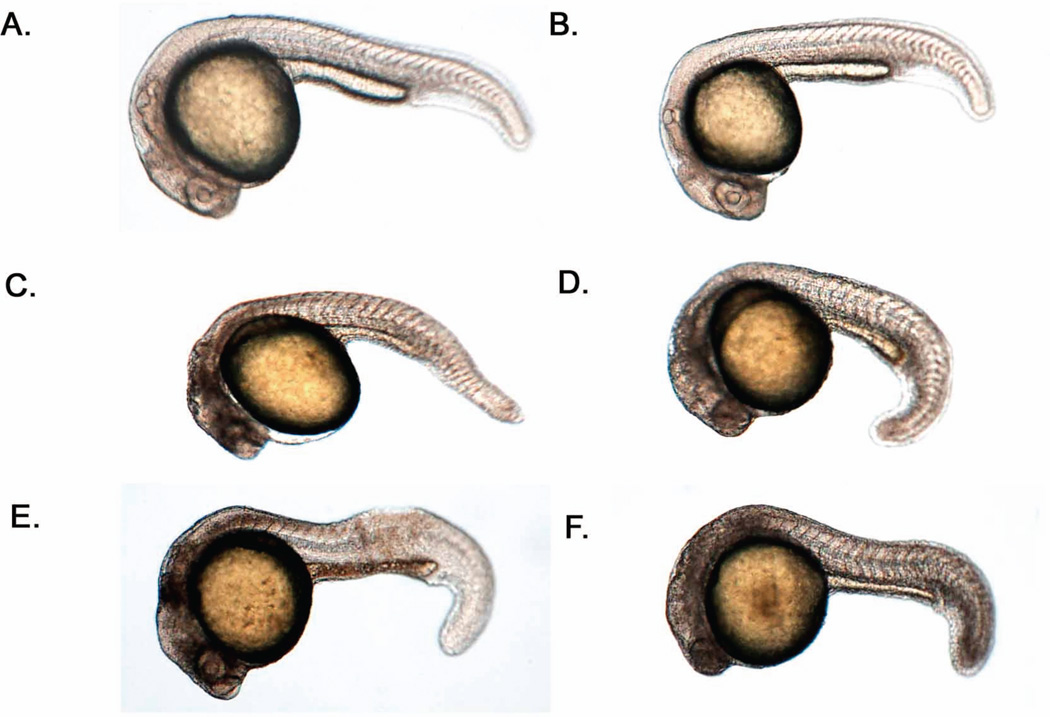
Given previous research reporting a role for MARCKS in mammalian and Xenopus laevis development, we hypothesized that both Marcksa and Marcksb play a role in the development of zebrafish. In order to test this hypothesis, we utilized anti-sense morpholinos (MOs) to evaluate Marcksa and Marcksb function in the zebrafish embryo. MAT and MATU are translation-blocking morpholinos designed to target Marcksa with MAT binding at the AUG start codon and MATU binding upstream of the AUG start codon. MBT and MBS morpholinos are designed to target Marcksb and are translation-blocking (binding at the AUG start codon) and splice-blocking morpholinos, respectively. Single cell zebrafish embryos were injected with various doses of these MOs and phenotypes were characterized at 24, 48, 72 and 96 hpf. Injection of MOs designed to target either Marcksa or Marcksb produced embryos with a similar range of phenotpyes that were classified into one of four phenotypic groups; normal, mild, moderate and severe (Fig. 3). The mild phenotype fish have a slight curve to their full-length tail whereas moderate phenotype fish have a full-length tail with a severe curve or twist. Severe phenotype fish have a cropped or absent tail, with the tail not extending far beyond the yolk sac. Mild, moderate and severe phenotyped embryos all appear to have abnormal brain development, evidenced by the lack of normal eye and neural tissue architecture. We observed increased numbers of mild, moderate and severe phenotypes in both Marcksa and Marcksb targeted embryos relative to control injected and wild type, non-injected embryos (Figs. 4 and 5). MAT and MATU injected embryos had similar numbers of mild, moderate and severe phenotyped embryos relative to control injected and wild type, non-injected (Fig. 4, Tables 1 and 2) embryos. Similarly, MBT and MBS injected embryos had similar numbers of abnormal (mild, moderate and severe) phenotyped embryos relative to control injected and wild type, non-injected (Fig. 5, Tables 3 and 4) embryos. Although we cannot directly confirm knockdown of either Marcksa or Marcksb protein level due to the lack of a zebrafish MARCKS specific antibody, our data does suggest specificity of the MOs as the same phenotypes were observed in Marcksa (MAT and MATU) and Marcksb (MBT and MBS) targeted embryos.
Figure 3.
Phenotypic characterization of Marcksa and Marcksb targeted zebrafish morphants. Representative examples of normal, mild, moderate and severe phenotypes are depicted at 24 hpf (A), 48 hpf (B), 72 hpf (C) and 96 hpf (D).
Figure 4.
Phenotypic quantification of zebrafish Marcksa targeted morphants. Embryos were either not injected (WT) or injected at the single cell stage with 0.5, 2 or 4 ng MAT (A–D), 2, 4, or 6 ng MATU (E–H) or respective dose of control MO (CONT). Phenotypic characterization was quantified at 24 hpf (A, E), 48 hpf (B, F), 72 hpf (C, G) and 96 hpf (D, H) and phenotypes were classified into five phenotypes: normal, mild, moderate, severe and dead. The data is pooled from three separate experiments with a total of 120 embryos injected in the MAT and MATU groups and 110 embryos injected in the control injected or WT groups. Data is represented as the percentage of injected embryos.
Figure 5.
Phenotypic quantification of zebrafish Marcksb targeted morphants. Embryos were either not injected (WT) or injected at the single cell stage with 2, 4, or 6 ng MBT (A–D), MBS (E–H) or control MO (A–H). Phenotypic characterization was quantified at 24 hpf (A, E), 48 hpf (B, F), 72 hpf (C, G) and 96 hpf (D, H) and phenotypes were classified into five phenotypes: normal, mild, moderate, severe and dead. The data is pooled from three separate experiments with a total of 120 embryos injected in the MBT and MBS groups and 110 embryos injected in the control injected or WT groups. Data is represented as the percentage of injected embryos.
Table 1.
Increased mortality and abnormal (sum of mild, moderate and severe) phenotypes are observed in MAT injected embryos1.
| MO | MAT | Control MO | WT | |||||
|---|---|---|---|---|---|---|---|---|
| ng | 0.5 | 2 | 4 | 0.5 | 2 | 4 | - | |
| 24 hpf | Dead | 10.67± 3.51 |
10.67± 2.08 |
20.67± 9.022 |
8.67± 1.53 |
10.00± 7.00 |
10.33± 6.66 |
1.67± 2.89 |
| Normal | 19.67± 9.454 |
7.00± 7.554,5 |
0.33± 0.584,5 |
27.67± 7.51 |
27.33± 8.34 |
25.33± 10.41 |
35.00± 5.00 |
|
| Abnormal | 9.67± 6.35 |
22.33± 8.082,3 |
19.00± 8.542,3 |
0.00± 1.00 |
0.67± 1.15 |
1.00± 1.00 |
0.00± 0.00 |
|
| 48 hpf | Dead | 11.00± 3.00 |
12.67± 2.53 |
29.00± 11.532,3 |
8.33± 3.06 |
8.67± 8.15 |
10.67± 7.10 |
2.33± 2.52 |
| Normal | 26.67± 5.03 |
6.67± 6.114,5 |
0.00± 0.004,5 |
27.33± 7.51 |
27.67± 8.33 |
26.00± 10.15 |
34.33± 4.04 |
|
| Abnormal | 2.33± 2.10 |
20.67± 8.622,3 |
11.00± 11.532,3 |
0.00± 1.00 |
0.33± 0.58 |
0.33± 0.58 |
0.00± 0.00 |
|
| 72 hpf | Dead | 15.33± 8.082 |
14.00± 4.58 |
31.67± 11.152,3 |
8.33± 3.06 |
8.67± 8.15 |
10.67± 7.10 |
2.33± 2.52 |
| Normal | 22.67± 9.02 |
3.67± 4.734,5 |
0.00± 0.004,5 |
27.33± 7.51 |
27.67± 8.33 |
25.67± 10.26 |
34.33± 4.04 |
|
| Abnormal | 2.00± 2.00 |
22.33± 9.292,3 |
8.33± 11.15 |
0.00± 1.00 |
1.00± 1.73 |
0.33± 0.58 |
0.00± 0.00 |
|
| 96 hpf | Dead | 15.33± 8.082 |
16.33± 3.062 |
32.33± 10.792,3 |
8.33± 3.06 |
8.67± 8.15 |
10.67± 7.10 |
2.33± 2.52 |
| Normal | 23.33± 8.08 |
6.00± 5.204,5 |
0.00± 0.004,5 |
27.33± 7.51 |
27.67± 8.33 |
25.67± 10.26 |
34.33± 4.04 |
|
| Abnormal | 1.33± 2.31 |
17.67± 5.512,3 |
7.67± 10.792 |
0.00± 1.00 |
0.33± 0.58 |
0.33± 0.58 |
0.00± 0.00 |
|
Data represented as average ± standard deviation with statistical significance set at (p<0.05).
Denotes a significant increase relative to wild type
Denotes a significant increase relative to control MO injected
Denotes a significant decrease relative to wild type
Denotes a significant decrease relative to control MO injected.
Table 2.
Increased mortality and abnormal (sum of mild, moderate and severe) phenotypes are observed in MATU injected embryos1.
| MO | MATU | Control MO | WT | |||||
|---|---|---|---|---|---|---|---|---|
| ng | 2 | 4 | 6 | 2 | 4 | 6 | - | |
| 24 hpf | Dead | 18.33± 17.04 |
17.00± 8.54 |
26.67± 12.70 |
8.00± 6.56 |
10.33± 7.57 |
13.57± 10.02 |
10.00± 8.19 |
| Normal | 16.67± 13.45 |
15.00± 9.85 |
5.33± 6.66 |
31.00± 7.21 |
29.00± 8.72 |
26.00± 9.85 |
30.00± 8.19 |
|
| Abnormal | 5.00± 4.00 |
8.00± 6.24 |
8.00± 6.08 |
1.00± 1.00 |
0.67± 1.15 |
0.33± 0.58 |
0.00± 0.00 |
|
| 48 hpf | Dead | 20.33± 15.31 |
19.00± 7.21 |
28.33± 10.69 |
8.00± 6.56 |
10.33± 7.57 |
13.67± 10.02 |
10.00± 8.19 |
| Normal | 17.00± 14.00 |
14.67± 9.29 |
7.67± 9.81 |
31.33± 7.09 |
29.00± 8.72 |
26.33± 10.02 |
30.00± 8.19 |
|
| Abnormal | 2.67± 2.08 |
6.33± 4.93 |
4.00± 1.00 |
0.67± 0.58 |
0.67± 1.15 |
0.00± 0.00 |
0.00± 0.00 |
|
| 72 hpf | Dead | 21.67± 15.14 |
22.00± 7.00 |
29.67± 9.29 |
8.00± 6.56 |
10.33± 7.57 |
13.67± 10.02 |
10.00± 8.19 |
| Normal | 11.33± 13.32 |
14.00± 7.94 |
8.00± 9.54 |
31.00± 7.55 |
29.00± 8.72 |
26.33± 10.02 |
30.00± 8.19 |
|
| Abnormal | 2.33± 1.15 |
4.00± 2.65 |
2.33± 0.58 |
1.00± 1.00 |
0.67± 1.15 |
0.00± 0.00 |
0.00± 0.00 |
|
| 96 hpf | Dead | 22.00± 15.72 |
22.00± 7.00 |
29.67± 9.29 |
8.00± 6.56 |
10.33± 7.57 |
13.67± 10.02 |
10.00± 8.19 |
| Normal | 15.67± 13.80 |
14.00± 7.94 |
8.00± 9.54 |
31.00± 7.55 |
29.00± 8.72 |
26.33± 10.02 |
30.00± 8.19 |
|
| Abnormal | 2.33± 2.08 |
4.00± 2.65 |
2.33± 0.58 |
1.00± 1.00 |
0.67± 1.15 |
0.00± 0.00 |
0.00± 0.00 |
|
Data represented as average ± standard deviation with statistical significance set at (p<0.05).
Denotes a significant increase relative to wild type
Denotes a significant increase relative to control MO injected
Denotes a significant decrease relative to wild type
Denotes a significant decrease relative to control MO injected.
Table 3.
Increased mortality and abnormal (sum of mild, moderate and severe) phenotypes are observed in MBT injected embryos1.
| MO | MBT | Control MO | WT | |||||
|---|---|---|---|---|---|---|---|---|
| ng | 2 | 4 | 6 | 2 | 4 | 6 | - | |
| 24 hpf | Dead | 15.33± 9.872 |
20.33± 11.242 |
22.33± 9.612,3 |
10.00± 7.00 |
10.33± 6.66 |
9.67± 4.73 |
1.67± 2.89 |
| Normal | 3.00± 5.204,5 |
0.00± 0.004,5 |
0.00± 0.004,5 |
27.33± 8.34 |
25.33± 10.41 |
25.67± 7.37 |
35.00± 5.00 |
|
| Abnormal | 18.67± 1.152,3 |
19.67± 11.242,3 |
17.67± 9.612,3 |
0.67± 1.15 |
1.00± 1.00 |
1.33± 1.16 |
0.00± 0.00 |
|
| 48 hpf | Dead | 17.33± 9.302 |
27.00± 7.002,3 |
29.00± 5.302,3 |
8.67± 8.15 |
10.67± 7.10 |
9.67± 4.73 |
2.33± 2.52 |
| Normal | 8.67± 8.084,5 |
0.00± 0.004,5 |
0.00± 0.004,5 |
27.67± 8.33 |
26.00± 10.15 |
26.33± 6.66 |
34.33± 4.04 |
|
| Abnormal | 13.33± 4.732,3 |
13.00± 7.002,3 |
11.00± 5.302,3 |
0.33± 0.58 |
0.33± 0.58 |
0.67± 1.15 |
0.00± 0.00 |
|
| 72 hpf | Dead | 20.00± 12.122 |
34.00± 3.612,3 |
35.33± 2.082,3 |
8.67± 8.15 |
10.67± 7.10 |
10.00± 5.29 |
2.33± 2.52 |
| Normal | 8.00± 7.214,5 |
0.00± 0.004,5 |
0.00± 0.004,5 |
27.67± 8.33 |
25.67± 10.26 |
26.33± 6.66 |
34.33± 4.04 |
|
| Abnormal | 12.00± 6.082,3 |
6.00± 3.60 |
4.67± 2.08 |
1.00± 1.73 |
0.33± 0.58 |
0.33± 0.58 |
0.00± 0.00 |
|
| 96 hpf | Dead | 22.00± 13.122,3 |
34.67± 2.902,3 |
37.00± 1.732,3 |
8.67± 8.15 |
10.67± 7.10 |
10.33± 5.86 |
2.33± 2.52 |
| Normal | 8.67± 6.814,5 |
0.00± 0.004,5 |
0.00± 0.004,5 |
27.67± 8.33 |
25.67± 10.26 |
26.00± 6.93 |
34.33± 4.04 |
|
| Abnormal | 9.33± 7.512,3 |
5.30± 2.89 |
3.00± 1.73 |
0.33± 0.58 |
0.33± 0.58 |
0.67± 1.15 |
0.00± 0.00 |
|
Data represented as average ± standard deviation with statistical significance set at (p<0.05).
Denotes a significant increase relative to wild type
Denotes a significant increase relative to control MO injected
Denotes a significant decrease relative to wild type
Denotes a significant decrease relative to control MO injected.
Table 4.
Increased mortality and abnormal (sum of mild, moderate and severe) phenotypes are observed in MBS injected embryos1.
| MO | MBS | Control MO | WT | |||||
|---|---|---|---|---|---|---|---|---|
| ng | 2 | 4 | 6 | 2 | 4 | 6 | - | |
| 24 hpf | Dead | 11.67± 1.53 |
26.67± 12.662 |
37.67± 2.082,3 |
8.33± 6.03 |
8.33± 6.03 |
10.67± 5.51 |
3.23± 3.21 |
| Normal | 19.33± 8.14 |
0.67± 1.154,5 |
0.33± 0.584,5 |
27.67± 10.69 |
28.00± 8.89 |
25.33± 3.51 |
36.33± 3.06 |
|
| Abnormal | 9.00± 6.93 |
12.67± 12.90 |
2.00± 1.73 |
0.67± 0.58 |
0.33± 0.58 |
0.67± 1.15 |
0.33± 0.58 |
|
| 48 hpf | Dead | 13.00± 1.73 |
30.67± 11.932,3 |
38.67± 1.152,3 |
8.33± 6.03 |
8.33± 8.50 |
11.00± 6.00 |
3.33± 3.21 |
| Normal | 23.00± 2.00 |
0.67± 1.154,5 |
0.33± 0.584,5 |
27.67± 10.69 |
28.00± 8.89 |
25.00± 4.00 |
36.33± 3.06 |
|
| Abnormal | 4.00± 1.00 |
8.33± 12.74 |
1.00± 1.00 |
0.67± 0.58 |
0.33± 0.58 |
0.67± 1.15 |
0.33± 0.58 |
|
| 72 hpf | Dead | 13.67± 2.52 |
33.33± 8.142,3 |
38.67± 1.152,3 |
8.33± 6.03 |
8.33± 8.50 |
11.00± 6.00 |
3.33± 3.21 |
| Normal | 23.33± 2.89 |
1.00± 1.004,5 |
0.33± 0.584,5 |
27.67± 10.69 |
28.00± 8.89 |
25.00± 4.00 |
36.33± 3.06 |
|
| Abnormal | 3.00± 1.73 |
5.67± 8.96 |
1.00± 1.00 |
0.67± 0.58 |
0.33± 0.58 |
0.67± 1.15 |
0.33± 0.58 |
|
| 96 hpf | Dead | 13.67± 2.52 |
33.33± 8.142,3 |
38.67± 1.152,3 |
8.33± 6.03 |
8.33± 8.50 |
11.00± 6.00 |
3.33± 3.21 |
| Normal | 23.33± 2.89 |
1.00± 1.004,5 |
0.33± 0.584,5 |
27.67± 10.69 |
28.00± 8.89 |
25.00± 4.00 |
36.33± 6.06 |
|
| Abnormal | 3.00± 1.73 |
5.67± 8.96 |
1.00± 1.00 |
0.67± 0.58 |
0.33± 0.58 |
0.67± 1.15 |
0.33± 0.58 |
|
Data represented as average ± standard deviation with statistical significance set at (p<0.05).
Denotes a significant increase relative to wild type
Denotes a significant increase relative to control MO injected
Denotes a significant decrease relative to wild type
Denotes a significant decrease relative to control MO injected.
Besides abnormal phenotypes, we also observed increased mortality in Marcksa and Marcksb targeted embryos. We observed increased mortality in MAT and MATU injected embryos relative to control injected and wild type, non-injected (Fig. 4, Tables 1 and 2). This same observation of increased mortality was also observed in MBT and MBS injected embryos (Fig. 5, Tables 3 and 4). Since we did not observe 100% mortality in either Marcksa or Marcksb MO injected zebrafish embryos at any of the doses indicated, we asked if co-injection of Marcksa and Marcksb MOs would result in 100% mortality. Two ng combinations of MAT, MBT or control MOs (4 ng of MO injected total) were co-injected into 1-cell zebrafish embryos and mortality was determined at 24 hpf. Embryos co-injected with 2 ng MAT + 2 ng MBT had an increased mortality compared to embryos co-injected with 2 ng MAT + 2 ng control MO, 2 ng MBT + 2 ng control MO or non-injected, wild type embryos (Table 5). The increase in mortality in the MAT + MBT co-injected embryos was statistically significant compared to MBT + control MO co-injected and wild type (non injected embryos), but was not statistically increased compared to MAT + control co-injected embryos (Table 5). Interestingly, when we injected 2 ng of MAT or MBT alone, we observed 40.8±7.6% and 55.0±32.8% mortality, respectively. Further, when we injected 4 ng MAT or MBT alone, we observed 80.8±26.9% and 86.5±7.4% mortality, respectively (Figs. 4 and 5). Given that the mortality in the MAT and MBT co-injected group was not 100% and that the mortality was similar to the mortality in the 4 ng MAT or MBT alone injected groups suggests that Marcksa and Marcksb may play an additive role in zebrafish development. However, we cannot determine if Marcksa and Marcksb play redundant or non-redundant roles because we are unable to determine protein expression levels and localization.
Table 5.
Increased mortality is observed at 24 hpf in zebrafish embryos co-injected with Marcksa and Marcksb morpholinos1.
| Treatment Group | % Mortality |
|---|---|
| 2 ng MAT + 2 ng MBT | 97.1 ± 5.0 |
| 2 ng MAT + 2 ng CONT | 83.1 ± 11.7 |
| 2 ng MBT + 2 ng CONT | 58.2 ± 20.12 |
| Wild type, non-injected | 10.7 ± 11.72 |
Data is representative of average ± SEM
Denoting a statistically significant decrease in % mortality relative to 2 ng MAT + 2 ng MBT group (p<0.05)
Marcksa and Marcksb are required for neural development and retinal histogenesis in zebrafish
Our phenotypic characterization of Marcksa and Marcksb targeted zebrafish revealed that both proteins might be involved in neural development. With both Marcksa and Marcksb targeting MOs, we observed neural abnormalities that were not present in the control injected or wild type non-injected embryos (Fig. 3). Recently, Ekker and colleagues have shown that MOs can elicit off target effects, resulting in the activation of the p53 pathway and a “neural dead” phenotype (Robu et al., 2007). Therefore, to confirm that the neural phenotype that we observed in Marcksa and Marcksb targeted embryos was not due to off target effects, we performed p53 MO co-injection experiments. Because we observed the same phenotypes with MAT and MATU as well as MBT and MBS morpholinos, for the remainder of experiments we chose to use MAT and MBT to target Marcksa and Marcksb, respectively. For the p53 co-injection experiments, we injected the following groups: 2ng MAT + 4 ng p53 MO, 2 ng MBT + 4 ng p53 MO, 2 ng MAT + 4 ng control MO and 2 ng MBT + 4 ng control MO. Observations of the co-injected groups were made at 24 hpf (Fig. 6). In the MAT + control MO and MBT + control MO groups the same neural phenotype (characterized by a lack of normal eye and neural tissue architecture) was observed (Fig. 6). Similarly, in the MAT + p53 MO and MBT + p53 MO groups the neural phenotype was once again observed (Fig. 6). These findings confirm that the neural phenotypes observed in Marcksa and Marcksb targeted embryos are true phenotypes. Further, it suggests that Marcks is involved in the neural development of zebrafish.
Figure 6.
Marcksa and Marcksb are involved in normal zebrafish neural development. Embryos were co-injected in the single cell stage with MAT or MBT MOs along with p53 or control MOs. Embryos were analyzed at 24 hpf for the presence of the neural dead phenotype. Representative embryos are shown; wild type, non-injected (A), 4 ng p53 MO + 2 ng control MO (B), 2 ng MBT + 4 ng p53 MO (C), 2 ng MAT + 4 ng p53 MO (D), 2 ng MBT + 4 ng control MO (E), 2 ng MAT + 4 ng control MO (F).
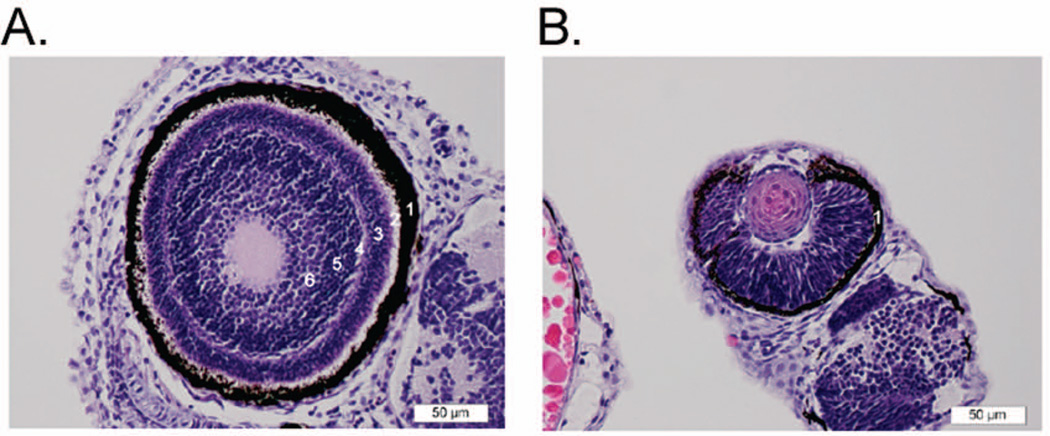
Histopathological analysis further revealed neural abnormalities in both Marcksa and Marcksb deficient zebrafish. MAT and MBT embryos were euthanized 72 hpf, fixed and routinely processed, and multiple sections stained with hematoxylin & eosin (H & E) were examined microscopically. The observed neural abnormality consists of irregular retinal layering in both Marcksa and Marcksb targeted embryos, whereas control injected and wild type embryos have normal retinas (Fig. 7). While the outermost layer, the retinal pigment epithelium appears normal in Marcksa and Marcksb targeted zebrafish, the other layers essentially consist of a mass of poorly organized neural cell nuclei with no clear separation into normal retinal layers (Fig. 7B). In addition, the overall size of the eyes in these fish appears smaller (Fig. 7) and interestingly, we did observe a correlation between the severity of the retinal layering and the severity of the phenotypic characterization of the embryo (data not shown). Taken together, our data indicate that Marcks is involved in the neural development of zebrafish.
Figure 7.
Marcks is involved in retinal histogenesis of zebrafish. (A) Normal eye from 72 hpf wild type zebrafish embryo. Here, the layers of the retina are relatively well defined. Starting from the outermost layer and working toward the center of the eye: 1) the dark brown/black retinal pigment epithelium; 2) layer of rods and cones; 3) outer nuclear layer (dark blue/purple); 4) outer plexiform layer; 5) inner nuclear layer; 6) inner plexiform layer. The ganglion cell and nerve fiber layers are less distinct in this section. The plexiform layers consist mainly of synapses of the various sensory neural cells. (B) Eye from 72 hpf MAT injected zebrafish embryo (moderate phenotype). The retina consists of a mass of poorly organized neural cell nuclei with no clear separation into normal retinal layers. Only the retinal pigment epithelium forms a distinct layer (1). This morphology is consistent in both the MAT and MBT injected fish, and is most severe in the severe phenotype fish (data not shown).
Marcksa and Marcksb are required for gill formation and skeletal muscle cell morphology in developing zebrafish
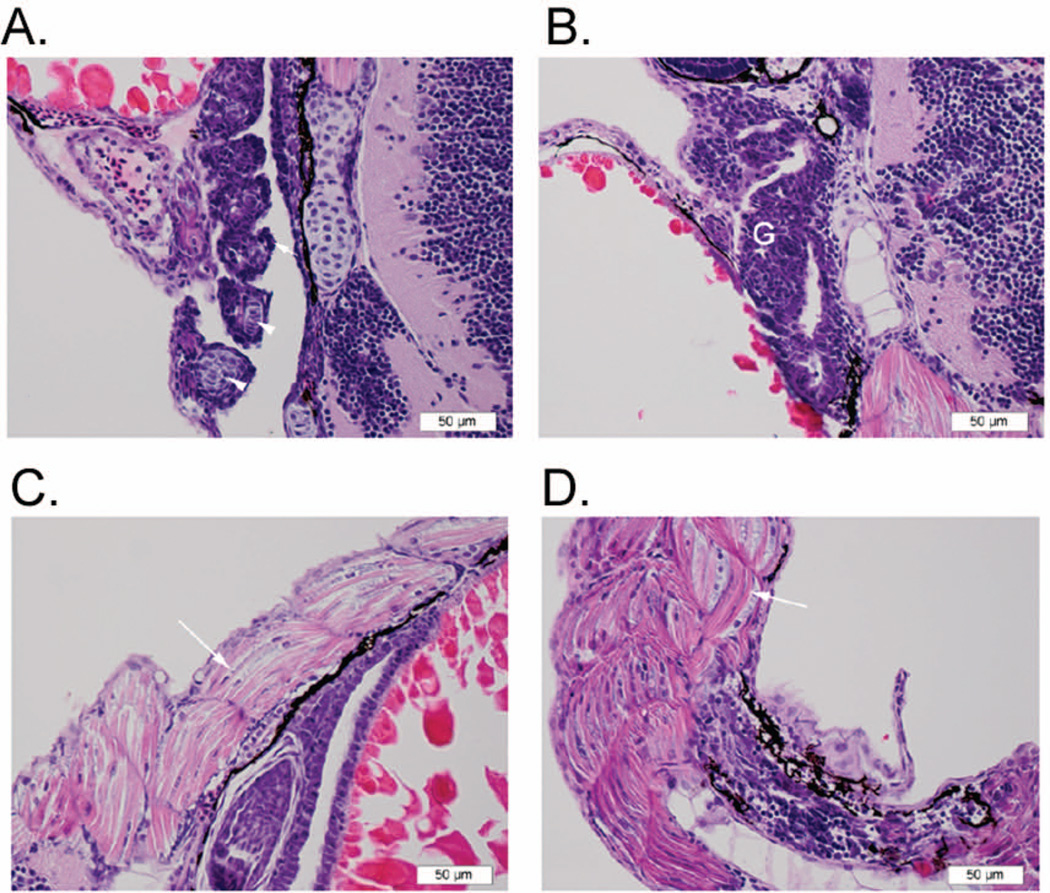
Histopathology on 72 hpf Marcksa and Marcksb targeted zebrafish embryos also revealed abnormal gill formation. The gills in these specimens consist of jumbled layers of epithelial cells with no discernable separation into lamellar units that would normally be separated by a core of cartilage (Fig. 8 A and B). Further, consistent with the curved or twisted tail phenotype, histopathology revealed that the skeletal muscle in the tails of Marcksa and Marcksb targeted zebrafish embryos contains numerous curved/crescent-shaped fibers and plumper, more numerous nuclei as compared to control injected and wild type, non-injected embryos (Fig. 8 C and D). Again, a correlation between the abnormalities in gill formation and muscle cell morphology and the severity of the phenotypic characterization of the embryo was observed (data not shown).
Figure 8.
Marcks targeting zebrafish results in abnormal gill development and muscle cell morphology. (A) Normal gill tissue from 72 hpf wild type zebrafish embryo. The primary lamellae (filaments) of the developing gill are shown in cross section, with normal supporting chondrocytes (arrow heads) at the center and a covering of squamous epithelial cells (arrow). (B) Gill tissue (G) from 72 hpf MAT injected zebrafish embryo (moderate phenotype). In this section, the gill is comprised of a mass of numerous, poorly organized epithelial cells with no clear separation into primary lamellae and no discernable central supporting chrondrocytes. This morphology is consistent in both MAT and MBT injected fish, and is most severe in the severe phenotype fish (data not shown). (C) Skeletal muscle fibers (arrow) from the tail of 72 hpf control MO injected zebrafish are relatively straight, with thin, regularly spaced nuclei. (D) Skeletal muscle from the tail of 72 hpf MAT injected zebrafish (moderate phenotype) contains numerous curved/crescent-shaped fibers (arrow) and plumper, more numerous nuclei as compared to control and wild type fish. This morphology is consistent in both the MAT and MBT fish, and is most severe in the severe phenotype fish (data not shown).
Discussion
MARCKS has been identified as a protein involved in mammalian and Xenopus development and in the present study we demonstrate a role for MARCKS in zebrafish development. Zebrafish express two MARCKS genes, marcksa and marcksb; both of which are expressed throughout early development and have similar homology to MARCKS expressed by other vertebrate species. Targeted disruption of Marcksa or Marcksb proteins in zebrafish results in increased mortality as well as several other developmental abnormalities. To our knowledge, this is the first analysis for the role of Marcks in zebrafish development. The role of MARCKS in the embryonic and neural development of other species has been previously examined (Stumpo et al., 1995, Hamada et al., 2003, Higo et al., 2004, Iioka et al., 2004) and our results support this research and offers further evidence for the role of MARCKS in developmental biology.
One of the most noticeable phenotypes observed in Marcks targeted zebrafish was increased mortality. An increased mortality in Marcksa or Marcksb targeted zebrafish was observed in all doses relative to wild type non-injected and control MO injected embryos and mortality increased as the embryos aged (Figs. 4 – 5, Tables 1–4). Further, we observed nearly complete (97.1 ± 5.0%) mortality at 24 hpf when embryos were co-injected 2ng of MAT and MBT morpholinos (Table 5). The increased mortality in Marcks targeted zebrafish is consistent with previous data in a mouse model. Disruption of MARCKS in mice (Marcks−/−) is embryonic lethal and mouse pups that survive birth die shortly thereafter (Stumpo et al., 1995). Interestingly, MARCKS heterozygotes (Marcks+/−) appeared normal and were capable of reproduction (Stumpo et al., 1995) and homozygous non-myristoylated MARCKS transgenic mice had an increased postnatal survival rate, with 25% of the mouse pups surviving the perinatal period (Swierczynski et al., 1996).
Besides increased mortality, we observed anatomical abnormalities in Marcksa or Marcksb targeted zebrafish embryos. The abnormalities observed could be divided into four phenotypic groups: normal, mild, moderate and severe. Mild phenotyped embryos had a slightly curved tail wheras moderate phenotyped embryos have a more severe curve or twist to the tail. Severe phenotyped embryos had truncated or cropped tails, with tails not extending beyond the yolk sac (Fig. 3). Although we could not confirm knockdown of either Marcksa or Marcksb by western blot due to the lack of an antibody specific for zebrafish Marcks, we did observe the same phenotypic characteristics in zebrafish when injected with two separate MOs targeting the same gene. As shown in figure 4 and tables 1 and 2, MAT and MATU, both of which target Marcksa, result in the same phenotypes (mild, moderate and severe) in a dose dependent manner and this same observation was also made when embryos were injected with MBT and MBS (Fig. 5, Tables 3 and 4), both of which target Marcksb. We acknowledge that this does not confirm knockdown of Marcksa or Marcksb, but it does, however, suggest specificity of the MOs designed for targeting Marcksa and Marcksb and that the phenotypes observed are real.
The phenotypes that we observed in Marcksa and Marcksb targeted zebrafish occurred as early as 24 hpf, suggesting that Marcks may play a role in the early development of zebrafish and potentially during gastrulation. In Xenopus laevis, knockdown of MARCKS leads to defective gastrulation, which is the result of impaired involution of the mesoderm and failed blastopore closure. MARCKS deficiencies in Xenopus were also associated with impaired convergent extension (Iioka et al., 2004). Convergent extension results in body axis elongation via polarized cell movements that converge medio-laterally and extends anterior-posteriorly (Keller, 2002, Schlessinger et al., 2009). Previous studies have shown the phenotype of convergent extension defective zebrafish embryos (Liu et al., 2009, van Eekelen et al., 2010) and the curved, twisted and truncated tails (mild, moderate and severe phenotypes, respectively) observed in the current study (Fig. 3) suggest that Marcks may also be involved in zebrafish convergent extension.
Other abnormalities observed in Marcks targeted zebrafish were neural abnormalities, as shown by co-injection of either Marcksa or Marcksb MOs with the p53 MO (Fig. 6). Studies have revealed that some MOs can elicit off target effects which are characterized by neural abnormalities. This phenotype is due to activation of the p53 pathway and studies have revealed that co-injection with a p53 MO ameliorate the off target effects and subsequent neural abnormalities (Robu, et al., 2007). Given that we still observed neural abnormalities when we co-injected either Marcksa + p53 MO or Marcksb + p53 MO suggests that MARCKS is involved in the neural development of zebrafish. Consistent with our findings, MARCKS deficiencies in mice also result in neural abnormalities, including exencephaly resulting from a failure in neural tube closure (Stumpo et al., 1995, Blackshear et al., 1996, Zolessi and Arruti, 2001). MARCKS deficiencies in non-exencephalic mice resulted in the failure of fusion of the cerebral hemispheres and agenesis of the corpus callosum and other forebrain commisures (Stumpo et al., 1995). In the studies presented herein, we did not observe defects in neural tube formation in Marcksa or Marcksb targeted zebrafish embryos by histopathologic evaluation. This was surprising given that there has been a large amount of literature describing the neural tube abnormalities associated with MARCKS deficiencies in mice. Zebrafish neural tube formation occurs prior to 24 hpf at the 20–25 somites stage (Lowery and Sive, 2004) and thus it is possible that the high mortality rate observed in MARCKS MO injected embryos is due to defects in neural tube formation. However, supporting a role for Marcksa and Marcksb in other aspects of neural development, we observed abnormal retinal histogenesis in MARCKS targeted zebrafish embryos (Fig. 7), and this data correlates with data that previously established MARCKS involvement in the histogenesis of the retina (Stumpo et al., 1995, Zolessi and Arruti, 2004). Specifically, it was found that the thick non-nuclear layer (layer of Chievitz) was absent in MARCKS homozygous deficient mice retinas (Stumpo et al., 1995). Taken together, our data suggests that Marcks plays a role in the neural development of zebrafish, specifically retinal histogenesis, a finding consistent with observations in other species.
Besides neural and retinal abnormalities, we also observed abnormal gill formation and muscle cell morphology (Fig. 8). The observed abnormal gill formation could certainly contribute to the increased mortality in Marcksa and Marcksb targeted embryos (Figs. 4 and 5, Tables 1–4). The role of MARCKS in respiratory physiology has been well studied as previous investigation has demonstrated that MARCKS is involved in mucin secretion in the respiratory epithelium (Singer et al., 2004, Park et al., 2008). However, to our knowledge, a role for MARCKS in respiratory development has yet to be demonstrated, making our observation of abnormal gill formation in Marcks targeted zebrafish a novel finding. This finding also suggests the need for further research on the role of MARCKS in respiratory development of both mammalian and non-mammalian vertebrates. The abnormal muscle morphology observed in Marcksa or Marcksb targeted zebrafish embryos supports the argument that MARCKS may play a role during in vivo myogenesis. Interestingly, myocyte adhesion and spreading (Disatnik et al., 2002, Disatnik et al., 2004) as well as migration (Dedieu et al., 2004) are associated with MARCKS function in vitro, as is fusion of embryonic myoblasts (Kim et al., 2002). Given these observations, zebrafish may be a suitable model for further studying the role of MARCKS in both respiratory and muscle development.
One potential process that is altered in MARCKS targeted zebrafish is cell migration. Cell migration is critical to development biology, especially during the morphogenetic movements of gastrulation (Chuai et al., 2009, Roszko et al., 2009, Wang and Steinbeisser, 2009) and recent evidence suggests a role for MARCKS in gastrulation (Iioka et al., 2004). MARCKS function is also required for mesenchymal and leukocyte cell migration. The growth hormone, platelet derived growth factor-BB (PDGF-BB) results in activation of PKCδ, which in turns phosphorylates MARCKS leading to migration of hepatic stellate cells (Rombouts et al., 2008). MARCKS silencing is also associated with inhibition of vascular smooth muscle cell migration (Monahan et al., 2009) and inhibition of calpain, a protease of which MARCKS is a substrate, leads to an accumulation of MARCKS and inhibition of myoblast migration (Dedieu et al., 2003, Dedieu et al., 2004). We have demonstrated a role for the myristoylated amino-terminus of MARCKS in the regulation of neutrophil migration in vitro (Eckert et al., 2009) and MARCKS has also been associated with the establishment of polarity of T lymphocytes (Friedl and Brocker, 2000). Taken together, MARCKS function is required for migration of diverse cell populations, which may explain the variety of developmental abnormalities observed in Marcks targeted zebrafish.
The experiments presented here describe a useful in vivo model to study MARCKS function. Zebrafish are a well established model for developmental biology and are becoming increasingly popular in other fields, including innate immunology (Mathias et al., 2006, Renshaw et al., 2006, Jima et al., 2009). Given the results of these experiments, we now have a model to study MARCKS function in vivo, increasing our ability to better understand the mechanisms by which MARCKS acts.
Acknowledgements
The authors would like to thank Aaron Garner and Amy Heffelfinger for assistance with microinjections and zebrafish husbandry. The authors would also like to thank Dr. Jonathan Horowitz for assistance with microscopy. Finally, the authors would like to thank Dr. Nanette Nascone-Yoder for helpful discussions and review of this manuscript.
Sequence from this article have been deposited with the GenBank and ZFIN databases under accession numbers GU563328 and ZDB-GENE-050522-145 (marcksa), and GU563329 and ZDB-GENE-030131-1921 (marcksb).
Literature Cited
- Aderem A. The MARCKS brothers: A family of protein kinase C substrates. Cell. 1992;71:713–716. doi: 10.1016/0092-8674(92)90546-o. [DOI] [PubMed] [Google Scholar]
- Ali N, Macala LJ, Hayslett JP. Identification and characterization of MARCKS in xenopus laevis. Biochem Biophys Res Commun. 1997;234:143–146. doi: 10.1006/bbrc.1997.6604. [DOI] [PubMed] [Google Scholar]
- Allen LH, Aderem A. A role for MARCKS, the alpha isozyme of protein kinase C and myosin I in zymosan phagocytosis by macrophages. J Exp Med. 1995;182:829–840. doi: 10.1084/jem.182.3.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbuzova A, Schmitz AA, Vergeres G. Cross-talk unfolded: MARCKS proteins. Biochem J. 2002;362:1–12. doi: 10.1042/0264-6021:3620001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackshear PJ. The MARCKS family of cellular protein kinase C substrates. J Biol Chem. 1993;268:1501–1504. [PubMed] [Google Scholar]
- Blackshear PJ, Lai WS, Tuttle JS, Stumpo DJ, Kennington E, Nairn AC, Sulik KK. Developmental expression of MARCKS and protein kinase C in mice in relation to the exencephaly resulting from MARCKS deficiency. Brain Res Dev Brain Res. 1996;96:62–75. doi: 10.1016/0165-3806(96)00097-1. [DOI] [PubMed] [Google Scholar]
- Chuai M, Dormann D, Weijer CJ. Imaging cell signalling and movement in development. Semin Cell Dev Biol. 2009;20:947–955. doi: 10.1016/j.semcdb.2009.09.001. [DOI] [PubMed] [Google Scholar]
- Dedieu S, Mazeres G, Poussard S, Brustis JJ, Cottin P. Myoblast migration is prevented by a calpain-dependent accumulation of MARCKS. Biol Cell. 2003;95:615–623. doi: 10.1016/j.biolcel.2003.09.005. [DOI] [PubMed] [Google Scholar]
- Dedieu S, Poussard S, Mazeres G, Grise F, Dargelos E, Cottin P, Brustis JJ. Myoblast migration is regulated by calpain through its involvement in cell attachment and cytoskeletal organization. Exp Cell Res. 2004;292:187–200. doi: 10.1016/j.yexcr.2003.08.014. [DOI] [PubMed] [Google Scholar]
- Disatnik MH, Boutet SC, Lee CH, Mochly-Rosen D, Rando TA. Sequential activation of individual PKC isozymes in integrin-mediated muscle cell spreading: A role for MARCKS in an integrin signaling pathway. J Cell Sci. 2002;115:2151–2163. doi: 10.1242/jcs.115.10.2151. [DOI] [PubMed] [Google Scholar]
- Disatnik MH, Boutet SC, Pacio W, Chan AY, Ross LB, Lee CH, Rando TA. The bi-directional translocation of MARCKS between membrane and cytosol regulates integrin-mediated muscle cell spreading. J Cell Sci. 2004;117:4469–4479. doi: 10.1242/jcs.01309. [DOI] [PubMed] [Google Scholar]
- Eckert RE, Neuder LE, Park J, Adler KB, Jones SL. Myristoylated alanine-rich C-kinase substrate (MARCKS) protein regulation of human neutrophil migration. Am J Respir Cell Mol Biol. 2010;42:586–594. doi: 10.1165/rcmb.2008-0394OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallman M, Gullberg M, Hellberg C, Andersson T. Complement receptor-mediated phagocytosis is associated with accumulation of phosphatidylcholine-derived diglyceride in human neutrophils. involvement of phospholipase D and direct evidence for a positive feedback signal of protein kinase. J Biol Chem. 1992;267:2656–2663. [PubMed] [Google Scholar]
- Friedl P, Brocker EB. T cell migration in three-dimensional extracellular matrix: Guidance by polarity and sensations. Dev Immunol. 2000;7:249–266. doi: 10.1155/2000/56473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada H, Zhang YL, Kawai A, Li F, Hibino Y, Hirashima Y, Kurimoto M, Hayashi N, Kato I, Endo S, Hiraga K. Development-associated myristoylated alanine-rich C kinase substrate phosphorylation in rat brain. Childs Nerv Syst. 2003;19:152–158. doi: 10.1007/s00381-002-0713-x. [DOI] [PubMed] [Google Scholar]
- Hartwig JH, Thelen M, Rosen A, Janmey PA, Nairn AC, Aderem A. MARCKS is an actin filament crosslinking protein regulated by protein kinase C and calcium-calmodulin. Nature. 1992;356:618–622. doi: 10.1038/356618a0. [DOI] [PubMed] [Google Scholar]
- Heemskerk FM, Chen HC, Huang FL. Protein kinase C phosphorylates Ser152, Ser156 and Ser163 but not Ser160 of MARCKS in rat brain. Biochem Biophys Res Commun. 1993;190:236–241. doi: 10.1006/bbrc.1993.1036. [DOI] [PubMed] [Google Scholar]
- Higo N, Oishi T, Yamashita A, Murata Y, Matsuda K, Hayashi M. Northern blot and in situ hybridization analyses for the development of myristoylated alanine-rich c-kinase substrate mRNA in the monkey cerebral cortex. Neuroscience. 2004;129:167–177. doi: 10.1016/j.neuroscience.2004.07.039. [DOI] [PubMed] [Google Scholar]
- Iioka H, Ueno N, Kinoshita N. Essential role of MARCKS in cortical actin dynamics during gastrulation movements. J Cell Biol. 2004;164:169–174. doi: 10.1083/jcb.200310027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jima DD, Shah RN, Orcutt TM, Joshi D, Law JM, Litman GW, Trede NS, Yoder JA. Enhanced transcription of complement and coagulation genes in the absence of adaptive immunity. Mol Immunol. 2009;46:1505–1516. doi: 10.1016/j.molimm.2008.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller R. Shaping the vertebrate body plan by polarized embryonic cell movements. Science. 2002;298:1950–1954. doi: 10.1126/science.1079478. [DOI] [PubMed] [Google Scholar]
- Kim SS, Kim JH, Kim HS, Park DE, Chung CH. Involvement of the theta-type protein kinase C in translocation of myristoylated alanine-rich C kinase substrate (MARCKS) during myogenesis of chick embryonic myoblasts. Biochem J. 2000;347(Pt 1):139–146. [PMC free article] [PubMed] [Google Scholar]
- Kim SS, Kim JH, Lee SH, Chung SS, Bang OS, Park D, Chung CH. Involvement of protein phosphatase-1-mediated MARCKS translocation in myogenic differentiation of embryonic muscle cells. J Cell Sci. 2002;115:2465–2473. doi: 10.1242/jcs.115.12.2465. [DOI] [PubMed] [Google Scholar]
- Kumar S, Nei M, Dudley J, Tamura K. MEGA: A biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform. 2008;9:299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Li Y, Martin LD, Spizz G, Adler KB. MARCKS protein is a key molecule regulating mucin secretion by human airway epithelial cells in vitro. J Biol Chem. 2001;276:40982–40990. doi: 10.1074/jbc.M105614200. [DOI] [PubMed] [Google Scholar]
- Liu JX, Hu B, Wang Y, Gui JF, Xiao W. Zebrafish eaf1 and eaf2/u19 mediate effective convergence and extension movements through the maintenance of wnt11 and wnt5 expression. J Biol Chem. 2009;284:16679–16692. doi: 10.1074/jbc.M109.009654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowery LA, Sive H. Stratagies of vertebrate neurulation and a re-evalulation of teleost neural tube formation. Mechanisms of Development. 2004;121:1189–1197. doi: 10.1016/j.mod.2004.04.022. [DOI] [PubMed] [Google Scholar]
- Mathias JR, Perrin BJ, Liu TX, Kanki J, Look AT, Huttenlocher A. Resolution of inflammation by retrograde chemotaxis of neutrophils in transgenic zebrafish. J Leukoc Biol. 2006;80:1281–1288. doi: 10.1189/jlb.0506346. [DOI] [PubMed] [Google Scholar]
- Monahan TS, Andersen ND, Martin MC, Malek JY, Shrikhande GV, Pradhan L, Ferran C, LoGerfo FW. MARCKS silencing differentially affects human vascular smooth muscle and endothelial cell phenotypes to inhibit neointimal hyperplasia in saphenous vein. FASEB J. 2009;23:557–564. doi: 10.1096/fj.08-114173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myat MM, Anderson S, Allen LA, Aderem A. MARCKS regulates membrane ruffling and cell spreading. Curr Biol. 1997;7:611–614. doi: 10.1016/s0960-9822(06)00262-4. [DOI] [PubMed] [Google Scholar]
- Neuder LE, Keener JM, Eckert RE, Trujillo JC, Jones SL. Role of p38 MAPK in LPS induced pro-inflammatory cytokine and chemokine gene expression in equine leukocytes. Vet Immunol Immunopathol. 2009;129:192–199. doi: 10.1016/j.vetimm.2008.11.006. [DOI] [PubMed] [Google Scholar]
- Park J, Fang S, Crews AL, Lin KW, Adler KB. MARCKS regulation of mucin secretion by airway epithelium in vitro: Interaction with chaperones. Am J Respir Cell Mol Biol. 2008;39:68–76. doi: 10.1165/rcmb.2007-0139OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel J, Kligman D. Purification and characterization of an mr 87,000 protein kinase C substrate from rat brain. J Biol Chem. 1987;262:16686–16691. [PubMed] [Google Scholar]
- Renshaw SA, Loynes CA, Trushell DM, Elworthy S, Ingham PW, Whyte MK. A transgenic zebrafish model of neutrophilic inflammation. Blood. 2006;108:3976–3978. doi: 10.1182/blood-2006-05-024075. [DOI] [PubMed] [Google Scholar]
- Robu ME, Larson JD, Nasevicius A, Beiraghi S, Brenner C, Farber SA, Ekker SC. P53 activation by knockdown technologies. PLoS Genet. 2007;3:e78. doi: 10.1371/journal.pgen.0030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roszko I, Sawada A, Solnica-Krezel L. Regulation of convergence and extension movements during vertebrate gastrulation by the Wnt/PCP pathway. Semin Cell Dev Biol. 2009;20:986–997. doi: 10.1016/j.semcdb.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarlett CO, Blackshear PJ. Neuroanatomical development in the absence of PKC phosphorylation of the myristoylated alanine-rich C-kinase substrate (MARCKS) protein. Brain Res Dev Brain Res. 2003;144:25–42. doi: 10.1016/s0165-3806(03)00155-x. [DOI] [PubMed] [Google Scholar]
- Schlessinger K, Hall A, Tolwinski N. Wnt signaling pathways meet rho GTPases. Genes Dev. 2009;23:265–277. doi: 10.1101/gad.1760809. [DOI] [PubMed] [Google Scholar]
- Seykora JT, Myat MM, Allen LA, Ravetch JV, Aderem A. Molecular determinants of the myristoyl-electrostatic switch of MARCKS. J Biol Chem. 1996;271:18797–18802. doi: 10.1074/jbc.271.31.18797. [DOI] [PubMed] [Google Scholar]
- Singer M, Martin LD, Vargaftig BB, Park J, Gruber AD, Li Y, Adler KB. A MARCKS-related peptide blocks mucus hypersecretion in a mouse model of asthma. Nat Med. 2004;10:193–196. doi: 10.1038/nm983. [DOI] [PubMed] [Google Scholar]
- Song JC, Hrnjez BJ, Farokhzad OC, Matthews JB. PKC-epsilon regulates basolateral endocytosis in human T84 intestinal epithelia: Role of F-actin and MARCKS. Am J Physiol. 1999;277:C1239–49. doi: 10.1152/ajpcell.1999.277.6.C1239. [DOI] [PubMed] [Google Scholar]
- Strausberg RL, Feingold EA, Grouse LH, Derge JG, Klausner RD, Collins FS, Wagner L, Shenmen CM, Schuler GD, Altschul SF, Zeeberg B, Buetow KH, Schaefer CF, Bhat NK, Hopkins RF, Jordan H, Moore T, Max SI, Wang J, Hsieh F, Diatchenko L, Marusina K, Farmer AA, Rubin GM, Hong L, Stapleton M, Soares MB, Bonaldo MF, Casavant TL, Scheetz TE, Brownstein MJ, Usdin TB, Toshiyuki S, Carninci P, Prange C, Raha SS, Loquellano NA, Peters GJ, Abramson RD, Mullahy SJ, Bosak SA, McEwan PJ, McKernan KJ, Malek JA, Gunaratne PH, Richards S, Worley KC, Hale S, Garcia AM, Gay LJ, Hulyk SW, Villalon DK, Muzny DM, Sodergren EJ, Lu X, Gibbs RA, Fahey J, Helton E, Ketteman M, Madan A, Rodrigues S, Sanchez A, Whiting M, Madan A, Young AC, Shevchenko Y, Bouffard GG, Blakesley RW, Touchman JW, Green ED, Dickson MC, Rodriguez AC, Grimwood J, Schmutz J, Myers RM, Butterfield YS, Krzywinski MI, Skalska U, Smailus DE, Schnerch A, Schein JE, Jones SJ, Marra MA Mammalian Gene Collection Program Team. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci U S A. 2002;99:16899–16903. doi: 10.1073/pnas.242603899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpo DJ, Bock CB, Tuttle JS, Blackshear PJ. MARCKS deficiency in mice leads to abnormal brain development and perinatal death. Proc Natl Acad Sci U S A. 1995;92:944–948. doi: 10.1073/pnas.92.4.944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaram M, Cook HW, Byers DM. The MARCKS family of phospholipid binding proteins: Regulation of phospholipase D and other cellular components. Biochem Cell Biol. 2004;82:191–200. doi: 10.1139/o03-087. [DOI] [PubMed] [Google Scholar]
- Swierczynski SL, Siddhanti SR, Tuttle JS, Blackshear PJ. Nonmyristoylated MARCKS complements some but not all of the developmental defects associated with MARCKS deficiency in mice. Dev Biol. 1996;179:135–147. doi: 10.1006/dbio.1996.0246. [DOI] [PubMed] [Google Scholar]
- Tanabe A, Kamisuki Y, Hidaka H, Suzuki M, Negishi M, Takuwa Y. PKC phosphorylates MARCKS Ser159 not only directly but also through RhoA/ROCK. Biochem Biophys Res Commun. 2006;345:156–161. doi: 10.1016/j.bbrc.2006.04.082. [DOI] [PubMed] [Google Scholar]
- Taylor JS, Braasch I, Frickey T, Meyer A, Van de Peer Y. Genome duplication, a trait shared by 22000 species of ray-finned fish. Genome Res. 2003;13:382–390. doi: 10.1101/gr.640303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JS, Van de Peer Y, Braasch I, Meyer A. Comparative genomics provides evidence for an ancient genome duplication event in fish. Philos Trans R Soc Lond B Biol Sci. 2001;356:1661–1679. doi: 10.1098/rstb.2001.0975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse B, Thisse C. Fast Release Clones: A High Throughput Expression Analysis. ZFIN Direct Data Submission. 2004 http://zfin.org.
- Tzlil S, Murray D, Ben-Shaul A. The “electrostatic-switch” mechanism: Monte carlo study of MARCKS-membrane interaction. Biophys J. 2008;95:1745–1757. doi: 10.1529/biophysj.108.132522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Eekelen M, Runtuwene V, Overvoorde J, den Hertog J. RPTPalpha and PTPepsilon signaling via Fyn/Yes and RhoA is essential for zebrafish convergence and extension cell movements during gastrulation. Dev Biol. 2010;340:626–639. doi: 10.1016/j.ydbio.2010.02.026. [DOI] [PubMed] [Google Scholar]
- Wang Y, Steinbeisser H. Molecular basis of morphogenesis during vertebrate gastrulation. Cell Mol Life Sci. 2009;66:2263–2273. doi: 10.1007/s00018-009-0018-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijer CJ. Collective cell migration in development. J Cell Sci. 2009;122:3215–3223. doi: 10.1242/jcs.036517. [DOI] [PubMed] [Google Scholar]
- Weimer JM, Yokota Y, Stanco A, Stumpo DJ, Blackshear PJ, Anton ES. MARCKS modulates radial progenitor placement, proliferation and organization in the developing cerebral cortex. Development. 2009;136:2965–2975. doi: 10.1242/dev.036616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarmola EG, Edison AS, Lenox RH, Bubb MR. Actin filament cross-linking by MARCKS: Characterization of two actin-binding sites within the phosphorylation site domain. J Biol Chem. 2001;276:22351–22358. doi: 10.1074/jbc.M101457200. [DOI] [PubMed] [Google Scholar]
- Zolessi FR, Arruti C. Apical accumulation of MARCKS in neural plate cells during neurulation in the chick embryo. BMC Dev Biol. 2001;1:7. doi: 10.1186/1471-213X-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolessi FR, Arruti C. MARCKS in advanced stages of neural retina histogenesis. Dev Neurosci. 2004;26:371–379. doi: 10.1159/000082279. [DOI] [PubMed] [Google Scholar]
- Zolessi FR, Duran R, Engstrom U, Cervenansky C, Hellman U, Arruti C. Identification of the chicken MARCKS phosphorylation site specific for differentiating neurons as ser 25 using a monoclonal antibody and mass spectrometry. J Proteome Res. 2004;3:84–90. doi: 10.1021/pr034066f. [DOI] [PubMed] [Google Scholar]