Abstract
We propose that microbial diversity must be viewed in light of gene flow and selection, which define units of genetic similarity, and of phenotype and ecological function, respectively. Here, we discuss to what extent ecological and genetic units overlap to form cohesive populations in the wild, based on recent evolutionary modeling and on evidence from some of the first microbial populations studied with genomics. These show that if recombination is frequent and selection moderate, ecologically adaptive mutations or genes can spread within populations independently of their original genomic background (gene-specific sweeps). Alternatively, if the effect of recombination is smaller than selection, genome-wide selective sweeps should occur. In both cases, however, distinct units of overlapping ecological and genotypic similarity will form if microgeographic separation, likely involving ecological tradeoffs, induces barriers to gene flow. These predictions are supported by (meta)genomic data, which suggest that a ‘reverse ecology’ approach, in which genomic and gene flow information is used to make predictions about the nature of ecological units, is a powerful approach to ordering microbial diversity.
Keywords: population genomics, ecological differentiation, reverse ecology, gene flow, selective sweeps, mosaic sympatric speciation
Introduction and motivation
It is often said that species are fundamental units of ecology because they comprise individuals that are phenotypically and hence ecologically more similar to each other than to other species [1,2]. This notion was extended in Mayr’s biological species concept [3], which states that species are reproductively isolated units, implying that adaptive mutations can spread within a species leaving other co-existing species unaffected. Although recent evidence has shown that reproductive boundaries can be leaky [4–6], species are still regarded as congruent genetic and ecological units for sexual eukaryotes, even if hybrids and intermediate forms are common [7]. For bacteria and archaea, however, the situation has been marred by several complicating factors that question whether such units can be defined.
In addressing whether we can identify genetically and ecologically congruent units, we need to take into account the peculiarities of bacterial and archaeal evolution, i.e., the varying modes and rates of genetic exchange. In these organisms, incorporation of new genetic material is always unidirectional and leads either to gene conversion by homologous recombination or gene addition by non-homologous recombination (see Glossary). (In fact, the distinction might not be so clear: there is mounting evidence that homologous recombination is often involved in gene addition and loss [8–11]). Importantly, the rates and bounds of this gene transfer can vary considerably. While some lineages follow a highly clonal mode of evolution, in others, rates of recombination can differ by several orders of magnitude. Regardless of the overall rate of gene flow, genetic material can, in principle, be incorporated from distantly related organisms. This variation in genetic exchange and its effect on genotypic integrity and ecological adaptation is at the heart of the debate about what constitutes ecological and genetic units for bacteria and archaea.
In particular, horizontal gene transfer (HGT) among distantly related organisms can create genotypes that vary in properties of ecological relevance by acquiring functions, such as antibiotic resistance or nitrogen fixation, that distinguish them from otherwise closely related genotypes [4,12]. At the same time, the recipient genotype has also become ecologically similar, in at least one niche dimension, to the organism from which it acquired the novel pathway. In fact, such functional differentiation is observed among closely related environmental isolates [13] and, in combination with high gene turnover, has been taken as evidence that gene acquisition and loss is so high as to quickly erode any niche association of lineages [12]. By extension, the very notion of a lineage has been questioned on the same grounds – with the consequence that nearly each genotype might represent its own, independent ecological unit [14] that can only be recognized by the functional genes it carries [15].
In recent years, however, analysis of environmental isolates and metagenomes has shown that microbial communities consist of genotypic clusters of closely related organisms and that these can display cohesive environmental associations and dynamics that clearly distinguish them from other such clusters co-existing in the same samples. Despite also showing evidence for extensive gene flow, genetically distinguishable clusters have been observed among closely related environmental and pathogenic isolates by multilocus sequence analysis and genomics [1,16,17], and by metagenomics [18–20]. Moreover, cohesive ecological dynamics and associations have been demonstrated for a growing number of cases, including for vibrios, sulfate-reducing bacteria and cyanobacteria, as well for organisms represented in several marine, freshwater and acid-mine drainage community metagenomes. These observations suggest congruence of genotypic and ecological units and are, in principle, consistent with the notion of populations as locally co-existing members of a species. As we will discuss below, selection and recombination are paramount in shaping and maintaining such units, although the effects of biogeography, on both local [20,21] and global [22,23] scales may also come into play.
The idea that genotypic clusters should be rapidly eroded by HGT might in part be an artifact of early comparative studies of quite anciently diverged genomes. In these, only a fraction of genes in the core genome showed phylogenetic congruence, and the flexible genome seemed to be completely unrelated [12,24]. Moreover, we often call organisms closely related if their 16S rRNA genes, which are commonly used as taxonomic markers, show few percent nucleotide differences, yet such difference may indicate millions of years of separate evolution with associated large genome changes [25]. But even as closely related genomes (e.g., identical in 16S rRNA genes) began to be sequenced, these usually were not isolated from the same habitat and hence were not part of the same populations of interacting genotypes. This means that the effect of environmental selection might not be easily disentangled from genetic divergence due to geographic separation [26]. For example, in the marine cyanobacterium Prochlorococcus, populations in the Atlantic contain genes responsible for efficient phosphorus acquisition that are absent from populations in the Pacific [27]. Hence these genes are part of the core genome of Atlantic populations but would be judged flexible genes if closely related isolates were compared from both ocean regions. We therefore believe that an important step forward will be to emphasize population-thinking in microbiology by assembling genomic datasets that represent clusters of close relatives co-occurring in the same environment – as only these will allow interpretation of how environmental selection acts on genomes from within the same population.
The challenge is then to develop an understanding of how genotypic clusters originate and are maintained, and whether they are selectively optimized to occupy sufficiently different niches to co-exist with other clusters. Importantly, any such attempt needs to take into account the considerable genotypic diversity encountered in environmental populations, which often consist of genomes differing by a considerable fraction of their gene content and displaying large allelic diversity even if most of their genes suggest close relationships [26].
In this review, we begin by discussing the extent to which ecological and genetic units overlap, and under what circumstances genetic units can be used as a proxy for ecological units. We argue that, while it is essential to sequence populations of microbial genomes and record ecological metadata, a powerful alternative is represented by a ‘reverse ecology’ approach in which genomic and gene flow information is used to make predictions about the nature of ecological units (Box 1). What distinguishes reverse ecology from the broader field of ecological genomics is its focus on simultaneously predicting ecological and genetic units, rather than mapping ecological data onto pre-defined genetic units. These predictions can then be tested using ecological metadata and experimental follow-up. Then we describe in detail two examples of reverse ecology applied to different closely related, sympatric, natural microbial populations. We synthesize conclusions from these examples, along with data from more distantly related genomic comparisons and evolutionary models, and propose a process of speciation that can operate under different regimes of selection and recombination (Table 1). Early stages of the speciation process are driven by either gene-specific or genome-wide selective sweeps as microbes adapt to new sympatric niches, with either high or low levels of recombination between niches, respectively. At more advanced stages, if barriers to gene flow between niches emerge, distinct units of microbial diversity come into focus and can be potentially be recognized (Box 2). As we have reasoned previously, units can be defined operationally in cases where both genotypic and phenotypic variance is much greater between than within such units [28]. We wish to make a strong distinction between this process of speciation – which we define as any stage of the dynamic process of ecological and genetic differentiation – and the concept of species, which we are not attempting to address. Speciation need not proceed to completion, and recognizable units of genotypic and ecological similarity will often contain abundant genetic and phenotypic diversity within them. We conclude by briefly highlighting the potential for reverse ecology to identify natural units of microbial diversity, and for genome-wide association studies (GWAS) to identify mutations and genes underlying ecologically relevant traits.
Box 1. How to perform a reverse ecology population genomic study.
The goal of the reverse ecology approach is to determine whether a sample of closely related, sympatric genome sequences constitute one or more genotypic units, and to test how these units might differ in their ecology either by mapping of these clusters onto environmental gradients or patches, or by laboratory tests. The sampling scheme need not be entirely unbiased. For example, isolates should be intentionally chosen to be closely-related. They could also be chosen from two or more hypothesized niches or phenotypic groups in order to test whether these groups behave as separate genotypic units (Box 2), and to uncover the genes or mutations that might contribute to their ecological differences. Isolates should, however, be sampled from the same geographic location in order to reduce the effects of allopatric divergence and focus on the effects of local selection and recombination. Some a priori information – perhaps from a previous phylogenetic or metagenomic survey – may also be required in order to select a subset of closely related populations from the community.
Choose a genomic or metagenomic approach. Whole-genome sequencing of cultured isolates or isolated (but uncultured) single cells is preferable because it reveals information about how genes and mutations are linked within genomes, facilitating inferences about recombination events among genomes. Metagenomic sequencing has the advantage of sampling more individuals within an environment than are generally possible to isolate, but linkage information will be limited by the sequencing read length and quality of the assembly. Most importantly, unbiased metagenomic sequencing will only provide an appropriate population genomic dataset for populations that are relatively abundant in the sampled environment. The power of metagenomic data can be boosted significantly if they are gathered as a time series. Although such data sets are currently rare and potentially challenging to collect, they can follow the speciation process (Table 1) in real time, and potentially catch selective sweeps and niche-specifying events in action. Fine-grained time series might also follow shifting ecolological conditions over time, revealing independent behaviors of different clusters.
Assemble genome sequences. Complete genome sequences are more readily assembled from isolates, but assembly can also be attempted on metagenomic data, taking care to guard against or account for different individuals being co-assembled into a single genome.
Align genome sequences and define core and flexible components. Here, particular care must be taken to only define these categories for organisms that co-occur and hence have the potential to be connected by contemporary gene flow and be subject to consistent environmental selection.
Evaluate phylogenetic signals in single nucleotide polymorphisms (SNPs) found in the core genome. Standard phylogenetic methods can be used to build a core genome-wide phylogeny, and the average impact of recombination can be measured by assessing linkage disequilibrium between SNPs. Specific recombination events and breakpoints can then be identified using methods such as BratNextGen [77], ClonalFrame/ClonalOrigin [78], and STARRInIGHTS [55]. These analyses will reveal the number of major genotypic units (well-supported monophyletic groups), and whether these units are supported genome-wide (consistent with mostly clonal evolution) or in ‘islands’ or ‘continents’ of the genome.
If populations were hypothesized a priori based on an ecological axis of interest, assess whether these presumed populations correspond to genotypic clusters or not. If genome-wide diversity is clustered according to ecology, this suggests that stable clusters have formed (Table 1, Stages 4–5). If there is little or no phylogenetic clustering according to ecology, the hypothesized populations likely constitute a single, phenotypically diverse population. In this case, certain (flexible) genes or (core) mutations that associate with ecology might be identified by GWAS (Figure 3). If there is a preference for recombination within rather than between ecological groups, the single population might be on a trajectory towards speciation (Table 1, Stage 3).
If populations were not hypothesized a priori (a ‘purer’ reverse ecology approach), assess how many phylogenetic groups were identified. If phylogenetic groupings are supported genome-wide, this suggests stable differentiation (Table 1, Stages 4–5), the ecological basis of which remains unknown but can be tested by phenotypically characterizing representative isolates from each group and/or mapping genotypic clusters onto environmental samples. If groupings are not supported genome-wide, genomic regions containing the bulk of the phylogenetic signal, or signals of positive selection, frequent recombination, or dense polymorphism can be functionally annotated to generate hypotheses about their possible ecological roles.
Table 1.
Stages of microbial speciation under different rates of selection and homologous recombinationa
| (A) r/s ≫ 1 | (B) r/s ≪ 1 | |
|---|---|---|
| Stage 1 | New niche-specifying variant(s) acquired by mutation, homologous recombination or HGT | |
| Stage 2 | Ecological separation: new variant spreads in new niche by recombination | Ecological separation: new variant spreads in new niche by clonal expansion |
| Stage 3 | Genetic separation driven by genome- wide depression in recombination between new and ancestral niches | Genetic separation driven by periodic selection and drift |
| Stage 4 | Genetic separation maintained by genetic barriers to recombination, including sequence divergence and epistasis; otherwise lineages may merge back together | Genetic separation maintained by further periodic selection and drift events; lineages are permanently separate |
| Stage 5 | Lineages remain ecologically and genetically distinct (at both adaptive and neutral loci, genome-wide) until extinction | |
The relative influence of selection [s, the average fitness difference experienced by a niche-specifying (adaptive) allele in different niches] and recombination (r, the recombination rate per locus per generation) is expressed as the r/s ratio. The stages represent rough, potentially overlapping and potentially terminal steps (e.g. Stage 2 need not lead to Stage 3).
Box 2. Challenges in identifying natural units of microbial diversity.
To determine whether two hypothesized units are indeed distinct, one must reject the hypothesis that they are both part of the same unit. This means that they must differ in at least one ecological dimension, and must show more genome-wide cohesion within than between units. The cohesion could be due to higher rates of recombination within than between populations, or due to independent genome-wide selective sweeps occurring in each population, without significant recombination. Therefore, natural units of genome-wide and ecological similarity can be produced under different regimes of selection and recombination (Table 1). Importantly, no absolute cutoff (for either genetic or ecological similarity) is necessary to define these units.
One challenge to overcome in identifying natural units is that some degree of recombination is to be expected between separate units, which might exchange ‘globally adaptive’ genes or alleles, while remaining separate elsewhere in the genome [79]. For example, different species of Campylobacter exchange genes of certain functions only, while remaining distinct throughout most of the genome [80,81]. In Streptococcus, cross-species exchange is often accompanied by positive selection (high dN/dS in the exchanged genes [47]). This suggests that biased cellular functions and positive selection might be general features of globally adaptive genes, allowing them to be recognized and excluded from phylogenetic or recombination-based tests for separation between units.
A second challenge is that a single cohesive population may still contain significant phenotypic and genotypic variation, but this variation will be restricted to only a relatively small fraction of the core and flexible genome. For example, genes under diversifying or frequency-dependent selection within a single cohesive populations might be mistaken for niche-specifying genes driving adaptation between populations. With careful application of population-genetic tests for natural selection, combined with phenotypic characterization of these genes, it is possible (if challenging) to distinguish between these two scenarios.
Defining genetic and ecological units
Ecological units, in the most basic sense, denote groups of organisms with common ecological functions. It is immediately obvious that this definition represents an abstraction by the observer and is hence subject to individual preferences of how finely one wishes to demarcate units [29]. For example, does the acquisition of an antibiotic resistance gene generate a new ecological unit or simply a variant within an existing unit? Do all sulfate-reducing bacteria represent one ecological unit since they all carry out a common, highly relevant environmental function? In other words, is an ecotype (defined here as ecologically completely equivalent genotypes) the right unit, or should we define ecological units more broadly? To understand the genetic basis of ecological preferences, microbiologists will generally make educated guesses about important and measurable dimensions of niche space (e.g., host preference and ability to grow on a particular carbon source) and embark on a population genomics study. Similar to classical, trait-based taxonomy, this approach is potentially subject to arbitrary weighing of phenotypes in order to define an ecological unit. An alternative approach is to avoid a priori guesses as much as possible, and sample closely related microbes, identify genomic units among them, and make hypotheses about their ecological differences (if any) based on the predicted or experimentally validated effects of these genomic differences, or, as we will argue later, based on patterns of gene flow. We refer to this as a reverse ecology approach [30–32] (Box 1). If these genomic units correspond to natural populations, this approach also provides the opportunity to test hypotheses about the evolutionary mechanism creating and maintaining diversity within and between such genomic units.
Genotypes are in principle easier to delineate than ecological types because a cell’s genome can be measured by sequencing, whereas it is not clear how many phenotypic properties have to be measured before a cell’s ecology is exhaustively captured. However, defining genetic units suffers from similar problems as for ecological units because it is not clear what measure of similarity to use and where to draw the bounds. Genotypes can be grouped into units based on various measures of genomic similarity: DNA hybridization assays, percent similarity in a marker gene, average nucleotide identity (ANI) across the genome, or the proportion of shared genes [33]. These are all convenient measures, but they all require that we decide on a cutoff value to divide units, often referred to as operational taxonomic units (OTUs).
An alternative is to search for natural genetic units based on the mechanisms capable of clustering genetic diversity, namely migration, mutation, recombination and selection. As detailed in Box 1, this can be done with a relatively limited sample of genomes from the same environment if one focuses on a defined taxonomic group. We exclude from this review patterns of biogeography that arise due to allopatric speciation, and we refer the reader to excellent recent work on the topic [20,34,35]. While allopatric speciation is conceptually more straightforward, sympatric speciation is thought to be more common in microbial populations [84]. We therefore focus on recombination and selection in sympatric settings.
Modeling the interplay of selection and recombination
In answering how genotypic clusters originate and are maintained, it is critical to evaluate the interplay of recombination and selection, both of which can vary widely. But while recombination rates can be measured to some extent, the magnitude of selection is difficult to assess directly in the wild, so that we have to rely on reasonable guesses. Below, we give an overview of current knowledge of recombination rates, and then show how mathematical models that explicitly incorporate recombination have been used to explore (i) the probability that clusters arise in sympatry due to neutral processes, and (ii) the effect of different recombination and selection rates on the spread of adaptive loci or alleles within and across populations.
As noted above, homologous recombination rates can vary tremendously in different lineages of bacteria and archaea, with some evolving in a highly clonal fashion while others are considered sexual, with recombination rates up to 10-fold higher than mutation rates [36], resulting in >10-fold more polymorphism from recombination than mutation [37]. It is, however, likely that most measured recombination rates are underestimates since typical analyses allow inference of recombination only when highly polymorphic segments of DNA are observed. Hence these measured rates might give a fairly accurate picture of recombination between but not within clusters. Moreover, experimental observations have suggested that the frequency of recombination drops exponentially with sequence divergence due to the requirement of a 20 bp stretch of identical DNA sequence for efficient initiation of recombination [38,39]. Such a rapid drop in frequency should limit efficient exchange of DNA to closely related genomes, as expected within genotypic clusters, and might play a role in maintaining the cohesion of clusters. Although such relationships have been demonstrated for several, divergent groups of bacteria, in some archaea, the requirement for short, identical DNA stretches seems to be absent [40,41], even though environmental observations support decreased rates of recombination across clusters [42,43]. Moreover, recent comparison of very closely related genomes has also shown that very little sequence similarity appears to be required for integration of long stretches of highly divergent DNA (including single nucleotide changes and structural variants) into the genome [9,11] although the mechanisms remain unclear. These recent results show that much remains to be learned about how recombination proceeds in different groups of bacteria and archaea, making mathematical models an important tool to explore potential outcomes, given reasonable assumptions about the importance of recombination relative to mutation and selection.
Whether genotypic clusters can arise neutrally in sympatry was addressed with a simple computational model starting with a single population that evolves by mutation and varying degrees of recombination, but in the absence of selection [44]. Without recombination, clonal clusters emerge by random mutation, but quickly drift to extinction. Because these clusters are short lived, they accumulate very little sequence diversity and would be hard to recognize in samples of microbes. When recombination rates become more frequent than mutation rates, however, clusters no longer emerge, and the population remains homogenous. A critical parameter in this model is the decline in the rate of homologous recombination with sequence divergence. Separate clusters are only formed if the rate of decline of recombination with mutational divergence is unrealistically high compared to those observed experimentally [9,11,38,39]. Hence the model suggests that natural selection should be required to produce stable genotypic clusters, and that neutral cluster formation is extremely unlikely – a prediction that is borne out in long-term microbial experimental evolution studies. These studies have provided evidence that most fixed mutations tend to be adaptive, not neutral [45], and that formation of new genotypic clusters might involve adaptation to using novel ressources [46].
Building on these results, a model was developed that includes one or more loci under selection, conferring adaptation to either of two sympatric niches, which are completely geographically overlapping, ensuring frequent mixing of all genotypes and an equal probability of sharing genes [47,48]. In this sympatric simulation (symsim) model, niche adaptation is encoded by genes or alleles already segregating within, or recently horizontally transferred into the population, with de novo adaptive mutation assumed to be negligible. The symsim model readily describes the simple case in which niches correspond to two different carbon sources dissolved in a single well-mixed aquatic environment. With rates of recombination much higher than selection (r/s ≫ 1), diversity at any neutral locus was unaffected by a selective sweep of an adaptive locus [47]. Conversely, when selection coefficients are much higher than recombination rates (r/s ≪ 1), an adaptive allele will generally sweep to fixation on a single genetic background, homogenizing neutral variation, as in the Stable Ecotype model (Box 3). However, even with r/s ≪ 1, given enough time before any further selective events, and assuming that the two niches remain sympatric, neutral alleles will eventually become randomly distributed across genotypes, with only adaptive alleles being selectively maintained [48]. Moreover, when the selective coefficient is distributed across more than one adaptive locus, this reduces the effective strength of selection and results in even stronger homogenization of neutral loci (and even to some extent, adaptive loci) across niches.
Box 3. The Stable Ecotype model.
The Stable Ecotype model of speciation, as developed by Cohan, invokes a prominent role for natural selection to form and maintain separate genetic clusters. It also provides an appealing mechanistic link between ecological and genetic units. In its basic form, an ecotype can be understood as the domain of competitive superiority of an adaptive mutant [82]. When an adaptive mutant arises within an ecotype population, it outcompetes its neighbors, purging diversity in a periodic selection event. Importantly, diversity is not purged in other ecotypes, which compete in independent niches. Ecotypes are also subject to neutral mutation and drift, which, along with periodic selection events, result in separate clusters of ecological and genetic diversity. The model states that observed rates of recombination are not high enough to unlink adaptive and neutral loci in the genome. Therefore, periodic selection is predicted to purge diversity genome-wide.
The Stable Ecotype model has never been directly observed in nature. Reasons for this might include recombination rates being underestimated, and niche complementarity maintaining multiple genotypes within a population. Support for the Stable Ecotype model has come from experimental evolution studies, in which diversity can only be generated by mutation within a restricted population, without the possibility for recombination with distant relatives. In these studies, adaptive mutations increase in frequency, eventually reaching fixation on a single genomic background, along with neutral ‘hitchhiking’ mutations [45]. Some mutations may found a new ecotype by allowing colonization of a new niche [83], followed by successive genome-wide sweeps in the new ecotype.
Hence the model shows that although adaptation spreads differently in the two regimes of recombination vs. selection, the eventual outcome is similar, i.e., recombination will eventually homogenize perfectly sympatric genotypes even if they carry niche-specific adaptations. The important consequence is that some kind of micro-geographic separation between niches, akin to the ‘mosaic sympatry’ described by Mallet [7], might be required to reduce gene flow between niche-adapted genotypes before clusters of selectively neutral genome-wide diversity may develop. Mosaic sympatry essentially means that niches are distributed patchily, without being completely allopatric [7]. This situation might readily describe many microbial environments, such as soil, oceans and animal hosts, where resources are distributed in small-scale patches, but patches may be short-lived and colonizing populations may mix frequently because of the need to recolonize new patches [28]. Barriers to gene flow might also arise to due to incompatible restriction modification or competence peptide systems yielding a form of mosaic sympatry, although empirical evidence that either system actually promotes speciation is lacking [16, 85]. Overall, there is a growing consensus that bacterial speciation generally takes place in sympatric or mosaic sympatric settings [84].
Taken together, these models suggest, first, that in the absence of selection, neither clonal nor sexual populations will split into stable, sympatric genotypic clusters due to neutral processes. Second, selection on niche-specifying variants should be accompanied or followed by habitat separation for genetic exchange to be reduced across the genome. With r/s ≪ 1, stable clusters of ecological and genome-wide similarity can develop quickly (as in the Stable Ecotype model, Box 3), and can remain distinct if gene-flow is impeded by habitat partitioning. With r/s ≫ 1, genotypic clusters of distinct ecology would take longer to establish since the gradual accumulation of sequence diversity by the interplay of population-specific mutation and recombination is required for distinct genetic clusters to emerge [49].
Important further predictions of these models are, first, that if we observe co-occurring genotypic clusters, these should be ecologically distinct (even if they are closely-related). This prediction has largely been supported by surveys of genetic diversity in the wild that identified clusters with overlapping genetic and ecological similarity [18,20,50–53]. Second, for a new niche-specifying gene or allele to induce habitat separation, there must be some form of tradeoff that reduces its success in the former habitat while increasing it in the new [15]. In the absence of such tradeoffs, an ecological generalist might evolve that is successful in both habitats and remains cohesive by gene flow. As demonstrated below, we have recently detected two nascent populations that appear to have evolved an ecological tradeoff explaining their distribution [54].
Genomics of nascent clusters
As suggested by Wiedenbeck and Cohan [15], detailed investigations of the very early stages of ecological differentiation – whether or not it proceeds to completion – are essential to understanding the interplay of recombination and selection in generating ecological and genetic units. We discuss two such snapshots of slightly different stages in this dynamic process.
In the first, 20 Vibrio cyclitrophicus genomes with identical 16S rRNA genes were sequenced and found to share >99% amino acid identity genome-wide. Despite being so genetically similar, two separate groups with distinct ecological preferences were recognized: isolates associated with organic particles and those free-living in coastal ocean water [55]. These distinct lifestyles are made possible by the patchy distribution of resources in the ocean, which might promote a form of mosaic sympatry. In the second investigation, Cadillo-Quiroz et al. [56] sequenced 12 genomes of the archaeon Sulfolobus islandicus from a hotspring in Kamchatka, Russia. These studies were similar in that both sampled closely related isolates from the same geographic location, without apparent barriers to genetic exchange (either a single hotspring or a single bucket of seawater), and from groups of bacteria or archaea with relatively high rates of recombination [37,42]. The studies differed in that the first study had an a priori notion of ecological association for the two Vibrio populations due to the sequencing of a gene under potential environmental selection [50,55], while Cadillo-Quiroz et al. took a purer reverse ecology approach (Box 1), identifying two phylogenetic groups based on overall genomic similarity, then investigating recombination rates within and between groups, and characterizing phenotypic differences between them.
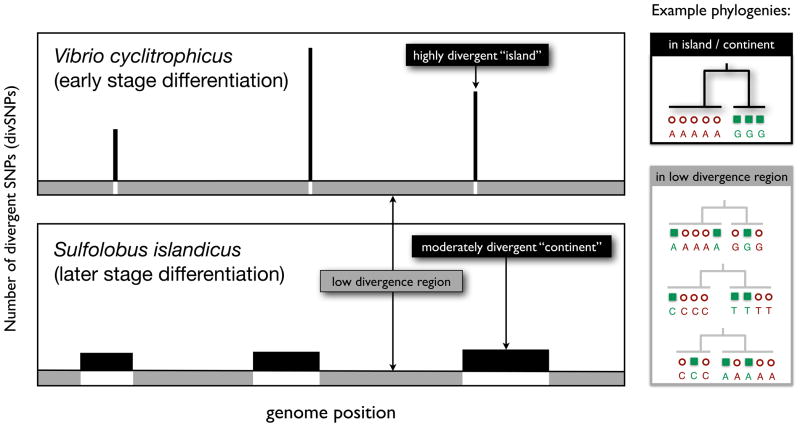
Both studies identified distinct regions of the genome containing single nucleotide polymorphisms (SNPs) clearly dividing the two groups of isolates. In [55], these were referred to as ecoSNPs, because they were fixed genetic differences between groups with previously known ecological associations. Here, we refer to them more inclusively as ‘divergent SNPs’ (divSNPs) – a term that can also be applied to the Sulfolobus populations since in these association with ecological differentiation is still unclear. In the Vibrio genomes, the divSNPs were localized in densely clustered ‘islands’, whereas divSNPs were both more numerous and more broadly dispersed across ‘continents’ of the Sulfolobus genomes (Figure 1), likely reflecting a more advanced stage of differentiation (Table 1). (Here we use the terms ‘islands’ and ‘continents’ in a metaphorical sense, not in a biogeographical sense). Many of the divSNPs in the Sulfolobus continents are probably not directly involved in ecological adaptation, and might be hitchhiking with putative adaptive variants. The extent of this divergence hitchhiking [57] is much smaller in the Vibrio islands, which are rich in ecologically relevant genes, such as those involved in stress responses, attachment and biofilm formation [55]. Outside of these islands or continents, both studies found poorly resolved phylogenetic separation – in fact, a plethora of distinct and conflicting phylogenies across the genome – and shared genetic diversity between groups, indicated by low fixation indices (FST). This provided evidence for a history of rampant recombination among all sympatric isolates, not just those sharing an ecological preference.
Figure 1. Genomic islands and continents of speciation.
Based on data from aligned core genomes of Vibrio cyclitrophicus [55] (upper panel) and Sulfolobus islandicus [56] (lower panel), the distribution of divergent SNPs (divSNPs) fixed between populations is plotted along the genome as black bars. Vibrio and Sulfolobus contain, respectively, 725 divSNPs distributed over ~1% of the genome, and 4232 divSNPs over ~36% of the genome. Scale is approximate for divSNPs (y-axis) and genome position (x-axis). The y-axis is not to scale for SNPs rejecting differentiation between populations (regions of the genome shown in grey).
Do these observations support genome-wide or gene-specific selective sweeps (Figure 2), and what are the implications for ecological/genetic units? One possibility is that genome-wide sweeps did occur in each habitat, but the clonal frames were gradually eroded by recombination of neutral loci between habitats, leaving behind islands or continents as the only traces of the ancient clonal divergence. Modeling suggests that this gradual erosion would require several thousand generations [48], over which time the islands or continents of divSNPs (containing the habitat-specific alleles) would accumulate polymorphism within populations. Yet, in the Vibrio genomes, most of the habitat-specific alleles show very low polymorphism and high synonymous divergence between habitats. This suggests their recent acquisition by recombination from more distant relatives, rather than being the remnant of a more ancient genome-wide sweep. Moreover, a genome-wide sweep would not explain the presence of the same habitat-specific allele in different clonal frames, which was observed to be the case at the RpoS/RTX locus [55]. These observations show that niche-specifying genes or alleles may reside in different genotypes that are otherwise homogenized by gene flow (Figure 2).
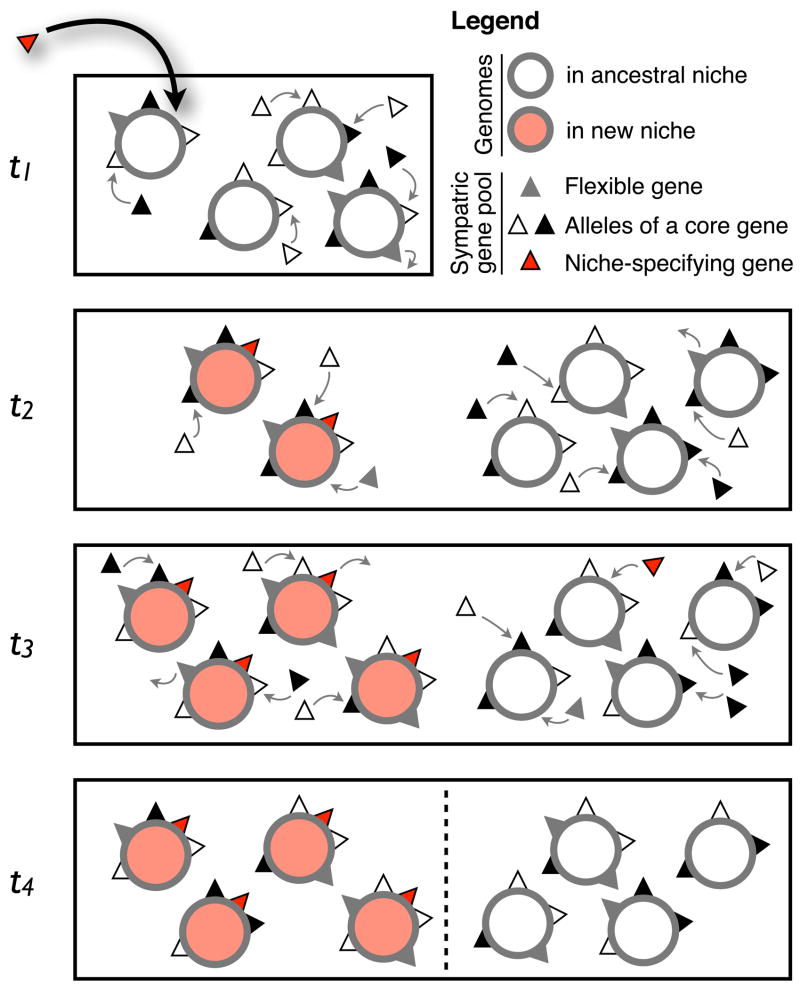
Figure 2. Ecological differentiation via a gene-specific sweep.
This illustration follows the basic steps of the symsim model [48]. At the first time point (t1), a niche-specifying gene (red triangle) arrives into a homogeneously recombining population occupying a single niche. Between t1 and t2 (as between all time points), recombination occurs at random from a sympatric pool (1–2 events per genome, illustrated as arrows), then genomes reproduce clonally, and are culled to a carrying capacity of 4 genomes per niche. Because the niche-specifying gene confers a selective advantage in the new niche, genomes that contain it grow exponentially until the carrying capacity is reached at t3. Other genomes are culled at random, because the rest of the gene pool is neutral to fitness. By t4, the gene-specific sweep is complete: the niche- specifying gene is in perfect association with the new niche, but all other genes are randomly distributed across niches. At this point, barriers to recombination between niches (dashed line) may or may not emerge. (Note: recombination events at t4 are not shown for purposes of clarity, but this does not mean they do not occur).
Genome-wide sweeps were not as firmly excluded in the Sulfolobus populations, although deemed unlikely based on the relatively high inferred recombination rates among populations [56]. As discussed above, archaea like Sulfolobus, which lack mismatch repair machinery, generally show very little reduction in recombination as sequence divergence increases, suggesting weak barriers to recombination between incipient clusters, favoring gene-specific sweeps. Hence very strong divergent selection would be required for the populations to have diverged before much recombination occurred between them, yielding an effectively genome-wide selective sweep. Even in the absence of selection, however, the populations could have diverged in allopatry before re-encountering each other in the same hotspring. Either way, the resulting clonal frame would be observable as large continents of divergence between populations (Figure 1), interrupted by recombination events following the clonal divergence (a scenario not excluded by Cadillo-Quiroz et al.).
While the Sulfolobus populations appear to behave as two distinct genetic units, at first glance, the two ecological populations identified in V. cyclitrophicus are contained within a single genotypic cluster that appears thoroughly mixed by recombination at all except the divSNP-containing loci (Figure 1). This picture of the vibrios as a single gene flow unit changes, however, when inferred recombination events are separated into more ancient and more recent ones – those that have presumably occurred before and after the ecological split, respectively. Such analysis shows that the more recent events are biased to occur among genotypes within either of the two habitats, whereas more ancient events connect all genotypes. This suggests an evolutionary trajectory, most likely induced by microhabitat separation, from a single freely recombining population towards two increasingly separate gene pools. The same trend was observed in both the core and flexible components of the population genomes of vibrios and Sulfolobus alike, and, if projected into the future, might lead to the evolution of clearly distinct genotypic clusters.
As predicted in the models described above, the habitat separation of the two nascent Vibrio populations appears to be associated with an ecological tradeoff. While one population specializes in organic particle exploitation through strong attachment and growth in biofilms, the other population only rarely attaches yet is specialized for dispersal by rapidly detecting and swimming toward new particles, implying that it can better exploit short-lived nutrient patches [54]. Based on their genetic distinctness, we would also predict the ecological distinctness of the two Sulfolobus populations. Indeed, Cadillo-Quiroz et al. [56] went on to show that the two populations differed in growth characteristics in the lab, suggesting distinct niches. It is not yet clear whether these growth differences are relevant to fitness in the wild, and further research will be needed to exclude the possibility that they evolved as a consequence of neutral divergence in allopatry. However, assuming that sequence divergence does not present a significant barrier to gene flow, the preference for recombination within rather than between Sulfolobus populations is likely driven at least in part by differences in ecological associations. Hence, these examples of closely related populations suggest that, in recombining microbes, gene flow barriers may help maintain established units (Sulfolobus), and may initiate formation of new units (Vibrio). The Vibrio example, in particular, further suggests that genes can spread in a population specific manner and, perhaps, initiate microgeographic structure.
This model also helps explain previous findings in genomic and metagenomic surveys that have found location-specific genes or alleles in genotypic clusters that are broadly distributed but seem otherwise phylogenetically ‘well-mixed’ in neutral genes across the genome [27,58–61]. While such mixing may be expected in microbes separated by only a few microns in a biofilm [59], it is perhaps more surprising that Vibrio cholerae from different continents [61], Prochloroccous from different oceans [27], or even haloarchaea separated by a few hundred kilometers [58] remain cohesive at neutral loci. This may be explained by at least a few genotypes being mixed across geographic distances in every generation, allowing allopatric populations to remain homogenous outside of a few environment-specific loci under location-specific selection (e.g., Prochlorococcus phosphorus utilization genes discussed above). In other cases, geographic distance has been correlated to significant divergence between populations [23] but such divergence need not be permanent once gene flow barriers are removed. For example, clonal divergence was reported between Leptospirillum populations that had been separated for ~1000 years [20,21]. However, once the separation ended due to commercial mining activities, it took only ~150 years (~100,000 bacterial generations) for the incipient populations to become well-mixed by recombination of neutral loci across the genome [21].
In summary, these considerations suggest that gene-specific, rather than genome-wide (clonal) selective sweeps may be more common in nature than previously thought. As we discuss below, such gene-specific mechanisms begin with poor mapping between ecological preference and neutral genetic diversity, but eventually result in tight ecological and genetic units.
Stages in the speciation spectrum
The snapshots described above suggest a gradual process by which a new niche becomes accessible when novel genes or alleles arise by mutation or HGT in an ancestral population (Table 1, Stage 1). With sufficiently high recombination rates relative to selection, this niche-specifying variant will spread in a gene-specific sweep (Figure 2, Stage 2A). If the new and ancestral niches remain fully sympatric, with no barriers to recombination between them, the process will stop here, as in the symsim model described above [48]. However, if the new niche is also somehow associated with barriers to recombination (perhaps due to a reduced encounter rate with genomes in the ancestral niche), genetic separation will begin to occur at neutral loci throughout the genome (Stage 3). These ecological barriers might later be reinforced by genetic barriers, as sequence divergence accumulates between lineages, eventually inhibiting recombination genome-wide (Stage 4). Genetic isolation may also develop more quickly if the capacity for recombination is transiently lost, either genetically (e.g., [62]) or physiologically (e.g., by modulating expression of recombination and mismatch repair machinery).
Whether the early stages of speciation involve gene-specific or genome-wide selective sweeps will depend on the r/s ratio, but both regimes can eventually lead to the same end products of overlapping genotypic and ecological units (Table 1). The Sulfolobus populations may be in an intermediate regime, with a low enough r/s ratio to allow clonal sweeps at Stage 2, but sufficient recombination to generate the patterns of gene flow observed at Stages 3 and 4. Based on the number of conflicting phylogenetic signals in the genome, flexible genome diversity, and presence of niche-specific genes in multiple different clonal frames, it appears that the vibrios are firmly in the r/s ≫ 1 regime. Yet this this seems at odds with experimentally estimated recombination rates, which appear to be generally much lower than even moderate selection coefficients [15].
What factors might keep r/s so high – astoundingly high, in fact, compared to our expectations? One possibility is that genome-wide selective sweeps are slowed by negative frequency-dependent selection [26], imposed on traits involved in susceptibility to phage predation (e.g., surface structures [63]) or in social interactions within microbial populations (e.g., siderophore or antibiotic production [10,64]). Another possibility is that genome-wide sweeps are slowed by clonal interference (e.g., [65]), allowing more time for recombination to occur before all diversity is purged. Further research will be needed to distinguish between these possibilities.
We propose that the Sulfolobus lineages are approximately at Stage 3 or 4, while the vibrios are at around Stage 2 or 3. As a result, potential niche-specifying genes or alleles are much more readily pinpointed in the Vibrio islands than the Sulfolobus continents. Whether the nascent Vibrio lineages will persist cannot be predicted but we note that 3 years after the initial sampling, the same populations with the same set of habitat-specific flexible genes were observed once again, suggesting a reasonably stable association between ecological units and selected parts of the genome [66]. However, we note that at Stage 2, the two nascent populations cannot be differentiated from a single population with the putative niche-specifying genes under balancing or negative frequency-dependent selection within the population [26,67]. Only with ecological information that demonstrates poor habitat overlap and/or reduced recombination throughout the genome (Stages 3–4) can we reasonably consider the population to be splitting in two.
As this split occurs, lineages might eventually become permanently separate, forming distinct ecological and genome-wide sequence clusters (at neutral loci across the genome, interrupted by occasional exchange between lineages), until one or both go extinct (Stage 5). Importantly, this long-term result of formation of congruent ecological and genetic clusters is expected under both high and low r/s ratios. Therefore, comparing microbial genomes that have already reached this stage is not expected to be informative about the relative influence of selection and recombination at early stages. From a practical standpoint, once lineages have diverged to Stage 5, they should be easily recognizable as distinct ecological and genetic units. For example, environmental and gut-associated groups of Escherichia coli have distinct ecologies and have diverged genetically throughout the genome [17]. It therefore appears to be justifiable not to group these lineages together into the same species (Box 2).
We stress that, just because these stages of speciation can be defined, it does not mean that all populations that start at Stage 1 will make it to Stage 5. In fact, the intermediates may be more numerous than the end products. As James Mallet [7] wrote, “speciation appears to be easy; the intermediate stages are all around us.” If this is the case, many more examples should be forthcoming from across the microbial world. Studied using the framework of population genomics and reverse ecology (Box 1), they will test the generality of the speciation process that we propose based on current data.
Variation within a cohesive population
Our discussion thus far has focused on how to define and delimit the boundaries between internally cohesive microbial populations. Cohesive populations may nevertheless contain high levels of genotypic (and to some extent, phenotypic) diversity within them. How can this be explained? First, as discussed earlier, niche-specifying variants (genes or alleles) may come with a fitness tradeoff, such that they are adaptive in one niche but not another (indeed, one might even define them as such). In a genetically cohesive population that spans two niches, different niche-specifying variants will be maintained in each niche, leading to variation at the level of the entire population. (In fact, without knowledge of habitat specificity and tendency toward within-habitat recombination, the vibrios could be thought of in this way: as a single cohesive population, with diversity at the level of niche-specifying variants that have failed to sweep through the entire population because of some tradeoff). Second, frequency-dependent selection might maintain diversity in a subset of genes involved in niche complementarity, social interactions, and predator-prey interactions [26]. A relatively high proportion of genes in the flexible genome may be involved in such interactions. It has been argued previously that many genes occurring at intermediate frequency within genomes are involved in predation evasion by varying surface antigenicity [26,63]. Moreover, intermediate frequency genes may be involved in frequency dependent interactions such as public good production and cheating as well as niche-complementation [26]. This may also explain some phenotypic variation frequently observed among closely related genotypes. For example, any secreted compound, such as enzymes, antibiotics, or signaling molecules, can become a public good that may invite cheating given sufficiently stable population structure. Lastly, we should not forget that many genes, typically localized in genomic islands of high variation, appear to have such high turnover within populations that a high fraction might be (nearly) neutral to bacterial fitness [68–70]. Similarly, if genome-wide selective sweeps do not periodically reduce diversity, substantial allelic diversity will be preserved through speciation. In other words, allelic diversity will be much older than the population itself [71]. Importantly, interpretation of such microevolutionary changes in the context of selection and population dynamics requires that sympatric genomes (i.e., from the same population) are sampled.
Another portion of genes that are generally considered as part of the flexible genome may actually be part of the core genome of local populations and hence be under purifying selection. The example of Prochlorococcus populations in the Atlantic and Pacific given earlier in this review falls into this category. Another recent example is Campylobacter jejuni strains that were isolated from both cattle and chickens, but the genome-wide phylogeny provided little evidence for host preference [72]. In other words, host-switching is relatively rapid and long-term host preferences have not been established. However, a gene cluster involved in vitamin B5 biosynthesis is universally present in cattle isolates, but mostly absent in chicken isolates. This gene cluster appears to provide a selective advantage in B5-depleted environments, which might include the cattle gut [72]. An ecological trait is therefore associated with variation in a single gene cluster, but not with diversity across the entire genome. The gene cluster can be thought of as a niche-specifying variant, and the cattle- and chicken-associated isolates could be placed at Stage 1 of the differentiation spectrum (Table 1). This by no means guarantees that Stage 1 will proceed to Stage 2 and onward to genome-wide divergence. Rather, phenotypic diversity in host preference might be thought of as part of the shared ecology of all C. jejuni. Regardless, these examples highlight the importance of considering allele frequencies (in the core and flexible genome) in the context of carefully sampled populations (Box 1).
Whether or not niche-specifying variants trigger further differentiation, they can provide insights into the mechanisms of niche adaptation, and can be identified by properly designed GWAS. An appropriate microbial GWAS should account for the degree of recombination or clonality in the population of interest [73]. Especially in highly clonal populations, associations should be based on a convergence criterion [74,75], in which phenotypes of interest are acquired independently in different lineages (Figure 3). Mutations, alleles or genes that are repeatedly associated with these phenotypic transitions can then be identified, and the statistical significance of their associations assessed relative to a neutral model [76]. In highly recombining populations, convergence tests are still justified [47,67], but might lack power relative to approaches that take into account rapid recombination. For example, in Vibrio, the flexible genome turns over very rapidly [55,61,69], such that associations between habitat preference and flexible genes are unlikely to be maintained by vertical descent, but rather by habitat-specific selective pressures. Similar reasoning was used to identify E. coli flexible genes associated with environmental or gut-associated lifestyles [17].
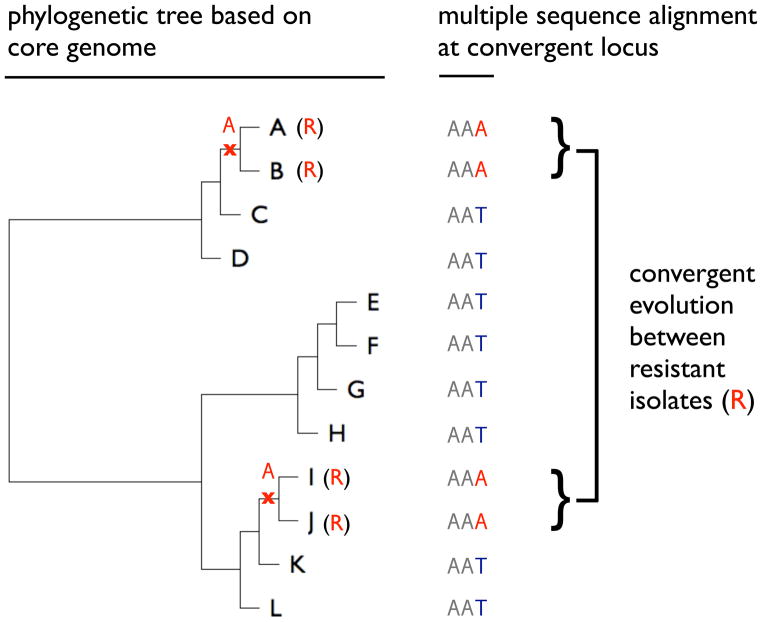
Figure 3. Evolutionary convergence as the basis for GWAS.
In this simplified example, the genome-wide core phylogeny has been inferred for a sample of mostly clonal bacterial isolates (A–L). A convergent (homoplasic) SNP is identified in 4 genomes. Importantly, this corresponds to only 2 independent mutation events (T→A, indicated in the phylogenetic tree by the red “X” with an “A” above it), which associate perfectly with 2 independent transitions from the antibiotic sensitive to resistant (R) state. The significance of the association can be assessed by calculating a P-value by resampling from the genome-wide distribution of mutations and phenotypic states (resistant or sensitive) on the phylogeny. By failing to account for population structure (e.g. the phylogenetic information), 4 events would be counted, thereby overestimating the significance of the association. GWAS can also be performed considering entire genes, instead of individual nucleotide sites, as targets of convergent mutations. See [72,76] for examples.
Concluding remarks and prospects for reverse ecology
As we have outlined in this review, when genotypic clusters can be detected, they should be ecologically differentiated from other such clusters. Although this does not preclude some level of ecological diversity within these clusters due to the potential of acquisition of novel, niche-specifying genes, such diversity should be relatively minor because selection can only maintain a limited number of ecologically divergent loci within the same, genetically mixed population [48]. Hence a reverse ecology strategy, in which genotypic clusters among co-existing microbes are identified as a first step toward identifying ecologically cohesive populations, is potentially easier than the forward approach, which is to map marker genes onto many environmental samples in the hopes of finding significant ecological associations.
As genome sequencing becomes more and more broadly accessible because of decreased cost and increased throughput, it will become feasible to sequence sufficient numbers of closely related genomes from the same environmental samples, either in the form of isolates or single-cell genomes. Moreover, improved coverage and assembly techniques will also allow increased identification of genotypic clusters from metagenomic samples. Once these genomes are available, they can serve two purposes. First, they can be used to delineate clusters, and second, they can help build hypotheses of environmental differentiation by searching for genes of potential ecological relevance. In that way, some guess as to the population’s niche can be made before engaging in the exercise of mapping the cluster onto environmental samples and identifying correlations with biotic and abiotic environmental metadata. We stress that this exercise must consider the fine structure of the environment since microbial habitats and interactions often occur at small spatial (micro- to millimeters) and temporal scales (minutes to days) [28]. Given sufficient environmental and genomic sampling, GWAS can provide valuable further insights as to the causes of allele and gene diversity within and between populations (Box 4).
Box 4. Outstanding questions.
How frequently does speciation happen in sympatry, and how long-lived are genotypic/phenotypic units, on average?
What types of ecological tradeoffs are associated with mosaic sympatry?
Does the relative importance of gene-specific and genome-wide selective sweeps vary across different natural microbial populations and/or habitats?
What evolutionary or ecological mechanisms depress selection rates in the environment, hence delaying or preventing genome-wide selective sweeps?
Why do genomes, even within ecologically cohesive populations, display such large gene-content variation?
Enabled by our still-evolving knowledge of microbial diversity, the combined toolkits of population genomics, reverse ecology, and GWAS will no doubt continue to enrich and expand our understanding as we move from descriptive to more mechanistic models of the ecological and genetic structure of microbes in the wild.
Highlights.
Mathematical models and genomic data provide new insights into microbial speciation.
Adaptive genes can either spread within populations or trigger genome-wide sweeps.
Formation of new genotypic clusters in sympatry requires (microgeographic) gene-flow barriers.
if arisen in sympatry, genotypic clusters represent congruent ecological and genetic units.
Acknowledgments
We thank Libusha Kelly, Rex Malmstrom, Gabriel Perron and Rachel Whitaker for their insightful comments. Funding for MFP was provided by National Science Foundation grant DEB 0821391, National Institute of Environmental Health Sciences grant P30-ES002109, the Moore Foundation and the Broad Institute’s SPARC program. Funding for BJS was provided by the Natural Sciences and Engineering Research Council of Canada and the Canada Research Chairs program.
Glossary
- Allopatric
a set of sampled isolates or genomes from different geographic areas, where barriers to migration and gene flow are significant
- Clonal frame
the portion of the genome transmitted by vertical (clonal) evolution, unimpacted by HGT. Mutations in the clonal frame should all fall parsimoniously on a single phylogenetic tree
- Core genome
the portion of the genome that is present (or in practice, that can be aligned) in all of a given set of sequenced isolates or metagenomes
- Flexible genome
the set of genes or DNA that is present in only a fraction of a given set of sequenced isolates or metagenomes
- Gene-specific selective sweep
the process in which an adaptive gene or allele (possibly a niche-specifying variant) spreads in a population by recombination faster than by clonal expansion. The result is that the adaptive variant is present in more than a single clonal background, and that diversity is not purged genome-wide
- Genome-wide association study (GWAS)
a technique commonly used in eukaryotic genetics to identify genomic variants that are associated with a phenotype of interest. In highly structured populations (e.g., clonal microbes), it is essential to correct for false associations due to phylogenetic structure
- Genome-wide selective sweep
the process in which an adaptive gene or allele (possibly a niche-specifying variant) spreads in a population by clonal expansion of the genome that first acquired it. The result is that diversity is purged genome-wide, and that the adaptive variant is linked in the same clonal frame as the rest of the genome
- Horizontal gene transfer (HGT)
the incorporation of foreign DNA into a genome. Incorporation can be mediated by either homologous recombination or non-homologous recombination of DNA that enters a cell via transformation, transduction or conjugation. In bacteria and archaea, all gene transfer is horizontal (i.e., always unidirectional)
- Homologous recombination
a mechanism of DNA integration requiring at least short tracts of identity between the genome and the foreign DNA, mediated by RecA and mismatch-repair machinery. The integrated DNA can result in single-nucleotide changes and in some cases, addition or loss of relatively long stretch of DNA including entire genes
- Metagenome
the total set of all the genomic DNA in a particular environment or sample
- Negative frequency dependent selection
a type of natural selection that favors rare phenotypes in a population
- Niche
a specific set of ecological parameters (environments, resources, physical and chemical characteristics, biotic interactions etc.) to which an organism is adapted. This does not necessarily imply (but does not exclude) physical separation between niches
- Niche-specifying variant
a mutation, gene or allele that allows a cell to be part of a particular niche. These variants are under positive selection within the particular niche, but not outside it
- Non-homologous recombination
integration of DNA with no homologous allele already present in the genome, often mediated by phage and integrative elements. This results in the acquisition of entirely new genes
- Population
a group of individuals sharing genetic and ecological similarity, and co-existing in a sympatric setting
- Positive selection
a type of natural selection that favors variants conferring a fitness advantage, causing them to increase in frequency in a population
- Sympatric
a set of sampled isolates or genomes from the same geographic area, where barriers to migration and gene flow are low or non-existent
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gevers D, et al. Opinion: re-evaluating prokaryotic species. Nature Reviews Microbiology. 2005;3:733–739. doi: 10.1038/nrmicro1236. [DOI] [PubMed] [Google Scholar]
- 2.Cohan FM, Koeppel AF. The origins of ecological diversity in prokaryotes. Current Biology. 2008;18:1024–1034. doi: 10.1016/j.cub.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 3.Mayr E. Systematics and the Origin of Species. Columbia University Press; 1942. [Google Scholar]
- 4.Syvanen M. Evolutionary implications of horizontal gene transfer. Annu Rev Genet. 2012;46:341–358. doi: 10.1146/annurev-genet-110711-155529. [DOI] [PubMed] [Google Scholar]
- 5.Danchin EGJ, Rosso MN. Lateral gene transfers have polished animal genomes: lessons from nematodes. Front Cell Infect Microbiol. 2012;2:27–27. doi: 10.3389/fcimb.2012.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schönknecht G, et al. Horizontal gene acquisitions by eukaryotes as drivers of adaptive evolution. Bioessays. 2013;36:9–20. doi: 10.1002/bies.201300095. [DOI] [PubMed] [Google Scholar]
- 7.Mallet J. Hybridization, ecological races and the nature of species: empirical evidence for the ease of speciation. Philos Trans R Soc Lond, B, Biol Sci. 2008;363:2971–2986. doi: 10.1098/rstb.2008.0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Vries J, Wackernagel W. Integration of foreign DNA during natural transformation of Acinetobacter sp. by homology-facilitated illegitimate recombination. Proc Natl Acad Sci USA. 2002;99:2094–2099. doi: 10.1073/pnas.042263399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mell JC, et al. Transformation of natural genetic variation into Haemophilus influenzae genomes. PLoS Pathog. 2011;7:e1002151. doi: 10.1371/journal.ppat.1002151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cordero OX, et al. Public good dynamics drive evolution of iron acquisition strategies in natural bacterioplankton populations. Proc Natl Acad Sci USA. 2012;109:20059–20064. doi: 10.1073/pnas.1213344109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Croucher NJ, et al. A high-resolution view of genome-wide pneumococcal transformation. PLoS Pathog. 2012;8:e1002745. doi: 10.1371/journal.ppat.1002745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doolittle WF, Papke RT. Genomics and the bacterial species problem. Genome Biol. 2006;7:116. doi: 10.1186/gb-2006-7-9-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hahn MW, Pockl M. Ecotypes of planktonic Actinobacteria with identical 16S rRNA genes adapted to thermal niches in temperate, subtropical, and tropical freshwater habitats. Applied and Environmental Microbiology. 2005;71:766–773. doi: 10.1128/AEM.71.2.766-773.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doolittle WF, Zhaxybayeva O. On the origin of prokaryotic species. Genome Research. 2009;19:744–756. doi: 10.1101/gr.086645.108. [DOI] [PubMed] [Google Scholar]
- 15.Wiedenbeck J, Cohan FM. Origins of bacterial diversity through horizontal genetic transfer and adaptation to new ecological niches. FEMS Microbiology Reviews. 2011;35:957–976. doi: 10.1111/j.1574-6976.2011.00292.x. [DOI] [PubMed] [Google Scholar]
- 16.Hanage WP, et al. Fuzzy species among recombinogenic bacteria. BMC Biol. 2005;3:6. doi: 10.1186/1741-7007-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luo C, et al. Genome sequencing of environmental Escherichia coli expands understanding of the ecology and speciation of the model bacterial species. Proc Natl Acad Sci USA. 2011;108:7200–7205. doi: 10.1073/pnas.1015622108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Konstantinidis KT, Delong EF. Genomic patterns of recombination, clonal divergence and environment in marine microbial populations. The ISME Journal. 2008;2:1052–1065. doi: 10.1038/ismej.2008.62. [DOI] [PubMed] [Google Scholar]
- 19.Oh S, et al. Metagenomic insights into the evolution, function, and complexity of the planktonic microbial community of Lake Lanier, a temperate freshwater ecosystem. Applied and Environmental Microbiology. 2011;77:6000–6011. doi: 10.1128/AEM.00107-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Denef VJ, et al. AMD biofilms: using model communities to study microbial evolution and ecological complexity in nature. The ISME Journal. 2010;4:599–610. doi: 10.1038/ismej.2009.158. [DOI] [PubMed] [Google Scholar]
- 21.Simmons SL, et al. Population genomic analysis of strain variation in Leptospirillum group II bacteria involved in acid mine drainage formation. Plos Biol. 2008;6:1427–1442. doi: 10.1371/journal.pbio.0060177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Whitaker RJ, et al. Geographic barriers isolate endemic populations of hyperthermophilic archaea. Science. 2003;301:976–978. doi: 10.1126/science.1086909. [DOI] [PubMed] [Google Scholar]
- 23.Reno ML, et al. Biogeography of the Sulfolobus islandicus pan-genome. Proc Natl Acad Sci USA. 2009;106:8605–8610. doi: 10.1073/pnas.0808945106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Welch RA, et al. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc Natl Acad Sci USA. 2002;99:17020–17024. doi: 10.1073/pnas.252529799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kettler GC, et al. Patterns and implications of gene gain and loss in the evolution ofProchlorococcus. PLoS Genet. 2007;3:2515–2528. doi: 10.1371/journal.pgen.0030231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cordero OX, Polz MF. Explaining microbial genomic diversity in light of evolutionary ecology. Nature Review Microbiology. 2014 doi: 10.1038/nrmicro3218. in press. [DOI] [PubMed] [Google Scholar]
- 27.Coleman ML, Chisholm SW. Ecosystem-specific selection pressures revealed through comparative population genomics. Proc Natl Acad Sci USA. 2010;107:18634–18639. doi: 10.1073/pnas.1009480107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Polz MF, et al. Patterns and mechanisms of genetic and phenotypic differentiation in marine microbes. Philos Trans R Soc Lond, B, Biol Sci. 2006;361:2009–2021. doi: 10.1098/rstb.2006.1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jax K. Ecological units: definitions and application. Q Rev Biol. 2006;81:237–258. doi: 10.1086/506237. [DOI] [PubMed] [Google Scholar]
- 30.Li YF, et al. “Reverse ecology” and the power of population genomics. Evolution. 2008;62:2984–2994. doi: 10.1111/j.1558-5646.2008.00486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levy R, Borenstein E. Reverse ecology: from systems to environments and back. Advances in experimental medicine and biology. 2012;751:329–345. doi: 10.1007/978-1-4614-3567-9_15. [DOI] [PubMed] [Google Scholar]
- 32.Ellison CE, et al. Population genomics and local adaptation in wild isolates of a model microbial eukaryote. Proc Natl Acad Sci USA. 2011;108:2831–2836. doi: 10.1073/pnas.1014971108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Konstantinidis KT, Tiedje JM. Towards a genome-based taxonomy for prokaryotes. Journal of Bacteriology. 2005;187:6258–6264. doi: 10.1128/JB.187.18.6258-6264.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hanson CA, et al. Beyond biogeographic patterns: processes shaping the microbial landscape. Nature Reviews Microbiology. 2012;10:497–506. doi: 10.1038/nrmicro2795. [DOI] [PubMed] [Google Scholar]
- 35.Nemergut DR, et al. Global patterns in the biogeography of bacterial taxa. Environ Microbiol. 2010;13:135–144. doi: 10.1111/j.1462-2920.2010.02315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith JM, et al. How clonal are bacteria? Proc Natl Acad Sci USA. 1993;90:4384–4388. doi: 10.1073/pnas.90.10.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vos M, Didelot X. A comparison of homologous recombination rates in bacteria and archaea. The ISME Journal. 2009;3:199–208. doi: 10.1038/ismej.2008.93. [DOI] [PubMed] [Google Scholar]
- 38.Vulić M, et al. Molecular keys to speciation: DNA polymorphism and the control of genetic exchange in enterobacteria. Proc Natl Acad Sci USA. 1997;94:9763–9767. doi: 10.1073/pnas.94.18.9763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Majewski JJ, Cohan FMF. DNA sequence similarity requirements for interspecific recombination inBacillus. Genetics. 1999;153:1525–1533. doi: 10.1093/genetics/153.4.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grogan DW, Stengel KR. Recombination of synthetic oligonucleotides with prokaryotic chromosomes: substrate requirements of the Escherichia coli/λRed and Sulfolobus acidocaldarius recombination systems. Molecular Microbiology. 2008;69:1255–1265. doi: 10.1111/j.1365-2958.2008.06356.x. [DOI] [PubMed] [Google Scholar]
- 41.Naor A, et al. Low species barriers in halophilic archaea and the formation of recombinant hybrids. Curr Biol. 2012;22:1444–1448. doi: 10.1016/j.cub.2012.05.056. [DOI] [PubMed] [Google Scholar]
- 42.Whitaker RJ, et al. Recombination shapes the natural population structure of the hyperthermophilic archaeon Sulfolobus islandicus. Mol Biol Evol. 2005;22:2354–2361. doi: 10.1093/molbev/msi233. [DOI] [PubMed] [Google Scholar]
- 43.Williams D, et al. Quantifying homologous replacement of loci between haloarchaeal species. Genome Biology and Evolution. 2012;4:1223–1244. doi: 10.1093/gbe/evs098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fraser C, et al. Recombination and the nature of bacterial speciation. Science. 2007;315:476–480. doi: 10.1126/science.1127573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barrick JE, et al. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature. 2009;461:1243–1247. doi: 10.1038/nature08480. [DOI] [PubMed] [Google Scholar]
- 46.Blount ZD, et al. Genomic analysis of a key innovation in an experimental Escherichia coli population. Nature. 2012;488:513–518. doi: 10.1038/nature11514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shapiro BJ, et al. Looking for Darwin’s footprints in the microbial world. Trends in Microbiology. 2009;17:196–204. doi: 10.1016/j.tim.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 48.Friedman J, et al. Sympatric speciation: when is it possible in bacteria? PLoS ONE. 2013;8:e53539. doi: 10.1371/journal.pone.0053539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Polz MF, et al. Horizontal gene transfer and the evolution of bacterial and archaeal population structure. Trends in Genetics. 2013;29:170–175. doi: 10.1016/j.tig.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hunt DE, et al. Resource partitioning and sympatric differentiation among closely related bacterioplankton. Science. 2008;320:1081–1085. doi: 10.1126/science.1157890. [DOI] [PubMed] [Google Scholar]
- 51.Koeppel A, et al. Identifying the fundamental units of bacterial diversity: a paradigm shift to incorporate ecology into bacterial systematics. Proc Natl Acad Sci USA. 2008;105:2504–2509. doi: 10.1073/pnas.0712205105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rocap G, et al. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature. 2003;424:1042–1047. doi: 10.1038/nature01947. [DOI] [PubMed] [Google Scholar]
- 53.Johnson ZI, et al. Niche partitioning among Prochlorococcus ecotypes along ocean-scale environmental gradients. Science. 2006;311:1737–1740. doi: 10.1126/science.1118052. [DOI] [PubMed] [Google Scholar]
- 54.Yawata Y, et al. A competition-dispersal trade-off ecologically differentiates recently speciated marine bacterioplankton populations. Proc Natl Acad Sci U S A. 2014 doi: 10.1073/pnas.1318943111. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shapiro BJ, et al. Population genomics of early events in the ecological differentiation of bacteria. Science. 2012;336:48–51. doi: 10.1126/science.1218198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cadillo-Quiroz H, et al. Patterns of gene flow define species of thermophilic archaea. Plos Biol. 2012;10:e1001265. doi: 10.1371/journal.pbio.1001265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Via S. Divergence hitchhiking and the spread of genomic isolation during ecological speciation-with-gene-flow. Philos Trans R Soc Lond, B, Biol Sci. 2012;367:451–460. doi: 10.1098/rstb.2011.0260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Papke RT, et al. Searching for species in haloarchaea. Proc Natl Acad Sci USA. 2007;104:14092–14097. doi: 10.1073/pnas.0706358104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Denef VJ, et al. Proteogenomic basis for ecological divergence of closely related bacteria in natural acidophilic microbial communities. Proc Natl Acad Sci USA. 2010;107:2383–2390. doi: 10.1073/pnas.0907041107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Burke C, et al. Bacterial community assembly based on functional genes rather than species. Proc Natl Acad Sci USA. 2011;108:14288–14293. doi: 10.1073/pnas.1101591108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Boucher Y, et al. Local mobile gene pools rapidly cross species boundaries to create endemicity within global Vibrio cholerae populations. mBio. 2011;2:e00335–10. doi: 10.1128/mBio.00335-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Katz LS, et al. Evolutionary dynamics of Vibrio cholerae O1 following a single-source introduction to Haiti. mBio. 2013;4:e00398–13. doi: 10.1128/mBio.00398-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rodriguez-Valera F, et al. Explaining microbial population genomics through phage predation. Nature Reviews Microbiology. 2009;7:828–836. doi: 10.1038/nrmicro2235. [DOI] [PubMed] [Google Scholar]
- 64.Cordero OX, et al. Ecological populations of bacteria act as socially cohesive units of antibiotic production and resistance. Science. 2012;337:1228–1231. doi: 10.1126/science.1219385. [DOI] [PubMed] [Google Scholar]
- 65.Lieberman TD, et al. Genetic variation of a bacterial pathogen within individuals with cystic fibrosis provides a record of selective pressures. Nature Genetics. 2013 doi: 10.1038/ng.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Szabó G, et al. Reproducibility of Vibrionaceae population structure in coastal bacterioplankton. The ISME Journal. 2013;7:509–519. doi: 10.1038/ismej.2012.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shapiro BJ. Signatures of natural selection and ecological differentiation in microbial genomes. Advances in experimental medicine and biology. 2014;781:339–359. doi: 10.1007/978-94-007-7347-9_17. [DOI] [PubMed] [Google Scholar]
- 68.Berg OG, Kurland CG. Evolution of microbial genomes: sequence acquisition and loss. Molecular biology and evolution. 2002;19:2265–2276. doi: 10.1093/oxfordjournals.molbev.a004050. [DOI] [PubMed] [Google Scholar]
- 69.Thompson JR, et al. Genotypic diversity within a natural coastal bacterioplankton population. Science. 2005;307:1311–1313. doi: 10.1126/science.1106028. [DOI] [PubMed] [Google Scholar]
- 70.Haegeman B, Weitz JS. A neutral theory of genome evolution and the frequency distribution of genes. Bmc Genomics. 2012;13:196–196. doi: 10.1186/1471-2164-13-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Castillo-Ramírez S, et al. The impact of recombination on dN/dS within recently emerged bacterial clones. PLoS Pathog. 2011;7:e1002129. doi: 10.1371/journal.ppat.1002129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sheppard SK, et al. Genome-wide association study identifies vitamin B5 biosynthesis as a host specificity factor inCampylobacter. Proc Natl Acad Sci USA. 2013;110:11923–11927. doi: 10.1073/pnas.1305559110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Falush D, Bowden R. Genome-wide association mapping in bacteria? Trends Microbiol. 2006;14:353–355. doi: 10.1016/j.tim.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 74.Sokurenko EV. Selection footprint in the FimH adhesin shows pathoadaptive niche differentiation in Escherichia coli. Mol Biol Evol. 2004;21:1373–1383. doi: 10.1093/molbev/msh136. [DOI] [PubMed] [Google Scholar]
- 75.Chattopadhyay S, et al. Tracking recent adaptive evolution in microbial species using TimeZone. Nat Protoc. 2013;8:652–665. doi: 10.1038/nprot.2013.031. [DOI] [PubMed] [Google Scholar]
- 76.Farhat MR, et al. Genomic analysis identifies targets of convergent positive selection in drug-resistant Mycobacterium tuberculosis. Nature Genetics. 2013;45:1183–1189. doi: 10.1038/ng.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Marttinen P, et al. Detection of recombination events in bacterial genomes from large population samples. Nucleic Acids Res. 2012;40:e6. doi: 10.1093/nar/gkr928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Didelot X, et al. Inference of homologous recombination in bacteria using whole-genome sequences. Genetics. 2010;186:1435–1449. doi: 10.1534/genetics.110.120121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Majewski J, Cohan FM. Adapt globally, act locally: the effect of selective sweeps on bacterial sequence diversity. Genetics. 1999;152:1459–1474. doi: 10.1093/genetics/152.4.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Caro-Quintero A, et al. Genomic insights into the convergence and pathogenicity factors of Campylobacter jejuni and Campylobacter coli species. Journal of Bacteriology. 2009;191:5824–5831. doi: 10.1128/JB.00519-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Caro-Quintero A, Konstantinidis KT. Bacterial species may exist, metagenomics reveal. Environ Microbiol. 2011;14:347–355. doi: 10.1111/j.1462-2920.2011.02668.x. [DOI] [PubMed] [Google Scholar]
- 82.Cohan FM. Bacterial species and speciation. Systematic Biology. 2001;50:513–524. doi: 10.1080/10635150118398. [DOI] [PubMed] [Google Scholar]
- 83.Koeppel AF, et al. Speedy speciation in a bacterial microcosm: new species can arise as frequently as adaptations within a species. The ISME Journal. 2013;7:1080–1091. doi: 10.1038/ismej.2013.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vos M. A species concept for bacteria based on adaptive divergence. Trends Microbiol. 2011;19:1–7. doi: 10.1016/j.tim.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 85.Cornejo OE, et al. Polymorphic competence peptides do not restrict recombination in Streptococcus pneumoniae. Mol Biol Evol. 2010;27:694–702. doi: 10.1093/molbev/msp287. [DOI] [PubMed] [Google Scholar]