Abstract
We characterized the full-length genomes of nine hepatitis C virus genotype 3 (HCV-3) isolates: QC7, QC8, QC9, QC10, QC34, QC88, NE145, NE274 and 811. To the best of our knowledge, NE274 and NE145 were the first full-length genomes for confirming the provisionally assigned subtypes 3d and 3e, respectively, whereas 811 represented the first HCV-3 isolate that had its extreme 3′ UTR terminus sequenced. Based on these full-length genomes, together with 42 references representing eight assigned subtypes and an unclassified variant of HCV-3, and 10 sequences of six other genotypes, a timescaled phylogenetic tree was reconstructed after an evolutionary analysis using a coalescent Bayesian procedure. The results indicated that subtypes 3a, 3d and 3e formed a subset with a common ancestor dated to ~202.89 [95 % highest posterior density (HPD): 160.11, 264.6] years ago. The analysis of all of the HCV-3 sequences as a single lineage resulted in the dating of the divergence time to ~457.81 (95 % HPD: 350.62, 587.53) years ago, whereas the common ancestor of all of the seven HCV genotypes dated to ~780.86 (95 % HPD: 592.15, 1021.34) years ago. As subtype 3h and the unclassified variant were relatives, and represented the oldest HCV-3 lineages with origins in Africa and the Middle East, these findings may indicate the ancestral origin of HCV-3 in Africa. We speculate that the ancestral HCV-3 strains may have been brought to South Asia from Africa by land and/or across the sea to result in its indigenous circulation in that region. The spread was estimated to have occurred in the era after Vasco da Gama had completed his expeditions by sailing along the eastern coast of Africa to India. However, before this era, Arabians had practised slave trading from Africa to the Middle East and South Asia for centuries, which may have mediated the earliest spread of HCV-3.
Introduction
Hepatitis C virus (HCV) is a blood-borne pathogen that infects an estimated >185 million people worldwide, which corresponds to 2.8 % of the global population (Alter, 1999). The infection is characterized by the establishment of chronic hepatitis in 70–85 % of the infected individuals, of whom >20 % subsequently develop cirrhosis, liver failure and hepatocellular carcinoma, (Hoofnagle, 2002; Zoulim et al., 2003). HCV is a cause of substantial morbidity and mortality, the rates of which are expected to increase over the next decades (Williams, 1999).
HCV possesses a positive-sense ssRNA genome of ~9600 nt in length and is classified into the Hepacivirus genus of the Flaviviridae family (Thiel et al., 2005). Based on a recent paper on its expanded classification, HCV has now been divided into seven genotypes and 67 subtypes, of which 10 subtypes and an unclassified novel variant have been assigned within genotype 3 (Smith et al., 2014). Different genotypes exhibit different geographical distribution patterns and are associated with different treatment outcomes. In general, genotypes 1–3 are distributed worldwide, whereas genotypes 4–7 are restricted to certain regions (Simmonds et al., 2005). Clinically, those infected with genotypes 2 and 3 present a higher rate of sustained virological responses to therapy with IFN-α and ribavirin than those infected with genotypes 1 and 4 (Feld & Hoofnagle, 2005; Manns et al., 2006). With respect to complications, the isolates of genotype 3 are linked to the development of more pronounced hepatocellular steatosis (Abid et al., 2005; Hourioux et al., 2007; Jackel-Cram et al., 2007; Negro, 2006; Rubbia-Brandt et al., 2000), an accelerated fibrosis progression rate (Bochud et al., 2009), an increased risk of developing hepatocellular carcinoma (Nkontchou et al., 2011) and an increased all-cause mortality (van der Meer et al., 2012). Although the mechanisms involved in HCV-3-associated pathogenesis are not fully understood, they may be due to the genetic variation of the virus.
With the completion of one of our recent studies, six subtypes (3a, 3b, 3g, 3h, 3i and 3k) in genotype 3 have been confirmed and their full-length genomes have been characterized (Lu et al., 2013). However, due to the availability of only partial sequences, four other subtypes (3c, 3d, 3e and 3f) have not yet been sequenced completely. In the present study, we determined the full-length genomes of subtypes 3d and 3e to partially fill this information gap.
Among the subtypes in genotype 3, both 3a and 3b are distributed worldwide and are often detected in developed countries – a phenomenon that is ascribed to their propensity of being transmitted via intravenous drug use (IDU) (Bourliere et al., 2002; McCaw et al., 1997; Pawlotsky et al., 1995; Silini et al., 1995). In contrast, other subtypes have been detected mainly in South Asia and its neighbouring regions, and thus show a higher genetic complexity that indicates their possible long-term indigenous circulation and origin in the regions (Hotta et al., 1994; Lole et al., 2003; Lu et al., 2013; Murphy et al., 2007; Okamoto et al., 1994; Panigrahi et al., 1996; Simmonds et al., 1996, 2005; Tokita et al., 1996; Valliammai et al., 1995). Although a limited number of subtype 3h isolates have been detected in countries that are geographically well separated, these were obtained exclusively from immigrants who originated in Africa and showed the longest genetic distances compared with all of the other assigned subtypes (Abid et al., 2000; Bernier et al., 1996; Colson et al., 2011; Corbet et al., 2003; Murphy et al., 2007). In addition to some other variants originating in the Middle East, two isolates of the provisionally proposed subtype 3l were identified recently in Iran (Amini et al., 2006). These are two lines of evidence that may challenge the commonly accepted belief that HCV-3 may have its origin in South Asia (Simmonds et al., 2005). Instead, the ancestral origin may have been in Africa and this genotype may have been brought to areas outside of Africa via the Middle East or across the sea to South Asia. To test this hypothesis, we determined the full-length genomes of four subtype 3a, two subtype 3b and one subtype 3h isolates to not only calculate a molecular clock for genotype 3, but also perform evolutionary analysis. Using the sequence data retrieved from the Los Alamos HCV database (http://www.hcv.lanl.gov), and determined in this and previous studies, a timescaled phylogenetic tree was reconstructed. Based on this tree, the ancestral origin of HCV-3 was estimated and a new hypothesis was suggested.
Results
Genome sequence and organization
The full-length genomes of nine HCV-3 isolates were characterized: NE145, NE274, 811, QC7, QC8, QC9, QC10, QC34 and QC88, each using 16–23 overlapping fragments. Of these isolates, 811 had a genome size of 9633 nt, beginning from the 5′ terminus and ending at the extreme 3′ end, with the latter obtained using 3′ RACE reverse transcription (RT)-PCR. Each of the other eight genomes spanned 9458–9465 nt from the 5′ UTR to the poly(U) track of the 3′ UTR (Table 1). These nine genomes each contained a single ORF of 9060–9069 nt. The 5′ UTRs were 339–340 nt in length, whereas the 3′ UTRs varied from 52 to 234 nt. The sizes of the 10 protein-coding regions were the same as those of the H77 genome with the exception of those of the E2 and NS5A regions, which were variable in length (Table 1).
Table 1. Patient information for the nine genotype 3 isolates and the variable number of nucleotides/amino acids in certain genomic regions.
| ID | Subtype | Origin | Full | ORF | 5′ UTR | E2 | NS5A | 3′ UTR |
| H77 | 1a | USA | 9646 | 9036/3011 | 341 | 1089/363 | 1344/448 | 269 |
| NE274 | 3d | Nepal | 9458 | 9066/3021 | 339 | 1104/368 | 1359/453 | 53 |
| NE145 | 3e | Nepal | 9458 | 9066/3021 | 340 | 1107/369 | 1356/452 | 52 |
| 811 | 3h | Somalia | 9633 | 9060/3019 | 339 | 1104/368 | 1353/451 | 234 |
| QC07 | 3a | Canada | 9462 | 9066/3021 | 339 | 1107/369 | 1356/452 | 57 |
| QC08 | 3a | Canada | 9458 | 9066/3021 | 339 | 1107/369 | 1356/452 | 53 |
| QC09 | 3a | Canada | 9465 | 9066/3021 | 339 | 1107/369 | 1356/452 | 60 |
| QC10 | 3a | Canada | 9464 | 9069/3022 | 339 | 1110/370 | 1356/452 | 56 |
| QC34 | 3b | Canada | 9458 | 9066/3021 | 339 | 1107/369 | 1356/452 | 53 |
| QC88 | 3b | Canada | 9463 | 9069/3022 | 339 | 1110/370 | 1356/452 | 55 |
The genotype 1 sequence H77 (GenBank accession number AF009606) is included for comparison.
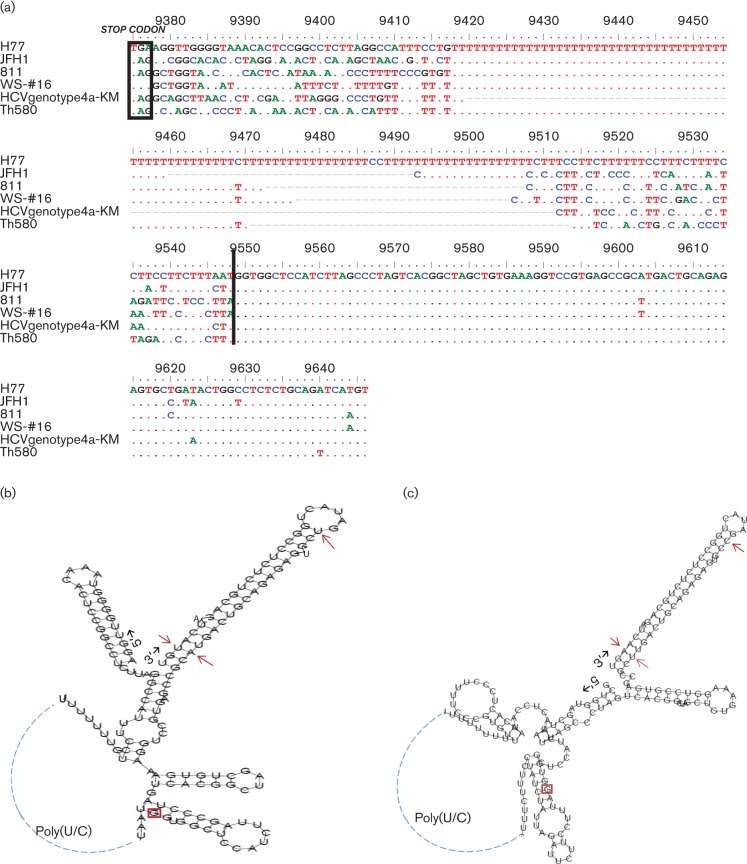
Fig. 1(a) shows an alignment of the 3′ UTR sequences from six isolates, which were selected to represent different genotypes of HCV. The 811 isolate shows a 3′ UTR that is 234 nt in length, consisting of a type-specific region immediately following the stop codon at the end of the NS5B region, a poly(U) stretch, a C(U)n-repeat and a highly conserved 98 nt region at the extreme 3′ end (Kolykhalov et al., 1996; Yamada et al., 1996). Using an online computer program (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi), we predicted the secondary structure for this 234 nt RNA sequence, which showed five typical hairpins, as reported previously (Fig. 1b). To obtain a better understanding of the structural similarity, we pairwise compared the 811 sequence in this region against that of the five other isolates, i.e. WS-#16, HCVgenotype4a-KM, JFH1, Th580 and H77, and revealed p distances of 0.225, 0.275, 0.298, 0.326 and 0.331, respectively. This finding indicated that the 811 sequence in this region was more closely related to WS-#16, which belongs to subtype 3a (Kolykhalov et al., 1996). The results also indicated that the 98 nt in the extreme 3′-terminus were highly conserved among the genotypes. Although a few substitutions were observed in the very last 46 nt, these did not appear to disturb the stability of the predicted secondary structure (Fig. 1c) (Blight & Rice, 1997).
Fig. 1.
(a) An alignment of the 3′ UTR sequences from the H77 (AF009606), JFH1 (AB047639), 811 (JF735126), WS-#16 (AF009075), HCVgenotype4a-KM (AB795432) and Th580 (D84262) isolates representing subtypes 1a, 2a, 3h, 3a, 4a and 6c, respectively. (GenBank accession numbers are given in parentheses.) The rectangle at the beginning of the alignment highlights the stop codons preceding each 3′ UTR. A vertical line in the middle marks the start of the highly conserved 98 bases at the extreme 3′ end. The ruler indicates the numbering of the H77 genome. The dots inside the alignment show the bases identical to those of the H77 isolate, whereas the short bars represent the absent bases. (b) The optimal RNA secondary structure of the 269 nt 3′ UTR of the H77 isolate was predicted, using an online computer program (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi), with a minimum free energy of −61.90 kcal mol−1. (c) The optimal RNA secondary structure of the 234 nt 3′ UTR of the 811 isolate was predicted with a minimum free energy of −55.10 kcal mol−1. In the highly conserved 98 nt of the extreme 3′ UTR (the 5′ starting nucleotide is boxed), the different nucleotides shown by 811 in comparison with H77 are indicated by arrows.
Genotype 3 classification
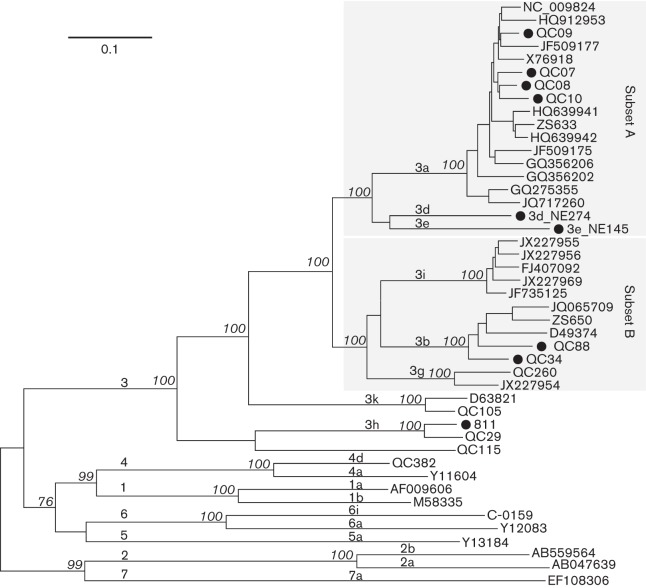
Fig. 2 depicts a maximum-likelihood (ML) tree that was reconstructed with the 45 full-length HCV genome sequences assembled in this study. The tree shows that the nine genomes determined in this study were all classified into genotype 3, but represented different subtypes. Based on the other sequences that were co-analysed, QC7, QC8, QC9 and QC10 can be classified into subtype 3a, QC34 and QC88 can be classified into subtype 3b, and 811 can be classified into subtype 3h. In contrast, NE145 and NE274 each led to a single branch, and both formed a distantly linked twin. In the tree, a total of eight subtypes and a novel subtype equivalent represented by the QC115 variant were classified under genotype 3. These formed two subsets and three independent subtypes. From the top of the tree, subset A contained 3a, 3d and 3e, and subset B contained 3i, 3b and 3g. Below and adjacent to these two subsets was the relatively distant subtype 3k, which used to be classified as genotype 10 because of its greater genetic differences from the members of the two subsets (Tokita et al., 1996). The base of the genotype 3 clade exhibited the divergent subtype 3h and the QC115 isolate, both of which showed the largest genetic differences from all other HCV-3 members. In general, when more than one sequence was grouped to form a subtype cluster, a bootstrap support of 100 % was shown. Based on this tree, pairwise p distances were calculated by comparing subtype 3h to the other seven assigned subtypes. The results showed distances ranging from 0.261 to 0.278; the largest was from the HCV-Tr/3b, NE145/3e and QC260/3g isolates, and the smallest was from the QC268/3i isolate. Apparently, these inter-subtype genome-wide distances were larger compared with those found among other genotypes. This finding was consistent with recently described results (Smith et al., 2014).
Fig. 2.
ML tree estimated using 45 full-length genome sequences. In addition to the nine isolates determined in this study (•), references from subtypes 1a, 1b, 2a, 2b, 3a, 3b, 3d, 3e, 3g, 3k, 3i, 3h, 4a, 4d, 5a, 6a, 6i, 7a and a novel HCV-3 variant QC115 were also included. Each branch is led by an isolate and denoted by the ID of the isolate (from this and our previous studies) or its GenBank accession number. The bootstrap supports are shown in italics. Bar, 0.1 nt substitutions per site. For descriptive purposes, the genotypes and subtypes are indicated above the related branches, and the two subsets A and B are highlighted by two shaded rectangles.
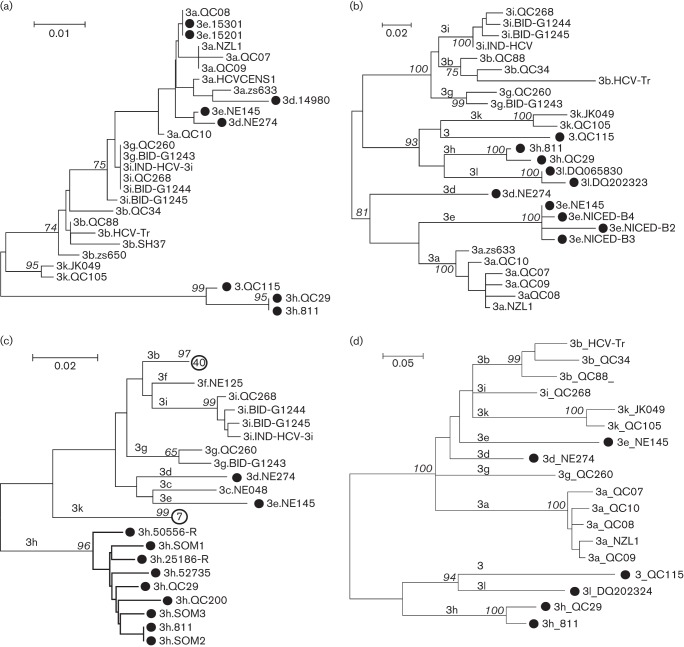
Fig. 3 presents four ML trees reconstructed using the partial sequences in the 5′ UTR, core, E1 and NS5B regions, respectively. These trees showed that NE274, NE145 and 811 each represented a distinct subtype: 3d, 3e and 3h, respectively. In the Los Alamos HCV database, two subtype 3d isolates, i.e. 14980 and NE274, were recorded. The 14980 (GenBank accession number AY766524) has a determined 5′ UTR sequence, but its sampling country was unknown. As it was more closely related to an isolate of 3a, i.e. ZS633 (Xu et al., 2013), than to NE274 in the 5′ UTR tree, 14980 may not represent a subtype 3d isolate. Six isolates corresponding to subtype 3e were archived. Two (15201 and 15301) showed 5′ UTR sequences identical to that of QC08/3a, whereas four (NICED-B2, NICED-B3, NICED-B4 and NE145) formed a solid 3e cluster based on their core region sequences (Chowdhury et al., 2003). With the exception of NE145, which was completely sequenced in this study, NICED-B2, NICED-B3 and NICED-B4 were all sampled in India, whereas 15201 and 15301 were of unknown origin. The full-length genome sequence was available for one subtype 3h isolate while partial E1 sequences were available for seven others; six of these were obtained from African immigrants (SOM1, SOM2, SOM3, QC29 and 52735 were obtained from Somalia, and QC200 was obtained from Congo) and two (25186-R and 50556-R) were detected in the UK, but the patients’ information is unknown. In addition, 8332488, which was obtained from an immigrant originating in Djibouti, a country that neighbours Somalia, was also classified into subtype 3h based on its cloned NS3 sequences (Colson et al., 2011). Two novel HCV-3 variants were detected recently in Iran and have been assigned provisionally as subtype 3l (Amini et al., 2006). The core sequences of both of these and the partial NS5B sequence of one of these were available in the Los Alamos HCV database, and were also analysed in this study. In the tree based on the core region sequences, both isolates were led by a single internal branch parallel to that of subtype 3h, whereas in the tree based on the NS5B sequences, the single sequence was more closely related to that of the QC115 isolate and those of subtype 3h than to any other sequences. Regardless, we observed that the 5′ UTR sequences poorly differentiated the subtypes within genotype 3.
Fig. 3.
Four ML trees estimated using partial sequences in the (a) 5′ UTR, (b) core, (c) E1 and (d) NS5B regions. Sequences of the following four subtypes, 3d, 3e, 3h, 3l and a novel variant QC115 are marked each with a black circle (•) to highlight their classifications. Each branch is led by an isolate, and denoted by the subtype and the ID of the isolate. However, in the E1 tree two hollow circles with a number inside are used to label two branches, which indicate the numbers of sequences led by these two branches. Bootstrap supports are shown in italics. Subtypes are indicated above the related branches. Bars, 0.1, 0.2 or 0.5 nt substitutions per site for different trees.
Evolutionary analysis
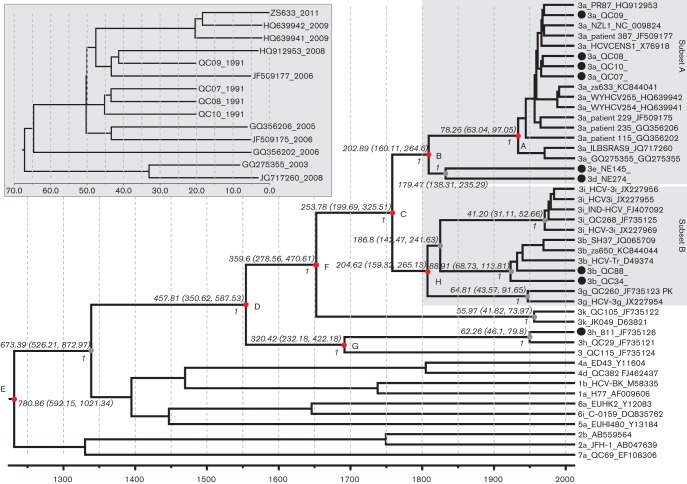
Based on the 14 full-length subtype 3a sequences, which exhibited sampling dates covering 22 years from 1991 to 2012, an evolutionary rate of 1.289×10−3±1.47×10−5 substitutions per site per year (mean±1.96 se) was estimated (see insert in Fig. 4). By increasing the se 14.14-fold and applying this rate in a normal distribution as the prior rate to a subsequent Markov chain Monte Carlo (MCMC) procedure under the uncorrelated log-normal clock model, we analysed the 45 full-length HCV genomes assembled in this study to yield the timescaled phylogenetic tree presented in Fig. 4. This tree showed a topology highly similar to that of the tree shown in Fig. 2, with the exception that all of the tips were aligned to the right end, and all of the nodes and branches were measured by grids corresponding to the timescale shown at the tree base. This scale indicates the year at which the different branches diverged. It was estimated that subtype 3a has a common ancestor dated to ~78.26 [95 % highest posterior density (HPD): 63.04, 97.05] years ago (point A), which is after the era in which the ancestral 3a strains diverged from the ancestor of both subtypes 3d and 3e ~202.89 (95 % HPD: 160.11, 264.6) years ago (point B). Considering both subsets A and B as a whole, the most recent common ancestor was dated to ~253.78 (95 % HPD: 199.69, 325.51) years ago (point C), whereas the analysis of genotype 3 as a single lineage dated the divergence time to ~457.81 (95 % HPD: 350.62, 587.53) years ago (point D). The inclusion of all of the analysed 45 HCV genomes representing genotypes 1–7 as a whole in the analysis resulted in the dating of the common ancestor to ~780.86 (95 % HPD: 592.15, 1021.34) years ago (point E). It was shown that both subtypes 3d and 3e were closer relatives to subtype 3a than to any other subtypes, and that these three may have represented a single epidemiological lineage during a certain period in the past. Similarly, the past co-origin of both subsets A and B was also indicated – both appeared to have diverged from each other ~100 years after the era when their ancestor diverged from subtype 3k (point F). Among all of the HCV-3 lineages, however, subtype 3h and the novel variant QC115 were the oldest, and both were separated from each other ~320.42 (95 % HPD: 232.18, 422.18) years ago (point G). In the tree, all of these major divergences showed full posterior values, indicating robust classification and reliable MCMC processing.
Fig. 4.
A timescaled phylogenetic tree estimated using the 45 sequences shown in Fig. 2. Each branch is led by an isolate and is denoted by the following format: subtype_ID_GenBank accession number. The branch lengths represent the evolutionary time that is measured by the grids corresponding to the timescale (years) shown at the tree base. To simplify the tree, only those posterior probability scores equal to 1 are shown in italics to the left of the related nodes; otherwise, the scores are <1. Eight time points are given after running the tMRCA function of beast software and these are indicated with the red circles A–G at the related internal nodes. These points measure the time and the estimated 95 % HPD, which are detailed on the circles to indicate the divergence of the related HCV-3 lineages. The two subsets A and B marked in Fig. 2 are also highlighted with two shaded rectangles. Another rectangle insert, which highlights the 14 dated subtype 3a full-length genomes that were used to estimate the molecular rate, is shown in the upper left of the tree.
Geographical distribution of HCV-3 isolates
By December 2011, the Los Alamos HCV database had archived a total of 7840 genotype 3 sequences. After removing those that did not include information regarding their sampling countries, those obtained from experimental animals, those representing various genomic regions of the same isolates and multiple clones from single subjects, a total of 5846 individual isolates were differentiated. These were obtained from different patients and their sampling countries were known. The information on their geographical distribution is summarized in Tables 2 and S1 (available in the online Supplementary Material). Table 2 shows that subtype 3a isolates had been identified worldwide and accounted for 88.6 % of the total 5846 isolates. In contrast, the numbers of other subtypes were remarkably decreased. With the exception of subtype 3b, which was found in 18 countries/regions and accounted for 8.96 % of the total 5846 isolates, the other subtypes each accounted for no more than 1.09 % of the isolates and were limited to certain regions. The results showed that the highest diversity of the lineages of HCV-3 was found in Asia (nine of the 10 subtypes plus one recombinant) followed by North America (six subtypes and an unclassified variant). Based on the number of isolates, however, the majority of the lineages were reported in Europe (2662/5846, 45.5 %) followed by Asia, America, Australia and then the other continents.
Table 2. Geographical distribution of HCV-3 subtypes.
| Subtype | No. of countries* in which subtype is detected | No. of isolates (n = 5846) | Percentage of isolates | |
| Into subtypes | Not into subtypes | |||
| 3a | 55 (worldwide) | 4632 | 545 | 88.6 |
| 3b | 18 (AU, BD, BR, CA, CN, DE, GB, HK, ID, IN, JP, KH, MM, MY, NP, PK, TH, VN) | 482 | 42 | 8.96 |
| 3a/3b | 1 (CN) | 1 | 0.0171 | |
| 3c | 1 (NP) | 1 | 0.0171 | |
| 3d | 1 (NP) | 1 | 0.0171 | |
| 3e | 1 (NP) | 1 | 0.0171 | |
| 3f | 2 (NP, PK) | 2 | 0.0342 | |
| 3 g | 2 (CA, IN) | 19 | 4 | 0.394 |
| 3h | 4(CA, DK, FR, SO) | 7 | 2 | 0.154 |
| 3i | 2 (CA, IN) | 14 | 0.239 | |
| 3k | 6 (CA, ES, ID, IN, NL, PK) | 49 | 16 | 1.11 |
| 3l | 1 (IR) | 2 | 0.0342 | |
| 3 | 6 (BR, CA, ID, IN, TH, TR) | 26 | 0.445 | |
| Total | 59 | 5211 | 635 | 100 |
Country codes officially assigned in ISO 3166-1: AU, Australia; BD, Bangladesh; BR, Brazil; CA, Canada; CN, China; DE, Germany; DK, Denmark; ES, Spain; FR, France; GB, Great Britain; HK, Hong Kong; ID, Indonesia; IN, India; IR, Iran; JP, Japan; KH, Cambodia; MM, Myanmar; MY, Malaysia; NL, Netherlands; NP, Nepal; PK, Pakistan; SO, Somalia; TH, Thailand; TR, Turkey; VN, Viet Nam (http://en.wikipedia.org/wiki/ISO_3166-1_alpha-2).
Discussion
In this study, the full-length genome sequences of nine genotype 3 isolates of HCV, denoted QC7, QC8, QC9, QC10, QC34, QC88, NE145, NE274 and 811, were characterized. These isolates represent subtypes 3a, 3b, 3d, 3e and 3h. In previous studies, partial sequences had been determined for these isolates, but their full-length genomes had not been obtained (Abid et al., 2000; Murphy et al., 2007; Tokita et al., 1994). By applying our modified approaches (Lu et al., 2005), these nine isolates were entirely sequenced in the present study not only for the confirmation of their subtype assignments within genotype 3, but also for the reconstruction of a timescaled phylogenetic tree to estimate the origin and divergence of this HCV genotype.
To the best of our knowledge, NE274 and NE145 represent the first full-length genomes of subtypes 3d and 3e, respectively. In a recent paper on the expanded HCV classification, both 3d and 3e were categorized into the provisionally assigned subtypes (Smith et al., 2014). As their full-length genomes were determined in this study, these two subtypes are now confirmed, which allowed us to fill the information gap. Although 811 represents the second full-length genome of subtype 3h, it is, to the best of our knowledge, the first HCV-3 full-length genome to have its extreme 3′ UTR sequence characterized. Based on this information, we were able to predict its secondary structure to show its similarity to other HCV genotypes. Together with the full-length genomes of subtypes 3g, 3h, 3i and 3k obtained in one of our recent studies (Lu et al., 2013), and those of four subtype 3a and two subtype 3b isolates, which were completely sequenced in this study, an expanded panel of HCV-3 full-length genomes is provided. These genomes represent all of the assigned subtypes within HCV-3 with the exception of 3c and 3f, which remain our future goals. To date, subtype 3c has three isolates archived in the Los Alamos HCV database: NE048 from Nepal, S48 from Singapore (Greene et al., 1995) and JN656627 from India. However, our reanalysis revealed that the latter two isolates can be reclassified into subtype 3a. Consequently, NE048 represents the only subtype 3c isolate that has been identified to date. Regarding subtype 3f, two isolates have been recorded: NE125 from Nepal (Tokita et al., 1994) and PK64 from Pakistan (Stuyver et al., 1996). Although we attempted to sequence the NE048/3c and NE125/3f isolates in the present study, we failed to provide their full-length genomes due to the limited number of samples with low viral titres. Therefore, our future studies will rely on more surveys to find additional 3c and 3f isolates in the regions in which these subtypes are indigenous.
Our recent studies have shown a very high degree of genetic diversity among the HCV-6 isolates found in South-East Asia, which therefore suggests South-East Asia as the region of origin for this oldest HCV lineage (Wang et al., 2013). However, to a lesser extent, the present study also revealed the high complexity of HCV-3 isolates in the Indian subcontinent and in countries around the Indian Ocean, where 10 of the 11 assigned HCV-3 subtypes have been detected (Table 2). Based on these data, we may conclude that the Indian subcontinent is the region of origin for HCV-3. In fact, this conclusion reflects a commonly accepted belief. Regardless, the number of HCV-3 isolates reported in the Los Alamos HCV database is highest in Europe (2662/5846 = 45.5 %) compared with those reported in other continents. This difference is observed because almost all of the European HCV-3 isolates belong to subtype 3a, with the exception of five that can be classified into other subtypes (Castillo et al., 2006; Colson et al., 2011; Corbet et al., 2003; Ross et al., 2000; van Doorn et al., 1994). In addition, subtype 3a strains have been found to be distributed worldwide because of their long-distance transmission via the IDU network. Furthermore, molecular epidemiological studies of HCV have been more frequently performed in Europe than in other continents, which has resulted in the determination of a large number of 3a sequences.
With the exception of 3a and 3b that are distributed worldwide, only a small number of the isolates of the other subtypes in genotype 3 have been detected in geographical regions outside South and South-East Asia. These include three 3g (QC260 and QC299 in Canada, BID-G1243 in Europe), 10 3i (QC100, QC102, QC125, QC238, QC268, QC270 and QC310 in Canada, BID-G1244, BID-G1245 and 17872-R in Europe), five 3k (DQ641982-85, HE974762 and NL96 in Europe, QC105 in Canada, KC118332 in Iran), nine 3h (QC29 and QC200 in Canada, SOM1, SOM2, SOM3, 52735, 25186-R, 50556-R and 8332488 in Europe) and two 3l isolates (DQ202323 and DQ065830 in Iran) (Abid et al., 2000; Amini et al., 2006; Bernier et al., 1996; Castillo et al., 2006; Colson et al., 2011; Corbet et al., 2003; Murphy et al., 2007; Newman et al., 2013; Samimi-Rad et al., 2013; van Doorn et al., 1994). Of these, five 3i isolates (QC100, QC102, QC125, QC238 and QC310) have been detected among Caucasians in Canada and these five isolates shared a unique insertion of 12 nt, which may indicate a single source of infection, likely via IDU (Murphy et al., 2007). With the exception of these five 3i isolates, the majority of the above-described non-3a/3b isolates were from immigrants originating in South Asia or Africa, although a few were detected in the Middle East. These findings appear to agree with the above-described and commonly accepted belief that HCV-3 may have its origin in South Asia and subsequently spread to other regions. However, the ancestral origin of HCV-3 could have been in Africa, as suggested by the timescaled phylogenetic tree reconstructed in the present study. The tree shows that subtype 3h and the branch led by the QC115 isolate represent the oldest HCV-3 lineages. As all of the 3h isolates had their origins in Africa, the tree may suggest that the ancestral origin of HCV-3 is in Africa. From Africa, the ancient HCV-3 strains may have spread to South Asia either across the sea or by land via the Middle East. In support of the latter hypothesis, the novel QC115 isolate was detected in an immigrant from the Middle East (Lu et al., 2013) and isolates of the newly proposed subtype 3l, which are more genetically related to subtype 3h and the Q115 isolate than to any other genotype 3 members, have been detected in Iran (Amini et al., 2006). These findings suggest that there could be additional genotype 3 lineages of HCV that have not been detected in the Middle East. Based on the timescaled tree, the period during which the spread of the ancestral HCV-3 strains from Africa to the Middle East and South Asia could have occurred was estimated. This period was dated to ~457 (95 % HPD: 351, 588) years ago, i.e. in the middle of the sixteenth century (1553, 95 % HPD: 1422, 1660). Coincidently, this time frame corresponds to the era during which Vasco da Gama completed his expeditions by sailing along the eastern coast of Africa to India (http://en.wikipedia.org/wiki/Vasco_da_Gama). However, before da Gama’s voyages, slave trading from Eastern Africa to the Middle East and South Asia had taken place for centuries (Gordon, 1989; http://www.arabslavetrade.com/). Since the seventh century, Arabian traders had brought Africans across the Indian Ocean from the coasts of the Red Sea in Eritrea and Ethiopia, from the Swahili Coast of present-day Kenya, Mozambique and Tanzania, and from elsewhere in South-East Africa to present-day Iraq, Iran, Kuwait, Turkey and other parts of the Middle East, and to Pakistan and India in South Asia. Such slave-trading activities may have mediated the spread of the earliest HCV-3 strains from Eastern Africa to South Asia and the islands of Indonesia. However, additional studies on the molecular epidemiology of HCV not only in Africa but, more importantly, in the Middle East are required to obtain more solid evidence in support of this hypothesis.
In this study, we analysed the geographical distribution patterns of HCV-3 only based on the sequences archived in the Los Alamos HCV database. Their percentages represented the frequencies in the database, but not in the population. These frequencies may not really reflect the epidemiological situation although they can be a good approximation. Therefore, these percentages should be extrapolated with caution.
Errors may exist in the evolutionary analysis. For example, in this study only a limited number of HCV-3 sequences, sampled during a very short period of time from 1991 to 2011, were used for the time calibration. Among these sequences there may exist some sampling or stochastic errors, whilst such a relatively small error may be magnified in the subsequent estimation of the past histories of HCV that have lasted for centuries or millennia. Other potential errors may involve the possible saturation of nucleotide substitution and different rates along branches or within genomic regions. All evolutionary analyses of the highly variable RNA viruses such as HCV are subjected to these errors. Theoretically, the resulting data should be adjusted; unfortunately, however, we still lack the knowledge for these adjustments.
Methods
Samples.
Three serum samples, NE274, NE145 and 811, were obtained from patients with chronic liver disease. Of these, both NE274 and NE145 were sampled in Nepal (Tokita et al., 1994). These were selected because partial sequences that had been determined previously in these two samples represented the provisionally assigned subtypes 3d and 3e. The 811 sample was collected from a Somalian refugee in Switzerland, in which one of the first partial 3h sequences was isolated (Abid et al., 2000). To yield the dated full-length genome sequences for the reconstruction of the ancestral relationship among genotype 3 isolates, six additional samples, denoted QC7, QC8, QC9, QC10, QC34 and QC88, were also included. These were obtained from patients living in the Canadian province of Quebec (Murphy et al., 2007). Among these, QC7, QC8, QC9 and QC10 were sampled in 1991 and represented subtype 3a isolates. QC34 and QC88 were sampled in 1995 and 2002, respectively, and represented subtype 3b isolates.
PCR amplification and sequencing.
From each 100 µl serum sample, the viral RNA was extracted using the QIAamp Viral RNA Mini kit (Qiagen) following the manufacturer’s protocol. The cDNA was synthesized using the RevertAid First-Strand cDNA Synthesis kit (Fermentas Life Science) under slightly modified conditions: the RNA pellet was dissolved in a random primer solution and incubated at 95 °C for 5 min before the other reagents were added. The resulting cDNA was used as a template to amplify the target sequences through conventional nested PCR. With the exception of the above-described protocols, all of the other procedures, such as the degenerate primers, the overlapping PCR strategies, the BigDye DNA sequencing method and the full-length genome sequence assembling, were the same as those reported previously (Li et al., 2012; Lu et al., 2013).
Amplification of the extreme 5′ end.
The extreme 5′ UTR termini of the HCV-3 isolates were highly conserved. To amplify such terminal sequences through conventional PCR, a set of 5′-end degenerate primers was designed based on the known 3a and 3k sequences (forward, 5′-ACCTGCCTCTTWCGAGGCGACACT-3′; inner reverse, 5′-TGGTGCACGGTCTACGAGACCT-3′; outer reverse, 5′-CTTTGAGGTTTAGGATTCGTGCTC-3′).
Amplification of the 3′ end.
For the QC7, QC8, QC9, QC10, QC34, QC88, NE145 and NE274 isolates, the 3′ termini were extended to the poly(U) tracks using conventional PCR (Li et al., 2012). However, for the 811 isolate, the extreme 3′ terminus was determined based on 3′ RACE RT-PCR, in which the RNA was polyadenylated using the A-Plus Poly(A) Polymerase Tailing kit (Epicenter Biotechnologies) and then extracted using Tri-pure (Lu et al., 2005). After the RNA pellet was resuspended, RT was performed and the cDNA was synthesized based on the poly(T) primer CACT35, which contained the 35mer (T). The resulting cDNA was then used to amplify the poly(T) ends using the CACT35 as the downstream primer and an 811-specific primer 5′-GCTATTTACTCCTGTGCCTACTCC-3′ (corresponding to nt 9332–9355 in the 811 genome) as the upstream primer. The PCR was performed using FastStart Taq DNA Polymerase (Roche) with the following strategy: a first cycle of denaturation at 95 °C for 3 min, 35 cycles of 95 °C for 30 s, 55 °C for 30 s and 72 °C for 30 s, and a final cycle of extension at 72 °C for 7 min. After amplification, the products were cloned into a TA vector and 10 clones were selected for DNA sequencing (Lu et al., 2008).
Inspection of genetic variations and phylogenetic analyses.
The nine full-length HCV genomes obtained were denoted according to the nucleotide numbering in the H77 genome. To reconstruct the ancestral relationship, an additional 36 full-length HCV genome sequences were retrieved from the Los Alamos HCV database (Kuiken et al., 2006). These not only represented genotype 3, but also genotypes 1, 2, 4, 5, 6 and 7. A total of 45 full-length genome sequences were thus assembled using the BioEdit program (Tippmann, 2004). These were aligned to display their genomic organization and to identify the genetic differences.
Based on the 45 full-length HCV genome sequences, ML phylogenetic trees were reconstructed using mega5 software (Tamura et al., 2011), in which the best trees were heuristically searched under the most appropriate substitution model GTR+I+Γ6 using the nearest-neighbour-interchange perturbation algorithms. To assess the statistical robustness of the phylogenetic groupings, bootstrap analyses were conducted with 500 replicates.
Evolutionary analysis.
Based on the 45 full-length HCV genome sequences assembled, an evolutionary timescale tree was further estimated using the MCMC algorithm implemented in the beast package (version 1.6.1) (Drummond & Rambaut, 2007). We recently analysed the full-length subtype 1a and 1b sequences, and identified that the relaxed log-normal clock model is better than the exponential and strict models (Yuan et al., 2013). Therefore, we used this model in combination with the GTR+I+Γ substitution model and the Bayesian skyline model in this study for the estimation of the timescale tree in the MCMC analysis, but we inputted an evolutionary rate of 1.289×10−3±1.47×10−5 substitutions per site per year as the prior rate, which was estimated directly using the 14 dated 3a sequences assembled in this study using a previously described method (Yuan et al., 2013). After these parameters were set, we ran the MCMC procedure for 300 million states and obtained a tree every 30 000 states. After discarding the first 10 % burn-in, the output was examined for convergence through a visual inspection of the chain length and by comparing the statistics of the effective sample size (ESS) using the Tracer program (http://tree.bio.ed.ac.uk). In this study, sufficient sampling was considered to have been achieved when all of the ESS numbers were ≥200. We used the TreeAnnotator program to reconstruct a tree from the resulting set of credible trees and the final tree was denoted the MCC (maximum clade credibility) tree. As a molecular clock was incorporated, the branch lengths and the tree node heights were marked in units of years. The phylogenetic structure was then displayed using the FigTree program, and the clades, lineages and internal node heights were indicated as necessary.
To further comprehend the genetic complexity of subtypes 3e, 3d, 3h and 3l, the related partial sequences available in the Los Alamos HCV database were retrieved to represent these four subtypes and analysed phylogenetically. Four datasets, denoted 5′ UTR, core, E1 and NS5B, were therefore assembled.
To exclude the recent viral recombination events (Lee et al., 2010; Legrand-Abravanel et al., 2007), rdp3 software (Martin et al., 2010) was run using the modified settings that we described previously (Lu et al., 2007). This analysis was only performed for the full-length genome sequences.
Geographical distribution of genotype 3 sequences.
A total of 7840 HCV isolates from the Los Alamos HCV database accessed on December, 2011 were classified as genotype 3. Of these, 5846 had known sampling countries and represented individual isolates. Based on these 5846 isolates, the correlation between subtypes and sampling countries was investigated.
Acknowledgements
The study described was supported by a grant from the National Institute of Allergy and Infectious Diseases (5 R01 AI080734-03A).
Footnotes
One supplementary table is available with the online version of this paper.
References
- Abid K., Quadri R., Veuthey A. L., Hadengue A., Negro F. (2000). A novel hepatitis C virus (HCV) subtype from Somalia and its classification into HCV clade 3. J Gen Virol 81, 1485–1493 [DOI] [PubMed] [Google Scholar]
- Abid K., Pazienza V., de Gottardi A., Rubbia-Brandt L., Conne B., Pugnale P., Rossi C., Mangia A., Negro F. (2005). An in vitro model of hepatitis C virus genotype 3a-associated triglycerides accumulation. J Hepatol 42, 744–751 10.1016/j.jhep.2004.12.034 [DOI] [PubMed] [Google Scholar]
- Alter M. J. (1999). Hepatitis C virus infection in the United States. J Hepatol 31 (Suppl. 1), 88–91 10.1002/hep.1840360704 [DOI] [PubMed] [Google Scholar]
- Amini S., Ahmadi Pour M. H., Azadmanesh K. (2006). The phylogenetic analysis of hepatitis C virus isolates obtained from two Iranian carriers revealed evidence for a new subtype of HCV genotype 3. Virus Genes 33, 271–278 [DOI] [PubMed] [Google Scholar]
- Bernier L., Willems B., Delage G., Murphy D. G. (1996). Identification of numerous hepatitis C virus genotypes in Montreal, Canada. J Clin Microbiol 34, 2815–2818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blight K. J., Rice C. M. (1997). Secondary structure determination of the conserved 98-base sequence at the 3′ terminus of hepatitis C virus genome RNA. J Virol 71, 7345–7352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochud P. Y., Cai T., Overbeck K., Bochud M., Dufour J. F., Müllhaupt B., Borovicka J., Heim M., Moradpour D. & other authors (2009). Genotype 3 is associated with accelerated fibrosis progression in chronic hepatitis C. J Hepatol 51, 655–666 10.1016/j.jhep.2009.05.016 [DOI] [PubMed] [Google Scholar]
- Bourlière M., Barberin J. M., Rotily M., Guagliardo V., Portal I., Lecomte L., Benali S., Boustière C., Perrier H. & other authors (2002). Epidemiological changes in hepatitis C virus genotypes in France: evidence in intravenous drug users. J Viral Hepat 9, 62–70 10.1046/j.1365-2893.2002.00319.x [DOI] [PubMed] [Google Scholar]
- Castillo I., Rodríguez-Iñigo E., López-Alcorocho J. M., Pardo M., Bartolomé J., Carreño V. (2006). Hepatitis C virus replicates in the liver of patients who have a sustained response to antiviral treatment. Clin Infect Dis 43, 1277–1283 10.1086/508198 [DOI] [PubMed] [Google Scholar]
- Chowdhury A., Santra A., Chaudhuri S., Dhali G. K., Chaudhuri S., Maity S. G., Naik T. N., Bhattacharya S. K., Mazumder D. N. (2003). Hepatitis C virus infection in the general population: a community-based study in West Bengal, India. Hepatology 37, 802–809 [DOI] [PubMed] [Google Scholar]
- Colson P., Gayet S., Gerolami R. (2011). NS3 protease of genotype 3 subtype h HCV identified in southeastern France. Antivir Ther 16, 615–619 10.3851/IMP1765 [DOI] [PubMed] [Google Scholar]
- Corbet S., Bukh J., Heinsen A., Fomsgaard A. (2003). Hepatitis C virus subtyping by a core-envelope 1-based reverse transcriptase PCR assay with sequencing and its use in determining subtype distribution among Danish patients. J Clin Microbiol 41, 1091–1100 10.1128/JCM.41.3.1091-1100.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A. J., Rambaut A. (2007). beast: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7, 214 10.1186/1471-2148-7-214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feld J. J., Hoofnagle J. H. (2005). Mechanism of action of interferon and ribavirin in treatment of hepatitis C. Nature 436, 967–972 [DOI] [PubMed] [Google Scholar]
- Gordon M. (1989). Slavery in the Arab World. New York: New Amsterdam Press [Google Scholar]
- Greene W. K., Cheong M. K., Ng V., Yap K. W. (1995). Prevalence of hepatitis C virus sequence variants in South-East Asia. J Gen Virol 76, 211–215 10.1099/0022-1317-76-1-211 [DOI] [PubMed] [Google Scholar]
- Hoofnagle J. H. (2002). Course and outcome of hepatitis C. Hepatology 36 (Suppl 1), S21–S29 10.1002/hep.1840360704 [DOI] [PubMed] [Google Scholar]
- Hotta H., Handajani R., Lusida M. I., Soemarto W., Doi H., Miyajima H., Homma M. (1994). Subtype analysis of hepatitis C virus in Indonesia on the basis of NS5b region sequences. J Clin Microbiol 32, 3049–3051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hourioux C., Patient R., Morin A., Blanchard E., Moreau A., Trassard S., Giraudeau B., Roingeard P. (2007). The genotype 3-specific hepatitis C virus core protein residue phenylalanine 164 increases steatosis in an in vitro cellular model. Gut 56, 1302–1308 10.1136/gut.2006.108647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackel-Cram C., Babiuk L. A., Liu Q. (2007). Up-regulation of fatty acid synthase promoter by hepatitis C virus core protein: genotype-3a core has a stronger effect than genotype-1b core. J Hepatol 46, 999–1008 10.1016/j.jhep.2006.10.019 [DOI] [PubMed] [Google Scholar]
- Kolykhalov A. A., Feinstone S. M., Rice C. M. (1996). Identification of a highly conserved sequence element at the 3′ terminus of hepatitis C virus genome RNA. J Virol 70, 3363–3371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiken C., Combet C., Bukh J., Shin-I T., Deleage G., Mizokami M., Richardson R., Sablon E., Yusim K. & other authors (2006). A comprehensive system for consistent numbering of HCV sequences, proteins and epitopes. Hepatology 44, 1355–1361 10.1002/hep.21377 [DOI] [PubMed] [Google Scholar]
- Lee Y. M., Lin H. J., Chen Y. J., Lee C. M., Wang S. F., Chang K. Y., Chen T. L., Liu H. F., Chen Y. M. (2010). Molecular epidemiology of HCV genotypes among injection drug users in Taiwan: full-length sequences of two new subtype 6w strains and a recombinant form_2b6w. J Med Virol 82, 57–68 10.1002/jmv.21658 [DOI] [PubMed] [Google Scholar]
- Legrand-Abravanel F., Claudinon J., Nicot F., Dubois M., Chapuy-Regaud S., Sandres-Saune K., Pasquier C., Izopet J. (2007). New natural intergenotypic (2/5) recombinant of hepatitis C virus. J Virol 81, 4357–4362 10.1128/JVI.02639-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Cao H., Lu L., Murphy D. (2012). Full-length sequences of 11 hepatitis C virus genotype 2 isolates representing five subtypes and six unclassified lineages with unique geographical distributions and genetic variation patterns. J Gen Virol 93, 1173–1184 10.1099/vir.0.038315-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lole K. S., Jha J. A., Shrotri S. P., Tandon B. N., Prasad V. G., Arankalle V. A. (2003). Comparison of hepatitis C virus genotyping by 5′ noncoding region- and core-based reverse transcriptase PCR assay with sequencing and use of the assay for determining subtype distribution in India. J Clin Microbiol 41, 5240–5244 10.1128/JCM.41.11.5240-5244.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L., Nakano T., He Y., Fu Y., Hagedorn C. H., Robertson B. H. (2005). Hepatitis C virus genotype distribution in China: predominance of closely related subtype 1b isolates and existence of new genotype 6 variants. J Med Virol 75, 538–549 10.1002/jmv.20307 [DOI] [PubMed] [Google Scholar]
- Lu L., Li C., Fu Y., Gao F., Pybus O. G., Abe K., Okamoto H., Hagedorn C. H., Murphy D. (2007). Complete genomes of hepatitis C virus (HCV) subtypes 6c, 6l, 6o, 6p and 6q: completion of a full panel of genomes for HCV genotype 6. J Gen Virol 88, 1519–1525 10.1099/vir.0.82820-0 [DOI] [PubMed] [Google Scholar]
- Lu L., Tatsunori N., Li C., Waheed S., Gao F., Robertson B. H. (2008). HCV selection and HVR1 evolution in a chimpanzee chronically infected with HCV-1 over 12 years. Hepatol Res 38, 704–716 10.1111/j.1872-034X.2008.00320.x [DOI] [PubMed] [Google Scholar]
- Lu L., Li C., Yuan J., Lu T., Okamoto H., Murphy D. G. (2013). Full-length genome sequences of five hepatitis C virus isolates representing subtypes 3g, 3h, 3i and 3k, and a unique genotype 3 variant. J Gen Virol 94, 543–548 10.1099/vir.0.049668-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manns M. P., Wedemeyer H., Cornberg M. (2006). Treating viral hepatitis C: efficacy, side effects, and complications. Gut 55, 1350–1359 10.1136/gut.2005.076646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D. P., Lemey P., Lott M., Moulton V., Posada D., Lefeuvre P. (2010). rdp3: a flexible and fast computer program for analyzing recombination. Bioinformatics 26, 2462–2463 10.1093/bioinformatics/btq467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaw R., Moaven L., Locarnini S. A., Bowden D. S. (1997). Hepatitis C virus genotypes in Australia. J Viral Hepat 4, 351–357 10.1046/j.1365-2893.1997.00060.x [DOI] [PubMed] [Google Scholar]
- Murphy D. G., Willems B., Deschênes M., Hilzenrat N., Mousseau R., Sabbah S. (2007). Use of sequence analysis of the NS5B region for routine genotyping of hepatitis C virus with reference to C/E1 and 5′ untranslated region sequences. J Clin Microbiol 45, 1102–1112 10.1128/JCM.02366-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negro F. (2006). Mechanisms and significance of liver steatosis in hepatitis C virus infection. World J Gastroenterol 12, 6756–6765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman R. M., Kuntzen T., Weiner B., Berical A., Charlebois P., Kuiken C., Murphy D. G., Simmonds P., Bennett P. & other authors (2013). Whole genome pyrosequencing of rare hepatitis C virus genotypes enhances subtype classification and identification of naturally occurring drug resistance variants. J Infect Dis 208, 17–31 10.1093/infdis/jis679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nkontchou G., Ziol M., Aout M., Lhabadie M., Baazia Y., Mahmoudi A., Roulot D., Ganne-Carrie N., Grando-Lemaire V. & other authors (2011). HCV genotype 3 is associated with a higher hepatocellular carcinoma incidence in patients with ongoing viral C cirrhosis. J Viral Hepat 18, e516–e522 10.1111/j.1365-2893.2011.01441.x [DOI] [PubMed] [Google Scholar]
- Okamoto H., Kojima M., Sakamoto M., Iizuka H., Hadiwandowo S., Suwignyo S., Miyakawa Y., Mayumi M. (1994). The entire nucleotide sequence and classification of a hepatitis C virus isolate of a novel genotype from an Indonesian patient with chronic liver disease. J Gen Virol 75, 629–635 10.1099/0022-1317-75-3-629 [DOI] [PubMed] [Google Scholar]
- Panigrahi A. K., Roca J., Acharya S. K., Jameel S., Panda S. K. (1996). Genotype determination of hepatitis C virus from northern India: identification of a new subtype. J Med Virol 48, 191–198 [DOI] [PubMed] [Google Scholar]
- Pawlotsky J. M., Tsakiris L., Roudot-Thoraval F., Pellet C., Stuyver L., Duval J., Dhumeaux D. (1995). Relationship between hepatitis C virus genotypes and sources of infection in patients with chronic hepatitis C. J Infect Dis 171, 1607–1610 10.1093/infdis/171.6.1607 [DOI] [PubMed] [Google Scholar]
- Ross R. S., Viazov S. O., Holtzer C. D., Beyou A., Monnet A., Mazure C., Roggendorf M. (2000). Genotyping of hepatitis C virus isolates using CLIP sequencing. J Clin Microbiol 38, 3581–3584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubbia-Brandt L., Quadri R., Abid K., Giostra E., Malé P. J., Mentha G., Spahr L., Zarski J. P., Borisch B. & other authors (2000). Hepatocyte steatosis is a cytopathic effect of hepatitis C virus genotype 3. J Hepatol 33, 106–115 10.1016/S0168-8278(00)80166-X [DOI] [PubMed] [Google Scholar]
- Samimi-Rad K., Asgari F., Nasiritoosi M., Esteghamati A., Azarkeyvan A., Eslami S. M., Zamani F., Magnius L., Alavian S. M., Norder H. (2013). Patient-to-patient transmission of hepatitis C at Iranian thalassemia centers shown by genetic characterization of viral strains. Hepat Mon 13, e7699 10.5812/hepatmon.7699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silini E., Bono F., Cividini A., Cerino A., Maccabruni A., Tinelli C., Bruno S., Bellobuono A., Mondelli M. (1995). Molecular epidemiology of hepatitis C virus infection among intravenous drug users. J Hepatol 22, 691–695 10.1016/0168-8278(95)80225-8 [DOI] [PubMed] [Google Scholar]
- Simmonds P., Mellor J., Sakuldamrongpanich T., Nuchaprayoon C., Tanprasert S., Holmes E. C., Smith D. B. (1996). Evolutionary analysis of variants of hepatitis C virus found in South-East Asia: comparison with classifications based upon sequence similarity. J Gen Virol 77, 3013–3024 10.1099/0022-1317-77-12-3013 [DOI] [PubMed] [Google Scholar]
- Simmonds P., Bukh J., Combet C., Deléage G., Enomoto N., Feinstone S., Halfon P., Inchauspé G., Kuiken C. & other authors (2005). Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology 42, 962–973 10.1002/hep.20819 [DOI] [PubMed] [Google Scholar]
- Smith D. B., Bukh J., Kuiken C., Muerhoff A. S., Rice C. M., Stapleton J. T., Simmonds P. (2014). Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: updated criteria and genotype assignment web resource. Hepatology 59, 318–327 10.1002/hep.26744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuyver L., Wyseur A., van Arnhem W., Hernandez F., Maertens G. (1996). Second-generation line probe assay for hepatitis C virus genotyping. J Clin Microbiol 34, 2259–2266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). mega5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28, 2731–2739 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel H. J., Collett M. S., Gould E. A., Heinz F. X., Houghton M., Meyers G., Purcell R. H., Rice C. M. (2005). Flaviviridae. In Virus Taxonomy: VIIIth Report of the International Committee on Taxonomy of Viruses, pp. 981–998 Edited by Fauquet C. M., Mayo, M., Maniloff J., Desselberger U., Ball L. A. San Diego, CA: Academic Press [Google Scholar]
- Tippmann H. F. (2004). Analysis for free: comparing programs for sequence analysis. Brief Bioinform 5, 82–87 10.1093/bib/5.1.82 [DOI] [PubMed] [Google Scholar]
- Tokita H., Shrestha S. M., Okamoto H., Sakamoto M., Horikita M., Iizuka H., Shrestha S., Miyakawa Y., Mayumi M. (1994). Hepatitis C virus variants from Nepal with novel genotypes and their classification into the third major group. J Gen Virol 75, 931–936 10.1099/0022-1317-75-4-931 [DOI] [PubMed] [Google Scholar]
- Tokita H., Okamoto H., Iizuka H., Kishimoto J., Tsuda F., Lesmana L. A., Miyakawa Y., Mayumi M. (1996). Hepatitis C virus variants from Jakarta, Indonesia classifiable into novel genotypes in the second (2e and 2f), tenth (10a) and eleventh (11a) genetic groups. J Gen Virol 77, 293–301 10.1099/0022-1317-77-2-293 [DOI] [PubMed] [Google Scholar]
- Valliammai T., Thyagarajan S. P., Zuckerman A. J., Harrison T. J. (1995). Diversity of genotypes of hepatitis C virus in southern India. J Gen Virol 76, 711–716 10.1099/0022-1317-76-3-711 [DOI] [PubMed] [Google Scholar]
- van der Meer A. J., Veldt B. J., Feld J. J., Wedemeyer H., Dufour J. F., Lammert F., Duarte-Rojo A., Heathcote E. J., Manns M. P. & other authors (2012). Association between sustained virological response and all-cause mortality among patients with chronic hepatitis C and advanced hepatic fibrosis. JAMA 308, 2584–2593 10.1001/jama.2012.144878 [DOI] [PubMed] [Google Scholar]
- van Doorn L. J., Kleter B., Stuyver L., Maertens G., Brouwer H., Schalm S., Heijtink R., Quint W. (1994). Analysis of hepatitis C virus genotypes by a line probe assay and correlation with antibody profiles. J Hepatol 21, 122–129 10.1016/S0168-8278(94)80148-7 [DOI] [PubMed] [Google Scholar]
- Wang H., Yuan Z., Barnes E., Yuan M., Li C., Fu Y., Xia X., Li G., Newton P. N. & other authors (2013). Eight novel hepatitis C virus genomes reveal the changing taxonomic structure of genotype 6. J Gen Virol 94, 76–80 10.1099/vir.0.047506-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams I. (1999). Epidemiology of hepatitis C in the United States. Am J Med 107 (Suppl 2), 2–9 10.1016/S0002-9343(99)00373-3 [DOI] [PubMed] [Google Scholar]
- Xu R., Tong W., Gu L., Li C., Fu Y., Lu L. (2013). A panel of full-length HCV genome sequences characterized in China representing 11 subtypes and two novel variants. Infect Genet Evol 20, 225–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada N., Tanihara K., Takada A., Yorihuzi T., Tsutsumi M., Shimomura H., Tsuji T., Date T. (1996). Genetic organization and diversity of the 3′ noncoding region of the hepatitis C virus genome. Virology 223, 255–261 10.1006/viro.1996.0476 [DOI] [PubMed] [Google Scholar]
- Yuan M., Lu T., Li C., Lu L. (2013). The evolutionary rates of HCV estimated with subtype 1a and 1b sequences over the ORF length and in different genomic regions. PLoS ONE 8, e64698 10.1371/journal.pone.0064698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoulim F., Chevallier M., Maynard M., Trepo C. (2003). Clinical consequences of hepatitis C virus infection. Rev Med Virol 13,57–68 10.1002/rmv.371 [DOI] [PubMed] [Google Scholar]