Abstract
Dengue virus (DENV) infection in humans can cause flu-like illness, life-threatening haemorrhagic fever or even death. There is no specific anti-DENV therapeutic or approved vaccine currently available, partially due to the possibility of antibody-dependent enhancement reaction. Small interfering RNAs (siRNAs) that target specific viral genes are considered a promising therapeutic alternative against DENV infection. However, in vivo, siRNAs are vulnerable to degradation by serum nucleases and rapid renal excretion due to their small size and anionic character. To enhance siRNA delivery and stability, we complexed anti-DENV siRNAs with biocompatible gold nanoparticles (AuNPs) and tested them in vitro. We found that cationic AuNP–siRNA complexes could enter Vero cells and significantly reduce DENV serotype 2 (DENV-2) replication and infectious virion release under both pre- and post-infection conditions. In addition, RNase-treated AuNP–siRNA complexes could still inhibit DENV-2 replication, suggesting that AuNPs maintained siRNA stability. Collectively, these results demonstrated that AuNPs were able to efficiently deliver siRNAs and control infection in vitro, indicating a novel anti-DENV strategy.
Introduction
Dengue virus (DENV; family Flaviviridae) is a positive-sense ssRNA virus. DENV is transmitted to humans primarily by the mosquito vectors, Aedes aegypti and Aedes albopictus (Xi et al., 2008). After DENV binds to host cellular receptors, it is endocytosed and uncoated within the endosome to release its RNA genome into the cytoplasm, where replication of the viral genome and assembly of virions occur (van der Schaar et al., 2008). The common symptoms of DENV infection in humans are characterized by fever, headache, joint and muscle pain, and measle-like rashes (Halstead, 2007; Wilder-Smith et al., 2010). Although asymptomatic in most cases, DENV infection can lead to serious disease, such as haemorrhagic fever and systemic shock syndrome, that can potentially be fatal. There are four serotypes of DENV (DENV-1–4) and illness can be produced by any serotype (Tomashek & Margolis, 2011). Although recovery from infection by one serotype provides lifelong immunity against that particular serotype, subsequent infection by other co-circulating serotypes may cause an antibody-dependent enhancement reaction, which increases the risk of developing severe dengue haemorrhagic fever, shock and even death (Halstead, 1982; Kliks et al., 1989). DENV infection has been reported in >100 countries, including the United States (Gubler, 2002; Ramos et al., 2008; Tomashek & Margolis, 2011). A recent report has estimated that 390 million people are infected with DENV annually, with 96 million people manifesting clinical symptoms of disease (Bhatt et al., 2013).
Since antibody-dependent enhancement hinders the development of an antibody-based vaccine, the development of therapeutics that target replication or expression of the viral genome may serve as a promising alternative to control DENV infection. Anti-host receptor (Alhoot et al., 2011) and anti-DENV (Stein et al., 2011; Subramanya et al., 2010) small interfering RNAs (siRNAs) have considerably advanced both clinical- and laboratory-based applications as potential DENV therapeutic alternatives (Idrees & Ashfaq, 2013). However, efficient delivery of siRNAs remains a great challenge for clinical applications. The most common siRNA delivery approach is with a lipid-based transfection reagent, whereby the negatively charged siRNA duplex binds to the cationic liposome transfection reagent, which allows for cell membrane endocytosis, micelle formation and intracellular release of the siRNA into the cytosol (Zuhorn et al., 2002). Although this method is successful for siRNA delivery in vitro, it is not an ideal candidate to use in vivo, as these chemically based transfection reagents are physiologically toxic and could induce cellular stress or aberrant transcriptional profiles (Torchilin, 2005; Zhong et al., 2008).
Recently, gold nanoparticles (AuNPs) have been suggested as promising vehicles to deliver siRNA both in vitro and in vivo (Conde et al., 2012, 2013; Mitra et al., 2013; Yang et al., 2013). Several advantages make them well suited for safe and efficient protection and transport of siRNA: (i) biocompatibility (Chithrani & Chan, 2007; Lewinski et al., 2008), (ii) ease of preparation and facile surface modification (Elbakry et al., 2009; Lee et al., 2011; Song et al., 2010), and (iii) high siRNA loading capacity. In this study, we constructed novel anti-DENV AuNP–siRNA nanoplexes and found that AuNPs are able to promote siRNA stability and efficiently deliver anti-DENV siRNAs in vitro.
Results
Antiviral siRNAs efficiently inhibit DENV replication
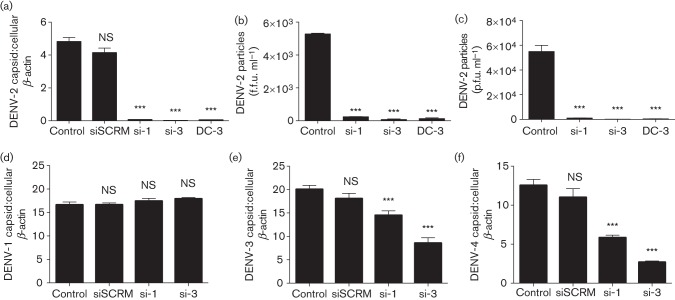
Two antiviral siRNAs (si-1 and si-3) were designed to target the most prevalent dengue serotype, DENV-2. Vero cells were transfected with siRNA (20 nM) using a Lipofectamine-based transfection reagent and subsequently infected with DENV-2 (m.o.i. 0.1). Sequence-scrambled siRNA (siSCRM) was used as a negative control and a previously described anti-DENV siRNA (DC-3) was used as a positive control (Stein et al., 2011). At 48 h post-infection, the cells were harvested for total RNA extraction, cDNA synthesis and subjected to real-time quantitative PCR (qPCR) for the measurement of DENV capsid gene and cellular β-actin gene expression. The qPCR results showed that si-1, si-3 and DC-3 significantly reduced DENV-2 capsid gene expression compared with the control, in which cells were treated with transfection reagent only (P<0.001, Fig. 1a). In contrast, siSCRM did not show any inhibitory effect (P = NS, Fig. 1a), suggesting that siRNA inhibition is sequence-specific. To test if infectious viral particles released from the infected cells were reduced with siRNA transfection, the media were quantified by a plaque-forming assay and an immunostaining assay, with the latter detecting viral envelope protein expression. As the DENV-2 genome shares some homology with other DENV serotypes, the antiviral effect of si-1 and si-3 (20 nM) was also evaluated against the replication of DENV-1, DENV-3 and DENV-4 (m.o.i. 0.1). qPCR analysis showed that si-1 and si-3 were able to inhibit DENV-3 and DENV-4 replication, but showed no inhibitory activity against DENV-1, compared with the control in which cells were treated with transfection reagent without siRNA (P<0.001 and P = NS, Fig. 1d–f). RNA sequence alignment analysis suggested that the lack of inhibition of si-1 and si-3 against DENV-1 replication might be due to lower homology between their targeting sequences (~50 and 70 %, respectively, data not shown). In summary, these results demonstrated that both si-1 and si-3 were able to significantly inhibit replication of DENV serotypes 2, 3 and 4.
Fig. 1.
Antiviral siRNAs inhibit replication of DENV-2, DENV-3 and DENV-4 serotypes. Vero cells were transfected with siRNAs (si-1 and si-3), siSCRM and a positive control siRNA (DC-3) using a Lipofectamine reagent for 48 h. Vero cells were then infected with DENV (m.o.i. 0.1) for an additional 48 h. (a) qPCR (mean unitless ratio of DENV-2 capsid : β-actin±sem) was performed 48 h post-DENV-2 infection. (b) Infectious DENV-2 viral particles (f.f.u. ml−1) in cell media were measured by an immunostaining assay. (c) Infectious DENV-2 viral particles (p.f.u. ml−1) in cell media were measured by a plaque assay. (d–f) qPCR assays were also performed for DENV-1 (d), DENV-3 (e) and DENV-4 (f) infection. All siRNA- and siSCRM-transfected samples were compared with DENV-infected controls with transfection reagent only (Control) and analysed using a two-tailed, Student’s t-test (n = 3; ***P<0.001; **P<0.005; NS, non-significant). The p.f.u. assay represents one experiment, and the qPCR and f.f.u. data represent three independent experiments.
Design and optimization of AuNP complexes for anti-DENV siRNA delivery in vitro
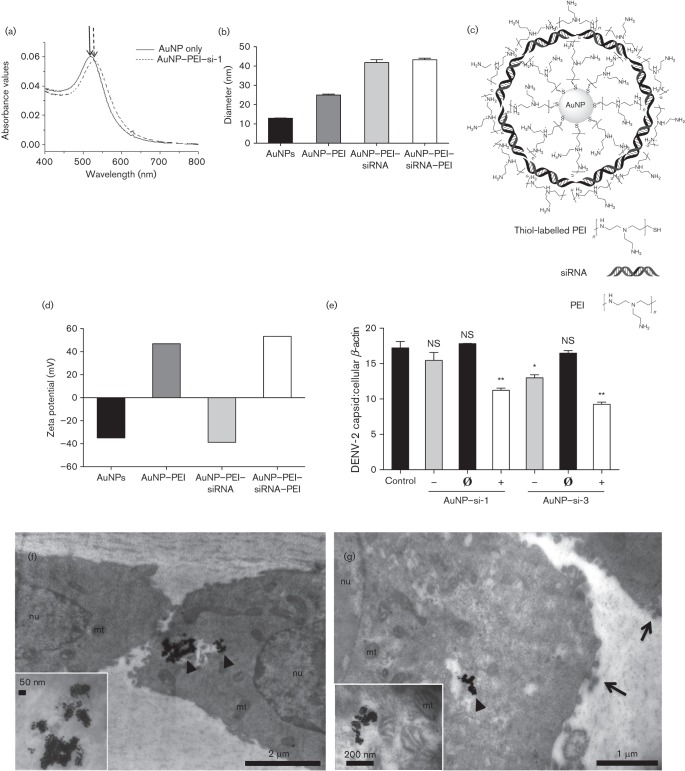
Recently, AuNPs have been designed and optimized as delivery vehicles for siRNA both in vitro and in vivo (Conde et al., 2013; Gilleron et al., 2013; Mitra et al., 2013; Yang et al., 2013; Zheng et al., 2012). Optimization of AuNPs has focused on certain properties that enhance siRNA delivery and reduce possible cytotoxicity. Such properties include surface charge, surface complexity and size of the AuNPs (de Jong & Borm, 2008). In this study, three AuNP–siRNA complexes (anionic, neutral and cationic) were prepared by a novel layer-by-layer (LbL) approach to deliver anti-DENV siRNA into Vero cells. To confirm each layer was successfully coated on the AuNPs, the AuNP complexes were characterized by UV-visible spectra, dynamic light scattering (DLS) and zeta potential. AuNPs and AuNP–polyethylenimine (PEI)–siRNAs were measured by UV-visible spectra analysis, which disclosed a shift in the surface plasmon resonance peaks with the addition of siRNA (Fig. 2a). DLS measured the hydrodynamic diameters of the different complexes: AuNPs (12.92 nm), AuNP–PEI (24.97 nm), AuNP–PEI–siRNA (41.79 nm) and AuNP–PEI–siRNA–PEI (43.25 nm), which indicated that the diameters increased with the addition of each layer (Fig. 2b). AuNPs were first coated with an inner layer of HS–PEI (thiol-labelled PEI) via gold–sulfur bonds, rendering the particles cationic. The negatively charged siRNAs were then coated on the AuNP–PEIs through electrostatic interactions, rendering the complexes anionic. Adjusting the concentration of the outer layer PEI with the AuNP–PEI–siRNA complexes (Fig. 2c) altered the overall charge from negative to positive. The zeta potential determined the surface charges of the complexes: AuNPs (−34.9 mV), AuNP–PEI (+46.9 mV), AuNP–PEI–siRNA (−38.8 mV) and AuNP–PEI–siRNA–PEI (+53.3 mV) (Fig. 2d, neutral charge data not shown). These results demonstrated the optimization of AuNPs and successful synthesis of AuNP–siRNA complexes.
Fig. 2.
Characterization of AuNP-conjugated siRNA complexes. AuNPs were prepared by a new LbL approach and were characterized by UV-visible spectrometry, DLS and zeta potential. (a) Absorbance peaks of the AuNP only (λmax = 512 nm, solid arrow) and the AuNP–PEI-si-1 complex (λmax = 525 nm, dashed arrow). (b) The diameters of the constructed AuNP complexes were measured using DLS (nm). (c) Schematic structure of an AuNP–siRNA complex. (d) The zeta potential (mV) of the constructed AuNP complexes. (e) AuNP–siRNA complexes (20 nM siRNA) with different charges (‘–’, −40 mV; ‘Ø’, 0 mV; ‘+’, +40 mV) were cultured with Vero cells for 48 h and subsequently infected with DENV-2 (m.o.i. 0.1) for an additional 48 h. qPCR analysis (mean unitless ratio of DENV-2 capsid : β-actin±sem) was performed to measure viral RNA replication 48 h post-infection. Control samples were with DENV-2 infection only (Control). (f, g) Cationic AuNP–si-1 complexes (80 nM siRNA) were cultured with Vero cells for 48 h and subsequently infected with DENV-2 (m.o.i. 0.1) for an additional 72 h. TEM was used to visualize AuNP–si-1 cellular localization (arrowheads, AuNP–siRNA complexes; mt, mitochondria; nu, nucleus; arrows, budding DENV-2). All qPCR data were analysed with a two-tailed Student’s t-test (n = 3, * P<0.05, **P<0.005). The qPCR data represent two independent experiments.
To test the effect of surface charge on siRNA delivery efficiency, we treated Vero cells with anionic (−40 mV), neutral (0 mV) and cationic (+40 mV) AuNP–siRNA complexes (20 nM) for 48 h, and subsequently infected them with DENV-2 (m.o.i. 0.1) for an additional 48 h. qPCR analysis showed that cationic AuNP–si-1 and AuNP–si-3 complexes significantly reduced DENV-2 replication (P<0.005, Fig. 2e), whilst anionic AuNP–si-3, but not AuNP–si-1 complexes, also inhibited DENV-2 replication compared with the control, in which cells were infected with DENV-2 only (P<0.05, Fig. 2e). In contrast, neither the neutrally charged AuNP–si-1 nor the AuNP–si-3 complex inhibited viral replication, suggesting inefficient delivery of siRNAs by neutral AuNP–siRNA complexes (Fig. 2e). In summary, these results showed that cationic AuNP–siRNA complexes could effectively deliver anti-DENV siRNAs into cells.
To confirm that cationic AuNP–siRNAs were able to enter cells, Vero cells were cultured with cationic AuNP–si-1 (80 nM) for 48 h and subsequently infected with DENV-2 (m.o.i. 0.1) for an additional 72 h. Ultra-thin sections embedded in epoxy resin were imaged by transmission electron microscopy (TEM). The TEM images showed that AuNPs localized within the cytoplasm of Vero cells (Fig. 2f, g, inserts), which implied AuNP complexes were able to deliver siRNAs into the cells.
Cationic AuNP–siRNA complexes are non-toxic to Vero cells
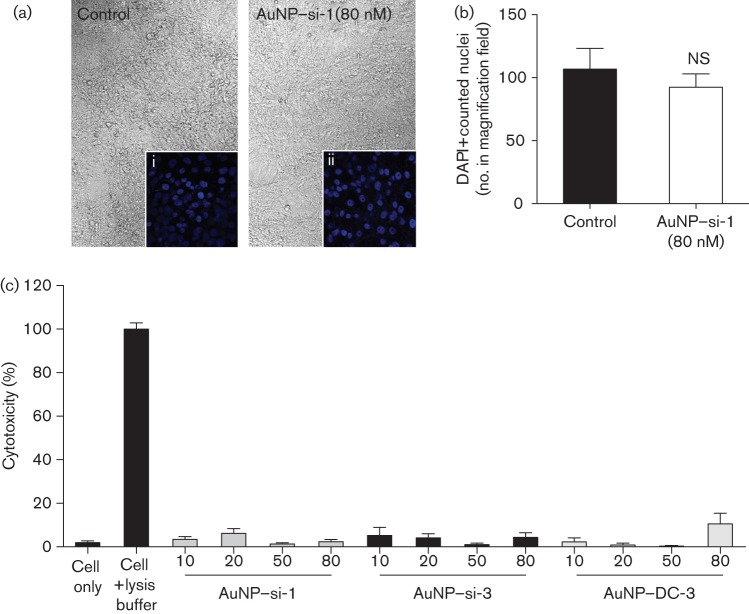
AuNPs have been noted as biocompatible vehicles for drug delivery (Connor et al., 2005; Shukla et al., 2005). However, they have also been suggested to have some cytotoxicity due to their cationic polarity and surface modifications (Gerber et al., 2013; Goodman et al., 2004). To exclude the possibility that inhibition of DENV-2 replication was due to the AuNP–siRNA complexes causing Vero cell death, we assessed the cytotoxicity of the AuNP–siRNA complexes when exposed to Vero cells. We incubated AuNP–siRNA complexes (+40 mV) with Vero cells under the same experimental conditions and concentrations that were used for the pre-treatment transfection assays, without DENV-2 infection. Cellular morphology was imaged, and cell number was quantified by using DAPI nuclear staining and examined under a confocal microscope. No morphological changes (Fig. 3a, i and ii) or cell number reduction (Fig. 3b) were observed with any AuNP–siRNA-treated samples compared with the untreated control. To further confirm this, cellular lactate dehydrogenase (LDH) released from broken cell membranes was also measured. The LDH cytotoxicity assay results showed no significant toxicity (≤6.5 % cytotoxicity), which was similar to the untreated cell control (2 % cytotoxicity, Fig. 3c). These data suggested that Vero cells exposed to AuNP–siRNA complexes had negligible cytotoxicity, which indicated that inhibition of DENV-2 replication was due to siRNA-mediated RNA interference.
Fig. 3.
Cationic AuNP–siRNA complexes are not toxic to Vero cells. Vero cells were cultured with or without AuNP–si-1 complexes (80 nM) under similar experimental conditions as for the pre-treatment assays. (a) Cell images were taken under a confocal microscope using differential interference contrast imaging (×40). DAPI-positive nuclear staining imaged under confocal microscopy: (i) control cells without AuNP–siRNA complexes (control) and (ii) cells with AuNP–si-1 complexes (×400). (b) DAPI-positive cell number was counted in the magnification fields in (a). (c) LDH released from lysed cells measured and reported as per cent cytotoxicity. All AuNP–siRNA complex-containing samples in the LDH assay were compared with cells without AuNP–siRNA complexes (Cell only) and analysed with a Bonferroni post hoc analysis (n = 3; NS, non-significant). The data in (b) represent three independent experiments and the data in (c) represent two independent experiments.
Cationic AuNP–siRNA complexes inhibit DENV-2 infection under both pre- and post-infection conditions
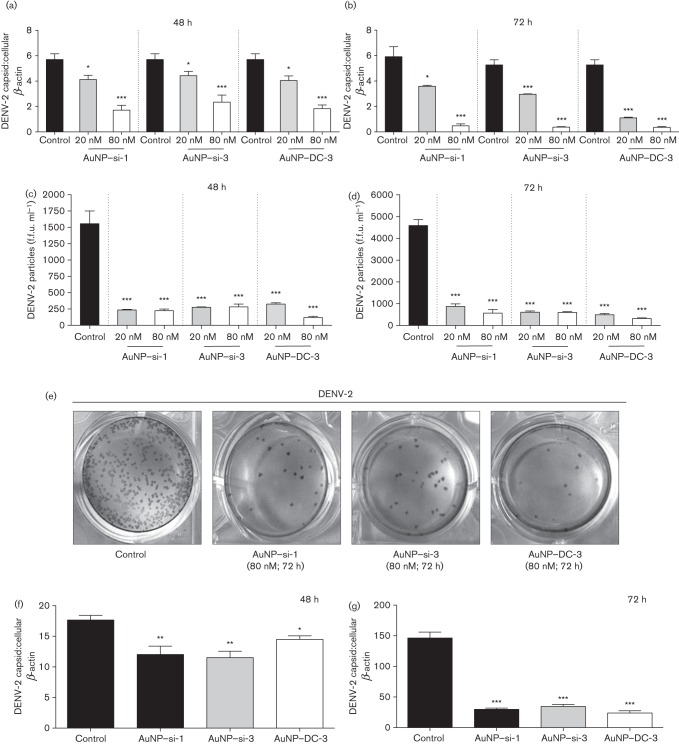
To further confirm that cationic AuNP–siRNA complexes inhibited DENV-2 replication, we pre-treated Vero cells with cationic AuNP–siRNAs (+40 mV) at 20 and 80 nM for 48 h, and subsequently infected them with DENV-2 (m.o.i. 0.1) for an additional 48 and 72 h, followed by qPCR analysis. The results showed that compared with DENV-2-only-infected controls, AuNP–si-1 (20 nM) reduced DENV replication approximately twofold at both 48 and 72 h (P<0.05, Fig. 4a, b) and AuNP–si-3 (20 nM) reduced DENV-2 replication approximately two- and three-fold at 48 and 72 h, respectively (P<0.05 and 0.001, Fig. 4a, b). In addition, cells pre-treated with either AuNP–si-1 or AuNP–si-3 at 80 nM further reduced DENV-2 replication, both approximately threefold at 48 h and sixfold at 72 h, respectively (P<0.001, Fig. 4a, b). Similar results were observed with AuNP–DC-3, whereby DENV-2 replication was reduced approximately two- and fivefold with 20 nM, and three- and fifteen-fold with 80 nM AuNP–siRNA complexes, at 48 h and 72 h, respectively (P<0.05 and 0.001, Fig. 4a, b). To test if AuNP–siRNA pre-treatment could inhibit production of infectious DENV particles, the viral-containing media were measured at 48 and 72 h by an immunostaining assay. AuNP–siRNAs were able to successfully reduce the DENV-2 titre from ~1500 (control) to <350 f.f.u. ml−1 (AuNP–siRNA pre-treated samples) at 48 h and ~4500 (control) to <900 f.f.u. ml−1 (AuNP–siRNA pre-treated samples) at 72 h (P<0.001, Fig. 4c–e). siSCRM-treated samples showed no difference compared with the control (data not shown). These results confirmed that cationic AuNP–si-1, AuNP–si-3 and AuNP–DC-3 pre-treatment inhibited DENV-2 replication.
Fig. 4.
Cationic AuNP–siRNA complexes inhibit DENV-2 replication and viral propagation in vitro. Cationic AuNP–siRNA complexes (20 and 80 nM siRNA) were added to Vero cell culture for 48 h and infected with DENV-2 (m.o.i. 0.1) for an additional 48 or 72 h. After infection, qPCR (mean unitless ratio of DENV-2 capsid : β-actin±sem) was performed to measure viral replication at (a) 48 and (b) 72 h. Infectious DENV-2-containing media collected at (c) 48 and (d) 72 h post-infection were measured by an immunostaining assay. (e) Visual representation of immunopositive foci. Vero cells were infected with DENV-2 (m.o.i. 0.1) for 6 h and AuNP–siRNAs (80 nM) were added for an additional 48 or 72 h. qPCR was performed at (f) 48 and (g) 72 h post-treatment. Data were compared with DENV-2-infected samples only (Control) with a two-tailed Student’s t-test (n = 3; *P<0.05; **P<0.005; ***P<0.001; NS, non-significant). The data represent three independent experiments.
We next asked if AuNP–siRNA complexes could still inhibit viral replication when the cells were infected with DENV prior to AuNP–siRNA treatment. To test this, we infected Vero cells with DENV-2 (m.o.i. 0.1) for 6 h at 37 °C, 5 % CO2, replaced the viral-containing media with fresh medium containing AuNP–si-1, AuNP–si-3 or AuNP–DC-3 at 80 nM and cultured the cells for an additional 48 or 72 h, followed by qPCR analysis. The qPCR results showed that all three AuNP–siRNAs significantly reduced DENV-2 replication at both 48 and 72 h post-infection compared with controls, without AuNP–siRNA treatment (Fig. 4f, g).
AuNPs protect siRNAs from enzymic degradation
AuNPs have been suggested to protect siRNAs from degradation by intracellular or extracellular RNase enzymes (Mitra et al., 2013). To test this, cationic AuNP–si-3 complexes (300 pmol siRNA) and naked si-3 (300 pmol) were treated with Riboshredder RNase blend (0.1 U/500 µl; Epicenter Biotechnologies) and the absorbance was monitored with an Agilent 8453 UV-Vis spectrophotometer. The naked si-3 control exhibited a rapid increase in the absorbance percentage after RNase treatment, which suggested rapid degradation of the naked si-3 (Fig. 5a). In contrast, the AuNP–si-3 sample did not increase the absorbance percentage within the same time period of RNase treatment, which indicated that the AuNP–si-3 complexes remained intact (Fig. 5a). To test if the RNase-treated AuNP–si-3 retained functionality to inhibit DENV-2 replication, we cultured Vero cells with the RNase-treated AuNP–si-3 (80 nM) for 48 h and infected them with DENV-2 (m.o.i. 0.1) for an additional 72 h, followed by qPCR analysis. The same volume of RNase-treated water served as a control. The qPCR results showed that RNase-treated AuNP–si-3 complexes still could inhibit DENV-2 replication compared with the control (P<0.001, Fig. 5b), which suggested that AuNPs maintained siRNA stability and retained siRNA functionality even after rigorous RNase treatment.
Fig. 5.
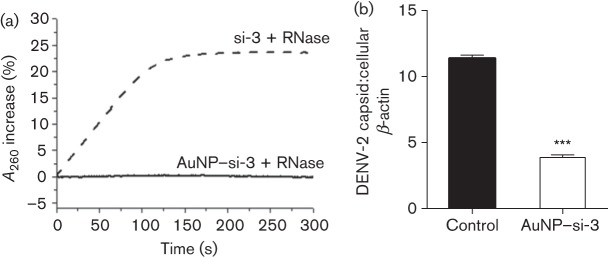
AuNP–siRNA complexes are resistant to nuclease degradation. AuNP–si-3 complexes (80 nM) were pre-treated with Riboshredder RNase blend and the change in absorbance at 260 nm was monitored over a timescale of 0–300 s. (a) Degradation of naked si-3 (dashed line) and AuNP–si-3 (solid line) with Riboshredder RNase blend (0.1 U/500 µl). RNase-treated AuNP–si-3 was cultured with Vero cells for 48 h and infected with DENV-2 (m.o.i. 0.1) for an additional 72 h. (b) qPCR (mean unitless ratio of DENV-2 capsid : β-actin±sem) was performed to measure DENV-2 replication at 72 h post-infection. qPCR data were compared with RNase-treated water only controls (Control) and analysed with a two-tailed Student’s t-test (n = 3; ***P<0.001). The data represent two independent experiments.
Discussion
AuNPs and their gold nanorod counterparts have been implicated as biocompatible carriers for siRNAs as novel treatment options in cancer (Mitra et al., 2013) and host gene regulation during human immunodeficiency virus infection (Reynolds et al., 2012). However, to the best of our knowledge, using AuNPs to deliver antiviral siRNAs to inhibit viral replication has not yet been investigated. In this study, we used DENV as a viral model to evaluate the feasibility of using AuNPs to deliver anti-DENV siRNA. We designed AuNP complexes that were encapsulated in a dual layer of the cationic polymer PEI, which has been suggested to enhance the conjugation of siRNAs to AuNPs and the binding/entry across negatively charged cell membranes (Thomas & Klibanov, 2003). In addition, the outer layer of PEI has been previously reported to act as a ‘proton sponge’, whereby it can buffer acidic compartments upon endosomal/lysosomal trafficking to effectively protect siRNA from intracellular degradation (Boussif et al., 1995; Zhou et al., 2013). This dual-layer modification makes these AuNP complexes an attractive candidate for siRNA transfection that differs from classical AuNP delivery vehicles for siRNA (Richards Grayson et al., 2006; Thomas & Klibanov, 2003; Urban-Klein et al., 2005). Our results demonstrated that the cationic AuNP–siRNA complexes were able to efficiently deliver siRNAs into cells and significantly inhibit DENV-2 replication when transfected prior to and after viral infection. The TEM images also provided visual evidence that cationic AuNP–siRNA complexes entered Vero cell cytoplasm, where DENV replicates its RNA genome and translates viral polyproteins. Although we did not investigate the cellular entry mechanisms of AuNP–siRNA complexes in this study, cationic AuNP–siRNA complexes may readily bind to the negatively charged cell membranes, which is an essential step in cellular uptake. As the AuNP–siRNA complexes are held together by electrostatic interactions, the siRNA can dissociate/release from the AuNPs upon cellular entry by factors that weaken their ionic interactions. It has been reported previously that low-molecular-mass salts can cause the dissociation of nanoplexes within cells due to electrostatic interactions (Zintchenko et al., 2008). Moreover, anionic components (such as negatively charged proteins) may compete with RNA binding to PEI, which could unpack AuNPs and release siRNA.
As lipid-based siRNA transfection reagents produce a high transfection efficiency, they are most commonly used in research; however, they also have some significant drawbacks. For example, lipid-based transfection reagents with siRNA complexes are unstable and need to be used immediately before transfection, which limits the use of these reagents as optimal therapeutics in the clinical setting (de Fougerolles, 2008; Gao & Huang, 2009). In addition, a high dose of the delivery vehicles in vivo (i.e. Lipofectamine-based) could lead to the induction of the cellular stress response or cell death (Breunig et al., 2007; Zhong et al., 2008). Furthermore, lipid-based transfection reagents that have been developed for in vivo studies, such as Invivofectamine 2.0 (Life Technologies), selectively target liver cells (Wang et al., 2013). However, DENV infection is not limited to the liver, but is systemic and ubiquitous during the infection process (Dalrymple & Mackow, 2011; Pham et al., 2012; Puccioni-Sohler et al., 2012), therefore a delivery vehicle that targets DENV-infected cells specifically would be an optimal therapeutic for siRNA delivery. In theory, AuNP–siRNA complexes would non-specifically target/enter various cell types in vivo. However, the outer layer of PEI can be tailored with various cell-targeting molecules or peptides that can specifically recognize DENV-infected cells. Future directions for improvement include the addition of a synthetic, dendritic cell-targeting peptide (Subramanya et al., 2010; Ye et al., 2011) that can direct the anti-DENV AuNP–siRNA complexes into dendritic cells, as DENV replicates efficiently within these cell types (Wu et al., 2000).
Although AuNPs have been suggested as biocompatible reagents, AuNPs can be cytotoxic in certain configurations, depending on size, surface charge and chemical complexity (de Jong & Borm, 2008; Lewinski et al., 2008). Previous reports suggested that cationic AuNPs were somewhat more cytotoxic than anionic AuNPs, but possessed better transfection ability by increased interaction with the cell membrane (Goodman et al., 2004; Thomas & Klibanov, 2003). However, the LDH cytotoxicity assay performed in this study showed that the cationic AuNP–siRNA complexes are not cytotoxic to Vero cells. Importantly, the AuNP–siRNA complexes can protect siRNAs from RNase digestion, suggesting AuNPs may prolong siRNA activity when used in vivo. Therefore, PEI–siRNA complexes conjugated to AuNPs may offer a possible solution to protect siRNAs from degradation when treating infection in vivo. In addition, a number of reports have introduced the use of AuNPs (Goel et al., 2007, 2009) and AuNP–siRNAs (Conde et al., 2013; Zheng et al., 2012) in vivo, showing promising results. More importantly, AuNPs have been implicated in a phase I clinical trial for cancer treatment with excellent prognosis (Libutti et al., 2010). However promising, variable surface modifications and size of AuNPs may play a prominent role in causing toxicity in vivo (Gerber et al., 2013; Zhang et al., 2011), and therefore additional testing of our complexes in vivo is required.
There are many questions that need to be answered before using AuNPs as delivery vehicles for siRNAs in clinical trials. We have noticed that the siRNA delivery efficiency of AuNPs is not as high as the lipid-based transfection reagent. We believe that improvements can be made through charge optimization and AuNP–siRNA complex outer layer modifications. In addition, specifying certain AuNP features, such as physiological dosage, biodistribution and fate of AuNPs in vivo, is warranted for further investigation. Nevertheless, this study provides several lines of evidence that cationic AuNPs can be used to deliver siRNAs against DENV infection in vitro, which can be further developed as novel therapeutics against DENV and other viral or non-viral diseases.
Methods
Viruses, cell culture and siRNA design.
DENV-1 (TUP-2180), DENV-3 (TUP-2342) and DENV-4 (TUP-2174) isolates were kindly provided by Dr John F. Anderson, and DENV-2 (VR-1584) was purchased from the ATCC. All virus strains were propagated one time in mosquito C6/36 cells (ATCC CRL-1660) and titrated using a Vero cell plaque assay as described previously (Bai et al., 2005). C6/36 cells were maintained at 28 °C, 5 % CO2 in Eagle’s minimum essential medium containing 10 % FBS. Vero cells (ATCC CCL-81) were maintained in Dulbecco’s modified Eagle’s medium (DMEM) containing 1 % l-glutamine, 1 % penicillin/streptomycin and 10 % FBS.
siRNAs were designed against DENV-2 (New Guinea strain C; GenBank accession number M29095.1) by using the Thermo Scientific siRNA designing tool (siDESIGN Center). si-1 that targeted non-structural region 1 (NS1), 5′-UGGCUAAAGUUGAGAGAAA-3′; si-3 that targeted non-structural region 5 (NS5), 5′-CCAAAGAGGUAGUGGACAA-3′; a si-3 scrambled control (siSCRM), 5′-GAUGGUACGACUAACAAGUGA-3′; and DC-3 that targeted the capsid region of DENV-2 (Stein et al., 2011) were synthesized either by Applied Biosystems or Integrated DNA Technologies. The siSCRM sequence was designed using InvivoGen’s Scramble siRNA Generator.
Preparation of AuNP–PEI–siRNA–PEI complexes.
AuNPs were synthesized by the reduction of chloroauric acid with subsequent addition of citrate as described previously (Huang & Shi, 2010; Storhoff et al., 1998). The preparation of AuNP–PEI–siRNA–PEI complexes followed the reported procedure (Elbakry et al., 2009) with significant modifications. HS–PEI (1.8 kDa) was synthesized by the conjugation of 12-mercaptododecanoic acid NHS ester (Sigma) with PEI (1 : 1 ratio). The HS–PEI was used to coat AuNPs to form AuNP–PEI complexes. siRNAs were conjugated to the AuNP–PEI (AuNP : siRNA = 1 : ~300) and an additional layer of PEI was added to make AuNP–PEI–siRNA–PEI complexes. The UV-visible spectra of the various AuNP complexes were measured using an Agilent 8453 UV-Vis spectrophotometer (Agilent Technologies), and DLS and zeta potential was measured by the Zetasizer Nano Range ZS instrument (Malvern).
siRNA transfection and AuNP–siRNA treatment.
siRNAs were transfected into Vero cells (8×104 cells per well in 24-well plates) with RNAiMAX Lipofectamine reagent (1 µl per well; Life Technologies) in OPTI-MEM medium (100 µl) for 30 min. DMEM containing 2 % l-glutamine and 2 % FBS was then added (500 µl per well) for 48 h at 37 °C, 5 % CO2. Following incubation, media was removed and cells were infected with DENV-1–4 (m.o.i. 0.1) for 1 h at room temperature. Viral-containing media was removed, and fresh medium (DMEM containing 1 % penicillin/streptomycin and 10 % FBS) was added and cultured for an additional 48 h.
AuNP–siRNAs were either added to Vero cells before or after DENV-2 infection. For pre-treatment assays, Vero cells were cultured with AuNP–siRNA complexes for 48 h, then infected with DENV-2 (m.o.i. 0.1) for 1 h at room temperature. After changing the media, cells were cultured for an additional 48 or 72 h. For post-treatment assays, Vero cells were infected with DENV-2 (m.o.i. 0.1) for 6 h at 37 °C, 5 % CO2, unattached virus was washed off, and fresh medium was added and cultured for an additional 48 or 72 h. Cell culture supernatants were collected at the respective time points and stored at −70 °C until ready to be tested.
qPCR assay.
Following DENV infection, Vero cells were collected for total RNA extraction with TRI Reagent (Molecular Research Center) and converted into cDNA using the iSCRIPT cDNA synthesis kit (Bio-Rad). qPCR assays were performed using iTAQ polymerase supermix for probe-based assays (Bio-Rad). DENV capsid gene and Vero cellular β-actin gene primer and probe sequences were adapted according to previous publications: DENV-1 (Sadon et al., 2008), DENV-2 (Mota & Rico-Hesse, 2009), DENV-3 and DENV-4 (Bai et al., 2008), and β-actin (Bai et al., 2005). All primers and probes were synthesized by Integrated DNA Technologies or Applied Biosystems.
Plaque-forming assays and immunostaining assays.
Plaque-forming assays were performed as described previously (Bai et al., 2005). Briefly, Vero cells were plated at 1×106 cells ml−1 and grown to 90–95 % confluence in six-well plates. DENV-2-containing media were added to the monolayer and incubated at room temperature for 1 h at 37 °C. The media were removed and cells were overlaid with 2 ml 1 % SeaPlaque agarose (Lonza) in PBS and incubated for 5–7 days. Following incubation, cells were stained with 4 % (v/v) Neutral Red (Sigma) and plaques were counted. Virus titre was recorded as p.f.u. ml−1.
In the immunostaining assays, Vero cells were plated to 90–95 % confluence, and DENV-2-containing media were added to the monolayer and incubated at room temperature for 1 h. The media were removed and cells were overlaid with fresh medium (OptiMEM-Glutamax containing 2 % FBS and 1 % methylcellulose; Sigma) and incubated for an additional 4 days. Overlay was removed and cells were washed with PBS then fixed with 4 % paraformaldehyde (PFA) in PBS for 15 min at room temperature. Cells were washed twice and blocked with 2 % normal goat serum (Life Technologies) with 0.4 % Triton X-100 for 1 h at room temperature and probed with monoclonal mouse anti-flavivirus glycoprotein E IgG antibody (4G2; 1 : 100) overnight at 4 °C (ATCC D1-4G2-4-15 HB-112). The cells were then washed and polyclonal goat anti-mouse HRP IgG (1 : 1000; KPL) was added for 2 h at room temperature. Immuno-positive cell foci were developed with TrueBlue peroxidase substrate (KPL) and counted using an Axiostar Plus light microscope (Zeiss) as reported previously (Bonafé et al., 2009). Virus titre was recorded as f.f.u. ml−1.
Confocal microscopy and TEM.
Vero cells (8×104 cells per well in 24-well plates) were cultured with or without AuNP–si-1 at 80 nM and DENV-2 (m.o.i. 0.1) as in pre-treatment assays. Following 72 h incubation, media were removed, and cells were washed with PBS and fixed in 4 % PFA in PBS for 15 min. Cells were mounted with Vectashield mounting medium containing DAPI (Vector Laboratories) to quantify cell number, and imaged by differential interference contrast and fluorescence with a confocal LSM 510 Microscope (Zeiss).
Vero cells were cultured with or without AuNP–si-1 at 80 nM and DENV-2 (m.o.i. 0.1) as in pre-treatment assays. Following 72 h incubation, cells were washed, fixed and embedded in epoxy resin ERL 4221 (Spurr, 1969). Ultra-thin (100 nm) serial sections were obtained on an Ultra-microtome MT-2B (Sorvall Porter-Blum). Sections were collected on Formvar-coated (5 % polyvinyl formal resin) copper grids and post-stained for 20 min in lead citrate (Sigma) followed by 20 min in 2 % aqueous uranyl acetate (Sigma). Using a Zeiss 900 transmission electron microscope, digital images were taken with a Gatan 785 ES1000W Erlangshen CCD camera.
LDH cytotoxicity assay.
Vero cells (1×104 cells per well in 96-well ELISA plates) were incubated with AuNP–siRNAs at serially diluted concentrations (10–80 nM siRNA = 34–277 pM AuNPs) for 48 h at 37 °C, 5 % CO2. The media were replaced with DMEM containing 10 % FBS and 1 % penicillin/streptomycin, followed by an additional 72 h incubation. After incubation, an LDH cytotoxicity assay was performed using a LDH Cytotoxicity Detection KitPLUS (Roche Diagnostics) and analysed using an ELx808 ultramicroplate reader (BioTek) at 450 nm. Per cent cytotoxicity was calculated according to the user’s manual.
Statistical analyses.
Data were compared with either a Student’s t-test or a two-way ANOVA and Bonferroni post hoc analysis. All statistical analyses were done by using GraphPad Prism software (version 6.0).
Acknowledgements
We thank Ms Baobin Kang for assistance with confocal microscopy and Dr Kenneth Curry for assistance with sample preparation for TEM. This work was supported in part by the University of Southern Mississippi New Faculty Start-up Funding (F. B.), the Mississippi INBRE is funded by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health (grant P20GM103476) and by a National Institutes of Health grant (R15CA152822) to F. H.
Reference
- Alhoot M. A., Wang S. M., Sekaran S. D. (2011). Inhibition of dengue virus entry and multiplication into monocytes using RNA interference. PLoS Negl Trop Dis 5, e1410. 10.1371/journal.pntd.0001410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai F., Wang T., Pal U., Bao F., Gould L. H., Fikrig E. (2005). Use of RNA interference to prevent lethal murine west nile virus infection. J Infect Dis 191, 1148–1154 10.1086/428507 [DOI] [PubMed] [Google Scholar]
- Bai Z., Liu L., Tu Z., Yao L., Liu J., Xu B., Tang B., Liu J., Wan Y. & other authors (2008). Real-time PCR for detecting circulating dengue virus in the Guangdong Province of China in 2006. J Med Microbiol 57, 1547–1552 10.1099/jmm.0.2008/003418-0 [DOI] [PubMed] [Google Scholar]
- Bhatt S., Gething P. W., Brady O. J., Messina J. P., Farlow A. W., Moyes C. L., Drake J. M., Brownstein J. S., Hoen A. G. & other authors (2013). The global distribution and burden of dengue. Nature 496, 504–507 10.1038/nature12060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonafé N., Rininger J. A., Chubet R. G., Foellmer H. G., Fader S., Anderson J. F., Bushmich S. L., Anthony K., Ledizet M. & other authors (2009). A recombinant West Nile virus envelope protein vaccine candidate produced in Spodoptera frugiperda expresSF+ cells. Vaccine 27, 213–222 10.1016/j.vaccine.2008.10.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boussif O., Lezoualc’h F., Zanta M. A., Mergny M. D., Scherman D., Demeneix B., Behr J. P. (1995). A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc Natl Acad Sci U S A 92, 7297–7301 10.1073/pnas.92.16.7297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breunig M., Lungwitz U., Liebl R., Goepferich A. (2007). Breaking up the correlation between efficacy and toxicity for nonviral gene delivery. Proc Natl Acad Sci U S A 104, 14454–14459 10.1073/pnas.0703882104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chithrani B. D., Chan W. C. (2007). Elucidating the mechanism of cellular uptake and removal of protein-coated gold nanoparticles of different sizes and shapes. Nano Lett 7, 1542–1550 10.1021/nl070363y [DOI] [PubMed] [Google Scholar]
- Conde J., Ambrosone A., Sanz V., Hernandez Y., Marchesano V., Tian F., Child H., Berry C. C., Ibarra M. R. & other authors (2012). Design of multifunctional gold nanoparticles for in vitro and in vivo gene silencing. ACS Nano 6, 8316–8324 10.1021/nn3030223 [DOI] [PubMed] [Google Scholar]
- Conde J., Tian F., Hernández Y., Bao C., Cui D., Janssen K. P., Ibarra M. R., Baptista P. V., Stoeger T., de la Fuente J. M. (2013). In vivo tumor targeting via nanoparticle-mediated therapeutic siRNA coupled to inflammatory response in lung cancer mouse models. Biomaterials 34, 7744–7753 10.1016/j.biomaterials.2013.06.041 [DOI] [PubMed] [Google Scholar]
- Connor E. E., Mwamuka J., Gole A., Murphy C. J., Wyatt M. D. (2005). Gold nanoparticles are taken up by human cells but do not cause acute cytotoxicity. Small 1, 325–327 10.1002/smll.200400093 [DOI] [PubMed] [Google Scholar]
- Dalrymple N., Mackow E. R. (2011). Productive dengue virus infection of human endothelial cells is directed by heparan sulfate-containing proteoglycan receptors. J Virol 85, 9478–9485 10.1128/JVI.05008-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Fougerolles A. R. (2008). Delivery vehicles for small interfering RNA in vivo. Hum Gene Ther 19, 125–132 10.1089/hum.2008.928 [DOI] [PubMed] [Google Scholar]
- de Jong W. H., Borm P. J. (2008). Drug delivery and nanoparticles:applications and hazards. Int J Nanomedicine 3, 133–149 10.2147/IJN.S596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbakry A., Zaky A., Liebl R., Rachel R., Goepferich A., Breunig M. (2009). Layer-by-layer assembled gold nanoparticles for siRNA delivery. Nano Lett 9, 2059–2064 10.1021/nl9003865 [DOI] [PubMed] [Google Scholar]
- Gao K., Huang L. (2009). Nonviral methods for siRNA delivery. Mol Pharm 6, 651–658 10.1021/mp800134q [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber A., Bundschuh M., Klingelhofer D., Groneberg D. A. (2013). Gold nanoparticles: recent aspects for human toxicology. J Occup Med Toxicol 8, 32. 10.1186/1745-6673-8-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilleron J., Querbes W., Zeigerer A., Borodovsky A., Marsico G., Schubert U., Manygoats K., Seifert S., Andree C. & other authors (2013). Image-based analysis of lipid nanoparticle-mediated siRNA delivery, intracellular trafficking and endosomal escape. Nat Biotechnol 31, 638–646 10.1038/nbt.2612 [DOI] [PubMed] [Google Scholar]
- Goel R., Swanlund D., Coad J., Paciotti G. F., Bischof J. C. (2007). TNF-alpha-based accentuation in cryoinjury–dose, delivery, and response. Mol Cancer Ther 6, 2039–2047 10.1158/1535-7163.MCT-06-0676 [DOI] [PubMed] [Google Scholar]
- Goel R., Shah N., Visaria R., Paciotti G. F., Bischof J. C. (2009). Biodistribution of TNF-alpha-coated gold nanoparticles in an in vivo model system. Nanomedicine (Lond) 4, 401–410 10.2217/nnm.09.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman C. M., McCusker C. D., Yilmaz T., Rotello V. M. (2004). Toxicity of gold nanoparticles functionalized with cationic and anionic side chains. Bioconjug Chem 15, 897–900 10.1021/bc049951i [DOI] [PubMed] [Google Scholar]
- Gubler D. J. (2002). The global emergence/resurgence of arboviral diseases as public health problems. Arch Med Res 33, 330–342 10.1016/S0188-4409(02)00378-8 [DOI] [PubMed] [Google Scholar]
- Halstead S. B. (1982). Immune enhancement of viral infection. Prog Allergy 31, 301–364 [PubMed] [Google Scholar]
- Halstead S. B. (2007). Dengue. Lancet 370, 1644–1652 10.1016/S0140-6736(07)61687-0 [DOI] [PubMed] [Google Scholar]
- Huang F., Shi Y. (2010). Synthesis of symmetrical thiol-adenosine conjugate and 5′ thiol-RNA preparation by efficient one-step transcription. Bioorg Med Chem Lett 20, 6254–6257 10.1016/j.bmcl.2010.08.097 [DOI] [PubMed] [Google Scholar]
- Idrees S., Ashfaq U. A. (2013). RNAi: antiviral therapy against dengue virus. Asian Pac J Trop Biomed 3, 232–236 10.1016/S2221-1691(13)60057-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliks S. C., Nisalak A., Brandt W. E., Wahl L., Burke D. S. (1989). Antibody-dependent enhancement of dengue virus growth in human monocytes as a risk factor for dengue hemorrhagic fever. Am J Trop Med Hyg 40, 444–451 [DOI] [PubMed] [Google Scholar]
- Lee M. Y., Park S. J., Park K., Kim K. S., Lee H., Hahn S. K. (2011). Target-specific gene silencing of layer-by-layer assembled gold-cysteamine/siRNA/PEI/HA nanocomplex. ACS Nano 5, 6138–6147 10.1021/nn2017793 [DOI] [PubMed] [Google Scholar]
- Lewinski N., Colvin V., Drezek R. (2008). Cytotoxicity of nanoparticles. Small 4, 26–49 10.1002/smll.200700595 [DOI] [PubMed] [Google Scholar]
- Libutti S. K., Paciotti G. F., Byrnes A. A., Alexander H. R., Jr, Gannon W. E., Walker M., Seidel G. D., Yuldasheva N., Tamarkin L. (2010). Phase I and pharmacokinetic studies of CYT-6091, a novel PEGylated colloidal gold-rhTNF nanomedicine. Clin Cancer Res 16, 6139–6149 10.1158/1078-0432.CCR-10-0978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra M., Kandalam M., Rangasamy J., Shankar B., Maheswari U. K., Swaminathan S., Krishnakumar S. (2013). Novel epithelial cell adhesion molecule antibody conjugated polyethyleneimine-capped gold nanoparticles for enhanced and targeted small interfering RNA delivery to retinoblastoma cells. Mol Vis 19, 1029–1038 [PMC free article] [PubMed] [Google Scholar]
- Mota J., Rico-Hesse R. (2009). Humanized mice show clinical signs of dengue fever according to infecting virus genotype. J Virol 83, 8638–8645 10.1128/JVI.00581-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham A. M., Langlois R. A., TenOever B. R. (2012). Replication in cells of hematopoietic origin is necessary for Dengue virus dissemination. PLoS Pathog 8, e1002465. 10.1371/journal.ppat.1002465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puccioni-Sohler M., Orsini M., Soares C. N. (2012). Dengue: a new challenge for neurology. Neurol Int 4, e15. 10.4081/ni.2012.e15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos M. M., Mohammed H., Zielinski-Gutierrez E., Hayden M. H., Lopez J. L., Fournier M., Trujillo A. R., Burton R., Brunkard J. M. & other authors (2008). Epidemic dengue and dengue hemorrhagic fever at the Texas-Mexico border: results of a household-based seroepidemiologic survey, December 2005. Am J Trop Med Hyg 78, 364–369 [PubMed] [Google Scholar]
- Reynolds J. L., Law W. C., Mahajan S. D., Aalinkeel R., Nair B., Sykes D. E., Yong K. T., Hui R., Prasad P. N., Schwartz S. A. (2012). Nanoparticle based galectin-1 gene silencing, implications in methamphetamine regulation of HIV-1 infection in monocyte derived macrophages. J Neuroimmune Pharmacol 7, 673–685 10.1007/s11481-012-9379-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards Grayson A. C., Doody A. M., Putnam D. (2006). Biophysical and structural characterization of polyethylenimine-mediated siRNA delivery in vitro. Pharm Res 23, 1868–1876 10.1007/s11095-006-9009-2 [DOI] [PubMed] [Google Scholar]
- Sadon N., Delers A., Jarman R. G., Klungthong C., Nisalak A., Gibbons R. V., Vassilev V. (2008). A new quantitative RT-PCR method for sensitive detection of dengue virus in serum samples. J Virol Methods 153, 1–6 10.1016/j.jviromet.2008.06.023 [DOI] [PubMed] [Google Scholar]
- Shukla R., Bansal V., Chaudhary M., Basu A., Bhonde R. R., Sastry M. (2005). Biocompatibility of gold nanoparticles and their endocytotic fate inside the cellular compartment: a microscopic overview. Langmuir 21, 10644–10654 10.1021/la0513712 [DOI] [PubMed] [Google Scholar]
- Song W. J., Du J. Z., Sun T. M., Zhang P. Z., Wang J. (2010). Gold nanoparticles capped with polyethyleneimine for enhanced siRNA delivery. Small 6, 239–246 10.1002/smll.200901513 [DOI] [PubMed] [Google Scholar]
- Spurr A. R. (1969). A low-viscosity epoxy resin embedding medium for electron microscopy. J Ultrastruct Res 26, 31–43 10.1016/S0022-5320(69)90033-1 [DOI] [PubMed] [Google Scholar]
- Stein D. A., Perry S. T., Buck M. D., Oehmen C. S., Fischer M. A., Poore E., Smith J. L., Lancaster A. M., Hirsch A. J. & other authors (2011). Inhibition of dengue virus infections in cell cultures and in AG129 mice by a small interfering RNA targeting a highly conserved sequence. J Virol 85, 10154–10166 10.1128/JVI.05298-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storhoff J. J., Elghanian R., Mucic R. C., Mirkin C. A., Letsinger R. L. (1998). One-pot colorimetric differentiation of polynucleotides with single base imperfections using gold nanoparticles probes. J Am Chem Soc 120, 1959–1964 10.1021/ja972332i [DOI] [Google Scholar]
- Subramanya S., Kim S. S., Abraham S., Yao J., Kumar M., Kumar P., Haridas V., Lee S. K., Shultz L. D. & other authors (2010). Targeted delivery of small interfering RNA to human dendritic cells to suppress dengue virus infection and associated proinflammatory cytokine production. J Virol 84, 2490–2501 10.1128/JVI.02105-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M., Klibanov A. M. (2003). Conjugation to gold nanoparticles enhances polyethylenimine’s transfer of plasmid DNA into mammalian cells. Proc Natl Acad Sci U S A 100, 9138–9143 10.1073/pnas.1233634100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomashek K. M., Margolis H. S. (2011). Dengue: a potential transfusion-transmitted disease. Transfusion 51, 1654–1660 10.1111/j.1537-2995.2011.03269.x [DOI] [PubMed] [Google Scholar]
- Torchilin V. P. (2005). Recent advances with liposomes as pharmaceutical carriers. Nat Rev Drug Discov 4, 145–160 10.1038/nrd1632 [DOI] [PubMed] [Google Scholar]
- Urban-Klein B., Werth S., Abuharbeid S., Czubayko F., Aigner A. (2005). RNAi-mediated gene-targeting through systemic application of polyethylenimine (PEI)-complexed siRNA in vivo. Gene Ther 12, 461–466 10.1038/sj.gt.3302425 [DOI] [PubMed] [Google Scholar]
- van der Schaar H. M., Rust M. J., Chen C., van der Ende-Metselaar H., Wilschut J., Zhuang X., Smit J. M. (2008). Dissecting the cell entry pathway of dengue virus by single-particle tracking in living cells. PLoS Pathog 4, e1000244. 10.1371/journal.ppat.1000244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Yu B., Ren W., Mo X., Zhou C., He H., Jia H., Wang L., Jacob S. T. & other authors (2013). Enhanced hepatic delivery of siRNA and microRNA using oleic acid based lipid nanoparticle formulations. J Control Release 172, 690–698 10.1016/j.jconrel.2013.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilder-Smith A., Ooi E. E., Vasudevan S. G., Gubler D. J. (2010). Update on dengue: epidemiology, virus evolution, antiviral drugs, and vaccine development. Curr Infect Dis Rep 12, 157–164 10.1007/s11908-010-0102-7 [DOI] [PubMed] [Google Scholar]
- Wu S. J., Grouard-Vogel G., Sun W., Mascola J. R., Brachtel E., Putvatana R., Louder M. K., Filgueira L., Marovich M. A. & other authors (2000). Human skin Langerhans cells are targets of dengue virus infection. Nat Med 6, 816–820 10.1038/77553 [DOI] [PubMed] [Google Scholar]
- Xi Z., Ramirez J. L., Dimopoulos G. (2008). The Aedes aegypti Toll pathway controls dengue virus infection. PLoS Pathog 4, e1000098. 10.1371/journal.ppat.1000098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Yang H., Wu J., Meng Z., Xing R., Tian A., Tian X., Guo L., Zhang Y. & other authors (2013). No overt structural or functional changes associated with PEG-coated gold nanoparticles accumulation with acute exposure in the mouse heart. Toxicol Lett 222, 197–203 10.1016/j.toxlet.2013.07.018 [DOI] [PubMed] [Google Scholar]
- Ye C., Abraham S., Wu H., Shankar P., Manjunath N. (2011). Silencing early viral replication in macrophages and dendritic cells effectively suppresses flavivirus encephalitis. PLoS ONE 6, e17889. 10.1371/journal.pone.0017889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. D., Wu D., Shen X., Liu P. X., Yang N., Zhao B., Zhang H., Sun Y. M., Zhang L. A., Fan F. Y. (2011). Size-dependent in vivo toxicity of PEG-coated gold nanoparticles. Int J Nanomedicine 6, 2071–2081 10.2147/IJN.S21657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D., Giljohann D. A., Chen D. L., Massich M. D., Wang X. Q., Iordanov H., Mirkin C. A., Paller A. S. (2012). Topical delivery of siRNA-based spherical nucleic acid nanoparticle conjugates for gene regulation. Proc Natl Acad Sci U S A 109, 11975–11980 10.1073/pnas.1118425109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y. Q., Wei J., Fu Y. R., Shao J., Liang Y. W., Lin Y. H., Liu J., Zhu Z. H. (2008). [Toxicity of cationic liposome Lipofectamine 2000 in human pancreatic cancer Capan-2 cells]. Nan Fang Yi Ke Da Xue Xue Bao 28, 1981–1984 (in Chinese). [PubMed] [Google Scholar]
- Zhou J., Shum K. T., Burnett J. C., Rossi J. J. (2013). Nanoparticle-based delivery of RNAi therapeutics: progress and challenges. Pharmaceuticals 6, 85–107 10.3390/ph6010085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zintchenko A., Philipp A., Dehshahri A., Wagner E. (2008). Simple modifications of branched PEI lead to highly efficient siRNA carriers with low toxicity. Bioconjug Chem 19, 1448–1455 10.1021/bc800065f [DOI] [PubMed] [Google Scholar]
- Zuhorn I. S., Kalicharan R., Hoekstra D. (2002). Lipoplex-mediated transfection of mammalian cells occurs through the cholesterol-dependent clathrin-mediated pathway of endocytosis. J Biol Chem 277, 18021–18028 10.1074/jbc.M111257200 [DOI] [PubMed] [Google Scholar]