SUMMARY
Lysine acetylation is a major posttranslational modification involved in a broad array of physiological functions. Here, we provide an organ-wide map of lysine acetylation sites from 16 rat tissues analyzed by high-resolution tandem mass spectrometry. We quantify 15,474 modification sites on 4,541 proteins and provide the data set as a web-based database. We demonstrate that lysine acetylation displays site-specific sequence motifs that diverge between cellular compartments, with a significant fraction of nuclear sites conforming to the consensus motifs G-AcK and AcK-P. Our data set reveals that the subcellular acetylation distribution is tissue-type dependent and that acetylation targets tissue-specific pathways involved in fundamental physiological processes. We compare lysine acetylation patterns for rat as well as human skeletal muscle biopsies and demonstrate its general involvement in muscle contraction. Furthermore, we illustrate that acetylation of fructose-bisphosphate aldolase and glycerol-3-phosphate dehydrogenase serves as a cellular mechanism to switch off enzymatic activity.
INTRODUCTION
Lysine acetylation is a reversible posttranslational modification involved in multiple cellular processes, where an acetyl group is transferred to the epsilon-amino group of an internal lysine residue of a protein. The importance of lysine acetylation is well appreciated in the context of nuclear histone modifications (Strahl and Allis, 2000), but the regulatory implications of the modification extend beyond gene regulation. Changes in cellular lysine acetylation status can alter metabolic enzyme activity and provide a mechanism for the cell to adapt to metabolic changes (Rodgers et al., 2008; Wang et al., 2010; Zhao et al., 2010), for instance by regulating key enzymes of the tricarboxylic acid cycle, the urea cycle, and fatty acid oxidation (Ahn et al., 2008; Hirschey et al., 2010; Kim et al., 2006; Nakagawa et al., 2009). The physiological importance of dynamic acetylation as a regulatory mechanism of metabolism has been highlighted in recent studies of the liver (Zhao et al., 2010), where the distribution of lysine acetylation sites changes under conditions of either acute fasting (Yang et al., 2011) or caloric restriction (Schwer et al., 2009). An overlap of 70% has been reported for acetylation sites identified from mouse and human liver tissues, whereas the overlap with data from cell lines was only 14% (Zhao et al., 2010). Because lysine acetylation sites are evolutionarily conserved (Wang et al., 2010; Weinert et al., 2011), this suggests that acetylation patterns vary depending on the particular cellular functions to be performed. To explore physiologically relevant lysine acetylation substrates, it is pivotal to have knowledge of which proteins are acetylated in vivo and to know the tissue distribution of the modified sites. The value of creating extensive, tissue-specific maps of protein lysine acetylation sites is further underscored by the significant role of posttranslational modifications in the evolution and phenotypic composition of vertebrates, as indicated by the significant selective pressure gene regulatory elements near posttranslational-modifying enzymes have come under in placental animals (Lowe et al., 2011). Here, we combined an efficient method for protein extraction from tissue samples with lysine-acetylated peptide immunoprecipitation and high-accuracy tandem mass spectrometric (MS/MS) measurements, which allowed us to generate an atlas of lysine acetylation sites in 16 different rat tissues. Our acetylome data set expands the current number of lysine acetylation sites by 4-fold and the number of acetylated proteins by 2-fold (Choudhary et al., 2009; Wang et al., 2010; Weinert et al., 2011). We achieved this by improving the antibody-based enrichment protocol significantly, such that 40% of all tandem mass spectra identify lysine-acetylated peptides. We map ~3,000–6,000 acetylation sites in most tissues by analyzing just three strong cation exchange (SCX) fractions with single 3 hr tandem mass spectrometric runs. We provide tissue-specific evidence that lysine acetylation is comparable to phosphorylation in cellular prevalence. Our data set highlights tissue-specific pathways involving lysine-acetylated proteins, which stresses the importance of mapping protein modifications in the physiologically relevant tissue. Among the findings, we observe that almost all proteins involved in striated muscle contraction are acetylated, and we confirm this finding in human skeletal muscle biopsies. Not only do we find that specific proteins and sites are differentially acetylated across tissues, we also show that the distribution of lysine-acetylated proteins in subcellular compartments is tissue specific. Our large data set allows us to delineate lysine acetylation sequence motifs, and contrary to what has been previously known, we show that there are subcellular compartment-specific sequence motifs. We provide easy access to all data via a searchable online database.
RESULTS
Identification of 15,474 Lysine Acetylation Sites from 4,541 Proteins
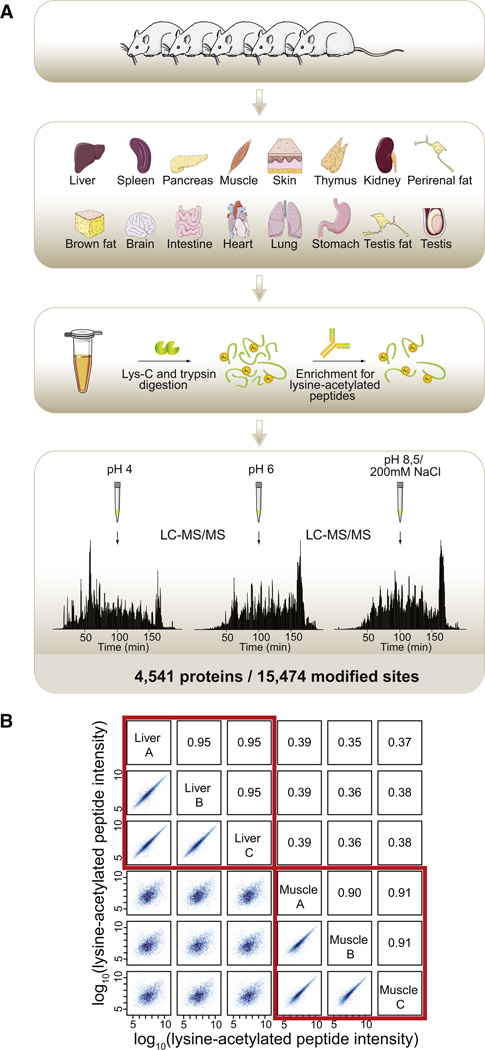
A total of 16 different organs and tissues were dissected from Sprague-Dawley albino rats (SPRD; Taconic, Denmark) to map lysine acetylation sites of proteins across tissues. The tissues were brain, heart, muscle, lung, kidney, liver, stomach, pancreas, spleen, thymus, intestine, skin, testis, testis fat, perirenal fat, and brown fat (Figure 1A). Organs from five rats were pooled to account for biological variation. Protein extracts were digested in solution in a urea buffer with sequential steps of Lys-C and trypsin incubations. The resulting peptides were desalted and enriched for lysine-acetylated peptides by immunoprecipitation followed by fractionation by microscale SCX chromatography (Weinert et al., 2011). The three resulting SCX fractions were analyzed by 3 hr nanoflow liquid chromatography tandem mass spectrometry (LC-MS/MS) gradients on a LTQ-Orbitrap Velos mass spectrometer (Olsen et al., 2009) with all tandem mass spectra recorded in the orbitrap analyzer at high resolution using the higher-energy collisional dissociation (HCD) technology (Olsen et al., 2007). Peptide sequences were identified by Mascot, and lysine-acetylated proteins were quantified using the MaxQuant software suite’s label-free algorithm based on peptide-extracted ion chromatograms (XICs). In total 1,060,605 high-resolution HCD-MS/MS events were collected resulting in 265,034 peptide-spectrum matches at a false discovery rate below 0.01. A total of 105,904 HCD spectra identified lysine-acetylated peptides, which corresponds to 40% of all MS/MS identifications. The data set covered 62,553 unique modification-specific peptides, of which 19,965 were lysine acetylated, matching to 15,474 lysine acetylation sites from 4,541 proteins.
Figure 1. Workflow for Acetylome Analysis of Rat Tissues.
(A) A total of 16 tissues were isolated from 5 male rats; the tissues were snap frozen, heat inactivated, homogenized, and solubilized. The extracted proteins were digested with endoproteinase Lys-C and trypsin, and lysine-acetylated peptides were enriched by immunoprecipitation. The acetylated peptide mixtures were fractionated by SCX in a STAGE tip, and three pH elutions per tissue were analyzed by high-resolution LC-MS/MS on a LTQ-Orbitrap Velos instrument resulting in identification of a total of 15,474 lysine acetylation sites from 4,541 proteins.
(B) For liver and muscle samples, results from three technical replicates prepared from the tissue homogenates are shown. Logarithmized intensities for acetylated peptides were plotted against each other and shown on the left side of the diagonal with the corresponding Pearson correlation coefficients given on the right side of the diagonal. Technical replicates of the same tissue are highly correlated.
See also Figures S1 and S2.
To facilitate the ease of searching the data set for modifications on particular proteins of interest, we have created an online database, the CPR PTM Resource, where we have recorded the entire tissue-specific lysine acetylation data set. At the CPR PTM Resource, information on the tissue distribution of posttranslational modifications on a given protein is provided along with topological protein information imported from other sources. To further allow for easy meta-analysis of the data set for the scientific community, we provide 60 raw files as well as 22,789 annotated lysine-acetylated peptide HCD-MS/MS spectra as a resource for download via TRANCHE (see Extended Experimental Procedures). Tables with all modification-specific peptides (Table S1), identified lysine acetylation sites (Table S2), and proteins (Table S3) are also provided both in the Extended Experimental Procedures and at the CPR PTM Resource website. Evaluation of the high-quality mass spectrometry (MS) and MS/MS data is shown in Figure S1. Figure S2 illustrates our successful enrichments of lysine-acetylated peptides and peptide fractionation, which were instrumental factors, for allowing us to expand the current number of lysine acetylation sites by 4-fold.
Distribution of Lysine Acetylation Sites across Tissues
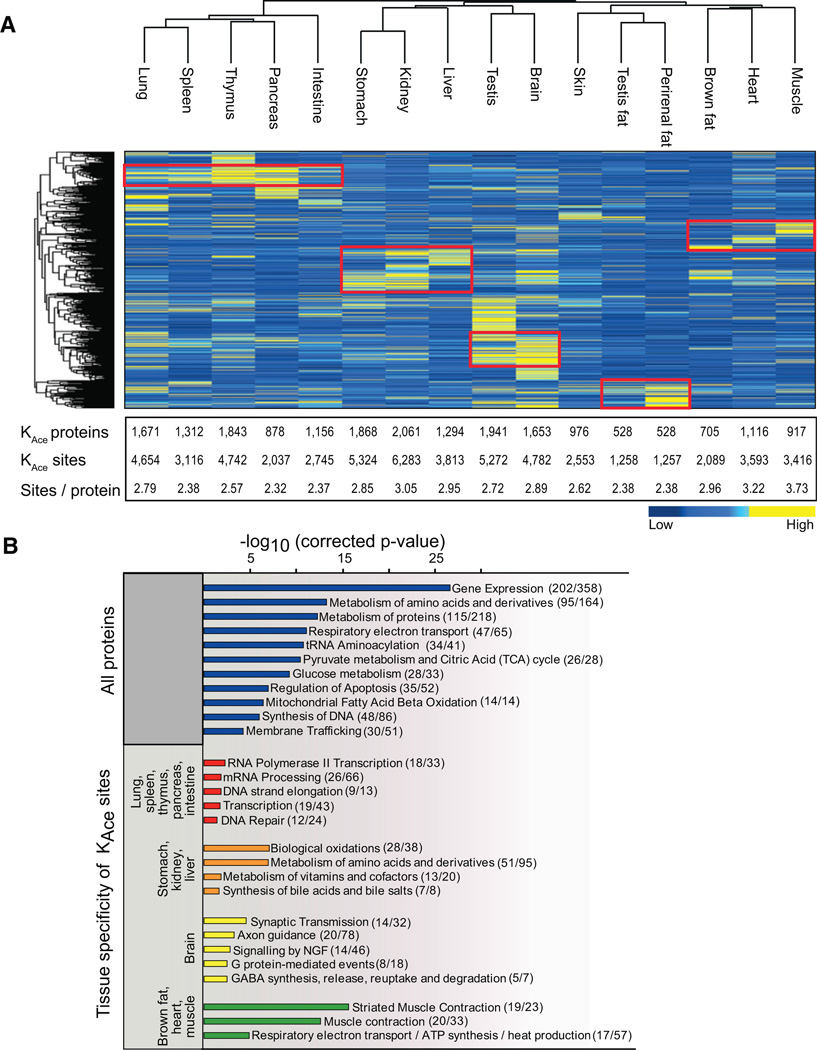
For individual tissues there is excellent technical reproducibility of the entire experimental workflow (Figure 1B). Figure 1B summarizes three technical replicates for two different tissue types: liver, which is known to have high abundance of lysine-acetylated proteins; and skeletal muscle, which has not previously been studied in the context of lysine acetylation. The correlation analysis of lysine-acetylated peptide MS signal intensities between technical replications prepared from the same tissue reveals Pearson coefficients < R > = 0.95 for liver and < R > = 0.91 for muscle. Importantly, there is poor correlation between muscle and liver tissues: < R > = 0.37. The high reproducibility of the MS data collected from a given tissue enables us to perform relative label-free quantification of lysine acetylation sites between tissues. We evaluated the tissue distribution of lysine-acetylated proteins by hierarchical clustering analysis of normalized acetylated protein intensities derived from summed MS signal intensities from each tissue (Figure 2A). The acetylated proteins are color coded according to their normalized intensities, which are a measure for their relative abundances in each tissue (de Godoy et al., 2008; Malmström et al., 2009). Protein acetylation patterns vary greatly across tissues, but functionally related tissues cluster together, as for instance the major energy-consuming tissues (heart, muscle, and brown fat) and organs containing lymphoid tissue (lung, spleen, thymus, pancreas, and intestine). Because we identify most high-abundant proteins to be acetylated, this clustering profile is likely partly due to similarities in protein expression patterns, in addition to similarities in acetylation patterns. For most tissues we identify on the order of 1,000 lysine-acetylated proteins, but a few tissues display significantly more acetylated proteins. For example in kidney there are 2,061 acetylated proteins containing 6,283 modified residues (Figure 2A). To explore the differences underlying the tissue-specific patterns of acetylated proteins in each tissue cluster, we performed gene ontology (GO) (Ashburner et al., 2000) and pathway (Reactome) (Croft et al., 2011) enrichment analyses. Testing which pathways are enriched in the entire data set of 4,541 acetylated proteins identifies the main pathways previously recognized to be regulated by lysine acetylation such as gene expression, protein metabolism, the citric acid (TCA) cycle, and apoptosis (Figure 2B). However, focusing on the acetylated proteins that are specifically enriched for each of the tissue clusters identifies tissue-specific pathways, which for the most part have not previously been shown to involve lysine acetylation. In general the tissue-specific pathways correlate well with the known physiological roles of the tissues, which possibly reflect that the high-abundant acetylated proteins are expressed in a tissue-specific manner. For instance the cluster with the major energy-consuming tissues is significantly enriched for proteins involved in striated muscle contraction as well as respiratory electron transport. Conversely, the cluster with organs containing lymphoid tissue is significantly enriched for proteins involved in various aspects of transcription and translation. The acetylated proteins that are more abundant in the brain than in other tissues are primarily involved in neuronal signal transmission, whereas the specific acetylated proteins of the stomach, liver, and kidney are enriched for proteins involved in metabolism of amino acids and vitamins (Figure 2B). Thus, lysine acetylation is targeting tissue-specific biological processes, and it is involved in diverse cellular functions across tissues.
Figure 2. Tissue Distribution of Lysine-Acetylated Proteins.
(A) Hierarchical clustering of the 16 investigated tissues and the identified acetylated proteins based on label-free quantification on their summed MS peptide signal intensities. Low-intensity proteins are depicted in blue, and high-intensity proteins are depicted in yellow. Protein clusters of highly abundant acetylated proteins are highlighted by red boxes. The table summarizes the number of lysine-acetylated proteins and sites identified in each tissue as well as the average number of acetylation sites per protein.
(B) Pathway enrichment analysis for all identified acetylated proteins as well as for acetylated proteins enriched in the main tissue clusters compared to all other tissues. Logarithmized corrected p values for significant overrepresentation are shown. In parenthesis we indicate how many proteins in each pathway we identify to be acetylated.
See also Figure S3.
As a starting point for deciphering tissue-specific molecular networks of lysine-acetylated proteins, we generated protein-protein interaction networks based on proteins that are acetylated in a tissue-specific manner. We identified interaction partners using the InWeb database of quality-controlled protein-protein interactions (Lage et al., 2007, 2008, 2010) and applied a network-building algorithm to build protein-interaction networks with quality thresholds optimized by permutation tests (Bergholdt et al., 2007; Lage et al., 2010). Five of the protein-interaction networks of tissue-specific acetylated proteins interact significantly (0.0261 ≤ adj. p ≤ 0.0467, adjusted for multiple testing using a Bonferroni correction), indicating that proteins with tissue-specific acetylation patterns have a strong tendency to directly interact or are part of connected tissue-specific pathways. The high connectivity in interaction space of acetylated proteins is clearly illustrated in the network based on proteins specifically acetylated in brain (Figure S3). We provide all networks in flat file and Cytoscape session formats as a resource for the community through the CPR PTM Resource web page.
Lysine Acetylation in Rat and Human Striated Muscle Contraction
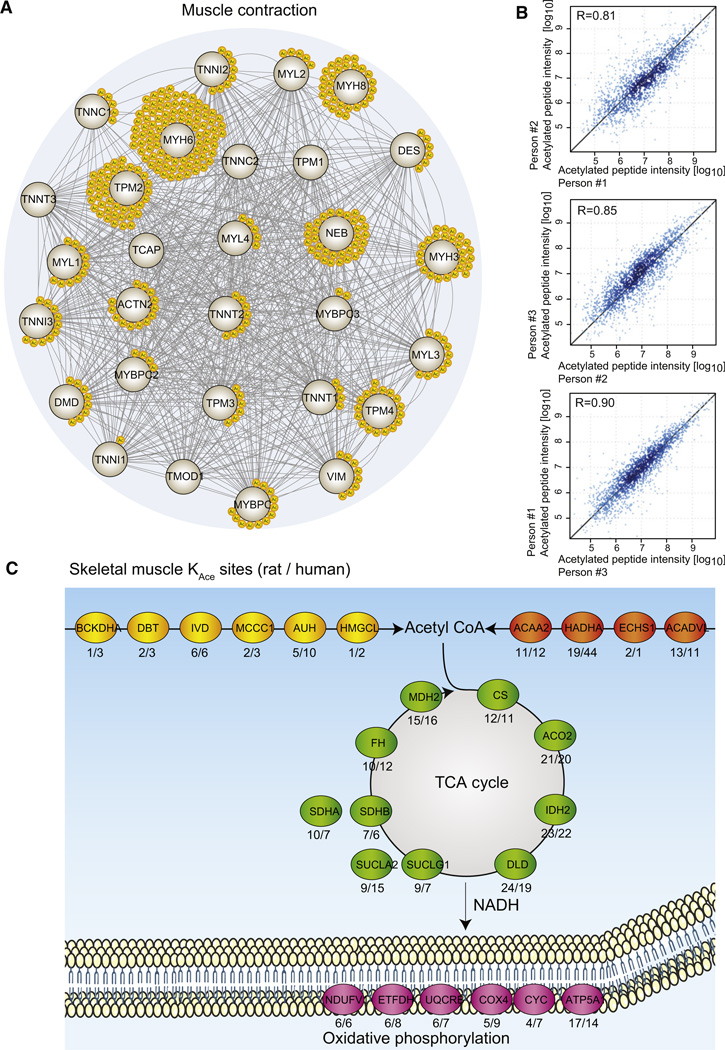
To investigate one of the tissue-specific roles of lysine acetylation further, we decided to focus on striated muscle contraction. More than 80% of the proteins involved in striated muscle contraction are acetylated (Figure 3A). To ensure that this finding is of general physiological relevance, we performed similar experiments on human skeletal muscle biopsies from three age-matched athletes. From these samples we assess the biological variation of lysine acetylation site abundance. Pearson correlation coefficients range from 0.81 to 0.90 (Figure 3B), thus indicating that it will be possible to perform quantitative lysine acetylation analyses of human samples, and not only from inbred animals. From the human skeletal muscle samples, we identify a total of 941 acetylated proteins containing 2,811 acetylation sites (Table S4). The lysine acetylation sites identified in human skeletal muscle confirm the acetylation pattern of proteins involved in muscle contraction. In muscle the cellular compartment with the highest abundance of lysine acetylation is the mitochondria, and in rat as well as in human muscle samples, we identify lysine acetylation sites on proteins involved in all enzymatic steps of the major mitochondrial metabolic pathways (Figure 3C). Thus, proteins involved in muscle contraction are hyperacetylated as are the metabolic enzymes generating ATP, which collectively points to an important role of lysine acetylation in skeletal muscle physiology.
Figure 3. Lysine Acetylation in Muscle Contraction.
(A) All proteins associated with striated muscle contraction were extracted from the Reactome pathway database, and a protein-protein interaction network was visualized with STRING (Szklarczyk et al., 2011). Each yellow circle represents a unique lysine acetylation site identified from rat skeletal or cardiac muscle samples.
(B) Correlation plots of lysine-acetylated peptide intensities from skeletal muscle samples from three human individuals. The Pearson correlation coefficient is provided in each plot.
(C) Four major mitochondrial metabolic pathways are depicted with the most acetylated protein identified in skeletal muscle for each enzymatic step in the pathways. The metabolic pathways of amino acid catabolism and fatty acid metabolism generate acetyl-CoA that is feeded into the TCA cycle. The TCA cycle generates NADH and FADH2, which serve as electron donors in the respiratory chain, ultimately resulting in the formation of ATP. All enzymes are represented by their gene names, and below each enzyme the number of identified lysine acetylation sites is given for rat and human muscle samples, respectively (rat/human).
Functional Analysis of Site-Specific Acetylation Sites on Glycolytic Enzymes
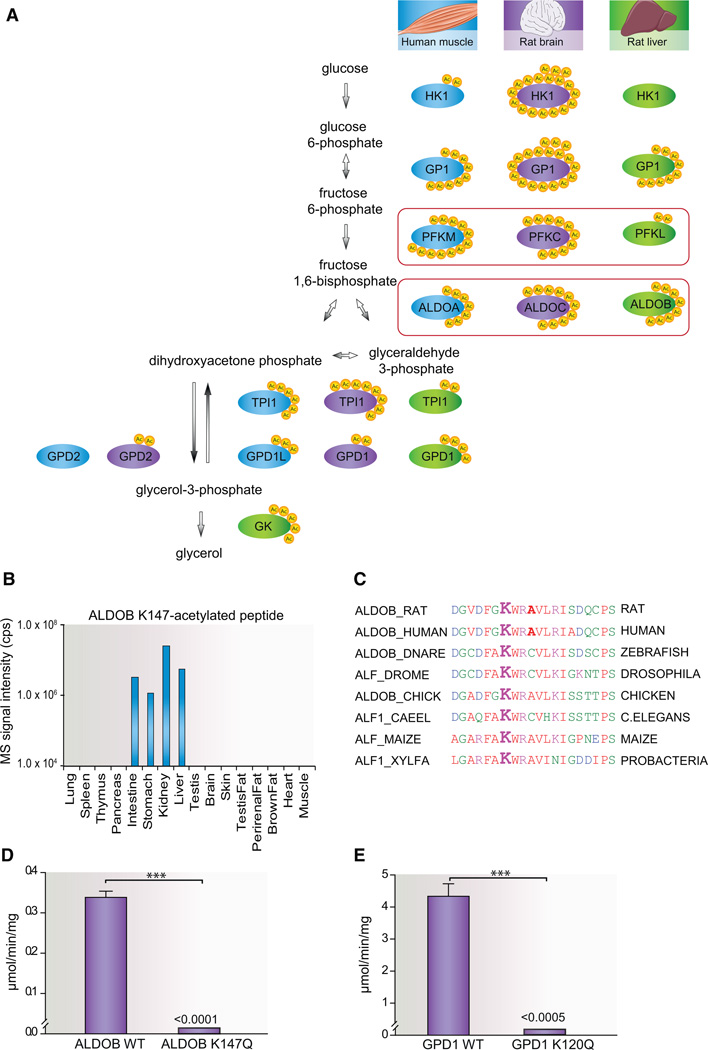
In accordance with the increasing evidence that supports lysine acetylation as a major cellular mechanism that regulates glucose metabolism (Wang et al., 2010; Yang et al., 2011; Zhao et al., 2010), we identify multiple acetylation sites on enzymes involved in the glycerol synthesis pathway. Interestingly, there is a tissue-specific distribution of sites between the enzymes in human skeletal muscle, rat brain and rat liver (Figure 4A). We identified all three genetically distinct and tissue-specific isozymes of fructose-bisphosphate aldolase known in mammals, with specific acetylated peptides from ALDOA in human muscle, ALDOB in liver, and ALDOC in brain. Aldolase deficiency in humans leads to fructose intolerance, and two-thirds of patients suffering from hereditary fructose intolerance have a common missense mutation in the ALDOB gene resulting in an amino acid substitution A150P (Davit-Spraul et al., 2008). This mutation is in close proximity of lysine 147 (K147), which we find to be abundantly acetylated in intestine, stomach, kidney, and liver (Figure 4B). To estimate the occupancy of this lysine acetylation site, we modified a method we previously developed to calculate phosphorylation site occupancy from SILAC-labeled samples (Olsen et al., 2010). Our approach relies on the label-free quantification measurements and uses the information contained in the normalized peptide intensities for a given acetylated peptide and its corresponding nonacetylated peptide combined with relative iBAQ estimated protein abundances between tissues (Schwanhäusser et al., 2011). Our occupancy calculations show that K147 is acetylated on ~33% of the ALDOB proteins in the liver and on ~20% of the proteins in the stomach, thus indicating a potential physiological role. K147 as well as the surrounding amino acids is highly conserved from vertebrates to probacteria (Figure 4C). Because K147 furthermore is important for substrate binding (St-Jean et al., 2009), we decided to investigate functional effects of acetylation of this residue on ALDOB enzymatic activity. We expressed full-length human ALDOB in a wild-type and K147Q acetylation mimetic mutant and measured the enzymatic activity of the purified proteins. Wild-type ALDOB efficiently converts fructose-1,6-bisphosphate to glyceraldehyde-3-phosphate and dihydroxyacetone phosphate at a rate of 0.34 ± 0.01 µmol/min/mg. Strikingly, the K147Q mutation appears to abolish the enzymatic activity of ALDOB because this mutant shows no detectable activity in our assay (Figure 4D). Hence, the wild-type enzyme is at least three orders of magnitude more active than the K147Q mutant. This finding highlights that site-specific acetylation can function as an efficient cellular mechanism to switch off the enzymatic activity of important regulatory proteins. To investigate if this regulatory switch is restricted to aldolases, or if lysine acetylation of substrate binding sites is a general mechanism to inhibit enzymes in the glycogen synthesis pathway, we tested the function of an equivalent lysine acetylation site we identified on glycerol-3-phosphate dehydrogenase (GPD1). K120 on GPD1 is predicted to serve as a substrate binding site (Ou et al., 2006), and we identified this residue to be acetylated. Analogous to the functional test of ALDOB, we purified wild-type GPD1 as well as the K120Q mutant and assayed their enzymatic activity (Figure 4E). The activity of wild-type enzyme was measured to 4.4 ± 0.4 µmol/min/mg, whereas the acetylation mimicking mutated enzyme has activity levels below the detection limit. The difference in activity between the wild-type and mutant GPD1 is more than three orders of magnitude, thus again providing evidence for lysine acetylation as a mechanism for switching off enzymatic activity.
Figure 4. Functional Analysis of Site-Specific Acetylation Sites on Glycolytic Enzymes.
(A) Tissue distribution of lysine acetylation sites on enzymes involved in glycerol synthesis. For each enzymatic step, enzymes from human skeletal muscle (blue), rat brain (purple), and rat liver (green) are visualized with the number of lysine acetylation sites identified (yellow). For aldolase (ALDOA, ALDOB, and ALDOC) and phosphofructosekinase (PFKM, PFKC, and PFKL), tissue-specific isozymes were identified and are boxed in red color.
(B) Tissue distribution of the mass spectrometric signal intensities of the ALDOB K147-acetylated peptide DGVDFGK(ac)WARAVLR.
(C) Conservation of rat ALDOB K147 across species from human to probacteria and plants.
(D) Catalytic activity of ALDOB WT and ALDOB K147Q toward fructose-1,6-bisphosphate assayed by downstream NADH oxidation, respectively (n = 3, mean ± SEM). The activity of ALDOB K174Q was below the assay detection limits of approximately 1 × 10−4 µmol/min/mg.
(E) Enzymatic activity of GPD1 WT and GPD1 K120Q toward dihydroxyacetone phosphate measured by NADH oxidation, respectively (n = 3, mean ± SEM). The activity of GPD1 K120Q was below the assay detection limit of approximately 5 × 10−4 µmol/min/mg.
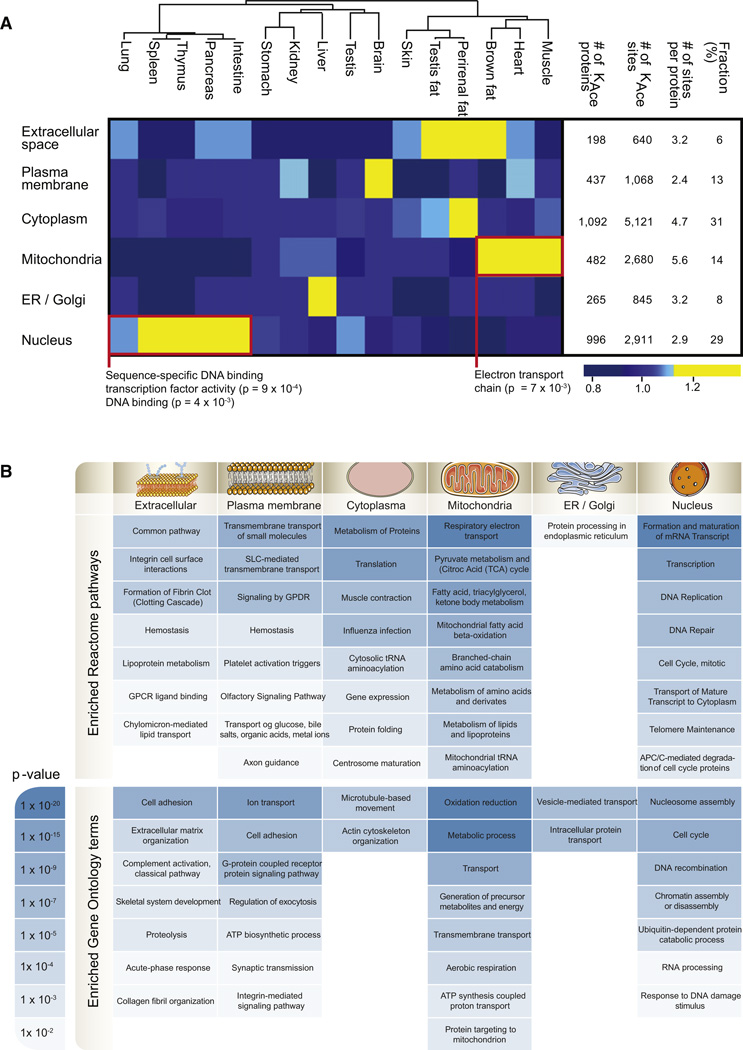
Cellular Compartment Distribution of Lysine-Acetylated Proteins
To investigate the distribution of acetylated proteins across cellular compartments, we evaluated the proteins according to their GO cellular compartment annotations. The majority of acetylated proteins reside in either the cytoplasm or the nucleus, which each contain ~30% of all the acetylated proteins. The mitochondria and the plasma membrane both account for ~15% of modified proteins, and the endoplasmic reticulum or Golgi apparatus (ER/Golgi) as well as the extracellular space harbor ~5% (Figure 5A). Two tissue atlases of phosphoproteins have been published, one for mouse and one for rat (Huttlin et al., 2010; Lundby et al., 2012), so to compare the distribution of phosphoproteins to that of acetylated proteins, we exploited the rat data set we have generated. Interestingly, the subcellular distribution of acetylated proteins is markedly different from the distribution of phosphorylated proteins (Figure S4). The fraction of modified proteins residing in the mitochondria is more than 3-fold higher for lysine-acetylated proteins compared to phosphorylated proteins. On the contrary the fraction of protein substrates localized to the plasma membrane is more than 2-fold higher for phosphorylation compared to acetylation. However, the subcellular distribution of lysine-acetylated proteins is tissue dependent and possibly reflects the distribution of unmodified proteins. For example the brain accounts for the largest fraction of acetylated membrane proteins, which explains why pathway enrichment analysis of all acetylated plasma membrane proteins emphasizes neurophysiological functions (Figure 5B). Alike, the three adipose tissues are abundant in acetylated proteins associated with the extracellular space, which is likely associated to adipose tissues being major endocrine organs (Kratchmarova et al., 2002; Saltiel, 2001). Organs containing lymphoid tissue (spleen, thymus, pancreas, lung, and intestine) contain a higher fraction of acetylated nuclear proteins than any of the other tissues, and these nuclear proteins are significantly enriched for sequence-specific DNA binding transcription factor activity (p = 9 × 10−4). Focusing on the proteins underlying this enrichment, we discover that lysine acetylation is significantly more abundant on 82 of 129 modified transcription factors in these tissues (Table S5). This observation likely reflects that lymphoid-containing tissues have high self-renewal rates (Pellettieri and Sánchez Alvarado, 2007) and, therefore, require high transcriptional activity.
Figure 5. Cellular Compartment Distribution of Acetylated Proteins across Tissues.
(A) All lysine-acetylated proteins were grouped based on their subcellular localization, and for each tissue the fraction of identified acetylated proteins per compartment was calculated. The deviation from the median was visualized as a heatmap according to the indicated color scale. For the clusters encircled by a red box, pathway enrichment analysis was performed, and the protein processes underlying significant overrepresentation is displayed. The number of acetylated proteins and sites per cellular compartment is provided in the table together with the average number of acetylation sites per protein per compartment and the fraction of acetylated proteins per compartment.
(B) GO and pathway enrichment analyses were made for each subcellular compartment, and enriched Reactome pathways and GO terms for biological processes are listed with their corresponding p values color coded according to the scale.
See also Figures S4 and S5.
At the global level we identify an average of 2.8 lysine acetylation sites per protein per tissue (Figure 2A), but as evident from Figure 5A, this number greatly varies between cellular compartments. Mitochondrial proteins exhibit the greatest number of acetylation sites with an average of 5.6 sites per protein. This may explain why many previous studies primarily identified acetylation sites from mitochondrial proteins, although this is not the compartment with the greatest number of acetylated proteins. To test if the tendency toward multiacetylated proteins in the mitochondria is biased by protein abundance, we estimated the relative protein abundance for all proteins using the iBAQ method. Analyzing the number of acetylation sites per protein as function of protein abundance revealed that mitochondrial proteins have significantly more modified sites at all protein abundance levels (Figure S5). The tissues with the largest fraction of acetylated mitochondrial proteins are muscle, heart, and brown fat. More than 35% of all identified acetylation sites in these tissues are mitochondrial, with a significant overrepresentation of proteins involved in the electron transport chain (p = 7 × 10−3). Thus, the result likely reflects the high-energy demand and oxidative capacity of these particular tissues.
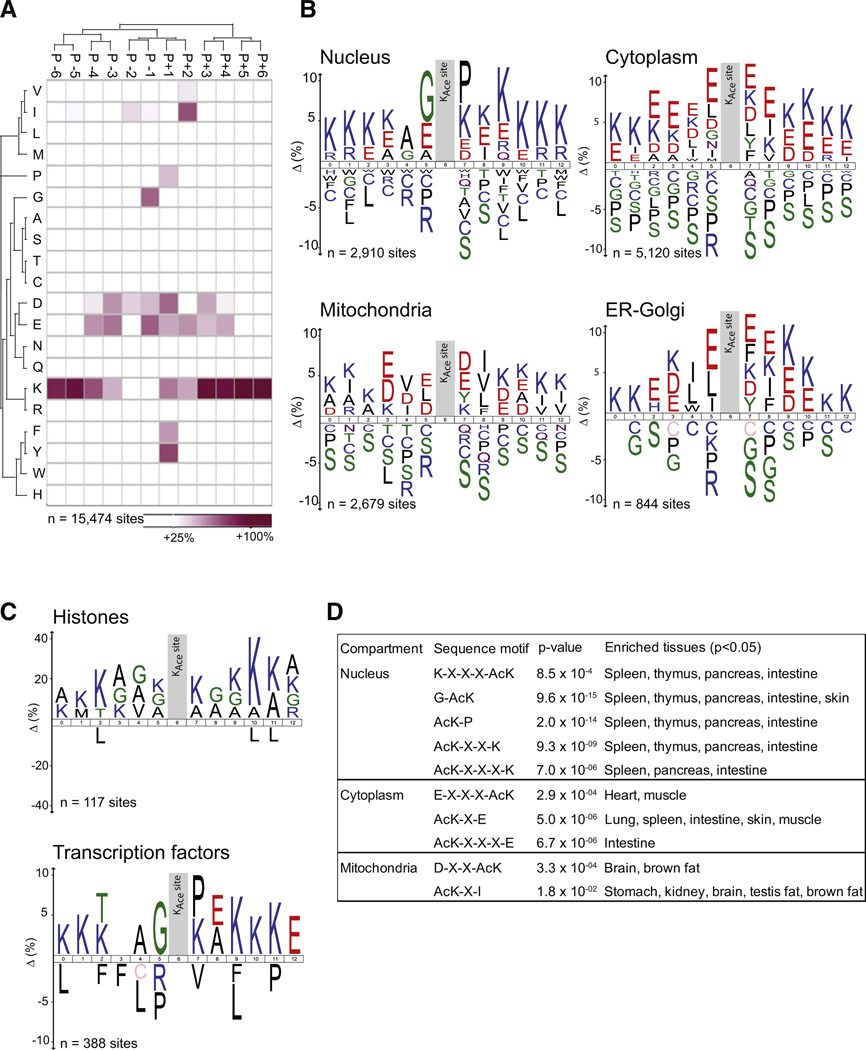
Lysine Acetylation Sequence Motifs Are Specific for Cellular Compartments
We explored our data set for lysine acetylation site-specific sequence motifs by analyzing the 12 residues flanking the modified site for overrepresentation of specific amino acids relative to the proteome background distribution. Analysis of all identified acetylation sites reveals general preferences for specific amino acid residues at particular positions surrounding the acetylated lysines (Figure 6A). As also previously reported (Weinert et al., 2011), we find that lysine acetylations preferentially occur in lysine-rich regions, with a tendency toward negatively charged residues in the immediate surroundings of the modified site. Position-specific preferences include glycine residues at amino acid position −1 relative to the acetylated residue, proline/phenylalanine/tyrosine residues at amino acid position +1, and valine/isoleucine residues at amino acid position +2. We next investigated if these sequence preferences are similar across all cellular compartments. Interestingly, the global sequence motif map we find appears to be a merged picture of compartment-specific motifs (Figure S6). To visualize compartment-specific sequence motifs, we analyzed all lysine acetylation sites from proteins localized to a particular subcellular compartment relative to the rat protein database using ice-Logo (Colaert et al., 2009) (Figure 6B). This analysis reveals that the sequence motifs indeed differ for subcellular compartments. The preference for lysine-rich regions is general to all compartments, but on cytoplasmic proteins there is a clear preference for glutamate residues at all positions in the vicinity of the acetylation site. Mitochondrial proteins have a preference for negatively charged residues in the immediate vicinity of the acetylation site, but they additionally show a strong preference for hydrophobic residues (V/I/L/F) at position +2. Proteins that reside in either ER/Golgi also have the general preference for negatively charged residues but further favor hydrophobic residues (I/L) at positions −1 and −2. The most distinct sequence motif is evident on nuclear proteins, where there is a strong preference for glycine residues in position −1 and proline residues in position +1. Because this nuclear sequence motif differs substantially from the previously reported motif for lysine acetylation of histones (KX-X-X-AcK-X-X-X-K) (Kim et al., 2006), we next analyzed sites identified from histones separately. This analysis indeed confirms that the preferred sequence motif for histones differs from other nuclear proteins and that it conforms to the known motif with lysines at positions ±4 (Figure 6C). Another group of nuclear proteins of particular interest is transcription factors, and because we have identified a total of 388 acetylation sites on transcription factors, we next tested if these contain a particular sequence motif. We find that transcription factors display a similar motif as the other nuclear proteins (G-AcK-P), which thus differs for the one found for histones (Figure 6C). The identified compartment-specific sequence motifs are summarized in Figure 6D.
Figure 6. Sequence Motifs for Lysine Acetylation Sites across Cellular Compartments.
(A) Heatmap indicating overrepresentation of amino acids in positions from −6 to +6 from the acetylated lysine residue based on all identified acetylation sites compared to the overall proteome amino acid frequency distribution.
(B) Sequence logos for acetylation sites identified on proteins residing in the nucleus, cytosol, mitochondria, or ER-Golgi.
(C) Sequence logos for subsets of the nuclear proteins. Sites identified from histones and transcription factors were analyzed separately.
(D) Table summarizing sequence motifs found for compartment-specific lysine acetylation sites with sequence motif and tissue enrichment p values indicated.
See also Figure S6.
DISCUSSION
The importance of tissue-specific mapping of posttranslational modifications of proteins is underscored by the substantial differences we find for lysine acetylation patterns across tissues. Our data set provides a rich resource of candidates for hypothesis generation and subsequent mechanism-focused studies to elucidate the tissue-specific protein networks that are controlled by regulatory site-specific lysine acetylation. In addition the data set provides evidence that site-specific lysine-acetylated proteins are involved in multiple physiological functions with which the modification has not previously been associated. For example we show that almost all proteins involved in muscle contraction are acetylated both in rat striated muscle as well as in human skeletal muscle. In particular we find that multiple enzymes involved in ATP generation are hyper-acetylated in skeletal muscle mitochondria, for many of which it is known that the enzymatic activity of their bacterial orthologs is regulated by lysine acetylation (Wang et al., 2010). Because acetylation of muscle LIM protein is moreover known to enhance calcium sensitivity of myofilaments in cardiomyocytes (Gupta et al., 2008), our findings strongly imply that muscle-contractile proteins are regulated by lysine acetylation in addition to the well-established role of phosphorylation in muscle contraction. This notion is further supported by our demonstration that site-specific acetylation of two glycolytic enzymes, ALDOB and GPD1, serves as a regulatory mechanism for switching off their enzymatic activity. It is intriguing to speculate that lysine acetylation of substrate binding sites may be a general cellular mechanism to control metabolic enzyme activity in a manner analogous to phosphorylation-dependent regulation of protein kinases and signal transducers. In contrast to the activating effects of phosphorylation, acetylation could function to switch off enzymatic activity, adding another layer of posttranslational regulatory control to dynamic cellular processes.
For more than a decade, it has been speculated that the extent of lysine acetylation matches that of phosphorylation (Kouzarides, 2000). With this data set we provide evidence that this is indeed the case. Intriguingly, despite the observation that phosphorylation and acetylation both have broad cellular scopes, we have shown that they have distinct subcellular substrate patterns. On average the fraction of lysine-acetylated proteins that reside in the mitochondria is three times greater than that of phosphorylated proteins. Conversely, the fraction of phosphorylated proteins at the plasma membrane is twice as high as that of acetylated proteins. However, the subcellular distribution of lysine acetylation sites varies across tissues. Lysine acetylation is in general abundant in mitochondria, a large fraction of mitochondrial proteins carry the modification, and the average number of modified sites per protein is high, but the modification is particularly prevalent in mitochondria of energy-generating tissues. In brown fat more than 50% of the acetylation sites we identified reside on mitochondrial proteins. Among the sites are three modifications on the uncoupling protein 1 (ucp1), which is essential for the tissue’s ability to dissipate energy as heat. Our data also reveal that the amino acids flanking the modified sites in brown fat mitochondria exhibit two significant sequence motifs: D-X-X-AcK and AcK-X-I.
Lysine acetylation has not previously been discussed in the context of secreted proteins, but we identify about 200 extracellular proteins that carry the modification. These proteins were primarily identified in adipose tissues, which likely reflect that adipose tissues are major endocrine organs (Kratchmarova et al., 2002; Saltiel, 2001). The functional roles of these lysine acetylation sites remain to be elucidated, as is also the case for the sites on the more than 400 plasma membrane proteins that we here show carry the modification. ER preparations purified from a human cell line were recently investigated in the context of lysine acetylation (Pehar et al., 2012). A total of 143 acetylated proteins were identified, which is about half the number reported herein. Two proteins highlighted in the study, GRP78 and CALR, were reported to harbor six and five acetylation sites, respectively. Although we have better coverage in our study, identifying 20 and 13 sites, respectively, a couple of sites reported by Pehar et al. (2012) were not covered in our data set. Artificial acetyl group transfers can be introduced in vitro by nonenzymatic reactions (Dormeyer et al., 2005) or by changes to a protein’s C terminus (Pasheva et al., 2004). To prevent false positives, nonenzymatic acetylation should therefore be considered in the evaluation of in vitro studies of acetylated peptides. Pehar et al. (2012) investigated ERs purified from cells overexpressing an ER membrane acetyl-CoA transporter, which led to increased ER acetylation compared to endogenous levels. For GRP78 and CALR their acetylation profiles were further investigated by analyses of overexpressed C terminus-tagged proteins. The differences between the results of Pehar and colleagues and the results in this study could therefore either be a result of false positives introduced by the high levels of acetyl-CoA in their cell system, a change in the C terminus of the two proteins, or simply reflect that our compendium is not yet comprehensive.
The highest fractions of nuclear acetylation sites were identified in the lymphoid-containing organs, which are known to have high mitotic activity. The acetylation level of K19 and K24 on histone H3 correlates with transcriptional activity, and intriguingly, these particular sites are most abundant in the lymphoid tissues, such as thymus and spleen, that primarily consist of dividing cells with active cell-cycle machinery. There is further evidence in our data set for this observation by a significant overrepresentation of lysine acetylation-modified transcription factors in the same lymphoid tissues. For a few transcription factors, such as p53, RELA, and STAT3, it is already known that they are functionally regulated by lysine acetylation (Gu and Roeder, 1997; Hoberg et al., 2006; Wang et al., 2005). Our identification of a total of 388 acetylation sites on transcription factors indicates that lysine acetylation may be a more ubiquitous mechanism of transcription factor regulation than previously appreciated. The notion that the modified sites are more prevalent in mitotic compared to postmitotic tissues, such as brain and heart muscle, further favors a functional role of these sites. In the lymphoid tissues we identify significant nuclear sequence motifs for lysine acetylation sites. We show that although lysine acetylation sites on histones exhibit the motif K-X-X-X-AcK-X-X-X-K, sites on nuclear proteins in general and on transcription factors, in particular, exhibit preference for the motif G-AcK-P. A glycine at position −1 has previously been demonstrated to be part of a recognition motif for the nuclear CBP/p300 lysine acetyl transferase (KAT) complex (Bannister et al., 2000), and a high-resolution crystal structure of the KAT domain of the STAGA transcription coactivator-complex member GCN5 bound to either coenzyme A or a histone H3 peptide also revealed binding specificity for a random coil structure with this motif (Rojas et al., 1999), thus supporting the functional relevance of glycine preceding lysine as part of a nuclear recognition motif.
Our finding that lysine acetylation sequence motifs are compartment specific favors the debated existence of subcellular compartment-specific KATs (Sadoul et al., 2011). Although a mitochondrial KAT is further supported by our identification of acetylation sites on four proteins encoded by mitochondrial DNA, we cannot rule out the possibility that the sequence motifs are due to compartment-specific protein expression patterns. The observed sequence preferences could also in part be mediated by lysine deacetylates (HDACs). HDAC inhibitors are promising therapeutic agents for treatment of a variety of cancers (Marks et al., 2001). Nevertheless, the underlying molecular mechanisms and effects of the clinically administered HDAC inhibitors are largely unknown. It would not only be of high medical relevance to probe patient tumor samples for aberrant acetylation patterns, but delineating downstream in vivo targets of HDAC inhibitors is of equal importance. Therefore, we are confident that the methodology we have developed and applied to investigate lysine acetylation sites in tissue samples will open new avenues for large-scale investigations of lysine acetylation patterns in disease tissues and for phenotyping samples from patients with cancer.
EXPERIMENTAL PROCEDURES
See Extended Experimental Proceduresfor detailed Experimental Procedures.
Rat Tissues and Peptide Extraction
Organs were quickly dissected from five Sprague-Dawley rats and snap frozen. Following heat inactivation tissues were homogenized and sonicated, proteins were acetone precipitated, resuspended in urea, and the concentration was determined. Proteins were reduced, alkylated, and digested with endoproteinase Lys-C followed by trypsin. Samples were desalted, and acetylated peptides were enriched using agarose-conjugated acetyl lysine antibody and separated by SCX fractionation before loading onto in-house packed C18 STAGE tips.
MS
Eluted peptide mixtures were reconstituted in 2% MeCN, 0.5% AcOH, 0.1% TFA, and separated by online reversed-phase C18 nanoscale liquid chromatography on a 15 cm × 75 µm column packed with ReproSil-Pur C18-AQ 3 µm resin. A nanoflow EASY-nLC system (Proxeon Biosystems, Odense, Denmark) was connected through a nano-electrospray ion source to the mass spectrometer. Peptides were separated by a linear gradient of increasing acetonitrile in 0.5% acetic acid for 180 min with a flow rate of 250 nl/min. The MS/MS was performed on a LTQ Orbitrap Velos mass spectrometer (Thermo Electron, Bremen, Germany) using a top10 HCD fragmentation method. Fullscan MS spectra were acquired at a target value of 1e6 and a resolution of 30,000, and the HCD-MS/MS spectra were recorded at a target value of 5e4 and with a resolution of 7,500 using normalized collision energy of 40%. Raw MS files were processed using the MaxQuant software (ver.1.0.14.7 and v.1.2.0.29). Precursor MS signal intensities were determined, and HCD MS/MS spectra were deisotoped and filtered such that only the ten most-abundant fragments per 100 m/z range were retained. Acetylated proteins were identified using the Mascot search algorithm. HCD-MS/MS spectra were searched with fixed modification of Carbamidomethyl-Cysteine and variable modifications of oxidation (M), acetylation (protein N-term), Gln- > pyro-Glu, and acetylation (K). Initial precursor ion tolerance was 7 ppm, MS/MS tolerance 0.02 Da, and strict tryptic specificity with maximum two missed cleavages were required. Label-free quantification and validation were performed in MaxQuant. Acetylated peptides were filtered based on Mascot score, PTM (Andromeda) score, precursor mass accuracy, peptide length, and summed protein score to achieve an estimated FDR <0.01 based on forward and reversed identifications. Hierarchical clustering was performed in Perseus (Max-Planck Institute of Biochemistry, Department of Proteomics and Signal Transduction, Munich) using Euclidian distance and average linkage clustering. Statistical evaluation of enriched Reactome pathways and GO terms for biological processes identified with the innate DB web tool (Lynn et al., 2008) were performed using a hypergeometric test and correcting for multiple testing by applying a Benjamini-Hochberg false discovery rate of 0.01. Sequence motif analysis was performed with ice-Logo using percent difference as scoring system and applying a significance cutoff of 0.01.
Enzymatic Experiments
The coding sequences for human ALDOB and GPD1were cloned into pDEST-15, point mutations were introduced, and constructs were transformed into Rosetta cells. Recombinant proteins were extracted and incubated with glutathione Sepharose beads. The concentrations of purified proteins were determined, and the amino acid sequences were checked by MS. Enzymatic activities were determined as initial velocity measurements by monitoring NADH decrease as a function of time at 340 nm absorbance reads on a Biotek Synergy H4 reader.
Supplementary Material
ACKNOWLEDGMENTS
We thank all members of the Department for Proteomics at the Center for Protein Research for scientific discussions and Dr. Nikolai Nordsborg for help with collecting human samples. The Center for Protein Research is partly supported by a generous donation from the Novo Nordisk Foundation. This work was supported by the Seventh Framework Programme of the European Union (Contract no. 262067-PRIME-XS) and the research career programme Sapere Aude from The Danish Council for Independent Research for Medical Sciences (to A.L. and J.V.O.). A.L. and J.V.O. designed all of the experiments, A.S. and C.L. coordinated animal and human sample handling, respectively, A.L. completed all sample preparation, A.L. performed MS analysis, B.T.W. provided input on SCX fractionation, C.D.K. provided input on Q Exactive measurements, K.L. generated protein interaction networks, D.B.B.-J. and T.S. performed enzyme experiments, A.D. made the MySQL database, A.L. and J.V.O. performed data analysis, B.T.W. and C.C. provided input on manuscript, and A.L. and J.V.O. wrote the paper.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes Extended Experimental Procedures, six figures, raw MS data, and five tables and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2012.07.006.
WEB RESOURCES
The URLs for data presented herein are as follows:
CPR PTM Resource, http://cpr1.sund.ku.dk/cgi-bin/PTM.pl
Mascot, http://www.matrixscience.com
Online Mendelian Inheritance in Man (OMIM), http://omim.org/
Reflect, http://reflect.cbs.dtu.dk/index.html
TRANCHE, http://www.proteomecommons.org
REFERENCES
- Ahn BH, Kim HS, Song S, Lee IH, Liu J, Vassilopoulos A, Deng CX, Finkel T. A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis. Proc. Natl. Acad. Sci. USA. 2008;105:14447–14452. doi: 10.1073/pnas.0803790105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. The Gene Ontology Consortium. Gene ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Miska EA, Görlich D, Kouzarides T. Acetylation of importin-alpha nuclear import factors by CBP/p300. Curr. Biol. 2000;10:467–470. doi: 10.1016/s0960-9822(00)00445-0. [DOI] [PubMed] [Google Scholar]
- Bergholdt R, Størling ZM, Lage K, Karlberg EO, Olason PI, Aalund M, Nerup J, Brunak S, Workman CT, Pociot F. Integrative analysis for finding genes and networks involved in diabetes and other complex diseases. Genome Biol. 2007;8:R253. doi: 10.1186/gb-2007-8-11-r253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- Colaert N, Helsens K, Martens L, Vandekerckhove J, Gevaert K. Improved visualization of protein consensus sequences by iceLogo. Nat. Methods. 2009;6:786–787. doi: 10.1038/nmeth1109-786. [DOI] [PubMed] [Google Scholar]
- Croft D, O’Kelly G, Wu G, Haw R, Gillespie M, Matthews L, Caudy M, Garapati P, Gopinath G, Jassal B, et al. Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Res. 2011;39(Database issue):D691–D697. doi: 10.1093/nar/gkq1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davit-Spraul A, Costa C, Zater M, Habes D, Berthelot J, Broué P, Feillet F, Bernard O, Labrune P, Baussan C. Hereditary fructose intolerance: frequency and spectrum mutations of the aldolase B gene in a large patients cohort from France—identification of eight new mutations. Mol. Genet. Metab. 2008;94:443–447. doi: 10.1016/j.ymgme.2008.05.003. [DOI] [PubMed] [Google Scholar]
- de Godoy LM, Olsen JV, Cox J, Nielsen ML, Hubner NC, Fröhlich F, Walther TC, Mann M. Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature. 2008;455:1251–1254. doi: 10.1038/nature07341. [DOI] [PubMed] [Google Scholar]
- Dormeyer W, Ott M, Schnölzer M. Probing lysine acetylation in proteins: strategies, limitations, and pitfalls of in vitro acetyltransferase assays. Mol. Cell. Proteomics. 2005;4:1226–1239. doi: 10.1074/mcp.M500047-MCP200. [DOI] [PubMed] [Google Scholar]
- Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- Gupta MP, Samant SA, Smith SH, Shroff SG. HDAC4 and PCAF bind to cardiac sarcomeres and play a role in regulating myofilament contractile activity. J. Biol. Chem. 2008;283:10135–10146. doi: 10.1074/jbc.M710277200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschey MD, Shimazu T, Goetzman E, Jing E, Schwer B, Lombard DB, Grueter CA, Harris C, Biddinger S, Ilkayeva OR, et al. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 2010;464:121–125. doi: 10.1038/nature08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoberg JE, Popko AE, Ramsey CS, Mayo MW. IkappaB kinase alpha-mediated derepression of SMRT potentiates acetylation of RelA/p65 by p300. Mol. Cell. Biol. 2006;26:457–471. doi: 10.1128/MCB.26.2.457-471.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, Villén J, Haas W, Sowa ME, Gygi SP. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 2010;143:1174–1189. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Acetylation: a regulatory modification to rival phosphorylation? EMBO J. 2000;19:1176–1179. doi: 10.1093/emboj/19.6.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratchmarova I, Kalume DE, Blagoev B, Scherer PE, Podtelejnikov AV, Molina H, Bickel PE, Andersen JS, Fernandez MM, Bunkenborg J, et al. A proteomic approach for identification of secreted proteins during the differentiation of 3T3-L1 preadipocytes to adipocytes. Mol. Cell. Proteomics. 2002;1:213–222. doi: 10.1074/mcp.m200006-mcp200. [DOI] [PubMed] [Google Scholar]
- Lage K, Karlberg EO, Størling ZM, Olason PI, Pedersen AG, Rigina O, Hinsby AM, Tümer Z, Pociot F, Tommerup N, et al. A human phenome-interactome network of protein complexes implicated in genetic disorders. Nat. Biotechnol. 2007;25:309–316. doi: 10.1038/nbt1295. [DOI] [PubMed] [Google Scholar]
- Lage K, Hansen NT, Karlberg EO, Eklund AC, Roque FS, Donahoe PK, Szallasi Z, Jensen TS, Brunak S. A large-scale analysis of tissue-specific pathology and gene expression of human disease genes and complexes. Proc. Natl. Acad. Sci. USA. 2008;105:20870–20875. doi: 10.1073/pnas.0810772105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lage K, Møllgård K, Greenway S, Wakimoto H, Gorham JM, Workman CT, Bendsen E, Hansen NT, Rigina O, Roque FS, et al. Dissecting spatio-temporal protein networks driving human heart development and related disorders. Mol. Syst. Biol. 2010;6:381. doi: 10.1038/msb.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe CB, Kellis M, Siepel A, Raney BJ, Clamp M, Salama SR, Kingsley DM, Lindblad-Toh K, Haussler D. Three periods of regulatory innovation during vertebrate evolution. Science. 2011;333:1019–1024. doi: 10.1126/science.1202702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundby A, Secher A, Lage K, Nordsborg NB, Dmytriyev A, Lundby C, Olsen JV. Quantitative maps of protein phosphorylation sites across 14 different rat organs and tissues. Nat. Commun. 2012 doi: 10.1038/ncomms1871. Published online June 6, 2012. http://dx.doi.org/10.1038/ncomms1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn DJ, Winsor GL, Chan C, Richard N, Laird MR, Barsky A, Gardy JL, Roche FM, Chan TH, Shah N, et al. InnateDB: facilitating systems-level analyses of the mammalian innate immune response. Mol. Syst. Biol. 2008;4:218. doi: 10.1038/msb.2008.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmström J, Beck M, Schmidt A, Lange V, Deutsch EW, Aebersold R. Proteome-wide cellular protein concentrations of the human pathogen Leptospira interrogans. Nature. 2009;460:762–765. doi: 10.1038/nature08184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks P, Rifkind RA, Richon VM, Breslow R, Miller T, Kelly WK. Histone deacetylases and cancer: causes and therapies. Nat. Rev. Cancer. 2001;1:194–202. doi: 10.1038/35106079. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Lomb DJ, Haigis MC, Guarente L. SIRT5 Deacetylates carbamoyl phosphate synthetase 1 and regulates the urea cycle. Cell. 2009;137:560–570. doi: 10.1016/j.cell.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen JV, Macek B, Lange O, Makarov A, Horning S, Mann M. Higher-energy C-trap dissociation for peptide modification analysis. Nat. Methods. 2007;4:709–712. doi: 10.1038/nmeth1060. [DOI] [PubMed] [Google Scholar]
- Olsen JV, Schwartz JC, Griep-Raming J, Nielsen ML, Damoc E, Denisov E, Lange O, Remes P, Taylor D, Splendore M, et al. A dual pressure linear ion trap Orbitrap instrument with very high sequencing speed. Mol. Cell. Proteomics. 2009;8:2759–2769. doi: 10.1074/mcp.M900375-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen JV, Vermeulen M, Santamaria A, Kumar C, Miller ML, Jensen LJ, Gnad F, Cox J, Jensen TS, Nigg EA, et al. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal. 2010;3:ra3. doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- Ou X, Ji C, Han X, Zhao X, Li X, Mao Y, Wong LL, Bartlam M, Rao Z. Crystal structures of human glycerol 3-phosphate dehydrogenase 1 (GPD1) . J. Mol. Biol. 2006;357:858–869. doi: 10.1016/j.jmb.2005.12.074. [DOI] [PubMed] [Google Scholar]
- Pasheva E, Sarov M, Bidjekov K, Ugrinova I, Sarg B, Lindner H, Pashev IG. In vitro acetylation of HMGB-1 and-2 proteins by CBP: the role of the acidic tail. Biochemistry. 2004;43:2935–2940. doi: 10.1021/bi035615y. [DOI] [PubMed] [Google Scholar]
- Pehar M, Lehnus M, Karst A, Puglielli L. Proteomic assessment shows that many ER-resident proteins are targeted by N{varepsilon}-lysine acetylation in the lumen of the organelle and predicts broad biological impact. J. Biol. Chem. 2012;287:22436–22440. doi: 10.1074/jbc.C112.362871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellettieri J, Sánchez Alvarado A. Cell turnover and adult tissue homeostasis: from humans to planarians. Annu. Rev. Genet. 2007;41:83–105. doi: 10.1146/annurev.genet.41.110306.130244. [DOI] [PubMed] [Google Scholar]
- Rodgers JT, Lerin C, Gerhart-Hines Z, Puigserver P. Metabolic adaptations through the PGC-1 alpha and SIRT1 pathways. FEBS Lett. 2008;582:46–53. doi: 10.1016/j.febslet.2007.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas JR, Trievel RC, Zhou J, Mo Y, Li X, Berger SL, Allis CD, Marmorstein R. Structure of Tetrahymena GCN5 bound to coenzyme A and a histone H3 peptide. Nature. 1999;401:93–98. doi: 10.1038/43487. [DOI] [PubMed] [Google Scholar]
- Sadoul K, Wang J, Diagouraga B, Khochbin S. The tale of protein lysine acetylation in the cytoplasm. J. Biomed. Biotechnol. 2011;2011:970382. doi: 10.1155/2011/970382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltiel AR. You are what you secrete. Nat. Med. 2001;7:887–888. doi: 10.1038/90911. [DOI] [PubMed] [Google Scholar]
- Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- Schwer B, Eckersdorff M, Li Y, Silva JC, Fermin D, Kurtev MV, Giallourakis C, Comb MJ, Alt FW, Lombard DB. Calorie restriction alters mitochondrial protein acetylation. Aging Cell. 2009;8:604–606. doi: 10.1111/j.1474-9726.2009.00503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Jean M, Blonski C, Sygusch J. Charge stabilization and entropy reduction of central lysine residues in fructose-bisphosphate aldolase. Biochemistry. 2009;48:4528–4537. doi: 10.1021/bi8021558. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39(Database issue):D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Zhang Y, Yang C, Xiong H, Lin Y, Yao J, Li H, Xie L, Zhao W, Yao Y, et al. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science. 2010;327:1004–1007. doi: 10.1126/science.1179687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Cherukuri P, Luo J. Activation of Stat3 sequence-specific DNA binding and transcription by p300/CREB-binding protein-mediated acetylation. J. Biol. Chem. 2005;280:11528–11534. doi: 10.1074/jbc.M413930200. [DOI] [PubMed] [Google Scholar]
- Weinert BT, Wagner SA, Horn H, Henriksen P, Liu WR, Olsen JV, Jensen LJ, Choudhary C. Proteome-wide mapping of the Drosophila acetylome demonstrates a high degree of conservation of lysine acetylation. Sci. Signal . 2011;4:ra48. doi: 10.1126/scisignal.2001902. [DOI] [PubMed] [Google Scholar]
- Yang L, Vaitheesvaran B, Hartil K, Robinson AJ, Hoopmann MR, Eng JK, Kurland IJ, Bruce JE. The fasted/fed mouse metabolic acetylome: N6-acetylation differences suggest acetylation coordinates organ-specific fuel switching. J. Proteome Res. 2011;10:4134–4149. doi: 10.1021/pr200313x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, et al. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.