Abstract
Most aspects of mammalian function display circadian rhythms driven by an endogenous clock. The circadian clock is operated by genes and comprises a central clock in the brain that responds to environmental cues and controls subordinate clocks in peripheral tissues via circadian output pathways. The central and peripheral clocks coordinately generate rhythmic gene expression in a tissue-specific manner in vivo to couple diverse physiological and behavioral processes to periodic changes in the environment. However, as the world industrialized, activities that disrupt endogenous homeostasis with external circadian cues have increased. This change in lifestyle has been linked to increased risk of diseases in all aspects of human health, including cancer. Studies in humans and animal models have revealed that cancer development in vivo is closely associated with the loss of circadian homeostasis in energy balance, immune function and aging that are supported by cellular functions important for tumor suppression including cell proliferation, senescence, metabolism and DNA damage response. The clock controls these cellular functions both locally in cells of peripheral tissues and at the organismal level via extracellular signaling. Thus, the hierarchical mammalian circadian clock provides a unique system to study carcinogenesis as a deregulated physiological process in vivo. The asynchrony between host and malignant tissues in cell proliferation and metabolism also provides new and exciting options for novel anti-cancer therapies.
Introduction
Circadian rhythms in physiological and behavioral processes in plants and animals have been known since fourth century BC. These rhythms were originally attributed to a passive response of organisms to diurnal changes in external light cues, but were later discovered to be generated by an endogenous clock in all species studied1. In mammals, circadian rhythms are generated by a central clock located in the hypothalamus suprachiasmatic nucleus (SCN) that constantly synchronizes with environmental cues via circadian input pathways and controls the peripheral clocks through circadian output pathways2,3.
Both central and peripheral clocks are operated by the same set of circadian genes expressed in all tissues studied. The molecular clockwork in mammals has been described in detail in several recent reviews4–6. Briefly, it is based on autoregulatory transcriptional feedback loops driven by the heterodimer of bHLH-PAS transcription factors BMAL1/CLOCK or BMAL1/NPAS2 that activate their downstream transcriptional repressor targets Cryptochrome (Cry1,2) and Period (Per1–3) at the beginning of a circadian day. The accumulation of PER and CRY proteins in the cytoplasm at the end of a circadian day, controlled by the SCF (Skp1-Cullin-F-box protein) E3 ubiquitin ligase complexes, casein kinase 1ε/δ (CK1ε/δ) and AMP kinase (AMPK), leads to the formation of a PER/CRY repressor complex that translocates into the nucleus at the beginning of a circadian night to inhibit the activity of BMAL1/CLOCK or BMAL1/NPAS2 heterodimers and recruit the transcriptional termination complex to the Per and Cry genes7. In addition, the transcription of Bmal1 is alternatively regulated by its own transcription targets, the nuclear receptors Rev-erbα/β (the repressors) and Rorα (the activator)8–10. The multiple interlocked autoregulatory feedback loops result in a robust circadian variation in the expression and activity of Bmal1 over a 24 hour period, providing a driving force for circadian oscillation of the molecular clockwork.
The circadian regulators also target clock-controlled genes to generate circadian rhythms in all major cellular processes in both SCN neurons and peripheral organs, resulting in a rhythmic expression of 3–10% of all mRNAs expressed in a given tissue due to time-dependent interactions between the circadian regulators with specific gene promoter sequences, transcription factors, or transcriptional initiation, elongation and termination complexes, as well as the key factors controlling chromatin remodeling7,11–15. The clock-controlled genes usually do not share overlapping expression patterns between tissues, suggesting a key role for the circadian clock in controlling tissue-specific function in vivo. Clock-controlled genes expressed in all tissues studied include the key regulators of cell proliferation, metabolism, senescence and DNA damage response16–23.
The molecular clock in SCN neurons and peripheral tissues can be entrained or phase-shifted by cellular signaling. The most potent circadian time cue for the SCN clock is light which is received by a subset of melanopsin-expressing retinal ganglion cells and transmitted directly to the SCN neurons via the retinohypothalamic tract (RHT). Upon activation, the RHT produces neurotransmitters that activate a cascade of signal transduction events leading to circadian phase resetting24,25. Although the SCN clock on its own is capable of generating autonomic circadian outputs, the constant coupling of the central clock with environmental cues provides a survival advantage by synchronizing daily physiology and behavior with local time cues25,26. A shift in environmental cues, such as traveling across several times zones on an aircraft, induces a phase-shift in the central clock and the subsequent SCN-controlled phase-shift in peripheral tissues via circadian output pathways to reestablish the endogenous circadian homeostasis to the new local time. The number of days needed to fully adjust to the new time zone is dependent on the number of time zones crossed during the trip. Constant back-and-forth phase-shifts of environmental light cues resulted from rotating work schedules or chronic jet-lag, disrupts endogenous circadian homeostasis by uncoupling the central and peripheral clock coordination27–29.
The best studied circadian output pathways include the autonomic nervous system (ANS) and neuroendocrine system (NES) that control all aspects of mammalian physiology as well as the peripheral clocks via cellular signaling. The rhythmic activities of these systems provide a mechanism for the central clock to control peripheral tissues directly and indirectly via peripheral clocks24,29–33.
Dramatic changes in lifestyles since the industrial revolution due to increased use of artificial lighting, nightshift working schedules, or rapid long-distance transmeridian travelling have led to frequent disruption of endogenous circadian homeostasis in modern societies. These changes in lifestyle are coupled with a significant increase in the risk of diseases in all aspects of human health including cancer.
Circadian dysfunction promotes cancer development in humans
Circadian disruption is an independent cancer risk factor for humans
Recent epidemiology studies have linked circadian disruption to increased susceptibility of cancer development in all key organ systems in humans. The cancers observed from these studies included breast, ovarian, lung, pancreatic, prostate, colorectal and endometrial cancers, non-Hodgkin's lymphoma (NHL), osteosarcoma, acute myeloid leukemia (AML), head and neck squamous cell carcinoma, and hepatocellular carcinoma34–49. Circadian dysfunction-induced cancer risk increases with the number of years, the frequency of rotating work schedules, and the number of hours per week working at night among human night-shift workers45,46,50–52. Together, these findings suggest that loss of circadian homeostasis could be an independent cancer risk factor for humans53. Due to the prevalence of night shift-work schedules in modern societies, the World Health Organization’s International Agency for Research on Cancer (IARC) listed "shiftwork that involves circadian disruption" as a probable carcinogen in 2007.
Circadian disruption is associated with poor prognosis and early mortality of cancer patients
Loss of circadian homeostasis not only promotes cancer development, but is also associated with poor performance to anticancer treatments and early mortality among cancer patients. After adjusting for other factors that might affect survival, circadian rhythm in salivary and serum cortisol levels as well as daily rest/activity patterns are used as independent prognosis factors for survival and therapeutic response of patients with metastatic breast, lung and colorectal cancers54–61.
Disruption of the molecular clockworks in human cancers
Ample evidence has linked dysfunction of the molecular clock with pathogenesis of human cancers (Table 1). The mechanisms of dysregulation of the core circadian genes in human cancers discovered to date include epigenetic silencing by promoter methylation, deregulation at the transcriptional and post-transcriptional levels, and structural variations of clock proteins due to circadian gene polymorphisms.
Table 1.
Deregulation of the Core Clock Genes in Human Cancers
| Cancer Type | Circadian genes Deregulated | Deregulated Targets | Cellular Functions Affected | References |
|---|---|---|---|---|
| Breast cancer | Bmal1, Clock, Cry1, Cry2, Per1, Per2, Per3, and Npas2 | BCCIP, BCL2, BRAC1, ERα, Estrogen, EXO1, cAMP, CDKN1A, Cortisol, CyclinD1, c-ERBB2, GADD45A, HERC5, Melatonin, MCM5, MSH2, p21, p38, p53, PARP, PKA, PPP1R15A, SIRT1, SUMO1, TERT, TIP60, and UBA1 | Apoptosis, cell cycle control, chromatin remodeling, DNA damage repair, and teleomere length | 38, 39, 42, 44,45, 48–50, 53, 55, 63, 66, 74, 80, 92, 96, 99, 100, 102, 104–107 |
| Acute Lymphocytic Leukemia (ALL) | Bmal1 and Clock | Catalase, c-MYC, and p300 | Cell cycle control and chromatin remodeling | 78 |
| Acute Myeloid Leukemia (AML) | Bmal1, Per1, Per2, and Per3 | Catalase, c-MYC, and p300 | Cell cycle control and chromatin remodeling | 78 |
| Chronic Lymphocytic Leukemia (CLL) | Cry1 and Per2 | ZAP70 | Cell cycle control, chromatin remodeling, and DNA damage repair | 86, 91 |
| Chronic Myeloid Leukemia (CML) | Bmal1, Cry1, Cry2, Per1, Per2, and Per3 | c-MYC, CyclinB1, and p53 | Apoptosis, cell cycle control, and chromatin remodeling | 75,94 |
| Colorectal cancer | Clock, Per1, Per2, and Per3 | Cortisol, ATM, EGFR, ER-β, EXO1, IL-6, MSH2, p53, PARP, TGFα, and TNFα | Chromatin remodeling and DNA damage repair | 34, 43, 54, 58, 62, 82, 98, 101, 102 |
| Endometrial cancer | Cry1, Per1, Per2, and Per3 | Melatonin | Chromatin remodeling | 36, 42, 76 |
| Glioma | Cry1,Cry2, Per1, Per2, and Per3 | N/A | Apoptosis and cell cycle control | 64, 65, 90 |
| Head and neck squamous cell carcinoma | Bmal1, CKIε, Cry1, Cry2, Per1, Per2, Per3, and Tim | TIP60 | N/A | 52, 70, 85, 104 |
| Hepatocellular carcinoma | Cry2, Per1, Per2, Per3, and Tim | CDC2, CyclinB1, EZH2, GR, IGF-1, and WEE1 | Cell cycle control, chromatin remodeling | 47, 71 |
| Lung cancer | Clock, Per1, Per2, and Per3 | Cortisol and TIP60 | Cell cycle control, chromatin remodeling, DNA damage repair | 43, 56, 57, 72, 98, 104 |
| Malignant pleural mesothelioma | Bmal1, Cry2, Per1, Per3, Npas2, Rev-erbα, Rev-erbβ and Tim | CASP3, CyclinB, CyclinE, p21WAF1/CIP1, and WEE1 | Apoptosis, cell cycle control, chromatin remodeling, and deregulated chemotherapy drug response | 83, 87 |
| Non-Hodgkin's Lymphoma (NHL) | Npas2 | DMC1, EXO1, and MSH2 | Cell cycle control, DNA damage repair, and immune deficiency | 37, 79, 102 |
| Diffuse large B-cell Lymphoma | Bmal1 and Clock | Catalase, c-MYC, and p300 | Chromatin remodeling | 78 |
| Osteosarcoma | CK1ε and Per2 | CASP3, CyclinB, and CyclinA2 | Apoptosis and cell cycle control | 40, 88 |
| Ovarian cancer | Bmal1, CK1ε, Clock, Cry1, Cry2, Per1, Per2, and Per3 | Cortisol | Apoptosis, cell cycle control and deregulated chemotherapy drug response | 39, 84, 103 |
| Pancreatic cancer | Bmal1, CK1ε, Clock, Cry1, Cry2, DEC1, Per1, Per2, Per3, Tim, and Tipin | BCL-XL, CDC2, CyclinB1, TNF-α, and USP30 | Apoptosis, cell cycle control and chromatin remodeling | 68, 69, 77, 89 |
| Prostate cancer | Bmal1, CK1ε, Clock, Cry1, Cry2, Npas2, Per1, Per2, and Per3 | Melatonin, SIRT1, and TIP60 | Apoptosis, cell cycle control, DNA damage repair and transactivation of AR | 35, 43, 45, 67, 73, 81, 97, 104 |
When compared to normal host tissues, decreased expression and polymorphism of the core circadian genes Per1, Per2 and Per3 are frequently found in human breast, endometrial, prostate, pancreatic, colorectal and non-small cell lung cancers (NSCLC), as well as hepatocellular carcinoma, neck squamous cell carcinoma, glioma, AML and chronic myeloid lymphoma (CML)62–73. In CML, breast, endometrial and NSCLC, this deregulation is often linked to hypermethylation of CpG islands or aberrant acetylation in the promoters of Per genes, which leads to gene silencing72,74–77. Other core circadian genes are also frequently deregulated or silenced in human cancers. For example, the epigenetic inactivation of Bmal1 is often linked to hematologic malignancies including NHL, diffuse large B-cell lymphoma, acute lymphocytic leukemia (ALL) and AML, whereas polymorphisms in Clock, Cry1, Cry2 and Npas2 gene are frequently found associated with increased risk or recurrence of NHL, AML, endometrial ovarian, colorectal and breast cancers76,78–82. In most studies examining the role of the molecular clock in human cancers, deregulation or polymorphism of multiple or all core circadian genes is observed. For example, deregulation or polymorphism of Per1, Per2 and Per3, Clock, Bmal1, Cry1, Cry2, Clock, Npas2, and/or CK1ε are frequently found in human CML, prostate, pancreatic and epithelial ovarian cancers, leukemia, pleural mesothelioma, hepatocellular carcinoma, glioma and neck squamous cell carcinoma69,75,81,83–90. Based on these discoveries, a combined deregulation of Cry1 and Bmal1, or Cry1 and Per2, has been suggested as a negative prognostic marker for epithelial ovarian cancer and CML, respectively84,86,91.
Deregulation of the core circadian genes in human cancers is closely associated with a constitutive activation of intracellular inflammatory and oncogenic signaling pathways including constitutive activation of p38, c-Myc, NF-kB, Bcl-XL, and protein kinase A (PKA)68,78,92–94, aberrant chromatin remodeling, deregulation of inflammatory cytokines, catalase, Tip60, telomerase, PARP [poly (ADP-ribose) polymerase], Sirt1 and p30078,92,93,95–97, over-expression of ERα, G1 and S-phase cyclins and suppression of tumor suppressors ATM, p53, p21 and Wee183,94,98–100. Deregulation of the molecular clock is correlated with the loss of control in cell proliferation, metabolism, DNA replication and repair, senescence, apoptosis, DNA damage response and increased drug resistance in all types of human cancer cells studied (Table 1)68,78,83,88,94,96,98,99,101–107.
Central clock dysfunction increases cancer risk in humans
In the hierarchical organization of the mammalian circadian clock, the peripheral clock can only sense changes in environmental light cues via central clock-controlled circadian output pathways. Thus, central clock dysfunction induced by frequent back-and-forth phase-shifts of environmental cues may play a key role in promoting cancer development among human night-shift workers by disrupting the homeostasis of neuroendocrine function95,108–110. This hypothesis is supported by the facts that visually impaired people who are insensitive to changes in environmental circadian light cues and largely or completely depend on a free-running endogenous clock to organize their daily physiology display a lower cancer risk compared to the general population111–113.
In summary, ample evidence obtained from human studies suggests that the mammalian circadian clock plays a key role in tumor suppression. Therefore, disruption of circadian homeostasis of mammalian physiology is a novel risk factor for cancer (Table 1).
Circadian disruption promotes cancer development in animal models
The central clock suppresses tumor initiation and progression in animal models
The pioneer studies to investigate the role of circadian disruption in cancer development using experimental animal models started in late 1960s. These studies demonstrate that disruption of circadian endocrine rhythms either by constant light exposure or pinealectomy increases spontaneous and carcinogen-induced mammary gland and hepatocellular carcinogenesis in rodents114–116. Similar experiments conducted in recent years have also shown that disruption of circadian homeostasis by a short period of back-and-forth or consecutive phase advance shifts of environmental light cues, or by constant light exposure significantly accelerate tumor growth in animals117–121. Compared to sham-operated animals, mice lacking a central clock due to surgical ablation of the SCN were unable to maintain circadian rhythmicity in locomotor activity, body temperature and immune function. This loss of circadian homeostasis in SCN-lesioned mice is coupled with a significant decrease in survival time due to increased rate of tumor growth compared to control tumor bearing mice with an intact SCN122. Together, these studies agree with the findings from human studies in that circadian dysfunction increases the risk of cancer by demonstrating that disruption of the central circadian clock promotes cancer development and progression in rodents.
Variation in cancer phenotypes reported for circadian gene-mutant mouse models
The role for mammalian circadian genes in cancer genetics was first reported in 2002 in a study showing that mice expressing a mutant PER2 (Per2m/m) defective in PER2-mediated protein/protein interactions due to a 85 amino acids in-frame deletion in the PAS domain of Per2 gene display multiple tumor-prone phenotypes including increased spontaneous and γ-radiation-induced lymphoma, hyperplastic growth in salivary and preputial glands, resistance to radiation-induced apoptosis in thymocytes, and deregulation of key tumor suppressors, cyclins and proto-oncogenes, such as p53, Gadd45α, Cyclin D1, Cyclin A, c-Myc and Mdm2123,124. The same study also shows that Per2-null (Per2−/−) mice display similar cancer prone phenotypes as Per2m/m mice (Supplemental data)124.
In contrast, other reports indicate that mice deficient in other core circadian genes either lack neoplastic phenotypes or are tumor resistant. For example, mice lacking Bmal1 (Bmal1−/−) show significant reduced lifespan and premature aging, but not spontaneous tumor development125. Hepatocytes in mice lacking both Cry1 and Cry2 (Cry1−/−;Cry2−/−) proliferated slower than wild-type (Wt) hepatocytes in the first 72 hours immediately after partial hepatectomy126. Ablation of both Cry1 and Cry2 reduced cancer risk for p53-null mice127. Clock-mutant (Clockm/m) and Cry1−/−;Cry2−/− mice did not show predisposition to cancer in response to a low dose of γ-irradiation128,129. Furthermore, MEFs isolated from Cry1−/−;Cry2−/− mice do not show deficiencies in γ-radiation-induced cell cycle arrest, whereas Clockm/m MEFs show lower levels of DNA synthesis and cell proliferation than wild-type controls19,128. Together, the studies described above led to the conclusion that the cancer-prone phenotypes discovered in Per2m/m and Per2−/− mice are the result of loss of a ”none-clock function” of the Per2 gene but not the function of the mammalian circadian clock6.
The molecular clock suppresses tumor development in mice
We e suggest that a more detailed analysis of the available information supports a direct role for the molecular clock in tumor suppression in mice.
First, the observation of a temporary slowdown in Cry1−/−;Cry2−/− hepatocyte proliferation immediately after partial hepatectomy cannot predict whether Cry1−/−;Cry2−/− mice are tumor-resistant under normal physiological conditions. Surgical stress caused by partial hepatectomy can suppress the growth of hepatocellular carcinoma in mouse livers until the third post-operative day130. In fact, compared to wild-type controls, most mouse models prone to spontaneous hepatocellular carcinoma show an initial delay in hepatocyte proliferation after partial hepatectomy. For example, compared to wild-type controls, mice lacking the nuclear receptor FXR display a delay in hepatocyte proliferation after partial hepatectomy until the 9th post-operative days. However, Fxr−/− mice quickly regain the ability of rapid hepatocyte proliferation and develop malignant liver tumors after 12 months of age131,132. Since Cry1−/−;Cry2−/− mice show significantly dampened Bmal1 expression and deregulation of the Mitogen-activated Protein Kinase/Extracellular Signal-regulated Kinase (MAPK/ERK) pathway in the liver133–135, it would be important to examine whether the temporary delay in hepatocyte proliferation in Cry1−/−;Cry2−/− mice after partial hepatectomy is caused by deficiencies in cellular signaling essential for G1 cell cycle initiation136,137.
Second, the conclusions that Per-mutants are cancer-prone but Cry-mutants are tumor resistant are confounded by significant differences in the phenotypes of control wild-type and p53-null mice but not Per- and Cry-mutant mice in different studies29,124,127,128. For example, the conclusion that a single 4-Gy sublethal dose of γ-irradiation led to a similar rate of decline in the survival of irradiated wild-type and Cry1−/−;Cry2−/− mice was based on an unusual sensitivity of wild-type mice to γ-irradiation128, which was not observed in the earlier study indicating increased sensitivity of the Per2-mutant mice to the same 4 Gy sublethal γ-radiation124.
Similarly, discrepancies on the effects of Per or Cry gene ablation on the survival and tumor developing rate of p53-null mice can also be attributed to a significant difference in the average and maximal lifespans of p53-null mice29,127, which vary from 15 to 30 and 28 to 60 weeks of age, respectively, as reported in different studies138–145. In accordance with our studies demonstrating that chronic back-and-forth jet-lag significantly accelerated tumor development and reduced survival of p53-null mice29, mice harboring both a mutant p53 correspondent to the p53R175H hotspot mutation in humans and a mutant Per2 allele (Per2S662G)146, which leads to a short and phase-advanced behavior rhythm among human patients suffering from familial advanced sleep phase syndrome due to a single serine to glycine mutation within the CKIε binding region in PER2147, also display increased tumor incidence and decreased survival compared to p53R175H mice148. These findings suggest strongly that circadian dysfunction cooperates with loss of p53 to promote tumor development.
Different research teams have also independently reached the same conclusion that Per- and Cry-mutant mice display a similar neoplastic growth of osteoblasts in bone149,150, and that disruption of Period genes increases cancer risk in mice29,124,148,151–153. Therefore, it would be important to verify the role of Per and Cry genes in cancer risk since Per and Cry genes are both indispensable for operating the same negative loop in the molecular clock and display the same deregulated behavioral phenotypes123,154,155. In addition, despite of a high Per2 mRNA expression, PER2 protein is not detected in peripheral tissues of Cry1−/−;Cry2−/− mice156. Indeed, when Per- and Cry-mutant mice of the same mouse strain were studied under the exact same conditions, the two mouse models display the same increased rate of tumor development in the skeletal, immune, reproductive and digestive systems when kept in steady-state 12 hour light/12 hour dark (24 hour LD) condition, in response to a sub-lethal dose of γ-radiation, or treated with chronic jet-lag after γ-radiation29. Together, the evidence obtained from studying mice lacking Per or Cry under the same conditions strongly argues that as found in human studies, the Cryptochrome genes also function in tumor suppression in rodents.
Third, Clockm/m mice show a significant decrease in survival compared to wild-type controls at 80 weeks of age after a sub-lethal dose of γ-radiation. No significant difference in tumor incidence or the rate of radiation-induced apoptosis between wild-type and Clockm/m splenocytes was reported. The decrease in the survival of irradiated Clockm/m mice was attributed to accelerated aging but not tumor development129. Since only the apoptotic response of in vitro cultured Clockm/m splenocytes to a lethal not a sub-lethal dose of γ-radiation was studied, and that the similar aging phenotypes displayed by irradiated Clockm/m mice are also commonly observed in other irradiated circadian gene-mutant mouse models29,124,128,129, a role for Clock in tumor suppression cannot be ruled out without examining the cancer risk of Clock−/− and Clock−/−;Npas2−/− mice as well as irradiated Clockm/m mice after 80 weeks of age. This is because aging is considered a primary risk factor for cancer157, and the mammalian CLOCK may play a direct role in DNA damage repair after γ-radiation158. In addition, CLOCK and NPAS2 play overlapping roles in the molecular clock159,160. Unlike Per2m/m and Per2−/− mice that show a similar deregulated circadian phenotype123,124,155,161, Clockm/m and Clock−/− mice display different circadian behavioral phenotypes and patterns of deregulation of gene expression in somatic cells162–165.
Fourth, the observations of aggressive aging phenotypes and lack of tumor incidence among Bmal1-null mice are not sufficient to exclude a role for Bmal1 in tumor suppression. When kept in stable 24 hour LD cycles, most spontaneous tumors are identified after 50 weeks of age in circadian gene-mutant mice29, which is an age most Bmal1−/− mice would not live to125. After being treated with a sublethal dose of γ-radiation, although the maximal life-span of Bmal1−/− mice was further decreased, about 6–7% of them developed malignant lymphomas before or at about 40 weeks of age. This rate of tumor development in irradiated Bmal1−/− mice is very similar to the reported rate of tumor development among irradiated Per- and Cry-mutant mice29,124,128. In addition, Bmal1+/− mice that have a similar life-span as Per- and Cry-mutants display the same rate of spontaneous and radiation-induced tumor development in the same organ systems as Per- and Cry-mutant mice29.
Bmal1-null mice also display a delay in anagen progression and decreased cell proliferation in secondary hair germ cells, which is coupled with increased G1 cell cycle block as shown by decreased levels of RB phosphorylation, increased expression of cyclin-dependent kinase inhibitors p21WAF1/CIP1 and p16Ink4A, and accelerated epidermal aging166. In contrast, mice lacking Bmal1 only in keratinocytes show constitutive elevation of cell proliferation and intracellular redox levels as well as deregulated UVB-induced DNA damage in the epidermis at linear growth age after wean22. However, in a different study, the same keratinocyte-specific Bmal1−/− mouse model was reported to display aging phenotypes in the skin starting from 10 months of age, an age most Bmal1-null mice could not survive to125. The decreased regeneration of keratinocyte-specific Bmal1−/− hair germ cells reported in this study cannot be rescued by overexpressing oncogenic Sos, a Ras activator167, suggesting an early onset of replicative or cellular senescence (explained later in this review). Together with the findings that Bmal1-null mice display normal skin regeneration and aggressive hyperplastic growth in bone at a young age as well as increased lymphoma development after γ-radiation, and that targeted silencing of Bmal1 in tumors induces immune suppression and accelerated tumor growth in mice29,125,149,168, the studies described above suggest that cellular senescence resulted from hyperplastic growth, oncogenic activation and accumulated free radical-induced DNA damage is intrinsic to Bmal1−/− somatic cells. However, in tumors and somatic tissues that can overcome the barriers of cellular senescence and ROS-induced apoptosis, loss of Bmal1 only accelerates tumor initiation and growth22,29,166,168. Bmal1−/− mice are specially distinct from other circadian gene-mutant mouse models in that they lack circadian homeostasis even when kept in 24 hour LD conditions169. This severe disruption of endogenous homeostasis at the organismal level may also contribute to increased senescence at the organismal level. However, if Bmal1-null mice can overcome aggressive aging, they are likely cancer prone.
Cellular based studies using mouse primary cells lacking core circadian genes
The role of core circadian genes in controlling cell proliferation and DNA damage response has also been studied using various types of primary cells isolated from different circadian gene mouse models in vitro19,29,124,128,129,149,170. The results obtained from these studies should be explained with caution. For example, primary MEFs cultured in vitro behave very differently from somatic cells in tissues prone to tumor development. MEFs are known to be resistant to sublethal doses of γ-radiation induced apoptosis regardless of genotypes, and therefore should not display a high rate of apoptotic death after a sublethal dose of γ-radiation in the absence of aberrant oncogenic activation128,171,172. In addition, the cell cycle clock is different from the molecular clock in that it does not free-run (Figure 1)173,174. Therefore, the serum shock protocol for setting free-running status of the molecular clock in cultured MEFs is not suitable for studying the role of a core circadian gene in cell cycle control because this protocol requires confluent cell culture condition and only provides growth factor-containing serum for a few hours at the initial serum shocking stage, which leads to uncoupling of the cell cycle clock from the molecular clock due to growth arrest induced by contact inhibition and lack of proper extracellular signals to induce immediate early genes essential for G1 cell cycle progression after the first day of the experiment19,175,176.
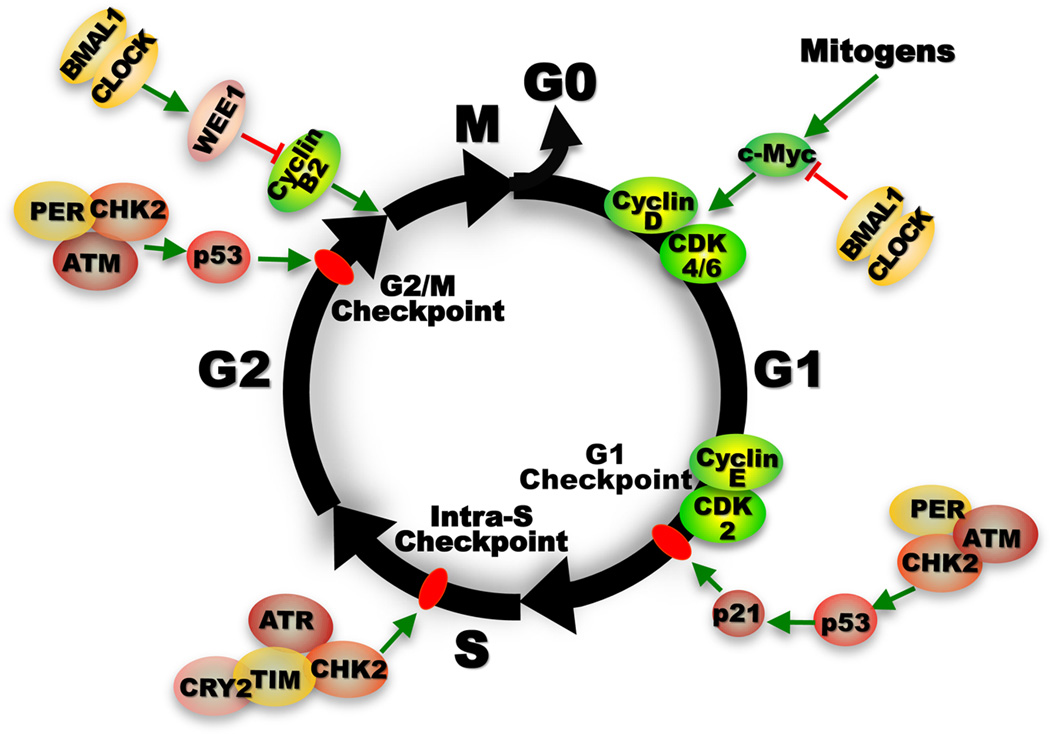
Figure 1. Control of cell proliferation by the molecular clock.
Unlike the molecular clock, the cell cycle does not free run before passing the G1/S phase transition. The initiation of cell cycle progression is strictly controlled by extracellular mitogenic signals that transiently activate immediate early genes such as c-Myc, which then induces Cyclin D leading to activation of CyclinD/CDK4/6 complex that in turn activates E2F-dependent Cyclin E expression by suppressing tumor suppressor RB (not shown). The interaction of Cyclin E with CDK2 allows G1 to S phase transition. G1 is the longest phase in the cell cycle during which most biosynthesis essential for supporting cell cycle progression occurs. c-MYC or E2F oncogenic activation induced elevation of G1 Cyclin expression or genomic DNA damage leads to activation of G1 checkpoint mediated by p16Ink4A and p21WAF1/CIP1, controlled by RB and p53 respectively. P16Ink4A disrupts Cyclin D/CDK4/6 complex (not shown), whereas p21WAF1/CIP1 disrupts Cyclin E/CDK2 interaction. The activation G1 checkpoint leads cells to pause before entering S-phase to repair damaged DNA or exit cell cycle to enter G0 phase (non-dividing status). Under certain conditions, excessive DNA damage or uncontrolled oncogenic signaling can both activate RB and/or p53 tumor suppression pathways leading to cellular senescence. DNA damage induced by UV radiation leads to activation of ATR/CHK1-mediated intra-S checkpoint that couples DNA damage repair with replication. Whereas double-stranded DNA damage induced by γ-radiation activates ATM/CHK2-mediated G1/S and G2/M checkpoints to prevent damaged cells to enter S or mitotic (M) phase. Prolonged G2/M checkpoint arrest is also often associated with p53-mediated apoptosis. G2/M transition is also regulated by WEE1, a kinase that phosphorylates and inactivates the Cyclin B1/Cell Division Cycle 2 (CDC2) complex essential for G2/M transition. Upon the completion of mitosis, cells either enter the next cell cycle stimulated by extracellular mitogen, or withdraw from cell cycle to enter the G0 phase in the absence of mitogenic signals174,234. The molecular clock functions in all phases of the cell cycle to prevent neoplastic growth. At the early G1 phase, the BMAL1/CLOCK heterodimer down regulates Myc transcription to prevent its overexpression29,124,149. PER1 directly interacts with ATM and CHK2 to control G1 checkpoint in response to double-strand DNA damage98. In the S phase, CRY2/TIM complex directly interacts with ATR/CHK1 to control intra-S checkpoint242. In the G2 phase, PER-mediated ATM/CHK2/p53 signaling in response to DNA double-strand breaks and BMAL1/CLOCK activated Wee1 expression both lead to activation of G2/M checkpoint to prevent inappropriate M phase entry98,126.
The choice of cell types and in vitro cell culture conditions used in a study may lead to different conclusions on the role of a gene in cell cycle control. For example, under confluent culture condition, serum-shocked differentiated skeletal muscle cells and hepatocytes from Bmal1−/− mice show high level expression of p21WAF1/CIP1, leading to the conclusion that loss of Bmal1 decreases the rate of cell proliferation177. However, when a human RNAi library targeting 8,000 human genes was studied to identify modulators of p53 function using the BJ-TERT-tsLT cells under subconfluent condition, which were originally isolated from normal human diploid foreskin fibroblasts, Bmal1 was identified as a novel positive regulator of the tumor suppressor p53. Inhibition of Bmal1 expression in this system led to loss of p53-mediated G1 cell cycle arrest at least in part due to an inability to activate the p53 target p21WAF1/CIP1 100.
In summary, genetic studies using various circadian gene-mutant mouse models strongly suggest that as found in human studies, both positive and negative loops of the molecular clock function in tumor suppression in rodents (Table 2).
Table 2.
Phenotypes of Circadian Gene-mutant mouse models
| Circadian Genes |
Mouse Models | Circadian Behavior Phenotypes |
Disease Phenotype | Key Targets Affected | References |
|---|---|---|---|---|---|
| Bmal1 | Bmal1-heterozygous | Normal | Premature aging and cancer prone | N/A | 29 |
| Bmal1-null | Arrhythmic | Premature aging, metabolic syndrome, immune deficiency, caner prone, deregulated drug response | CDC2, CyclinB1, CyclinD1, CyclinE, p21WAF1/CIP1, p16Ink4a, p53, RB, and WEE1 | 23, 29,124,125, 149,166,168, 169, 410, 424, 472 | |
| Keratinocyte-specific Bmal1-null | Normal | Hyperproliferation and deregulated UVB DNA damage of hair cells at a young age | DAB2, DKK3, LEF1, p16, SMAD3, TGFBR2, and WNT10a | 22 | |
| Keratinocyte-specific Bmal1/oncogenic Sos | Normal | Resistance to cutaneous squamous tumors, senescence of hair cells at 10 months of age | CD34+ and Integrin-α6 | 167 | |
| Macrophage-specific Bmal1-null | Normal | Immune deficiency | IL-6 | 405 | |
| Clock | Clock-mutant (Clock Δ19) | Lengthened period & arrhythmic in constant darkness | Metabolic syndrome, premature aging, and deregulated drug response | AKT1, ATR1, Cdk2, Chk1, Chk2, CyclinD3, CyclinE1, EGF, ERα, JAK2, p21, p27, PBEF, TGFβ, and WEE1 | 19,129, 163, 166, 407, 423–425, 471 |
| Clock-null | Shortened period & failure of phase delay/advance | Premature aging and immune deficiency | NF-κB and WEE1 | 162, 164 | |
| Cry | Cry1-null | Shortened period | Cancer prone | N/A | 29, 154, |
| Cry2-null | Lengthened period | Cancer prone | N/A | 29, 150, 154 | |
| Cry1; Cry2 double knockout | Arrhythmic | Cancer prone, immune deficiency, premature aging, metabolic syndrome, cell cycle control, neuroendocrine deficiency, and deregulated drug and DNA damage response | IL-6, MAPK/ERK, TNFα, WEE1, and XPA, GR and Gsα | 29, 93, 126,134, 149, 154, 170, 353, 377–378, 474 | |
| Tumor resistant | 127,128 | ||||
| Per | Per1-null | Unstable period length | Immune deficiency | IFNγ, Perforin, Granzyme B | 155,406 |
| Per2-null | Shortened period & arrhythmic in constant darkness | Cancer prone, premature aging, immune deficiency, and deregulated drug response | c-MYC, CyclinD1, CyclinA, GADD45α, IFNγ, IL-1β, LY49C, MDM2, and p53 | 29, 124,161, 409, | |
| Per2 mutant (Per2Brdm1) | Shortened period & arrhythmic in constant darkness | Cancer prone, vascular senescence, premature aging, immune and neuroendocrine deficiency, and metabolic syndrome | AKT, β-catenin, CAR, c-MYC, CyclinD1, CyclinA, GADD45α, IFNγ, IL-1, IL-6, IL-10, MDM2, p53, and NKG2D, | 29,123,124,149–153, 155, 322, 395, 405, 409, 411 | |
| Per2 mutant (Per2S662G) | Shortened period & phase-advance rhythm | Cancer prone | CyclinD and p21 | 146, 148 | |
| Per1; Per2 double knockout | Arrhythmic | Premature aging, cancer prone and metabolic syndrome | AP1, ATM, c-MYC, CyclinD1, CyclinE, MDM2, and p53 | 29, 149, 167 | |
| Rev-erb | Rev-Erbα-null | Shortened period | Metabolic syndrome and immune deficiency | CCL2, CXCL6, CXCL11, IL-6, IL-19, and IL-10 | 9, 407 |
| Rev-Erbα; Rev-erbβ double knockout | Arrhythmic | Metabolic syndrome | POR, PPARα and SCO2 | 9 | |
| Rorα | Rorα-null | Shortened period | Metabolic syndrome and cerebellar ataxia | TUBα8 and RASD1 | 10 |
The role of the mammalian circadian clock in tumor suppression
Cancer is a multifactorial disease in vivo. Its initiation and progression need various manifestations of abnormal physiological conditions in vivo. As the master regulator of mammalian physiology, the circadian clock acts at the molecular, cellular, tissue/organ and organismal levels to suppress tumor development by maintaining homeostasis of physiology.
The role of peripheral clock in tumor suppression
In cells of peripheral tissues, the clock orchestrates diverse cellular functions in a diurnal oscillating pattern via generation of a network of gene expression at the transcriptional and post-transcriptional levels9,178–180. Disruption of the circadian profiles of this gene expression network leads to loss of homeostasis in cell/tissue function, a key mechanism in circadian dysfunction-induced diseases. Recent studies have revealed that in both human and experimental animal models, circadian disruption specially increases the risk of cancers in the immune, digestive and reproductive systems that need daily cell proliferation to support their functions. These findings highlight the importance of circadian control of cell cycle progression in both homeostasis of tissue function and tumor suppression in vivo. In peripheral tissues, the molecular clock suppresses neoplastic growth by controlling cell proliferation, metabolism, senescence and DNA damage response.
Control of cell proliferation by the molecular clock
Both cell cycle and molecular clocks are operated by interlocked feedback loops of genes that display periodic and sequential phases of activation and repression at the transcription, post-transcription and post-translational levels181. However, although both are considered as “intracellular clocks”, a cell cycle clock is fundamentally different from the molecular clock. First, unlike the molecular clock, the cell cycle clock does not free-run in normal somatic cells. Therefore, the activities of the peripheral clock and cell cycle clock can be separated in peripheral tissues in the absence of proper extracellular mitogenic signals174,182–184. Second, the period of a cell cycle clock is not always fixed as 24 hours throughout one’s lifespan. It can vary from only a few hours in early embryogenesis to 24 hours in rapid renewing somatic tissues in adult life or indefinitely long due to cellular senescence185–188. Since tumor cells often display the properties of de-differentiation and rapid self-renewal with a cell cycle period shorter than 24 hours, loss of circadian coupling of cell cycle progression in adult life may play a key role in the initiation of neoplastic growth in vivo.
Circadian variation of mitotic activity in normal human tissues has been described since 1938189. The uncoupling of mitotic rhythm between normal host tissue and metastasizing cancer was first reported in 1940190. It is now well-known that cell proliferation in all rapidly renewing mammalian tissues studied follow a diurnal oscillating rhythm under normal physiological conditions but is altered in tumors78,101,191–197. The coupling of cell proliferation rhythm between host and tumor has been observed in slow-growing tumors that show considerably higher levels of DNA synthesis and mitotic indices than host tissues throughout a 24 hour period197–199. Whereas an ultradian rhythm less than 24 hour in cell proliferation is often found in metastasizing cancers199–202.
Genome-wide studies have identified a number of genes controlling the key steps of initiation, progression and checkpoint functions of the cell cycle clock as clock-controlled genes7,11–14. These genes encode proto-oncogenes, tumor suppressors, caspases, cyclins, transcription factors and ubiquitin-associated factors essential for regulating cell proliferation and death16–23. Clock-controlled cell cycle regulators are also expressed in all circadian gene-mutant mouse models studied except that they display significant changes in expression profiles and amplitudes over a 24 hour period, which is coupled with loss of cell cycle control in adult tissues of mutant mice22,124,126,149,166,167. Thus, cell cycle clock can function independent of the molecular clock. However, the control of the expression of key cell cycle genes by an intact molecular clock is indispensable for coupling the rate of cell proliferation to diurnal changes in mammalian physiology in vivo.
Both positive and negative loops of the molecular clock are involved in circadian control of cell cycle gene expression in peripheral tissues. The best studied clock-controlled cell cycle regulators include oncogenes c-Myc, Mdm2 and β-catenin, Cyclins D1, B and A, and tumor suppressors p53, Wee1 and p21WAF1/CIP198,124,126,149,166,177. c-Myc is an immediate early responsive gene to diverse cell proliferation stimuli. It plays a key role in G1 cell cycle initiation as well as cell growth and death (Figure 1)203. The expression of c-Myc is tightly controlled in somatic cells in response to diverse stimuli at the transcriptional level via multiple cis-acting sequences in its proximal promoter region204. Deregulation of c-Myc in cooperation with loss of function in p53 promotes neoplastic transformation, tumor initiation, maintenance and metastasis205. In addition to clock-controlled chromatin remodeling, BMAL1/CLOCK and BMAL1/NPAS2 directly regulate c-Myc transcription via the two E-boxes in the P1 promoter of c-Myc124,149,206. Disruption of circadian rhythm leads to deregulation of c-Myc which is coupled with increased neoplastic growth in mice, suggesting that the control of G1 cell cycle initiation is one of the key mechanisms for circadian control of tumor suppression29,124,149. Many key cell cycle regulators, such as Cdk4, Itga6, Wnt3, LHx2, Tcf4, Sox 9, Smad7 and Wee1 are also directly regulated at the transcriptional level by BMAL1/CLOCK heterodimer via E-box-mediated interaction in the gene promoters126,167,206. p53 and MDM2 are controlled indirectly by the Per genes via the tumor suppressor ataxia-telangiectasia mutated (ATM) at the post-transcriptional level29,98. Bmal1 has also been reported to regulate p53-p21WAF1/CIP1 signaling, although the molecular mechanism of this regulation is still not clear83,100,177. The rhythmic expression of cell cycle genes and tumor suppressor p53 is synchronized with the oscillation patterns of the core circadian genes in normal somatic tissues in both humans and mouse models29,124,126,207–209.
The core circadian regulators also regulate the activity of β-Catenin via directly controlling intracellular signaling pathways. Constitutive activation of the core circadian regulator CKIε mimics the effect of WNT signaling, resulting in cytoplasmic accumulation of β-Catenin and its subsequent nuclear localization210,211. In the nucleus, β-Catenin interacts with transcription factors of the T-cell-specific transcription factor/lymphoid enhancer factor-1 (TCF/LEF) family to regulate transcription and promote tumorigenesis212,213. Genes activated by β-Catenin/TCF/LEF include members of the AP1 transcription family, c-Myc and Cyclin D1214–219. Interestingly, the molecular clock itself also responds to β-Catenin nuclear localization to regulate the expression of genes via BMAL1/CLOCK-mediated transcriptional regulation167. Aberrant activation of β-Catenin also disrupts the molecular clock to promote neoplastic transformation by inducing β-transducin repeat-containing protein (β-TrCP)-mediated PER2 degradation151.
In the absence of WNT signaling, β-Catenin is destabilized by glycogen synthase kinase-3β (GSK3β), a functional homologue of the core circadian gene Shaggy in the fruit fly, which functions in regulating the period length of circadian cycles by indirectly controlling PER stability and nuclear entry220,221. Recent studies have revealed that deregulation of GSK3β promotes tumor cell survival, proliferation, invasion and resistance to chemo- and radiation therapy in humans by inhibiting p53 and RB tumor suppressors, inducing intracellular NF-κB signaling, Cyclin D1 over-expression and local chronic inflammation222,223. The activity of GSK3β exhibits robust circadian rhythm in both SCN and peripheral tissues, suggesting that GSK3β may also indirectly target PER2 in the mammalian molecular clock224. Together, the evidence discussed above suggests that the molecular clock couples cell proliferation with mammalian daily physiology by rhythmically pacing the key cell proliferation and tumor suppression pathways at the cellular level. Since the molecular clockworks operate as interlocked feedback loops, disruption of either a positive or negative loop would disrupt the stability of the molecular clock leading to loss of control in the circadian homeostasis of cell cycle progression29,135,156,225,226. Indeed, deregulation of both positive and negative loops of molecular clock frequently occurs in the same type of tumor in humans76,78–81,83,84,86,88,94,227,228.
Control of DNA-damage response by the molecular clock
An average human being contains about 1014 cells, each receives tens of thousands DNA lesions every day. If not repaired, these lesions induce harmful mutations that could affect the survival of cells or even an organism229. DNA damage activates genes encoding key enzymes in DNA repair machinery and cell cycle checkpoints that pause cell cycle progression to give the cell time to repair damaged DNA before continuing to divide230. When DNA damage exceeds the capacity of the cell to repair, efficient elimination of damaged cells by apoptosis or necrosis plays a key role in suppressing tumor development231.
DNA damage response in mammals is mainly controlled by two master kinases, ATM and ataxia telangiectasia and Rad3-related (ATR)232,233. ATM/ATR targets the protein kinases CHK1 and CHK2, which together with ATM and ATR suppress cyclin-dependent kinase (CDK) activity and activate CKIs such as p21WAF1/CIP1 in a p53-dependent manner205,234. Inhibition of CDKs and activation of CKIs lead to arrest of cell cycle progression at the G1/S, intra-S or G2/M checkpoints to allow DNA damage repair235,236. ATM/ATR signaling also enhances DNA repair by transcriptionally and post-transcriptionally activating DNA-repair genes and by recruiting repair factors to the sites of damage. Activation of p53 by ATM/ATR signaling in response to genomic DNA damage often leads to p53-mediated apoptosis (Figure 1)237.
A large number of key players in DNA replication, recombination and repair has been identified as clock-controlled genes in mice7,11–14. Some of these genes, such as Xeroderma pigmentosum A (XPA) that plays an essential role in nucleotide excision repair, are deregulated in Cry1−/−;Cry2−/− mice, which correlates with a dampened circadian rhythm in nucleotide excision repair after UVB irradiation in Cry1−/−;Cry2−/− epidermis238,239. The evidence of direct involvement of Cryptochromes in DNA damage repair in mammals is still missing, although Cryptochromes are structurally related to evolutionarily conserved DNA photolyases that catalyze light-dependent DNA repair in plants240. However, mammalian CLOCK may play a direct role in DNA damage repair because it directly locates to the sites of γ-radiation-induced DNA double-strand breaks158. PER1 was found directly interacting with ATM and CHK2 in response to γ-radiation-induced DNA damage. Overexpression of PER1 in human cancer cells sensitizes cells to radiation-induced apoptosis by activation of Myc-mediated pro-apoptotic pathways98. The human CRY2 has been reported to interact with ATR and CHK1 to regulate intra-S checkpoint function in UV-induced DNA damage response via Timeless (TIM), a natural partner of PER in Drosophila and is necessary for maintaining the robustness of the mammalian central clock241,242. The role for BMAL1 in DNA damage response is shown by a recent study in which knock-down Bmal1 expression abolishes γ-radiation-induced p53 activation as well as p53-dependent p21WAF1/CIP1 induction in human colorectal carcinoma cells100. All core circadian genes studied are activated by exogenous DNA damage agents such as γ-radiation in peripheral tissues following a time-dependent profile over a 24 hour period in mice. Wild-type mouse thymocytes display a time-dependent response to γ-radiation-induced apoptosis in vivo, while loss of function in Per2 leads to resistance to radiation-induced apoptosis in thymocytes throughout a 24 hour period and increased risk of radiation-induced lymphoma in Per2m/m and Per2−/− mice29,124,243,244. Thus, the potentiation of the molecular clock to respond to DNA damage agents varies at different times during a day and plays a key role in determining the outcomes of DNA damage response.
Control of cell metabolism by the molecular clock
Cancer is classically considered as a disease originated from genetic and epigenetic abnormalities that lead to uncontrolled cell growth and division, and formation of metastasizing tumors. The prevalence of metabolic syndromes worldwide and its coherence to cancer has led to a renewed interest in the Warburg effect, which describes an essential role of deregulation of cell metabolism in cancer initiation. In 1956, Otto Warburg observed that normal quiescent somatic cells metabolize glucose to CO2 and H2O via a low rate of glycolysis followed by oxidation of pyruvate in TCA cycle in mitochondria. However, cancer cells predominantly use glucose to produce energy through a high rate of glycolysis followed by lactic acid fermentation in the cytosol even in the presence of abundance oxygen. Warburg predicted that this metabolic reprogram is a fundamental cause of cancer245. Studies in the past decade have revealed that cancer cells display an array of metabolic abnormalities and that both oncoproteins and tumor suppressors can influence the switch between aerobic glycolysis and the use of TCA cycle to generate ATP246–251. The predominant use of the aerobic glycolysis pathway in cancer cells not only leads to a high level of intracellular reactive oxygen species (ROS), a major source of endogenous DNA damage agents promoting cancer and aging, but also the accumulation of intermediate products including purines, pyrimidines, nonessential amino acids and free fatty acids that can be used for anabolic synthesis, cell growth and division249,252–254.
The mammalian circadian clock is a master regulator of metabolic homeostasis both at the organismal and peripheral tissue levels6,255–258. In peripheral tissues, the molecular clock regulates nutrient uptake, metabolism, energy storage, mitochondria biosynthesis and intracellular redox levels by targeting key metabolic genes including those controlling the Warburg effect, such as glucose-6-phosphatase, pyruvate kinase and glucose transporter 2 (GLUT)17,20,179,259,260. It also responds to food cues and nutrient sensors to shift metabolic balance independent of SCN clock function12,261–266. The peripheral clock also indirectly controls Warburg switch by regulating the expression of oncoproteins and tumor suppressors. For example, p53 inhibits aerobic glycolysis and decreases intracellular ROS levels by suppressing GLUT1–4 and fructose-2,6-bisphosphate, a critical substrate of aerobic glycolysis, via the tumor suppressor p53-induced glycolysis and apoptosis regulator (TIGAR)267,268, and promotes the use of TCA cycle by inhibiting glycolic enzyme phosphoglycerol mutatase269. Whereas c-MYC stimulates biosynthesis to support cell growth and proliferation via upregulating lipogenetic, glycolytic and mitochondrial genes270–273, and increases glutamine uptake to compensate for the progressive loss of glucose as a mitochondrial substrate in cancers cells due to Warburg effect274. Myc also upregulates mitochondrial biosynthesis by regulating genes including nuclear respiratory factor 1 (NRF1) and the transcription factor A, mitochondrial (TFAM) as well as cytochrome C to stimulate the production of ROS275–279. Although elevated ROS leads to Myc-mediated apoptosis in normal somatic cells280, loss of function or deregulation of p53 in combination of Myc oncogenic activation results in the metabolic switch to support cell proliferation, apoptotic resistance and neoplastic transformation254,281–285.
The molecular clock not only controls metabolic homeostasis by regulating nuclear receptors and their co-activators and suppressors as well as chromatin remodeling in metabolically active peripheral tissues6,255–258,286, but is also regulated by nuclear receptors REV-ERBα/β, RORα and PPARγ coactivator PGC-1α that play an active role in energy metabolism, adipogenesis and lipid storage in peripheral tissues, and by nutrient sensors such as adenosine monophosphate-activated protein kinase (AMPK)8–10,265,287. Therefore, disruption of the molecular clockworks would inevitably shift the homeostasis of metabolism and energy balance in peripheral tissues to provide an intracellular environment that favors tumor initiation and progression.
Control of cell senescence by the molecular clock
Aging is frequently associated with disruption of circadian rhythmicity in humans and animal models288–290. Premature aging is commonly observed among circadian gene-mutant mouse models. Among them, Bmal1−/− mice display the most aggressive aging phenotypes leading to a significantly reduced lifespan125. Other circadian gene-mutant mouse models including mice carrying a mutated Clock (Clockm/m) or lacking Per or Cry genes (Cry1−/−;Cry2−/−, Per1−/−;Per2−/−, Per2m/m or Per2−/−) also display premature aging phenotypes which become more evident in response to DNA damage agents such as γ-radiation29,124,128,129. These premature aging phenotypes of circadian gene-mutant mouse models are closely related to deregulation of cell proliferation and DNA damage response in vivo.
Aging is an universal process for all multi-cell organisms on earth, which is measured chronologically by biological changes over time and is accompanied with a dramatic increase in the risk of various diseases at the mid-point of the life span291. Although aging is marked by progressive degeneration of tissue and cell function in vivo, it is also coupled with increased risk of neoplastic growth in renewable tissues in mammals, leading to the conclusion that aging is a primary risk factor of cancer292,293. The mechanisms linking aging to cancer including immunosenescence and age-related endocrine dysfunction at the organismal level as well as telomere shortening, reproductive cell cycle and accumulation of DNA damage over a lifetime294–299. These adverse age-related pathological changes have led to the conclusions that if humans live long enough, they would all eventually develop cancer300,301.
Throughout the life span, mammals need continuous cell proliferation to support their daily physiology. However, the inherent limitations in DNA repair mechanisms inevitably lead to accumulation of errors in genomic DNA, which often results in replicative or cellular senescence, a direct cause of aging302–306. Senescence refers to a state of permanent and irreversible withdrawal from cell cycle resulted from accumulation of cellular damages including DNA lesions, oncogenic activation, and/or over-expression of tumor suppressors157. Since cancer cells do not have a limited replicative life span, cellular senescence is often used to enforce the idea that it suppresses cancer development307. However, mounting evidence suggests that cellular senescence also promotes cancer initiation and progression. Senescent cells are still metabolically active and often show changes in chromatin organization and gene expression, leading to secretion of proinflammatory cytokines, proteases and growth factors. The paracrine activities of senescent cells have been found to stimulate proliferation and migration of neighboring cells and promote the development of metastatic tumors308–311. The activation of Myc, Ras/MAPK oncogenic signaling and/or p53/p21WAF1/CIP1 and pRB/p16INK4a tumor suppressing pathways are established molecular mechanisms for cellular senescence312–315. Under certain conditions, suppression or even a subtle change in the expression of these tumor suppressors could lead to senescent cells to rapidly regain the ability to proliferate316.
Increased oxidative stress in circadian gene mutant mice may lead to a higher level accumulation of DNA damage which induces early cellular senescence to promote aging125,289,317. The modulators of the molecular clock, such as NAD-dependent deacetylase sirtuin-1 (SIRT1) may play a role in bridging aging and cancer-prone phenotypes found in circadian gene-mutant mice6. SIRT1 is a class III histone deacetylase that promotes cell survival by inhibiting apoptosis and cellular senescence in mammals318. SIRT1 expression follows a robust circadian expression in vivo. It directly interacts with CLOCK and deacetylates BMAL1 to regulate the activity of the molecular clock12,13, and also plays a role in regulating cell proliferation and apoptosis by deacetylating key tumor suppressors and oncoproteins including p53, β-Catenin and DNA repair protein KU70319–321. Both Bmal1 and Per2 are found to inhibit cellular senescence in vivo possibly by distinct mechanisms. Per2-mutation leads to AKT-dependent vascular senescence that impairs endothelial progenitor cell function, while loss of Bmal1 promotes senescence in vivo via a p53-independent mechanism23,322. The increased cellular senescence found in circadian gene-mutant mice may be explained by their inherent high risk of deregulation of oncogenic and tumor suppression pathways, high intracellular levels of redox and its associated accumulation of genomic DNA damage, and abnormal internal physiological environments that promote oncogenic extracellular signaling. These abnormalities together with lack of proper control of gene expression could increase paracrine activities and loss of senescence surveillance, which not only leads to increased local tissue damage and inflammation that stimulate cell regeneration but also the possibility of re-entering cell cycle of senescent cells. Since cancer is a clonogenic disease in vivo, one or a few premalignant cells that successfully escape senescence surveillance would be sufficient enough to promote cancer development in vivo205,323.
Tumor suppression in vivo is a clock-controlled physiological function
Although an intact molecular clock can provide self-sustained circadian oscillations in peripheral tissues, peripheral organs rely on daily entrainment signals from the central pacemaker to maintain the synchrony of the internal physiology. Disruption of central clock function leads to phase desynchrony among peripheral tissues182. Such desynchronization of the internal circadian homeostasis is closely related to increased risk of tumor development and accelerates tumor progression in both humans and experimental animal models34–49,54–59,117–122. The key entrainment signals from the central clock include extracellular signaling controlled by the autonomic nervous and neuroendocrine systems (ANS and NES)24. Loss of homeostasis of ANS and NES disrupts circadian homeostasis of cancer immune surveillance and energy balance as well as G1 cell cycle progression in renewable peripheral tissues, which synergistically promote tumor development.
The mechanisms of SCN control of peripheral tissues have been discussed in detail in several recent reviews24,30–33,258,324. Briefly, via direct and indirect targeting, the SCN clock controls brain centers especially those in the hypothalamus including the paraventricular nucleus (PVN), arcuate nucleus (ARC), dorsomedial hypothalamus (DMH), lateral hypothalamus (LHA) and endocrine neurons producing corticotropin-releasing hormone (CRH), thyrotropin-releasing hormone (TRH) and gonadotropin-releasing hormone (GnRH). The CRH, TRH and GnRH control the activity of NES via the hypothalamic-pituitary-adrenal (HPA) and hypothalamic-pituitary-gonadal (HPG) axes, while ARC, DMH, LHA and PVN control energy expenditure and food-intake in response to both central and peripheral signals. The autonomic paraventricular neurons (aPVN) directly project to the preganglionic parasympathatic and sympathetic neurons in the brainstem nuclei, dorsal motor nucleus of the vagus (DMV), and intermediolateral cell columns (IML) of the spinal cord to control parasympathatic and sympathetic nervous systems. The SCN control of aPVN neurons leads to a robust circadian oscillation in the function of the autonomic nervous system (ANS) in vivo. The example of SCN control of NES is demonstrated by the rhythmic activity of HPA axis. The SCN pacemaker indirectly generates circadian oscillation of Adrenocorticotropic hormone (ACTH)-controlled corticosterone production from the adrenal gland into the blood via controlling hypothalamic CRH endocrine neurons324. In vivo, the ANS innervates all peripheral tissues except skeletal muscle through G-protein-coupled transmembrane-receptor (GPCR) in a tissue and/or cell type-specific manner325–327. Hormones produced by the pineal gland, HPA and HPG axes, such as melatonin, glucocorticoids, and oestrogen target a wide range of peripheral tissues especially the immune, metabolically active and reproductive organs328–335. The rhythmic intracellular signaling generated by neurotransmitters and hormones plays an essential role in maintaining homeostasis of tissue microenvironment (Figure 2).
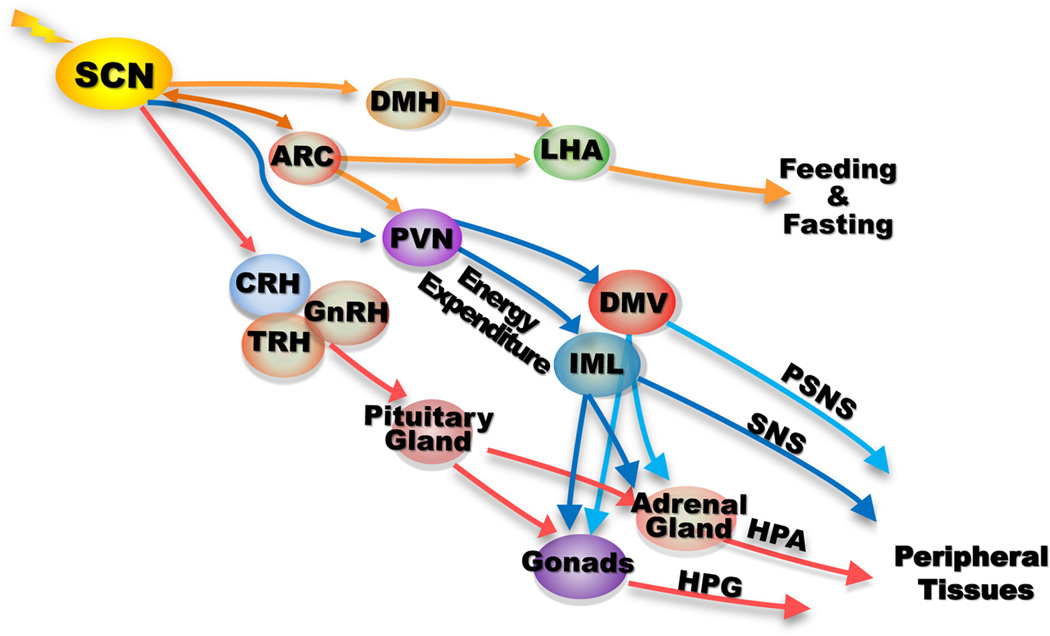
Figure 2. Peripheral control by the SCN pacemaker.
The SCN clock targets a variety of brain centers within the hypothalamus to control homeostasis of endogenous physiology. It controls nutrient intake and energy expenditure by targeting the brain energy homeostasis center arcuate nucleus (ARC) and catabolic center paraventricular nucleus (PVN) directly, and feeding and satiety center LHA indirectly via ARC and dorsomedial hypothalamus (DMH). It also controls the neuroendocrine system (NES) by directly targeting the corticotropin-releasing hormone (CRH), thyrotropin-releasing hormone (TRH) and gonadotropin-releasing hormone (GnRH) neurons that control the adrenal and gonadal glands via pituitary glands. The SCN pacemaker directly targets the autonomic paraventricular (aPVN) neurons that project to the preganglionic parasympathatic and sympathetic neurons in the dorsal motor nucleus of the vagus (DMV) and intermediolateral cell columns (IML) of the spinal cord to control parasympathatic and sympathetic nervous systems (PSNS and SNS). Both PSNS and SNS also crosstalk with the HPA and HPG axes by directly innervating the adrenal and gonadal glands. The NES and ANS innervate all peripheral tissues in vivo to generate circadian rhythm of internal physiology by controlling extracellular signaling and peripheral clock activity24,30–33,258,324.
Control of G1 cell cycle progression in peripheral tissues by the central pacemaker
In the central pacemaker, light stimuli activates a cascade of intracellular signal transduction pathways in the SCN neurons to phase-shift the center pacemaker including the MAPK, ERK, Protein Kinase C alpha (PKCα), Calcium/Calmodulin-dependent Protein Kinases II (CaM kinases II), c-Jun N-terminal kinase (JNK), c-AMP-Protein Kinase A (PKA) and Nitric Oxide (NO)/c-GMP pathways that differentially regulate the expression of the immediate early genes c-Fos and JunB and the core circadian genes Per1 and Per2 in a time-dependent manner336–345. The peripheral clocks do not directly respond to light stimuli, but are instead synchronized by cyclic changes in the levels of neurotransmitters, growth factors, cytokines and hormones in tissue microenvironment324,346,347. Despite the sensitivity of the peripheral clock to non-SCN cues such as food cues, metabolites, or fluctuation in body temperature, the central pacemaker plays a dominant role in peripheral clock control to maintain the integrity of the internal physiology24.
The best understood intracellular signaling pathways for entraining the peripheral clock by ANS and NES include glucocorticoid and beta-adrenergic receptor (ADRβ)-mediated activation of the molecular clock. The interaction of glucocorticoid with glucocorticoid receptor (GR) in the cytoplasm stimulates GR nuclear localization and activation of GR-mediated transcription via Glucocorticoid-Responsive Elements (GREs) in gene promoters348,349. GREs are found in the regulatory regions of Per1, Per2, Bmal1 and Cry1 genes350–352, and both CRY1 and 2 directly interact with GR in a ligand-dependent fashion to modulate the transcriptional activity of GR353. Administration of dexamethasone, a glucocorticoid analog, phase-shifts the molecular clock in cultured rat-1 fibroblasts as well as in mouse livers. Although unable to phase-shift the SCN clock, dexamethasone can resynchronize about 60% of the circadian transcriptome in the livers of SCN-lesioned mice via at least in part directly phase-shifting the molecular clock31,350. Adrenalectomy results in deregulation of the core clock genes, desynchronization and dampening of the molecular clock in multiple peripheral tissues354.
The SNS directly innervates all peripheral tissues by releasing norepinephrine (NE) to target adrenergic receptors (ADRs) on cell membranes. It also controls the production and secretion of epinephrine (EPI) from the adrenal medulla, which then target all cells expressing ADRs in the body via blood circulation355. ADRβ2 is the best studied ADR that responds to ligand binding by activating cAMP Response Element-Binding Protein (CREB) which then interacts with the cAMP Response Element (CRE) in promoters to regulate gene transcription. Per1 and Per2 both contain CREs in the promoters and are among the immediate early genes activated by CREB in cultured primary osteoblasts, NIH3T3 cells and mouse liver slides in response to administration of isoproterenol, a synthetic agonist of ADRβ2, EPI or NE in vitro and in mouse livers after intraperitoneal injection of adrenalines in vivo29,149,356. Loss of function in Per1 and Per2 or sympathectomy abolishes SNS-induced peripheral clock activation in affected tissues in rodents29,149,357–360.
The activation of ADRβ is followed by β-arrestin-mediated receptor desensitization. As multifunctional scaffold proteins, the interaction of β-arrestins with ADRβ leads to activation of other signal transduction pathways including the MAPK, Ras/ERK, JNK, nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB), β-Catenin, CaM kinases II, Phosphatidylinositide 3-Kinases/Protein Kinase B/Mammalian Target of Rapamycin (PI3K/AKT/mTOR), Janus kinase 3/Signal Transducer and Activator of Transcription (JAK3/STAT), insulin-like growth factor 1 (IGF-1) and MDM2-p53 pathways134,361–365. Glucocorticoid signaling also crosstalks with pathways controlled by NF-κB, β-Catenin, PI3K and epidermal growth factor receptor (EGFR)366–370. The pathways stimulated by glucocorticoid and ADRβ signaling not only modulate the molecular clock at transcriptional and post-translational levels but also cell proliferation, apoptosis and metabolism in a tissue and cell type-specific manner, leading to coupling of peripheral clock activity with tissue-specific functions in vivo (Figure 3)246,252,254,316,371–374.
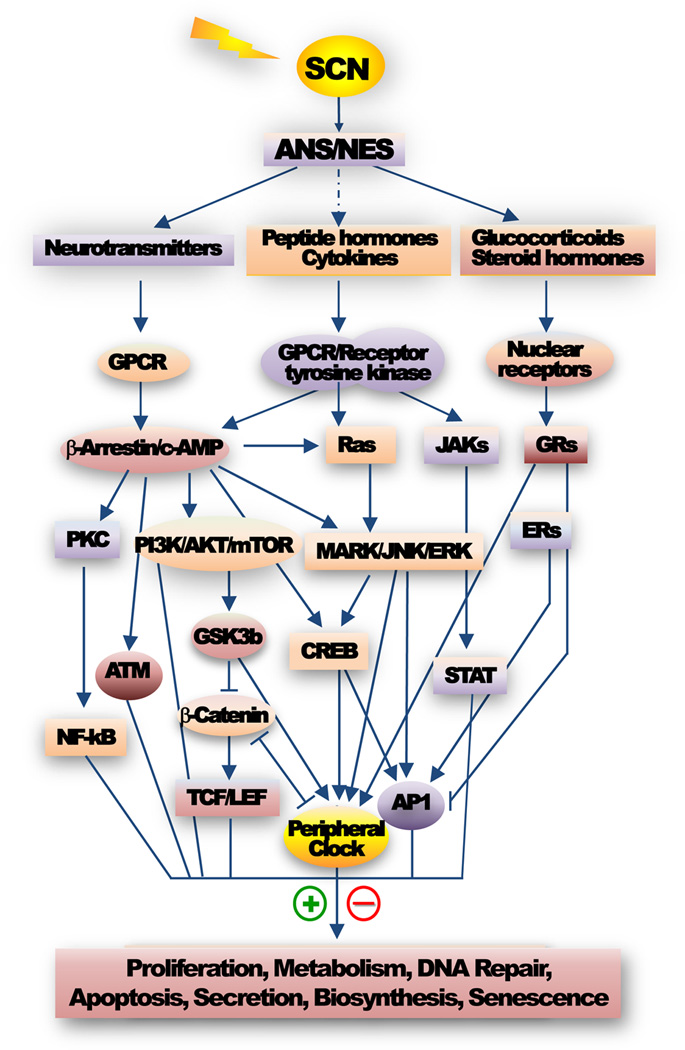
Figure 3. Circadian control of intracellular signaling.
The central pacemaker controlled autonomic nervous and neuroendocrine systems (ANS and NES) rhythmically signal to all of their target tissues. The resulting circadian rhythm in peripheral tissue function also generates local and/or circulating signaling molecules that rhythmically act on their targets. Together, these extracellular signals including neurotransmitters, steroid hormones, peptide hormones, chemokines, growth factors and cytokines activate intracellular signaling mediated by G-protein coupled receptors (GPCRs), tyrosine kinase receptors, integrins (not shown), and nuclear receptors in a tissue and cell-type specific manner. These same intracellular signaling pathways also activate the peripheral clock. The coordinated activities of the central and peripheral clocks orchestrate the complicated extracellular and intracellular signaling to maintain tissue homeostasis by controlling a network of gene expression. Disruption of the central clock-controlled extracellular signaling or mutations in core circadian genes both abolish peripheral clock activity leading to loss of circadian homeostasis in peripheral tissues. The representative intracellular signaling pathways directly or indirectly controlled by the central clock shown in the figure include the c-AMP/PKA/CREB/AP1, Ras/MARK/JNK/ERK and PI3K/AKT/β-Catenin/TCF/LEF pathways essential for c-Myc activation and cell cycle progression478–480, the PI3K/AKT/mTOR signaling controlling biosynthesis and drug resistance451,481, the GPCR/ATM signaling for p53 activation29, the GPCR/PKC/NF-κB pathway that regulates stress and immune response482, the JAK/STAT pathway controlling apoptotic response483, and the GR and ERα signaling pathways cross-talking with the AP1 signaling484,485. These signaling pathways also control the expression and function of circadian genes leading to a coupled activation of the molecular clock with tissue-specific function in vivo including cell proliferation, metabolism, apoptosis, DNA repair, biosynthesis, secretion and senescence371–374.
The initiation of G1 cell cycle progression in vivo is strictly controlled by extracellular signals that activate proto-oncogenes c-Myc and/or E2f via intracellular signaling include c-AMP-PKA, MAPK, Ras/ERK, JNK, β-Catenin, and/or PI3K pathways in a cell type-specific manner174. Loss of homeostasis in HPA axis and ANS signaling is frequently associated with increased risk of cancer375,376. The role of central clock in controlling cell cycle progression in peripheral tissues can be explained by a model obtained from studying circadian control of Myc transcriptional activation. In cultured primary osteoblasts, isoproterenol-mediated ADRβ signaling stimulates cell cycle progression via a coupled activation of peripheral clock, cell cycle clock and p53 controlled by immediate early genes including Per1 and Per2, Ap1 and Myc, and ATM, respectively. Activation of Myc leads to cell cycle progression, while activation of peripheral clock prevents Myc overexpression and also stimulates ATM-mediated p53 induction. Loss of function in the Per genes, or elevated concentration of isoproterenol, prevents the activation of peripheral clock and ATM but not AP1-controlled Myc activation, leading to suppression of p53 induction and uncontrolled osteoblast proliferation due to Myc overexpression. The PER proteins may be directly involved in SNS-stimulated ATM activation since PER1 has been found to interact with ATM in response to γ-radiation98, whereas CRY proteins may prevent uncontrolled c-AMP-PKA-CREB-AP1-c-Myc signaling in response to ADRβ activation by directly inhibiting the Gs alpha subunit (Gsα) essential for activating adenylate cyclase377. In vivo, Per- and Cry-mutant mice both show uncontrolled SNS signaling and display a phenotype of neoplastic growth of osteoblasts in bone29,149,150,377,378. Disruption of homeostasis of SNS signaling by chronic jetlag is associated with the disruption of peripheral clock function, suppression of p53 and Myc oncogenic activation, which is coupled with increased tumor development in all mouse models studied29. Together with previous reports on increased tumor development and progression in rodent models treated with constant light exposure, pinealectomy, chronic jet-lag, or SCN-lesion, these findings provide an explanation on how circadian dysfunction induces tumor development in the absence of gene mutations117–122, which is especially relevant for developing novel strategies for cancer prevention in the modern world in which frequent disruption of endogenous circadian homeostasis due to lifestyle change is associated with a dramatic increase in the risk of sporadic cancers (Figure 4)379.
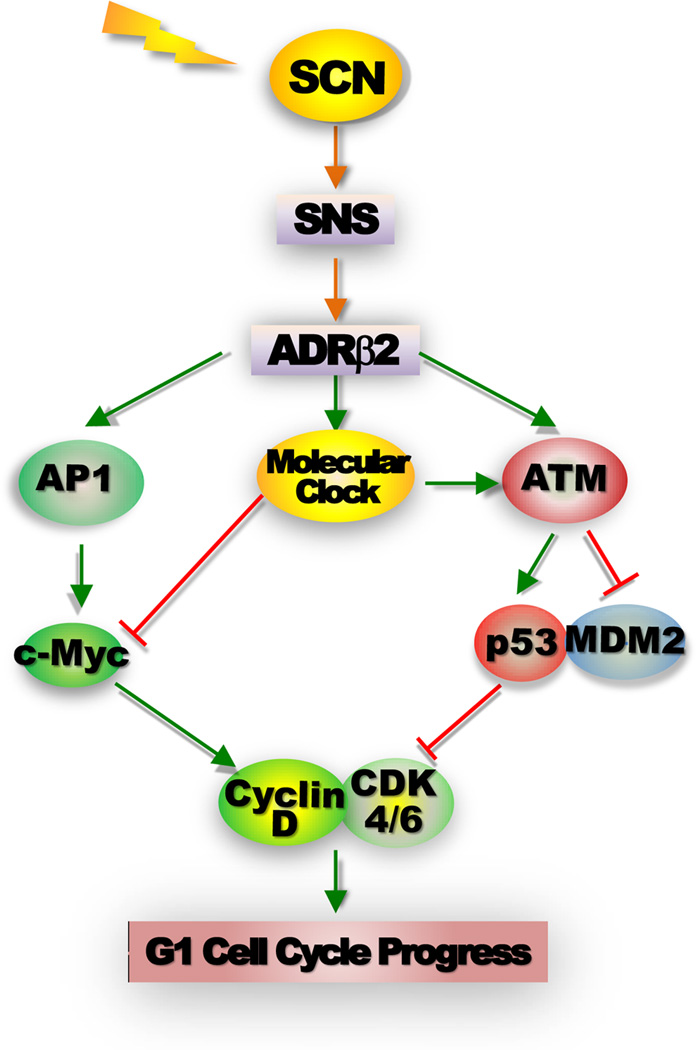
Figure 4. Control of G1 cell cycle progression by the peripheral clock and the SNS.
The activation of the β-adrenergic receptor 2 (ADRβ2) by SNS signaling leads a coupled induction of Ap1 and Period genes via CREB-mediated transcriptional regulation, which in turn activates AP1-controlled Myc induction and Myc-dependent G1 cell cycle progression as well as peripheral clock that prevents Myc overexpression via BMAL1/CLOCK-mediated transcriptional regulation. The activation of ADRβ2 intracellular signaling and the peripheral clock also synergistically activate ATM, which induces p53 by blocking p53-MDM2 interaction to provide an additional mechanism for preventing MYC oncogenic activation. Disruption of the central clock-SNS-peripheral clock axis in mice by chronic jet-lag or ablation of Per genes suppresses peripheral clock activation in response to ADRβ2 activation and abolishes ATM-mediated p53 induction but had no effect on Ap1-Myc signaling. Together these events lead to uncontrolled G1 cell cycle progression and neoplastic growth of osteoblasts both in vitro and in vivo29,149.
Circadian control of cancer immune surveillance
The current concept of cancer immunoediting is based on the evidence of sequential steps of elimination of transformed cells in vivo by the immune system380. When transformed cells accumulated above a threshold, they are recognized by lymphocytes including Nature Killing T (NKT), Nature Killing (NK) and gamma deltaT (γδT) cells that are stimulated by transformed cells to produce interferon γ (IFN-γ). This triggers a cascade of innate immunity including the induction of chemokines CXCL9, 10 and 11 to block neovascularization in the tumor and the recruitment of NK cells, dendritic cells, macrophages and other immune effector cells to the tumor site. The anti-proliferative effects of IFN-γ on transformed cells and the cytocidal activities of macrophages and NK cells result in the death of tumor cells which are ingested by dendritic cells and trafficked to the draining lymph node, where the tumor specific CD4+ and CD8+ T-lymphocytes are developed. These tumor-specific T-lymphocytes are then directed to the tumor site along a chemokine gradient, where they act together with NK cells and activated macrophages to recognize and destroy tumor cells380,381. Mice deficient in cancer immunoediting display a significantly higher risk of spontaneous tumor development in the immune, digestive, respiration and reproductive organs382–386. Cancer immunoediting is usually abolished by cancer-induced immunosuppression in human cancer patients380,381.
The mechanisms of cancer immunosuppression include deregulation or loss of expression of cancer cell surface markers leading to lack of recognition of transformed cells by cytotoxic T lymphocytes, resistance to cell death induced by cytotoxic T lymphocytes due to deregulation of apoptotic factors and death receptors, production of immunosuppressive factors including free radicals, cytokines and growth factors that negatively affect cancer immunoediting by impeding the proliferation and/or function of CD4+ and CD8+ T cells. The immunosuppressive microenvironment in tumors also stimulates the generation and/or promotion of immunosuppressive cells such as type 2 macrophages, myeloid-derived suppressor cells, immature dendritic cells and regulatory T lymphocytes380,381,387,388.
Both primary and secondary lymphoid organs including thymus, spleen and lymph nodes are intensively innervated by ANS and NES389–394. Under normal physiological conditions, the production of cytokines and cytolytic factors, proliferation of leukocytes, activities of NK cells, and redistribution of T and B lymphocytes, dendritic cells, leukocytes and macrophages to lymphoid organs all follow a robust circadian rhythm in vivo395–403. Disruption of circadian homeostasis is closely related to immune suppression404. Ablation or deregulation of the core circadian genes Per1, Per2, Bmal1, Rev-erbα, or Clock in mice induces an array of abnormalities in the immune system, including deregulation of proinflammatory cytokines, cytotoxic receptors, immunoregulatory genes and NK and mast cell activities, and inhibition of B lymphocyte differentiation164,397,405–411. Mice lacking both Cry1 and Cry2 display constitutive elevation of proinflammatory cytokines and are prone to chronic inflammation, a common underlying mechanism for cancer93. Importantly, the immune function is not controlled at the cell autonomous level in vivo. Consecutive phase advance shifts of environmental light cues disrupt the molecular clock and circadian homeostasis of NK cell function in rats412. Ablation of sympathetic innervation abolishes the circadian oscillation of cytokines and cytolytic factors in splenocytes and NK cells359, haematopoietic stem cell trafficking and the expression of chemokine CXCL12360. Adoptive transfer of Bmal1−/− bone marrow deficient in B lymphocyte differentiation to lethally irradiated BALB/cRag2−/− recipients that are unable to generate mature B lymphocytes due to lack of V(D)J recombination activating gene 2 (Rag2) resulted in normal T and B lymphocyte differentiation from Bmal1−/− bone marrow. However, reciprocal transfer of BALB/c Rag2−/− bone marrow to lethally irradiated Bmal1−/− mice did not lead to normal B lymphocyte development, suggesting that the SCN control of tissue microenvironment in bone marrow plays a dominant role in lymphocyte precursor proliferation and differentiation in vivo410.
The central pacemaker not only controls the function of immune system but can also be modulated by immune products such as proinflammatory cytokines Interleukin 1 and 6 (IL-1 and IL-6), Tumor Necrosis Factor-α (TNF-α) and anti-inflammatory drugs, which alter intracellular expression of Bmal1, Npas2, Cry1 and/or Per2413–415 and the activity of the SCN clock and HPA axis at the organismal level416,417. Such modulation provides a circadian-paced immune-regulatory feedback loop that acts as an internal cue to the central clock to control the homeostasis of internal physiology.
Circadian homeostasis of energy balance suppresses tumor development
Human nightshift workers show a coupled increase in the risk of metabolic syndromes and cancers29,46,418–422. Disruption of circadian rhythm in mouse models also induces metabolic syndromes and whole body, local and/or organ-specific adiposity6,255–258,423–425, which is coupled with a significantly higher risk of tumor development in the digestive system29. As discovered from cancer epidemiological studies, deregulation, epigenetic silencing and/or polymorphisms of core circadian genes including Per2, Bmal1, Cry2 and Clock are frequently associated with increased risk of obesity and metabolic syndromes in humans426–431. Together, these findings strongly argue that disruption of circadian homeostasis of energy balance is an important cancer risk factor.
Energy homeostasis in vivo is maintained by the reciprocal interaction of peripheral adiposity signals and the central nervous system432,433, which are both under close surveillance of the circadian clock258,434. Although beyond the scope of this review, the evidence of direct and indirect interactions between the SCN pacemaker and the brain energy homeostasis center ARC, feeding and satiety center LHA and catabolic center PVN suggest that although the central circadian clock does not directly respond to food cues435, it plays an active role in the brain circuits of energy balance436–441. Together with its role in modulating daily activity of NES and ANS that controls the production and circulating levels of most if not all peripheral adiposity signals as well as metabolic balance in peripheral tissues33,442–448, the central pacemaker acts as a master regulator of energy homeostasis in vivo (Figure 2).
Although the biological mechanisms linking obesity and cancer are still not well-understood, obesity-induced deregulation of extracellular signaling, angiogenesis and chronic inflammation are considered as the major pathways promoting cancer development449–451. Obesity leads to increased production of metabolic hormones from the fat and livers of obese subjects, such as Leptin and bioavailable IGF-1, which stimulate biosynthesis, cell growth, proliferation and survival via activation of intracellular signaling pathways including the PI3K/AKT/mTOR, MAPK and JAK3/STAT pathways that are often abrrantly activated in tumors452–454. Obesity also promotes chronic inflammation by increasing circulating free fatty acids, proinflammatory cytokines IL-1β, IL-6, TNF-α and monocyte chemoattractant protein (MCP)-1, stimulating type 2 macrophages polarization, and activating NF-κB-mediated proinflammatory and pro-proliferation pathways450,455–457. Such changes in tissue microenvironment lead to tissue damage due to increased cell necrosis induced by constant stress of increasing in biosynthesis, oxidative stress and DNA damage, which in turn stimulates tissue regeneration that needs active cell proliferation to support. However, the deregulation of multiple oncogenic and tumor suppression pathways as well as immunosuppression due to circadian disruption would result in increased risk of neoplastic growth that accelerate cancer development in obese subjects458–460.
Anticancer chronotherapy
Chemotherapy using one or more cytotoxic drugs in conjunction with radiation therapy or surgery is one of the most common procedures for anticancer therapy. Since chemotherapeutic drugs indiscriminately target both cancerous cells and normal host cells in renewable tissues, these drugs often generate intolerable side effects that impair the treatment461. The “targeted” anticancer therapy developed in recent years is aimed at blocking the growth of cancer cells via monoclonal antibodies recognizing tumor-specific antigens on tumor cell surface or small molecules that block intracellular tyrosine-kinase signaling including MARK/ERK, JAK, PI3K, CDKs, estrogen receptor (ER), epithelial growth factor receptor (EGFR) and vascular endothelial growth factor receptor (VEGFR) controlled pathways462. However, since these signaling pathways are also important for mediating neuroendocrine functions in host tissues, these small inhibitors also potentially increase the risk of fatal side effects among cancer patients463. Thus, to maximally increase tumor targeting efficiency and protect normal host tissues are the biggest challenges for successful anticancer treatments.
Tumors are derived from host tissues after regaining the properties of rapid self-renewing and de-differentiation phenotypes. Most biological processes supporting tumor growth are also essential for normal physiological functions of the host except that the biological processes in normal host tissues are properly controlled and integrated with the daily physiology. Thus, anticancer treatments would inevitably have adverse effect on patients. Biological processes determining the efficiency of anticancer drugs including drug absorption, distribution, intracellular metabolism and elimination all follow circadian rhythms in the host464. In addition, once inside the tumor cells, the function of a cytotoxic drug is largely determined by the circadian phase of cell proliferation in the tumors, which even in the advanced stage tumors is not temporally disorganized192,197–201,465. Therefore, to apply anticancer treatment at a selected time during the day based on circadian variation in host internal physiology and the asynchronies in cell proliferation and drug metabolic rhythms between normal and malignant tissues could maximize drug toxicity to tumors and increase the efficiency of anti-cancer treatment466.
Studies using mouse models have revealed that both host tolerability and drug efficacy are affected by circadian timing, and that the best therapeutic index is achieved by coupling the time for drug delivery with host endogenous circadian rhythms467. These results have been extrapolated to randomized clinic trials of patients undergoing treatment for advanced-stage cancers including metastatic ovarian, lung, colorectal, and breast cancers using conventional chemotherapeutic drugs58,468,469. The results of these studies have revealed that current procedures of anticancer chronotherapy indeed leads to better therapeutic outcomes and is especially more beneficial to patients who still maintain the endogenous circadian rhythmicity. The improved therapeutic index is shown by reduced drug toxicity, improved tumor response rate and the duration of the response, and decreased frequency of tumor metastasis. However, although chronotherapy has been shown to significantly increase the survival time for children with ALL470, it does not increase the long-term survival of patients with metastasizing cancers471, suggesting that anti-cancer chronotherapy is still at its initial stage of practice and needs further improvement. The facts that the molecular clock directly responds to genotoxic insults and that mice deficient in the Clock, Bmal1 and Cry genes all respond differently to anti-cancer drugs compared to wild-type controls suggest that the response to anticancer treatment is a complicated clock-controlled physiological function in vivo124,243,244,472. Further investigation of the mechanisms of cancer chronotherapy, especially the mechanisms controlling the response of the circadian clock to anticancer treatment, the consequence of such response to host physiology and tumor biology, the effect of tumor development on host circadian homeostasis and the ability of the clock to respond to anticancer treatment would contribute greatly to improve the current anticancer chronotherapy.
Conclusions
The rate of cancer continues to rise as more people live to an old age and changes in lifestyle affect more developing countries473. Cancer is usually not detected at the early stage due to lack of obvious symptoms474,475. However, the fact that only 5–10% of all cancers diagnosed are caused by inheritated genetic mutations suggests that an unhealthy lifestyle is the major risk factor for cancer. Therefore, cancer is preventable379. Mounting evidence obtained from recent studies suggests that disruption of endogenous circadian homeostasis is a novel and independent risk factor for cancer. In addition, as a master regulator of mammalian physiology, disruption of the clock likely manifests cancer development and progression induced by previously identified exogenous and endogenous cancer risk factors including diet choices, tobacoo and alcohol usage, viral infection, air polutions, aging, endocrine dysfunction, metabolic syndormes and immnue deficiencies476,477. Although still at the initial stage, recent advancements strongly suggest that both tumor suppression and response to anticancer treatments are clock-controlled physiological functions in vivo. Therefore, the mammalian circadian clock provides an invaluable and exciting system to study the mechanism of cancer and anticancer chronotherapy at the molecular, cellular, tissue organ and organismal levels in vivo. Such studies will have a significant impact on human health in the future by improving both cancer prevention and treament.
Aknowledgments
The authors would like to thank D. D. Moore for input in preparing the manuscript. This work is supported by the grants from USDA/ARS (6250-51000-055) and NIH/NCI (R01 CA137019-01A) to L. Fu and the training grant award from NIH/NIDDK to N. M. Kettner (5T32DK007696-20).
Literature cited
- 1.Young MW, Kay SA. Time zones: a comparative genetics of circadian clocks. Nat Rev Genet. 2001 Sep;2(9):702–715. doi: 10.1038/35088576. [DOI] [PubMed] [Google Scholar]
- 2.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002 Aug 29;418(6901):935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 3.Buijs RM, Scheer FA, Kreier F, et al. Organization of circadian functions: interaction with the body. Prog Brain Res. 2006;153:341–360. doi: 10.1016/S0079-6123(06)53020-1. [DOI] [PubMed] [Google Scholar]
- 4.Guo H, Brewer JM, Lehman MN, Bittman EL. Suprachiasmatic regulation of circadian rhythms of gene expression in hamster peripheral organs: effects of transplanting the pacemaker. J Neurosci. 2006 Jun 14;26(24):6406–6412. doi: 10.1523/JNEUROSCI.4676-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takahashi JS, Hong HK, Ko CH, McDearmon EL. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat Rev Genet. 2008 Oct;9(10):764–775. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sahar S, Sassone-Corsi P. Metabolism and cancer: the circadian clock connection. Nat Rev Cancer. 2009 Dec;9(12):886–896. doi: 10.1038/nrc2747. [DOI] [PubMed] [Google Scholar]
- 7.Padmanabhan K, Robles MS, Westerling T, Weitz CJ. Feedback regulation of transcriptional termination by the mammalian circadian clock PERIOD complex. Science. 2012 Aug 3;337(6094):599–602. doi: 10.1126/science.1221592. [DOI] [PubMed] [Google Scholar]
- 8.Preitner N, Damiola F, Lopez-Molina L, et al. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002 Jul 26;110(2):251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 9.Cho H, Zhao X, Hatori M, et al. Regulation of circadian behaviour and metabolism by REV-ERB-alpha and REV-ERB-beta. Nature. 2012 May 3;485(7396):123–127. doi: 10.1038/nature11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sato TK, Panda S, Miraglia LJ, et al. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron. 2004 Aug 19;43(4):527–537. doi: 10.1016/j.neuron.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 11.Duong HA, Robles MS, Knutti D, Weitz CJ. A molecular mechanism for circadian clock negative feedback. Science. 2011 Jun 17;332(6036):1436–1439. doi: 10.1126/science.1196766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakahata Y, Kaluzova M, Grimaldi B, et al. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008 Jul 25;134(2):329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Asher G, Gatfield D, Stratmann M, et al. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008 Jul 25;134(2):317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- 14.Feng D, Liu T, Sun Z, et al. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science. 2011 Mar 11;331(6022):1315–1319. doi: 10.1126/science.1198125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stratmann M, Suter DM, Molina N, Naef F, Schibler U. Circadian Dbp transcription relies on highly dynamic BMAL1-CLOCK interaction with E boxes and requires the proteasome. Mol Cell. 2012 Oct 26;48(2):277–287. doi: 10.1016/j.molcel.2012.08.012. [DOI] [PubMed] [Google Scholar]
- 16.Akhtar RA, Reddy AB, Maywood ES, et al. Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr Biol. 2002 Apr 2;12(7):540–550. doi: 10.1016/s0960-9822(02)00759-5. [DOI] [PubMed] [Google Scholar]
- 17.Duffield GE, Best JD, Meurers BH, Bittner A, Loros JJ, Dunlap JC. Circadian programs of transcriptional activation, signaling, and protein turnover revealed by microarray analysis of mammalian cells. Curr Biol. 2002 Apr 2;12(7):551–557. doi: 10.1016/s0960-9822(02)00765-0. [DOI] [PubMed] [Google Scholar]
- 18.Hughes ME, DiTacchio L, Hayes KR, et al. Harmonics of circadian gene transcription in mammals. PLoS Genet. 2009 Apr;5(4):e1000442. doi: 10.1371/journal.pgen.1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller BH, McDearmon EL, Panda S, et al. Circadian and CLOCK-controlled regulation of the mouse transcriptome and cell proliferation. Proc Natl Acad Sci U S A. 2007 Feb 27;104(9):3342–3347. doi: 10.1073/pnas.0611724104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Panda S, Antoch MP, Miller BH, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002 May 3;109(3):307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 21.Storch KF, Lipan O, Leykin I, et al. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002 May 2;417(6884):78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- 22.Geyfman M, Kumar V, Liu Q, et al. Brain and muscle Arnt-like protein-1 (BMAL1) controls circadian cell proliferation and susceptibility to UVB-induced DNA damage in the epidermis. Proc Natl Acad Sci U S A. 2012 Jul 17;109(29):11758–11763. doi: 10.1073/pnas.1209592109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khapre RV, Kondratova AA, Susova O, Kondratov RV. Circadian clock protein BMAL1 regulates cellular senescence in vivo. Cell Cycle. 2011 Dec 1;10(23):4162–4169. doi: 10.4161/cc.10.23.18381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mohawk JA, Green CB, Takahashi JS. Central and peripheral circadian clocks in mammals. Annu Rev Neurosci. 2012;35:445–462. doi: 10.1146/annurev-neuro-060909-153128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Golombek DA, Rosenstein RE. Physiology of circadian entrainment. Physiol Rev. 2010 Jul;90(3):1063–1102. doi: 10.1152/physrev.00009.2009. [DOI] [PubMed] [Google Scholar]
- 26.Do MT, Yau KW. Intrinsically photosensitive retinal ganglion cells. Physiol Rev. 2010 Oct;90(4):1547–1581. doi: 10.1152/physrev.00013.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davidson AJ, Castanon-Cervantes O, Leise TL, Molyneux PC, Harrington ME. Visualizing jet lag in the mouse suprachiasmatic nucleus and peripheral circadian timing system. Eur J Neurosci. 2009 Jan;29(1):171–180. doi: 10.1111/j.1460-9568.2008.06534.x. [DOI] [PubMed] [Google Scholar]
- 28.Kennaway DJ, Owens JA, Voultsios A, Varcoe TJ. Functional central rhythmicity and light entrainment, but not liver and muscle rhythmicity, are Clock independent. Am J Physiol Regul Integr Comp Physiol. 2006 Oct;291(4):R1172–R1180. doi: 10.1152/ajpregu.00223.2006. [DOI] [PubMed] [Google Scholar]
- 29.Lee S, Donehower LA, Herron AJ, Moore DD, Fu L. Disrupting circadian homeostasis of sympathetic signaling promotes tumor development in mice. PLoS One. 2010;5(6):e10995. doi: 10.1371/journal.pone.0010995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barclay JL, Tsang AH, Oster H. Interaction of central and peripheral clocks in physiological regulation. Prog Brain Res. 2012;199:163–181. doi: 10.1016/B978-0-444-59427-3.00030-7. [DOI] [PubMed] [Google Scholar]
- 31.Balsalobre A, Brown SA, Marcacci L, et al. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science. 2000 Sep 29;289(5488):2344–2347. doi: 10.1126/science.289.5488.2344. [DOI] [PubMed] [Google Scholar]
- 32.Richter HG, Torres-Farfan C, Rojas-Garcia PP, Campino C, Torrealba F, Seron-Ferre M. The circadian timing system: making sense of day/night gene expression. Biol Res. 2004;37(1):11–28. doi: 10.4067/s0716-97602004000100003. [DOI] [PubMed] [Google Scholar]
- 33.Kalsbeek A, Bruinstroop E, Yi CX, Klieverik LP, La Fleur SE, Fliers E. Hypothalamic control of energy metabolism via the autonomic nervous system. Ann N Y Acad Sci. 2010 Nov;1212:114–129. doi: 10.1111/j.1749-6632.2010.05800.x. [DOI] [PubMed] [Google Scholar]
- 34.Schernhammer ES, Laden F, Speizer FE, et al. Night-shift work and risk of colorectal cancer in the nurses' health study. J Natl Cancer Inst. 2003 Jun 4;95(11):825–828. doi: 10.1093/jnci/95.11.825. [DOI] [PubMed] [Google Scholar]
- 35.Kubo T, Ozasa K, Mikami K, et al. Prospective cohort study of the risk of prostate cancer among rotating-shift workers: findings from the Japan collaborative cohort study. Am J Epidemiol. 2006 Sep 15;164(6):549–555. doi: 10.1093/aje/kwj232. [DOI] [PubMed] [Google Scholar]
- 36.Viswanathan AN, Hankinson SE, Schernhammer ES. Night shift work and the risk of endometrial cancer. Cancer Res. 2007 Nov 1;67(21):10618–10622. doi: 10.1158/0008-5472.CAN-07-2485. [DOI] [PubMed] [Google Scholar]
- 37.Lahti TA, Partonen T, Kyyronen P, Kauppinen T, Pukkala E. Night-time work predisposes to non-Hodgkin lymphoma. Int J Cancer. 2008 Nov 1;123(9):2148–2151. doi: 10.1002/ijc.23566. [DOI] [PubMed] [Google Scholar]
- 38.Stevens RG. Working against our endogenous circadian clock: Breast cancer and electric lighting in the modern world. Mutat Res. 2009 Nov-Dec;680(1–2):106–108. doi: 10.1016/j.mrgentox.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 39.Touitou Y, Bogdan A, Levi F, Benavides M, Auzeby A. Disruption of the circadian patterns of serum cortisol in breast and ovarian cancer patients: relationships with tumour marker antigens. Br J Cancer. 1996 Oct;74(8):1248–1252. doi: 10.1038/bjc.1996.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Panzer A. Melatonin in osteosarcoma: an effective drug? Med Hypotheses. 1997 Jun;48(6):523–525. doi: 10.1016/s0306-9877(97)90123-7. [DOI] [PubMed] [Google Scholar]
- 41.Skibola CF, Holly EA, Forrest MS, et al. Body mass index, leptin and leptin receptor polymorphisms, and non-hodgkin lymphoma. Cancer Epidemiol Biomarkers Prev. 2004 May;13(5):779–786. [PubMed] [Google Scholar]
- 42.Viswanathan AN, Schernhammer ES. Circulating melatonin and the risk of breast and endometrial cancer in women. Cancer Lett. 2009 Aug 18;281(1):1–7. doi: 10.1016/j.canlet.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kloog I, Haim A, Stevens RG, Portnov BA. Global co-distribution of light at night (LAN) and cancers of prostate, colon, and lung in men. Chronobiology international. 2009 Jan;26(1):108–125. doi: 10.1080/07420520802694020. [DOI] [PubMed] [Google Scholar]
- 44.Knutsson A, Alfredsson L, Karlsson B, et al. Breast cancer among shift workers: results of the WOLF longitudinal cohort study. Scand J Work Environ Health. 2012 Sep 24; doi: 10.5271/sjweh.3323. [DOI] [PubMed] [Google Scholar]
- 45.Bhatti P, Mirick DK, Davis S. Invited commentary: Shift work and cancer. Am J Epidemiol. 2012 Nov 1;176(9):760–763. doi: 10.1093/aje/kws311. discussion 764–765. [DOI] [PubMed] [Google Scholar]
- 46.Haus EL, Smolensky MH. Shift work and cancer risk: Potential mechanistic roles of circadian disruption, light at night, and sleep deprivation. Sleep Med Rev. 2012 Nov 5; doi: 10.1016/j.smrv.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 47.Buzzelli G, Dattolo P, Pinzani M, Brocchi A, Romano S, Gentilini P. Circulating growth hormone and insulin-like growth factor-I in nonalcoholic liver cirrhosis with or without superimposed hepatocarcinoma: evidence of an altered circadian rhythm. Am J Gastroenterol. 1993 Oct;88(10):1744–1748. [PubMed] [Google Scholar]
- 48.Davis S, Mirick DK, Stevens RG. Night shift work, light at night, and risk of breast cancer. J Natl Cancer Inst. 2001 Oct 17;93(20):1557–1562. doi: 10.1093/jnci/93.20.1557. [DOI] [PubMed] [Google Scholar]
- 49.Rafnsson V, Tulinius H, Jonasson JG, Hrafnkelsson J. Risk of breast cancer in female flight attendants: a population-based study (Iceland) Cancer Causes Control. 2001 Feb;12(2):95–101. doi: 10.1023/a:1008983416836. [DOI] [PubMed] [Google Scholar]
- 50.Schernhammer ES, Laden F, Speizer FE, et al. Rotating night shifts and risk of breast cancer in women participating in the nurses' health study. J Natl Cancer Inst. 2001 Oct 17;93(20):1563–1568. doi: 10.1093/jnci/93.20.1563. [DOI] [PubMed] [Google Scholar]
- 51.Stevens RG, Hansen J, Costa G, et al. Considerations of circadian impact for defining 'shift work' in cancer studies: IARC Working Group Report. Occup Environ Med. 2011 Feb;68(2):154–162. doi: 10.1136/oem.2009.053512. [DOI] [PubMed] [Google Scholar]
- 52.Pukkala E, Aspholm R, Auvinen A, et al. Cancer incidence among 10,211 airline pilots: a Nordic study. Aviat Space Environ Med. 2003 Jul;74(7):699–706. [PubMed] [Google Scholar]
- 53.Keith LG, Oleszczuk JJ, Laguens M. Circadian rhythm chaos: a new breast cancer marker. Int J Fertil Womens Med. 2001 Sep-Oct;46(5):238–247. [PubMed] [Google Scholar]
- 54.Rich T, Innominato PF, Boerner J, et al. Elevated serum cytokines correlated with altered behavior, serum cortisol rhythm, and dampened 24-hour rest-activity patterns in patients with metastatic colorectal cancer. Clin Cancer Res. 2005 Mar 1;11(5):1757–1764. doi: 10.1158/1078-0432.CCR-04-2000. [DOI] [PubMed] [Google Scholar]
- 55.Sephton SE, Sapolsky RM, Kraemer HC, Spiegel D. Diurnal cortisol rhythm as a predictor of breast cancer survival. J Natl Cancer Inst. 2000 Jun 21;92(12):994–1000. doi: 10.1093/jnci/92.12.994. [DOI] [PubMed] [Google Scholar]
- 56.Kim KS, Kim YC, Oh IJ, Kim SS, Choi JY, Ahn RS. Association of worse prognosis with an aberrant diurnal cortisol rhythm in patients with advanced lung cancer. Chronobiology international. 2012 Oct;29(8):1109–1120. doi: 10.3109/07420528.2012.706767. [DOI] [PubMed] [Google Scholar]
- 57.Sephton SE, Lush E, Dedert EA, et al. Diurnal cortisol rhythm as a predictor of lung cancer survival. Brain, behavior, and immunity. 2012 Aug 3; doi: 10.1016/j.bbi.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 58.Innominato PF, Giacchetti S, Bjarnason GA, et al. Prediction of overall survival through circadian rest-activity monitoring during chemotherapy for metastatic colorectal cancer. Int J Cancer. 2012 Dec 1;131(11):2684–2692. doi: 10.1002/ijc.27574. [DOI] [PubMed] [Google Scholar]
- 59.Wu HS, Davis JE, Natavio T. Fatigue and disrupted sleep-wake patterns in patients with cancer: a shared mechanism. Clin J Oncol Nurs. 2012 Apr;16(2):E56–E68. doi: 10.1188/12.CJON.E56-E68. [DOI] [PubMed] [Google Scholar]
- 60.Levi F, Filipski E, Iurisci I, Li XM, Innominato P. Cross-talks between circadian timing system and cell division cycle determine cancer biology and therapeutics. Cold Spring Harb Symp Quant Biol. 2007;72:465–475. doi: 10.1101/sqb.2007.72.030. [DOI] [PubMed] [Google Scholar]
- 61.Mormont MC, Waterhouse J, Bleuzen P, et al. Marked 24-h rest/activity rhythms are associated with better quality of life, better response, and longer survival in patients with metastatic colorectal cancer and good performance status. Clin Cancer Res. 2000 Aug;6(8):3038–3045. [PubMed] [Google Scholar]
- 62.Mostafaie N, Kallay E, Sauerzapf E, et al. Correlated downregulation of estrogen receptor beta and the circadian clock gene Per1 in human colorectal cancer. Mol Carcinog. 2009 Jul;48(7):642–647. doi: 10.1002/mc.20510. [DOI] [PubMed] [Google Scholar]
- 63.Winter SL, Bosnoyan-Collins L, Pinnaduwage D, Andrulis IL. Expression of the circadian clock genes Per1 and Per2 in sporadic and familial breast tumors. Neoplasia. 2007 Oct;9(10):797–800. doi: 10.1593/neo.07595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xia HC, Niu ZF, Ma H, et al. Deregulated expression of the Per1 and Per2 in human gliomas. Can J Neurol Sci. 2010 May;37(3):365–370. doi: 10.1017/s031716710001026x. [DOI] [PubMed] [Google Scholar]
- 65.Fujioka A, Takashima N, Shigeyoshi Y. Circadian rhythm generation in a glioma cell line. Biochemical and biophysical research communications. 2006 Jul 21;346(1):169–174. doi: 10.1016/j.bbrc.2006.05.094. [DOI] [PubMed] [Google Scholar]
- 66.Chen ST, Choo KB, Hou MF, Yeh KT, Kuo SJ, Chang JG. Deregulated expression of the PER1, PER2 and PER3 genes in breast cancers. Carcinogenesis. 2005 Jul;26(7):1241–1246. doi: 10.1093/carcin/bgi075. [DOI] [PubMed] [Google Scholar]
- 67.Jung-Hynes B, Huang W, Reiter RJ, Ahmad N. Melatonin resynchronizes dysregulated circadian rhythm circuitry in human prostate cancer cells. J Pineal Res. 2010 Aug;49(1):60–68. doi: 10.1111/j.1600-079X.2010.00767.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Oda A, Katayose Y, Yabuuchi S, et al. Clock gene mouse period2 overexpression inhibits growth of human pancreatic cancer cells and has synergistic effect with cisplatin. Anticancer Res. 2009 Apr;29(4):1201–1209. [PubMed] [Google Scholar]
- 69.Pogue-Geile KL, Lyons-Weiler J, Whitcomb DC. Molecular overlap of fly circadian rhythms and human pancreatic cancer. Cancer Lett. 2006 Nov 8;243(1):55–57. doi: 10.1016/j.canlet.2005.11.049. [DOI] [PubMed] [Google Scholar]
- 70.Hsu CM, Lin SF, Lu CT, Lin PM, Yang MY. Altered expression of circadian clock genes in head and neck squamous cell carcinoma. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 2012 Feb;33(1):149–155. doi: 10.1007/s13277-011-0258-2. [DOI] [PubMed] [Google Scholar]
- 71.Lin YM, Chang JH, Yeh KT, et al. Disturbance of circadian gene expression in hepatocellular carcinoma. Mol Carcinog. 2008 Dec;47(12):925–933. doi: 10.1002/mc.20446. [DOI] [PubMed] [Google Scholar]
- 72.Gery S, Komatsu N, Kawamata N, et al. Epigenetic silencing of the candidate tumor suppressor gene Per1 in non-small cell lung cancer. Clin Cancer Res. 2007 Mar 1;13(5):1399–1404. doi: 10.1158/1078-0432.CCR-06-1730. [DOI] [PubMed] [Google Scholar]
- 73.Cao Q, Gery S, Dashti A, et al. A role for the clock gene per1 in prostate cancer. Cancer Res. 2009 Oct 1;69(19):7619–7625. doi: 10.1158/0008-5472.CAN-08-4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kuo SJ, Chen ST, Yeh KT, et al. Disturbance of circadian gene expression in breast cancer. Virchows Arch. 2009 Apr;454(4):467–474. doi: 10.1007/s00428-009-0761-7. [DOI] [PubMed] [Google Scholar]
- 75.Yang MY, Chang JG, Lin PM, et al. Downregulation of circadian clock genes in chronic myeloid leukemia: alternative methylation pattern of hPER3. Cancer Sci. 2006 Dec;97(12):1298–1307. doi: 10.1111/j.1349-7006.2006.00331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shih MC, Yeh KT, Tang KP, Chen JC, Chang JG. Promoter methylation in circadian genes of endometrial cancers detected by methylation-specific PCR. Mol Carcinog. 2006 Oct;45(10):732–740. doi: 10.1002/mc.20198. [DOI] [PubMed] [Google Scholar]
- 77.Suzuki T, Sato F, Kondo J, et al. Period is involved in the proliferation of human pancreatic MIA-PaCa2 cancer cells by TNF-alpha. Biomed Res. 2008 Apr;29(2):99–103. doi: 10.2220/biomedres.29.99. [DOI] [PubMed] [Google Scholar]
- 78.Taniguchi H, Fernandez AF, Setien F, et al. Epigenetic inactivation of the circadian clock gene BMAL1 in hematologic malignancies. Cancer Res. 2009 Nov 1;69(21):8447–8454. doi: 10.1158/0008-5472.CAN-09-0551. [DOI] [PubMed] [Google Scholar]
- 79.Zhu Y, Leaderer D, Guss C, et al. Ala394Thr polymorphism in the clock gene NPAS2: a circadian modifier for the risk of non-Hodgkin's lymphoma. Int J Cancer. 2007 Jan 15;120(2):432–435. doi: 10.1002/ijc.22321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dai H, Zhang L, Cao M, et al. The role of polymorphisms in circadian pathway genes in breast tumorigenesis. Breast Cancer Res Treat. 2011 Jun;127(2):531–540. doi: 10.1007/s10549-010-1231-2. [DOI] [PubMed] [Google Scholar]
- 81.Zhu Y, Stevens RG, Hoffman AE, et al. Testing the circadian gene hypothesis in prostate cancer: a population-based case-control study. Cancer Res. 2009 Dec 15;69(24):9315–9322. doi: 10.1158/0008-5472.CAN-09-0648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhou F, He X, Liu H, et al. Functional polymorphisms of circadian positive feedback regulation genes and clinical outcome of Chinese patients with resected colorectal cancer. Cancer. 2012 Feb 15;118(4):937–946. doi: 10.1002/cncr.26348. [DOI] [PubMed] [Google Scholar]
- 83.Elshazley M, Sato M, Hase T, et al. The circadian clock gene BMAL1 is a novel therapeutic target for malignant pleural mesothelioma. Int J Cancer. 2012 Dec 15;131(12):2820–2831. doi: 10.1002/ijc.27598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tokunaga H, Takebayashi Y, Utsunomiya H, et al. Clinicopathological significance of circadian rhythm-related gene expression levels in patients with epithelial ovarian cancer. Acta Obstet Gynecol Scand. 2008;87(10):1060–1070. doi: 10.1080/00016340802348286. [DOI] [PubMed] [Google Scholar]
- 85.Hsu CM, Lin PM, Wang YM, Chen ZJ, Lin SF, Yang MY. Circulating miRNA is a novel marker for head and neck squamous cell carcinoma. Tumour Biol. 2012 Dec;33(6):1933–1942. doi: 10.1007/s13277-012-0454-8. [DOI] [PubMed] [Google Scholar]
- 86.Lewintre EJ, Martin CR, Ballesteros CG, et al. Cryptochrome-1 expression: a new prognostic marker in B-cell chronic lymphocytic leukemia. Haematologica. 2009 Feb;94(2):280–284. doi: 10.3324/haematol.13052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Roe OD, Anderssen E, Helge E, et al. Genome-wide profile of pleural mesothelioma versus parietal and visceral pleura: the emerging gene portrait of the mesothelioma phenotype. PLoS One. 2009;4(8):e6554. doi: 10.1371/journal.pone.0006554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yang WS, Stockwell BR. Inhibition of casein kinase 1-epsilon induces cancer-cell-selective, PERIOD2-dependent growth arrest. Genome Biol. 2008;9(6):R92. doi: 10.1186/gb-2008-9-6-r92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Relles D, Sendecki J, Chipitsyna G, Hyslop T, Yeo CJ, Arafat HA. Circadian Gene Expression and Clinicopathologic Correlates in Pancreatic Cancer. J Gastrointest Surg. 2012 Dec 20; doi: 10.1007/s11605-012-2112-2. [DOI] [PubMed] [Google Scholar]
- 90.Luo Y, Wang F, Chen LA, et al. Deregulated expression of cry1 and cry2 in human gliomas. Asian Pac J Cancer Prev. 2012;13(11):5725–5728. doi: 10.7314/apjcp.2012.13.11.5725. [DOI] [PubMed] [Google Scholar]
- 91.Eisele L, Prinz R, Klein-Hitpass L, et al. Combined PER2 and CRY1 expression predicts outcome in chronic lymphocytic leukemia. Eur J Haematol. 2009 Oct;83(4):320–327. doi: 10.1111/j.1600-0609.2009.01287.x. [DOI] [PubMed] [Google Scholar]
- 92.Blask DE, Hill SM, Dauchy RT, et al. Circadian regulation of molecular, dietary, and metabolic signaling mechanisms of human breast cancer growth by the nocturnal melatonin signal and the consequences of its disruption by light at night. J Pineal Res. 2011 Oct;51(3):259–269. doi: 10.1111/j.1600-079X.2011.00888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Narasimamurthy R, Hatori M, Nayak SK, Liu F, Panda S, Verma IM. Circadian clock protein cryptochrome regulates the expression of proinflammatory cytokines. Proc Natl Acad Sci U S A. 2012 Jul 31;109(31):12662–12667. doi: 10.1073/pnas.1209965109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sun CM, Huang SF, Zeng JM, et al. Per2 inhibits k562 leukemia cell growth in vitro and in vivo through cell cycle arrest and apoptosis induction. Pathol Oncol Res. 2010 Sep;16(3):403–411. doi: 10.1007/s12253-009-9227-0. [DOI] [PubMed] [Google Scholar]
- 95.Blask DE. Melatonin, sleep disturbance and cancer risk. Sleep Med Rev. 2009 Aug;13(4):257–264. doi: 10.1016/j.smrv.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 96.Xiang S, Coffelt SB, Mao L, Yuan L, Cheng Q, Hill SM. Period-2: a tumor suppressor gene in breast cancer. J Circadian Rhythms. 2008;6:4. doi: 10.1186/1740-3391-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jung-Hynes B, Schmit TL, Reagan-Shaw SR, Siddiqui IA, Mukhtar H, Ahmad N. Melatonin, a novel Sirt1 inhibitor, imparts antiproliferative effects against prostate cancer in vitro in culture and in vivo in TRAMP model. J Pineal Res. 2011 Mar;50(2):140–149. doi: 10.1111/j.1600-079X.2010.00823.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gery S, Komatsu N, Baldjyan L, Yu A, Koo D, Koeffler HP. The circadian gene per1 plays an important role in cell growth and DNA damage control in human cancer cells. Mol Cell. 2006 May 5;22(3):375–382. doi: 10.1016/j.molcel.2006.03.038. [DOI] [PubMed] [Google Scholar]
- 99.Gery S, Virk RK, Chumakov K, Yu A, Koeffler HP. The clock gene Per2 links the circadian system to the estrogen receptor. Oncogene. 2007 Dec 13;26(57):7916–7920. doi: 10.1038/sj.onc.1210585. [DOI] [PubMed] [Google Scholar]
- 100.Mullenders J, Fabius AW, Madiredjo M, Bernards R, Beijersbergen RL. A large scale shRNA barcode screen identifies the circadian clock component ARNTL as putative regulator of the p53 tumor suppressor pathway. PLoS One. 2009;4(3):e4798. doi: 10.1371/journal.pone.0004798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Brandi G, Calabrese C, Pantaleo MA, et al. Circadian variations of rectal cell proliferation in patients affected by advanced colorectal cancer. Cancer Lett. 2004 May 28;208(2):193–196. doi: 10.1016/j.canlet.2003.11.015. [DOI] [PubMed] [Google Scholar]
- 102.Hoffman AE, Zheng T, Ba Y, Zhu Y. The circadian gene NPAS2, a putative tumor suppressor, is involved in DNA damage response. Mol Cancer Res. 2008 Sep;6(9):1461–1468. doi: 10.1158/1541-7786.MCR-07-2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Klevecz RR, Shymko RM, Blumenfeld D, Braly PS. Circadian gating of S phase in human ovarian cancer. Cancer Res. 1987 Dec 1;47(23):6267–6271. [PubMed] [Google Scholar]
- 104.Miyamoto N, Izumi H, Noguchi T, et al. Tip60 is regulated by circadian transcription factor clock and is involved in cisplatin resistance. The Journal of biological chemistry. 2008 Jun 27;283(26):18218–18226. doi: 10.1074/jbc.M802332200. [DOI] [PubMed] [Google Scholar]
- 105.Hoffman AE, Zheng T, Ba Y, et al. Phenotypic effects of the circadian gene Cryptochrome 2 on cancer-related pathways. BMC Cancer. 2010;10:110. doi: 10.1186/1471-2407-10-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yang X, Wood PA, Ansell C, Hrushesky WJ. Circadian time-dependent tumor suppressor function of period genes. Integr Cancer Ther. 2009 Dec;8(4):309–316. doi: 10.1177/1534735409352083. [DOI] [PubMed] [Google Scholar]
- 107.Yang X, Wood PA, Ansell CM, et al. The circadian clock gene Per1 suppresses cancer cell proliferation and tumor growth at specific times of day. Chronobiology international. 2009 Oct;26(7):1323–1339. doi: 10.3109/07420520903431301. [DOI] [PubMed] [Google Scholar]
- 108.Morris CJ, Aeschbach D, Scheer FA. Circadian system, sleep and endocrinology. Mol Cell Endocrinol. 2012 Feb 5;349(1):91–104. doi: 10.1016/j.mce.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Boersma GJ, Salton SR, Spritzer PM, Steele CT, Carbone DL. Models and mechanisms of metabolic regulation: genes, stress, and the HPA and HPG axes. Horm Metab Res. 2012 Jul;44(8):598–606. doi: 10.1055/s-0032-1311576. [DOI] [PubMed] [Google Scholar]
- 110.Wirth M, Burch J, Violanti J, et al. Shiftwork duration and the awakening cortisol response among police officers. Chronobiology international. 2011 May;28(5):446–457. doi: 10.3109/07420528.2011.573112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Feychting M, Osterlund B, Ahlbom A. Reduced cancer incidence among the blind. Epidemiology. 1998 Sep;9(5):490–494. [PubMed] [Google Scholar]
- 112.Kliukiene J, Tynes T, Andersen A. Risk of breast cancer among Norwegian women with visual impairment. Br J Cancer. 2001 Feb 2;84(3):397–399. doi: 10.1054/bjoc.2000.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pukkala E, Verkasalo P, Ojamo M, Rudanko SL. Response to the letter by Maria Feychting and Anders Ahlbom (Cancer Causes and Control 10: 637, 1999.) Why is the cancer pattern so different among visually impaired persons in Finland and Sweden? Cancer Causes Control. 2000 Jan;11(1):99–100. doi: 10.1023/a:1008994724025. [DOI] [PubMed] [Google Scholar]
- 114.Hamilton T, Sneddon A. Endocrine differences between rats bearing simple and malignant mammary tumours induced by 9,10-dimethyl-1,2-benzanthracene. Br J Cancer. 1968 Jun;22(2):324–329. doi: 10.1038/bjc.1968.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.van den Heiligenberg S, Depres-Brummer P, Barbason H, Claustrat B, Reynes M, Levi F. The tumor promoting effect of constant light exposure on diethylnitrosamine-induced hepatocarcinogenesis in rats. Life Sci. 1999;64(26):2523–2534. doi: 10.1016/s0024-3205(99)00210-6. [DOI] [PubMed] [Google Scholar]
- 116.Shah PN, Mhatre MC, Kothari LS. Effect of melatonin on mammary carcinogenesis in intact and pinealectomized rats in varying photoperiods. Cancer Res. 1984 Aug;44(8):3403–3407. [PubMed] [Google Scholar]
- 117.Li JC, Xu F. Influences of light-dark shifting on the immune system, tumor growth and life span of rats, mice and fruit flies as well as on the counteraction of melatonin. Biol Signals. 1997 Mar-Apr;6(2):77–89. doi: 10.1159/000109112. [DOI] [PubMed] [Google Scholar]
- 118.Wu M, Zeng J, Chen Y, et al. Experimental chronic jet lag promotes growth and lung metastasis of Lewis lung carcinoma in C57BL/6 mice. Oncol Rep. 2012 May;27(5):1417–1428. doi: 10.3892/or.2012.1688. [DOI] [PubMed] [Google Scholar]
- 119.Filipski E, Subramanian P, Carriere J, Guettier C, Barbason H, Levi F. Circadian disruption accelerates liver carcinogenesis in mice. Mutat Res. 2009 Nov-Dec;680(1–2):95–105. doi: 10.1016/j.mrgentox.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 120.Logan RW, Zhang C, Murugan S, et al. Chronic shift-lag alters the circadian clock of NK cells and promotes lung cancer growth in rats. Journal of immunology. 2012 Mar 15;188(6):2583–2591. doi: 10.4049/jimmunol.1102715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yasuniwa Y, Izumi H, Wang KY, et al. Circadian disruption accelerates tumor growth and angio/stromagenesis through a Wnt signaling pathway. PLoS One. 2010;5(12):e15330. doi: 10.1371/journal.pone.0015330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Filipski E, King VM, Li X, et al. Host circadian clock as a control point in tumor progression. J Natl Cancer Inst. 2002 May 1;94(9):690–697. doi: 10.1093/jnci/94.9.690. [DOI] [PubMed] [Google Scholar]
- 123.Zheng B, Larkin DW, Albrecht U, et al. The mPer2 gene encodes a functional component of the mammalian circadian clock. Nature. 1999 Jul 8;400(6740):169–173. doi: 10.1038/22118. [DOI] [PubMed] [Google Scholar]
- 124.Fu L, Pelicano H, Liu J, Huang P, Lee C. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell. 2002 Oct 4;111(1):41–50. doi: 10.1016/s0092-8674(02)00961-3. [DOI] [PubMed] [Google Scholar]
- 125.Kondratov RV, Kondratova AA, Gorbacheva VY, Vykhovanets OV, Antoch MP. Early aging and age-related pathologies in mice deficient in BMAL1, the core component of the circadian clock. Genes Dev. 2006 Jul 15;20(14):1868–1873. doi: 10.1101/gad.1432206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Matsuo T, Yamaguchi S, Mitsui S, Emi A, Shimoda F, Okamura H. Control mechanism of the circadian clock for timing of cell division in vivo. Science. 2003 Oct 10;302(5643):255–259. doi: 10.1126/science.1086271. [DOI] [PubMed] [Google Scholar]
- 127.Ozturk N, Lee JH, Gaddameedhi S, Sancar A. Loss of cryptochrome reduces cancer risk in p53 mutant mice. Proc Natl Acad Sci U S A. 2009 Feb 24;106(8):2841–2846. doi: 10.1073/pnas.0813028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gauger MA, Sancar A. Cryptochrome, circadian cycle, cell cycle checkpoints, and cancer. Cancer Res. 2005 Aug 1;65(15):6828–6834. doi: 10.1158/0008-5472.CAN-05-1119. [DOI] [PubMed] [Google Scholar]
- 129.Antoch MP, Gorbacheva VY, Vykhovanets O, et al. Disruption of the circadian clock due to the Clock mutation has discrete effects on aging and carcinogenesis. Cell Cycle. 2008 May 1;7(9):1197–1204. doi: 10.4161/cc.7.9.5886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Barbeito CG, Garcia MN, Flamini MA, Andrini LB, Badran AF. Effect of partial and sham hepatectomy on the growth of a hepatocellular carcinoma. J Exp Clin Cancer Res. 2001 Mar;20(1):153–158. [PubMed] [Google Scholar]
- 131.Huang W, Ma K, Zhang J, et al. Nuclear receptor-dependent bile acid signaling is required for normal liver regeneration. Science. 2006 Apr 14;312(5771):233–236. doi: 10.1126/science.1121435. [DOI] [PubMed] [Google Scholar]
- 132.Yang F, Huang X, Yi T, Yen Y, Moore DD, Huang W. Spontaneous development of liver tumors in the absence of the bile acid receptor farnesoid X receptor. Cancer Res. 2007 Feb 1;67(3):863–867. doi: 10.1158/0008-5472.CAN-06-1078. [DOI] [PubMed] [Google Scholar]
- 133.Doi M, Hirayama J, Sassone-Corsi P. Circadian regulator CLOCK is a histone acetyltransferase. Cell. 2006 May 5;125(3):497–508. doi: 10.1016/j.cell.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 134.Tsuchiya Y, Minami I, Kadotani H, Todo T, Nishida E. Circadian clock-controlled diurnal oscillation of Ras/ERK signaling in mouse liver. Proceedings of the Japan Academy. Series B, Physical and biological sciences. 2013;89(1):59–65. doi: 10.2183/pjab.89.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Oster H, van der Horst GT, Albrecht U. Daily variation of clock output gene activation in behaviorally arrhythmic mPer/mCry triple mutant mice. Chronobiology international. 2003 Jul;20(4):683–695. doi: 10.1081/cbi-120022408. [DOI] [PubMed] [Google Scholar]
- 136.Hoshino R, Chatani Y, Yamori T, et al. Constitutive activation of the 41-/43-kDa mitogen-activated protein kinase signaling pathway in human tumors. Oncogene. 1999 Jan 21;18(3):813–822. doi: 10.1038/sj.onc.1202367. [DOI] [PubMed] [Google Scholar]
- 137.Zou Y, Bao Q, Kumar S, Hu M, Wang GY, Dai G. Four waves of hepatocyte proliferation linked with three waves of hepatic fat accumulation during partial hepatectomy-induced liver regeneration. PLoS One. 2012;7(2):e30675. doi: 10.1371/journal.pone.0030675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lawler J, Miao WM, Duquette M, Bouck N, Bronson RT, Hynes RO. Thrombospondin-1 gene expression affects survival and tumor spectrum of p53-deficient mice. Am J Pathol. 2001 Nov;159(5):1949–1956. doi: 10.1016/S0002-9440(10)63042-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Yamamoto M, Tsukamoto T, Sakai H, et al. p53 knockout mice (−/−) are more susceptible than (+/−) or (+/+) mice to N-methyl-N-nitrosourea stomach carcinogenesis. Carcinogenesis. 2000 Oct;21(10):1891–1897. doi: 10.1093/carcin/21.10.1891. [DOI] [PubMed] [Google Scholar]
- 140.Tyner SD, Venkatachalam S, Choi J, et al. p53 mutant mice that display early ageing-associated phenotypes. Nature. 2002 Jan 3;415(6867):45–53. doi: 10.1038/415045a. [DOI] [PubMed] [Google Scholar]
- 141.Oda H, Zhang S, Tsurutani N, et al. Loss of p53 is an early event in induction of brain tumors in mice by transplacental carcinogen exposure. Cancer Res. 1997 Feb 15;57(4):646–650. [PubMed] [Google Scholar]
- 142.Hursting SD, Perkins SN, Phang JM. Calorie restriction delays spontaneous tumorigenesis in p53-knockout transgenic mice. Proc Natl Acad Sci U S A. 1994 Jul 19;91(15):7036–7040. doi: 10.1073/pnas.91.15.7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Nozawa H, Oda E, Nakao K, et al. Loss of transcription factor IRF-1 affects tumor susceptibility in mice carrying the Ha-ras transgene or nullizygosity for p53. Genes Dev. 1999 May 15;13(10):1240–1245. doi: 10.1101/gad.13.10.1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Liu G, McDaniel TK, Falkow S, Karlin S. Sequence anomalies in the Cag7 gene of the Helicobacter pylori pathogenicity island. Proc Natl Acad Sci U S A. 1999 Jun 8;96(12):7011–7016. doi: 10.1073/pnas.96.12.7011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Donehower LA, Harvey M, Slagle BL, et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992 Mar 19;356(6366):215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 146.Xu Y, Toh KL, Jones CR, Shin JY, Fu YH, Ptacek LJ. Modeling of a human circadian mutation yields insights into clock regulation by PER2. Cell. 2007 Jan 12;128(1):59–70. doi: 10.1016/j.cell.2006.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Toh KL, Jones CR, He Y, et al. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science. 2001 Feb 9;291(5506):1040–1043. doi: 10.1126/science.1057499. [DOI] [PubMed] [Google Scholar]
- 148.Gu X, Xing L, Shi G, et al. The circadian mutation PER2(S662G) is linked to cell cycle progression and tumorigenesis. Cell Death Differ. 2012 Mar;19(3):397–405. doi: 10.1038/cdd.2011.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Fu L, Patel MS, Bradley A, Wagner EF, Karsenty G. The molecular clock mediates leptin-regulated bone formation. Cell. 2005 Sep 9;122(5):803–815. doi: 10.1016/j.cell.2005.06.028. [DOI] [PubMed] [Google Scholar]
- 150.Maronde E, Schilling AF, Seitz S, et al. The clock genes Period 2 and Cryptochrome 2 differentially balance bone formation. PLoS One. 2010;5(7):e11527. doi: 10.1371/journal.pone.0011527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Yang X, Wood PA, Ansell CM, et al. Beta-catenin induces beta-TrCP-mediated PER2 degradation altering circadian clock gene expression in intestinal mucosa of ApcMin/+ mice. J Biochem. 2009 Mar;145(3):289–297. doi: 10.1093/jb/mvn167. [DOI] [PubMed] [Google Scholar]
- 152.Wood PA, Yang X, Taber A, et al. Period 2 mutation accelerates ApcMin/+ tumorigenesis. Mol Cancer Res. 2008 Nov;6(11):1786–1793. doi: 10.1158/1541-7786.MCR-08-0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Miyazaki K, Wakabayashi M, Hara Y, Ishida N. Tumor growth suppression in vivo by overexpression of the circadian component, PER2. Genes Cells. 2010 Apr 1;15(4):351–358. doi: 10.1111/j.1365-2443.2010.01384.x. [DOI] [PubMed] [Google Scholar]
- 154.van der Horst GT, Muijtjens M, Kobayashi K, et al. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999 Apr 15;398(6728):627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- 155.Zheng B, Albrecht U, Kaasik K, et al. Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell. 2001 Jun 1;105(5):683–694. doi: 10.1016/s0092-8674(01)00380-4. [DOI] [PubMed] [Google Scholar]
- 156.Tanioka M, Yamada H, Doi M, et al. Molecular clocks in mouse skin. J Invest Dermatol. 2009 May;129(5):1225–1231. doi: 10.1038/jid.2008.345. [DOI] [PubMed] [Google Scholar]
- 157.Campisi J. Aging, Cellular Senescence, and Cancer. Annu Rev Physiol. 2012 Nov 8; doi: 10.1146/annurev-physiol-030212-183653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Cotta-Ramusino C, McDonald ER, 3rd, Hurov K, Sowa ME, Harper JW, Elledge SJ. A DNA damage response screen identifies RHINO, a 9-1-1 and TopBP1 interacting protein required for ATR signaling. Science. 2011 Jun 10;332(6035):1313–1317. doi: 10.1126/science.1203430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.DeBruyne JP, Weaver DR, Reppert SM. CLOCK and NPAS2 have overlapping roles in the suprachiasmatic circadian clock. Nat Neurosci. 2007 May;10(5):543–545. doi: 10.1038/nn1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Bertolucci C, Cavallari N, Colognesi I, et al. Evidence for an overlapping role of CLOCK and NPAS2 transcription factors in liver circadian oscillators. Mol Cell Biol. 2008 May;28(9):3070–3075. doi: 10.1128/MCB.01931-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Bae K, Jin X, Maywood ES, Hastings MH, Reppert SM, Weaver DR. Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron. 2001 May;30(2):525–536. doi: 10.1016/s0896-6273(01)00302-6. [DOI] [PubMed] [Google Scholar]
- 162.Debruyne JP, Noton E, Lambert CM, Maywood ES, Weaver DR, Reppert SM. A clock shock: mouse CLOCK is not required for circadian oscillator function. Neuron. 2006 May 4;50(3):465–477. doi: 10.1016/j.neuron.2006.03.041. [DOI] [PubMed] [Google Scholar]
- 163.Asher G, Schibler U. A CLOCK-less clock. Trends Cell Biol. 2006 Nov;16(11):547–549. doi: 10.1016/j.tcb.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 164.Spengler ML, Kuropatwinski KK, Comas M, et al. Core circadian protein CLOCK is a positive regulator of NF-kappaB-mediated transcription. Proc Natl Acad Sci U S A. 2012 Sep 11;109(37):E2457–E2465. doi: 10.1073/pnas.1206274109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Antoch MP, Song EJ, Chang AM, et al. Functional identification of the mouse circadian Clock gene by transgenic BAC rescue. Cell. 1997 May 16;89(4):655–667. doi: 10.1016/s0092-8674(00)80246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Lin KK, Kumar V, Geyfman M, et al. Circadian clock genes contribute to the regulation of hair follicle cycling. PLoS Genet. 2009 Jul;5(7):e1000573. doi: 10.1371/journal.pgen.1000573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Janich P, Pascual G, Merlos-Suarez A, et al. The circadian molecular clock creates epidermal stem cell heterogeneity. Nature. 2011 Dec 8;480(7376):209–214. doi: 10.1038/nature10649. [DOI] [PubMed] [Google Scholar]
- 168.Zeng ZL, Wu MW, Sun J, et al. Effects of the biological clock gene Bmal1 on tumour growth and anti-cancer drug activity. J Biochem. 2010 Sep;148(3):319–326. doi: 10.1093/jb/mvq069. [DOI] [PubMed] [Google Scholar]
- 169.Bunger MK, Wilsbacher LD, Moran SM, et al. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell. 2000 Dec 22;103(7):1009–1017. doi: 10.1016/s0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Gaddameedhi S, Reardon JT, Ye R, Ozturk N, Sancar A. Effect of circadian clock mutations on DNA damage response in mammalian cells. Cell Cycle. 2012 Sep 15;11(18):3481–3491. doi: 10.4161/cc.21771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Maclean KH, Keller UB, Rodriguez-Galindo C, Nilsson JA, Cleveland JL. c-Myc augments gamma irradiation-induced apoptosis by suppressing Bcl-XL. Mol Cell Biol. 2003 Oct;23(20):7256–7270. doi: 10.1128/MCB.23.20.7256-7270.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Yuan ZM, Huang Y, Ishiko T, Kharbanda S, Weichselbaum R, Kufe D. Regulation of DNA damage-induced apoptosis by the c-Abl tyrosine kinase. Proc Natl Acad Sci U S A. 1997 Feb 18;94(4):1437–1440. doi: 10.1073/pnas.94.4.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Marks PA, Rifkind RA, Gambari R, Epner E, Chen ZX, Banks J. Commitment to terminal differentiation and the cell cycle. Curr Top Cell Regul. 1982;21:189–203. doi: 10.1016/b978-0-12-152821-8.50012-8. [DOI] [PubMed] [Google Scholar]
- 174.Massague J. G1 cell-cycle control and cancer. Nature. 2004 Nov 18;432(7015):298–306. doi: 10.1038/nature03094. [DOI] [PubMed] [Google Scholar]
- 175.Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998 Jun 12;93(6):929–937. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- 176.Skehan P. Control models of cell cycle transit, exit, and arrest. Biochem Cell Biol. 1988 Jun;66(6):467–477. doi: 10.1139/o88-059. [DOI] [PubMed] [Google Scholar]
- 177.Grechez-Cassiau A, Rayet B, Guillaumond F, Teboul M, Delaunay F. The circadian clock component BMAL1 is a critical regulator of p21WAF1/CIP1 expression and hepatocyte proliferation. The Journal of biological chemistry. 2008 Feb 22;283(8):4535–4542. doi: 10.1074/jbc.M705576200. [DOI] [PubMed] [Google Scholar]
- 178.Koike N, Yoo SH, Huang HC, et al. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012 Oct 19;338(6105):349–354. doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Masri S, Zocchi L, Katada S, Mora E, Sassone-Corsi P. The circadian clock transcriptional complex: metabolic feedback intersects with epigenetic control. Ann N Y Acad Sci. 2012 Aug;1264(1):103–109. doi: 10.1111/j.1749-6632.2012.06649.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Reddy AB, Karp NA, Maywood ES, et al. Circadian orchestration of the hepatic proteome. Curr Biol. 2006 Jun 6;16(11):1107–1115. doi: 10.1016/j.cub.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 181.Hunt T, Sassone-Corsi P. Riding tandem: circadian clocks and the cell cycle. Cell. 2007 May 4;129(3):461–464. doi: 10.1016/j.cell.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 182.Yoo SH, Yamazaki S, Lowrey PL, et al. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci U S A. 2004 Apr 13;101(15):5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Schibler U, Sassone-Corsi P. A web of circadian pacemakers. Cell. 2002 Dec 27;111(7):919–922. doi: 10.1016/s0092-8674(02)01225-4. [DOI] [PubMed] [Google Scholar]
- 184.Nagoshi E, Saini C, Bauer C, Laroche T, Naef F, Schibler U. Circadian gene expression in individual fibroblasts: cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell. 2004 Nov 24;119(5):693–705. doi: 10.1016/j.cell.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 185.Florian MC, Geiger H. Concise review: polarity in stem cells, disease, and aging. Stem Cells. 2010 Sep;28(9):1623–1629. doi: 10.1002/stem.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.de Boer BA, van den Berg G, Soufan AT, et al. Measurement and 3D-visualization of cell-cycle length using double labelling with two thymidine analogues applied in early heart development. PLoS One. 2012;7(10):e47719. doi: 10.1371/journal.pone.0047719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Rubin P, Williams JP, Devesa SS, Travis LB, Constine LS. Cancer genesis across the age spectrum: associations with tissue development, maintenance, and senescence. Semin Radiat Oncol. 2010 Jan;20(1):3–11. doi: 10.1016/j.semradonc.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 188.Shackleton M. Normal stem cells and cancer stem cells: similar and different. Semin Cancer Biol. 2010 Apr;20(2):85–92. doi: 10.1016/j.semcancer.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 189.Cooper ZKaS. Mitotic rhythm in human epidermis. Proc. Soc. Exp. Biol. Med. 1938;(39):323–324. [Google Scholar]
- 190.Dublin WB, Gregg RO, Broders AC. Mitosis in specimens removed during day and night from carcinoma of large intestine. Arch. Pathol. 1940;(30):893–895. [Google Scholar]
- 191.Smaaland R, Laerum OD, Lote K, Sletvold O, Sothern RB, Bjerknes R. DNA synthesis in human bone marrow is circadian stage dependent. Blood. 1991 Jun 15;77(12):2603–2611. [PubMed] [Google Scholar]
- 192.Buchi KN, Moore JG, Hrushesky WJ, Sothern RB, Rubin NH. Circadian rhythm of cellular proliferation in the human rectal mucosa. Gastroenterology. 1991 Aug;101(2):410–415. doi: 10.1016/0016-5085(91)90019-h. [DOI] [PubMed] [Google Scholar]
- 193.Marra G, Anti M, Percesepe A, et al. Circadian variations of epithelial cell proliferation in human rectal crypts. Gastroenterology. 1994 Apr;106(4):982–987. doi: 10.1016/0016-5085(94)90757-9. [DOI] [PubMed] [Google Scholar]
- 194.Sothern RB, Smaaland R, Moore JG. Circannual rhythm in DNA synthesis (S-phase) in healthy human bone marrow and rectal mucosa. FASEB J. 1995 Mar;9(5):397–403. doi: 10.1096/fasebj.9.5.7896010. [DOI] [PubMed] [Google Scholar]
- 195.Rydell R, Wennerberg J, Willen R. Circadian variations in cell cycle phase distribution in a squamous cell carcinoma xenograft; effects of cisplatin and fluorouracil treatment. In Vivo. 1990 Nov-Dec;4(6):385–389. [PubMed] [Google Scholar]
- 196.Rotstein J, Sarma DS, Farber E. Sequential alterations in growth control and cell dynamics of rat hepatocytes in early precancerous steps in hepatocarcinogenesis. Cancer Res. 1986 May;46(5):2377–2385. [PubMed] [Google Scholar]
- 197.Izquierdo JN. Increased cell proliferation with persistence of circadian rhythms in hamster cheek pouch neoplasms. Cell Tissue Kinet. 1977 Jul;10(4):313–322. doi: 10.1111/j.1365-2184.1977.tb00299.x. [DOI] [PubMed] [Google Scholar]
- 198.Hrushesky WJ, Lannin D, Haus E. Evidence for an ontogenetic basis for circadian coordination of cancer cell proliferation. J Natl Cancer Inst. 1998 Oct 7;90(19):1480–1484. doi: 10.1093/jnci/90.19.1480. [DOI] [PubMed] [Google Scholar]
- 199.Echave Llanos JM, Nash RE. Mitotic circadian rhythm in a fast-growing and a slow-growing hepatoma: mitotic rhythm in hepatomas. J Natl Cancer Inst. 1970 Mar;44(3):581–585. [PubMed] [Google Scholar]
- 200.Klevecz RR, Braly PS. Circadian and ultradian rhythms of proliferation in human ovarian cancer. Chronobiology international. 1987;4(4):513–523. doi: 10.3109/07420528709078543. [DOI] [PubMed] [Google Scholar]
- 201.Klevecz RR, Braly PS. Circadian and ultradian cytokinetic rhythms of spontaneous human cancer. Ann N Y Acad Sci. 1991;618:257–276. doi: 10.1111/j.1749-6632.1991.tb27248.x. [DOI] [PubMed] [Google Scholar]
- 202.Smaaland R, Lote K, Sothern RB, Laerum OD. DNA synthesis and ploidy in non-Hodgkin's lymphomas demonstrate intrapatient variation depending on circadian stage of cell sampling. Cancer Res. 1993 Jul 1;53(13):3129–3138. [PubMed] [Google Scholar]
- 203.Dang CV. MYC on the path to cancer. Cell. 2012 Mar 30;149(1):22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Levens D. You Don't Muck with MYC. Genes Cancer. 2010 Jun 1;1(6):547–554. doi: 10.1177/1947601910377492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Evan GI, Vousden KH. Proliferation, cell cycle and apoptosis in cancer. Nature. 2001 May 17;411(6835):342–348. doi: 10.1038/35077213. [DOI] [PubMed] [Google Scholar]
- 206.Repouskou A, Sourlingas TG, Sekeri-Pataryas KE, Prombona A. The circadian expression of c-MYC is modulated by the histone deacetylase inhibitor trichostatin A in synchronized murine neuroblastoma cells. Chronobiology international. 2010 Jun;27(4):722–741. doi: 10.3109/07420521003786800. [DOI] [PubMed] [Google Scholar]
- 207.Bjarnason GA, Jordan R. Circadian variation of cell proliferation and cell cycle protein expression in man: clinical implications. Prog Cell Cycle Res. 2000;4:193–206. doi: 10.1007/978-1-4615-4253-7_17. [DOI] [PubMed] [Google Scholar]
- 208.Bjarnason GA, Jordan RC, Sothern RB. Circadian variation in the expression of cell-cycle proteins in human oral epithelium. Am J Pathol. 1999 Feb;154(2):613–622. doi: 10.1016/S0002-9440(10)65306-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Bjarnason GA, Jordan RC, Wood PA, et al. Circadian expression of clock genes in human oral mucosa and skin: association with specific cell-cycle phases. Am J Pathol. 2001 May;158(5):1793–1801. doi: 10.1016/S0002-9440(10)64135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Casagolda D, Del Valle-Perez B, Valls G, et al. A p120-catenin-CK1epsilon complex regulates Wnt signaling. J Cell Sci. 2010 Aug 1;123(Pt 15):2621–2631. doi: 10.1242/jcs.067512. [DOI] [PubMed] [Google Scholar]
- 211.Lee E, Salic A, Kirschner MW. Physiological regulation of [beta]-catenin stability by Tcf3 and CK1epsilon. J Cell Biol. 2001 Sep 3;154(5):983–993. doi: 10.1083/jcb.200102074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Anastas JN, Moon RT. WNT signalling pathways as therapeutic targets in cancer. Nat Rev Cancer. 2013 Jan;13(1):11–26. doi: 10.1038/nrc3419. [DOI] [PubMed] [Google Scholar]
- 213.Valenta T, Hausmann G, Basler K. The many faces and functions of beta-catenin. EMBO J. 2012 Jun 13;31(12):2714–2736. doi: 10.1038/emboj.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Batlle E, Henderson JT, Beghtel H, et al. Beta-catenin and TCF mediate cell positioning in the intestinal epithelium by controlling the expression of EphB/ephrinB. Cell. 2002 Oct 18;111(2):251–263. doi: 10.1016/s0092-8674(02)01015-2. [DOI] [PubMed] [Google Scholar]
- 215.van de Wetering M, de Lau W, Clevers H. WNT signaling and lymphocyte development. Cell. 2002 Apr;109(Suppl):S13–S19. doi: 10.1016/s0092-8674(02)00709-2. [DOI] [PubMed] [Google Scholar]
- 216.van de Wetering M, Sancho E, Verweij C, et al. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell. 2002 Oct 18;111(2):241–250. doi: 10.1016/s0092-8674(02)01014-0. [DOI] [PubMed] [Google Scholar]
- 217.Tetsu O, McCormick F. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999 Apr 1;398(6726):422–426. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- 218.Crawford HC, Fingleton B, Gustavson MD, et al. The PEA3 subfamily of Ets transcription factors synergizes with beta-catenin-LEF-1 to activate matrilysin transcription in intestinal tumors. Mol Cell Biol. 2001 Feb;21(4):1370–1383. doi: 10.1128/MCB.21.4.1370-1383.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Mann B, Gelos M, Siedow A, et al. Target genes of beta-catenin-T cell-factor/lymphoid-enhancer-factor signaling in human colorectal carcinomas. Proc Natl Acad Sci U S A. 1999 Feb 16;96(4):1603–1608. doi: 10.1073/pnas.96.4.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Ahmed Y, Hayashi S, Levine A, Wieschaus E. Regulation of armadillo by a Drosophila APC inhibits neuronal apoptosis during retinal development. Cell. 1998 Jun 26;93(7):1171–1182. doi: 10.1016/s0092-8674(00)81461-0. [DOI] [PubMed] [Google Scholar]
- 221.Martinek S, Inonog S, Manoukian AS, Young MW. A role for the segment polarity gene shaggy/GSK-3 in the Drosophila circadian clock. Cell. 2001 Jun 15;105(6):769–779. doi: 10.1016/s0092-8674(01)00383-x. [DOI] [PubMed] [Google Scholar]
- 222.Shimasaki T, Kitano A, Motoo Y, Minamoto T. Aberrant glycogen synthase kinase 3beta in the development of pancreatic cancer. J Carcinog. 2012;11:15. doi: 10.4103/1477-3163.100866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Shimura T. Acquired radioresistance of cancer and the AKT/GSK3beta/cyclin D1 overexpression cycle. J Radiat Res. 2011;52(5):539–544. doi: 10.1269/jrr.11098. [DOI] [PubMed] [Google Scholar]
- 224.Iitaka C, Miyazaki K, Akaike T, Ishida N. A role for glycogen synthase kinase-3beta in the mammalian circadian clock. The Journal of biological chemistry. 2005 Aug 19;280(33):29397–29402. doi: 10.1074/jbc.M503526200. [DOI] [PubMed] [Google Scholar]
- 225.Kondratov RV, Shamanna RK, Kondratova AA, Gorbacheva VY, Antoch MP. Dual role of the CLOCK/BMAL1 circadian complex in transcriptional regulation. FASEB J. 2006 Mar;20(3):530–532. doi: 10.1096/fj.05-5321fje. [DOI] [PubMed] [Google Scholar]
- 226.Doi M, Takahashi Y, Komatsu R, et al. Salt-sensitive hypertension in circadian clock-deficient Cry-null mice involves dysregulated adrenal Hsd3b6. Nature medicine. 2010 Jan;16(1):67–74. doi: 10.1038/nm.2061. [DOI] [PubMed] [Google Scholar]
- 227.Hua H, Wang Y, Wan C, et al. Circadian gene mPer2 overexpression induces cancer cell apoptosis. Cancer Sci. 2006 Jul;97(7):589–596. doi: 10.1111/j.1349-7006.2006.00225.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Relles D, Sendecki J, Chipitsyna G, Hyslop T, Yeo CJ, Arafat HA. Circadian gene expression and clinicopathologic correlates in pancreatic cancer. J Gastrointest Surg. 2013 Mar;17(3):443–450. doi: 10.1007/s11605-012-2112-2. [DOI] [PubMed] [Google Scholar]
- 229.Lindahl T, Barnes DE. Repair of endogenous DNA damage. Cold Spring Harb Symp Quant Biol. 2000;65:127–133. doi: 10.1101/sqb.2000.65.127. [DOI] [PubMed] [Google Scholar]
- 230.Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004 Nov 18;432(7015):316–323. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- 231.Thompson CB. Apoptosis in the pathogenesis and treatment of disease. Science. 1995 Mar 10;267(5203):1456–1462. doi: 10.1126/science.7878464. [DOI] [PubMed] [Google Scholar]
- 232.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003 Jan 30;421(6922):499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 233.Cimprich KA, Shin TB, Keith CT, Schreiber SL. cDNA cloning and gene mapping of a candidate human cell cycle checkpoint protein. Proc Natl Acad Sci U S A. 1996 Apr 2;93(7):2850–2855. doi: 10.1073/pnas.93.7.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009 Oct 22;461(7267):1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.el-Deiry WS, Tokino T, Velculescu VE, et al. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993 Nov 19;75(4):817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 236.Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993 Nov 19;75(4):805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 237.Rich T, Allen RL, Wyllie AH. Defying death after DNA damage. Nature. 2000 Oct 12;407(6805):777–783. doi: 10.1038/35037717. [DOI] [PubMed] [Google Scholar]
- 238.Kang TH, Lindsey-Boltz LA, Reardon JT, Sancar A. Circadian control of XPA and excision repair of cisplatin-DNA damage by cryptochrome and HERC2 ubiquitin ligase. Proc Natl Acad Sci U S A. 2010 Mar 16;107(11):4890–4895. doi: 10.1073/pnas.0915085107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Gaddameedhi S, Selby CP, Kaufmann WK, Smart RC, Sancar A. Control of skin cancer by the circadian rhythm. Proc Natl Acad Sci U S A. 2011 Nov 15;108(46):18790–18795. doi: 10.1073/pnas.1115249108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Chaves I, Pokorny R, Byrdin M, et al. The cryptochromes: blue light photoreceptors in plants and animals. Annu Rev Plant Biol. 2011 Jun;62:335–364. doi: 10.1146/annurev-arplant-042110-103759. [DOI] [PubMed] [Google Scholar]
- 241.Barnes JW, Tischkau SA, Barnes JA, et al. Requirement of mammalian Timeless for circadian rhythmicity. Science. 2003 Oct 17;302(5644):439–442. doi: 10.1126/science.1086593. [DOI] [PubMed] [Google Scholar]
- 242.Unsal-Kacmaz K, Mullen TE, Kaufmann WK, Sancar A. Coupling of human circadian and cell cycles by the timeless protein. Mol Cell Biol. 2005 Apr;25(8):3109–3116. doi: 10.1128/MCB.25.8.3109-3116.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Oklejewicz M, Destici E, Tamanini F, Hut RA, Janssens R, van der Horst GT. Phase resetting of the mammalian circadian clock by DNA damage. Curr Biol. 2008 Feb 26;18(4):286–291. doi: 10.1016/j.cub.2008.01.047. [DOI] [PubMed] [Google Scholar]
- 244.Gamsby JJ, Loros JJ, Dunlap JC. A phylogenetically conserved DNA damage response resets the circadian clock. J Biol Rhythms. 2009 Jun;24(3):193–202. doi: 10.1177/0748730409334748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Warburg O. On the origin of cancer cells. Science. 1956 Feb 24;123(3191):309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 246.Ferreira LM, Hebrant A, Dumont JE. Metabolic reprogramming of the tumor. Oncogene. 2012 Sep 6;31(36):3999–4011. doi: 10.1038/onc.2011.576. [DOI] [PubMed] [Google Scholar]
- 247.Hsu PP, Sabatini DM. Cancer cell metabolism: Warburg and beyond. Cell. 2008 Sep 5;134(5):703–707. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
- 248.Kroemer G, Pouyssegur J. Tumor cell metabolism: cancer's Achilles' heel. Cancer Cell. 2008 Jun;13(6):472–482. doi: 10.1016/j.ccr.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 249.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009 May 22;324(5930):1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Tennant DA, Duran RV, Boulahbel H, Gottlieb E. Metabolic transformation in cancer. Carcinogenesis. 2009 Aug;30(8):1269–1280. doi: 10.1093/carcin/bgp070. [DOI] [PubMed] [Google Scholar]
- 251.Bensaad K, Vousden KH. p53: new roles in metabolism. Trends in cell biology. 2007 Jun;17(6):286–291. doi: 10.1016/j.tcb.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 252.Levine AJ, Puzio-Kuter AM. The control of the metabolic switch in cancers by oncogenes and tumor suppressor genes. Science. 2010 Dec 3;330(6009):1340–1344. doi: 10.1126/science.1193494. [DOI] [PubMed] [Google Scholar]
- 253.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011 Feb;11(2):85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 254.Schulze A, Harris AL. How cancer metabolism is tuned for proliferation and vulnerable to disruption. Nature. 2012 Nov 15;491(7424):364–373. doi: 10.1038/nature11706. [DOI] [PubMed] [Google Scholar]
- 255.Huang W, Ramsey KM, Marcheva B, Bass J. Circadian rhythms, sleep, and metabolism. The Journal of clinical investigation. 2011 Jun;121(6):2133–2141. doi: 10.1172/JCI46043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Maury E, Ramsey KM, Bass J. Circadian rhythms and metabolic syndrome: from experimental genetics to human disease. Circ Res. 2010 Feb 19;106(3):447–462. doi: 10.1161/CIRCRESAHA.109.208355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Bass J, Takahashi JS. Circadian integration of metabolism and energetics. Science. 2010 Dec 3;330(6009):1349–1354. doi: 10.1126/science.1195027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Bass J. Circadian topology of metabolism. Nature. 2012 Nov 15;491(7424):348–356. doi: 10.1038/nature11704. [DOI] [PubMed] [Google Scholar]
- 259.Koike N, Yoo SH, Huang HC, et al. Transcriptional Architecture and Chromatin Landscape of the Core Circadian Clock in Mammals. Science. 2012 Aug 30; doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Garbarino-Pico E, Green CB. Posttranscriptional regulation of mammalian circadian clock output. Cold Spring Harb Symp Quant Biol. 2007;72:145–156. doi: 10.1101/sqb.2007.72.022. [DOI] [PubMed] [Google Scholar]
- 261.Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela F, Schibler U. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000 Dec 1;14(23):2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Stokkan KA, Yamazaki S, Tei H, Sakaki Y, Menaker M. Entrainment of the circadian clock in the liver by feeding. Science. 2001 Jan 19;291(5503):490–493. doi: 10.1126/science.291.5503.490. [DOI] [PubMed] [Google Scholar]
- 263.Rutter J, Reick M, Wu LC, McKnight SL. Regulation of clock and NPAS2 DNA binding by the redox state of NAD cofactors. Science. 2001 Jul 20;293(5529):510–514. doi: 10.1126/science.1060698. [DOI] [PubMed] [Google Scholar]
- 264.Ramsey KM, Yoshino J, Brace CS, et al. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009 May 1;324(5927):651–654. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Lamia KA, Sachdeva UM, DiTacchio L, et al. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science. 2009 Oct 16;326(5951):437–440. doi: 10.1126/science.1172156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Yin L, Wu N, Curtin JC, et al. Rev-erbalpha, a heme sensor that coordinates metabolic and circadian pathways. Science. 2007 Dec 14;318(5857):1786–1789. doi: 10.1126/science.1150179. [DOI] [PubMed] [Google Scholar]
- 267.Dang CV. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol Cell Biol. 1999 Jan;19(1):1–11. doi: 10.1128/mcb.19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Bensaad K, Tsuruta A, Selak MA, et al. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006 Jul 14;126(1):107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 269.Matoba S, Kang JG, Patino WD, et al. p53 regulates mitochondrial respiration. Science. 2006 Jun 16;312(5780):1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- 270.Fernandez PC, Frank SR, Wang L, et al. Genomic targets of the human c-Myc protein. Genes Dev. 2003 May 1;17(9):1115–1129. doi: 10.1101/gad.1067003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Orian A, Grewal SS, Knoepfler PS, Edgar BA, Parkhurst SM, Eisenman RN. Genomic binding and transcriptional regulation by the Drosophila Myc and Mnt transcription factors. Cold Spring Harb Symp Quant Biol. 2005;70:299–307. doi: 10.1101/sqb.2005.70.019. [DOI] [PubMed] [Google Scholar]
- 272.Li Z, Van Calcar S, Qu C, Cavenee WK, Zhang MQ, Ren B. A global transcriptional regulatory role for c-Myc in Burkitt's lymphoma cells. Proc Natl Acad Sci U S A. 2003 Jul 8;100(14):8164–8169. doi: 10.1073/pnas.1332764100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Cawley S, Bekiranov S, Ng HH, et al. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 2004 Feb 20;116(4):499–509. doi: 10.1016/s0092-8674(04)00127-8. [DOI] [PubMed] [Google Scholar]
- 274.Gao P, Tchernyshyov I, Chang TC, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009 Apr 9;458(7239):762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Cam H, Balciunaite E, Blais A, et al. A common set of gene regulatory networks links metabolism and growth inhibition. Mol Cell. 2004 Nov 5;16(3):399–411. doi: 10.1016/j.molcel.2004.09.037. [DOI] [PubMed] [Google Scholar]
- 276.Morrish F, Giedt C, Hockenbery D. c-MYC apoptotic function is mediated by NRF-1 target genes. Genes Dev. 2003 Jan 15;17(2):240–255. doi: 10.1101/gad.1032503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Hansson A, Hance N, Dufour E, et al. A switch in metabolism precedes increased mitochondrial biogenesis in respiratory chain-deficient mouse hearts. Proc Natl Acad Sci U S A. 2004 Mar 2;101(9):3136–3141. doi: 10.1073/pnas.0308710100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Vafa O, Wade M, Kern S, et al. c-Myc can induce DNA damage, increase reactive oxygen species, and mitigate p53 function: a mechanism for oncogene-induced genetic instability. Mol Cell. 2002 May;9(5):1031–1044. doi: 10.1016/s1097-2765(02)00520-8. [DOI] [PubMed] [Google Scholar]
- 279.Tanaka H, Matsumura I, Ezoe S, et al. E2F1 and c-Myc potentiate apoptosis through inhibition of NF-kappaB activity that facilitates MnSOD-mediated ROS elimination. Mol Cell. 2002 May;9(5):1017–1029. doi: 10.1016/s1097-2765(02)00522-1. [DOI] [PubMed] [Google Scholar]
- 280.K CS, Carcamo JM, Golde DW. Antioxidants prevent oxidative DNA damage and cellular transformation elicited by the over-expression of c-MYC. Mutat Res. 2006 Jan 29;593(1–2):64–79. doi: 10.1016/j.mrfmmm.2005.06.015. [DOI] [PubMed] [Google Scholar]
- 281.Pelengaris S, Khan M, Evan G. c-MYC: more than just a matter of life and death. Nat Rev Cancer. 2002 Oct;2(10):764–776. doi: 10.1038/nrc904. [DOI] [PubMed] [Google Scholar]
- 282.Breckenridge DG, Shore GC. Regulation of apoptosis by E1A and Myc oncoproteins. Crit Rev Eukaryot Gene Expr. 2000;10(3–4):273–280. doi: 10.1615/critreveukargeneexpr.v10.i3-4.50. [DOI] [PubMed] [Google Scholar]
- 283.Blyth K, Terry A, O'Hara M, et al. Synergy between a human c-myc transgene and p53 null genotype in murine thymic lymphomas: contrasting effects of homozygous and heterozygous p53 loss. Oncogene. 1995 May 4;10(9):1717–1723. [PubMed] [Google Scholar]
- 284.Elson A, Deng C, Campos-Torres J, Donehower LA, Leder P. The MMTV/c-myc transgene and p53 null alleles collaborate to induce T-cell lymphomas, but not mammary carcinomas in transgenic mice. Oncogene. 1995 Jul 6;11(1):181–190. [PubMed] [Google Scholar]
- 285.Lowe SW, Cepero E, Evan G. Intrinsic tumour suppression. Nature. 2004 Nov 18;432(7015):307–315. doi: 10.1038/nature03098. [DOI] [PubMed] [Google Scholar]
- 286.Pan X, Zhang Y, Wang L, Hussain MM. Diurnal regulation of MTP and plasma triglyceride by CLOCK is mediated by SHP. Cell Metab. 2010 Aug 4;12(2):174–186. doi: 10.1016/j.cmet.2010.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Liu C, Li S, Liu T, Borjigin J, Lin JD. Transcriptional coactivator PGC-1alpha integrates the mammalian clock and energy metabolism. Nature. 2007 May 24;447(7143):477–481. doi: 10.1038/nature05767. [DOI] [PubMed] [Google Scholar]
- 288.Hofman MA, Swaab DF. Living by the clock: the circadian pacemaker in older people. Ageing research reviews. 2006 Feb;5(1):33–51. doi: 10.1016/j.arr.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 289.Kondratov RV. A role of the circadian system and circadian proteins in aging. Ageing research reviews. 2007 May;6(1):12–27. doi: 10.1016/j.arr.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 290.Asai M, Yoshinobu Y, Kaneko S, et al. Circadian profile of Per gene mRNA expression in the suprachiasmatic nucleus, paraventricular nucleus, and pineal body of aged rats. J Neurosci Res. 2001 Dec 15;66(6):1133–1139. doi: 10.1002/jnr.10010. [DOI] [PubMed] [Google Scholar]
- 291.Lachman ME, Agrigoroaei S. Promoting functional health in midlife and old age: long-term protective effects of control beliefs, social support, and physical exercise. PLoS One. 2010;5(10):e13297. doi: 10.1371/journal.pone.0013297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Balducci L, Ershler WB. Cancer and ageing: a nexus at several levels. Nat Rev Cancer. 2005 Aug;5(8):655–662. doi: 10.1038/nrc1675. [DOI] [PubMed] [Google Scholar]
- 293.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA: a cancer journal for clinicians. 2010 Sep-Oct;60(5):277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 294.Rose MR. The evolutionary Biology of Aging. 1991 [Google Scholar]
- 295.G W. Pleiotropy, Natural Selection, and Senescence. Evolution. 1957;11:398–411. [Google Scholar]
- 296.Atwood CS, Bowen RL. The reproductive-cell cycle theory of aging: an update. Exp Gerontol. 2011 Feb-Mar;46(2–3):100–107. doi: 10.1016/j.exger.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 297.Strehler BL. Genetic instability as the primary cause of human aging. Exp Gerontol. 1986;21(4–5):283–319. doi: 10.1016/0531-5565(86)90038-0. [DOI] [PubMed] [Google Scholar]
- 298.Pawelec G, Derhovanessian E, Larbi A. Immunosenescence and cancer. Critical reviews in oncology/hematology. 2010 Aug;75(2):165–172. doi: 10.1016/j.critrevonc.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 299.Anisimov VN, Sikora E, Pawelec G. Relationships between cancer and aging: a multilevel approach. Biogerontology. 2009 Aug;10(4):323–338. doi: 10.1007/s10522-008-9209-8. [DOI] [PubMed] [Google Scholar]
- 300.Lee DJ, Cha EK, Dubin JM, et al. Novel therapeutics for the management of castration-resistant prostate cancer (CRPC) BJU international. 2012 Apr;109(7):968–985. doi: 10.1111/j.1464-410X.2011.10643.x. [DOI] [PubMed] [Google Scholar]
- 301.Minelli A, Bellezza I, Conte C, Culig Z. Oxidative stress-related aging: A role for prostate cancer? Biochim Biophys Acta. 2009 Apr;1795(2):83–91. doi: 10.1016/j.bbcan.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 302.Best BP. Nuclear DNA damage as a direct cause of aging. Rejuvenation research. 2009 Jun;12(3):199–208. doi: 10.1089/rej.2009.0847. [DOI] [PubMed] [Google Scholar]
- 303.Espejel S, Martin M, Klatt P, Martin-Caballero J, Flores JM, Blasco MA. Shorter telomeres, accelerated ageing and increased lymphoma in DNA-PKcs-deficient mice. EMBO Rep. 2004 May;5(5):503–509. doi: 10.1038/sj.embor.7400127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.de Boer J, Andressoo JO, de Wit J, et al. Premature aging in mice deficient in DNA repair and transcription. Science. 2002 May 17;296(5571):1276–1279. doi: 10.1126/science.1070174. [DOI] [PubMed] [Google Scholar]
- 305.Dolle ME, Busuttil RA, Garcia AM, et al. Increased genomic instability is not a prerequisite for shortened lifespan in DNA repair deficient mice. Mutat Res. 2006 Apr 11;596(1–2):22–35. doi: 10.1016/j.mrfmmm.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 306.Cao L, Li W, Kim S, Brodie SG, Deng CX. Senescence, aging, and malignant transformation mediated by p53 in mice lacking the Brca1 full-length isoform. Genes Dev. 2003 Jan 15;17(2):201–213. doi: 10.1101/gad.1050003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Sager R. Senescence as a mode of tumor suppression. Environmental health perspectives. 1991 Jun;93:59–62. doi: 10.1289/ehp.919359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Noureddine H, Gary-Bobo G, Alifano M, et al. Pulmonary artery smooth muscle cell senescence is a pathogenic mechanism for pulmonary hypertension in chronic lung disease. Circ Res. 2011 Aug 19;109(5):543–553. doi: 10.1161/CIRCRESAHA.111.241299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Parrinello S, Coppe JP, Krtolica A, Campisi J. Stromal-epithelial interactions in aging and cancer: senescent fibroblasts alter epithelial cell differentiation. J Cell Sci. 2005 Feb 1;118(Pt 3):485–496. doi: 10.1242/jcs.01635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Krtolica A, Parrinello S, Lockett S, Desprez PY, Campisi J. Senescent fibroblasts promote epithelial cell growth and tumorigenesis: a link between cancer and aging. Proc Natl Acad Sci U S A. 2001 Oct 9;98(21):12072–12077. doi: 10.1073/pnas.211053698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Liu D, Hornsby PJ. Senescent human fibroblasts increase the early growth of xenograft tumors via matrix metalloproteinase secretion. Cancer Res. 2007 Apr 1;67(7):3117–3126. doi: 10.1158/0008-5472.CAN-06-3452. [DOI] [PubMed] [Google Scholar]
- 312.Guney I, Wu S, Sedivy JM. Reduced c-Myc signaling triggers telomere-independent senescence by regulating Bmi-1 and p16(INK4a) Proc Natl Acad Sci U S A. 2006 Mar 7;103(10):3645–3650. doi: 10.1073/pnas.0600069103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Campisi J, d'Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol. 2007 Sep;8(9):729–740. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- 314.Adams PD. Healing and hurting: molecular mechanisms, functions, and pathologies of cellular senescence. Mol Cell. 2009 Oct 9;36(1):2–14. doi: 10.1016/j.molcel.2009.09.021. [DOI] [PubMed] [Google Scholar]
- 315.Collins CJ, Sedivy JM. Involvement of the INK4a/Arf gene locus in senescence. Aging Cell. 2003 Jun;2(3):145–150. doi: 10.1046/j.1474-9728.2003.00048.x. [DOI] [PubMed] [Google Scholar]
- 316.Rufini A, Tucci P, Celardo I, Melino G. Senescence and aging: the critical roles of p53. Oncogene. 2013 Feb 18; doi: 10.1038/onc.2012.640. [DOI] [PubMed] [Google Scholar]
- 317.Hardeland R, Coto-Montes A, Poeggeler B. Circadian rhythms, oxidative stress, and antioxidative defense mechanisms. Chronobiol Int. 2003 Nov;20(6):921–962. doi: 10.1081/cbi-120025245. [DOI] [PubMed] [Google Scholar]
- 318.Longo VD, Kennedy BK. Sirtuins in aging and age-related disease. Cell. 2006 Jul 28;126(2):257–268. doi: 10.1016/j.cell.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 319.Vaziri H, Dessain SK, Ng Eaton E, et al. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001 Oct 19;107(2):149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 320.Jeong J, Juhn K, Lee H, et al. SIRT1 promotes DNA repair activity and deacetylation of Ku70. Experimental & molecular medicine. 2007 Feb 28;39(1):8–13. doi: 10.1038/emm.2007.2. [DOI] [PubMed] [Google Scholar]
- 321.Cho IR, Koh SS, Malilas W, et al. SIRT1 inhibits proliferation of pancreatic cancer cells expressing pancreatic adenocarcinoma up-regulated factor (PAUF), a novel oncogene, by suppression of beta-catenin. Biochem Biophys Res Commun. 2012 Jun 29;423(2):270–275. doi: 10.1016/j.bbrc.2012.05.107. [DOI] [PubMed] [Google Scholar]
- 322.Wang CY, Wen MS, Wang HW, et al. Increased vascular senescence and impaired endothelial progenitor cell function mediated by mutation of circadian gene Per2. Circulation. 2008 Nov 18;118(21):2166–2173. doi: 10.1161/CIRCULATIONAHA.108.790469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Kang TW, Yevsa T, Woller N, et al. Senescence surveillance of pre-malignant hepatocytes limits liver cancer development. Nature. 2011 Nov 24;479(7374):547–551. doi: 10.1038/nature10599. [DOI] [PubMed] [Google Scholar]
- 324.Kalsbeek A, van der Spek R, Lei J, Endert E, Buijs RM, Fliers E. Circadian rhythms in the hypothalamo-pituitary-adrenal (HPA) axis. Molecular and cellular endocrinology. 2012 Feb 5;349(1):20–29. doi: 10.1016/j.mce.2011.06.042. [DOI] [PubMed] [Google Scholar]
- 325.Rajagopal S, Rajagopal K, Lefkowitz RJ. Teaching old receptors new tricks: biasing seven-transmembrane receptors. Nature reviews. Drug discovery. 2010 May;9(5):373–386. doi: 10.1038/nrd3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Rosenbaum DM, Rasmussen SG, Kobilka BK. The structure and function of G-protein-coupled receptors. Nature. 2009 May 21;459(7245):356–363. doi: 10.1038/nature08144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Janig W, Habler HJ. Specificity in the organization of the autonomic nervous system: a basis for precise neural regulation of homeostatic and protective body functions. Prog Brain Res. 2000;122:351–367. doi: 10.1016/s0079-6123(08)62150-0. [DOI] [PubMed] [Google Scholar]
- 328.Hanaoka BY, Peterson CA, Horbinski C, Crofford LJ. Implications of glucocorticoid therapy in idiopathic inflammatory myopathies. Nature reviews. Rheumatology. 2012 Aug;8(8):448–457. doi: 10.1038/nrrheum.2012.85. [DOI] [PubMed] [Google Scholar]
- 329.Samarasinghe RA, Witchell SF, DeFranco DB. Cooperativity and complementarity: synergies in non-classical and classical glucocorticoid signaling. Cell Cycle. 2012 Aug 1;11(15):2819–2827. doi: 10.4161/cc.21018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Deblois G, Giguere V. Oestrogen-related receptors in breast cancer: control of cellular metabolism and beyond. Nat Rev Cancer. 2013 Jan;13(1):27–36. doi: 10.1038/nrc3396. [DOI] [PubMed] [Google Scholar]
- 331.Cui J, Shen Y, Li R. Estrogen synthesis and signaling pathways during aging: from periphery to brain. Trends in molecular medicine. 2013 Jan 21; doi: 10.1016/j.molmed.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Hasan KM, Rahman MS, Arif KM, Sobhani ME. Psychological stress and aging: role of glucocorticoids (GCs) Age (Dordr) 2012 Dec;34(6):1421–1433. doi: 10.1007/s11357-011-9319-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Coto-Montes A, Boga JA, Rosales-Corral S, Fuentes-Broto L, Tan DX, Reiter RJ. Role of melatonin in the regulation of autophagy and mitophagy: a review. Molecular and cellular endocrinology. 2012 Sep 25;361(1–2):12–23. doi: 10.1016/j.mce.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 334.Greene MW. Circadian rhythms and tumor growth. Cancer Lett. 2012 May 28;318(2):115–123. doi: 10.1016/j.canlet.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 335.Reiter RJ, Calvo JR, Karbownik M, Qi W, Tan DX. Melatonin and its relation to the immune system and inflammation. Ann N Y Acad Sci. 2000;917:376–386. doi: 10.1111/j.1749-6632.2000.tb05402.x. [DOI] [PubMed] [Google Scholar]
- 336.Xu CX, Wang C, Krager SL, Bottum KM, Tischkau SA. Aryl Hydrocarbon Receptor Activation Attenuates Per1 Gene Induction and Influences Circadian Clock Resetting. Toxicol Sci. 2013 Feb 1; doi: 10.1093/toxsci/kfs345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Zhu H, Vadigepalli R, Rafferty R, Gonye GE, Weaver DR, Schwaber JS. Integrative gene regulatory network analysis reveals light-induced regional gene expression phase shift programs in the mouse suprachiasmatic nucleus. PLoS One. 2012;7(5):e37833. doi: 10.1371/journal.pone.0037833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Jakubcakova V, Oster H, Tamanini F, et al. Light entrainment of the mammalian circadian clock by a PRKCA-dependent posttranslational mechanism. Neuron. 2007 Jun 7;54(5):831–843. doi: 10.1016/j.neuron.2007.04.031. [DOI] [PubMed] [Google Scholar]
- 339.Hainich EC, Pizzio GA, Golombek DA. Constitutive activation of the ERK-MAPK pathway in the suprachiasmatic nuclei inhibits circadian resetting. FEBS Lett. 2006 Dec 11;580(28–29):6665–6668. doi: 10.1016/j.febslet.2006.11.019. [DOI] [PubMed] [Google Scholar]
- 340.Hirota T, Fukada Y. Resetting mechanism of central and peripheral circadian clocks in mammals. Zoological science. 2004 Apr;21(4):359–368. doi: 10.2108/zsj.21.359. [DOI] [PubMed] [Google Scholar]
- 341.Shigeyoshi Y, Taguchi K, Yamamoto S, et al. Light-induced resetting of a mammalian circadian clock is associated with rapid induction of the mPer1 transcript. Cell. 1997 Dec 26;91(7):1043–1053. doi: 10.1016/s0092-8674(00)80494-8. [DOI] [PubMed] [Google Scholar]
- 342.Takeuchi J, Shannon W, Aronin N, Schwartz WJ. Compositional changes of AP-1 DNA-binding proteins are regulated by light in a mammalian circadian clock. Neuron. 1993 Nov;11(5):825–836. doi: 10.1016/0896-6273(93)90112-5. [DOI] [PubMed] [Google Scholar]
- 343.Obrietan K, Impey S, Storm DR. Light and circadian rhythmicity regulate MAP kinase activation in the suprachiasmatic nuclei. Nat Neurosci. 1998 Dec;1(8):693–700. doi: 10.1038/3695. [DOI] [PubMed] [Google Scholar]
- 344.Ding JM, Faiman LE, Hurst WJ, Kuriashkina LR, Gillette MU. Resetting the biological clock: mediation of nocturnal CREB phosphorylation via light, glutamate, and nitric oxide. J Neurosci. 1997 Jan 15;17(2):667–675. doi: 10.1523/JNEUROSCI.17-02-00667.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Hurst WJ, Mitchell JW, Gillette MU. Synchronization and phase-resetting by glutamate of an immortalized SCN cell line. Biochem Biophys Res Commun. 2002 Oct 18;298(1):133–143. doi: 10.1016/s0006-291x(02)02346-x. [DOI] [PubMed] [Google Scholar]
- 346.Kon N, Hirota T, Kawamoto T, Kato Y, Tsubota T, Fukada Y. Activation of TGF-beta/activin signalling resets the circadian clock through rapid induction of Dec1 transcripts. Nat Cell Biol. 2008 Dec;10(12):1463–1469. doi: 10.1038/ncb1806. [DOI] [PubMed] [Google Scholar]
- 347.Gerber A, Esnault C, Aubert G, Treisman R, Pralong F, Schibler U. Blood-Borne Circadian Signal Stimulates Daily Oscillations in Actin Dynamics and SRF Activity. Cell. 2013 Jan 31;152(3):492–503. doi: 10.1016/j.cell.2012.12.027. [DOI] [PubMed] [Google Scholar]
- 348.Lu NZ, Cidlowski JA. The origin and functions of multiple human glucocorticoid receptor isoforms. Ann N Y Acad Sci. 2004 Jun;1024:102–123. doi: 10.1196/annals.1321.008. [DOI] [PubMed] [Google Scholar]
- 349.Chung S, Son GH, Kim K. Adrenal peripheral oscillator in generating the circadian glucocorticoid rhythm. Ann N Y Acad Sci. 2011 Mar;1220:71–81. doi: 10.1111/j.1749-6632.2010.05923.x. [DOI] [PubMed] [Google Scholar]
- 350.Reddy AB, Maywood ES, Karp NA, et al. Glucocorticoid signaling synchronizes the liver circadian transcriptome. Hepatology. 2007 Jun;45(6):1478–1488. doi: 10.1002/hep.21571. [DOI] [PubMed] [Google Scholar]
- 351.Yamamoto T, Shimano H, Nakagawa Y, et al. SREBP-1 interacts with hepatocyte nuclear factor-4 alpha and interferes with PGC-1 recruitment to suppress hepatic gluconeogenic genes. J Biol Chem. 2004 Mar 26;279(13):12027–12035. doi: 10.1074/jbc.M310333200. [DOI] [PubMed] [Google Scholar]
- 352.So AY, Bernal TU, Pillsbury ML, Yamamoto KR, Feldman BJ. Glucocorticoid regulation of the circadian clock modulates glucose homeostasis. Proc Natl Acad Sci U S A. 2009 Oct 13;106(41):17582–17587. doi: 10.1073/pnas.0909733106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Lamia KA, Papp SJ, Yu RT, et al. Cryptochromes mediate rhythmic repression of the glucocorticoid receptor. Nature. 2011 Dec 22;480(7378):552–556. doi: 10.1038/nature10700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Pezuk P, Mohawk JA, Wang LA, Menaker M. Glucocorticoids as entraining signals for peripheral circadian oscillators. Endocrinology. 2012 Oct;153(10):4775–4783. doi: 10.1210/en.2012-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Furness JB. The organisation of the autonomic nervous system: peripheral connections. Auton Neurosci. 2006 Dec 30;130(1–2):1–5. doi: 10.1016/j.autneu.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 356.Shibata S. Neural regulation of the hepatic circadian rhythm. The anatomical record. Part A, Discoveries in molecular, cellular, and evolutionary biology. 2004 Sep;280(1):901–909. doi: 10.1002/ar.a.20095. [DOI] [PubMed] [Google Scholar]
- 357.Vujovic N, Davidson AJ, Menaker M. Sympathetic input modulates, but does not determine, phase of peripheral circadian oscillators. Am J Physiol Regul Integr Comp Physiol. 2008 Jul;295(1):R355–R360. doi: 10.1152/ajpregu.00498.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Takekida S, Yan L, Maywood ES, Hastings MH, Okamura H. Differential adrenergic regulation of the circadian expression of the clock genes Period1 and Period2 in the rat pineal gland. Eur J Neurosci. 2000 Dec;12(12):4557–4561. doi: 10.1046/j.0953-816x.2000.01324.x. [DOI] [PubMed] [Google Scholar]
- 359.Logan RW, Arjona A, Sarkar DK. Role of sympathetic nervous system in the entrainment of circadian natural-killer cell function. Brain Behav Immun. 2011 Jan;25(1):101–109. doi: 10.1016/j.bbi.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Mendez-Ferrer S, Lucas D, Battista M, Frenette PS. Haematopoietic stem cell release is regulated by circadian oscillations. Nature. 2008 Mar 27;452(7186):442–447. doi: 10.1038/nature06685. [DOI] [PubMed] [Google Scholar]
- 361.Kang J, Shi Y, Xiang B, et al. A nuclear function of beta-arrestin1 in GPCR signaling: regulation of histone acetylation and gene transcription. Cell. 2005 Dec 2;123(5):833–847. doi: 10.1016/j.cell.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 362.Tilley DG. G protein-dependent and G protein-independent signaling pathways and their impact on cardiac function. Circ Res. 2011 Jul 8;109(2):217–230. doi: 10.1161/CIRCRESAHA.110.231225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Reiter E, Ahn S, Shukla AK, Lefkowitz RJ. Molecular mechanism of beta-arrestin-biased agonism at seven-transmembrane receptors. Annu Rev Pharmacol Toxicol. 2012;52:179–197. doi: 10.1146/annurev.pharmtox.010909.105800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.DeFea KA. Beta-arrestins as regulators of signal termination and transduction: how do they determine what to scaffold? Cell Signal. 2011 Apr;23(4):621–629. doi: 10.1016/j.cellsig.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 365.Verkaar F, van Rosmalen JW, Blomenrohr M, et al. G protein-independent cell-based assays for drug discovery on seven-transmembrane receptors. Biotechnology annual review. 2008;14:253–274. doi: 10.1016/S1387-2656(08)00010-0. [DOI] [PubMed] [Google Scholar]
- 366.Ling J, Kumar R. Crosstalk between NFkB and glucocorticoid signaling: a potential target of breast cancer therapy. Cancer Lett. 2012 Sep 28;322(2):119–126. doi: 10.1016/j.canlet.2012.02.033. [DOI] [PubMed] [Google Scholar]
- 367.Grossmann C, Gekle M. Interaction between mineralocorticoid receptor and epidermal growth factor receptor signaling. Molecular and cellular endocrinology. 2012 Mar 24;350(2):235–241. doi: 10.1016/j.mce.2011.07.045. [DOI] [PubMed] [Google Scholar]
- 368.Schakman O, Gilson H, Kalista S, Thissen JP. Mechanisms of muscle atrophy induced by glucocorticoids. Hormone research. 2009 Nov;72(Suppl 1):36–41. doi: 10.1159/000229762. [DOI] [PubMed] [Google Scholar]
- 369.Mulholland DJ, Dedhar S, Coetzee GA, Nelson CC. Interaction of nuclear receptors with the Wnt/beta-catenin/Tcf signaling axis: Wnt you like to know? Endocr Rev. 2005 Dec;26(7):898–915. doi: 10.1210/er.2003-0034. [DOI] [PubMed] [Google Scholar]
- 370.Lang F, Perrotti N, Stournaras C. Colorectal carcinoma cells--regulation of survival and growth by SGK1. Int J Biochem Cell Biol. 2010 Oct;42(10):1571–1575. doi: 10.1016/j.biocel.2010.05.016. [DOI] [PubMed] [Google Scholar]
- 371.Fu L, Lee CC. The circadian clock: pacemaker and tumour suppressor. Nat Rev Cancer. 2003 May;3(5):350–361. doi: 10.1038/nrc1072. [DOI] [PubMed] [Google Scholar]
- 372.Rhind N, Russell P. Signaling pathways that regulate cell division. Cold Spring Harbor perspectives in biology. 2012;4(10) doi: 10.1101/cshperspect.a005942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Gallego M, Virshup DM. Post-translational modifications regulate the ticking of the circadian clock. Nat Rev Mol Cell Biol. 2007 Feb;8(2):139–148. doi: 10.1038/nrm2106. [DOI] [PubMed] [Google Scholar]
- 374.Cao R, Obrietan K. mTOR Signaling and Entrainment of the Mammalian Circadian Clock. Molecular and cellular pharmacology. 2010;2(4):125–130. doi: 10.4255/mcpharmacol.10.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Volden PA, Conzen SD. The influence of glucocorticoid signaling on tumor progression. Brain Behav Immun. 2012 Nov 16; doi: 10.1016/j.bbi.2012.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Cole SW. Nervous system regulation of the cancer genome. Brain Behav Immun. 2012 Dec 1; doi: 10.1016/j.bbi.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Zhang EE, Liu Y, Dentin R, et al. Cryptochrome mediates circadian regulation of cAMP signaling and hepatic gluconeogenesis. Nature medicine. 2010 Oct;16(10):1152–1156. doi: 10.1038/nm.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Ikeda H, Yong Q, Kurose T, et al. Clock gene defect disrupts light-dependency of autonomic nerve activity. Biochemical and biophysical research communications. 2007 Dec 21;364(3):457–463. doi: 10.1016/j.bbrc.2007.10.058. [DOI] [PubMed] [Google Scholar]
- 379.Anand P, Kunnumakkara AB, Sundaram C, et al. Cancer is a preventable disease that requires major lifestyle changes. Pharmaceutical research. 2008 Sep;25(9):2097–2116. doi: 10.1007/s11095-008-9661-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Dunn GP, Old LJ, Schreiber RD. The three Es of cancer immunoediting. Annual review of immunology. 2004;22:329–360. doi: 10.1146/annurev.immunol.22.012703.104803. [DOI] [PubMed] [Google Scholar]
- 381.Mlecnik B, Bindea G, Pages F, Galon J. Tumor immunosurveillance in human cancers. Cancer metastasis reviews. 2011 Mar;30(1):5–12. doi: 10.1007/s10555-011-9270-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Street SE, Cretney E, Smyth MJ. Perforin and interferon-gamma activities independently control tumor initiation, growth, and metastasis. Blood. 2001 Jan 1;97(1):192–197. doi: 10.1182/blood.v97.1.192. [DOI] [PubMed] [Google Scholar]
- 383.Street SE, Trapani JA, MacGregor D, Smyth MJ. Suppression of lymphoma and epithelial malignancies effected by interferon gamma. J Exp Med. 2002 Jul 1;196(1):129–134. doi: 10.1084/jem.20020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.van den Broek ME, Kagi D, Ossendorp F, et al. Decreased tumor surveillance in perforin-deficient mice. J Exp Med. 1996 Nov 1;184(5):1781–1790. doi: 10.1084/jem.184.5.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Shinkai Y, Rathbun G, Lam KP, et al. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 1992 Mar 6;68(5):855–867. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- 386.Shankaran V, Ikeda H, Bruce AT, et al. IFNgamma and lymphocytes prevent primary tumour development and shape tumour immunogenicity. Nature. 2001 Apr 26;410(6832):1107–1111. doi: 10.1038/35074122. [DOI] [PubMed] [Google Scholar]
- 387.Khong HT, Restifo NP. Natural selection of tumor variants in the generation of "tumor escape" phenotypes. Nature immunology. 2002 Nov;3(11):999–1005. doi: 10.1038/ni1102-999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Mellman I, Coukos G, Dranoff G. Cancer immunotherapy comes of age. Nature. 2011 Dec 22;480(7378):480–489. doi: 10.1038/nature10673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Scheiermann C, Kunisaki Y, Frenette PS. Circadian control of the immune system. Nature reviews. Immunology. 2013 Mar;13(3):190–198. doi: 10.1038/nri3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Bellinger DL, Millar BA, Perez S, et al. Sympathetic modulation of immunity: relevance to disease. Cellular immunology. 2008 Mar-Apr;252(1–2):27–56. doi: 10.1016/j.cellimm.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Sloan EK, Capitanio JP, Cole SW. Stress-induced remodeling of lymphoid innervation. Brain Behav Immun. 2008 Jan;22(1):15–21. doi: 10.1016/j.bbi.2007.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Srinivasan V, Pandi-Perumal SR, Brzezinski A, Bhatnagar KP, Cardinali DP. Melatonin, immune function and cancer. Recent patents on endocrine, metabolic & immune drug discovery. 2011 May;5(2):109–123. doi: 10.2174/187221411799015408. [DOI] [PubMed] [Google Scholar]
- 393.Bellavance MA, Rivest S. The neuroendocrine control of the innate immune system in health and brain diseases. Immunol Rev. 2012 Jul;248(1):36–55. doi: 10.1111/j.1600-065X.2012.01129.x. [DOI] [PubMed] [Google Scholar]
- 394.Chighizola C, Meroni PL. The role of environmental estrogens and autoimmunity. Autoimmunity reviews. 2012 May;11(6–7):A493–A501. doi: 10.1016/j.autrev.2011.11.027. [DOI] [PubMed] [Google Scholar]
- 395.Arjona A, Sarkar DK. Circadian oscillations of clock genes, cytolytic factors, and cytokines in rat NK cells. Journal of immunology. 2005 Jun 15;174(12):7618–7624. doi: 10.4049/jimmunol.174.12.7618. [DOI] [PubMed] [Google Scholar]
- 396.Arjona A, Boyadjieva N, Sarkar DK. Circadian rhythms of granzyme B, perforin, IFN-gamma, and NK cell cytolytic activity in the spleen: effects of chronic ethanol. J Immunol. 2004 Mar 1;172(5):2811–2817. doi: 10.4049/jimmunol.172.5.2811. [DOI] [PubMed] [Google Scholar]
- 397.Arjona A, Sarkar DK. Evidence supporting a circadian control of natural killer cell function. Brain, behavior, and immunity. 2006 Sep;20(5):469–476. doi: 10.1016/j.bbi.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 398.Arjona A, Sarkar DK. Are circadian rhythms the code of hypothalamic-immune communication? Insights from natural killer cells. Neurochemical research. 2008 Apr;33(4):708–718. doi: 10.1007/s11064-007-9501-z. [DOI] [PubMed] [Google Scholar]
- 399.Haus E, Smolensky MH. Biologic rhythms in the immune system. Chronobiol Int. 1999 Sep;16(5):581–622. doi: 10.3109/07420529908998730. [DOI] [PubMed] [Google Scholar]
- 400.Silver AC, Arjona A, Hughes ME, Nitabach MN, Fikrig E. Circadian expression of clock genes in mouse macrophages, dendritic cells, and B cells. Brain Behav Immun. 2012 Mar;26(3):407–413. doi: 10.1016/j.bbi.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Suzuki S, Toyabe S, Moroda T, et al. Circadian rhythm of leucocytes and lymphocytes subsets and its possible correlation with the function of the autonomic nervous system. Clin Exp Immunol. 1997 Dec;110(3):500–508. doi: 10.1046/j.1365-2249.1997.4411460.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Okamoto S, Ibaraki K, Hayashi S, Saito M. Ventromedial hypothalamus suppresses splenic lymphocyte activity through sympathetic innervation. Brain Res. 1996 Nov 11;739(1–2):308–313. doi: 10.1016/s0006-8993(96)00840-2. [DOI] [PubMed] [Google Scholar]
- 403.Maestroni GJ, Cosentino M, Marino F, et al. Neural and endogenous catecholamines in the bone marrow. Circadian association of norepinephrine with hematopoiesis? Exp Hematol. 1998 Nov;26(12):1172–1177. [PubMed] [Google Scholar]
- 404.Sephton S, Spiegel D. Circadian disruption in cancer: a neuroendocrine-immune pathway from stress to disease? Brain Behav Immun. 2003 Oct;17(5):321–328. doi: 10.1016/s0889-1591(03)00078-3. [DOI] [PubMed] [Google Scholar]
- 405.Arjona A, Sarkar DK. The circadian gene mPer2 regulates the daily rhythm of IFN-gamma. Journal of interferon & cytokine research : the official journal of the International Society for Interferon and Cytokine Research. 2006 Sep;26(9):645–649. doi: 10.1089/jir.2006.26.645. [DOI] [PubMed] [Google Scholar]
- 406.Logan RW, Wynne O, Levitt D, Price D, Sarkar DK. Altered Circadian Expression of Cytokines and Cytolytic Factors in Splenic Natural Killer Cells of Per1(−/−) Mutant Mice. Journal of interferon & cytokine research : the official journal of the International Society for Interferon and Cytokine Research. 2013 Feb 12; doi: 10.1089/jir.2012.0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Gibbs JE, Blaikley J, Beesley S, et al. The nuclear receptor REV-ERBalpha mediates circadian regulation of innate immunity through selective regulation of inflammatory cytokines. Proc Natl Acad Sci U S A. 2012 Jan 10;109(2):582–587. doi: 10.1073/pnas.1106750109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Wang X, Reece SP, Van Scott MR, Brown JM. A circadian clock in murine bone marrow-derived mast cells modulates IgE-dependent activation in vitro. Brain, behavior, and immunity. 2011 Jan;25(1):127–134. doi: 10.1016/j.bbi.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 409.Luo Y, Tian W, Cai L, et al. Expression profiling reveals a positive regulation by mPer2 on circadian rhythm of cytotoxicity receptors: Ly49C and Nkg2d. Chronobiology international. 2009 Dec;26(8):1514–1544. doi: 10.3109/07420520903553435. [DOI] [PubMed] [Google Scholar]
- 410.Sun Y, Yang Z, Niu Z, et al. MOP3, a component of the molecular clock, regulates the development of B cells. Immunology. 2006 Dec;119(4):451–460. doi: 10.1111/j.1365-2567.2006.02456.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Liu J, Malkani G, Shi X, et al. The circadian clock Period 2 gene regulates gamma interferon production of NK cells in host response to lipopolysaccharide-induced endotoxic shock. Infection and immunity. 2006 Aug;74(8):4750–4756. doi: 10.1128/IAI.00287-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.Logan RW, Sarkar DK. Circadian nature of immune function. Molecular and cellular endocrinology. 2012 Feb 5;349(1):82–90. doi: 10.1016/j.mce.2011.06.039. [DOI] [PubMed] [Google Scholar]
- 413.Tong X, Buelow K, Guha A, Rausch R, Yin L. USP2a protein deubiquitinates and stabilizes the circadian protein CRY1 in response to inflammatory signals. The Journal of biological chemistry. 2012 Jul 20;287(30):25280–25291. doi: 10.1074/jbc.M112.340786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Murphy BA, Vick MM, Sessions DR, Cook RF, Fitzgerald BP. Acute systemic inflammation transiently synchronizes clock gene expression in equine peripheral blood. Brain, behavior, and immunity. 2007 May;21(4):467–476. doi: 10.1016/j.bbi.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 415.Kouri VP, Olkkonen J, Kaivosoja E, et al. Circadian timekeeping is disturbed in rheumatoid arthritis at molecular level. PLoS One. 2013;8(1):e54049. doi: 10.1371/journal.pone.0054049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Ohdo S, Koyanagi S, Suyama H, Higuchi S, Aramaki H. Changing the dosing schedule minimizes the disruptive effects of interferon on clock function. Nat Med. 2001 Mar;7(3):356–360. doi: 10.1038/85507. [DOI] [PubMed] [Google Scholar]
- 417.Besedovsky H, del Rey A, Sorkin E, Dinarello CA. Immunoregulatory feedback between interleukin-1 and glucocorticoid hormones. Science. 1986 Aug 8;233(4764):652–654. doi: 10.1126/science.3014662. [DOI] [PubMed] [Google Scholar]
- 418.Pan A, Schernhammer ES, Sun Q, Hu FB. Rotating night shift work and risk of type 2 diabetes: two prospective cohort studies in women. PLoS Med. 2011 Dec;8(12):e1001141. doi: 10.1371/journal.pmed.1001141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Husse J, Hintze SC, Eichele G, Lehnert H, Oster H. Circadian clock genes Per1 and Per2 regulate the response of metabolism-associated transcripts to sleep disruption. PLoS One. 2012;7(12):e52983. doi: 10.1371/journal.pone.0052983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Schiavo-Cardozo D, Lima MM, Pareja JC, Geloneze B. Appetite-regulating hormones from the upper gut: disrupted control of xenin and ghrelin in night workers. Clin Endocrinol (Oxf) 2012 Dec 1; doi: 10.1111/cen.12114. [DOI] [PubMed] [Google Scholar]
- 421.Morris CJ, Yang JN, Scheer FA. The impact of the circadian timing system on cardiovascular and metabolic function. Prog Brain Res. 2012;199:337–358. doi: 10.1016/B978-0-444-59427-3.00019-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.Roenneberg T, Allebrandt KV, Merrow M, Vetter C. Social jetlag and obesity. Curr Biol. 2012 May 22;22(10):939–943. doi: 10.1016/j.cub.2012.03.038. [DOI] [PubMed] [Google Scholar]
- 423.Arble DM, Bass J, Laposky AD, Vitaterna MH, Turek FW. Circadian timing of food intake contributes to weight gain. Obesity. 2009 Nov;17(11):2100–2102. doi: 10.1038/oby.2009.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Marcheva B, Ramsey KM, Buhr ED, et al. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature. 2010 Jul 29;466(7306):627–631. doi: 10.1038/nature09253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425.Turek FW, Joshu C, Kohsaka A, et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005 May 13;308(5724):1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426.Milagro FI, Gomez-Abellan P, Campion J, Martinez JA, Ordovas JM, Garaulet M. CLOCK, PER2 and BMAL1 DNA methylation: association with obesity and metabolic syndrome characteristics and monounsaturated fat intake. Chronobiol Int. 2012 Nov;29(9):1180–1194. doi: 10.3109/07420528.2012.719967. [DOI] [PubMed] [Google Scholar]
- 427.Garcia-Rios A, Perez-Martinez P, Delgado-Lista J, et al. A Period 2 genetic variant interacts with plasma SFA to modify plasma lipid concentrations in adults with metabolic syndrome. J Nutr. 2012 Jul;142(7):1213–1218. doi: 10.3945/jn.111.156968. [DOI] [PubMed] [Google Scholar]
- 428.Gomez-Abellan P, Madrid JA, Lujan JA, et al. Sexual dimorphism in clock genes expression in human adipose tissue. Obesity surgery. 2012 Jan;22(1):105–112. doi: 10.1007/s11695-011-0539-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 429.Gomez-Abellan P, Hernandez-Morante JJ, Lujan JA, Madrid JA, Garaulet M. Clock genes are implicated in the human metabolic syndrome. Int J Obes (Lond) 2008 Jan;32(1):121–128. doi: 10.1038/sj.ijo.0803689. [DOI] [PubMed] [Google Scholar]
- 430.Williams SR, Zies D, Mullegama SV, Grotewiel MS, Elsea SH. Smith-Magenis syndrome results in disruption of CLOCK gene transcription and reveals an integral role for RAI1 in the maintenance of circadian rhythmicity. Am J Hum Genet. 2012 Jun 8;90(6):941–949. doi: 10.1016/j.ajhg.2012.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Garaulet M, Corbalan-Tutau MD, Madrid JA, et al. PERIOD2 variants are associated with abdominal obesity, psycho-behavioral factors, and attrition in the dietary treatment of obesity. Journal of the American Dietetic Association. 2010 Jun;110(6):917–921. doi: 10.1016/j.jada.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432.Morton GJ, Cummings DE, Baskin DG, Barsh GS, Schwartz MW. Central nervous system control of food intake and body weight. Nature. 2006 Sep 21;443(7109):289–295. doi: 10.1038/nature05026. [DOI] [PubMed] [Google Scholar]
- 433.Myers MG, Jr, Olson DP. Central nervous system control of metabolism. Nature. 2012 Nov 15;491(7424):357–363. doi: 10.1038/nature11705. [DOI] [PubMed] [Google Scholar]
- 434.Gonnissen HK, Hulshof T, Westerterp-Plantenga MS. Chronobiology, endocrinology, and energy- and food-reward homeostasis. Obesity reviews : an official journal of the International Association for the Study of Obesity. 2013 Feb 6; doi: 10.1111/obr.12019. [DOI] [PubMed] [Google Scholar]
- 435.Schibler U, Ripperger J, Brown SA. Peripheral circadian oscillators in mammals: time and food. J Biol Rhythms. 2003 Jun;18(3):250–260. doi: 10.1177/0748730403018003007. [DOI] [PubMed] [Google Scholar]
- 436.Verwey M, Amir S. Nucleus-specific effects of meal duration on daily profiles of Period1 and Period2 protein expression in rats housed under restricted feeding. Neuroscience. 2011 Sep 29;192:304–311. doi: 10.1016/j.neuroscience.2011.07.016. [DOI] [PubMed] [Google Scholar]
- 437.Guilding C, Hughes AT, Brown TM, Namvar S, Piggins HD. A riot of rhythms: neuronal and glial circadian oscillators in the mediobasal hypothalamus. Mol Brain. 2009;2:28. doi: 10.1186/1756-6606-2-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438.Mieda M, Williams SC, Richardson JA, Tanaka K, Yanagisawa M. The dorsomedial hypothalamic nucleus as a putative food-entrainable circadian pacemaker. Proc Natl Acad Sci U S A. 2006 Aug 8;103(32):12150–12155. doi: 10.1073/pnas.0604189103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.Wyse CA, Coogan AN. Impact of aging on diurnal expression patterns of CLOCK and BMAL1 in the mouse brain. Brain Res. 2010 Jun 14;1337:21–31. doi: 10.1016/j.brainres.2010.03.113. [DOI] [PubMed] [Google Scholar]
- 440.Abrahamson EE, Leak RK, Moore RY. The suprachiasmatic nucleus projects to posterior hypothalamic arousal systems. Neuroreport. 2001 Feb 12;12(2):435–440. doi: 10.1097/00001756-200102120-00048. [DOI] [PubMed] [Google Scholar]
- 441.Saper CB, Scammell TE, Lu J. Hypothalamic regulation of sleep and circadian rhythms. Nature. 2005 Oct 27;437(7063):1257–1263. doi: 10.1038/nature04284. [DOI] [PubMed] [Google Scholar]
- 442.Kalra SP, Kalra PS. Neuroendocrine control of energy homeostasis: update on new insights. Prog Brain Res. 2010;181:17–33. doi: 10.1016/S0079-6123(08)81002-3. [DOI] [PubMed] [Google Scholar]
- 443.Beaudry JL, Riddell MC. Effects of glucocorticoids and exercise on pancreatic beta-cell function and diabetes development. Diabetes/metabolism research and reviews. 2012 Oct;28(7):560–573. doi: 10.1002/dmrr.2310. [DOI] [PubMed] [Google Scholar]
- 444.Roberge C, Carpentier AC, Langlois MF, et al. Adrenocortical dysregulation as a major player in insulin resistance and onset of obesity. Am J Physiol Endocrinol Metab. 2007 Dec;293(6):E1465–E1478. doi: 10.1152/ajpendo.00516.2007. [DOI] [PubMed] [Google Scholar]
- 445.Thorens B. Brain glucose sensing and neural regulation of insulin and glucagon secretion. Diabetes, obesity & metabolism. 2011 Oct;13(Suppl 1):82–88. doi: 10.1111/j.1463-1326.2011.01453.x. [DOI] [PubMed] [Google Scholar]
- 446.Chandra R, Liddle RA. Recent advances in pancreatic endocrine and exocrine secretion. Current opinion in gastroenterology. 2011 Sep;27(5):439–443. doi: 10.1097/MOG.0b013e328349e2e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447.Gale SM, Castracane VD, Mantzoros CS. Energy homeostasis, obesity and eating disorders: recent advances in endocrinology. J Nutr. 2004 Feb;134(2):295–298. doi: 10.1093/jn/134.2.295. [DOI] [PubMed] [Google Scholar]
- 448.Gautron L, Elmquist JK. Sixteen years and counting: an update on leptin in energy balance. The Journal of clinical investigation. 2011 Jun;121(6):2087–2093. doi: 10.1172/JCI45888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 449.Vansaun MN. Molecular Pathways: Adiponectin and Leptin Signaling in Cancer. Clin Cancer Res. 2013 Jan 25; doi: 10.1158/1078-0432.CCR-12-0930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 450.Hursting SD, Dunlap SM. Obesity, metabolic dysregulation, and cancer: a growing concern and an inflammatory (and microenvironmental) issue. Ann N Y Acad Sci. 2012 Oct;1271:82–87. doi: 10.1111/j.1749-6632.2012.06737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 451.Vucenik I, Stains JP. Obesity and cancer risk: evidence, mechanisms, and recommendations. Ann N Y Acad Sci. 2012 Oct;1271:37–43. doi: 10.1111/j.1749-6632.2012.06750.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 452.Brugge J, Hung MC, Mills GB. A new mutational AKTivation in the PI3K pathway. Cancer Cell. 2007 Aug;12(2):104–107. doi: 10.1016/j.ccr.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 453.Boudny V, Kovarik J. JAK/STAT signaling pathways and cancer. Janus kinases/signal transducers and activators of transcription. Neoplasma. 2002;49(6):349–355. [PubMed] [Google Scholar]
- 454.Bodart JF. Extracellular-regulated kinase-mitogen-activated protein kinase cascade: unsolved issues. Journal of cellular biochemistry. 2010 Apr 1;109(5):850–857. doi: 10.1002/jcb.22477. [DOI] [PubMed] [Google Scholar]
- 455.Okwan-Duodu D, Umpierrez GE, Brawley OW, Diaz R. Obesity-driven inflammation and cancer risk: role of myeloid derived suppressor cells and alternately activated macrophages. American journal of cancer research. 2013;3(1):21–33. [PMC free article] [PubMed] [Google Scholar]
- 456.Smorlesi A, Frontini A, Giordano A, Cinti S. The adipose organ: white-brown adipocyte plasticity and metabolic inflammation. Obesity reviews : an official journal of the International Association for the Study of Obesity. 2012 Dec;13(Suppl 2):83–96. doi: 10.1111/j.1467-789X.2012.01039.x. [DOI] [PubMed] [Google Scholar]
- 457.Karin M. Nuclear factor-kappaB in cancer development and progression. Nature. 2006 May 25;441(7092):431–436. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- 458.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002 Dec 19–26;420(6917):860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459.Allavena P, Sica A, Garlanda C, Mantovani A. The Yin-Yang of tumor-associated macrophages in neoplastic progression and immune surveillance. Immunol Rev. 2008 Apr;222:155–161. doi: 10.1111/j.1600-065X.2008.00607.x. [DOI] [PubMed] [Google Scholar]
- 460.Koki A, Khan NK, Woerner BM, et al. Cyclooxygenase-2 in human pathological disease. Adv Exp Med Biol. 2002;507:177–184. doi: 10.1007/978-1-4615-0193-0_28. [DOI] [PubMed] [Google Scholar]
- 461.Khan Z, Pillay V, Choonara YE, du Toit LC. Drug delivery technologies for chronotherapeutic applications. Pharmaceutical development and technology. 2009;14(6):602–612. doi: 10.3109/10837450902922736. [DOI] [PubMed] [Google Scholar]
- 462.Firer MA, Gellerman G. Targeted drug delivery for cancer therapy: the other side of antibodies. Journal of hematology & oncology. 2012;5:70. doi: 10.1186/1756-8722-5-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 463.Lodish MB. Kinase Inhibitors: Adverse Effects Related to the Endocrine System. J Clin Endocrinol Metab. 2013 Feb 28; doi: 10.1210/jc.2012-4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 464.Philip AK, Philip B. Chronopharmaceuticals: hype or future of pharmaceutics. Current pharmaceutical design. 2011;17(15):1512–1516. doi: 10.2174/138161211796197151. [DOI] [PubMed] [Google Scholar]
- 465.Smaaland R. Circadian rhythm of cell division. Prog Cell Cycle Res. 1996;2:241–266. doi: 10.1007/978-1-4615-5873-6_23. [DOI] [PubMed] [Google Scholar]
- 466.Levi F, Okyar A, Dulong S, Innominato PF, Clairambault J. Circadian timing in cancer treatments. Annu Rev Pharmacol Toxicol. 2010;50:377–421. doi: 10.1146/annurev.pharmtox.48.113006.094626. [DOI] [PubMed] [Google Scholar]
- 467.Levi F, Focan C, Karaboue A, et al. Implications of circadian clocks for the rhythmic delivery of cancer therapeutics. Advanced drug delivery reviews. 2007 Aug 31;59(9–10):1015–1035. doi: 10.1016/j.addr.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 468.Innominato PF, Levi FA, Bjarnason GA. Chronotherapy and the molecular clock: Clinical implications in oncology. Advanced drug delivery reviews. 2010 Jul 31;62(9–10):979–1001. doi: 10.1016/j.addr.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 469.Giacchetti S, Dugue PA, Innominato PF, et al. Sex moderates circadian chemotherapy effects on survival of patients with metastatic colorectal cancer: a meta-analysis. Ann Oncol. 2012 Dec;23(12):3110–3116. doi: 10.1093/annonc/mds148. [DOI] [PubMed] [Google Scholar]
- 470.Rivard GE, Infante-Rivard C, Dresse MF, Leclerc JM, Champagne J. Circadian time-dependent response of childhood lymphoblastic leukemia to chemotherapy: a long-term follow-up study of survival. Chronobiol Int. 1993 Jun;10(3):201–204. doi: 10.3109/07420529309073888. [DOI] [PubMed] [Google Scholar]
- 471.Librodo P, Buckley M, Luk M, Bisso A. Chronotherapeutic drug delivery. Journal of infusion nursing : the official publication of the Infusion Nurses Society. 2012 Sep-Oct;35(5):329–334. doi: 10.1097/NAN.0b013e31826597d3. [DOI] [PubMed] [Google Scholar]
- 472.Gorbacheva VY, Kondratov RV, Zhang R, et al. Circadian sensitivity to the chemotherapeutic agent cyclophosphamide depends on the functional status of the CLOCK/BMAL1 transactivation complex. Proc Natl Acad Sci U S A. 2005 Mar 1;102(9):3407–3412. doi: 10.1073/pnas.0409897102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 473.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA: a cancer journal for clinicians. 2011 Mar-Apr;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 474.Verma M, Srivastava S. New cancer biomarkers deriving from NCI early detection research. Recent results in cancer research. Fortschritte der Krebsforschung. Progres dans les recherches sur le Cancer. 2003;163:72–84. doi: 10.1007/978-3-642-55647-0_7. discussion 264–266. [DOI] [PubMed] [Google Scholar]
- 475.Saunders NA, Simpson F, Thompson EW, et al. Role of intratumoural heterogeneity in cancer drug resistance: molecular and clinical perspectives. EMBO molecular medicine. 2012 Aug;4(8):675–684. doi: 10.1002/emmm.201101131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 476.Stein CJ, Colditz GA. Modifiable risk factors for cancer. Br J Cancer. 2004 Jan 26;90(2):299–303. doi: 10.1038/sj.bjc.6601509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 477.McPherson K, Steel CM, Dixon JM. ABC of breast diseases. Breast cancer-epidemiology, risk factors, and genetics. BMJ. 2000 Sep 9;321(7261):624–628. doi: 10.1136/bmj.321.7261.624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 478.Zhang JY, Selim MA. The role of the c-Jun N-terminal Kinase signaling pathway in skin cancer. American journal of cancer research. 2012;2(6):691–698. [PMC free article] [PubMed] [Google Scholar]
- 479.Romitti M, Ceolin L, Siqueira DR, Ferreira CV, Wajner SM, Maia AL. Signaling pathways in follicular cell-derived thyroid carcinomas (review) Int J Oncol. 2013 Jan;42(1):19–28. doi: 10.3892/ijo.2012.1681. [DOI] [PubMed] [Google Scholar]
- 480.Servillo G, Della Fazia MA, Sassone-Corsi P. Coupling cAMP signaling to transcription in the liver: pivotal role of CREB and CREM. Exp Cell Res. 2002 May 1;275(2):143–154. doi: 10.1006/excr.2002.5491. [DOI] [PubMed] [Google Scholar]
- 481.Rodon J, Dienstmann R, Serra V, Tabernero J. Development of PI3K inhibitors: lessons learned from early clinical trials. Nature reviews. Clinical oncology. 2013 Feb 12;10(3):143–153. doi: 10.1038/nrclinonc.2013.10. [DOI] [PubMed] [Google Scholar]
- 482.Diaz-Meco MT, Moscat J. The atypical PKCs in inflammation: NF-kappaB and beyond. Immunol Rev. 2012 Mar;246(1):154–167. doi: 10.1111/j.1600-065X.2012.01093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 483.Steelman LS, Pohnert SC, Shelton JG, Franklin RA, Bertrand FE, McCubrey JA. JAK/STAT, Raf/MEK/ERK, PI3K/Akt and BCR-ABL in cell cycle progression and leukemogenesis. Leukemia. 2004 Feb;18(2):189–218. doi: 10.1038/sj.leu.2403241. [DOI] [PubMed] [Google Scholar]
- 484.Dobrovolna J, Chinenov Y, Kennedy MA, Liu B, Rogatsky I. Glucocorticoid-dependent phosphorylation of the transcriptional coregulator GRIP1. Mol Cell Biol. 2012 Feb;32(4):730–739. doi: 10.1128/MCB.06473-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 485.Dahlman-Wright K, Qiao Y, Jonsson P, Gustafsson JA, Williams C, Zhao C. Interplay between AP-1 and estrogen receptor alpha in regulating gene expression and proliferation networks in breast cancer cells. Carcinogenesis. 2012 Sep;33(9):1684–1691. doi: 10.1093/carcin/bgs223. [DOI] [PubMed] [Google Scholar]