Abstract
IL-10 is a potent anti-inflammatory molecule that regulates excessive production of inflammatory cytokines during an infection or tissue damage. Dysregulation of IL-10 is associated with a number of autoimmune diseases, and so, understanding the mechanisms by which IL-10 gene expression is regulated remains an important area of study. Macrophages represent a major source of IL-10, which is generated in response to TLR signaling as a feedback mechanism to curtail inflammatory response. In this study, we identify a signaling pathway in murine bone marrow-derived macrophages in which activation of TLR4 by LPS induces the expression of IL-10 through the sequential induction of type I IFNs followed by induction and signaling through IL-27. We demonstrate that IL-27 signaling is required for robust IL-10 induction by LPS and type I IFNs. IL-27 leads directly to transcription of IL-10 through the activation of two required transcription factors, STAT1 and STAT3, which are recruited to the IL-10 promoter. Finally, through systematic functional promoter-reporter analysis, we identify three cis elements within the proximal IL-10 promoter that play an important role in regulating transcription of IL-10 in response to IL-27.
Innate immune cells respond to bacterial or viral infection by the rapid activation of proinflammatory cytokines that serve to initiate host defense against microbial invasion. However, excess proinflammatory cytokines give rise to systemic metabolic and hemodynamic disturbances that are harmful to the host. To avert these deleterious effects, IL-10 is produced by macrophages as a negative-feedback mechanism to dampen uncontrolled production of inflammatory cytokines and excessive inflammation during infection. IL-10 is a potent anti-inflammatory cytokine with a broad effect on both innate and adaptive immune systems.
Induction of innate immunity is mediated by diverse families of pattern recognition receptors that recognize microbial components termed pathogen-associated molecular patterns, which can be viewed as a molecular signature of the invading pathogens. TLRs are a major family of pattern recognition receptors that are mainly expressed by cells of the innate immune system (1, 2). TLRs can initiate distinct innate immune responses through recruitment of different MyD88 adaptor family members, primarily MyD88 and Toll/IL-1R domain-containing adaptor inducing IFN-β (TRIF) (3, 4). Currently, 11 TLRs have been cloned in mammals, and each receptor is involved in the recognition of a unique set of pathogen-associated molecular patterns (e.g., TLR4 recognizes LPS) (2, 5, 6). Although it is well established that IL-10 is induced in innate immune cells in response to TLR agonists like LPS, the molecular events responsible for upregulation of IL-10 remain to be elucidated. In addition, because LPS induction of IL-10 requires signaling through both feed-forward and feedback loops that may mitigate aspects of IL-10 regulation, it is difficult to identify transcription factors and cis elements within the IL-10 promoter.
We and others have identified two major signaling pathways mediated by TLR4 activation: the MyD88-dependent activation of NF-κB that results in the induction of inflammatory genes, such as TNF, IL-6, and IL-1β, and TRIF-dependent pathways involving the induction of type I IFNs and, subsequently, secondary-response genes induced by IFN-β in an autocrine/paracrine manner. The TRIF adaptor molecule has been shown to be indispensable for TLR4-mediated IL-10 activation (4, 7–10).
Classically, type I IFNs (IFN-β/α) bind to a cognate hetero-dimeric IFNR (IFN-αR) to activate the Jak–STAT pathway leading to expression of antiviral genes. In addition to their antiviral functions, type I IFNs are capable of exerting immunomodulatory effects on both innate and adaptive immune cells, in large part by inducing expression of IL-10 (11–14). Previously, we have demonstrated that type I IFN signaling is required for TLR-mediated induction of IL-10 (11, 15). In addition, we have demonstrated that type I IFNs are required for TLR-mediated induction of another anti-inflammatory cytokine, IL-27, in a dose-dependent manner (15).
IL-27 functions as a heterodimer composed of p28 and EBV-induced gene 3 (Ebi3), which have homologies to IL-12p35 and p40, respectively. The IL-27R complex consists of the unique subunit IL-27R (also referred to as TCCR and WSX-1) and the gp130 chain of IL-6R, which then activate transcription factors STAT1 and STAT3 via Jak-mediated phosphorylation (16, 17). IL-27 is produced by innate immune cells and has potent immune-suppressive effects on T cell immunity, including the inhibition of Th17 and Th1 differentiation, as well as in several infection models (15, 18–22). In addition, IL-27R–deficient mice develop excessive tissue inflammation in the context of infection or in autoimmune conditions (18, 23, 24). Importantly, although the underlying molecular mechanisms of IL-27–mediated immune suppression are not well understood, a number of studies have highlighted the importance of IL-27–mediated production of IL-10 to promote an anti-inflammatory state in lymphocytes (16, 25, 26). To date, the role of IL-27 in the regulation of IL-10 in macrophages is less understood.
In this study, we reveal a pathway in which LPS stimulation leads to induction of IL-10 in bone marrow-derived macrophages (BMDMs) through the sequential progression of type I IFN induction followed by IL-27 activation in an autocrine/paracrine manner. Importantly, induction of IL-10 by IL-27 in BMDMs is direct, allowing us to identify three cis regulatory elements through functional characterization of the murine IL-10 promoter. IL-27–mediated IL-10 expression is STAT3 and STAT1 dependent, and we demonstrate that both transcription factors are mobilized to the IL-10 promoter in vivo upon stimulation with LPS, type I IFNs, and IL-27.
Materials and Methods
Plasmids
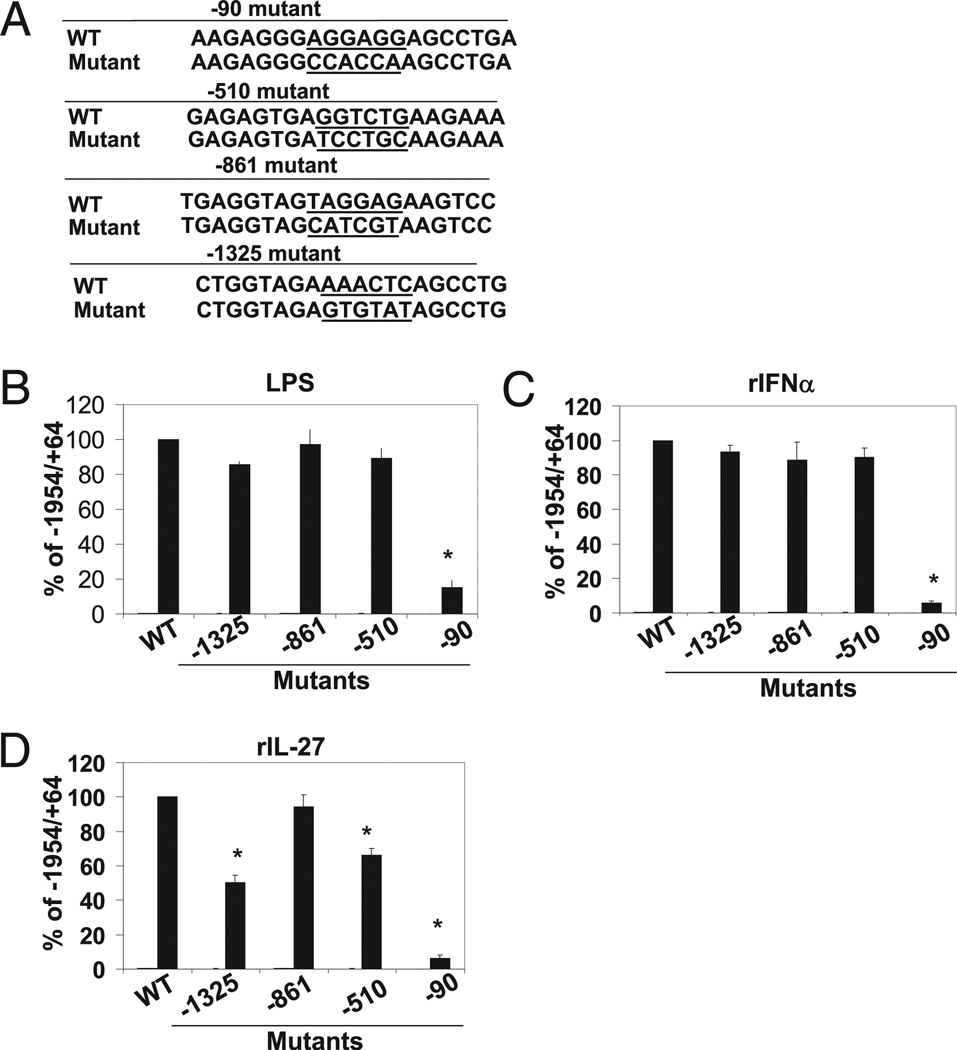
A ~2-kb fragment of the IL-10 promoter (−1954/+64) was amplified by PCR from mouse genomic DNA and subsequently cloned into the XhoI and HindIII sites of the pGL4.20 polylinker (Promega, Madison, WI). Promoter deletion mutants were amplified from the −1954/+64 promoter clone by PCR using an upstream primer containing an XhoI restriction site and a downstream primer containing a HindIII site. The PCR products were then inserted into the luciferase reporter vector pGL4.20. Substitution mutants were generated via a two-step PCR procedure using overlapping internal primers that contain a mutant sequence. All plasmids used in transient transfection assays were purified using an endotoxin-free purification system (Invitrogen Purelink, Invitrogen, Carlsbad, CA). Substitution mutants are presented in Fig. 5A.
FIGURE 5.
Mapping of critical LPS, IFN-α, and IL-27 response elements in IL-10 promoter by substitution mutant analysis. A, 6-nt mutants were constructed spanning across entirety of the regions −1541/−1232 and −595/−455 within the context of the −1954/+64 IL-10 promoter reporter construct. Constructs were transfected in RAW 264.7 cells and assessed for luciferase activity as described in B. Sequences of representative wild-type or substitution mutants are indicated. B, Empty pGL4.20, −1954/+64 IL10 promoter, and mutant IL-10 promoter constructs (−1954/+64 backbone) were transfected into RAW264.7 cells and left unstimulated or stimulated with LPS (1 µg/ml) (B), rIFN-α (1000 U/ml) (C), and rIL-27 (80 ng/ml) (D). Luciferase activity is displayed as percent of full-length wild-type (−1954/+64) IL-10 promoter activity normalized to renilla activity representing three independent experiments. Student t test was performed. *p < 0.05.
BMDM preparation and mice
Wild-type and IL-27R knockout (TCCR/WSX1−/−), STAT1−/− mice were on a C57BL/6 background (The Jackson Laboratory, Bar Harbor, ME). IFN-α Ro/o 129/Sv (IFNAR−/−) mice were from B&K Universal (Aldbrough, U.K.) and were backcrossed with C57BL/6 for six generations. Bones for macrophage-specific STAT3−/− (STAT3flox/floxLysCre+/−) and STAT3+/+ (STAT3flox/floxLysMCre−/−) were kindly provided by Dr. Bin Gao (National Institute of Alcohol Abuse and Alcoholism, Bethesda, MD) and generated as previously described (27). All mice were maintained and bred at the University of California, Los Angeles, Department of Laboratory Animal Medicine mouse facility (Los Angeles, CA) under specific pathogen-free conditions.
Murine BMDMs were generated by flushing bone marrow cells from the femurs and tibias of mice. These cells were cultured for 7 d in DMEM (Life Technologies, Rockville, MD) containing 10% FBS (Omega Scientigfic, Tarzana, CA), 1% penicillin/streptomycin (Life Technologies), and 10% conditioned media (CM) from L929 cells overexpressing M-CSF. CM was replaced on day 4 of differentiation and every 2 d thereafter. BMDMs were serum starved in DMEM, 1% FBS, and 1% penicillin/streptomycin overnight prior to cytokine stimulation.
Real-time quantitative PCR and ELISA
Post-stimulation, BMDMs were harvested in PBS and lysed in 500 µl TRIzol reagent (Invitrogen). RNA was isolated via chloroform extraction. cDNA was synthesized from 1 µg RNA per sample by reverse transcription using Iscript (Bio-Rad, Hercules, CA) and an oligo(dT) primer. The following primers were used: IL-10 forward: 5′-CGTCGGATCCGCCATGCCTGGCTCACCACTGCT-3′ and reverse: 5′-CGTCTCTAGATTAGCTTTTCATTTTGATCA-3′; IL-27p28 forward: 5′-CTCTGCTTCCTCGCTACCAC-3′ and reverse: 5′-GGGGCAGCTTCTTTTCTTCT-3′; EBI3 forward: 5′-TGAAACAGCTCTCGTGGCTCTA-3′ and reverse: 5′-GCCACGGGATACCGAGAA-3′; IFN-β forward: 5′-AGCTCCAAGAAAGGACGAACAT-3′ and reverse: 5′-GCCCTGTAGGTGAGGTTGATCT-3′; IFNα4 forward: 5′-CCTGTGTGATGCAGGAACC-3′ and reverse: 5′-TCACCTCCCAGGCACAGA-3′; and L32 forward: 5′-AAGCGAAACTGGCGGAAAC-3′ and reverse: 5′-TAACCGATGTTGGGCATCAG-3′.
Quantitative PCR (qPCR) was conducted using 95°C (5 min) to denature DNA strands followed by 95°C (30 s), 55°C (30 s), and 72°C (45 s) for 40 cycles followed by a 72°C 5-min extension. Analyses were done using the iCycler thermocycler (Bio-Rad). Relative transcript levels were normalized to L32 expression.
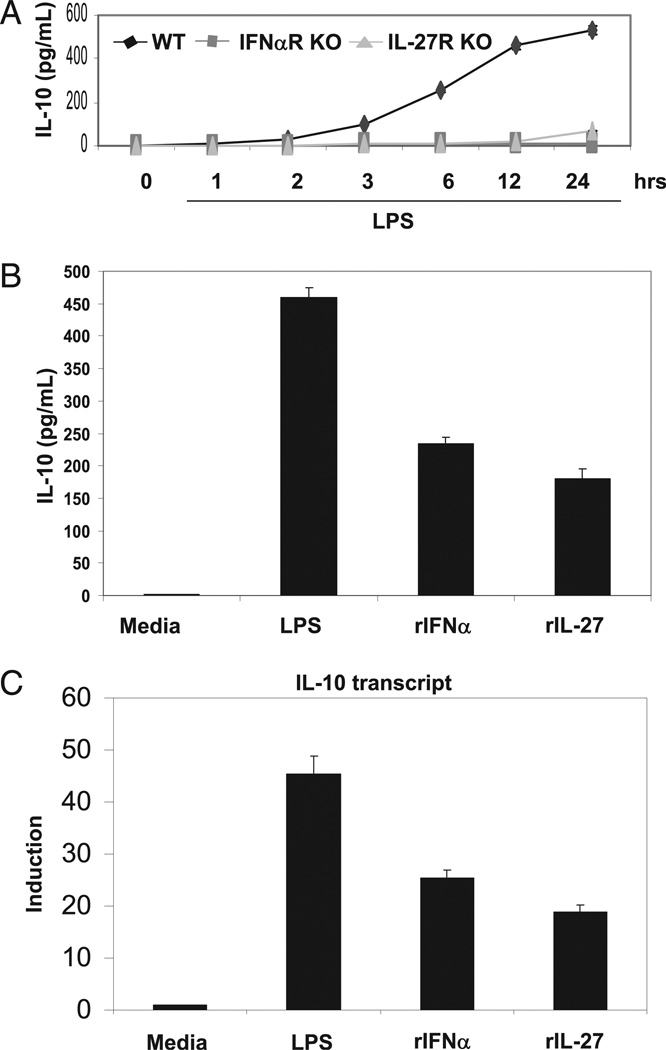
Murine IL-10 protein was measured from 0.5 × 106 BMDM cells cultured in a six-well plate in serum-starved conditions prior to cytokine treatment for indicated times (Fig. 1A, 1B). A total of 100 µl was then assayed using a murine IL-10 ELISA kit (BD Biosciences, San Jose, CA) according to the manufacturer’s instructions.
FIGURE 1.
Induction of IL-10 protein and mRNA in BMDMs. A, Total of 0.5 × 106 WT, IFN-αR KO, and IL-27R KO BMDMs were stimulated with LPS (50 ng/ml), and supernatant was collected at the indicated time points. IL-10 protein production was quantified via ELISA. B, Total of 0.5 × 106 BMDMs were unstimulated or stimulated with LPS (50 ng/ml), IFN-α (250 U/ml), or IL-27 (10 ng/ml), supernatant was collected, and IL-10 protein production was quantified via ELISA. C, Total of 0.5 × 106 BMDMs were unstimulated or stimulated with LPS (50 ng/ml), IFN-α (250U/ml), or IL-27 (10 ng/ml) for 4 h, RNA was harvested, and relative IL-10 transcript was detected via qPCR normalized to L32.
Cell lines, reagents, and transfection
The RAW264.7 murine macrophage cell line (American Type Culture Collection, Manassas, VA) was maintained in DMEM supplemented with 5% FBS and 1% penicillin/streptomycin (complete DMEM). LPS (Escherichia coli, strain O55:B5, Sigma-Aldrich, St. Louis, MO), murine rIFN-α4 (catalog number 12110-9, PBL InterferonSource, Piscataway, NJ), and rIL-27 (catalog number 1799-ML, R&D Systems, Minneapolis, MN) were used at the concentrations described in the figures. Murine rIFN-α4 and rIL-27 were both tested for endotoxin to <1 endotoxin unit/µg levels using the Limulus amebocyte lysate method. In the cycloheximide (CHX) experiments, BMDMs were pretreated with CHX (2 µg/ml) or ethanol for 15 min before cytokine stimulation.
RAW264.7 cells were transiently transfected using the FuGeneHD (Roche Diagnostic Systems, Somerville, NJ) reagent. Briefly, 2.5 × 106 cells were plated in a six-well plate. The following day, the cells were washed with PBS and transfected with 2.5 µg IL-10 promoter or empty firefly luciferase reporter plasmid and 100 ng Renilla luciferase reporter. DNA in serum-free DMEM (100 µl) were incubated at room temperature with FuGeneHD reagent at a 1:2 ratio of DNA to FuGeneHD for 15 min. Transfectant mixture was then added to each well drop-wise and incubated at 37°C for 4 h. Cells were washed with PBS and split into two wells in 2 ml complete DMEM. The cells in one of the two wells were stimulated with LPS (1 µg/ml), rIFN-α4 (1000 U/ml), or rIL-27 (80 ng/ml) 6 h posttransfection and incubated for 24 h. Luciferase activity was assayed using the Dual-Luciferase Reporter Assay kit (Promega) according to the manufacturer’s instructions. Luciferase activity was determined using 50 µl (of 200 µl total). Promoter activity is presented normalized to Renilla luciferase activity to control for transfection efficiency.
Immunoblot/chromatin immunoprecipitation
For Western blot analyses, cells were lysed in 50 mM Tris (pH 7.5), 140mM NaCl, 1% Igepal CA-630, 0.25% sodium deoxycholate, 1 mM EGTA, and 5 mM NaF. Protein was quantified using a BCA kit (Thermo Scientific, Waltham, MA) according to the manufacturer’s instructions. Proteins were denatured and separated on 10% SDS-PAGE, transferred to nitrocellulose membranes, and immunoblotted in 5% BSA or milk using standard methods. The following Abs were used at 1:1000 concentrations: anti-Sp1, anti-STAT1, anti-STAT2, and anti-STAT3 (Santa Cruz Biotechnology, Santa Cruz, CA), anti–phospho-STAT1 (Y701), anti–phospho-STAT3 (Y705) (Cell Signaling Technology, Beverly, MA), and c-Maf (Santa Cruz Biotechnology).
Chromatin immunoprecipitation (ChIP) was performed by cross-linking stimulated cells with formaldehyde (1% final concentration). Cells were lysed in cell lysis buffer (5 mM PIPES [pH 8], 85 mM KCl, 0.5% Nonidet P-40) on ice (5 min). Nuclei were lysed in nuclei lysis buffer (50 mM Tris-HCl [pH 8.1], 10 mM EDTA, and 1% SDS supplemented with protease inhibitors [Roche Diagnostic Systems]). Sonication was performed using a Diagenode Biorupter (Diagenode, Denville, NJ) for 8 min (30 s on/off) twice to generate sheared fragments of ~500 bp. A total of 100 µg chromatin was diluted to 300 µl in dilution buffer (16.7 mM Tris-HCl [pH 8.1], 167 mM NaCl, 1.2 mM EDTA, 0.01% SDS, and 1.1% Triton X-100 supplemented with protease inhibitors) and incubated with 5–10 µl anti–phospho-STAT1 (Cell Signaling Technology), anti–phospho-STAT3 (Cell Signaling Technology), STAT2 (Santa Cruz Biotechnology), c-Maf (Santa Cruz Biotechnology), or IgG1 (Santa Cruz Biotechnology) overnight at 4°C in the presence of 100 ng/ml sheared salmon sperm (Invitrogen). Immunocomplexes were recovered with 40 µl Protein A agarose/salmon sperm DNA beads (Upstate Biotechnology, Lake Placid, NY) at 4°C for 1 h. Five percent of input was collected for Ab normalization. Beads were washed twice in high-salt wash buffer (50 mM HEPES [pH 7.9], 0.5 M NaCl, 1 mM EDTA, 0.1% SDS, 1% Triton X-100, and 0.1% deoxycholate) and twice with TE buffer for 10 min room temperature. Beads were resuspended in 300 µl elution buffer (50 mM Tris-HCl [pH 8], 10 mM EDTA, and 1% SDS) supplemented with 20 µg proteinase K (Invitrogen) for 2 h at 55°C. DNA was reverse cross-linked at 65°C overnight. DNA was PCR purified (Invitrogen Purelink, Invitrogen), and immunoprecipitated DNA fragments were quantified via qPCR using the following primers: IL-10 proximal promoter forward: 5′- GGACCAAGAACAGGAGGT-3′ and reverse: 5′-ACTAAAAGTTGTATTTCC-3′, which amplifies 225 bp from −228 to −4 relative to the transcription start site; and IL-10 distal promoter forward: 5′-CCCCTTCCCTGTGCTTG-3′ and reverse: 5′-GAGAGGGTTACCACACCAGGG-3′, which amplifies 221 bp from −1353 to −1130 relative to the transcription start site.
Results
LPS, type I IFN, and IL-27 signaling induce IL-10 expression in BMDMs
Previously, we had demonstrated LPS mediated induction of IL-10 through a TRIF-dependent mechanism that involves the production and signaling of type I IFNs (11, 28–30). In light of a role of IL-27 as an anti-inflammatory agent and a potent inducer of IL-10, we asked whether IL-27 signaling was involved in LPS-mediated IL-10 induction. To address this question, we stimulated wild-type, IFN-αR–deficient, and IL-27R–deficient BMDMs with LPS and assessed IL-10 protein production over time via ELISA. As expected, LPS-mediated production of IL-10 was deficient in BMDMs lacking IFN-αR (Fig. 1A). Surprisingly, IL-27R–deficient BMDMs also exhibited significant defects in IL-10 production in response to LPS (Fig. 1A). We then confirmed IL-10 protein production in BMDMs in response to LPS, IFN-α, and IL-27 (Fig. 1B). To assess IL-10 expression at the transcriptional level, relative IL-10 mRNA production was assessed after 4 h of stimulation with LPS, IFN-α, and IL-27 by qPCR. Data are presented normalized to L32 mRNA levels (Fig. 1C). Importantly, relative to induction of proinflammatory cytokines like IL-12p40 and IL-6, IL-10 transcription in response to LPS appeared delayed, with detectable transcripts first appearing after 2 h of stimulation. In contrast, both IFN-α and IL-27 displayed faster kinetics of IL-10 transcription, with both able to generate detectable transcripts within 1 h of stimulation, even at low-dose concentration (data not shown).
IL-27 signaling occurs downstream of TLR4 and type I IFNs in LPS-mediated IL-10 gene expression
Previously, we showed that LPS-mediated IL-10 induction was type I IFN dependent (11). To confirm these results, we stimulated wild-type BMDMs with LPS for 2 h and then transferred the conditioned media (LPS-CM) from unstimulated and stimulated BMDMs to previously unstimulated wild-type and IFN-αR–deficient BMDMs for 3 h and monitored relative IL-10 induction by qPCR. Presumably, the LPS-CM at 2 h CM contains physiologically detectable levels of type I IFN protein, but not IL-27 protein, although this was not explicitly tested. As expected, LPS-CM was able to stimulate robust IL-10 transcript production in wild-type, whereas IL-10 induction in IFN-αR–deficient BMDMs was significantly abrogated (Fig. 2A, left panel). We chose the 2 h time point based on the relative kinetics of IFN-β versus IL-27p28 transcript expression (Fig. 3A). Detectable levels of IFN-β appear 1 h post LPS treatment and peak at ~3 h. In contrast, induction of IL-27p28 appears delayed relative to IFN-β, with robust mRNA detection occurring 3 h post LPS treatment and peaking at ~6 h.
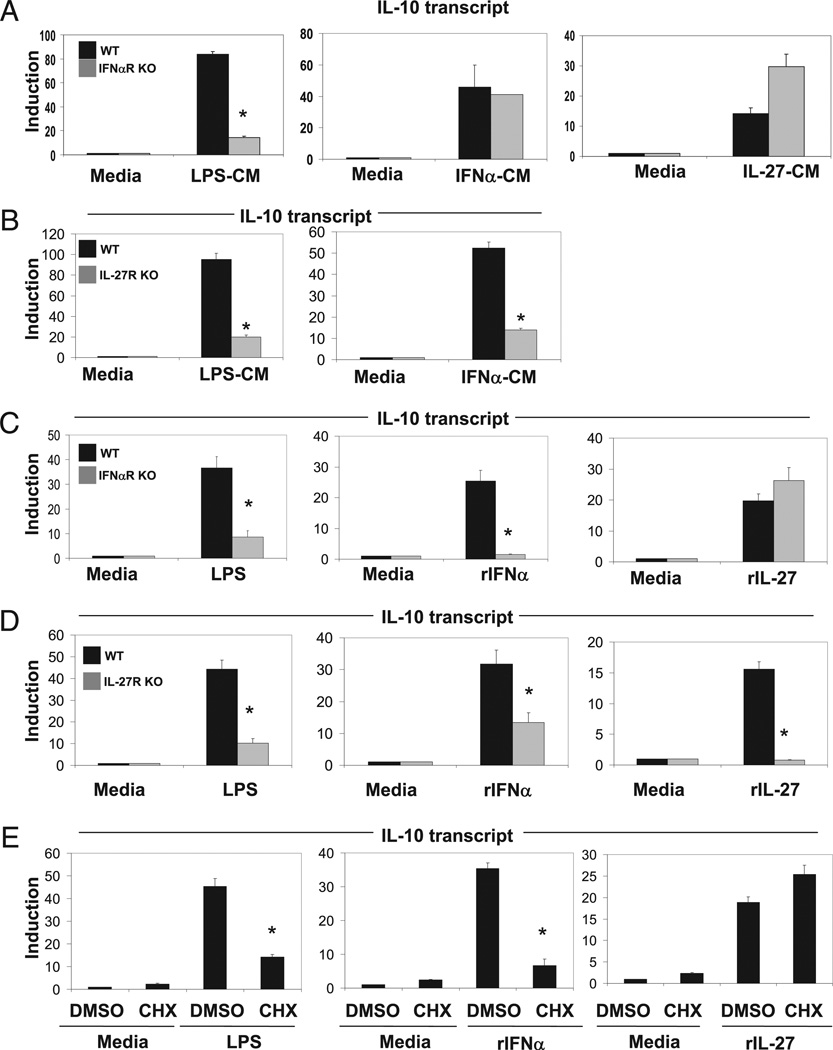
FIGURE 2.
LPS-mediated induction of IL-10 is type I IFN and IL-27 signaling dependent. A, Total of 0.5 × 106 WT were unstimulated or stimulated with LPS (50 ng/ml), rIFN-α (250 U/ml), or rIL-27 (10 ng/ml) for 2 h. CM were collected and transferred to 0.5 × 106 WT or IFN-αR−/− previously serum-starved (1% FBS) BMDMs in the absence of any stimulation. Cells were incubated with the indicated CM for 3 h, RNA was collected, and IL-10 transcript level was detected via qPCR normalized to L32. B, Total of 0.5 × 106 WT BMDMs were unstimulated or stimulated with LPS (50 ng/ml) or IFN-α (250 U/ml) for 2 h. CM were collected and transferred with 0.5 × 106 WT or IL27R−/− previously serum-starved BMDMs in the absence of any stimulation. Cells were incubated with the indicated CM for 3 h, and IL-10 expression was assessed as in A. Data from supernatant transfers (A, B) represent two independent experiments. C, Total of 0.5 × 106 WT or IFN-αR−/− BMDMs were unstimulated or stimulated with LPS (50 ng/ml), IFN-α (250 U/ml), or IL-27 (10 ng/ml) for 3 h. RNA was collected, and IL-10 transcript level was detected via qPCR normalized to L32. D, WT or IL27R−/− BMDMs were unstimulated or stimulated with LPS (50 ng/ml), IFN-α (250 U/ml), or IL-27 (10 ng/ml) for 3 h. RNA was collected, and IL-10 transcript level was detected via qPCR normalized to L32. E, Total of 0.5 × 106 WT BMDMs were pretreated with DMSO or CHX (2 µg/ml) and then stimulated with LPS (50 ng/ml), IFN-α (250 U/ml), or IL-27 (10 ng/ml) for 3 h. RNAwas collected, and IL-10 transcript level was detected via qPCR normalized to L32. Data from C and D represent mean and SD from three independent experiments. Student t test was performed. *p < 1 × 10−4.
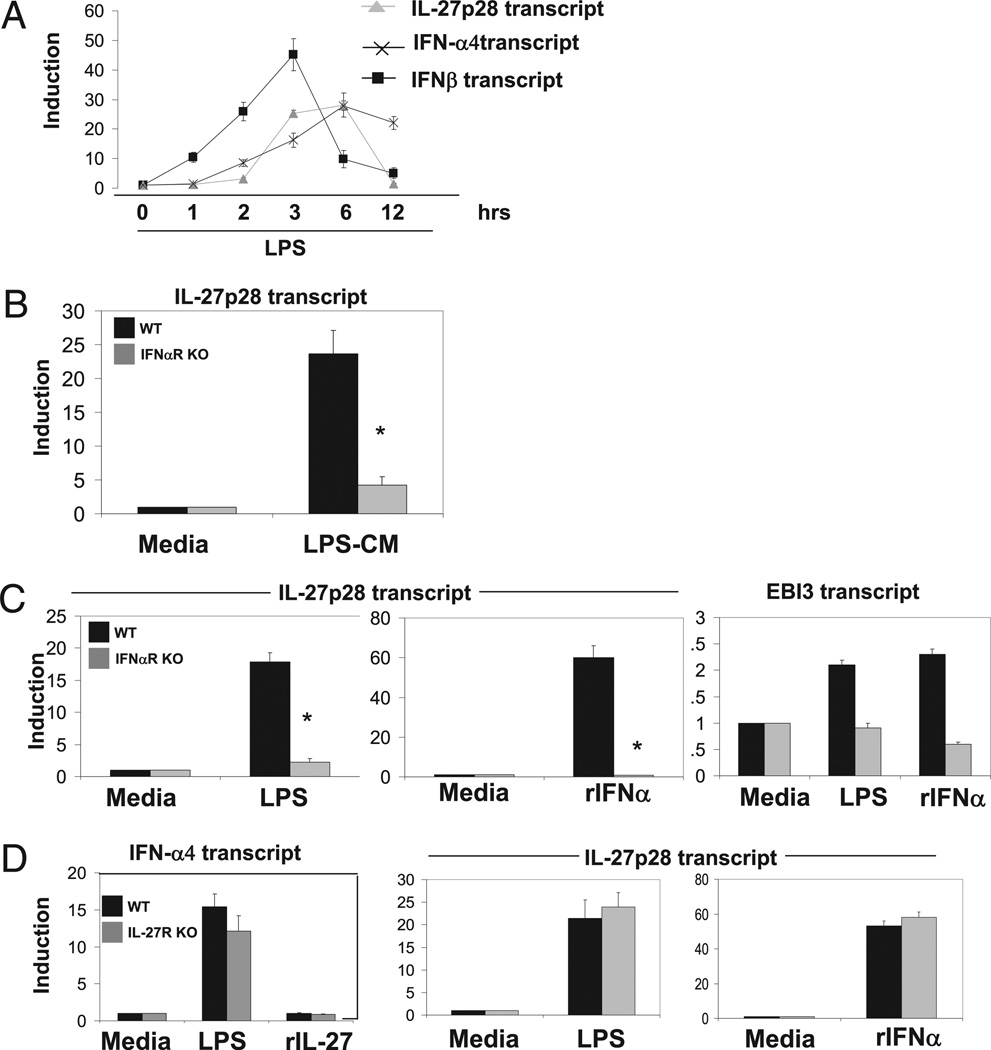
FIGURE 3.
LPS induction of IL-10 requires sequential induction of type I IFNs and IL-27, respectively. A, Total of 0.5 × 106 WT BMDMs were were stimulated with LPS (50 ng/ml) for the indicated time points. RNAwas collected, and IFN-β, IFN-α4, and IL-27p28 transcript level was detected via qPCR normalized to L32. B, Total of 0.5 × 106 WT were unstimulated or stimulated with LPS for 2 h. CM were collected and transferred to 0.5 × 106 WT or IFN-αR−/− previously serum-starved (1% FBS) BMDMs in the absence of any stimulation. Cells were incubated with the indicated CM for 3 h, RNA was collected, and IL-27p28 transcript level was detected via qPCR normalized to L32. C, Total of 0.5 × 106 WT or IFN-αR−/− BMDMs were unstimulated or stimulated with LPS (50 ng/ml) or IFN-α (250 U/ml) for 3 h. RNA was collected, and IL-27p28 or EBI3 transcript level was detected via qPCR normalized to L32. D, Total of 0.5 × 106 WT or IL-27R−/− BMDMs were unstimulated or stimulated with LPS (50 ng/ml), IFN-α (250 U/ml), or rIL-27 (10 ng/ml) for 3 h. RNA was collected, and IFN-α and IL-27p28 transcript level was detected via qPCR normalized to L32. Data represent mean and SD from three independent experiments. Student t test was performed. *p < 0.01.
We then performed an analogous experiment by stimulating wild-type BMDMs with IFN-α or IL-27 and then transferring the CM (IFN-α–CM, IL-27–CM) to previously unstimulated wild-type or IFN-αR–deficient BMDMs for 3 h and assessed IL-10 expression by qPCR. In contrast to the results from LPS-CM stimulation, both wild-type and IFN-αR–deficient BMDMs were able to generate similar levels of IL-10 induction in response to either IFN-α–CM or IL-27–CM (Fig. 2A, middle and right panels, respectively). We confirmed these results by stimulating wild-type and IFN-αR–deficient BMDMs directly with LPS, rIFN-α, and rIL-27. LPS induction of IL-10 in IFN-αR–deficient BMDMs was significantly abrogated compared with wild-type (Fig. 2C, left panel). IFN-α was unable to induce IL-10 in IFN-αR–deficient BMDMs due to lack of its receptor (Fig. 2C, middle panel). However, IL-10 was similarly induced in wild-type and IFN-αR–deficient BMDMs by rIL-27 stimulation (Fig. 2C, right panel). These data suggest that IL-27 can induce IL-10 expression independent of type I IFN signaling, whereas LPS and IFN-α cannot.
Next, we assessed the requirements for IL-27R signaling on LPS and IFN-α–mediated IL-10 gene expression. In this study, we treated previously unstimulated wild-type and IL-27R–deficient BMDMs with LPS-CM and IFN-α–CM for 3 h and monitored IL-10 expression by qPCR. Both LPS- and IFN-α–mediated induction of IL-10 were significantly decreased in IL-27R–deficient BMDMs compared with wild-type (Fig. 2B). We confirmed these results by inducing wild-type and IL-27R–deficient BMDMs with LPS, rIFN-α, and rIL-27 directly. Again, LPS and IFN-α were able to robustly induce IL-10 in IL-27R null BMDMs compared with wild-type (Fig. 2D, left and middle panels). IL-27 was unable to induce IL-10 expression in the IL-27R–deficient cells due to the absence of its cognate receptor (Fig. 2D, right panel). These data suggest that IL-27 signaling occurs downstream of TLR4 and type I IFN in the induction of IL-10 expression.
IL-27 induction of IL-10 is direct, whereas LPS- and IFN-α–mediated IL-10 expression require de novo protein synthesis
Our data suggest that both LPS and type I IFN induction of IL-10 occurs indirectly through subsequent signaling via IL-27. Previously, our laboratory and others have shown that LPS-mediated induction of IL-10 requires de novo synthesis and signaling by type I IFNs for robust induction of IL-10 transcripts (11). We next wanted to assess whether induction of IL-10 by IL-27 was direct or required subsequent de novo synthesis of a transcription factor or signaling molecule. To ascertain this, we pretreated wild-type BMDMs with CHX, a translation inhibitor, followed by stimulation with LPS, rIFN-α, or rIL-27. Inhibition of protein synthesis led to defects in LPS and IFN-α–mediated IL-10 induction (Fig. 2E, left and middle panels). In contrast, IL-27–induced IL-10 expression was unaffected by CHX treatment, suggesting that IL-27–mediated IL-10 transcription is direct (Fig. 2E, right panel).
LPS signaling leads to induction of both type I IFNs and IL-27p28 transcripts with induction of IL-27p28 displaying delayed kinetics of expression relative to IFN-β. Because both LPS- and IFN-α–mediated induction of IL-10 require IL-27 signaling and de novo protein synthesis, we propose that both LPS and IFNa induce expression of IL-27 and that subsequent IL-27 signaling leads to robust IL-10 gene expression. To test this, we assessed transcription of the IL-27p28 gene in WT and IFN-αR BMDMs treated with LPS-CM for 3 h. Expression of IL-27p28 by LPS-CM occurred in WT but not IFN-αR–deficient BMDMs (Fig. 3B). Induction of the second subunit of IL-27, EBI3, was not robust, and no significant differences between WT and IFN-αR KO were observed (data not shown). We confirmed these results by stimulating wild-type and IFN-αR–deficient BMDMs directly with LPS and rIFN-α. LPS-mediated induction of both IL-27p28 and EBI3 was type I IFN dependent (Fig. 3C). IFN-α was able to induce both IL27p28 and EBI3 in wild-type but not IFN-αR–deficient cells due to loss of its receptor. Interestingly, induction of IL-27p28 by LPS was abrogated in the presence of CHX, whereas IFN-α–mediated induction of IL-27p28 was not, suggesting that type I IFNs can induce IL-27 gene expression directly, whereas LPS requires de novo protein synthesis, presumably of type I IFNs (data not shown).
LPS induction of type I IFNs, IFN-β or IFN-α, is mediated primarily through the signaling by the TRIF adaptor molecule, leading to recruitment of the enhanceosome to the IFN-β promoter, resulting in gene induction (31, 32). Subsequent signaling of IFN-β leads to formation of the ISGF3 complex, leading to induction of multiple IFN-α genes as well as type I IFN-mediated antiviral gene targets in the murine genome (33). We propose that LPS-mediated induction of IL-27 requires de novo synthesis of type I IFNs and that subsequent IFN signaling leads directly to robust expression of the IL-27 heterodimeric subunits. To confirm that LPS-mediated induction of type I IFNs is IL-27 independent, we stimulated wild-type and IL-27R–deficient BMDMs and assessed expression of IFN-β. As expected, LPS-mediated induction of type I IFNs is independent of IL-27 signaling (Fig. 3D, left panel). Finally, both LPS and IFN-α were able to induce expression of IL-27p28 even in the absence of the IL-27R in BMDMs (Fig. 3D, middle and right panels). Thus, induction of IL-27 by LPS requires an intermediary step of type I IFN production. Taken together, we propose a signaling pathway by which LPS induces IL-10 gene expression in a two-step mechanism involving initial synthesis of type I IFNs followed by induction of IL-27, which then directly stimulates IL-10 production in an autocrine/paracrine manner.
Functional characterization of the IL-10 promoter reveals three cis regulatory elements mediating IL-27–mediated gene induction
Identifying functional elements within the IL-10 promoter has proven to be exceedingly difficult due to the complexity of signaling systems that induce IL-10 expression. Previous studies have correlated binding of several transcription factors to the IL-10 promoter with LPS-mediated gene induction in macrophages, including Sp1, Sp3, STAT1, STAT3, IFN regulatory factor (IRF) 1, AP-1, Maf, and NF-κB (34). However, to date, only a single Sp1 element upstream of the IL-10 core promoter appears to be essential for IL-10 promoter activity in response to LPS (30, 35). Our studies have revealed an autocrine/paracrine signaling loop by which LPS induces IL-10 expression through the sequential generation and signaling of type I IFNs and IL-27, respectively. Importantly, we have demonstrated that IL-27 can directly upregulate transcription of IL-10. Thus, studying IL-10 promoter activity in response to IL-27 eliminates some of the complexity of LPS signaling that can mask functional redundancy in the regulation of gene expression via multiple signaling pathways.
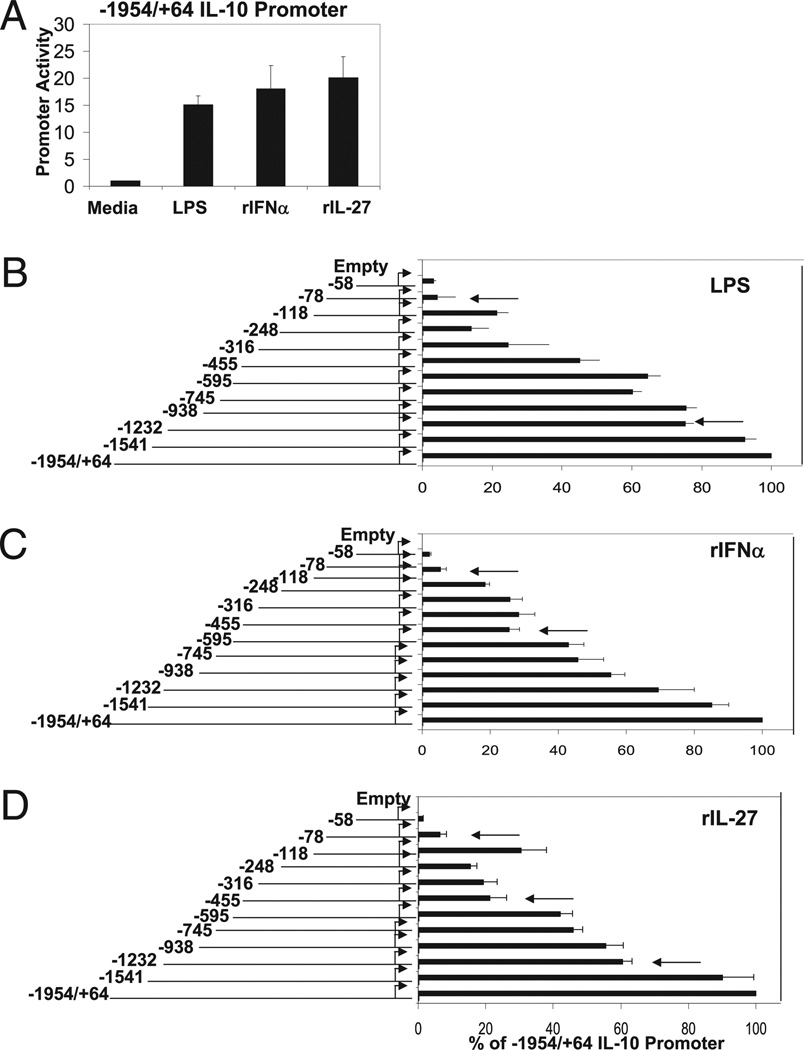
We cloned a fragment of the proximal IL-10 promoter extending from nucleotide −1954 to +64 relative to the +1 transcription start site (35), inserted into a firefly luciferase reporter vector. We then transiently transfected the promoter–reporter construct into the macrophage cell line RAW 264.7 to assess promoter activity. Importantly, RAW 264.7 cells exhibit similar gene profiles to BMDMs in response to LPS (28, 29). Following transfection, cells were stimulated with LPS, IFN-α, or IL-27 for 12 h. Each stimulant was able to induce robust IL-10 promoter activity compared with the empty luciferase vector. To make meaningful comparisons among LPS, IFN-α, and IL-27, we titrated each stimulatory agent and chose activation concentrations that resulted in a similar level of relative IL-10 promoter induction, normalized to renilla luciferase activity to control for transfection efficiency (Fig. 4A).
FIGURE 4.
Identification of LPS, IFN-α, and IL-27 response elements in IL-10 proximal promoter. A, RAW264.7 were transiently transfected with −1954/+64 IL-10 promoter ligated to firefly luciferase reporter (2.5 µg) and were unstimulated or titrated with stimulated with LPS, IFN-α, or IL-27. Stimulation with LPS (1 µg/ml), rIFN-α (1000 U/ml), or IL-27 (80 ng/ml) consistently gave equivalent firefly luciferase activities (normalized to renilla luciferase activity). Empty pGL4.20 or IL-10 promoter 5′ deletion mutants (as indicated) sharing +64 transcription start site were transfected into RAW264.7 cells and left untreated or stimulated with LPS (1 µg/ml) (B), rIFN-α (1000 U/ml) (C), or rIL-27 (80 ng/ml) (D). Luciferase activity displayed as percent of full-length (−1954/+64) IL-10 promoter activity normalized to renilla luciferase activity representing two independent experiments. Student t test was performed. Arrows indicate p < 0.05.
To identify DNA sequences that are necessary for LPS-, IFN-α–, or IL-27–induced promoter activity, a series of promoter mutants that contain successive deletions from the 5′ end were inserted upstream of the luciferase reporter gene. Each mutant shared the common +1 transcription start site. We then assessed the promoter activity in response to LPS, IFN-α, and IL-27 and measured relative induction as a percentage of the full-length −1954/+64 full-length IL-10 proximal promoter. Although some deletion constructs demonstrated slight defects in basal promoter activity, all were able to be robustly induced by LPS. We identified two major regions (indicated by arrows) located between −1232/−1541 and −78/−118 that, when deleted, led to defects in LPS-mediated promoter induction (Fig. 4B). IFN-α–mediated IL-10 promoter activity required elements within the −455/−595 and −78/−118 (Fig. 4C, indicated by arrows). Our analysis of IL-10 promoter activity in response to IL-27 revealed three major cis elements that appear to be important for promoter induction: −1232/−1541, −455/−595, and −78/−118 (Fig. 4D). Thus, although it appears that LPS, IFN-α, and IL-27 exert differential effects on the IL-10 promoter, they each share a common regulatory region located between −78/−118 relative the transcription start site that corresponds to a previously identified Sp1 binding site (30). Interestingly, there was an additional element located in a region between −118/−240 that may act as a negative regulatory element in response to IL-27 due to an increase in promoter activity when absent. With regards to this study, this putative negative regulatory element was not studied further. In addition, LPS and IL-27 share a common regulatory element located between −1232/−1541, whereas IFN-α and IL-27 appear to require a functional element located between −455/−595. These data emphasize that although LPS and IFN-α may induce IL-10 in distinct ways, they potentially share common regulatory elements within the IL-27 pathway, which they both require for robust IL-10 gene expression.
To further characterize the regulatory elements identified in our promoter-reporter analyses, we constructed 6-mer mutations across the entirety of the three regulatory regions identified in our promoter deletion analysis: −1232/−1541, −455/−595, and −78/−90 region as well as a neutral region, −745/−938, in the context of our full-length proximal promoter (−1954/+64) and assessed their function in response to LPS, IFN-α, and IL-27 using the same transfection/stimulation strategy described earlier. The sequences of four representative promoter mutations are displayed in Fig. 5A. None of the 6-mer mutant constructs displayed any significant defects in basal IL-10 promoter activity. Only the −78/−90 region, previously described as an Sp1 binding site, was universally required for IL-10 promoter activity in response to LPS, IFN-α, and IL-27 (Fig. 5B–D). No other scanning 6-mer mutations exerted any significant effects on LPS or IFN-α–mediated IL-10 induction in the context of the −1954/+64 promoter (Fig. 5B, 5C). However, we identified two cis regulatory elements located at −1324/−1319 and −510/−504 that led to significant defects in IL-27–mediated promoter activity (Fig. 5D).
IL-27–mediated IL-10 gene induction is regulated by STAT1 and STAT3
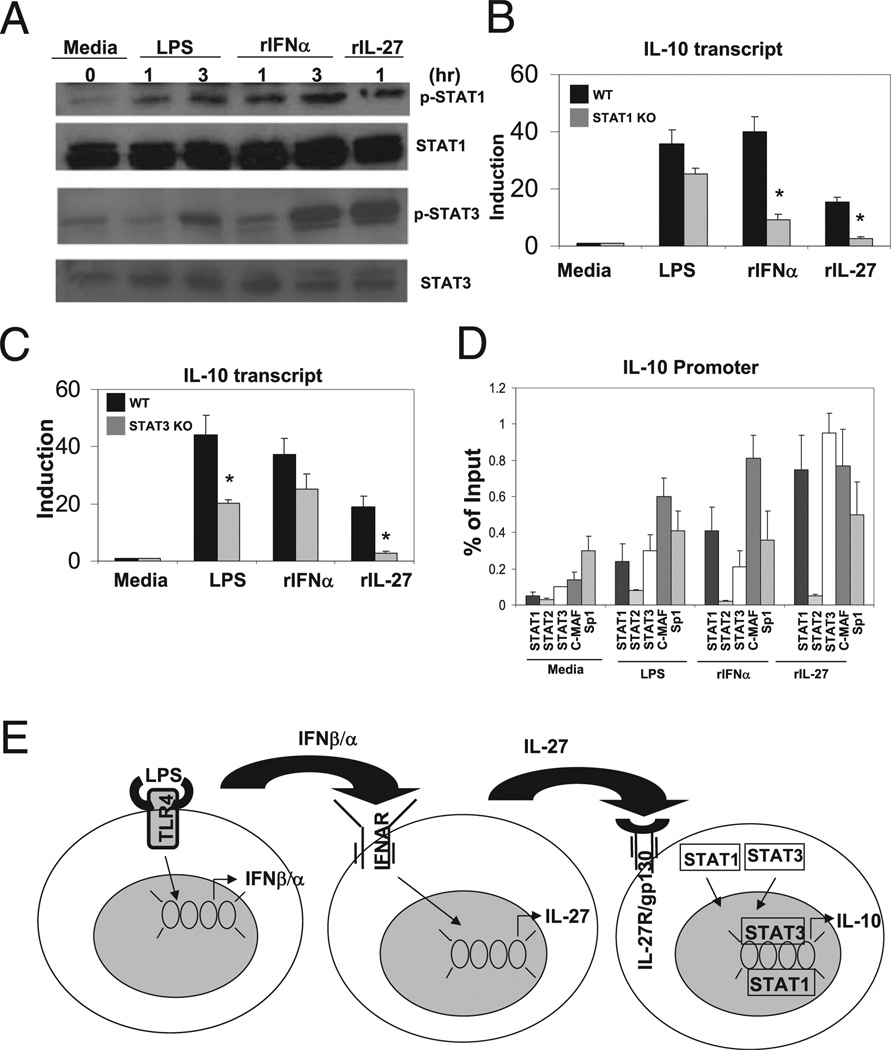
We next sought to identify potential transcription factors that mediate LPS-, IFN-α–, and IL-27–dependent IL-10 gene induction. A scan of the proximal IL-10 promoter using the program MatInspector to search for putative transcription factor binding sites, revealed that the −1324/−1319 and −510/−504 cis regulatory elements that mediate IL-27–dependent IL-10 promoter activity correspond to a putative STAT3 and a previously identified c-Maf binding motif (36, 37). IL-27 signaling through the heterodimeric receptor, IL-27R/gp130, is known to activate several members of the STAT transcription family, including STAT1, STAT2, STAT3, STAT4, and STAT5. To assess activation of the STAT family of transcription factors in macrophages, we stimulated BMDMs with LPS, IFN-α, and IL-27 at the indicated time points and assessed phosphorylation of STAT1, STAT2, STAT3, and STAT5 using specific phospho- Tyr Abs. LPS, IFN-α, and IL-27 were able to activate STAT1 and STAT3 phosphorylation (Fig. 6A), whereas LPS and IFN-α, but not IL-27, were able to activate STAT2 phosphorylation (data not shown). We also assessed induction of c-Maf by LPS, IFN-α, and IL-27; however, c-Maf appears to be constitutively expressed in BMDMs and not subject to regulation at the protein level at the time points measured (37).
FIGURE 6.
LPS, IFN-α, and IL-27 induction of IL-10 is STAT1 and STAT3 dependent. A, Total of 0.5 × 106 WT BMDMs were unstimulated or stimulated with LPS (50 ng/ml), IFN-α (250 U/ml), or IL-27 (10 ng/ml) for indicated time points. Cell lysate was collected and immune-blotted for STAT1 and STAT3 expression and STAT1 and STAT3 phosphorylation using specific Abs. B, Total of 0.5 × 106 WT and STAT1−/− BMDMs were unstimulated or stimulated with LPS (50 ng/ml), IFN-α (250 U/ml), or IL- 27 (10 ng/ml) for 3 h. RNA was harvested, and relative IL-10 transcript was detected via qPCR normalized to L32. Student t test was performed. *p < 1 × 103. C, Total of 0.5 × 106 macrophage-specific STAT3+/+ and STAT3−/− BMDMs were unstimulated or stimulated with LPS (50 ng/ml), IFN-α (250 U/ml), or IL-27 (10 ng/ml) for 3 h. RNA was harvested, and relative IL-10 transcript was detected via qPCR normalized to L32. Data represent two independent experiments. *p < 0.05. D, ChIP using Abs against STAT1, STAT2, STAT3, c-Maf, and Sp1 using primers specific to the IL-10 promoter. Transcription factor enrichment presented as percent of input representing two independent experiments. E, Model of LPS-mediated induction of IL-10 gene expression involves sequential induction and signaling type I IFN followed induction and signaling by IL-27 signaling in macrophages.
We next assessed the requirement for STAT1 and STAT3 in IL-10 induction. We stimulated wild-type and STAT1-deficient BMDMs with LPS, IFN-α, and IL-27 for 3 h and assessed IL-10 induction by qPCR. IL-27–mediated induction of IL-10 had an absolute requirement for STAT1, whereas IFN-α–mediated IL-10 induction exhibited partial but significant defects (Fig. 6B). Although there was a partial decrease in IL-10 induction in BMDMs deficient in STAT1 compared with wild-type in response to LPS, the difference was not statistically significant. Next, we stimulated wild-type and macrophage-specific STAT3-deficient BMDMs with LPS, IFN-α, and IL-27 and assessed IL-10 induction by qPCR. IL-27–mediated IL-10 induction was STAT3 dependent, whereas LPS and IFN-α displayed partial defects in IL-10 induction (Fig. 6C). Thus, we conclude that IL-27 induction of IL-10 expression requires STAT1 and STAT3.
Finally, we wanted to determine whether STAT1 and STAT3 are recruited to the IL-10 promoter or function mainly as signaling molecules. We expected that STAT1 and STAT3 could potentially directly regulate IL-10 gene expression because de novo synthesis of proteins is not required for IL-27–mediated IL-10 induction. We performed ChIP in BMDMs stimulated with LPS, IFN-α, and IL-27 using two sets of IL-10 promoter-specific promoters, one pair spanning the transcription start site and one pair to amplify the more distal regulatory region identified in our promoter activity analyses (−1324/−1319 bp). We show that both STAT1 and STAT3 are mobilized to the promoter in response to LPS, IFN-α, and IL-27, suggesting that they may directly regulate IL-10 promoter activity (Fig. 6D). C-Maf was recruited to the promoter by LPS, IFN-α, and IL-27 as well. Sp1 was shown to be constitutively bound at the IL-10 promoter with minimal enrichment upon stimulation, consistent with previous studies (23). In contrast, STAT2 is not recruited to the IL-10 promoter under any of the conditions tested, which is consistent with a nonessential role in IL-27–mediated IL-10 induction.
Discussion
The innate immune system is the first line of defense against infection. However, uncontrolled production of proinflammatory cytokines may initiate or exacerbate harmful inflammatory or autoimmune responses. IL-10 signaling represents one important source of such regulation. In macrophages, production of IL-10 is generated through TLR activation as a means of providing such a feedback response. In this study, we describe a signaling pathway that couples LPS stimulation of macrophages to IL-10 expression through the sequential generation and signaling of type I IFNs and IL-27, respectively. Our studies using LPS, IFN-α, and IL-27 CM to assess IL-10 expression suggest that the generation of signaling through these downstream gene products in response to LPS prior to IL-10 expression has physiological relevance in macrophage function.
In the past, identifying important transcription factors that mediate LPS upregulation of IL-10 has been difficult. A number of factors have been proposed to play a role in LPS-mediated IL-10 induction including IRFs, STATs, and NF-κB; however, to date, only one factor, Sp1, has proven to be essential (30, 34). One potential confounding element that may obscure the relative importance of specific transcription factors or regulatory motifs in mediating IL-10 induction by LPS is the multiple gene programs and autocrine/paracrine signaling pathways induced by LPS through TLR4. That is, although a specific transcription factor or cis regulatory element may play an important role in LPS-mediated IL-10 induction, its relative importance may be obscured through downstream events that provide compensatory or alternative means of gene activation. For example, we show that STAT1 is mobilized to the IL-10 locus in response to LPS, but we observe only partial and insignificant defects in IL-10 induction in macrophages deficient in STAT1 compared with wild-type. However, the role of STAT1 in IL-10 regulation is demonstrably clearer as we study its function in signaling pathways downstream of TLR4, like type I IFN and IL-27 (Fig. 6B). Because LPS-induced IL-10 expression indirectly occurs through the generation of secondary and tertiary signaling modules, the identification of important regulatory motifs may be obscured by these alternative or compensatory mechanisms.
In contrast, we demonstrate that IL-27–mediated IL-10 expression is direct in that it does not require the synthesis of a downstream signaling molecule or transcription factor prior to generation of IL-10 transcripts. Thus, we are more likely to identify functional cis elements within the IL-10 promoter in response to IL-27 unmitigated by additional downstream signaling events. Through promoter-reporter assays, we identify three major IL-27 response elements located at positions −1324/−1319, −510/−504, and −90/−78 relative to the transcription start site. The −90/−78 region is essential for LPS-, IFN-α–, and IL-27–mediated IL-10 promoter activity and corresponds to a previously identified Sp1 binding site (30). The −510/−504 site corresponds to a previously described c-Maf site (37). The −1324/−1319 and −510/−504 IL-27 responsive elements map larger promoter regions that correlate with LPS and IFN-α functional elements, respectively, from our promoter deletion studies, which is consistent with a downstream requirement for IL-27 signaling for LPS- and IFN-α–mediated IL-10 induction. Discrepancies between the relative roles for each element in IL-10 gene regulation may be due to the activation of alternative signaling pathways by LPS and IFN-α (34). For instance, it has been shown that maximal induction of IL-10 by LPS requires both MyD88 and TRIF adaptor molecules, whereas type I IFN production by LPS occurs solely through the TRIF signaling pathway (34).
We demonstrate that LPS, IFN-α, and IL-27 all activate and require both STAT1 and STAT3. Previous studies have suggested a role for both STAT1 and STAT3 in regulating of IL-10; however, direct binding of either factor to cis elements within the promoter have yet to be demonstrated (38). In this study, we show that both STAT1 and STAT3 as well as c-Maf are mobilized to the IL-10 promoter upon stimulation with LPS, IFN-α, and IL-27; however, we have not been able to map this binding to a specific region, though the −1319/−1324 region corresponds to a putative STAT3 site. Importantly, this site appears to be conserved from human to mouse. In addition, the site is physically distinct from a previously described IRF1/STAT3 composite site identified as a functional regulatory element in IFN-α–mediated IL-10 induction in human monocytes (38). Sp1 remains basally associated with the IL-10 promoter and is not enriched upon stimulation. It is possible that recruitment of STAT1 and STAT3 occurs only in the context of chromatin, as the IL-10 locus undergoes extensive epigenetic regulation (39–41).
In addition, it is important to note that whereas LPS, IFN-α, and IL-27 all exhibit STAT1- and STAT3-dependent activation of IL-10, the relative dependence on STAT1 and STAT3 differs among the three stimuli. Specifically, IL-27–mediated IL-10 induction exhibits an absolute requirement for STAT1 and STAT3 activity, whereas LPS and IFN-α display partial dependence on STAT3 and STAT1, respectively (Fig. 6B, 6C). Discrepancies between the relative importance of subsets of transcription factors, such as STAT1 and STAT3, as well as the relative use of cis elements within IL-10 promoter upon induction by LPS, IFN-α, and/or IL-27 highlight both the significance of the IL-27 pathway in LPS- and IFN-α–dependent IL-10 gene regulation, but also reveal a level of discordance in that both LPS and IFN-α appear to use alternative sets of signaling pathways and transcription factors to achieve maximal transcriptional induction of IL-10 (Fig. 6E) (34).
IL-27 has been implicated in both pro- and anti-inflammatory contexts. It is required for clearance of intracellular pathogens but also plays a role in inhibition of Th1/Th17 differentiation and confers a protective effect in the development of experimental allergic encephelomyeletis (15, 19, 42, 43). In this study, we show that IL-27 can contribute to a global anti-inflammatory state through the induction of IL-10 in macrophages. This is in contrast to one report in which IL-27 negatively regulates IL-10 induction in human monocytes and BMDMs (44). However, in that study, IL-27 downregulation of IL-10 occurred only when macrophages were preactivated with TLR2 or TLR4 agonists. How LPS and type I IFNs induce IL-27 expression remains to be elucidated and requires extensive promoter studies of the p28 and EBI3 loci that make up the functional Il-27 heterodimer. We show that LPS-mediated p28 induction is type I IFN dependent, consistent with studies of IL-27 regulation in human dendritic cells (45, 46). Induction of p28 expression appears to be NF-κB and IRF1 dependent (45). Further investigation into the regulation of IL-27 gene expression will undoubtedly reveal insights into the regulation of macrophage anti-inflammatory gene programs.
Acknowledgments
We thank Dr. Bin Gao (National Institute of Alcohol Abuse and Alcoholism) for providing bone marrow for macrophage-specific STAT3−/− (STAT3flox/flox LysCre+/−) and STAT3+/+ (STAT3flox/floxLysMCre−/−) used in this study. We also thank Dr. Steve Smale (MIMG, University of California, Los Angeles) for helpful feedback and members of the laboratory for engaging discussion.
Abbreviations used in this paper
- BMDM
bone marrow-derived macrophage
- ChIP
chromatin immunoprecipitation
- CHX
cycloheximide
- CM
conditioned media
- IRF
IFN regulatory factor
- qPCR
quantitative PCR
- TRIF
Toll/IL-1R domain-containing adaptor inducing IFN-β
Footnotes
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Meylan E, Tschopp J, Karin M. Intracellular pattern recognition receptors in the host response. Nature. 2006;442:39–44. doi: 10.1038/nature04946. [DOI] [PubMed] [Google Scholar]
- 2.Takeda K, Akira S. Toll-like receptors in innate immunity. Int. Immunol. 2005;17:1–14. doi: 10.1093/intimm/dxh186. [DOI] [PubMed] [Google Scholar]
- 3.Barton GM, Medzhitov R. Toll-like receptor signaling pathways. Science. 2003;300:1524–1525. doi: 10.1126/science.1085536. [DOI] [PubMed] [Google Scholar]
- 4.Kawai T, Akira S. TLR signaling. Cell Death Differ. 2006;13:816–825. doi: 10.1038/sj.cdd.4401850. [DOI] [PubMed] [Google Scholar]
- 5.Medzhitov R. Toll-like receptors and innate immunity. Nat. Rev. Immunol. 2001;1:135–145. doi: 10.1038/35100529. [DOI] [PubMed] [Google Scholar]
- 6.Kawai T, Akira S. TLR signaling. Semin. Immunol. 2007;19:24–32. doi: 10.1016/j.smim.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 7.Seth RB, Sun L, Chen ZJ. Antiviral innate immunity pathways. Cell Res. 2006;16:141–147. doi: 10.1038/sj.cr.7310019. [DOI] [PubMed] [Google Scholar]
- 8.Doyle SE, O’Connell RM, Miranda GA, Vaidya SA, Chow EK, Liu PT, Suzuki S, Suzuki N, Modlin RL, Yeh WC, et al. Toll-like receptors induce a phagocytic gene program through p38. J. Exp. Med. 2004;199:81–90. doi: 10.1084/jem.20031237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Severa M, Fitzgerald KA. TLR-mediated activation of type I IFN during antiviral immune responses: fighting the battle to win the war. Curr. Top. Microbiol. Immunol. 2007;316:167–192. doi: 10.1007/978-3-540-71329-6_9. [DOI] [PubMed] [Google Scholar]
- 10.Doyle SE, O’Connell R, Vaidya SA, Chow EK, Yee K, Cheng G. Toll-like receptor 3 mediates a more potent antiviral response than Toll-like receptor 4. J. Immunol. 2003;170:3565–3571. doi: 10.4049/jimmunol.170.7.3565. [DOI] [PubMed] [Google Scholar]
- 11.Chang EY, Guo B, Doyle SE, Cheng G. Cutting edge: involvement of the type I IFN production and signaling pathway in lipopolysaccharide-induced IL-10 production. J. Immunol. 2007;178:6705–6709. doi: 10.4049/jimmunol.178.11.6705. [DOI] [PubMed] [Google Scholar]
- 12.Rep MH, Schrijver HM, van Lopik T, Hintzen RQ, Roos MT, Adér HJ, Polman CH, van Lier RA. Interferon (IFN)-beta treatment enhances CD95 and interleukin 10 expression but reduces interferon-gamma producing T cells in MS patients. J. Neuroimmunol. 1999;96:92–100. doi: 10.1016/s0165-5728(98)00271-9. [DOI] [PubMed] [Google Scholar]
- 13.Aman MJ, Tretter T, Eisenbeis I, Bug G, Decker T, Aulitzky WE, Tilg H, Huber C, Peschel C. Interferon-alpha stimulates production of interleukin-10 in activated CD4+ T cells and monocytes. Blood. 1996;87:4731–4736. [PubMed] [Google Scholar]
- 14.Feng X, Yau D, Holbrook C, Reder AT. Type I interferons inhibit interleukin-10 production in activated human monocytes and stimulate IL-10 in T cells: implications for Th1-mediated diseases. J. Interferon Cytokine Res. 2002;22:311–319. doi: 10.1089/107999002753675730. [DOI] [PubMed] [Google Scholar]
- 15.Guo B, Chang EY, Cheng G. The type I IFN induction pathway constrains Th17-mediated autoimmune inflammation in mice. J. Clin. Invest. 2008;118:1680–1690. doi: 10.1172/JCI33342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stumhofer JS, Silver JS, Laurence A, Porrett PM, Harris TH, Turka LA, Ernst M, Saris CJ, O’Shea JJ, Hunter CA. Interleukins 27 and 6 induce STAT3-mediated T cell production of interleukin 10. Nat. Immunol. 2007;8:1363–1371. doi: 10.1038/ni1537. [DOI] [PubMed] [Google Scholar]
- 17.Yoshimura T, Takeda A, Hamano S, Miyazaki Y, Kinjyo I, Ishibashi T, Yoshimura A, Yoshida H. Two-sided roles of IL-27: induction of Th1 differentiation on naive CD4+ T cells versus suppression of proinflammatory cytokine production including IL-23-induced IL-17 on activated CD4+ T cells partially through STAT3-dependent mechanism. J. Immunol. 2006;177:5377–5385. doi: 10.4049/jimmunol.177.8.5377. [DOI] [PubMed] [Google Scholar]
- 18.Batten M, Kljavin NM, Li J, Walter MJ, de Sauvage FJ, Ghilardi N. Cutting edge: IL-27 is a potent inducer of IL-10 but not FoxP3 in murine T cells. J. Immunol. 2008;180:2752–2756. doi: 10.4049/jimmunol.180.5.2752. [DOI] [PubMed] [Google Scholar]
- 19.Stumhofer JS, Laurence A, Wilson EH, Huang E, Tato CM, Johnson LM, Villarino AV, Huang Q, Yoshimura A, Sehy D, et al. Interleukin 27 negatively regulates the development of interleukin 17-producing T helper cells during chronic inflammation of the central nervous system. Nat. Immunol. 2006;7:937–945. doi: 10.1038/ni1376. [DOI] [PubMed] [Google Scholar]
- 20.Kastelein RA, Hunter CA, Cua DJ. Discovery and biology of IL-23 and IL-27: related but functionally distinct regulators of inflammation. Annu. Rev. Immunol. 2007;25:221–242. doi: 10.1146/annurev.immunol.22.012703.104758. [DOI] [PubMed] [Google Scholar]
- 21.Li J, Gran B, Zhang GX, Rostami A, Kamoun M. IL-27 subunits and its receptor (WSX-1) mRNAs are markedly up-regulated in inflammatory cells in the CNS during experimental autoimmune encephalomyelitis. J. Neurol. Sci. 2005;232:3–9. doi: 10.1016/j.jns.2004.12.013. [DOI] [PubMed] [Google Scholar]
- 22.Hunter CA. New IL-12-family members: IL-23 and IL-27, cytokines with divergent functions. Nat. Rev. Immunol. 2005;5:521–531. doi: 10.1038/nri1648. [DOI] [PubMed] [Google Scholar]
- 23.Sonoda KH, Yoshimura T, Takeda A, Ishibashi T, Hamano S, Yoshida H. WSX-1 plays a significant role for the initiation of experimental autoimmune uveitis. Int. Immunol. 2007;19:93–98. doi: 10.1093/intimm/dxl125. [DOI] [PubMed] [Google Scholar]
- 24.Hölscher C, Hölscher A, Rückerl D, Yoshimoto T, Yoshida H, Mak T, Saris C, Ehlers S. The IL-27 receptor chain WSX-1 differentially regulates antibacterial immunity and survival during experimental tuberculosis. J. Immunol. 2005;174:3534–3544. doi: 10.4049/jimmunol.174.6.3534. [DOI] [PubMed] [Google Scholar]
- 25.Fitzgerald DC, Zhang GX, El-Behi M, Fonseca-Kelly Z, Li H, Yu S, Saris CJ, Gran B, Ciric B, Rostami A. Suppression of autoimmune inflammation of the central nervous system by interleukin 10 secreted by interleukin 27-stimulated T cells. Nat. Immunol. 2007;8:1372–1379. doi: 10.1038/ni1540. [DOI] [PubMed] [Google Scholar]
- 26.Awasthi A, Carrier Y, Peron JP, Bettelli E, Kamanaka M, Flavell RA, Kuchroo VK, Oukka M, Weiner HL. A dominant function for interleukin 27 in generating interleukin 10-producing anti-inflammatory T cells. Nat. Immunol. 2007;8:1380–1389. doi: 10.1038/ni1541. [DOI] [PubMed] [Google Scholar]
- 27.Horiguchi N, Wang L, Mukhopadhyay P, Park O, Jeong WI, Lafdil F, Osei-Hyiaman D, Moh A, Fu XY, Pacher P, et al. Cell type-dependent pro- and anti-inflammatory role of signal transducer and activator of transcription 3 in alcoholic liver injury. Gastroenterology. 2008;134:1148–1158. doi: 10.1053/j.gastro.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moore KJ, Labrecque S, Matlashewski G. Alteration of Leishmania donovani infection levels by selective impairment of macrophage signal transduction. J. Immunol. 1993;150:4457–4465. [PubMed] [Google Scholar]
- 29.Haskó G, Szabó C, Németh ZH, Kvetan V, Pastores SM, Vizi ES. Adenosine receptor agonists differentially regulate IL-10, TNF-alpha, and nitric oxide production in RAW 264.7 macrophages and in endotoxemic mice. J. Immunol. 1996;157:4634–4640. [PubMed] [Google Scholar]
- 30.Brightbill HD, Plevy SE, Modlin RL, Smale ST. A prominent role for Sp1 during lipopolysaccharide-mediated induction of the IL-10 promoter in macrophages. J. Immunol. 2000;164:1940–1951. doi: 10.4049/jimmunol.164.4.1940. [DOI] [PubMed] [Google Scholar]
- 31.Yamamoto M, Sato S, Mori K, Hoshino K, Takeuchi O, Takeda K, Akira S. Cutting edge: a novel Toll/IL-1 receptor domain-containing adapter that preferentially activates the IFN-βeta promoter in the Toll-like receptor signaling. J. Immunol. 2002;169:6668–6672. doi: 10.4049/jimmunol.169.12.6668. [DOI] [PubMed] [Google Scholar]
- 32.Panne D. The enhanceosome. Curr. Opin. Struct. Biol. 2008;18:236–242. doi: 10.1016/j.sbi.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 33.Levy DE, Kessler DS, Pine R, Darnell JE., Jr Cytoplasmic activation of ISGF3, the positive regulator of interferon-alpha-stimulated transcription, reconstituted in vitro. Genes and Dev. 1989;9:1362–1371. doi: 10.1101/gad.3.9.1362. [DOI] [PubMed] [Google Scholar]
- 34.Saraiva M, O’Garra A. The regulation of IL-10 production by immune cells. Nat. Rev. Immunol. 2010;10:170–181. doi: 10.1038/nri2711. [DOI] [PubMed] [Google Scholar]
- 35.Kim SJ, Onwuta US, Lee YI, Li R, Botchan MR, Robbins PD. The retinoblastoma gene product regulates Sp1-mediated transcription. Mol. Cell. Biol. 1992;12:2455–2463. doi: 10.1128/mcb.12.6.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- 37.Cao S, Liu J, Song L, Ma X. The protooncogene c-Maf is an essential transcription factor for IL-10 gene expression in macrophages. J. Immunol. 2005;174:3484–3492. doi: 10.4049/jimmunol.174.6.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ziegler-Heitbrock L, Lötzerich M, Schaefer A, Werner T, Frankenberger M, Benkhart E. IFN-alpha induces the human IL-10 gene by recruiting both IFN regulatory factor 1 and Stat3. J. Immunol. 2003;171:285–290. doi: 10.4049/jimmunol.171.1.285. [DOI] [PubMed] [Google Scholar]
- 39.Saraiva M, Christensen JR, Tsytsykova AV, Goldfeld AE, Ley SC, Kioussis D, O’Garra A. Identification of a macrophage-specific chromatin signature in the IL-10 locus. J. Immunol. 2005;175:1041–1046. doi: 10.4049/jimmunol.175.2.1041. [DOI] [PubMed] [Google Scholar]
- 40.Wang ZY, Sato H, Kusam S, Sehra S, Toney LM, Dent AL. Regulation of IL-10 gene expression in Th2 cells by Jun proteins. J. Immunol. 2005;174:2098–2105. doi: 10.4049/jimmunol.174.4.2098. [DOI] [PubMed] [Google Scholar]
- 41.Jones EA, Flavell RA. Distal enhancer elements transcribe intergenic RNA in the IL-10 family gene cluster. J. Immunol. 2005;175:7437–7446. doi: 10.4049/jimmunol.175.11.7437. [DOI] [PubMed] [Google Scholar]
- 42.Yoshida H, Miyazaki Y, Yoshiyuki M. Regulation of immune responses by interleukin-27. [Published erratum appears in 2009. Immunol. Rev. 227:283.] Immunol. Rev. 2008;226:234–247. doi: 10.1111/j.1600-065X.2008.00710.x. [DOI] [PubMed] [Google Scholar]
- 43.Schmidt C, Giese T, Ludwig B, Mueller-Molaian I, Marth T, Zeuzem S, Meuer SC, Stallmach A. Expression of interleukin-12-related cytokine transcripts in inflammatory bowel disease: elevated interleukin-23p19 and interleukin-27p28 in Crohn’s disease but not in ulcerative colitis. Inflamm. Bowel Dis. 2005;11:16–23. doi: 10.1097/00054725-200501000-00003. [DOI] [PubMed] [Google Scholar]
- 44.Kalliolias GD, Ivashkiv LB. IL-27 activates human monocytes via STAT1 and suppresses IL-10 production but the inflammatory functions of IL-27 are abrogated by TLRs and p38. J. Immunol. 2008;180:6325–6333. doi: 10.4049/jimmunol.180.9.6325. [DOI] [PubMed] [Google Scholar]
- 45.Liu J, Guan X, Ma X. Regulation of IL-27 p28 gene expression in macrophages through MyD88- and interferon-gamma-mediated pathways. J. Exp. Med. 2007;204:141–152. doi: 10.1084/jem.20061440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Remoli ME, Gafa V, Giacomini E, Severa M, Lande R, Coccia EM. IFN-βeta modulates the response to TLR stimulation in human DC: involvement of IFN regulatory factor-1 (IRF-1) in IL-27 gene expression. Eur. J. Immunol. 2007;37:3499–3508. doi: 10.1002/eji.200737566. [DOI] [PubMed] [Google Scholar]