SUMMARY
The p53-regulated long non-coding RNA lincRNA-p21 has been proposed to act in trans via several mechanisms ranging from repressing genes in the p53 transcriptional network to regulating mRNA translation and protein stability. To further examine lincRNA-p21 function we generated a conditional knockout mouse model. We find that lincRNA-p21 predominantly functions in cis to activate expression of its neighboring gene, p21. Mechanistically, we show that lincRNA-p21 acts in concert with hnRNP-K as a co-activator for p53-dependent p21 transcription. Additional phenotypes of lincRNA-p21 deficiency could be attributed to diminished p21 levels, including deregulated expression and altered chromatin state of some Polycomb target genes, defective G1/S checkpoint, increased proliferation rates, and enhanced reprogramming efficiency. These findings indicate that lincRNA-p21 affects global gene expression and influences the p53 tumor suppressor pathway by acting in cis as a locus-restricted co-activator for p53-mediated p21 expression.
INTRODUCTION
The p53 tumor suppressor pathway is activated in the presence of cellular stress, such as DNA damage and oncogenic signaling, and in turn coordinates the transcriptional response of hundreds of genes (Levine and Oren, 2009). Depending on the type of tissue and the nature of the stress signal, p53 activation can initiate multiple pathways that can lead to a temporary pause at a cell cycle checkpoint to allow for DNA repair, permanent growth arrest (senescence), or cell death (apoptosis) (Vousden and Prives, 2009). It is not clear what determines the outcome of p53 activation. Multiple phenomena, including the strength of p53 binding at the promoters of target genes and the dynamics of p53 oscillations, have been proposed to guide the transcriptional response leading to distinct cellular outcomes (Vousden and Prives, 2009; Purvis et al., 2012).
Based on the identification of mouse long non-coding RNAs (lncRNAs) that are directly induced by p53, recent studies have suggested that lncRNAs may provide an additional layer of transcriptional regulation in the p53 pathway (Guttman et al., 2009; Huarte et al., 2010). Among these, lincRNA-p21 has been proposed to promote apoptosis (Huarte et al., 2010). Other p53-regulated lncRNAs, including Pint and PANDA, have been found to antagonize p53 activity by promoting proliferation and by limiting the induction of pro-apoptotic genes (Hung et al., 2011; Marin-Bejar et al., 2013). In addition, lncRNAs expressed from p53-bound enhancer regions have been found to regulate checkpoint function (Melo et al., 2013a). These studies support a model in which p53-regulated lncRNAs fine-tune the p53 transcriptional response.
In recent years, significant insight has been gained into the numerous mechanisms by which lncRNAs function (Rinn and Chang, 2012). Some well-characterized nuclear lncRNAs, such as XIST and lncRNAs expressed from imprinted loci, have been shown to modulate gene expression in cis by acting as scaffolds for the recruitment of chromatin modifying complexes, notably the PRC2 complex, and by altering the chromatin structure of target genes (Lee and Bartolomei, 2013). Other cis-acting lncRNAs, such as enhancer RNAs (eRNAs), have been suggested to activate gene expression by locally regulating chromatin architecture (Melo et al., 2013b). In contrast, a growing class of lncRNAs, including lincRNA-p21, has been proposed to regulate gene expression in trans by directing the chromatin localization of protein binding partners (Fatica and Bozzoni, 2014). Finally, a class of cytosolic lncRNAs, including human lincRNA-p21, has been proposed to regulate mRNA translation and protein stability (Yang et al., 2014; Yoon et al., 2012).
Here, we have investigated the effects of lincRNA-p21 deficiency on the control of expression of p53 target genes and on the p53-dependent cellular response in cells, derived from a lincRNA-p21 conditional knockout mouse model. Our findings differ significantly from previous studies, which used RNAi to deplete lincRNA-p21 levels, and highlight the advantages of using a genetic system to study the function of low copy number, cis-acting lncRNAs. The data presented here reveal a novel mechanism for regulation in the p53 tumor suppressor network, whereby lincRNA-p21 acts as a transcriptional co-activator that locally reinforces p21 activation past a functional threshold.
RESULTS
Generation of lincRNA-p21 conditional knockout mice and cells
To investigate the role of lincRNA-p21 in vivo, we mutated the lincRNA-p21 locus using a conditional gene targeting strategy. The targeting design was directed at preventing expression of the lincRNA-p21 transcript by flanking the promoter region, which includes the p53 response element (p53RE) as well as the first exon of lincRNA-p21, with loxP sites (Fig. S1A, B). LincRNA-p21-deficient animals, generated by crossing founders to Deletor Cre transgenic animals (Lewandoski et al., 1997), are viable, born at Mendelian ratios, and display no significant abnormalities or predisposition to tumor development by the age of 18 months (Fig. S1A–E).
To characterize the function of lincRNA-p21, we isolated primary mouse embryonic fibroblasts (MEFs) from littermate E13.5 embryos with lincRNA-p21−/− and +/+ genotypes, as well as lincRNA-p21f/f; Rosa26-CreERT2 MEFs, in which lincRNA-p21 deletion could be induced by treatment with tamoxifen. Loss of lincRNA-p21 expression was confirmed by quantitative RT-PCR (qRT-PCR) and RNA-seq (Fig. S1F–H).
Deletion of lincRNA-p21 affects gene expression globally
To examine the role of lincRNA-p21 in the regulation of gene expression, we performed global expression analysis. RNA was collected from three independent pairs of littermate lincRNA-p21+/+ and lincRNA-p21−/− MEFs, harvested from untreated cells and at 24 hours following treatment with the DNA damage-inducing agent doxorubicin. To identify lincRNA-p21-responsive genes, we set a stringent classification in which high-confidence differentially expressed genes (FDR<0.05) were required to show consistent variation of 34% or more (|Log 2 Fold Change|>0.43) in all three lincRNA-p21-deficient samples compared to the corresponding littermate wild-type controls.
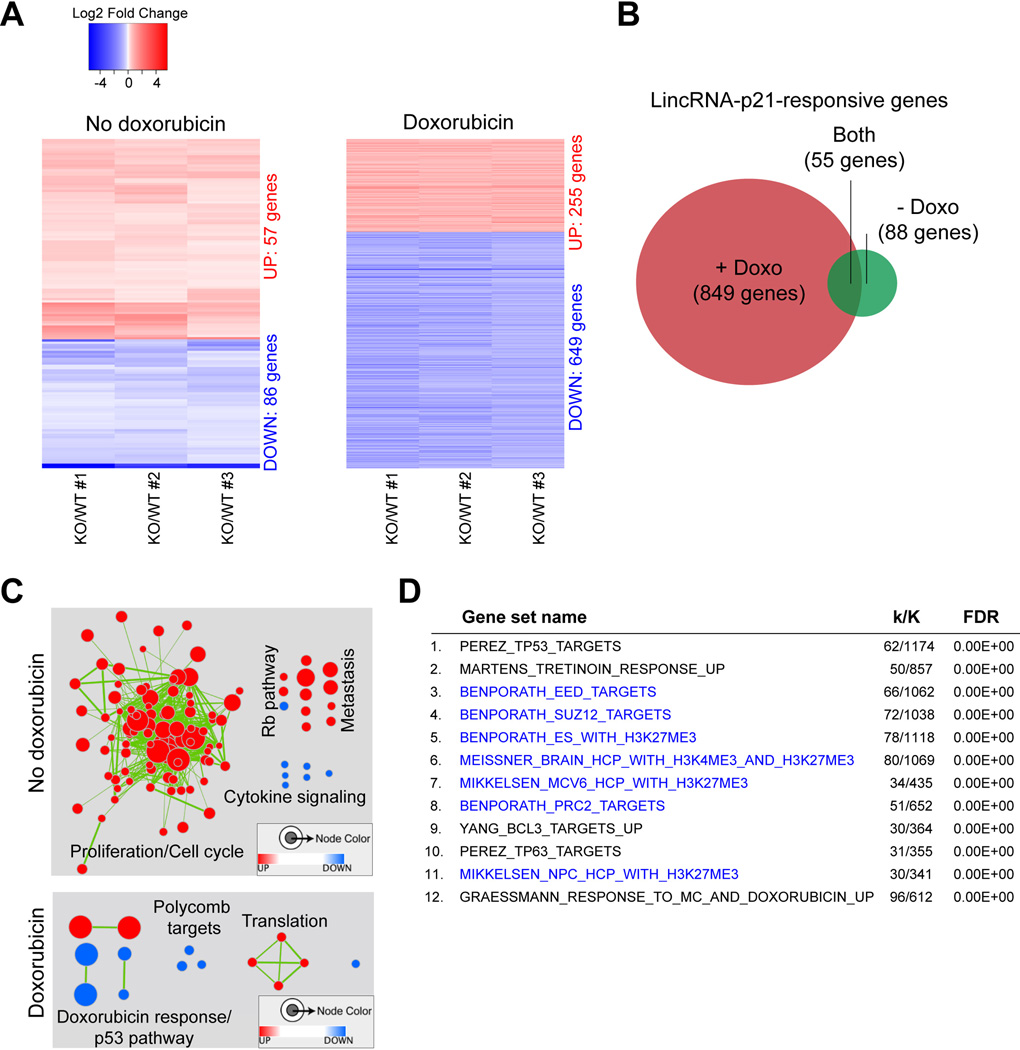
In the absence of DNA damage, we identified a set of 143 genes that were differentially expressed between lincRNA-p21-proficient and –deficient samples (Fig. 1A left and Table S1). To recognize statistically significant correlations with curated gene sets within the Molecular Signature Database (MSigDB), we performed gene set enrichment analysis (GSEA) and identified 122 gene sets affected by lincRNA-p21 loss (FDR<0.0001, Table S2) (Subramanian et al., 2005). Connectivity maps revealed a pattern associated with increased proliferation mediated by cell cycle regulators (Fig. 1C, top). We also noted that the cyclin-dependent kinase inhibitor p21 (also known as CDKN1a/Cip1/WAF1), located 16.7 Kb upstream of lincRNA-p21, was one of the differentially expressed genes. p21 is a direct target of p53 transcriptional regulation that is induced upon passaging of primary cell cultures as well as in response to cellular stress and acts to arrest cell cycle progression at the G1/S checkpoint (Deng et al., 1995; Brugarolas et al., 1995). p21 expression levels were reduced in all three lincRNA-p21-deficient MEF lines compared to wild-type controls (Fig. S2C and Table S1). The novel roles of lincRNA-p21 in cell cycle control and in promoting the expression of p21 will be examined in more detail below.
Figure 1. Loss of lincRNA-p21 affects gene expression globally.
(A) Heatmaps of differentially expressed genes (Table S1) in untreated (left) and doxorubicin (Doxo)-treated (right) pairs of littermate lincRNA-p21-proficient (WT) and – deficient (KO) MEFs, FDR<0.05, |Log2 Fold change KO/WT| >0.43 in all three biological replicates.
(B) Venn diagram of the overlap of the lincRNA-p21-reponsive genes in the presence and absence of doxorubicin (Doxo).
(C) Connectivity maps of gene sets based on GSEA of differentially expressed genes in (A) (Table S2, FDR<0.0001). Red nodes represent enrichment with loss of lincRNA-p21 and blue represents enrichment in wild-type cells. Node size reflects the number of genes in each set and edge thickness reflects the overlap between the sets.
(D) Top 12 categories of GSEA of 649 lincRNA-p21-responsive, doxorubicin-regulated genes using the CGP Collection of MSigDB. Gene sets related to the PRC2 pathway are highlighted in blue. k indicates the number of lincRNA-p21-responsive genes; K represents the number of genes included in a given gene set. See also Figures S1 and S2 and Tables 1 and 2.
Using the identical approach, we next singled out 904 genes whose expression levels were significantly altered in lincRNA-p21-deficient samples compared to wild-type controls following doxorubicin-induced DNA damage (Fig. 1A, right, B and Table S1). Enrichment map summaries of the differentially expressed genes revealed correlations with gene sets related to the doxorubicin response and the p53 pathway (Fig. 1C, bottom, FDR < 0.0001 and Table S2). Strikingly, GSEA also uncovered a negative correlation with multiple gene sets associated with the Polycomb Repressive Complex 2, PRC2 (Fig. 1C). In fact, pathway analysis using only the 649 genes that were downregulated in the doxorubicin-treated, lincRNA-p21-deficient samples, indicated a strong overlap with PRC2 function. 7 out of the top 12 MSigDB Chemical and Genetic Perturbations (CGP) categories represented gene sets whose promoters were either occupied by components of the PRC2 complex (Eed and Suz12) or carried the Polycomb chromatin signature (H3K27me3) in various cell types (Fig. 1D). The PRC2 complex, which plays an essential role in the epigenetic silencing of regulatory genes controlling differentiation during embryonic development (Aldiri and Vetter, 2012), has not previously been implicated in the DNA damage response or in the p53 pathway. However, multiple reports have shown important regulatory functions of lncRNAs in the PRC2 pathway (Davidovich et al., 2013; Kaneko et al., 2013; Khalil et al., 2009; Zhao et al., 2010).
Of note, comparison of the lincRNA-21-dependent genes identified here with the previously published lincRNA-p21-regulated gene set (Huarte et al., 2010) showed no significant overlap, despite the similarity in cell type (MEFs) and treatment (doxorubicin) (Fig. S2A–C). This discrepancy is likely due to the different methods used to downregulate lincRNA-p21 (potent genetic deletion versus RNAi-based post-transcriptional knockdown) and highlights the importance of complementary genetic approaches to functional studies.
LincRNA-p21 promotes the expression of a set of DNA damage-inducible, PRC2-target genes
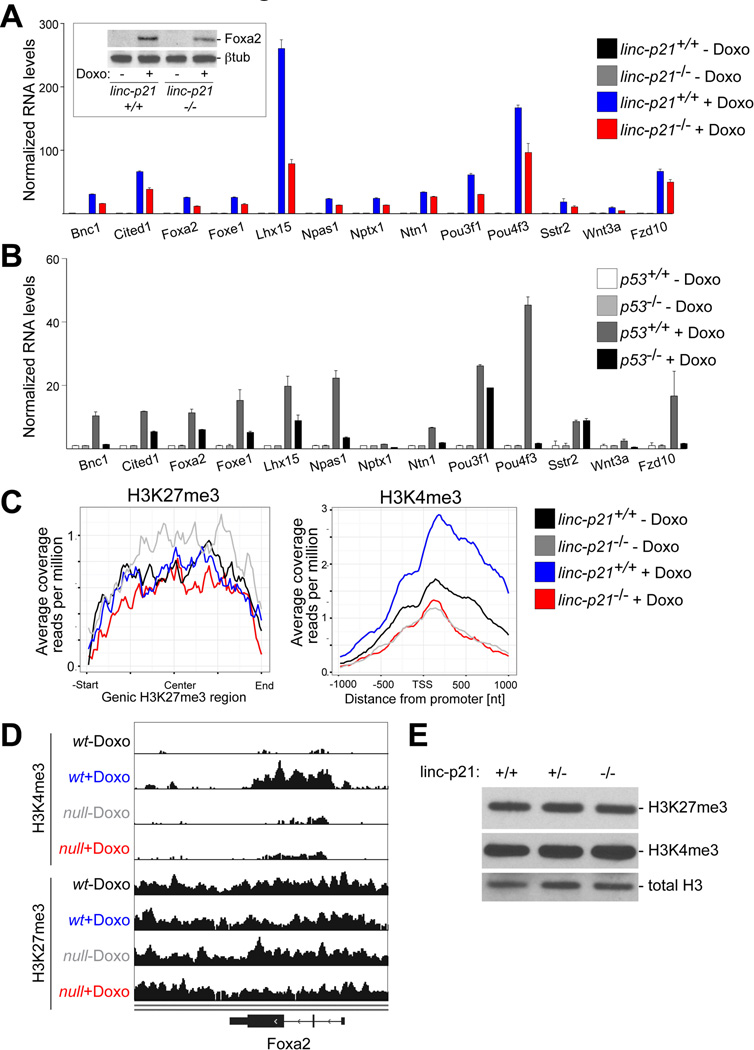
Since PRC2 targets (defined by Benporath_PRC2_Targets gene set) were significantly enriched in the lincRNA-p21-responsive gene set (Table S3), we investigated this connection further, anticipating that these studies might shed light on the role of lncRNAs in the regulation of Polycomb function. We observed that the majority of these genes were lineage-specific transcription factors, whose expression is expected to be repressed by Polycomb in primary MEFs and whose induction may be facilitating the activation of a wider transcriptional response. Indeed, the expression of several genes selected from this set was not detectable by qRT-PCR and by Western blotting in untreated samples (Fig. 2A). However, these genes became strongly upregulated at 24 hours following doxorubicin treatment and this induction was in part dependent on lincRNA-p21 (Fig. 2A and Fig. S3A, B). Global expression analysis and qRT-PCR validation indicated that the effect of lincRNA-p21 deficiency was limited to a subset of PRC2 target genes (Fig. S3B, C). Importantly, since lincRNA-p21 is a p53-regulated gene, we confirmed that the expression of this set of genes was also dependent on the p53 status (Fig. 2B), even though p53-binding sites were not identified in the promoters of these genes (data not shown). Moreover, induction of several of these genes could also be observed under other conditions that led to p53-dependent lincRNA-p21 expression, such as in response to oncogenic stress following restoration of p53 expression in a lung adenocarcinoma cell line (Fig. S3D) (Feldser et al., 2010). These data suggested an unanticipated connection between the p53-mediated stress response, lincRNA-p21, and Polycomb-regulated genes.
Figure 2. LincRNA-p21 promotes the expression and influences the chromatin state of a set of DNA damage-inducible, PRC2-target genes.
(A–B) qRT-PCR analysis of selected lincRNA-p21-responsive, PRC2 target genes in indicated MEFs and treatments. Data, replicated in >3 independent experiments, are represented mean±SEM of technical replicates. (Inset) Immunoblot analysis of Foxa2.
(C) Meta plots of the genic H3K27me3 (left) and promoter-associated H3K4me3 (right) ChIP-seq enrichment levels at lincRNA-p21-responsive, PRC2 target genes in indicated MEFs and treatments.
(D) Genome browser snapshot of Foxa2 locus, related to ChIP-seq in (C).
(E) Immunoblot analysis of H3K4me3 and H3K27me3 protein levels in whole cell extracts of doxorubicin-treated MEFs. Total H3 was used as a loading control. See also Figures S3, S4 and Table S4.
LincRNA-p21 influences the chromatin state of Polycomb target genes
We next sought to determine the mechanism by which lincRNA-p21 regulated this subset of PRC2 target genes. To test whether lincRNA-p21 regulated the chromatin state of its responsive Polycomb target genes, we performed H3K27me3 and H3K4me3 ChIP-seq in lincRNA-p21+/+ and −/− MEFs, harvested at 24 hours following mock or doxorubicin treatment. Meta analysis of the loci of the lincRNA-p21-regulated, PRC2 target gene set revealed that the PRC2-specific H3K27me3 modification was not affected by the doxorubicin treatment or by lincRNA-p21 deficiency (Fig. 2C and Fig. S4A). In contrast, whereas the activating H3K4me3 mark increased in wild-type cells in the presence of DNA damage, it remained unchanged following doxorubicin treatment in lincRNA-p21−/− MEFs (Fig. 2C, D, and Fig. S4A). Importantly, the effect of lincRNA-p21-deficiency did not lead to global changes in chromatin state (Fig. 2E and Fig. S4B) and not all loci subject to Polycomb regulation were affected (Fig. S4C). These data suggested that lincRNA-p21 status influenced the expression of a subset of DNA damage-inducible, PRC2 target genes by affecting positive regulators associated with transcriptional activation, which, in these cases, appeared to override the persistent H3K27me3 repressive mark.
LincRNA-p21 indirectly regulates PRC2 target genes via p21
We speculated that lincRNA-p21 may regulate the expression of the subset of PRC2 target genes by exerting a global effect on chromatin regulation. However, by RNA immunoprecipitation, we found no evidence for an interaction between lincRNA-p21 and components of the repressive PRC2 complex or other chromatin modifiers (Huarte et al., 2010 and data not shown). Moreover, the relatively low stability of lincRNA-p21 (half-life < 2 hours) and its low copy number (∼8 molecules/cell in the presence of DNA damage) were not consistent with a functional role as a widespread trans-regulator (Fig. S5A, B).
We next considered the possibility that lincRNA-p21 may directly bind at the promoters of PRC2 target genes and locally regulate the chromatin state at these sites. However, by single molecule RNA fluorescent in situ hybridization (RNA FISH), we did not observe co-localization between lincRNA-p21 RNA and a set of probes specific to the intron of the lincRNA-p21-responsive, PRC2 target gene Ntn1, designed to indicate the transcription site of this gene (Fig. S5C). These data suggested that lincRNA-p21 did not physically interact with the loci of PRC2 target genes.
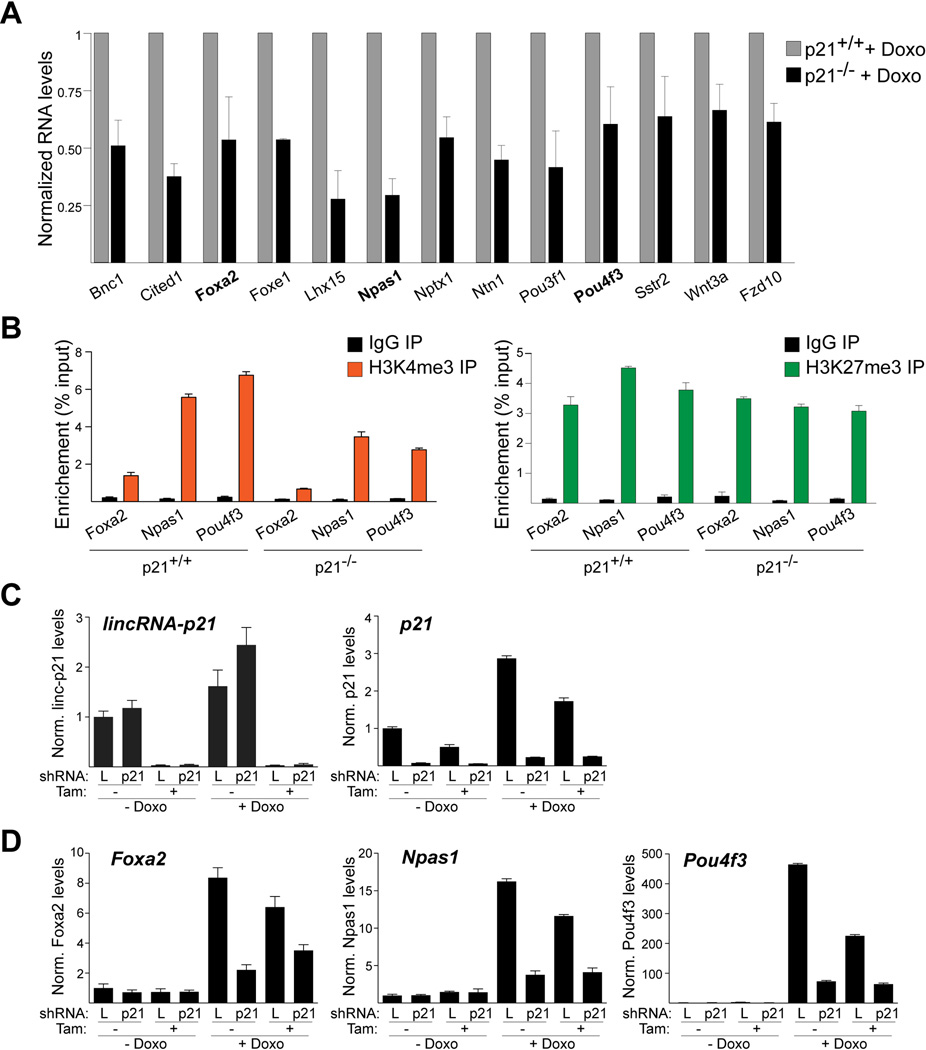
We therefore examined the possibility that lincRNA-p21 may affect the expression of PRC2 target genes indirectly. Since several reports have linked p21 and cellular differentiation (Missero et al., 1996; Steinman et al., 1994; Zhang et al., 1999), we speculated that the reduction of p21 levels observed by RNAseq in lincRNA-p21-deficient cells (Table S1) might contribute to the deregulation of the PRC2 target genes. Indeed, by qRT-PCR we found that all the examined lincRNA-p21-regulated, PRC2 target genes showed reduced induction levels following doxorubicin treatment in p21-deficient MEFs compared to wild-type controls (Fig. 3A). Moreover, by ChIP-qPCR we detected a reduction in the H3K4me3 chromatin mark and no change in the H3K27me3 chromatin modification at the promoters of Foxa2, Npas1, and Pou4f3, three genes randomly selected from the set of lincRNA-p21-responsive, PRC2 targets (Fig. 3B). These data indicated that lincRNA-p21 and p21 played similar roles in promoting the DNA damage-dependent induction of the identified set of PRC2 target genes.
Figure 3. LincRNA-p21 regulates PRC2 target genes through p21.
(A) Bar plot of normalized RNA levels of selected lincRNA-p21-regulated, PRC2 target genes in indicated cells, analyzed by qRT-PCR. Data are represented as mean±SEM, n=3.
(B) ChIP-qPCR analysis of the enrichment of H3K4me3 (left) and H3K27me3 (right) at the promoters of the indicated genes in doxorubicin-treated MEFs. Data, replicated in 2 independent experiments, are represented as mean±SEM of technical replicates.
(C) qRT-PCR analysis of lincRNA-p21 (left) and p21 (right) in a lincRNA-p21f/f; CreERT2 MEF line, infected with p21- or control luciferase (L)-specific shRNAs, harvested at 96 hours following mock treatment or tamoxifen (Tam)-mediated deletion of lincRNA-p21 and at 24 hours following mock treatment or doxorubicin (Doxo)-induced DNA damage. Data are represented as mean±SEM of technical replicates.
(D) qRT-PCR analysis of the effects of the treatments described in (C) on the expression levels of indicated lincRNA-p21-regulated, PRC2 target genes. Data were confirmed with an independent p21-targeting shRNA (not shown). See also Figure S5 and Table S4.
To establish whether lincRNA-p21 and p21 acted in the same pathway, we performed an epistasis experiment. We downregulated p21 in lincRNA-p21+/+ and −/−MEFs using a lentiviral short hairpin RNA (shRNA), which led to a greater than 90% knockdown of p21 (Fig. 3C). A luciferase-specific hairpin was used as a negative control. We found that both lincRNA-p21-deficiency and p21 knockdown led to diminished DNA damage-dependent induction of Foxa2, Npas1, and Pou4f3 (Fig. 3D). Importantly, combined loss of lincRNA-p21 and p21 did not further exacerbate the effect (Fig. 3D). Based on these findings, we concluded that the lincRNA-p21-dependent transcriptional induction of a subset of PRC2 target genes in response to genotoxic stress was mediated through its control of p21.
LincRNA-p21 activates the expression of p21
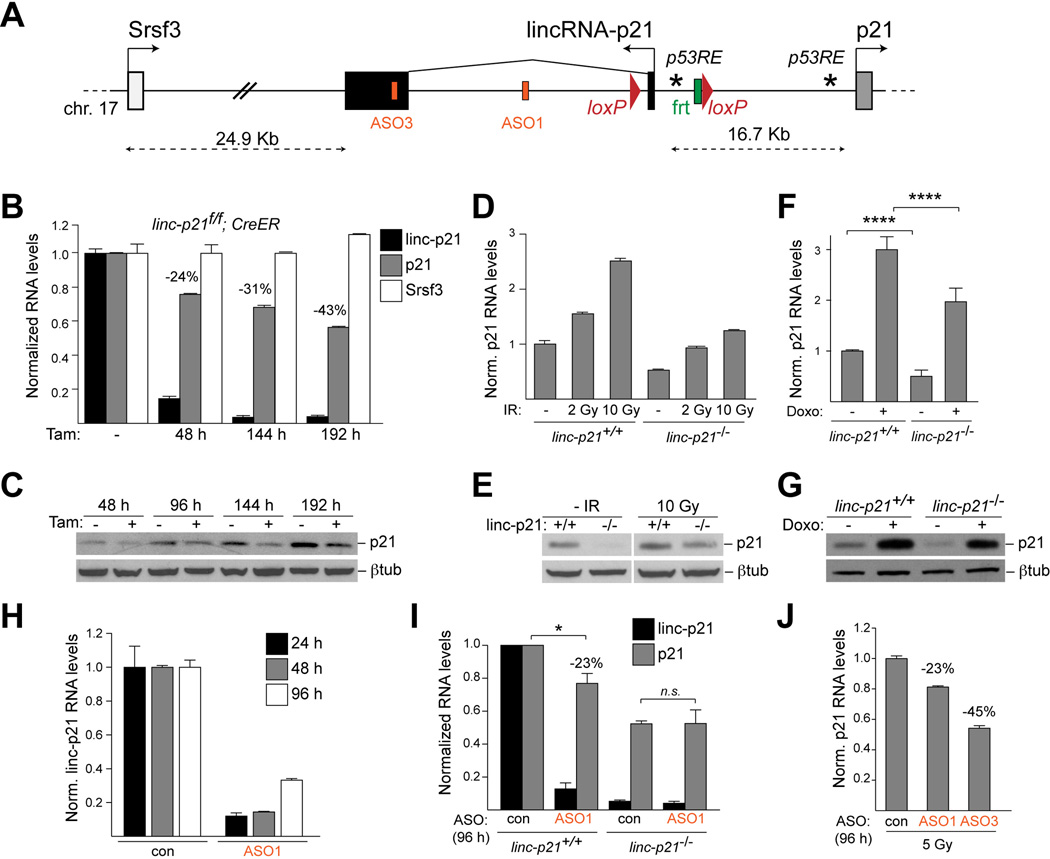
We therefore sought to dissect the role of lincRNA-p21 in the regulation of its neighboring gene, p21 (Fig. 4A). By qRT-PCR, we confirmed that acute, tamoxifen-mediated depletion of lincRNA-p21 from lincRNA-p21f/f; Rosa26-CreERT2 MEFs resulted in a reproducible 30–50% decrease of p21 RNA and protein levels over 2–3 cell divisions (Fig. 4B, C). The effect appeared specific to p21 since loss of lincRNA-p21 did not affect the expression of the gene distal to lincRNA-p21, Srsf3, and did not lead to a general defect in the activation of the p53 pathway (Fig. 4B and data not shown).
Figure 4. LincRNA-p21 RNA activates p21.
(A) Schematic of the lincRNA-p21/p21 locus, the targeting strategy, the positions of p53REs (*) in the promoters of lincRNA-p21 and p21, and the target sequences of lincRNA-p21-specific ASO1 and ASO3.
(B) qRT-PCR analysis of lincRNA-p21, p21, and Srsf3 RNA levels in indicated MEFs at indicated time-points after tamoxifen (Tam) treatment. Data, replicated in 2 independent experiments, are represented as mean±SEM of technical replicates.
(C) Immunoblot analysis of p21 protein levels in MEFs described in (B). β-tubulin was used as a loading control.
(D–E) qRT-PCR and immunoblot analysis of p21 in MEFs of indicated genotypes, harvested 8 hours following treatment with the indicated doses of γ-irradiation (IR). Data, replicated in 2 independent experiments, are represented as mean±SEM of technical replicates.
(F–G) qRT-PCR and immunoblot analysis of p21 in MEFs of indicated genotypes, harvested untreated or 8 hours following doxorubicin (Doxo) treatment. Data are represented as mean±SEM, n=8, p<0.0001, paired t-test.
(H) qRT-PCR analysis of lincRNA-p21 RNA levels at indicated time-points following transfection with the indicated control non-targeting or lincRNA-p21-specific ASOs. (I) qRT-PCR analysis of p21 RNA levels at 96 h following two consecutive ASO transfections, performed at a 48 h interval, in MEFs of indicated genotypes. Data are represented as mean±SEM, n=4, p=0.0318, paired t-test.
(J) qRT-PCR analysis of p21 RNA levels in ASO-treated wild-type MEFs, harvested 20 h post 5 Gy irradiation. Data are represented as mean±SEM of technical replicates. See also Figure S6 and Table S4.
We next asked whether the p53-dependent induction of p21 in response to DNA damage was affected by lincRNA-p21 deficiency. We observed that in lincRNA-p21−/−MEFs treated with γ-irradiation or doxorubicin, p21 was induced with the same kinetics as in wild-type cells but the total p21 RNA and protein levels remained significantly reduced by 30–50% compared to control cells (Fig. 4D–G and Fig. S6A). These data indicated that lincRNA-p21 positively regulates p21 levels in passaged primary MEFs and in response to genotoxic stress.
The region of deletion in lincRNA-p21-deficient cells is located 16.7 Kb upstream of the p21 transcription start site and contains binding sites for multiple transcription factors, including p53 (Huarte et al., 2010), as well as chromatin features that are associated with both promoter and enhancer regions. Therefore, it was formally possible that the observed defect in p21 expression could be due to deletion of a previously unidentified enhancer element located within the targeted region or due to loss of other non-coding transcripts expressed from the lincRNA-p21 locus. To address concerns with the specificity of the genetic model, we developed an independent approach to decrease lincRNA-p21 levels using antisense oligonucleotides (ASOs), which can mediate efficient co-transcriptional knockdown of nuclear RNAs in an RNase H-dependent manner (Vickers et al., 2003). Transfection with two independent lincRNA-p21-specific ASOs, ASO1 and ASO3, led to greater than 90% loss of lincRNA-p21 RNA levels at 24 hours following transfection, whereas ASO2 failed produce an efficient knockdown (Fig. 4A, H and Fig. S6B). The levels of inhibition diminished over time as MEFs underwent cell division and diluted the ASOs (Fig. 4H). Using this approach, we found that at 96 hours following transfection with ASO1 and ASO3, the levels of p21 were reproducibly reduced by 23±9% compared to cells treated with a control, non-targeting ASO (Fig. 4I and Fig. S6B). As a control for off-target effects, transfection of lincRNA-p21−/− MEFs with lincRNA-p21-specific ASOs did not further decrease p21 levels (Fig. 4I). We also observed that in the context of irradiation-induced DNA damage, transfection of wild-type MEFs with the two independent lincRNA-p21-specific ASOs led to a decrease in p21 levels (23% and 45% with ASO1 and ASO3, respectively) (Fig. 4J). Moreover, ASO-mediated lincRNA-p21 depletion in the context of oncogenic stress (Feldser et al., 2010) similarly affected the expression of p21 by 50% (Fig. S6C). Collectively, these experiments point to a specific role for the lincRNA-p21 transcript in the positive regulation of p21 expression.
LincRNA-p21 regulates p21 in cis
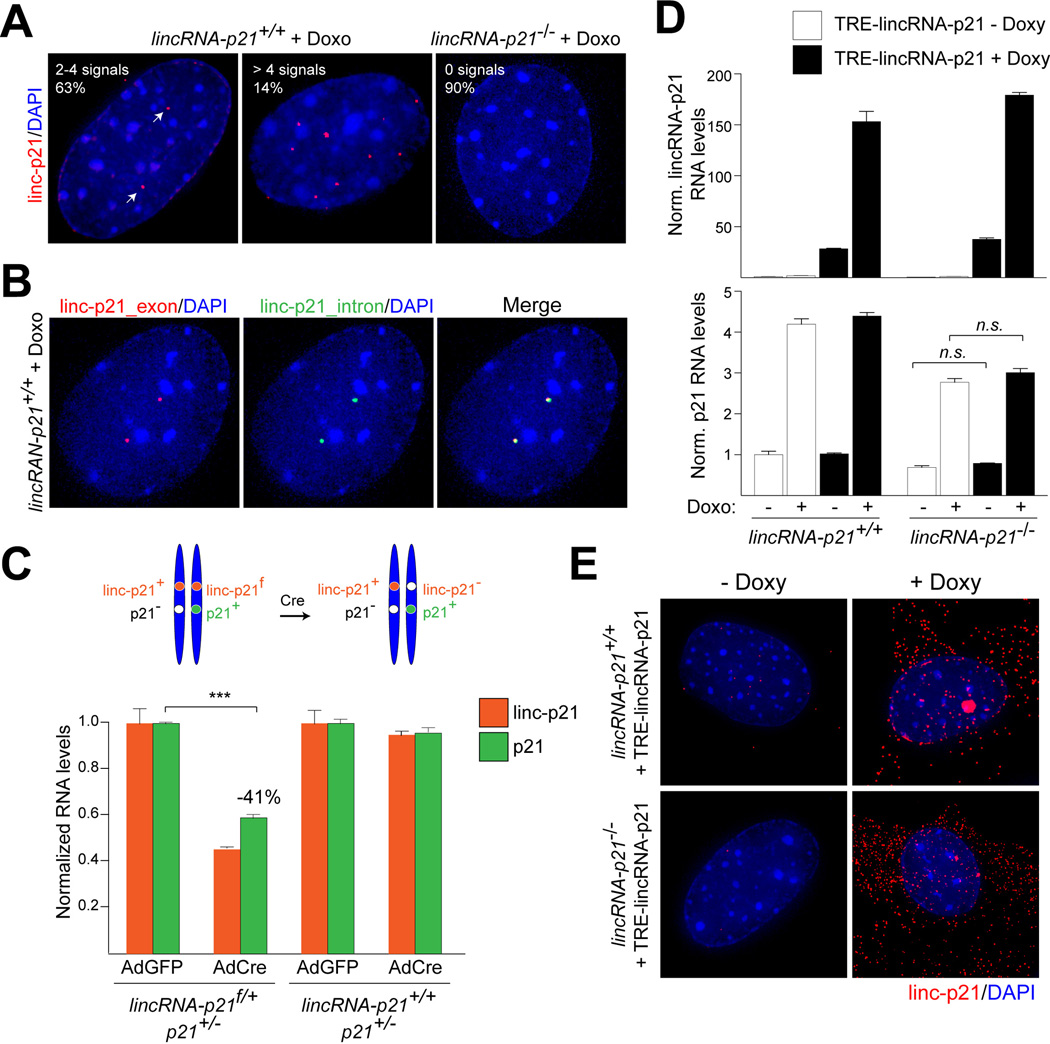
Recent data suggest that lncRNAs can regulate nearby genes via either cis- or transacting mechanisms (Fatica and Bozzoni, 2014). To visualize lincRNA-p21, we performed single molecule RNA FISH and observed that in wild-type MEFs treated with doxorubicin, lincRNA-p21 localized exclusively to the nucleus, with 63% of the cells containing 2 or 4 prominent lincRNA-p21 signals (Fig. 5A). As a negative control, over 90% of lincRNA-p21−/− cells contained no detectable signals (Fig. 5A). To test whether lincRNA-p21 was localized at or near its site of transcription, we designed RNA FISH probes complementary to the intron of lincRNA-p21. Since introns are removed co-transcriptionally and quickly degraded, we reasoned that the intron-specific RNA FISH signal should mark the genomic locus from which lincRNA-p21 is expressed. In accordance with a cis model, we observed perfect co-localization between lincRNA-p21 exonic and intronic signals in all wild-type cells, in which we could detect the short-lived intronic signal (Fig. 5B).
Figure 5. LincRNA-p21 regulates p21in cis.
(A) Single molecule RNA FISH detection of lincRNA-p21 with exon-specific probes in indicated MEFs (white arrows). DNA was counterstained with DAPI. % represents the fraction of cells with indicated number of signals per cell (n=150).
(B) Co-localization of exon- and intron-specific lincRNA-p21 signals using single molecule RNA FISH.
(C) (Top) Schematic. (Bottom) qRT-PCR analysis of lincRNA-p21 and p21 RNA levels in MEFs of indicated genotypes and treatments. Data are represented as mean±SEM, n=3, p<0.0001, paired t-test.
(D) qRT-PCR analysis of lincRNA-p21 (top) and p21 (bottom) RNA levels in MEFs of indicated genotypes, expressing TRE-lincRNA-p21, in the presence or absence of doxorubicin (Doxo) and doxycycline (Doxy). Data, replicated in 2 independent experiments, are represented as mean±SEM of technical replicates.
(E) Single molecule RNA-FISH detection of exogenously expressed lincRNA-p21 in (D).See also Table S4.
To further distinguish between cis and trans mechanisms of action, we crossed lincRNA-p21f/+ and p21−/− mice and isolated lincRNA-p21f/+; p21+/− MEFs, in which lincRNA-p21 and p21 mutations were in trans (Fig. 5C, top). In these cells, there was only one functional allele of p21, which was physically linked to a floxed allele of lincRNA-p21. We observed that upon adenoviral Cre-mediated deletion of the floxed allele of lincRNA-p21, the expression of p21 from the same chromosome diminished by 41±2% (Fig. 5C, bottom), which is comparable to the decrease of p21 levels in lincRNA-p21-deficient cells (Fig. 4B). Thus, the wild-type allele of lincRNA-p21 located in trans could not rescue p21 levels. In control experiments, we confirmed that the altered p21 expression required loss of lincRNA-p21 (Fig. 5C).
Consistent with a model in which lincRNA-p21 acts on p21 exclusively in cis, exogenous overexpression of lincRNA-p21 failed to rescue p21 levels in lincRNA-p21-deficient MEFs (Fig. 5D). This is likely because the majority of the overexpressed RNA did not properly localize to the endogenous locus and was exported to the cytoplasm (Fig. 5E).
LincRNA-p21 recruits hnRNP-K to promote p53-dependent transcription of p21
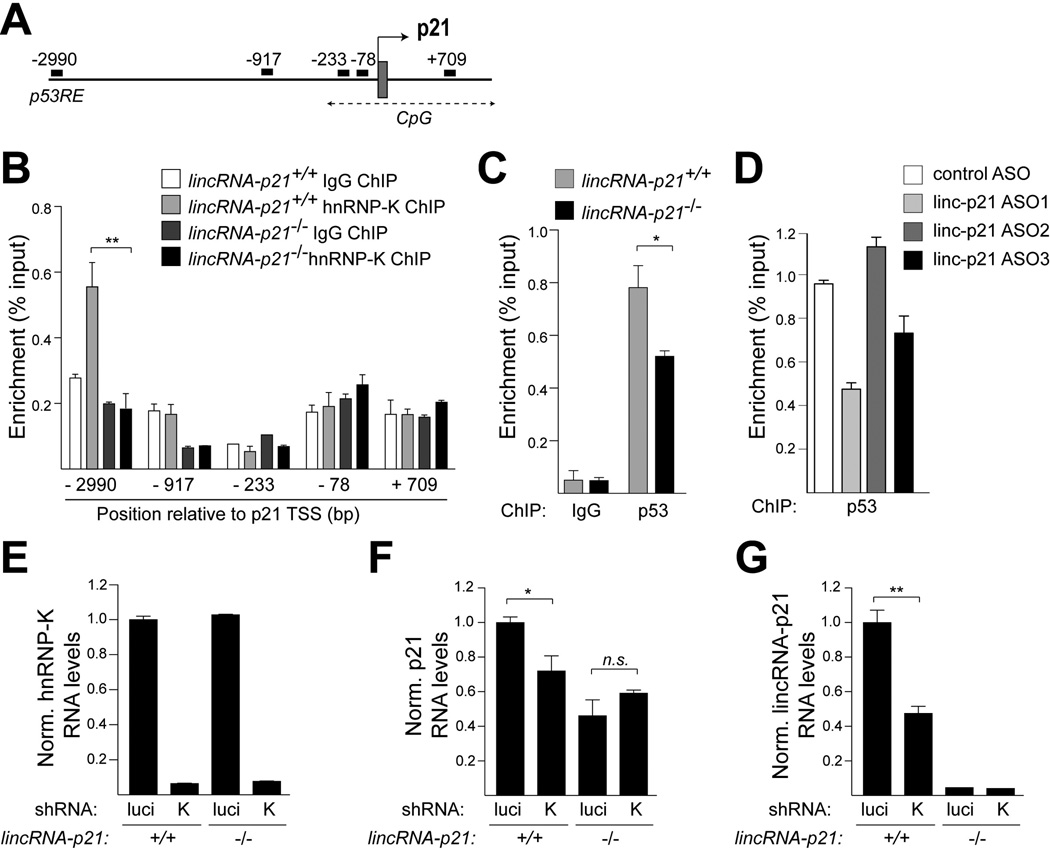
We next addressed the mechanism by which lincRNA-p21 promoted p21 expression. It has been established that p21 is a direct target of p53, but several transcriptional co-activators have been implicated in the regulation of p21 expression (Jung et al., 2010). In particular, studies in human cells have shown that one of these factors, hnRNP-K, binds to the p21 promoter in response to DNA damage and promotes p21 expression by recruiting p53 and/or by stabilizing p53 binding (Moumen et al., 2005). Since previous work had uncovered a direct interaction between lincRNA-p21 and hnRNP-K (Huarte et al., 2010), we speculated that lincRNA-p21 may be involved in mediating the co-activator function of hnRNP-K at the promoter of p21.
Consistent with previous data, we detected hnRNP-K and p53 at the p53RE in the p21 promoter by ChIP-qPCR in doxorubicin-treated, wild-type cells, despite the low efficiency of hnRNP-K immunoprecipitation (less than 1%) (Fig. 6A–C and Fig. S7A). We next asked whether lincRNA-p21 was required for the recruitment of hnRNP-K and p53 to the p21 promoter. ChIP-qPCR of hnRNP-K revealed that the binding of hnRNP-K at the p21 p53RE was reduced to background levels in lincRNA-p21-deficient MEFs (Fig. 6B). Moreover, as expected in the absence of proper localization of hnRNP-K at this site, we observed by ChIP-qPCR that the strength of p53 binding at the p21 promoter was diminished by 43±12% in lincRNA-p21-deficient cells compared to wild-type controls (Fig. 6C). Reduced binding of p53 at the p21 promoter was also observed in MEFs in which lincRNA-p21 RNA levels were downregulated by two independent lincRNA-p21-specific ASOs (ASO1 and ASO3) compared to a control, non-targeting ASO, whereas ASO2, which does not lead to efficient lincRNA-p21 downregulation (Fig. S6B), did not alter p53 binding (Fig. 6D). These data suggested that lincRNA-p21 RNA is required for the recruitment of hnRNP-K to the p53RE in the promoter of p21 and for the efficient binding of p53 at this site.
Figure 6. LincRNA-p21 acts with hnRNP-K as a transcriptional co-activator of p53-mediated expression of p21.
(A) Schematic of the positions of qPCR primers used for ChIP analysis, indicating the distance (base pairs) relative to the p21 transcription start site (TSS).
(B) A representative hnRNP-K ChIP-qPCR analysis in indicated MEF lines, harvested 8 hours following doxorubicin treatment. Data, replicated in 4 independent experiments, are represented as mean±SEM of technical replicates (n=4, p=0.0020, paired t-test).
(C) A representative p53 ChIP-qPCR analysis using the −2990 (p53RE) primer set in indicated MEF lines, harvested 8 hours following doxorubicin treatment. Data, replicated in 3 independent experiments, are represented as mean±SEM of technical replicates (n=3, p=0.043, paired t-test).
(D) ChIP-qPCR analysis of the relative p53 binding at the −2990 (p53RE) site of the p21 promoter in ASO-treated wild type MEFs. Data are represented as mean±SEM of technical replicates.
(E–G) qRT-PCR analysis of (E) hnRNP-K, (F) p21 (mean±SEM , n=4, p=0.0326, paired t-test), and (G) lincRNA-p21 (mean±SEM , n=3, p=0.0030, paired t-test) levels in indicated MEFs, expressing luciferase (luci) or hnRNP-K (K)-specific shRNAs, harvested at 8 hours following doxorubicin treatment. See also Figure S7A and Table S4.
In line with a model whereby lincRNA-p21 and hnRNP-K cooperate to promote p21 transcription, knockdown of hnRNP-K led to diminished p21 levels in doxorubicin-treated lincRNA-p21+/+ MEFs but not in lincRNA-p21−/− MEFs (Fig. 6E, F). Of note, hnRNP-K downregulation also resulted in reduced lincRNA-p21 levels, suggesting that hnRNP-K is required for the induction and/or stability of lincRNA-p21 (Fig. 6G)
LincRNA-p21 regulates the G1/S checkpoint, proliferation rates, and reprogramming efficiency
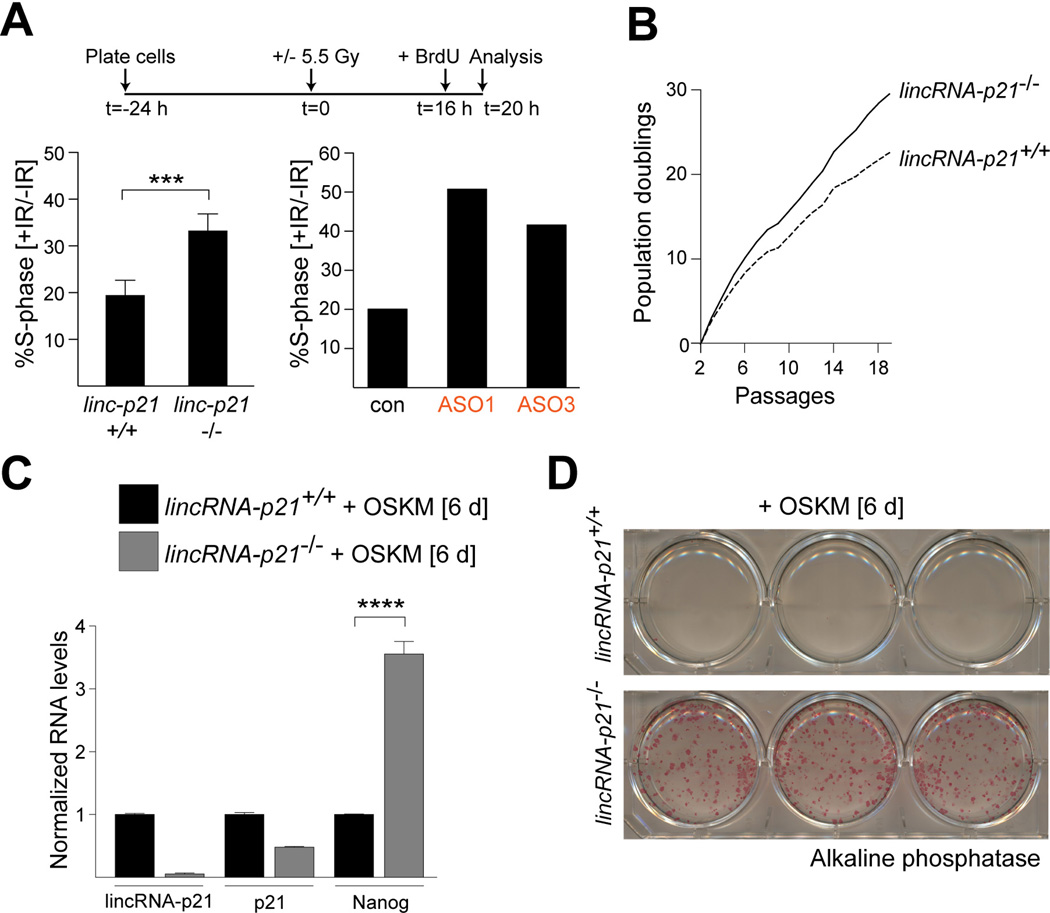
Finally, we investigated the physiological consequence of lincRNA-p21 deficiency in MEFs. Based on its role in regulating p21, which is a major mediator of checkpoint function in the p53 pathway, we anticipated that lincRNA-p21 deletion might affect the G1/S checkpoint. To test for this, we assayed the proportion of cells that entered S-phase following γ-irradiation in lincRNA-p21-proficient and -deficient cells. Compared to control cells, a significantly elevated fraction of lincRNA-p21−/− MEFs as well as MEFs transfected with lincRNA-p21-specific ASOs, continued to incorporate BrdU in the presence of DNA damage (Fig. 7A). This phenotype was comparable to the G1/S checkpoint defect observed in p21-deficient MEFs (Fig. S7B). We concluded that the G1/S checkpoint was indeed compromised in the absence of lincRNA-p21.
Figure 7. LincRNA-p21 controls the G1/S checkpoint, proliferation, and the induction of pluripotency.
(A) (Top) Schematic of the G1/S checkpoint assay. (Bottom, left) FACS analysis of the fraction of BrdU-positive cells in irradiated samples relative to untreated cells. Data are represented as mean±SEM, n=4, p=0.0060, paired t-test. (Bottom, right) FACS analysis of the fraction of BrdU-positive cells in wild type samples, transfected with the indicated ASOs.
(B) Proliferation of lincRNA-p21+/+ and −/− MEFs isolated from littermates.
(C) qRT-PCR analysis of lincRNA-p21, p21, and Nanog levels in RNA harvested from lincRNA-p21-proficient and –deficient cells at 6 days following infection with the reprogramming factors Oct4, Sox2, Klf4, and Myc (OSKM). Data are represented as mean±SEM, n=3, p=0.0004, paired t-test.
(D) Images of alkaline phosphatase-positive colonies stained 6 days following infection of MEFs with indicated genotypes with OSKM.
See also Figure S7B–F and Table S4.
Consistent with a defective checkpoint function, we reproducibly observed that lincRNA-p21-deficient MEFs proliferated faster compared to wild-type controls, which is similar to p21−/− cells (Fig. 7B and Fig. S7C). Moreover, we observed that lincRNA-p21-deficiency significantly increased the reprogramming efficiency of primary MEFs, as indicated by enhanced levels of the pluripotency marker Nanog and an increased number of alkaline phosphatase-positive colonies in lincRNA-p21−/− compared to littermate, wild-type MEFs (Fig. 7C, D).
In contrast, we did not find a role for lincRNA-p21 in other well-characterized functions of the p53 pathway in MEFs, including apoptosis, the ability to promote senescence following prolonged exposure to doxorubicin-induced DNA damage, and the capacity to inhibit cellular transformation (Fig. S7D–F). These data are consistent with the absence of these phenotypes in p21-deficient cells (Pantoja and Serrano, 1999). In sum, these data indicate that loss of lincRNA-p21 leads to cellular phenotypes similar to the consequences of p21 deletion in MEFs.
DISCUSSION
An emerging class of lncRNAs, including lincRNA-p21, has been proposed to regulate the expression of multiple genes in trans (Fatica and Bozzoni, 2014). For the most part, these studies rely on global gene expression profiling, which identifies hundreds, sometimes thousands, of differentially expressed genes in cells with RNAi-mediated knockdown or genetic depletion of lncRNAs. From a mechanistic perspective, however, it is important to differentiate direct targets of lncRNA regulation from genes that are indirectly affected by lncRNA loss. For instance, we found by global expression analysis that genetic deletion of lincRNA-p21 in primary MEFs led to the deregulation of close to one thousand genes. However, we present several lines of evidence that do not support widespread trans-regulatory activity of this lincRNA. First, single-molecule RNA FISH revealed that lincRNA-p21 did not interact with multiple distinct loci but was localized primarily at or near its transcription site. Next, we did not find evidence of nuclear domains near the lincRNA-p21 locus that cluster lincRNA-p21-regulated genes (Engreitz et al., 2013). Finally, the low copy number (∼8 lincRNA-p21 molecules/cell) does not support a global function for lincRNA-p21 as a protein-binding partner. For these reasons, we believe it unlikely that lincRNA-p21 has genome-wide regulatory functions.
The discrepancy with previous functional studies, which have proposed global nuclear and cytosolic trans functions for lincRNA-p21 (Huarte et al., 2010; Yang et al., 2014; Yoon et al., 2012), can likely be attributed to the different methods used to perform loss-of-function analyses (genetic deletion vs RNAi) and to the different cell types (MEFs vs human cancer cell lines). Importantly, a genetic deletion, rather than an RNAi approach, was required to unveil the cis function of lincRNA-p21 in the regulation of p21. Additional advantages of using a conditional knockout mouse model include specificity, efficiency, and opportunity to study the long-term consequences of lincRNA-p21 depletion in vitro and in vivo.
Using our genetic approach, we present compelling evidence that lincRNA-p21 activates the expression of its neighboring gene p21 by 30–50%. As several of the major phenotypes of lincRNA-p21-deficiency could be replicated by two independent lincRNA-p21-targeting ASOs, we conclude that this enhancer-like phenotype is mediated by the lincRNA-p21 RNA transcript and not through a genetic element located in the region of deletion. These data also argue against a role for another RNA species expressed from the lincRNA-p21 locus in these phenotypes and overcome concerns about non-specific consequences of altered chromatin architecture in mutant cells. Of note, the lincRNA-p21 locus does not bear the high H3K4me1/H3K4me3 ratio indicative of enhancer elements and lincRNA-p21 RNA has a half-life in the range of hours, rather than minutes, both of which suggest that lincRNA-p21 likely does not belong to the recently described class of eRNAs (Orom and Shiekhattar, 2011). However, we cannot exclude the possibility that lincRNA-p21 may arise from an enhancer region (Melo et al., 2013b).
Importantly, we formally show that lincRNA-p21 regulates p21 in cis. This conclusion is substantiated by a genetic experiment in which we place lincRNA-p21 and p21 mutations in trans as well as by the predominant two-dot nuclear pattern of lincRNA-p21 by RNA FISH and by the inability to rescue phenotypes of lincRNA-p21 deficiency by exogenous expression of lincRNA-p21. Consistent with our finding of cis regulation, the majority of the cellular phenotypes and expression changes observed in lincRNA-p21-deficient cells could be attributed to the role of lincRNA-p21 in activating p21, including defects in the G1/S cell cycle checkpoint and in proliferation control. Conversely, p21-independent functions of the p53 pathway in MEFs, including the abilities to promote apoptosis and senescence and the capacity to inhibit oncogene-mediated cellular transformation, were not affected by loss of lincRNA-p21. The lack of an overt organismal phenotype in lincRNA-p21-deficient mice is also in agreement with a cis regulatory model, given that p21-deficient animals develop normally and exhibit only mild susceptibility to tumor development with a long average latency (Martin-Caballero et al., 2001).
In further support of a cis regulatory model, we provide direct evidence that the regulation of a subset of PRC2 target genes by lincRNA-p21 is mediated through p21. This observation reveals a novel function of the p53 pathway. It remains to be determined what the physiological purpose of de-repression of PRC2 targets in response to prolonged genotoxic stress may be. One potential function could be the elimination of damaged cells by promoting terminal differentiation through the expression of multiple lineage-specific factors. This model is consistent with a growing body of literature that has linked p21 function in growth arrest with cellular differentiation and organism development (Munoz-Espin et al., 2013). Moreover, this phenomenon might be related to the increased reprogramming rates observed in p53-, p21-, and lincRNA-p21-deficient primary MEFs (Hanna et al., 2009; Kawamura et al., 2009). Whereas this phenotype was originally attributed to increased cellular proliferation rates, cell intrinsic factors were also acknowledged as potential modulators of the transition to a pluripotency state (Hanna et al., 2009).
The data presented here indicate that lincRNA-p21 acts in concert with hnRNP-K to promote p53-mediated expression of p21. Our findings support a model in which lincRNA-p21 and hnRNP-K act as transcriptional co-activators for local p53-mediated transcription. We propose that hnRNP-K binds directly to lincRNA-p21, which serves to promote the stability of lincRNA-p21 as well as to target hnRNP-K to the p21 locus. In the presence of lincRNA-p21 and hnRNP-K, p53 can carry out efficient induction of p21. These findings are consistent with previous studies in human cells that have implicated hnRNP-K as a transcriptional co-activator for p53-mediated expression of p21 (Moumen et al., 2005). An intriguing question is how lincRNA-p21 deficiency, which affects p21 expression levels by only 30–50%, leads to the range and extent of phenotypes that are comparable to complete loss of p21. We propose that lincRNA-p21-mediated reinforcement of activation may push p21 past a threshold for response and thus facilitate finer regulation of cellular responses.
It is important to understand the role of reinforcing p21 expression in cis. We speculate that this may be key in the context of the p53 tumor suppressor pathway, which is complex and whose outcome can be cell type- and context-specific (Vousden and Prives, 2009). p21 and hundreds of other genes are activated by p53 in response to stress and different targets have been shown to dictate different cellular outcomes (Levine and Oren, 2009). Thus, a function of lincRNA-p21 and hnRNP-K may be to locally amplify the execution of a specific p53-dependent transcription output, reliant on sustained p21 activation. The contribution of lincRNA-p21 is to recruit hnRNP-K in a locus-restricted manner and maintain p21 activation.
In summary, these data point to lincRNA-p21 as a key modulator of gene expression in the p53 pathway, influencing the activation and the chromatin state of hundreds of genes through its cis control of p21 expression. This work highlights the power of using defined genetic systems to dissect the contribution of lncRNAs to complex biological pathways. The model presented here has general implications for how low copy number, cis-acting lncRNAs may locally reinforce the specificity of broad transcriptional programs.
EXPERIMENTAL PROCEDURES
Mouse strains, cell culture, and treatments
LincRNA-p21 floxed and null mice and E13.5 MEFs were generated as described in Extended Experimental Procedures. Rosa26-CreERT2 (Ventura et al., 2007) and p21 mice (The Jackson Laboratory, stock number 003263) have previously been described .
To delete lincRNA-p21 in lincRNA-p21f/f; Rosa26-CreERT2 MEFs, cells were treated with 0.5 µM 4-OHT. To delete lincRNA-p21 in lincRNA-p21f/+; p21+/− MEFs, cells were infected 2 times at a 24-hour interval with Cre recombinase or control GFP adenovirus. To induce DNA damage, cells were treated with 0.5 µM doxorubicin or irradiated with 2 or 10 Gy of γ-irradiation and harvested at indicated time-points following treatment.
5–10-5 MOE gapmer oligonucleotides were designed and generously provided by Isis Pharmaceuticals (Table S4). 1 µM lincRNA-p21-specific or non-targeting control ASOs were transfected twice at a 48-hour interval into 1×106 lincRNA-p21+/+ and −/− MEFs using the Amaxa Mouse/Rat Hepatocyte Nucleofector Kit (Lonza) and the Nucleofector 2b Device (Lonza).
Constructs
Full-length mouse lincRNA-p21 cDNA (a gift from John Rinn) was expressed from the pSLIK lentiviral expression system (Shin et al., 2006). pLKO p21 and hnRNP-K hairpins were purchased from The RNA Consortium (TRC) collection (Thermo Scientific) (p21: clone IDs TRCN0000042583 (sh#1) and TRCN0000042584 (sh#2, used to replicate data); hnRNP-K: TRCN0000096825 (sh#4)). Lentivirus was produced as described in Extended Experimental Procedures.
RNA isolation and RT-qPCR
RNA was isolated with RNeasy Mini Kit (Qiagen) and reverse transcribed using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). SYBR Green PCR master mix (Applied Biosystems) was used for qPCR with primers listed in Table S4. Expression levels were calculated relative to GAPDH and normalized to control samples.
Western blotting
Western blotting was performed with whole cell lysates, using the following antibodies: p21 (clone F-5, sc-6246, Santa Cruz Biotechnology), hnRNP-K (ab70492, Abcam), H3K4me3 (ab8580, Abcam), H3K27me3 (ab6002, Abcam), and loading control β-tubulin (ab6046, Abcam).
Chromatin immunoprecipitation (ChIP)
ChIP was performed as described in Extended Experimental Procedures with the following antibodies: H3K4me3 (ab8580, Abcam), H3K27me3 (ab6002, Abcam), p53 (clone CM5, VP-P956, Vector Laboratories), hnRNP-K (ab70492, Abcam), and control IgG (ChIP grade, ab46540, Abcam). DNA was submitted for high-throughput sequencing (ChIP-seq) or used for qPCR analysis (ChIP-qPCR) using primers listed in Table S4. The data represent the percentage of input that was immunoprecipitated.
Single molecule RNA- Fluorescence in situ hybridization (RNA-FISH)
Single molecule RNA FISH was performed as previously described (Raj et al., 2008) with probes listed in Table S4.
G1/S checkpoint assay
G1/S checkpoint assay was performed as previously described (Brugarolas et al., 1995) with modifications (see Extended Experimental Procedures). The data represent the fraction of BrdU-positive cells in irradiated samples relative to non-irradiated samples for each cell line.
Supplementary Material
HIGHLIGHTS.
Murine knockout model reveals a role for lincRNA-p21 in activating p21 in cis
LincRNA-p21 acts with hnRNP-K as a transcriptional co-activator of p53
LincRNA-p21 promotes the expression of a set of Polycomb target genes via p21
LincRNA-p21 regulates the G1/S checkpoint, proliferation, and reprogramming
ACKNOWLEDGEMENTS
We thank John Rinn, Vasilena Gocheva and Thales Papagiannakopoulos for critical review of the manuscript. We are extremely grateful to Dirk Hockemeyer (UC Berkeley) for technical assistance and reagents related to the reprogramming experiment and to Jerry Ruth (Biosearch Technologies) for generously providing RNA FISH reagents. We are grateful to Aurora Burds O’Connor from the Rippel ES cell & Transgenics Facility of the Swanson Biotechnology Center. This work was supported by NIH (T.J.), the Howard Hughes Medical Institute and the Ludwig Center for Molecular Oncology at MIT. N.D. was supported by a Damon Runyon Fellowship Award. T.J. is the David H. Koch Professor of Biology and a Daniel K. Ludwig Scholar at MIT.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
All primary RNA-seq and ChIP-seq data are available at Gene Expression Omnibus (GSE52958).
DEDICATION
The authors wish to dedicate this paper to the memory of Officer Sean Collier, for his caring service to the MIT community and for his sacrifice.
REFERENCES
- Aldiri I, Vetter ML. PRC2 during vertebrate organogenesis: a complex in transition. Dev Biol. 2012;367(2):91–99. doi: 10.1016/j.ydbio.2012.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugarolas J, Chandrasekaran C, Gordon JI, Beach D, Jacks T, Hannon GJ. Radiation-induced cell cycle arrest compromised by p21 deficiency. Nature. 1995;377(6549):552–557. doi: 10.1038/377552a0. [DOI] [PubMed] [Google Scholar]
- Davidovich C, Zheng L, Goodrich KJ, Cech TR. Promiscuous RNA binding by Polycomb repressive complex 2. Nat Struct Mol Biol. 2013;20(11):1250–1257. doi: 10.1038/nsmb.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng C, Zhang P, Harper JW, Elledge SJ, Leder P. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell. 1995;82(4):675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- Engreitz JM, Pandya-Jones A, McDonel P, Shishkin A, Sirokman K, Surka C, Kadri S, Xing J, Goren A, Lander ES, Plath K, Guttman M. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science. 2013;341(6147):1237973. doi: 10.1126/science.1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15(1):7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- Feldser DM, Kostova KK, Winslow MM, Taylor SE, Cashman C, Whittaker CA, Sanchez-Rivera FJ, Resnick R, Bronson R, Hemann MT, Jacks T. Stage-specific sensitivity to p53 restoration during lung cancer progression. Nature. 2010;468(7323):572–575. doi: 10.1038/nature09535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN, Jaenisch R, Mikkelsen TS, Jacks T, Hacohen N, Bernstein BE, Kellis M, Regev A, Rinn JL, Lander ES. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458(7235):223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J, Saha K, Pando B, van Zon J, Lengner CJ, Creyghton MP, van Oudenaarden A, Jaenisch R. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature. 2009;462(7273):595–601. doi: 10.1038/nature08592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, Attardi LD, Regev A, Lander ES, Jacks T, Rinn JL. A Large Intergenic Noncoding RNA Induced by p53 Mediates Global Gene Repression in the p53 Response. Cell. 2010;142(3):409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, Horlings HM, Shah N, Umbricht C, Wang P, Wang Y, Kong B, Langerod A, Borresen-Dale AL, Kim SK, van de Vijver M, Sukumar S, Whitfield ML, Kellis M, Xiong Y, Wong DJ, Chang HY. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet. 2011;43(7):621–629. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung YS, Qian Y, Chen X. Examination of the expanding pathways for the regulation of p21 expression and activity. Cell Signal. 2010;22(7):1003–1012. doi: 10.1016/j.cellsig.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S, Son J, Shen SS, Reinberg D, Bonasio R. PRC2 binds active promoters and contacts nascent RNAs in embryonic stem cells. Nat Struct Mol Biol. 2013;20(11):1258–1264. doi: 10.1038/nsmb.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura T, Suzuki J, Wang YV, Menendez S, Morera LB, Raya A, Wahl GM, Izpisua Belmonte JC. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009;460(7259):1140–1144. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, Regev A, Lander ES, Rinn JL. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106(28):11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JT, Bartolomei MS. X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell. 2013;152(6):1308–1323. doi: 10.1016/j.cell.2013.02.016. [DOI] [PubMed] [Google Scholar]
- Levine AJ, Oren M. The first 30 years of p53: growing ever more complex. Nat Rev Cancer. 2009;9(10):749–758. doi: 10.1038/nrc2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin-Bejar O, Marchese FP, Athie A, Sanchez Y, Gonzalez J, Segura V, Huang L, Moreno I, Navarro A, Monzo M, Garcia-Foncillas J, Rinn JL, Guo S, Huarte M. Pint lincRNA connects the p53 pathway with epigenetic silencing by the Polycomb repressive complex 2. Genome Biol. 2013;14(9):R104. doi: 10.1186/gb-2013-14-9-r104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Caballero J, Flores JM, Garcia-Palencia P, Serrano M. Tumor susceptibility of p21(Waf1/Cip1)-deficient mice. Cancer Res. 2001;61(16):6234–6238. [PubMed] [Google Scholar]
- Melo CA, Drost J, Wijchers PJ, van de Werken H, de Wit E, Oude Vrielink JA, Elkon R, Melo SA, Leveille N, Kalluri R, de Laat W, Agami R. eRNAs are required for p53-dependent enhancer activity and gene transcription. Mol Cell. 2013a;49(3):524–535. doi: 10.1016/j.molcel.2012.11.021. [DOI] [PubMed] [Google Scholar]
- Melo CA, Leveille N, Agami R. eRNAs reach the heart of transcription. Cell Res. 2013b;23(10):1151–1152. doi: 10.1038/cr.2013.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missero C, Di Cunto F, Kiyokawa H, Koff A, Dotto GP. The absence of p21Cip1/WAF1 alters keratinocyte growth and differentiation and promotes ras-tumor progression. Genes Dev. 1996;10(23):3065–3075. doi: 10.1101/gad.10.23.3065. [DOI] [PubMed] [Google Scholar]
- Moumen A, Masterson P, O’Connor MJ, Jackson SP. hnRNP K: an HDM2 target and transcriptional coactivator of p53 in response to DNA damage. Cell. 2005;123(6):1065–1078. doi: 10.1016/j.cell.2005.09.032. [DOI] [PubMed] [Google Scholar]
- Munoz-Espin D, Canamero M, Maraver A, Gomez-Lopez G, Contreras J, Murillo-Cuesta S, Rodriguez-Baeza A, Varela-Nieto I, Ruberte J, Collado M, Serrano M. Programmed Cell Senescence during Mammalian Embryonic Development. Cell. 2013;155(5):1104–1118. doi: 10.1016/j.cell.2013.10.019. [DOI] [PubMed] [Google Scholar]
- Orom UA, Shiekhattar R. Long non-coding RNAs and enhancers. Curr Opin Genet Dev. 2011;21(2):194–198. doi: 10.1016/j.gde.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantoja C, Serrano M. Murine fibroblasts lacking p21 undergo senescence and are resistant to transformation by oncogenic Ras. Oncogene. 1999;18(35):4974–4982. doi: 10.1038/sj.onc.1202880. [DOI] [PubMed] [Google Scholar]
- Purvis JE, Karhohs KW, Mock C, Batchelor E, Loewer A, Lahav G. p53 dynamics control cell fate. Science. 2012;336(6087):1440–1444. doi: 10.1126/science.1218351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods. 2008;5(10):877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin KJ, Wall EA, Zavzavadjian JR, Santat LA, Liu J, Hwang JI, Rebres R, Roach T, Seaman W, Simon MI, Fraser ID. A single lentiviral vector platform for microRNA-based conditional RNA interference and coordinated transgene expression. Proc Natl Acad Sci U S A. 2006;103(37):13759–13764. doi: 10.1073/pnas.0606179103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinman RA, Hoffman B, Iro A, Guillouf C, Liebermann DA, el-Houseini ME. Induction of p21 (WAF-1/CIP1) during differentiation. Oncogene. 1994;9(11):3389–3396. [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, Newman J, Reczek EE, Weissleder R, Jacks T. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445(7128):661–665. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- Vickers TA, Koo S, Bennett CF, Crooke ST, Dean NM, Baker BF. Efficient reduction of target RNAs by small interfering RNA and RNase H-dependent antisense agents. A comparative analysis. J Biol Chem. 2003;278(9):7108–7118. doi: 10.1074/jbc.M210326200. [DOI] [PubMed] [Google Scholar]
- Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137(3):413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- Yang F, Zhang H, Mei Y, Wu M. Reciprocal regulation of HIF-1alpha and lincRNA-p21 modulates the Warburg effect. Mol Cell. 2014;53(1):88–100. doi: 10.1016/j.molcel.2013.11.004. [DOI] [PubMed] [Google Scholar]
- Yoon JH, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S, Huarte M, Zhan M, Becker KG, Gorospe M. LincRNA-p21 suppresses target mRNA translation. Mol Cell. 2012;47(4):648–655. doi: 10.1016/j.molcel.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Wong C, Liu D, Finegold M, Harper JW, Elledge SJ. p21(CIP1) and p57(KIP2) control muscle differentiation at the myogenin step. Genes Dev. 1999;13(2):213–224. doi: 10.1101/gad.13.2.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, Song JJ, Kingston RE, Borowsky M, Lee JT. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell. 2010;40(6):939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.