Significance
Obtaining cultures of microbes is essential for developing knowledge of bacterial genetics and physiology, but many microbes with potential biomedical significance identified from metagenomic studies have not yet been cultured due to the difficulty of identifying growth conditions, isolation, and characterization. We developed a microfluidics-based, genetically targeted approach to address these challenges. This approach corrects sampling bias from differential bacterial growth kinetics, enables the use of growth stimulants available only in small quantities, and allows targeted isolation and cultivation of a previously uncultured microbe from the human cecum that belongs to the high-priority group of the Human Microbiome Project’s “Most Wanted” list. This workflow could be leveraged to isolate novel microbes and focus cultivation efforts on biomedically important targets.
Keywords: microscale, anaerobe, aerobe, cultivate, metagenome
Abstract
This paper describes a microfluidics-based workflow for genetically targeted isolation and cultivation of microorganisms from complex clinical samples. Data sets from high-throughput sequencing suggest the existence of previously unidentified bacterial taxa and functional genes with high biomedical importance. Obtaining isolates of these targets, preferably in pure cultures, is crucial for advancing understanding of microbial genetics and physiology and enabling physical access to microbes for further applications. However, the majority of microbes have not been cultured, due in part to the difficulties of both identifying proper growth conditions and characterizing and isolating each species. We describe a method that enables genetically targeted cultivation of microorganisms through a combination of microfluidics and on- and off-chip assays. This method involves (i) identification of cultivation conditions for microbes using growth substrates available only in small quantities as well as the correction of sampling bias using a “chip wash” technique; and (ii) performing on-chip genetic assays while also preserving live bacterial cells for subsequent scale-up cultivation of desired microbes, by applying recently developed technology to create arrays of individually addressable replica microbial cultures. We validated this targeted approach by cultivating a bacterium, here referred to as isolate microfluidicus 1, from a human cecal biopsy. Isolate microfluidicus 1 is, to our knowledge, the first successful example of targeted cultivation of a microorganism from the high-priority group of the Human Microbiome Project’s “Most Wanted” list, and, to our knowledge, the first cultured representative of a previously unidentified genus of the Ruminococcaceae family.
This paper describes an integrated microfluidic workflow for genetically targeted cultivation and isolation of microorganisms. Microbes play critical functional roles in diverse environments ranging from soil and oceans to the human gut. The emergence of culture-independent techniques has provided insights into microbial ecology by revealing genetic signatures of uncultured microbial taxa (1–5). It also suggests that certain microbes may impact host phenotypes such as obesity, inflammation, and gastrointestinal integrity (6, 7). This explosion of sequencing data has presented new challenges and opportunities for microbial cultivation, which is critical for allowing direct access to microorganisms to test hypotheses experimentally, and is crucial for proper taxonomic classification, functional annotation of metagenomic sequences, and use of such microbes for environmental remediation, energy applications, and formulation of probiotics. However, a direct approach that cultivates, in a targeted fashion, microbes carrying genes of interest identified in metagenomic data sets remains mostly unexplored. As a result, for example, a list of the “Most Wanted” taxa that are urgently in need of cultivation has been issued by the Human Microbiome Project (HMP) from the National Institutes of Health. These microorganisms are highly prevalent and abundant in the human microbiome but poorly represented in cultured collections (2).
Most microbes do not grow using traditional cultivation methods and hence are referred to as “unculturable” (8–10). Although these microbes could be grown in their natural habitats (9), where effects such as cross-feeding (11) and microbe–host interactions (12, 13) are present, some biological samples, such as clinical biopsies, are often limited in quantity. This makes it challenging to set up cultivation experiments in large scale with these native media, but creates opportunities for miniaturized methods. Further, miniaturized methods that use compartmentalization can eliminate competition among species. Cultivation methods that use miniaturization and compartmentalization, including gel microdroplets (14), miniaturized Petri dishes (15), and microfluidics (16–19), have become increasingly promising as a basis for targeted microbial cultivation and isolation platforms, as they can limit the consumption of precious samples and also control the microenvironment around cells (20). We envisioned implementing targeted cultivation with microfluidics by focusing on two goals. The first goal is to efficiently identify cultivation conditions that support growth of target microbes. This can be accomplished by performing a genetic assay with target-specific primers or probes on the pooled microbial culture from a certain cultivation condition before isolation (21); however, designing specific probes based on short reads from high-throughput sequencing can be difficult. Moreover, it can be challenging to detect and cultivate slowly growing strains, as they often fall below the limit of detection, being outcompeted by rapidly growing strains in a complex community. A second goal of targeted cultivation is to focus isolation efforts on microbial targets of interest, thereby minimizing the effort associated with isolating off-target colonies. However, both PCR and fluorescence in situ hybridization (FISH) require access to genetic material, which is often not compatible with the goal of isolating and cultivating live cells. This paper addresses these challenges. In an accompanying paper (22), we describe the design, fabrication, and underlying physics of a microfluidic device to create arrays of individually addressable replica microbial cultures. Here, we integrate this device and additional devices and methods into a workflow for genetically targeted microbial cultivation, and validate this workflow by isolating a bacterium from the Most Wanted taxa.
Results and Discussion
Overview of Workflow for Genetically Targeted Microfluidics-Based Cultivation.
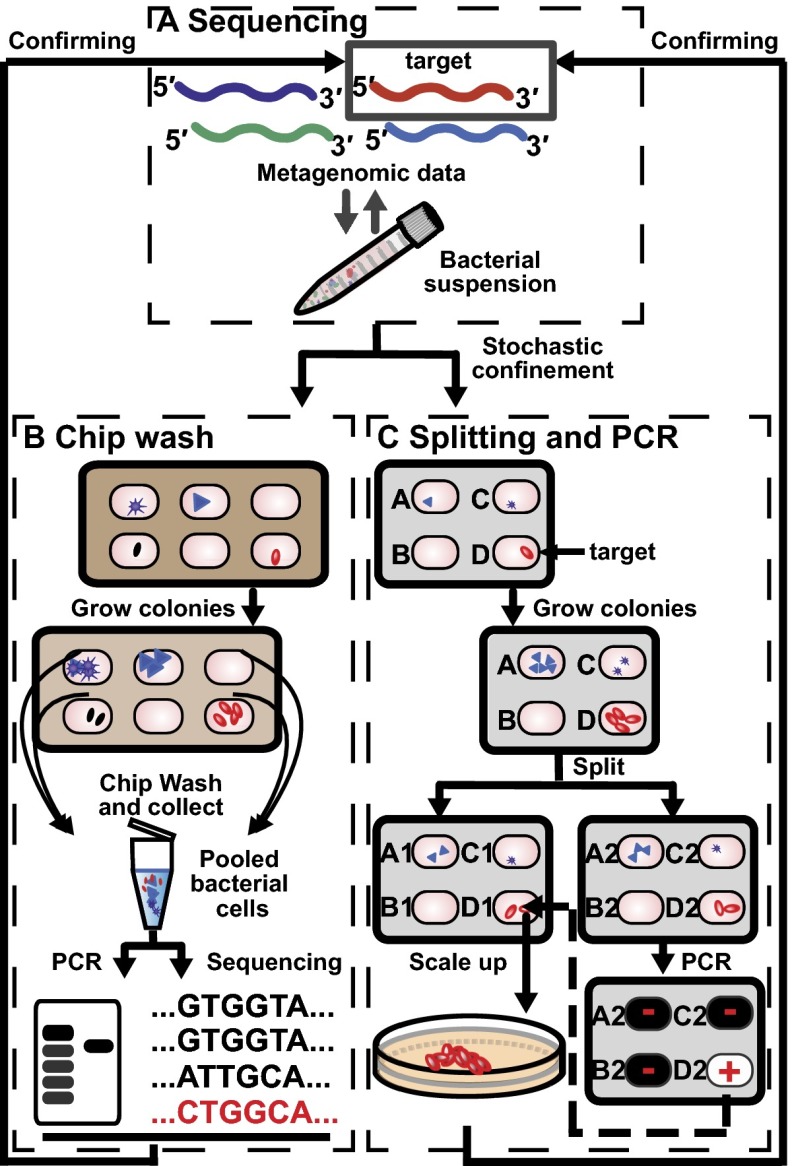
We envisioned isolating and cultivating microbial targets identified from metagenomic or 16S ribosomal RNA (16S rRNA) gene high-throughput sequencing studies by combining microfluidics with genetic assays (Fig. 1A). To address the goal of streamlining cultivation efforts using genetic assays, we created a general workflow with two major components: identification of cultivation conditions for the target organism (Fig. 1B) and isolation of the target (Fig. 1C). In both components, single bacterial cells from clinical samples are stochastically confined in nanoliter wells on a microfluidic device to promote the growth of microcolonies. This confinement can be useful for suppression of overgrowth from rapidly growing strains, in favor of slowly growing strains. In the first step, a “chip wash” method is used to monitor bacterial growth on a microfluidic device (Fig. 1B) under various conditions; miniaturization allows cultivation experiments that involve limited quantities of natural growth stimulants. In this method, microcolonies grown under each cultivation condition are collected into a single tube by washing the microwells after cultivation, analogously to the plate wash PCR method (21). DNA from the pooled cells is analyzed by sequencing, target-specific primers, or both, to determine whether the cultivation conditions for that chip allowed the growth of the target microorganism. This chip wash method can be repeated sequentially or in parallel until the growth conditions are identified. Then, the target organism is isolated and cultivated (Fig. 1C): The sample is cultivated on a separate microfluidic device, described in an accompanying paper (22), under the optimal condition identified during chip wash. After cultivation, this device splits each microcolony into two identical copies. We anticipate that multiple rounds of culture and splitting on the same device could be performed in a similar fashion. PCR is performed on the first copy to identify the compartment containing the target of interest, and then live cells can be retrieved from the corresponding well on the other half of the chip for scale-up cultivation.
Fig. 1.
Illustration representing the workflow for targeted cultivation and isolation of microbial organisms. (A) Microbial targets carrying genes of interest are identified by high-throughput sequencing of clinical samples. A representative sequence of the target is shown in red. To cultivate the target, the inoculum is suspended in cultivation medium and loaded onto a microfluidic device, enabling stochastic confinement of single cells and cultivation of individual species (represented by different shapes). (B) A chip wash method is used to monitor bacterial growth under different cultivation conditions. Cells are pooled en masse into a tube and DNA is extracted for genetic analysis such as sequencing and PCR. (C) The target can be isolated by growing the sample under the growth condition identified from the chip wash. The two halves of the device are separated, resulting in two copies of each colony. On one half of the chip, target colonies are identified using PCR. Then, the target colony on the other half of the chip is retrieved for a scale-up culture, after which sequencing is used to validate that the correct target has been isolated.
To implement this workflow, we relied on the SlipChip platform for three reasons (23). First, it can create thousands of miniaturized reactions without the need for bulky equipment. It can be used in the limited space of an anaerobic chamber, which is widely used to cultivate anaerobes that dominate the human gut microbiota. Second, SlipChip is compatible with PCR (24) and enzymatic assays (25). Third, compartmentalization on SlipChip is reversible and the microcolonies can be spatially indexed as described in an accompanying paper (22), which facilitates the retrieval of reagents and organisms from the device (24, 26).
Chip Wash Device.
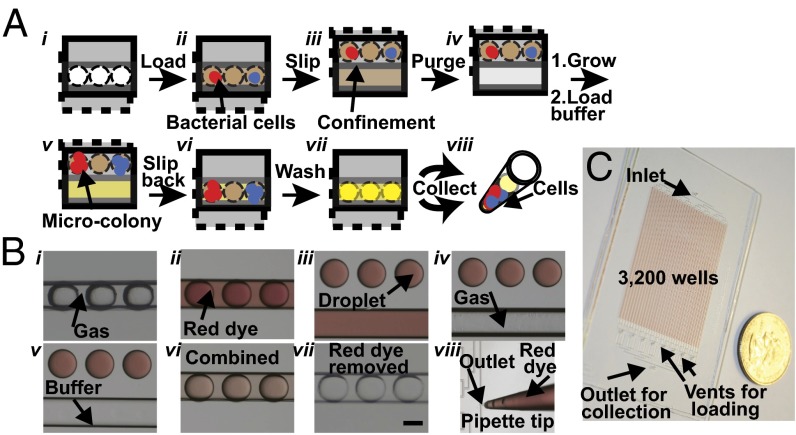
Fig. 2A shows the general workflow of a chip wash experiment. We designed a microfluidic device to perform up to 3,200 microbial cultivation experiments, each on a scale of ∼6 nL (Fig. 2C and SI Appendix). This device enables three capabilities: stochastic confinement of single cells from samples, microbial cultivation, and collection of cultivated cells. To confine single cells, a sample of bacteria suspended in cultivation medium is loaded into the channels and wells (Fig. 2A, ii). Slipping the bottom plate (dashed layer in Fig. 2A) upward enables stochastic confinement of bacterial cells in wells (Fig. 2 A, iii). To introduce gas into the channel and remove residual sample in the channel, the solution is purged from the channel by vacuum (Fig. 2A, iv). To cultivate microbes, the device is incubated and some of the single cells grow to microcolonies (Fig. 2A, v). After cultivation, the microchannel is loaded with buffer solution (Fig. 2A, v) to avoid the formation of gas bubbles. The presence of gas bubbles in a channel could increase flow resistance (27) and therefore slow down or stop the flow in that channel, resulting in inefficient washing in later steps. To allow collection of the microbial cells, the bottom plate is slipped back to overlay the wells with the channel (Fig. 2A, vi). A buffer solution is injected to flush the channel (Fig. 2A, vii) and is collected, from the outlet specifically designed for collection (Fig. 2 A, viii and C), in a pipette tip. The flow of fluid on SlipChip is controlled by positive pressure using a pipettor. This process of injection–collection is repeated three times. Immiscible oil is then injected to further displace the remaining aqueous phase. We used a red dye experiment to visualize the device operation described above (Fig. 2B), which allowed us to observe that the droplets remained intact during purging when gas was introduced into the channels. In addition, in the chip wash step, the solutions from the channel and the wells were merged and could be visualized by the originally colorless solutions from the channel turning red. The removal of red dye can be observed in Fig. 2B, vii as the solution in the channel turned back to colorless. To quantify the recovery efficiency of this method, a solution with a fluorescent dye was injected into the device and subsequently collected and quantified using a fluorospectrometer. We determined a recovery rate of 96% when comparing the fluorescence signal from the chip wash solution with the starting stock solution normalized to the same volume. A recovery rate of 83% was observed when Escherichia coli cells labeled with red fluorescent protein were used to quantify the recovery efficiency of bacterial cells.
Fig. 2.
Design and operation of the chip wash device. (A) Schematic drawings of the chip wash method illustrating device design for handling microbial cells. (B) Representative photographs showing device operation as visualized with red dye. See text for details. Scale bar in i–vii, 200 µm. (C) Photograph of 3,200 droplets generated and stored on the chip for chip wash, shown next to a US quarter.
Validating the Chip Wash Method with a Two-Species Model Community.
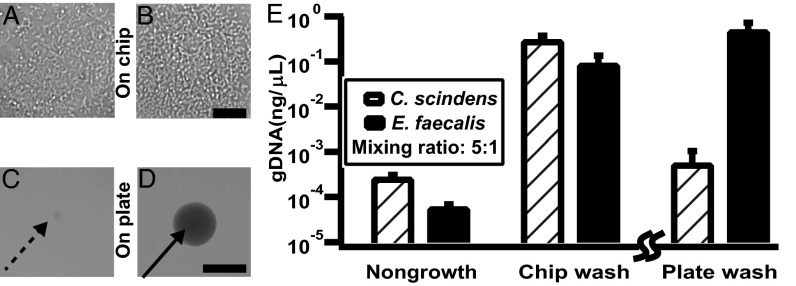
Having validated the device’s operation, we next tested the functionality of the chip wash method using a model community from the human gut microbiome (Fig. 3). First, we tested whether chip wash can detect microbial growth on SlipChip. We cultivated a mixture of Clostridium scindens and Enterococcus faecalis at a 5:1 ratio on the chip or agar plates. The genomic DNA of the starting inoculum and chip wash solution were extracted and quantified by quantitative PCR (qPCR). Cultivation on the chip followed by chip wash resulted in an ∼1,000-fold increase of DNA for each strain compared with DNA from the starting inoculum used as a nongrowth control (Fig. 3E), showing that chip wash can be used to detect microbial growth.
Fig. 3.
Validation of the chip wash method with a model community of C. scindens and E. faecalis. Samples were collected on day 1. (A and B) Representative optical microscopy of C. scindens (A) and E. faecalis (B) grown on SlipChip. (C and D) Representative photographs of C. scindens (C) and E. faecalis (D) grown on an agar plate. (E) Graph showing genomic DNA of C. scindens and E. faecalis recovered from nongrowth negative control, chip wash, and plate wash solutions. The nongrowth control and the chip wash experiments were performed using an identical procedure and can be directly compared. Because the plate wash experiment requires a different protocol, only the relative values can be compared (emphasized by the break in the axis). Error bars indicate SD (n = 3). Scale bar, 30 μm for A and B and 1 mm for C and D.
Second, we hypothesized that chip wash would detect, without bias, the growth of bacteria that grow at different rates but with similar carrying capacity, for the following reason. For the interest of detection, the optimal time for sampling is the late exponential phase or early stationary phase of the target to maximize the yield of biomass. A single cell growing on a plate starts at a density of ∼10 cfu mL−1 assuming the inoculation density is 300 cfu with 30 mL of medium, whereas a single cell growing in a 6-nL well starts at a density of ∼1.7 × 105 cfu mL−1. Typical carrying capacity of the media we used for gut anaerobes is ∼109 cfu mL−1; therefore, on the device the carrying capacity can be reached more rapidly, and for a larger range of growth rates, than on a plate. To test this hypothesis, we confirmed that under this particular cultivation condition, E. faecalis grew faster than C. scindens on agar plates, as observed from the difference in colony size on day 1 (Fig. 3 C and D). The cultivation medium has a similar carrying capacity for the two strains (SI Appendix). Consistent with the prediction, the two strains grew on the chip to a comparable density on day 1 (Fig. 3 A and B). As shown by the quantity of genomic DNA recovered from the two strains, sampling on day 1 by plate wash resulted in an ∼1,000-fold bias toward rapidly growing bacteria, whereas the chip wash method effectively corrected this bias, as the genomic DNA was comparable for each strain (Fig. 3E). This chip wash method provides an efficient way to detect slowly growing bacteria and is complementary to the plate wash method (21). Because we have shown that SlipChip is compatible with solutions used in membrane protein crystallization (28), we expect that SlipChip would be compatible with testing a wide range of growth media with different viscosities and surface tensions.
Using Splitting to Preserve Cultivar and Perform Genetic Assays.
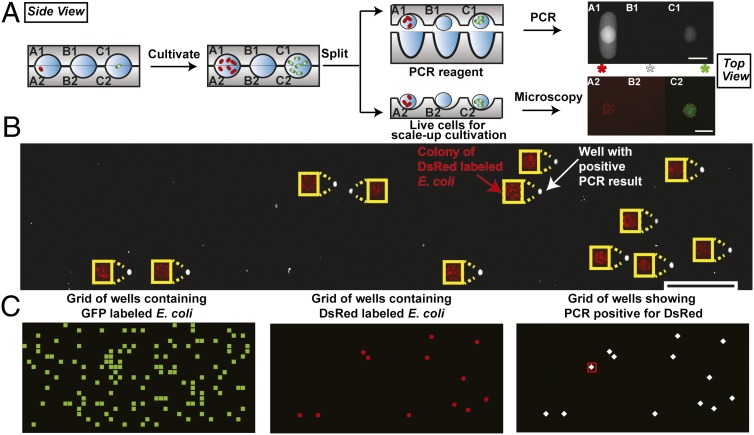
We next tested whether genetic assays could be used to identify and characterize microbes on the chip. We used a replica-SlipChip described in an accompanying paper (22) to split the microcolonies into two halves so that PCR could be performed with one of these halves and live microbes could be preserved on the other. To unambiguously establish the mapping from genotype to phenotype, we used E. coli cells expressing DsRed or GFP genes to ensure the genotype could be characterized by PCR, and the phenotype could be monitored by fluorescence microscopy (Fig. 4). We tested if this on-chip PCR approach could reliably distinguish the DsRed-labeled E. coli from the GFP-labeled E. coli. A mixture of E. coli cells labeled with GFP and DsRed proteins was loaded onto the chip, at final densities of 2 × 104 cfu mL−1 and 2 × 103 cfu mL−1, respectively. We assume that the cells are distributed in wells randomly and therefore that their distribution is governed by the Poisson statistics. We used a motile strain of E. coli to ensure uniform distribution of bacterial cells in both wells within 3 h of incubation. Individual cells were compartmentalized and cultivated, and then the chip was split into two daughter halves, each carrying a copy of the microcolonies (Fig. 4A). One chip was mixed with PCR reagents containing primers targeting the plasmid of DsRed and the other was imaged with a fluorescence microscope to check for the presence or absence of fluorescent proteins. We observed 125 wells that contained colonies with GFP E. coli and 12 wells containing DsRed E. coli. The wells showing PCR-positive matched the corresponding wells containing DsRed E. coli (Fig. 4 B and C); in contrast, blank wells that contained no bacteria and wells containing GFP-labeled E. coli were PCR-negative. We also noticed one well that showed increased fluorescence intensity in the PCR result, but no bacterial colony was detected in the other copy, which indicates that the well may have contained nongrowing cells or cell-free DNA from the solution. Many microbes residing in the human gut are not motile and might also adhere to surfaces. Therefore, we wanted to verify that this method would work with such organisms or whether active mixing inside SlipChip wells (25) would be required.
Fig. 4.
Cultivating pure microcolonies from a mixture and using PCR to identify specific microcolonies. Schematics show side views, whereas photographs show top views. (A) Schematic illustrating the cultivation of single cells from a mixture of E. coli expressing GFP and DsRed genes, as well as a method for splitting individual colonies. PCR was used to identify the E. coli expressing DsRed gene on one half of the split chip. The PCR reagents wells have an ellipsoidal cross-section from top view. An increase in fluorescence intensity indicated a positive result for PCR, and thus, the presence of the DsRed gene. Fluorescence microscopy identified wells that contained microbes expressing red and green fluorescent proteins, matching corresponding results in PCR. (B) To test the accuracy of the PCR assay, results from microscopy imaging (red), indicating E. coli colonies expressing DsRed gene, and PCR assay (white) were montaged with an offset to allow visualization without overlap. (C) Plot of a 20 × 50-well grid was used to represent the position of each well on the same device. Elements corresponding to wells were colored to highlight the presence of E. coli GFP colonies (green squares), E. coli DsRed colonies (red dots), and PCR positive results for DsRed (white diamonds). A red square in the third plot denotes a false positive result from PCR. The different shapes of markers used in C do not represent the shapes of wells. Scale bar, 200 µm for A, 2 mm for B. A 200-µm-wide yellow rectangle was used as scale bar for images showing DsRed expressing E. coli colonies in B. Note: schematics are not to scale; dimensions are provided in SI Appendix.
Identifying Cultivation Conditions for One of the Most Wanted Microbial Targets.
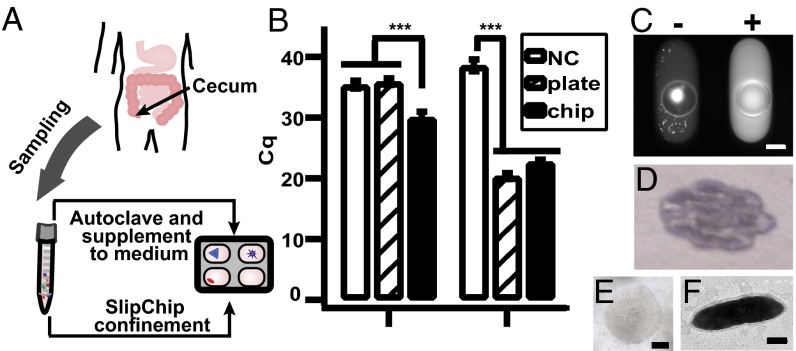
To test this workflow, we focused on isolating microbes from the human gut that belong to the high-priority groups of the Most Wanted list. The genus Oscillibacter is frequently observed in the Most Wanted list (2) and other sequencing data sets (29–31), but no human-associated member of this genus has been cultivated yet. To cultivate this genus, we collected samples from the human cecum using a brushing technique to obtain mucosa-associated microbes of high biomedical interest that may directly interact with the host (Fig. 5A). To identify microbial targets in the cultivar, we used 16S rRNA gene high-throughput sequencing with the V4 variable region (32) as a first screening. Reads were clustered to operational taxonomic units (OTUs) de novo with mothur software (33). First, the sample was cultured on agar plates in M2GSC medium (see SI Appendix for ingredients) and examined by plate wash. No OTU from the cultivar was classified as Oscillibacter. Next, miniaturization enabled by microfluidics allowed us to test if we could culture this genus by supplementing the medium with washing fluid from the sampling site in the human cecum. We obtained washing fluid by a lavage technique, autoclaved it, and spiked into the cultivation medium, referred to in this paper as M2LC (see SI Appendix for ingredients). The same amount of inoculum as plated on M2GSC agar was cultivated on SlipChip with M2LC medium and then chip wash was performed as described above. High-throughput sequencing of the V4 region of the 16S rRNA gene showed the successful cultivation of Oscillibacter on the chip (SI Appendix). We performed high-throughput sequencing of the V1V3 region of the 16S rRNA gene to test if the Oscillibacter recovered from chip wash belonged to the Most Wanted list. We were able to assign the reads classified as Oscillibacter to OTU_158_V1V3 (OTU158 for short, with an estimated ∼0.7% relative abundance in stool samples in the HMP dataset) from the list (SI Appendix) using a custom script (provided in SI Appendix) based on usearch software (34). PCR with OTU158-specific primers (OTU158P) confirmed that the target OTU indeed could be found in the cultivar (SI Appendix). OTU158P allowed us to validate results of the 16S survey conclusively by qPCR (Fig. 5B). We quantified the genomic DNA of OTU158 using OTU158P and total bacterial genomic DNA using 16S rRNA V4 universal primers. We observed genomic DNA of OTU158 from the chip wash experiment but not in the blank negative control or the plate wash experiment, whereas both plate wash and chip wash solutions had similar quantities of bacterial DNA that were higher than that of the blank negative control. Chip wash with M2GSC medium did not recover OTU158 (SI Appendix). We concluded that the M2LC medium with the washing fluid is an optimal condition to cultivate OTU158.
Fig. 5.
Targeted isolation of isolate microfluidicus 1 from SlipChip. (A) Illustration showing that mucosal biopsies obtained from the human cecum were used for stochastic confinement as well as supplemented into the medium to stimulate growth of microbes. (B) Identifying the cultivation condition of the microbial target OTU158 using qPCR. (Left) Graph showing that the use of target-specific primers revealed that the target was found in the chip wash solution (M2LC) but not in the blank negative control (NC) or the plate wash solution (M2GSC). (Right) Graph showing that the use of universal primers of 16S rRNA gene showed that both chip wash and plate wash solutions contained bacterial genomic DNA. A lower Cq value indicates higher concentration of DNA. Error bars indicate SD (n = 3). (C) Fluorescence microscopy photograph of on-chip colony PCR after the chip was split, showing a positive well (Right) for OTU158. A PCR negative well is shown on the left, as indicated by the low fluorescence intensity of the solution. The bright spot was presumably from cell material stained with SYBR Green. (D) Photograph of the first round of scaled-up culture of OTU158. (E) Microphotograph of a single colony of isolate microfluidicus 1. (F) Transmission electron microscopy image of a single OTU158 cell. Scale bar, 200 µm for C and E, and 0.5 µm for F.
Isolating OTU158 Using Replica-SlipChip.
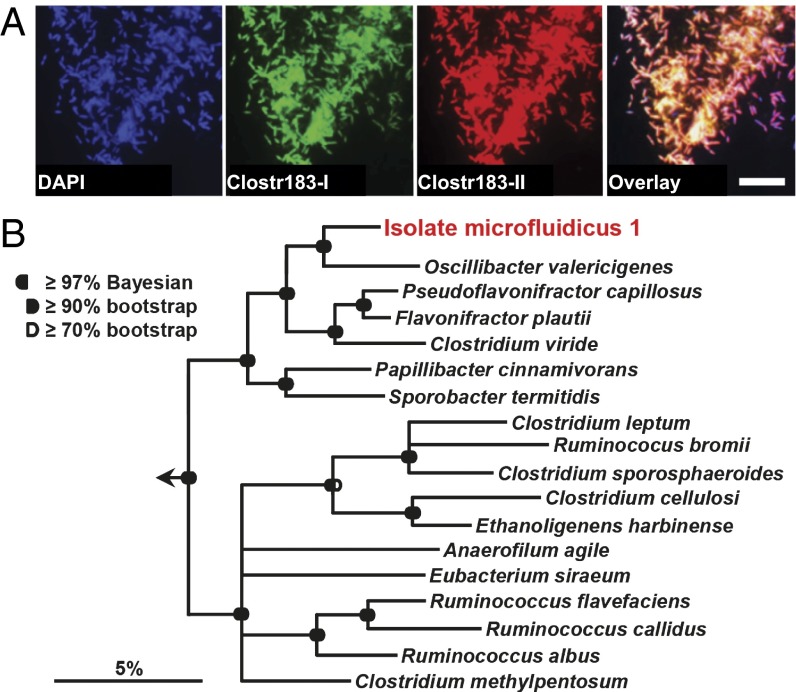
We further tested isolation and scale-up of microcolonies by cultivating the sample on the replica-SlipChip (22) with the M2LC medium containing the washing fluid from the sampling site. PCR was carried out with primers OTU158P targeting OTU158. We observed two positive wells (one is shown in Fig. 5C) from a single device with ∼500 microbial colonies (a negative PCR well is shown in Fig. 5C, Left). We scaled up the cultivar from one of the positive wells on an agar plate using the M2GSC medium. The intact scale-up culture after 3 d of incubation is shown in Fig. 5D. The culture contained multiple colonies, as shown in the picture, due to the presence of multiple cfus transferred from the same well of the chip. Although we did not observe the target from plate wash and chip wash experiments in the same medium, the cells could be scaled up on an agar plate with M2GSC medium. It is possible that the target grew in M2GSC medium but was outcompeted by rapidly growing strains in both plate wash and chip wash experiments, or that the target was in a dormant state until it was primed by washing fluid from the sampling site (35). Alternatively, the scaled-up colonies may represent a subpopulation of cells that can be cultivated with M2GSC, and the microcolony grown on the chip offered enough cells to cultivate these rare cells. We expect this observation can be understood as similar isolates are obtained using this method and as improved analytics are developed for quantitatively understanding the unculturable state of cells from environmental samples (10). Next, we performed colony PCR on this isolate with both species-specific and universal primers in bulk, and confirmed by Sanger sequencing that it was indeed the desired target. Although we observed that this was an almost pure culture (with some minor heterogeneity observed from chromatogram, shown in SI Appendix), we streaked the plates five times for purification to obtain single colonies (Fig. 5D) of target cells. This isolate, hereafter referred to in this paper as isolate microfluidicus 1, could then be routinely grown in bulk liquid culture to obtain enough biomass to initiate in vivo studies and whole genome sequencing. For example, the draft genome of this isolate was sequenced and assembled into 83 contigs comprising 3.4 Mbp sequences. We observed rod-shaped cells (Fig. 5F and SI Appendix) and two 16S rRNA gene types of 99.4% sequence identity to one another, each with 99% identical to OTU_158_V1V3 and OTU_896_V1V3 from the Most Wanted list (Table S1). Both OTUs are from the high-priority group classified as Oscillibacter, but their relative abundances differ by 20-fold in stool samples surveyed by the HMP (2). Although sequence heterogeneity among multiple 16S rRNA genes on the same genome is not uncommon (36), these two sequence types could either have been derived from a single strain or indicated the presence of two closely related strains. Therefore, we designed two oligonucleotide probes able to distinguish between the two sequence types and used them in FISH experiments (37, 38). All FISH-positive cells bound both sequence type-specific FISH probes (Clost183-I and Clost183-II, Fig. 6A), as well as the general probe mix EUB338I-III (SI Appendix), which specifically detects most members of the bacteria (39, 40). Together, these results demonstrate the presence of a single Ruminococcaceae species in the culture.
Fig. 6.
Phylogenetic affiliation of isolate microfluidicus 1 and validation of the purity of the culture by FISH. (A) Fluorescence images showing that both 16S rRNA types obtained from the culture are expressed within the same cells, demonstrating the presence of a single Ruminococcaceae species within the culture. Clostr183-I and Clostr183-II indicate FISH probes, each specific to a different sequence type. (Scale bar, 10 µm.) (B) 16S rRNA-based consensus tree demonstrating the positioning of isolate microfluidicus 1 within the Ruminococcaceae (Clostridia cluster IV). Please see SI Appendix for details.
Improved Taxonomic Assignment of the Isolate.
Short reads from 16S rRNA high-throughput sequencing may not be sufficient for assignment of taxonomy if the organisms are poorly represented in culture collections. Based on both 16S rRNA V4 and V1V3 high-throughput sequencing, the target was classified as Oscillibacter (see SI Appendix for Ribosomal Database Project classification). However, the pure culture suggests that isolate microfluidicus 1 is a member of a previously unidentified genus. The closest described relative for which a 16S rRNA sequence is available is Oscillibacter valericigenes, isolated from a Japanese clam (Corbicula) (41), which exhibits a sequence identity of 93.0% to the isolate of isolate microfluidicus 1. Phylogenetic analyses of the 16S rRNA of isolate microfluidicus 1 confirmed the unique positioning of this microbe within the Ruminococcaceae (42, 43) (Fig. 6B). These observations suggest that this highly sought (Table S2) bacterium may represent, to our knowledge, the first discovered species of an uncultured genus.
Materials and Methods
Sample Collection.
Brush and luminal cecum samples were collected from a healthy volunteer. Samples were transferred into an anaerobic chamber immediately after collection and homogenized in grants buffered saline solution (GBSS) supplemented with 5% DMSO by vortexing for 5 min. Aliquots of the samples were flash frozen with liquid nitrogen and preserved at −80 °C. Work with clinical samples for this project is approved by the Institutional Review Boards at California Institute of Technology and The University of Chicago, and by the Institutional Biosafety Committee.
SlipChip Cultivation.
The brush sample was serially diluted in GBSS buffer and then suspended in M2LC medium. This bacterial suspension was then loaded onto SlipChip designed for chip wash and incubated for 3 d.
Chip Wash.
The cultivar was collected into an Eppendorf pipette tip by flowing 90 µL PBS buffer three times and then 90 µL tetradecane into the SlipChip. The solution was then transferred into an Eppendorf tube. DNA was extracted using a QiaAmp kit following the manufacturer’s protocol and then used to prepare the library for high-throughput sequencing and PCR.
Isolation of Isolate Microfluidicus 1.
We used the replica-SlipChip to cultivate and split the microcolonies. One copy was used for colony PCR to identify the wells containing OTU158. The microcolony from the other copy was transferred on an M2GSC agar plate for scale-up culture.
Conclusions
In this paper, we describe an integrated microfluidic workflow for genetically targeted isolation of microbes, and validate it by successful isolation and cultivation of isolate microfluidicus 1 from the HMP's Most Wanted list. To our knowledge, this is the first example of targeted isolation of a high-priority member from the list, and is the first successful targeted cultivation from a complex biological sample of a previously uncultured taxon defined only by short reads from high-throughput sequencing of the 16S rRNA gene. We believe this genetically targeted workflow can become a general method beyond the isolate described in this paper, as in our preliminary experiments, an additional high-priority and three medium-priority organisms on the Most Wanted list have been isolated. We envision that the microfluidics-based workflow described in this paper will be useful for conclusively testing hypotheses generated from culture-independent studies by providing pure cultures of biomedically and environmentally significant microorganisms.
Supplementary Material
Acknowledgments
We thank Igor Antoshechkin and the Jacobs Genetics and Genomics Laboratory at California Institute of Technology for help with next-generation sequencing and assembly of the genome. We thank Alasdair McDowall from Prof. Grant Jensen’s lab for assistance with electron microscopy, Whitney Robles for contributions to writing and editing this manuscript, Prof. Tom Schmidt for discussions of 16S rRNA heterogeneity and experimental techniques, and for suggesting the creation of the 100 “Most Wanted” list at the HMP meeting, Prof. Jared Leadbetter for discussion on nomenclature of the isolate, Prof. Victoria Orphan for access to lab facilities, and Dionysios A. Antonopoulos for providing the anaerobic chamber to process the clinical samples. We thank Robert Edgar, J. Gregory Caporaso, Justin Kuczynski, William A. Walters, Hiroyuki Imachi, George M. Garrity, and Ashlee M. Earl for helpful discussions. Research reported in this publication was supported by the National Human Genome Research Institute of the National Institutes of Health under Award R01HG005826. R.H. was supported via an Erwin Schrӧdinger Postdoctoral Fellowship by the Austrian Science Fund (Fonds zur Förderung der Wissenschaftlichen Forschung; J 3162-B20).
Footnotes
Conflict of interest statement: R.F.I. has a financial interest in SlipChip Corporation.
This article is a PNAS Direct Submission.
Data deposition: The genome sequences reported in this paper have been deposited in the Joint Genome Institute's Integrated Microbial Genomes database, https://img.jgi.doe.gov/cgi-bin/w/main.cgi (accession no. 2545555870). The 16S rRNA gene sequences of isolate microfluidicus 1 reported in this paper have been deposited in the GenBank database (accession nos. KJ875866 and KJ875867).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1404753111/-/DCSupplemental.
References
- 1.Schloss PD, Handelsman J. Metagenomics for studying unculturable microorganisms: Cutting the Gordian knot. Genome Biol. 2005;6(8):229. doi: 10.1186/gb-2005-6-8-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fodor AA, et al. The “most wanted” taxa from the human microbiome for whole genome sequencing. PLoS ONE. 2012;7(7):e41294. doi: 10.1371/journal.pone.0041294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kennedy J, et al. Functional metagenomic strategies for the discovery of novel enzymes and biosurfactants with biotechnological applications from marine ecosystems. J Appl Microbiol. 2011;111(4):787–799. doi: 10.1111/j.1365-2672.2011.05106.x. [DOI] [PubMed] [Google Scholar]
- 4.Rooks DJ, McDonald JE, McCarthy AJ. Metagenomic approaches to the discovery of cellulases. Methods Enzymol. 2012;510:375–394. doi: 10.1016/B978-0-12-415931-0.00020-3. [DOI] [PubMed] [Google Scholar]
- 5.Reddy BVB, et al. Natural product biosynthetic gene diversity in geographically distinct soil microbiomes. Appl Environ Microbiol. 2012;78(10):3744–3752. doi: 10.1128/AEM.00102-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ridaura VK, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341(6150):1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frank DN, et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA. 2007;104(34):13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Staley JT, Konopka A. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu Rev Microbiol. 1985;39(1):321–346. doi: 10.1146/annurev.mi.39.100185.001541. [DOI] [PubMed] [Google Scholar]
- 9.Kaeberlein T, Lewis K, Epstein SS. Isolating “uncultivable” microorganisms in pure culture in a simulated natural environment. Science. 2002;296(5570):1127–1129. doi: 10.1126/science.1070633. [DOI] [PubMed] [Google Scholar]
- 10.Stewart EJ. Growing unculturable bacteria. J Bacteriol. 2012;194(16):4151–4160. doi: 10.1128/JB.00345-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.D’Onofrio A, et al. Siderophores from neighboring organisms promote the growth of uncultured bacteria. Chem Biol. 2010;17(3):254–264. doi: 10.1016/j.chembiol.2010.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giel JL, Sorg JA, Sonenshein AL, Zhu J. Metabolism of bile salts in mice influences spore germination in Clostridium difficile. PLoS ONE. 2010;5(1):e8740. doi: 10.1371/journal.pone.0008740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh S, et al. Cell extract-containing medium for culture of intracellular fastidious bacteria. J Clin Microbiol. 2013;51(8):2599–2607. doi: 10.1128/JCM.00719-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zengler K, et al. Cultivating the uncultured. Proc Natl Acad Sci USA. 2002;99(24):15681–15686. doi: 10.1073/pnas.252630999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ingham CJ, et al. The micro-Petri dish, a million-well growth chip for the culture and high-throughput screening of microorganisms. Proc Natl Acad Sci USA. 2007;104(46):18217–18222. doi: 10.1073/pnas.0701693104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eun Y-J, Utada AS, Copeland MF, Takeuchi S, Weibel DB. Encapsulating bacteria in agarose microparticles using microfluidics for high-throughput cell analysis and isolation. ACS Chem Biol. 2011;6(3):260–266. doi: 10.1021/cb100336p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin K, et al. Generation of larger numbers of separated microbial populations by cultivation in segmented-flow microdevices. Lab Chip. 2003;3(3):202–207. doi: 10.1039/b301258c. [DOI] [PubMed] [Google Scholar]
- 18.Park J, Kerner A, Burns MA, Lin XN. Microdroplet-enabled highly parallel co-cultivation of microbial communities. PLoS ONE. 2011;6(2):e17019. doi: 10.1371/journal.pone.0017019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu W, Kim HJ, Lucchetta EM, Du W, Ismagilov RF. Isolation, incubation, and parallel functional testing and identification by FISH of rare microbial single-copy cells from multi-species mixtures using the combination of chemistrode and stochastic confinement. Lab Chip. 2009;9(15):2153–2162. doi: 10.1039/b904958d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vincent ME, Liu W, Haney EB, Ismagilov RF. Microfluidic stochastic confinement enhances analysis of rare cells by isolating cells and creating high density environments for control of diffusible signals. Chem Soc Rev. 2010;39(3):974–984. doi: 10.1039/b917851a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stevenson BS, Eichorst SA, Wertz JT, Schmidt TM, Breznak JA. New strategies for cultivation and detection of previously uncultured microbes. Appl Environ Microbiol. 2004;70(8):4748–4755. doi: 10.1128/AEM.70.8.4748-4755.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma L, et al. 2014. Individually addressable arrays of replica microbial cultures enabled by splitting slipchips. Integr Biol, 10.1039/C4IB00109E.
- 23.Du W, Li L, Nichols KP, Ismagilov RF. SlipChip. Lab Chip. 2009;9(16):2286–2292. doi: 10.1039/b908978k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen F, et al. Nanoliter multiplex PCR arrays on a SlipChip. Anal Chem. 2010;82(11):4606–4612. doi: 10.1021/ac1007249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu W, Chen D, Du W, Nichols KP, Ismagilov RF. SlipChip for immunoassays in nanoliter volumes. Anal Chem. 2010;82(8):3276–3282. doi: 10.1021/ac100044c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Begolo S, Shen F, Ismagilov RF. A microfluidic device for dry sample preservation in remote settings. Lab Chip. 2013;13(22):4331–4342. doi: 10.1039/c3lc50747e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fuerstman MJ, et al. The pressure drop along rectangular microchannels containing bubbles. Lab Chip. 2007;7(11):1479–1489. doi: 10.1039/b706549c. [DOI] [PubMed] [Google Scholar]
- 28.Li L, Du W, Ismagilov R. User-loaded SlipChip for equipment-free multiplexed nanoliter-scale experiments. J Am Chem Soc. 2010;132(1):106–111. doi: 10.1021/ja908555n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Claesson MJ, et al. Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012;488(7410):178–184. doi: 10.1038/nature11319. [DOI] [PubMed] [Google Scholar]
- 30.Raman M, et al. Fecal microbiome and volatile organic compound metabolome in obese humans with nonalcoholic fatty liver disease. Clin Gastroenterol Hepatol. 2013;11(7):868–875, e1–e3. doi: 10.1016/j.cgh.2013.02.015. [DOI] [PubMed] [Google Scholar]
- 31.Lam YY, et al. Increased gut permeability and microbiota change associate with mesenteric fat inflammation and metabolic dysfunction in diet-induced obese mice. PLoS ONE. 2012;7(3):e34233. doi: 10.1371/journal.pone.0034233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Caporaso JG, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schloss PD, et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75(23):7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 35.Lewis K. Persister cells, dormancy and infectious disease. Nat Rev Microbiol. 2007;5(1):48–56. doi: 10.1038/nrmicro1557. [DOI] [PubMed] [Google Scholar]
- 36.Klappenbach JA, Saxman PR, Cole JR, Schmidt TM. Rrndb: The ribosomal RNA operon copy number database. Nucleic Acids Res. 2001;29(1):181–184. doi: 10.1093/nar/29.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Amann R, Fuchs BM. Single-cell identification in microbial communities by improved fluorescence in situ hybridization techniques. Nat Rev Microbiol. 2008;6(5):339–348. doi: 10.1038/nrmicro1888. [DOI] [PubMed] [Google Scholar]
- 38.Stoecker K, Daims H, Wagner M. Fluorescence in situ hybridization for the detection of prokaryotes. In: Osborn AM, Smith CJ, editors. Molecular Microbial Ecology. Abingdon, UK: Bios Advanced Methods; 2005. pp. 213–239. [Google Scholar]
- 39.Amann RI, et al. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56(6):1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Daims H, Brühl A, Amann R, Schleifer KH, Wagner M. The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: Development and evaluation of a more comprehensive probe set. Syst Appl Microbiol. 1999;22(3):434–444. doi: 10.1016/S0723-2020(99)80053-8. [DOI] [PubMed] [Google Scholar]
- 41.Iino T, Mori K, Tanaka K, Suzuki K, Harayama S. Oscillibacter valericigenes gen. nov., sp. nov., a valerate-producing anaerobic bacterium isolated from the alimentary canal of a Japanese corbicula clam. Int J Syst Evol Microbiol. 2007;57(Pt 8):1840–1845. doi: 10.1099/ijs.0.64717-0. [DOI] [PubMed] [Google Scholar]
- 42.Collins MD, et al. The phylogeny of the genus Clostridium: Proposal of five new genera and eleven new species combinations. Int J Syst Bacteriol. 1994;44(4):812–826. doi: 10.1099/00207713-44-4-812. [DOI] [PubMed] [Google Scholar]
- 43.Yutin N, Galperin MY. A genomic update on clostridial phylogeny: Gram-negative spore formers and other misplaced clostridia. Environ Microbiol. 2013;15(10):2631–2641. doi: 10.1111/1462-2920.12173. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.