Significance
The activity of the Hv1 proton channel has been shown to exacerbate neuronal death after cerebral hypoxia and promote tumor formation by highly metastatic breast cancer cells. Compounds able to inhibit excessive Hv1 activity could find applications as neuroprotective agents in ischemic stroke or anticancer drugs. In this work, we investigate how guanidine derivatives, like 2-guanidinobenzimidazole (2GBI), interact with Hv1 and inhibit proton conduction. We identify the binding site for 2GBI and determine the most likely orientation of the inhibitor inside the channel. We also show that a simple modification of 2GBI eliminates the requirement for the inhibitor to have direct access to the intracellular side of the channel to block the proton current.
Keywords: HVCN1 blocker, macrophage, microglial cell
Abstract
The voltage-gated proton channel Hv1 plays important roles in proton extrusion, pH homeostasis, and production of reactive oxygen species in a variety of cell types. Excessive Hv1 activity increases proliferation and invasiveness in cancer cells and worsens brain damage in ischemic stroke. The channel is composed of two subunits, each containing a proton-permeable voltage-sensing domain (VSD) and lacking the pore domain typical of other voltage-gated ion channels. We have previously shown that the compound 2-guanidinobenzimidazole (2GBI) inhibits Hv1 proton conduction by binding to the VSD from its intracellular side. Here, we examine the binding affinities of a series of 2GBI derivatives on human Hv1 channels mutated at positions located in the core of the VSD and apply mutant cycle analysis to determine how the inhibitor interacts with the channel. We identify four Hv1 residues involved in the binding: aspartate 112, phenylalanine 150, serine 181, and arginine 211. 2GBI appears to be oriented in the binding site with its benzo ring pointing to F150, its imidazole ring inserted between residue D112 and residues S181 and R211, and the guanidine group positioned in the proximity of R211. We also identify a modified version of 2GBI that is able to reach the binding site on Hv1 from the extracellular side of the membrane. Understanding how compounds like 2GBI interact with the Hv1 channel is an important step to the development of pharmacological treatments for diseases caused by Hv1 hyperactivity.
The Hv1 voltage-gated proton channel (also known as HVCN1 or voltage-sensor–only protein) regulates the production of superoxide and other reactive oxygen species by NADPH oxidase (NOX) enzymes in a variety of cell types, including microglial cells (1) and leukocytes (2). NOX activity causes membrane depolarization and intracellular accumulation of protons. Hv1 allows sustained NOX activity by repolarizing the membrane and extruding excess protons from the cell (3–5).
Hv1 has been shown to enhance brain damage in a mouse model of ischemic stroke through its NOX-modulating activity (1). The channel was also found overexpressed in many B-cell malignancies (6) and breast and colorectal cancer tissues (7, 8). High Hv1 activity was shown to increase invasiveness of breast cancer cells and be associated with shorter overall and recurrence-free survival in breast cancer patients (7). These findings highlight that excessive activity of the Hv1 channel can have serious pathological consequences in ischemic stroke and cancer and that small-molecule inhibitors targeting Hv1 could lead to the development of new neuroprotective or anticancer drugs.
The Hv1 protein is made of four membrane-spanning segments (S1–S4) (9, 10), and it is related to the voltage-sensing domains (VSDs) of other voltage-gated ion channels (11) and voltage-sensitive phosphatases (VSPs) (12). The inner end of the S4 segment is connected to a coiled-coil domain responsible for protein dimerization (13, 14). As a result, the channel is made of two VSD subunits, each containing a gated proton pore (15–17).
The block of voltage-gated sodium, potassium, and calcium channels by small molecules has been studied for decades. Its mechanism has been elucidated for many drugs, and in the majority of cases, the inhibitors were found to bind to different regions of the pore domain (18, 19). With the exception of peptide toxins (20, 21), not much is known about compounds interacting with VSDs (22), and only recently have there been successful attempts to produce small-molecule drugs that specifically target these domains in voltage-gated ion channels (23, 24).
We have recently shown that some guanidine derivatives have the ability to inhibit Hv1 activity and that one of these compounds, 2-guanidinobenzimidazole (2GBI), binds the channel's VSD only in the open conformation (25). We have also found that the binding site is within the proton permeation pathway and faces the cytoplasm.
Here, we explore the chemical space available to guanidine derivatives for Hv1 binding. We then use a mutation cycle analysis approach to identify the residues in the channel that contribute to the binding environment of 2GBI and establish the overall orientation of the blocker within the VSD in the open conformation. Our results suggest that residues D112, F150, S181, and R211 are located close to each other deep within the membrane and in the proximity of the intracellular vestibule of the VSD, where they can interact with the blocker. We discuss our binding model in the context of a recent crystal structure of the channel (26).
Results
Molecular Features of Guanidine Derivatives Regulating Hv1 Inhibition.
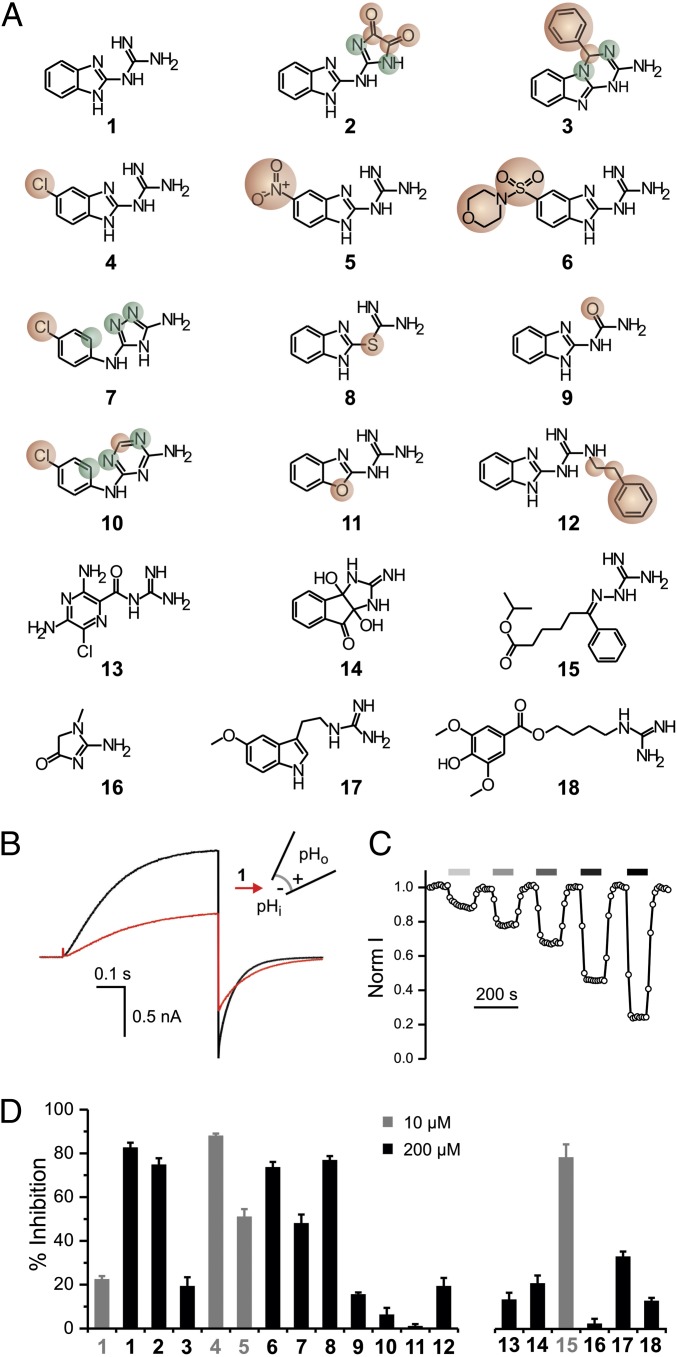
To understand what makes 2GBI (Fig. 1A, compound 1) an effective Hv1 inhibitor, we tested analogs 2–12 (Fig. 1A) for their ability to inhibit proton currents measured in inside-out patches from Xenopus oocytes expressing the human Hv1 channel. The analogs differed from 2GBI in selected molecular features, such as nature of heteroatoms, substituents, or ring connectivity. We also examined compounds 13–18 (Fig. 1A) to determine whether structures of guanidine derivatives unrelated to 2GBI could be compatible with tight binding.
Fig. 1.
Exploring Hv1 inhibition by guanidine derivatives. (A) Tested compounds: (1) 2GBI, (2) 2-(2-benzimidazolylamino)-imidazole-4,5-dione, (3) 4-phenyl-1,4-dihydro[1,3,5]triazino[1,2-a]benzimidazol-2-amine, (4) 5-chloro-2-guanidinobenzimidazole, (5) 5-nitro-2-guanidinobenzimidazole, (6) 1-[5-(morpholin-4-ylsulfonyl)-1H-benzimidazol-2-yl]guanidine, (7) N-(4-chlorophenyl)-4H-1,2,4-triazole-3,5-diamine, (8) S-1H-benzimidazol-2-yl-carbamothioate, (9) 1-(benzimidazol-2-yl)urea, (10) N-(4-chlorophenyl)-1,3,5-triazine-2,4-diamine, (11) 2-guanidino-benzoxazole, (12) 1-(benzimidazol-2-yl)-3-(2-phenylethyl)guanidine, (13) amiloride, (14) 3a,8a-dihydroxy-2-imino-2,3,3a,8a-tetrahydroindeno[1,2-d]imidazol-8(1H)-one, (15) isoproyl 6-(guanidinoimino)-6-phenylhexanoate, (16) creatinine, (17) 1-[2-(5-methoxy-1H-indol-3-yl)ethyl] guanidine, and (18) leonurine. Compounds 2–12 are structurally related to compound 1. Atoms or substituents differing from compound 1 are highlighted in red. Different connectivity from compound 1 is highlighted in green. (B) Proton currents measured in an inside-out patch from a Xenopus oocyte expressing human Hv1 WT in response to a depolarization to +120 mV from a holding potential of −80 mV. The black trace was recorded in the absence of inhibitor, and the red trace was recorded after the addition of 50 μM compound 1 (2GBI) in the bath solution. pHi = pHo = 6.0. (C) Example of a time course of inhibition of Hv1 channels exposed to increasing concentrations of 2GBI. Current was normalized to the value measured under bath perfusion in the absence of inhibitor. Horizontal bars of increasingly darker gray color indicate the presence of 5, 10, 20, 50, and 100 μM inhibitor in the bath. (D) Inhibition of Hv1 proton currents by the indicated compounds at 10 μM (gray) or 200 μM (black) concentration. Data are means ± SEMs (n ≥ 3). Currents were measured as in B.
Because 2GBI binds an intracellular receptor on the Hv1 channel (25), we added the guanidine derivatives to the intracellular side of the membrane patches (Fig. 1 B and C). The activity of different derivatives was compared at the final concentration of 200 µM (Fig. 1D, black bars), with the exception of compounds 4, 5, and 15, which were tested at the concentration of 10 µM (Fig. 1D, gray bars). The inhibition induced by compound 1 is reported in Fig. 1D at both concentrations for reference.
We found that, among the 2GBI analogs, two compounds with substitutions in the benzo ring (4, 5) were more effective than the reference compound when used at the same concentration. Three other analogs (2, 6, 8) showed activity similar to 2GBI, whereas the rest of the compounds were less effective. Among the guanidine derivatives unrelated to 2GBI, only compound 15 showed activity higher than 2GBI. Adding bulky hydrophobic groups to the guanidine moiety (compounds 3 and 12) impaired binding to the channel as did opening of the imidazole ring (compounds 7 and 10). Reducing the ability of the inhibitor to bind protons had different effects depending on the nature of the modification. Some of these modifications did not significantly affect the efficiency of binding (compounds 2 and 8), whereas others had quite a disruptive effect (compounds 9 and 11). Overall, these findings show that the activity of 2GBI can be increased by modifications of its benzo group and that the neutralization of its charge to increase membrane permeability is compatible with strong binding. In addition, the fact that compound 15 produced strong Hv1 inhibition suggests that the chemical space available for binding is not limited to molecules containing the benzimidazole group.
Binding Environment of Benzimidazole-Guanidine Compounds Within the Hv1 VSD.
We have previously shown that 2GBI inhibits the Hv1 channel by blocking its proton conduction pathway when the activation gate is open (25). However, how does 2GBI block the channel? What parts of the VSD make up the binding site, and how is the inhibitor molecule oriented inside it? Similar questions were previously addressed in a study of the Shaker potassium channel, where a mutant cycle analysis approach was used to map the binding site of the inhibitor agitoxin (27). Here, we use a similar approach to identify Hv1 residues interacting with 2GBI and investigate specific parts of the inhibitor that contribute to channel-blocker interactions.
The interaction between a specific residue (i) in a channel and a specific part (j) of a bound inhibitor can be detected by modifying i and j by either mutagenesis or chemical synthesis and then, measuring the dissociation constants (Kd values) for the four different combinations of modified and unmodified i and j (Fig. S1). The free energy of coupling between i and j [ΔΔGo(ij)] can be then calculated from the measured Kd values (Methods) (28). The size of the ΔΔGo(ij) quantifies the strength of the i − j interaction, whereas its sign can indicate whether the interaction is stabilizing or destabilizing.
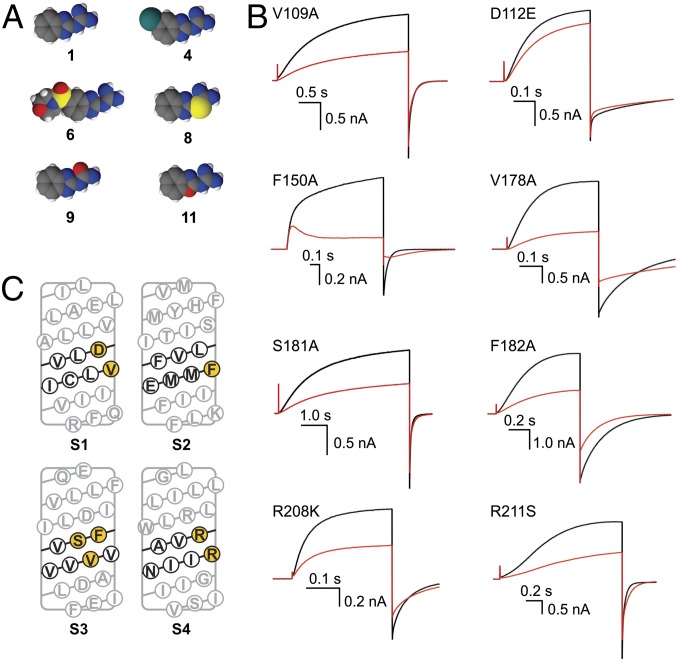
Among 2GBI analogs shown in Fig. 1A, we chose for the mutant cycle analysis five compounds that differ from the reference for individual substitutions at distinct locations within the 2GBI molecule (Fig. 2A). On the channel, we investigated mutations of eight residues shown in Fig. 2C. In the S2 helix, we chose position 150, because we found earlier that the nature of its side chain strongly modulates the binding efficiency of both 2GBI and compound 11 (25). Residues V109 and D112 in S1 and residue R211 in S4 have been previously proposed to face the proton conduction pathway (29–31), and in our structural models of Hv1, they are predicted to be in the vicinity of F150 (32). Another study recently proposed that R208 faces D112 in the core of the VSD in the open state (33). Therefore, we included positions 109, 112, 208, and 211 in our analysis. Because there is not clear experimental evidence as to whether specific S3 positions participate in proton conduction, we also included residues V178, S181, and F182 in the analysis. In our Hv1 models, these residues are predicted to be on the side of the S3 helix facing the center of the VSD and located in the middle of the transmembrane plane (32). Individual amino acid substitutions were selected as explained in Methods.
Fig. 2.
Selection of molecular features to be analyzed in channel and inhibitor. (A) Compounds selected for the mutant cycle analysis (space-filling representation). (B) Examples of current inhibitions observed with the indicated Hv1 mutants in the presence of 2GBI (red traces) in the bath solution (1 μM for the F150A mutant and 50 μM for the other channels). pHi = pHo = 6.0. Currents were measured in inside-out patches on depolarization to +120 mV from a holding potential of −80 mV like in Fig. 1B. (C) Sequence of Hv1 transmembrane segments S1–S4. Residues expected to be at the same transmembrane depth as F150 are shown in black. Black residues predicted to face the core of the VSD are highlighted in yellow.
We tested 2GBI and its five analogs on inside-out patches from oocytes expressing either the WT channel or one of its eight mutants. Fig. 1B shows an example of current traces from Hv1 WT in the absence of 2GBI and the presence of a 50-µm intracellular inhibitor. Fig. 2B shows equivalent traces for the individual mutants. The effects of the mutations on the channel’s voltage dependence of activation (represented as conductance vs. voltage relationship) are shown in Fig. S2.
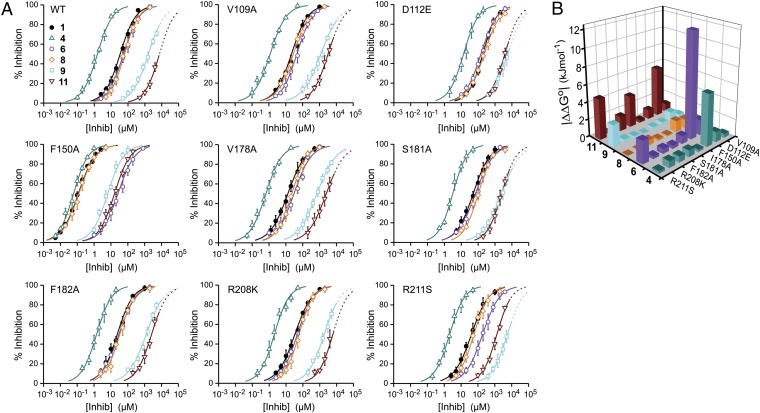
From measurements like the one shown in Fig. 1C, we determined the dose–response of inhibition for a total of 54 channel–inhibitor combinations (symbols in Fig. 3A). Each dose–response was fitted with the Hill equation (curves in Fig. 3A), and the apparent dissociation constants and Hill coefficients were determined, which are reported in Methods (Table S1). The Kd values for the WT channel and the mutants were then used to calculate the ΔΔGo of coupling for each combination of residue substitution and inhibitor modification. Fig. S1 shows two examples of individual mutant cycle analysis. In one case, the mutant channel F150A is paired to compound 6, and the combination produces a large ΔΔGo. In the other case, F150A is paired with compound 9, and the combination produces a ΔΔGo ∼ 0. These results indicate that phenylalanine 150 interacts with the part of the 2GBI molecule modified in compound 6 but not modified in compound 9.
Fig. 3.
Mutant cycle analysis. (A) Dose–response curves for the inhibition of WT and mutant channels produced by the indicated compounds (Fig. 2A). Reductions in proton current produced by increasing concentrations of inhibitor were measured like in Fig. 1C. Each point represents the average inhibition from three to six measurements ± SD. Curved lines are Hill fits. Fit parameters are reported in Table S1. Highest tested concentrations and relative inhibitions are reported in Table S3. Dotted lines for compounds 9 and 11 represent extrapolations into a concentration range where the drugs have low solubility (Methods). Inhibition of Hv1 WT and F150A by compounds 1 and 11 was reported in a previous study (25); in that study, the inhibitors were referred to as 2GBI and GBOZ, respectively (25). (B) Coupling free energy values for the indicated combinations of channel mutations and inhibitor modifications. Hv1 WT and compound 1 are the references. The absolute values of ΔΔGo were calculated from the apparent dissociation constants reported in Table S1 (Methods and Fig. S1).
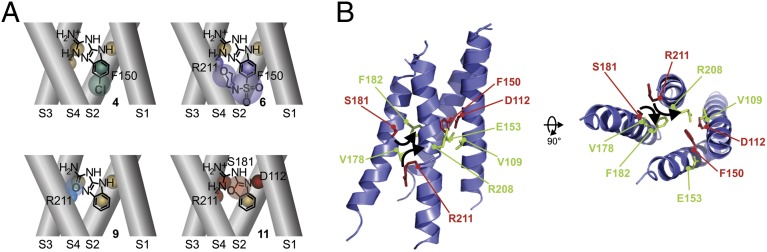
The absolute values of the ΔΔGo are reported in Fig. 3B, and their relative values, expressed as percentages of the total binding energy for 2GBI to Hv1 WT (−25 kJ/mol), are reported in Table S2. The combinations D112E-11, F150A-4, F150A-6, S181A-11, R211S-6, R211S-9, and R211S-11 had relative |ΔΔGo| values ≥ 10% of the 2GBI binding energy. The other 33 combinations had relative |ΔΔGo| values < 10%. The combinations showing large coupling energy were all consistent with the binding environment shown in Fig. 4A, in which the bound inhibitor is surrounded by D112, F150, S181, and R211 (spheres with different colors in Fig. 4A).
Fig. 4.
Interpretation of channel–inhibitor interactions. (A) Interpretation of the thermodynamic couplings calculated in Fig. 3B. Simple coupling occur between F150A and compound 4 and between R211S and compound 9 (Left). Simultaneous perturbation of multiple interactions must be invoked to explain the coupling between compounds 6 and 11 with the binding environment (Right). (B) Investigated Hv1 residues mapped on the structure of the Hv1-CiVSP chimera (26). Helix S0 and cytoplasmic coiled-coil domain are not shown for clarity. In Right, the VSD is viewed from the intracellular side of the membrane. Amino acid identity refers to human Hv1. Residues involved in 2GBI binding are shown in red. The other residues are shown in green. Black arrows indicate the rearrangements proposed to occur during activation to bring residues S181 and R211 in a position compatible with 2GBI binding (close to F150 and D112).
When the interaction between a channel mutant and a modified 2GBI molecule is different from the interaction between the WT channel and 2GBI (high ΔΔGo), the mutated position is indicated with the same color of the modified inhibitor (Fig. 4A). Otherwise, it is represented in yellow (small ΔΔGo) (Fig. 4A). Binding of compounds 4 and 6 were differentially perturbed in the F150A mutant. This finding is consistent with an interaction between F150 and the benzo ring of 2GBI. For compound 6, the binding to R211S was also perturbed, possibly because of the large size of the substituent (Fig. 4A, Upper Right).
Binding of compound 9 was differentially perturbed in the R211S mutant, indicating that the guanidine moiety of 2GBI is close to R211. We probed this association by also analyzing the binding of WT and R211S channels with compound 2. In this 2GBI derivative, the guanidine moiety is modified by cyclization rather than heteroatom substitution, like in compound 9 (Fig. S3). Both these modifications are expected to reduce the ability of the guanidine moiety to become protonated and donate hydrogen bonds. Accordingly, we found that the ΔΔGo values for combinations R211S-2 and R211S-9 were very similar (Fig. S3 and Table S2).
For compound 11, differential perturbations were measured in the D112E, S181A, and R211S mutants. This finding can be explained by assuming that compound 11 sits in the binding site with the orientation of the oxazole ring shown in Fig. 4A, Lower Right, with the ring oxygen facing R211 and S181 and the ring nitrogen facing D112.
To further constrain the possible orientations of 2GBI, we tested whether its guanidine moiety could interact with glutamate 153, the only negatively charged residue other than D112 located in the neighborhood of F150. We examined the binding of 2GBI and compound 9 to WT and E153C channels and found a ΔΔGo ∼ 0 (Fig. S4). This result excludes orientations of the inhibitor that bring the guanidine group in contact with E153.
Enabling Hv1 Inhibition by Extracellular Benzimidazole-Guanidine Compounds.
We have previously found that 2GBI does not produce a reduction in proton current when present in the extracellular medium (25). Because the binding site on the channel is intracellular, the compound must partition into the membrane to reach it from outside the cell. 2GBI is too polar to do this effectively (predicted logP for nonprotonated 2GBI = 0.52). We reasoned that modifications that make 2GBI less polar should improve membrane partitioning. The modification in compound 4 seemed particularly promising, because it reduces the overall polarity of the molecule (predicted logP nonprotonated 4 = 1.12), increasing, at the same time, the apparent affinity for the channel. Other modifications that resulted in more pronounced reductions in polarity (compounds 9, 11, and 12) negatively affected binding affinity.
We tested compound 4 (ClGBI) in outside-out patches from oocytes expressing Hv1 WT and found that the inhibitor can block the channel from the extracellular side of the membrane (Fig. S5 A and B). The apparent Kd and Hill coefficient for ClGBI extracellular binding were 26.3 ± 2.2 µM and 1.0 ± 0.12, respectively.
We then measured inhibition by ClGBI of native proton currents in microglial BV2 cells as well as monocyte/macrophage cells RAW264.7 and THP-1 (Fig. S5 C–E). These cells were previously shown to express high levels of proton channels (34–36). We found that the effect of the inhibitor was very similar in the native channels compared with recombinant Hv1 expressed in oocytes (Fig. S5F). These findings suggest that optimization of guanidine derivatives based on the 2GBI scaffold can lead to useful Hv1 inhibitors.
Discussion
2GBI was previously shown to act as an open channel blocker of Hv1 (25). Based on the results of the mutant cycle analysis, we can conclude that, when the VSD of Hv1 opens, 2GBI gets access to the innermost part of the intracellular vestibule and is surrounded by residues D112 in S1, F150 in S2, S181 in S3, and R211 in S4 (Fig. 4A). We propose a model of binding in which one of the nitrogen atoms of the imidazole ring interacts with D112 (stabilizing interaction), whereas the other points to S181 and R211. The benzo ring of 2GBI interacts with F150 (destabilizing interaction), and its guanidine moiety points to R211. The insertion of the imidazole ring between D112 and R211/S181 combined with the nonequivalent interactions between the two nitrogen atoms of the ring and the surrounding protein provide a simple explanation for why a single heteroatom replacement in compound 11 is coupled to three different positions in the channel located on opposite sides of the binding site (Fig. 4A, Lower Right). Previous studies have proposed the existence of water wires inside the open Hv1 channel (32, 37). Therefore, some of the observed interactions between 2GBI and the channel may be mediated by water molecules in the VSD.
Aspartate at position 112 has been shown to be the selectivity filter of Hv1 (29). By interacting directly with this residue, 2GBI could change the value of its pKa to values incompatible with effective proton permeation. Alternatively, the inhibitor could physically break the hydrogen-bonded chain required to deliver protons from the intracellular vestibule of the VSD to D112. An earlier study proposed that, in the open state, D112 and R211 are close enough to interact electrostatically (30). Our results are consistent with this conclusion. Other studies on the VSDs of voltage-gated potassium channels have concluded that the conserved phenylalanine corresponding to F150 interacts with different S4 arginines in different conformations (38, 39). Our findings indicate that R211 is close to F150 in the open state, which is consistent with other studies in which the accessibility of the S4 arginine to the intracellular solution was assessed in the closed and open conformations (33, 40).
Recently, the crystal structure of a chimera between Hv1 and the Ciona intestinalis voltage-sensitive phosphatase (CiVSP) has been solved (26). Although the channel seems to be in a nonconducting closed conformation, a relatively wide intracellular vestibule extends deep inside the VSD and contacts F150. The existence of a similar intracellular vestibule was recently reported in an Hv1 model based on the crystal structure of the VSD of CiVSP (41, 42). This vestibule is likely to be the pathway that 2GBI follows to reach its binding site from the intracellular medium. The crystallized channel also shows a small cavity on top of F150 and D112 followed by an extracellular constriction (26). If this constriction is maintained in the open state, it can explain why 2GBI is not able to reach its binding site from the extracellular medium.
We mapped the residues involved in 2GBI binding on the crystal structure of the Hv1-CiVSP chimera and show them in red in Fig. 4B. F150 and D112 are located close to each other and in the proximity of the intracellular vestibule, a position consistent with our 2GBI binding model. However, the residues in S3 and S4 facing F150 and D112 are F182 and R208, two of the positions tested that do not participate in 2GBI binding (green in Fig. 4B). This finding suggests that, when the channel transitions from the closed conformation (represented in the structure) to the open conformation capable of binding 2GBI, residues F182 and R208 change their positions with respect to F150 and D112. If, in the open conformation, S181 takes the place of F182 and R211 takes the place of R208 (black arrows in Fig. 4B), all four residues involved in 2GBI binding can face the center of the VSD and interact with the inhibitor. This structural rearrangement is compatible with the mechanism of voltage-driven activation of CiVSP proposed by Li et al. (42).
The high polarity of 2GBI limits its potential as an Hv1 inhibitor because of its inefficient partitioning into the membrane. Appending a hydrophobic group to the guanidine moiety could be a way to increase the lipophilic nature of the molecule. However, our results indicate that there is no space in the binding site to accommodate such a group. Accordingly, a 2GBI analog with this kind of modification proved to be a poor Hv1 blocker (compound 12 in Fig. 1 A and D). However, substituents at the benzo ring of 2GBI seem to be much better tolerated (compounds 4–6 in Fig. 1 A and D), and the 5-chloro substitution resulted in an increase in apparent binding affinity. In our binding model, the benzo ring is located in a region of the intracellular vestibule that is wide enough to accommodate large groups. The walls of the vestibule could provide an interacting surface for larger 2GBI analogs, resulting in stronger binding.
The ability of ClGBI to inhibit both recombinant and native proton channels from the extracellular medium (Fig. S5) suggests that guanidine derivatives based on the 2GBI scaffold could be optimized to reduce the pathological effects of excessive Hv1 activity in ischemic stroke and cancer.
Methods
Hv1 Expression.
Recombinant human Hv1 channel was subcloned in the pGEMHE vector (43) as previously described (25). Single-point mutations were introduced with standard PCR techniques. Plasmids were linearized with NheI restriction enzyme (New England Biolabs) before in vitro transcription. RNA synthesis was carried out with a T7 mMessage mMachine Transcription Kit (Ambion). cRNAs were injected in Xenopus oocytes (50 nL/cell, 0.3–1.5 µg/µL) 1–3 d before the electrophysiological measurements.
Cultured Cells.
Stages V and VI oocytes were either prepared from Xenopus laevis (NASCO) using well-established methods or purchased from Ecocyte Bioscience. Cells were kept at 18 °C in ND96 medium containing 96 mM NaCl, 2 mM KCl, 1.8 mM CaCl2, 1 mM MgCl2, 10 mM Hepes, 5 mM pyruvate, and 100 µg/mL gentamycin (pH 7.2). BV-2 and THP-1 cells were gifts from Heike Wulff (University of California, Davis, CA) and Albert Zlotnik (University of California, Irvine, CA), respectively. RAW264.7 cells were from ATCC (TIB-71). BV-2 and RAW264.7 cells were maintained in DMEM supplemented with 10% (wt/vol) FBS. THP-1 cells were maintained in RPMI-1640 medium supplemented with 10% (wt/vol) FBS and 50 µM 2-mercaptoethanol. Cells were kept at 37 °C in a 5% (vol/vol) CO2 incubator.
Tested Compounds.
2-GBI, 2-(2-benzimidazolylamino)-imidazole-4,5-dione, 5-chloro-2-guanidino-benzimidazole, 5-nitro-2-guanidinobenzimidazole, N-(4-chlorophenyl)-4H-1,2,4-triazole-3,5-diamine, 1-(benzimidazol-2-yl)urea, N-(4-chlorophenyl)-1,3,5-triazine-2,4-diamine hydrochloride, amiloride hydrochloride, isoproyl 6-(guanidinoimino)-6-phenylhexanoate nitrate, and 1-[2-(5-methoxy-1H-indol-3-yl)ethyl]guanidine hydroiodide were from Sigma-Aldrich. 1-[5-(Morpholin-4-ylsulfonyl)-1H-benzimidazol-2-yl]guanidine was from Enamine. 4-Phenyl-1,4-dihydro[1,3,5]triazino[1,2-a]benzimidazol-2-amine, S-1H-benzimidazol-2-yl-carbamothioate, 2-guanidinobenzoxazole, 1-(benzimidazol-2-yl)-3-(2-phenylethyl) guanidine, and 3a,8a-dihydroxy-2-imino-2,3,3a,8a-tetrahydroindeno[1,2-d]imidazol-8(1H)-one were from Vitas-M Laboratory. Creatinine was from Acros Organics, and leonurine was from Combi-Blocks. All of the guanidine derivatives used were at the highest purity commercially available. The compounds were directly dissolved in the recording solutions at the desired final concentrations or prepared as 100 mM stock solutions in DMSO. The bath chamber was perfused under gravity by manifold connected to a VC-6 Perfusion Valve System (Warner Instruments) controlled by the pClamp software by Transistor-Transistor Logic (TTL) signal. Predicted logP values were calculated with ALOGPS (Virtual Computational Chemistry Laboratory; www.vcclab.org) (44).
Electrophysiological Measurements.
Hv1 proton currents were measured in inside-out or outside-out patches from oocytes using an Axopatch 200B Amplifier controlled by pClamp10 software through an Axon Digidata 1440A (Molecular Devices). The intracellular solution contained 100 mM MES, 30 mM tetraethylammonium (TEA) methanesulfonate, 5 mM TEA chloride, and 5 mM EGTA adjusted to pH 6.0 with TEA hydroxide. For recordings carried out in the absence of pH gradient, extracellular and intracellular solutions had the same composition. For measurements performed in the presence of a pH gradient (pHi = 6.0, pHo = 7.5) the extracellular solution contained 100 mM Hepes, 40 mM TEA methanesulfonate, and 5 mM TEA chloride adjusted to pH 7.5 with TEA hydroxide.
Channel inhibition was determined by isochronal current measurements at the end of a depolarization pulse at +120 mV. The duration of the pulse was different for different channels (Fig. 2B) to account for the difference in time required to reach steady-state activation. An example of time course of inhibition is reported in Fig. 1C. At +120 mV (and pHi = pHo = 6.0), WT and mutant channels are maximally open, with the exception of R211S (Fig. S2). For this mutant under the experimental conditions used, only ∼50% of channels are open at +120 mV. Because the inhibitor binds Hv1 only in the open state and it does not change the channel’s voltage dependence of activation (25), the assessment of inhibition is not affected by the presence of closed channels. We interpret the change in conductance vs. voltage relationship produced by the R211S mutation as the direct result of a missing gating charge as previously shown (45).
Native proton currents in BV-2, RAW264.7, and THP-1 were measured in whole-cell configuration with an intracellular solution containing: 90 mM TEA methanesulfonate, 100 mM MES, 2 mM MgCl2, and 2 mM EGTA adjusted to pH 6.0 with TEA hydroxide. The extracellular solution contained 85 mM TEA methanesulfonate, 100 mM Hepes, 3 mM CaCl2, and 1 mM EGTA adjusted to pH 7.5 with TEA hydroxide.
All measurements were performed at 22 °C ± 2 °C. Pipettes had 1.5–4 MΩ access resistance. Current traces were filtered at 1 kHz, sampled at 5 kHz, and analyzed with Clampfit10.2 (Molecular Devices) and Origin8.1 (OriginLab).
Mutant Cycle Analysis.
Doses–responses of channel inhibition in Fig. 3A and Figs. S3, S4, and S5B were fitted by the Hill equation:
where [B] is the concentration of inhibitor B, Kd is the apparent dissociation constant, and h is the Hill coefficient. Fit parameters are reported in the text and Table S1. In some dose–response curves, because of limits in solubility of inhibitors 9 and 11, we could test maximal concentrations that produced less than 80% inhibition. In these cases, the Hill coefficient was constrained between 0.80 and 1.00 for the fitting, and the curves were extrapolated to 100% inhibition (dotted lines in Fig. 3A).
ΔΔGo(ij) values relative to position i in the channel and part j of the inhibitor were calculated according to the equation
where 0 stands for either a mutation in the channel at position i or a change in structural feature in the inhibitor at position j. For instance, the mutation (i → 0) in Hv1 can be F150A, and the change in the inhibitor (j → 0) can be a chlorine atom replacing the hydrogen at position 5 in the benzo ring of 2GBI (compound 4).
Mapping Tested Residues on the Hv1-CiVSP Chimera.
In the VSD of the chimera, residues E149–F171 of mouse Hv1/VSOP are replaced with residues D164–L188 of CiVSP. Tested residues in human Hv1 (V109, D112, F150, E153, V178, S181, F182, R208, and R211) correspond to residues V105, D108, F146, D149, V174, S177, F178, R204, and R207 of the chimera, respectively. Representation of the structure shown in Fig. 4B was made in PyMOL (Schrödinger).
Supplementary Material
Acknowledgments
We thank H. Wulff and V. Singh for providing useful information about the tested compounds and the gift of BV-2 cells. We thank A. Zlotnik for providing THP-1 cells. We also thank O. Yifrach, D. J. Tobias, and J. A. Freites for helpful discussion. We thank M. M. Pathak and the other members of the laboratory of F.T. for insightful comments on the manuscript. This work was supported by National Institutes of Health Grant R01GM098973.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 9673.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1324012111/-/DCSupplemental.
References
- 1.Wu LJ, et al. The voltage-gated proton channel Hv1 enhances brain damage from ischemic stroke. Nat Neurosci. 2012;15(4):565–573. doi: 10.1038/nn.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DeCoursey TE. Voltage-gated proton channels: Molecular biology, physiology, and pathophysiology of the H(V) family. Physiol Rev. 2013;93(2):599–652. doi: 10.1152/physrev.00011.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DeCoursey TE, Morgan D, Cherny VV. The voltage dependence of NADPH oxidase reveals why phagocytes need proton channels. Nature. 2003;422(6931):531–534. doi: 10.1038/nature01523. [DOI] [PubMed] [Google Scholar]
- 4.Ramsey IS, Ruchti E, Kaczmarek JS, Clapham DE. Hv1 proton channels are required for high-level NADPH oxidase-dependent superoxide production during the phagocyte respiratory burst. Proc Natl Acad Sci USA. 2009;106(18):7642–7647. doi: 10.1073/pnas.0902761106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.El Chemaly A, et al. VSOP/Hv1 proton channels sustain calcium entry, neutrophil migration, and superoxide production by limiting cell depolarization and acidification. J Exp Med. 2010;207(1):129–139. doi: 10.1084/jem.20091837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Capasso M, et al. HVCN1 modulates BCR signal strength via regulation of BCR-dependent generation of reactive oxygen species. Nat Immunol. 2010;11(3):265–272. doi: 10.1038/ni.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y, Li SJ, Wu X, Che Y, Li Q. Clinicopathological and biological significance of human voltage-gated proton channel Hv1 protein overexpression in breast cancer. J Biol Chem. 2012;287(17):13877–13888. doi: 10.1074/jbc.M112.345280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y, Wu X, Li Q, Zhang S, Li SJ. Human voltage-gated proton channel hv1: A new potential biomarker for diagnosis and prognosis of colorectal cancer. PLoS ONE. 2013;8(8):e70550. doi: 10.1371/journal.pone.0070550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramsey IS, Moran MM, Chong JA, Clapham DE. A voltage-gated proton-selective channel lacking the pore domain. Nature. 2006;440(7088):1213–1216. doi: 10.1038/nature04700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sasaki M, Takagi M, Okamura Y. A voltage sensor-domain protein is a voltage-gated proton channel. Science. 2006;312(5773):589–592. doi: 10.1126/science.1122352. [DOI] [PubMed] [Google Scholar]
- 11.Yu FH, Catterall WA. The VGL-chanome: A protein superfamily specialized for electrical signaling and ionic homeostasis. Sci STKE. 2004;2004(253):re15. doi: 10.1126/stke.2532004re15. [DOI] [PubMed] [Google Scholar]
- 12.Murata Y, Iwasaki H, Sasaki M, Inaba K, Okamura Y. Phosphoinositide phosphatase activity coupled to an intrinsic voltage sensor. Nature. 2005;435(7046):1239–1243. doi: 10.1038/nature03650. [DOI] [PubMed] [Google Scholar]
- 13.Li SJ, et al. The role and structure of the carboxyl-terminal domain of the human voltage-gated proton channel Hv1. J Biol Chem. 2010;285(16):12047–12054. doi: 10.1074/jbc.M109.040360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujiwara Y, et al. The cytoplasmic coiled-coil mediates cooperative gating temperature sensitivity in the voltage-gated H(+) channel Hv1. Nat Commun. 2012;3:816. doi: 10.1038/ncomms1823. [DOI] [PubMed] [Google Scholar]
- 15.Lee SY, Letts JA, Mackinnon R. Dimeric subunit stoichiometry of the human voltage-dependent proton channel Hv1. Proc Natl Acad Sci USA. 2008;105(22):7692–7695. doi: 10.1073/pnas.0803277105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tombola F, Ulbrich MH, Isacoff EY. The voltage-gated proton channel Hv1 has two pores, each controlled by one voltage sensor. Neuron. 2008;58(4):546–556. doi: 10.1016/j.neuron.2008.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koch HP, et al. Multimeric nature of voltage-gated proton channels. Proc Natl Acad Sci USA. 2008;105(26):9111–9116. doi: 10.1073/pnas.0801553105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hille B. Ion Channels of Excitable Membranes. 3rd Ed. Sunderland, MA: Sinauer; 2001. [Google Scholar]
- 19.Wulff H, Castle NA, Pardo LA. Voltage-gated potassium channels as therapeutic targets. Nat Rev Drug Discov. 2009;8(12):982–1001. doi: 10.1038/nrd2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swartz KJ, MacKinnon R. An inhibitor of the Kv2.1 potassium channel isolated from the venom of a Chilean tarantula. Neuron. 1995;15(4):941–949. doi: 10.1016/0896-6273(95)90184-1. [DOI] [PubMed] [Google Scholar]
- 21.Wang J, et al. Mapping the receptor site for alpha-scorpion toxins on a Na+ channel voltage sensor. Proc Natl Acad Sci USA. 2011;108(37):15426–15431. doi: 10.1073/pnas.1112320108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Börjesson SI, Elinder F. An electrostatic potassium channel opener targeting the final voltage sensor transition. J Gen Physiol. 2011;137(6):563–577. doi: 10.1085/jgp.201110599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li P, et al. The gating charge pathway of an epilepsy-associated potassium channel accommodates chemical ligands. Cell Res. 2013;23(9):1106–1118. doi: 10.1038/cr.2013.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peretz A, et al. Targeting the voltage sensor of Kv7.2 voltage-gated K+ channels with a new gating-modifier. Proc Natl Acad Sci USA. 2010;107(35):15637–15642. doi: 10.1073/pnas.0911294107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hong L, Pathak MM, Kim IH, Ta D, Tombola F. Voltage-sensing domain of voltage-gated proton channel Hv1 shares mechanism of block with pore domains. Neuron. 2013;77(2):274–287. doi: 10.1016/j.neuron.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takeshita K, et al. X-ray crystal structure of voltage-gated proton channel. Nat Struct Mol Biol. 2014;21(4):352–357. doi: 10.1038/nsmb.2783. [DOI] [PubMed] [Google Scholar]
- 27.Hidalgo P, MacKinnon R. Revealing the architecture of a K+ channel pore through mutant cycles with a peptide inhibitor. Science. 1995;268(5208):307–310. doi: 10.1126/science.7716527. [DOI] [PubMed] [Google Scholar]
- 28.Horovitz A, Fersht AR. Strategy for analysing the co-operativity of intramolecular interactions in peptides and proteins. J Mol Biol. 1990;214(3):613–617. doi: 10.1016/0022-2836(90)90275-Q. [DOI] [PubMed] [Google Scholar]
- 29.Musset B, et al. Aspartate 112 is the selectivity filter of the human voltage-gated proton channel. Nature. 2011;480(7376):273–277. doi: 10.1038/nature10557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berger TK, Isacoff EY. The pore of the voltage-gated proton channel. Neuron. 2011;72(6):991–1000. doi: 10.1016/j.neuron.2011.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morgan D, et al. Peregrination of the selectivity filter delineates the pore of the human voltage-gated proton channel hHV1. J Gen Physiol. 2013;142(6):625–640. doi: 10.1085/jgp.201311045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wood ML, et al. Water wires in atomistic models of the Hv1 proton channel. Biochim Biophys Acta. 2012;1818(2):286–293. doi: 10.1016/j.bbamem.2011.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kulleperuma K, et al. Construction and validation of a homology model of the human voltage-gated proton channel hHV1. J Gen Physiol. 2013;141(4):445–465. doi: 10.1085/jgp.201210856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Song JH, Marszalec W, Kai L, Yeh JZ, Narahashi T. Antidepressants inhibit proton currents and tumor necrosis factor-α production in BV2 microglial cells. Brain Res. 2012;1435:15–23. doi: 10.1016/j.brainres.2011.11.041. [DOI] [PubMed] [Google Scholar]
- 35.DeCoursey TE, Cherny VV. Voltage-activated proton currents in human THP-1 monocytes. J Membr Biol. 1996;152(2):131–140. doi: 10.1007/s002329900092. [DOI] [PubMed] [Google Scholar]
- 36.Sakai H, et al. Increases in intracellular pH facilitate endocytosis and decrease availability of voltage-gated proton channels in osteoclasts and microglia. J Physiol. 2013;591(Pt 23):5851–5866. doi: 10.1113/jphysiol.2013.263558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramsey IS, et al. An aqueous H+ permeation pathway in the voltage-gated proton channel Hv1. Nat Struct Mol Biol. 2010;17(7):869–875. doi: 10.1038/nsmb.1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tao X, Lee A, Limapichat W, Dougherty DA, MacKinnon R. A gating charge transfer center in voltage sensors. Science. 2010;328(5974):67–73. doi: 10.1126/science.1185954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lacroix JJ, Bezanilla F. Control of a final gating charge transition by a hydrophobic residue in the S2 segment of a K+ channel voltage sensor. Proc Natl Acad Sci USA. 2011;108(16):6444–6449. doi: 10.1073/pnas.1103397108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gonzalez C, Koch HP, Drum BM, Larsson HP. Strong cooperativity between subunits in voltage-gated proton channels. Nat Struct Mol Biol. 2010;17(1):51–56. doi: 10.1038/nsmb.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Q, Wanderling S, Perozo E. The resting state of human proton channel from functional and structural determinations. Biophys J. 2014;106(2):745a. [Google Scholar]
- 42.Li Q, et al. Structural mechanism of voltage-dependent gating in an isolated voltage-sensing domain. Nat Struct Mol Biol. 2014;21(3):244–252. doi: 10.1038/nsmb.2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liman ER, Tytgat J, Hess P. Subunit stoichiometry of a mammalian K+ channel determined by construction of multimeric cDNAs. Neuron. 1992;9(5):861–871. doi: 10.1016/0896-6273(92)90239-a. [DOI] [PubMed] [Google Scholar]
- 44.Tetko IV, et al. Virtual computational chemistry laboratory—design and description. J Comput Aided Mol Des. 2005;19(6):453–463. doi: 10.1007/s10822-005-8694-y. [DOI] [PubMed] [Google Scholar]
- 45.Gonzalez C, Rebolledo S, Perez ME, Larsson HP. Molecular mechanism of voltage sensing in voltage-gated proton channels. J Gen Physiol. 2013;141(3):275–285. doi: 10.1085/jgp.201210857. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.