Significance
All three domains of life—Eukarya, Bacteria, and Archaea—are known to inhabit the marine water column from surface waters to great depth. Planktonic marine Archaea are comprised of two dominant groups—the Thaumarchaeaota and Euryarchaeota. The Thaumarchaeota contain characteristic biomarker lipids known as tetraethers that are thought to be diagnostic for this group, and are used as paleotemperature proxies, since these lipids are well preserved in marine sediments. In this study, we show that planktonic Euryarchaeota produce the same types of archaeal tetraether lipids as do Thaumarchaeaota. Our results have important implications for environmental surveys of marine Archaea, and the use of their lipids for interpretation of the sedimentary record.
Keywords: euryarchaea, glycerol dialkyl glycerol tetraether, oceanography, environmental genomics, TEX86 index
Abstract
Archaea are ubiquitous in marine plankton, and fossil forms of archaeal tetraether membrane lipids in sedimentary rocks document their participation in marine biogeochemical cycles for >100 million years. Ribosomal RNA surveys have identified four major clades of planktonic archaea but, to date, tetraether lipids have been characterized in only one, the Marine Group I Thaumarchaeota. The membrane lipid composition of the other planktonic archaeal groups—all uncultured Euryarchaeota—is currently unknown. Using integrated nucleic acid and lipid analyses, we found that Marine Group II Euryarchaeota (MG-II) contributed significantly to the tetraether lipid pool in the North Pacific Subtropical Gyre at shallow to intermediate depths. Our data strongly suggested that MG-II also synthesize crenarchaeol, a tetraether lipid previously considered to be a unique biomarker for Thaumarchaeota. Metagenomic datasets spanning 5 y indicated that depth stratification of planktonic archaeal groups was a stable feature in the North Pacific Subtropical Gyre. The consistent prevalence of MG-II at depths where the bulk of exported organic matter originates, together with their ubiquitous distribution over diverse oceanic provinces, suggests that this clade is a significant source of tetraether lipids to marine sediments. Our results are relevant to archaeal lipid biomarker applications in the modern oceans and the interpretation of these compounds in the geologic record.
Early cultivation-independent molecular surveys led to the discovery of planktonic marine Euryarchaeota (1) and Thaumarchaeota (formerly called Crenarchaeota) (1, 2), and have since been used to describe the abundance and ecological distributions of archaeal groups in diverse ocean biomes (3). Metagenomic analyses and the isolation of several marine Thaumarchaeota have provided further insight into their physiology and biogeochemistry. Distinctive archaeal tetraether membrane lipids (SI Appendix, Fig. S1) have also been reported throughout the oceans (4). These compounds, collectively referred to as glycerol dialkyl glycerol tetraethers (GDGTs), have been useful tracers of archaeal biomass (5) and, via their isotopic composition, have provided new information about archaeal community carbon metabolism (6–8). GDGTs are relatively recalcitrant; they can be exported with little alteration to marine sediments, where their distributions have been exploited to develop proxies for reconstructing sea surface temperature (9) and terrigenous organic matter input (10). On the basis of GDGT abundances in a black shale dated at 112 million years (11), it was suggested that archaea have been significant members of marine ecosystems since at least the Mesozoic Era.
Although tetraether lipids are used with increasing frequency in paleoceanography and microbial ecology, their specific taxonomic sources in the water column are not well constrained. Of the four groups of planktonic archaea identified in the oceans, representatives of only one—the Marine Group I (MG-I) Thaumarchaeota (1, 2, 12)—have been isolated in pure culture. All MG-I strains isolated to date are chemolithoautotrophic, fixing inorganic carbon via energy obtained from the oxidation of ammonia to nitrite (13). Recent evidence suggests that MG-I also contribute to the flux of potent greenhouse gases nitrous oxide (14) and methane (15) from the water column to the atmosphere. The membrane lipid assemblage of MG-I includes GDGTs with zero through four cyclopentyl moieties and crenarchaeol, a GDGT containing one cyclohexyl and four cyclopentyl moieties (16). Crenarchaeol has been considered uniquely diagnostic for Thaumarchaeota (17) and, by extension, has been postulated as a biomarker for archaeal nitrification (18).
In addition to MG-I Thaumarchaeota, three other groups of archaea—all Euryarchaeota—inhabit the marine water column: Groups II (1), III (19), and IV (20). Of these, Marine Group II (MG-II) are the most abundant and frequently detected, often but not exclusively in near-surface waters. MG-II inhabit diverse oceanic provinces including the oligotrophic North Pacific Subtropical Gyre (21, 22), coastal California (23–25), the North Sea (26), Arctic (27, 28) and Antarctic (29–31) waters, the coastal Mediterranean Sea (32), the eastern tropical South Pacific oxygen minimum zone (33), waters surrounding a tropical atoll (34), the deep North Atlantic (35), and the East China Sea (36). The potential contribution of this cosmopolitan group to the marine tetraether lipid pool has been debated (37–40), but the lack of cultivated representatives of MG-II has precluded direct analysis of their membrane lipids, and incomplete knowledge of the genetic basis of archaeal tetraether lipid biosynthesis limits the ability of metagenomic studies to address this question.
We conducted parallel characterization of planktonic archaeal lipids and archaeal community DNA composition to constrain the taxonomic origins of GDGTs in marine plankton and their distribution in the marine water column. Metagenomic analyses, quantitative polymerase chain reaction (qPCR) of Thaumarchaeota marker genes, and small subunit ribosomal RNA (SSU rRNA) gene amplicon pyrosequencing were used to characterize the overall archaeal community composition in size-fractionated suspended particulate matter (SPM). Parallel measurements of archaeal tetraether lipids from the same filter samples enabled us to associate GDGTs with particular members of the planktonic archaeal community.
Results and Discussion
SPM for this study was collected in the North Pacific Subtropical Gyre (NPSG) in September 2011 using in situ pumps deployed at depths ranging from 11 to 559 m (SI Appendix, Figs. S2 and S3). SPM was fractionated into two size classes (0.3–3 µm and 3–57 µm) in an effort to separate the archaeal community into free-living and particle-associated/detrital components. Lipids and DNA (SI Appendix, Fig. S4) were extracted from the same filters.
Depth Distributions of Archaeal Phyla.
We characterized archaeal communities in our sample set using two independent techniques: metagenomics and pyrosequencing of amplicons of the V1−V3 region of the SSU rRNA gene in DNA. Counts of euryarchaeotal and thaumarchaeotal 16S rRNAs and functional gene sequences in metagenomic datasets (Table 1) indicated that Euryarchaeota outnumbered Thaumarchaeota at depths above 157 m in the 0.3- to 3-µm size fraction and in the 3- to 57-µm size fraction throughout the entire profile. Notably, at 83 m depth, the archaeal population was comprised nearly exclusively of MG-II.
Table 1.
Archaeal community composition and relative abundances of crenarchaeol and GDGTs with zero to three cyclopentyl moieties in suspended particulate matter samples
| Size, µm | z, m | % Eury | Relative abundance of GDGT cores | |||||
| rDNA | MGA | 0 | 1 | 2 | 3 | Cren | ||
| 0.3–3 | 83 | 97.2 | 94.4 | 22.4 | 8.2 | 8.1 | 0.1 | 61.1 |
| 131 | 74.4 | 66.7 | 21.8 | 8.3 | 12.1 | 6.0 | 51.7 | |
| 131* | 74.4 | 66.7 | 19.6 | 7.5 | 10.6 | 4.3 | 58.1 | |
| 157 | 35.3 | 26.4 | 19.1 | 6.0 | 10.0 | 3.9 | 61.0 | |
| 231 | 47.0 | 44.7 | 25.9 | 7.0 | 13.2 | 0.9 | 53.0 | |
| 331 | 36.2 | 28.9 | 40.3 | 5.6 | 8.6 | 0.0 | 45.5 | |
| 450 | 20.1 | 12.8 | 9.8 | 7.4 | 68.8 | 0.0 | 14.0 | |
| 559* | ND | ND | 25.5 | 10.3 | 13.7 | 0.4 | 50.1 | |
| 3–57 | 83 | 100.0 | 96.1 | 38.6 | 12.8 | 1.2 | 0.0 | 47.4 |
| 131 | 85.3 | 87.9 | 17.5 | 6.7 | 8.6 | 3.3 | 63.9 | |
| 157 | 75.0 | 74.3 | 18.0 | 6.7 | 11.4 | 3.6 | 60.3 | |
| 231 | 86.3 | 78.5 | 19.7 | 5.8 | 14.3 | 1.9 | 58.3 | |
| 331 | 71.0 | 66.1 | 21.2 | 5.0 | 16.8 | 0.7 | 56.3 | |
| 450 | 85.2 | 60.9 | 26.3 | 7.3 | 11.2 | 1.5 | 53.7 | |
Archaeal community composition was determined from MGA and 16S rDNA gene representation in metagenomic datasets. % Eury, % Euryachaeotal; Cren, crenarchaeol; MGA, metagenomic read annotations; z, depth.
Hydrolysates of total lipid extracts.
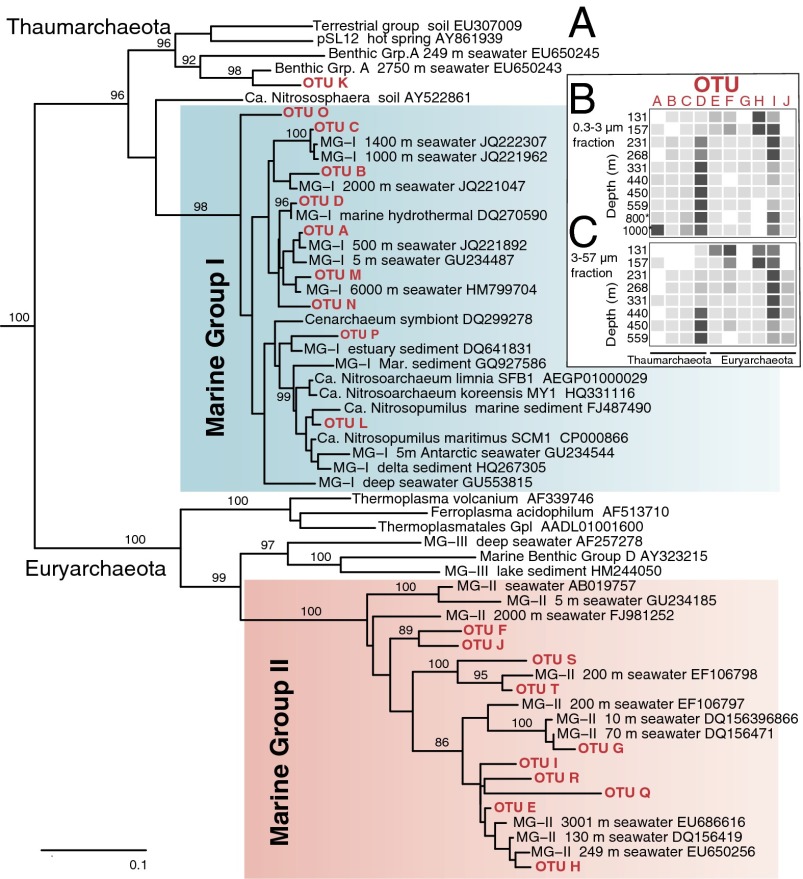
The 16S rRNA amplicon data provided a more detailed taxonomic breakdown of MG-I and MG-II phylotypes in the different size fractions and depths. We focused on samples from eight depths that yielded >1,000 amplicons in each size fraction (SI Appendix, Figs. S5 and S6 and Table S2), and the 0.22- to 1.6-µm size fraction from 800 m and 1000 m. In total, this sample set contained 323,924 archaeal amplicon sequences after curation and quality control. Archaeal rRNA sequences were clustered into operational taxonomic units (OTUs) using the software program QIIME (41) and taxonomy assigned through comparison with the ARB-SILVA SSU reference database using BLASTn.
The 10 most prevalent archaeal OTUs across the entire dataset (each representing >2,000 amplicon sequences and, in total, 82% of archaeal amplicons) were assigned to either the Thaumarchaeota (primarily MG-I) or MG-II Euryarchaeota (Fig. 1). Among the Thaumarchaeota, OTU D was predominant in both size fractions of SPM and represented a higher percent of total archaeal OTUs with increasing depth. At 1,000 m, OTU A had a higher relative abundance than OTU D, and OTU C rose in abundance, consistent with previous reports of genetic diversity of Thaumarchaeota in depth profiles (22).
Fig. 1.
Archaeal representation in different sample depths and size fractions. (A) Phylogenetic analysis (maximum likelihood; see SI Appendix) of the top 20 OTUs recovered by rRNA PCR (labeled in orange; 97% similarity level) that represent the majority of sequences across the dataset. (B and C) The depth and size class distribution of the 10 most prevalent archaeal OTUs. Darkest gray represents the most abundant OTU in a sample.
In sharp contrast to Thaumarchaeota, MG-II Euryarchaeota represented a much higher proportion of total archaeal OTUs at shallower depths, and the MG-II population was not dominated by a single phylotype (Fig. 1). Four OTUs—E, F, H, and I—were abundant in the 131- and 157-m samples, and OTU I became more prominent with depth. Such intragroup stratification may reflect niche partitioning, since some MG-II photic zone ecotypes encode the light-driven proton pump proteorhodopsin (42, 43), whereas mesopelagic MG-II ecotypes tend to lack the proteorhodopsin gene (42). Patterns of MG-II OTU abundance also varied between size classes; at 131 m, for instance, OTU F was more highly represented in the 3- to 57-µm fraction than in the 0.3- to 3-µm fraction.
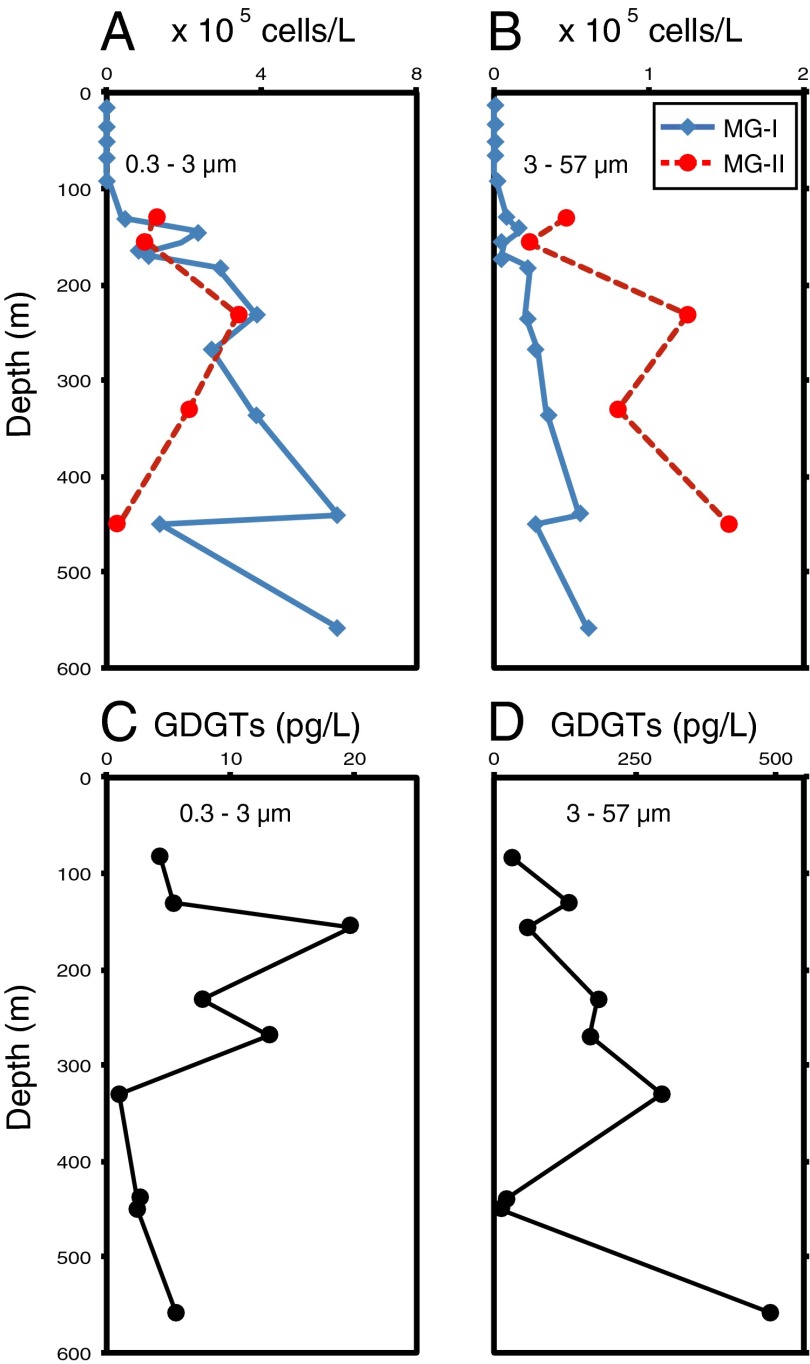
The near-binary nature of the archaeal community in the sample set enabled us to estimate MG-II cell densities using MG-I rRNA qPCR data and the relative abundances of MG-I and MG-II determined from metagenomes (Eq. 1 and Fig. 2 A and B). Overall, MG-I and MG-II distributions were similar to those reported in a previous NPSG study (22). Two independent metrics we report here also support the dominance of MG-II in the upper ocean: At 83 m, MG-I were not detectable by qPCR (Fig. 2 A and B), and Euryarchaeota comprised >94% of the 16S rRNA and archaeal metagenomic read annotations (MGAs) (Table 1).
Fig. 2.
Inferred cell densities of Thaumarchaeota and Euryarchaeota and core GDGT lipid concentrations in small (A and C) and large (B and D) size fractions of suspended particulate matter in the NPSG. MG-I cell densities (solid blue line) were determined by qPCR, and were below limits of detection at depths <100 m. MG-II cell densities (dashed red line) were inferred from archaeal community composition as determined from MG-I cell densities using SSU rRNA gene representation in metagenomic datasets (Eq. 1). Mean values of replicate GDGT analyses are shown; error bars indicating the range of measurements are smaller than the data points. Note variable scales on x axes.
Both MG-I and MG-II DNAs were more abundant in the 0.3- to 3-µm than in the 3- to 57-µm size fraction, consistent with a predominantly free-living existence (FISH) (21, 24). Both size class MG-I profiles (Fig. 2 A and B) showed a gradual increase in cell density with depth, and suggest a small percentage (10–15%) of a predominantly free-living thaumarchaeotal population was captured on the large filters.
MG-II depth profiles showed more pronounced differences between size fractions, with MG-II cells captured on the large filter ranging from 2% to 90% of the total MG-II detected at a given depth. This variability may be related to differences in cell size and the extent of particle association between ecotypes, as MG-II have been reported to be pleomorphic and larger than MG-I cells (44).
Tetraether Lipid Distributions and Partitioning.
Core GDGT molecules (SI Appendix, Fig. S1) are composed of two isoprenoidal C40 hydrocarbon cores linked to glycerol by ether bonds at each end. Many GDGTs contain internal cyclopentyl moieties, and crenarchaeol has an additional six-membered ring (SI Appendix, Fig. S1). In intact polar lipid (IPL) GDGTs, glycerol is linked to polar head groups, often hexose moieties. Because polar head groups have been found to degrade rapidly after cell death (45), IPLs in sediments have been proposed as indicators of live biomass. It has been presumed that only a small proportion of GDGTs in live cells exist as free core GDGTs, but how the relative proportion of these lipids varies with environmental conditions, taxa, or growth states, is not well constrained.
We measured core GDGTs in lipid extracts using high-performance liquid chromatography−atmospheric pressure chemical ionization (APCI) mass spectrometry (46, 47). In the small size fraction samples, free core GDGT concentrations were 1–20 pg/L and peaked at 157 m. Free core GDGTs (Fig. 2 C and D) were most abundant in the large size fraction, where their concentrations ranged from 16 to 490 pg/L. They reached an upper water column maximum at 331 m, but the highest concentration was measured at 559 m. Similar partitioning, with a higher proportion of core GDGTs detected in the larger size fraction, was reported in a study of size-fractionated SPM collected in Puget Sound (48). Notably, concentrations of core GDGTs (including crenarchaeol) in the 83-m samples dominated by MG-II (94–100% Euryarchaeota) were similar to those in deeper (>300 m) samples that had high MG-I representation (Fig. 2 C and D).
Intact Polar GDGTs.
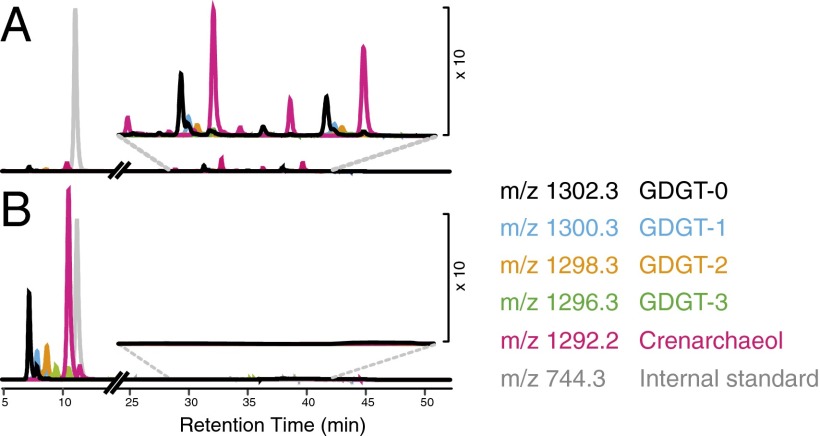
Several series of very late eluting peaks were prominent in extracted ion chromatograms of masses corresponding to GDGTs 0–3 and crenarchaeol in the small size fraction samples from 131 and 559 m (Fig. 3 and SI Appendix, Fig. S7). Within each series, the order of retention times was identical to that of commonly detected free GDGTs. Because they eluted at the stage of analysis at which HPLC eluents are most polar, we postulated that these peaks represented IPL GDGTs that lost their head groups during in-source fragmentation in the mass spectrometer. A hydrolysis experiment supported the IPL GDGT hypothesis: After the extracts were treated with hydrochloric acid (49), the late-eluting peaks disappeared and the peak areas of core GDGTs increased (Fig. 3 and SI Appendix, Fig. S7). This difference was most pronounced in the 131-m sample, in which concentrations of IPL-bound GDGTs were 123 pg/L, exceeding those of free core GDGTs (5 pg/L) by more than an order of magnitude. Euryarchaeotal DNAs were predominant in this sample (74% euryarchaeotal DNAs, 26% thaumarchaotal). At 559 m, the depth at which Thaumarchaeota reached a maximum, IPL-bound GDGT concentrations were 21 pg/L. Comparisons with samples in which IPL GDGTs had previously been reported using established techniques (50) suggested a large contribution from monoglycosyl GDGTs (SI Appendix, Fig. S1) at both 131 and 559 m. Identification of monoglycosyl crenarchaeol was confirmed on an accurate-mass quadrupole time-of-flight mass spectrometer (51) (SI Appendix, Fig. S8).
Fig. 3.
HPLC-APCI-MS composite extracted ion chromatograms (EIC) of total lipid extract (TLE) from 131 m, 0.3–3 µm size fraction sample before (A) and after (B) acid hydrolysis. Colored traces are EICs of m/z values of individual core GDGTs and the hydrolysis-resistant internal standard. Late eluting peaks in A represent three putative series of core GDGTs released from IPL GDGTs by fragmentation in the APCI source. After acid hydrolysis, late-eluting peaks disappeared and the peak area of core GDGTs increased, supporting this interpretation.
Marine Euryarchaeota Contain Crenarchaeol and Other Tetraether Lipids.
Two lines of convergent evidence indicate that MG-II Euryarchaeota are a major source of tetraether lipids in the NPSG and, by extrapolation, throughout the open ocean and in coastal waters where Euryarchaeota are found (3, 23–25). First, core GDGTs—including crenarchaeol—were detected in both SPM size fractions throughout the profile, most notably at depths at which MG-II dominated the archaeal population (Fig. 2). The 83-m samples composed nearly exclusively of Euryarchaeota (95–100%) contained as much core GDGTs as deeper samples with higher MG-I representation (Fig. 2 C and D). Because the turnover time of cells in the upper water column is on the order of a few days (52), the free core GDGTs we measured must have been derived from living or recently living archaea found at these depths. IPL-bound GDGTs (53–55) provide a second line of evidence for a MG-II contribution to the tetraether lipid pool. Concentrations of these lipids at 559 m (dominated by MG-I) were less than one-fifth those measured at 131 m (dominated by MG-II). It therefore appears unlikely that MG-I, less abundant at 131 m than at 559 m, could be the sole source of the much higher IPL GDGT concentrations detected in the shallower sample.
Similar distributions of MG-I and MG-II were reported in a previous metagenomic study using samples collected in 2002–2004 (22); this continuity suggests that the pattern of MG-II dominance in the upper open ocean is not ephemeral. To improve the temporal resolution of the historical record, we queried datasets from four metagenomic profiles and one metatranscriptomic profile generated from the NPSG against a comprehensive database that included a draft MG-II metagenome (43). Subsequent analysis of the relative contributions of Euryarchaeota and Thaumarchaeota to total protein-coding reads (SI Appendix, Fig. S9) revealed a pattern similar to that seen in the amplicon dataset. Thus, fosmid, amplicon, and pyrosequenced metagenomic datasets from samples spanning nearly a decade all indicated that euryarchaeotal dominance in the upper ocean is a persistent feature of the NPSG.
Our finding that Euryarchaeota are a major source of GDGTs in the water column is consistent with the observation that tetraether lipid biosynthesis—a broadly but not universally distributed trait among Euryarchaeota—is common among members of the Thermoplasmatales (56), the most closely related cultivars to MG-II. Moreover, biosynthesis of crenarchaeol by MG-II was previously proposed on the basis of water column GDGT and DNA distributions (37–40). A role for the cyclohexyl moiety in maintaining tetraether membrane fluidity at low temperatures, possibly enabling the expansion of thermophilic MG-I from hydrothermal environments to the open ocean, was postulated (17) but later contradicted by reports of crenarchaeol in hot springs (57, 58) and thermophilic MG-I isolates (18, 59).
Although crenarchaeol biosynthesis has been confirmed in MG-I isolates (e.g., ref. 16), determining its sources—and sources of other tetraether lipids—in environments in which multiple archaeal groups coexist has been difficult. Studies examining correlations between crenarchaeol and MG-I SSU rRNA or amoA gene transcript copy numbers in the water column may have been complicated by an early focus on core rather than IPL GDGTs, and the use of filters with different pore sizes for collection of lipids and DNA, and for FISH counts. The absence of an apparent correlation between community composition and the relative proportions of individual GDGTs detected in this study (Table 1) suggests that it may not be possible to structurally differentiate marine planktonic GDGT pools in the case of mixed MG-I and MG-II archaeal populations.
Geological and Ecological Implications.
Together with the widespread predominance of MG-II in the epipelagic zone, where exported GDGT signals have been reported to originate (60, 61), our evidence that MG-II are a source of tetraether lipids suggests that a significant proportion of GDGTs delivered to sediments are likely synthesized by Euryarchaeota. This finding has important implications for past and future oceanographic and organic geochemical studies, and may necessitate a reinterpretation of archaeal tetraether lipid distributions in the geologic record.
The paleotemperature proxy TEX86 (TetraEther indeX of tetraethers containing 86 carbon atoms) relies on an empirical relationship between sea surface temperature (SST) and relative abundances of GDGTs in sediments that have been presumed to be derived from planktonic Thaumarchaeota (9, 62). Although TEX86 is now widely used in paleoceanography and core top calibration studies have found correlations between temperatures inferred using it and SSTs to be strong (62), mechanisms underlying the proxy are not well understood (63). One critical but as yet unresolved issue is how SST signals can be closely reflected in the membrane lipid composition of planktonic Thaumarchaeota, which are typically less abundant in epipelagic waters, where much exported organic matter originates. Tetraether lipid biosynthesis by MG-II provides a possible solution: If temperature affects the membrane lipid composition of MG-II Euryarchaeota, MG-II GDGTs are more likely than those of deeper-dwelling MG-I Thaumarchaeota to reflect sea surface temperatures. Significant physiological differences between the groups, however, may complicate this scenario. Diverse heterotrophic MG-II populations may be influenced by very different ecological factors than those that impact chemolithoautotrophic MG-I populations. Heterotrophic MG-II populations likely respond to changes in organic matter availability and primary productivity, and their abundances have been observed to vary seasonally in a variety of habitats (24, 64).
Because TEX86 relies on relative rather than absolute abundances of GDGTs to estimate SST, it may in theory be immune to changes in GDGT flux or changes in community composition, provided that temperature is the only factor influencing archaeal membrane lipid composition in the ocean. This assumption is probably too simplistic, because the tetraether lipid composition of cultured archaea is known to vary with factors such as pH (65).
Analyses of one sample in this study (440 m, small size fraction) underscored the fact that our understanding of factors influencing relative proportions of GDGTs in the water column remains incomplete. This deep sample showed a GDGT pattern atypical of marine waters (SI Appendix, Fig. S10), with GDGT-2 more abundant than GDGT-0 or crenarchaeol—yet the archaeal community was dominated by the same MG-I OTU as at other depths with similar in situ temperatures. Neither temperature nor community composition seem likely explanations for this unusual GDGT signal, although the full genetic diversity of MG-I ecotypes is not entirely reflected in rRNA data.
A mixed biological origin for GDGTs detected in marine sediments may confound efforts to use universally applicable paleoceanographic tools. A recent attempt to detect marine carbon isotope excursions by measuring the carbon isotopic composition (δ13C) of GDGTs (66) made the assumption that these compounds were synthesized by autotrophic Thaumarchaeota and argued that the δ13C of GDGTs must covary with that of the dissolved inorganic carbon (DIC) substrate. If, however, heterotrophic Euryarchaeota also contributed to the GDGT pool (as now appears likely), the isotopic signal would be mixed, reflecting the δ13C of their organic carbon substrates as well as that of DIC.
Conclusions
The results presented should help guide future studies of planktonic archaea and their lipids in the marine water column and sediments. The enrichment and isolation of representative strains of MG-II would greatly aid attempts to understand its ecology, physiology, and membrane lipid composition, but further cultivation-independent studies are also likely to yield new information about this important group of marine plankton.
Isotopically labeled tracer experiments have high potential for probing MG-II populations. Early microcosm studies using 13C labeled bicarbonate amendments (67) were valuable in predicting the physiology of MG-I communities before any representatives were isolated. More recently, 13C tracer experiments have been successful in attributing the production of specific IPL classes to their autotrophic/heterotrophic bacterial sources (68). Similar experiments using labeled organic substrates may prove equally informative regarding the physiology and membrane lipid composition of MG-II Euryarchaeota.
In addition to better-constrained GDGT sources, a more complete knowledge of export processes and residence times will be necessary to interrelate the distributions of GDGT-containing archaea in the water column with their lipid profiles in the sedimentary record. Sediment trap studies, coupled with lipid and nucleic acid analyses of SPM and sediments, may reveal the depths from which exported GDGTs originate. Such investigations, conducted in both open ocean and coastal environments, in transects, profiles, and time series, should also help constrain the balance of euryarchaeotal/thaumarchaeotal lipids exported to the seafloor in different oceanic provinces and seasons. Further analyses of size-fractionated SPM, combining archaeal community and lipid composition measurements with compound-specific radiocarbon analyses and thorium-based measures of particle flux, may also yield more insight into the mechanisms and timescales of GDGT export.
Materials and Methods
Sample Collection.
Size-fractionated SPM samples (0.3–3 µm and 3–57 µm) were collected in September 2011 (SI Appendix, Figs. S2 and S3; details in SI Appendix) near Hawaii Ocean Time-series Station ALOHA using in situ pumps fitted with glass fiber filters.
DNA Methods.
DNA was extracted from filter sections corresponding to 20–70 L of seawater (SI Appendix, Fig. S4). The qPCR assays used primers MGI_751F; 5′-GTCTACCAGAACAYGTTC and MGI_956R; 5′-HGGCGTTGACTCCAATTG and generally followed the program of Mincer et al. (24) (details in SI Appendix). We amplified the V1−V3 variable region of the archaeal SSU rRNA gene using the primers 20F 5′-TCCGGTTGATCCYGCCRG-3′ (23) and a barcoded reverse primer 519R, 5′-GGTDTTACCGCGGCKGCT-3′ (69). Pooled amplicons were sequenced using the 454 Genome Sequencer (Roche; details in SI Appendix). We used AmpliconNoise (70) to remove 454 sequencing errors, QIIME (41) to demultiplex the dataset and cluster sequences into OTUs at the 97% identity level using OTU-picking scripts based on uclust (71), and the Ribosomal Database Project classifier to assign taxonomy for 4,900 OTUs. A matrix of predominant OTUs was generated; darkest gray indicates the most abundant OTU in a single sample (Fig. 1). A maximum likelihood tree of the 20 most abundant OTUs was constructed in ARB (72) and visualized in ITOL (73) (see SI Appendix).
Metagenomic data and analyses.
A subset of DNA samples were sequenced on an Illumina MiSeq Sequencer. Historical contributions of MG-I and MG-II to total protein-coding reads were determined from metagenomic datasets generated from SPM profiles collected on Hawaii Ocean Time-series (HOT) research cruises 179 [March 2006 (74)], 186 [October 2006 (75)], 194 (August 2007), and 215 (September 2009) and a metatranscriptomic dataset from HOT 186.
Illumina datasets.
Reads were curated with Trimmomatic (76) and joined using PandaSeq (77); unjoined read pairs were retained and tracked as single records. Trimmed reads were filtered using SortMeRNA (78) to identify those containing rRNA sequences (SI Appendix); taxonomy was determined by searching against SILVA release 115 using lastal (79).
454 datasets.
Duplicate reads (100% identity) were removed and FASTA files divided into rRNA and non-rRNA reads by using BLASTn (top hit, e-values <0.0001, bit scores >50) to compare with a reference database comprised of combined SILVA release 106 SSU and LSU databases and a 5S database (80). Using BLASTx or LAST (79), non-rRNA reads were compared with a reference database (SI Appendix) and their taxonomic affiliation determined in MEGAN (81). The proportion of thaumarchaeal and euryarchaeal reads was calculated as a percent of total reads assigned at or below phylum level.
Lipid extraction, hydrolysis, and analysis.
Lipids were extracted from glass fiber filters containing SPM following a modified Bligh−Dyer protocol (50) (details in SI Appendix). A C46 tetraether (82), which lacks a polar head group and is unmodified by acid hydrolysis, was used as an internal standard. Aliquots of TLE were subjected to acid hydrolysis to cleave head groups of intact polar GDGTs, converting them to core GDGTs (50) (SI Appendix). Core GDGTs were analyzed by high-performance liquid chromatography−APCI mass spectrometry (47, 48) (details in SI Appendix). IPLs in TLEs of the 0.3- to 3-µm size fraction of SPM from 131 and 559 m were analyzed by ultra-high-performance liquid chromatography−electrospray ionization mass spectrometry(51) (SI Appendix).
Cell density inference.
We inferred MG-II cell densities based on qPCR and community composition (SI Appendix) using Eq. 1, where n = number and f = fraction:
| [1] |
Supplementary Material
Acknowledgments
We thank the captain and crew of the R/V Kilo Moana, BioLINCs chief scientists Julie Robidart and Sam Wilson; John Waterbury for help with sampling; Jessica Bryant, Florence Schubotz, and Asuncion Martinez for advice on analytical methods, data processing, and helpful discussions; Tsultrim Palden and Rachel Barry for sequencing; and Carolyn Colonero for laboratory assistance. This work was supported by a Grants 492.01 and 3777 from the Gordon and Betty Moore Foundation (to E.F.D.), a gift from the Agouron Institute (to E.F.D. and R.E.S.), and National Science Foundation Science and Technology Center Award EF0424599 (to E.F.D.). This work is a contribution of the Center of Microbial Ecology: Research and Education.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper has been deposited in the NCBI sequence read archive, www.ncbi.nlm.nih.gov/sra (accession no. SRP042245, SRX553024, SRX553023, SRX556067, and SRX556088).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1409439111/-/DCSupplemental.
References
- 1.DeLong EF. Archaea in coastal marine environments. Proc Natl Acad Sci USA. 1992;89(12):5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fuhrman JA, McCallum K, Davis AA. Novel major archaebacterial group from marine plankton. Nature. 1992;356(6365):148–149. doi: 10.1038/356148a0. [DOI] [PubMed] [Google Scholar]
- 3.DeLong EF. Oceans of archaea. Am Soc Microbiol News. 2003;69(10):503–511. [Google Scholar]
- 4.Schouten S, Hopmans E, Sinninghe Damsté J. The organic geochemistry of glycerol dialkyl glycerol tetraether lipids: A review. Org Geochem. 2013;54:19–61. [Google Scholar]
- 5.Sinninghe Damsté JS, et al. Distribution of membrane lipids of planktonic Crenarchaeota in the Arabian Sea. Appl Environ Microbiol. 2002;68(6):2997–3002. doi: 10.1128/AEM.68.6.2997-3002.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wuchter C, Schouten S, Boschker HTS, Sinninghe Damsté JS. Bicarbonate uptake by marine Crenarchaeota. FEMS Microbiol Lett. 2003;219(2):203–207. doi: 10.1016/S0378-1097(03)00060-0. [DOI] [PubMed] [Google Scholar]
- 7.Pearson A, McNichol AP, Benitez-Nelson BC, Hayes JM, Eglinton TI. Origins of lipid biomarkers in Santa Monica Basin surface sediment: A case study using compound-specific Δ14C analysis. Geochim Cosmochim Acta. 2001;65(18):3123–3137. [Google Scholar]
- 8.Ingalls AE, et al. Quantifying archaeal community autotrophy in the mesopelagic ocean using natural radiocarbon. Proc Natl Acad Sci USA. 2006;103(17):6442–6447. doi: 10.1073/pnas.0510157103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schouten S, Hopmans EC, Schefuß E, Sinninghe Damsté JS. Distributional variations in marine crenarchaeotal membrane lipids: A new tool for reconstructing ancient sea water temperatures? Earth Planet Sci Lett. 2002;204(1-2):265–274. [Google Scholar]
- 10.Hopmans EC, et al. A novel proxy for terrestrial organic matter in sediments based on branched and isoprenoid tetraether lipids. Earth Planet Sci Lett. 2004;224(1-2):107–116. [Google Scholar]
- 11.Kuypers MM, et al. Massive expansion of marine archaea during a mid-Cretaceous oceanic anoxic event. Science. 2001;293(5527):92–95. doi: 10.1126/science.1058424. [DOI] [PubMed] [Google Scholar]
- 12.Brochier-Armanet C, Boussau B, Gribaldo S, Forterre P. Mesophilic Crenarchaeota: Proposal for a third archaeal phylum, the Thaumarchaeota. Nat Rev Microbiol. 2008;6(3):245–252. doi: 10.1038/nrmicro1852. [DOI] [PubMed] [Google Scholar]
- 13.Könneke M, et al. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature. 2005;437(7058):543–546. doi: 10.1038/nature03911. [DOI] [PubMed] [Google Scholar]
- 14.Santoro AE, Buchwald C, McIlvin MR, Casciotti KL. Isotopic signature of N2O produced by marine ammonia-oxidizing archaea. Science. 2011;333(6047):1282–1285. doi: 10.1126/science.1208239. [DOI] [PubMed] [Google Scholar]
- 15.Metcalf WW, et al. Synthesis of methylphosphonic acid by marine microbes: A source for methane in the aerobic ocean. Science. 2012;337(6098):1104–1107. doi: 10.1126/science.1219875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schouten S, et al. Intact membrane lipids of “Candidatus Nitrosopumilus maritimus,” a cultivated representative of the cosmopolitan mesophilic group I Crenarchaeota. Appl Environ Microbiol. 2008;74(8):2433–2440. doi: 10.1128/AEM.01709-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Damsté JS, Schouten S, Hopmans EC, van Duin AC, Geenevasen JA. Crenarchaeol: The characteristic core glycerol dibiphytanyl glycerol tetraether membrane lipid of cosmopolitan pelagic crenarchaeota. J Lipid Res. 2002;43(10):1641–1651. doi: 10.1194/jlr.m200148-jlr200. [DOI] [PubMed] [Google Scholar]
- 18.de la Torre JR, Walker CB, Ingalls AE, Könneke M, Stahl DA. Cultivation of a thermophilic ammonia oxidizing archaeon synthesizing crenarchaeol. Environ Microbiol. 2008;10(3):810–818. doi: 10.1111/j.1462-2920.2007.01506.x. [DOI] [PubMed] [Google Scholar]
- 19.Fuhrman JA, Davis AA. Widespread Archaea and novel Bacteria from the deep sea as shown by 16S rRNA gene sequences. Mar Ecol Prog Ser. 1997;150:275–285. [Google Scholar]
- 20.López-García P, Moreira D, López-López A, Rodríguez-Valera F. A novel haloarchaeal-related lineage is widely distributed in deep oceanic regions. Environ Microbiol. 2001;3(1):72–78. doi: 10.1046/j.1462-2920.2001.00162.x. [DOI] [PubMed] [Google Scholar]
- 21.Karner MB, DeLong EF, Karl DM. Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature. 2001;409(6819):507–510. doi: 10.1038/35054051. [DOI] [PubMed] [Google Scholar]
- 22.DeLong EF, et al. Community genomics among stratified microbial assemblages in the ocean’s interior. Science. 2006;311(5760):496–503. doi: 10.1126/science.1120250. [DOI] [PubMed] [Google Scholar]
- 23.Massana R, Murray AE, Preston CM, DeLong EF. Vertical distribution and phylogenetic characterization of marine planktonic Archaea in the Santa Barbara Channel. Appl Environ Microbiol. 1997;63(1):50–56. doi: 10.1128/aem.63.1.50-56.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mincer TJ, et al. Quantitative distribution of presumptive archaeal and bacterial nitrifiers in Monterey Bay and the North Pacific Subtropical Gyre. Environ Microbiol. 2007;9(5):1162–1175. doi: 10.1111/j.1462-2920.2007.01239.x. [DOI] [PubMed] [Google Scholar]
- 25.Ottesen EA, et al. Pattern and synchrony of gene expression among sympatric marine microbial populations. Proc Natl Acad Sci USA. 2013;110(6):E488–E497. doi: 10.1073/pnas.1222099110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herfort L, et al. Variations in spatial and temporal distribution of Archaea in the North Sea in relation to environmental variables. FEMS Microbiol Ecol. 2007;62(3):242–257. doi: 10.1111/j.1574-6941.2007.00397.x. [DOI] [PubMed] [Google Scholar]
- 27.Bano N, Ruffin S, Ransom B, Hollibaugh JT. Phylogenetic composition of Arctic Ocean archaeal assemblages and comparison with Antarctic assemblages. Appl Environ Microbiol. 2004;70(2):781–789. doi: 10.1128/AEM.70.2.781-789.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Galand PE, Casamayor EO, Kirchman DL, Potvin M, Lovejoy C. Unique archaeal assemblages in the Arctic Ocean unveiled by massively parallel tag sequencing. ISME J. 2009;3(7):860–869. doi: 10.1038/ismej.2009.23. [DOI] [PubMed] [Google Scholar]
- 29.Murray AE, et al. Seasonal and spatial variability of bacterial and archaeal assemblages in the coastal waters near Anvers Island, Antarctica. Appl Environ Microbiol. 1998;64(7):2585–2595. doi: 10.1128/aem.64.7.2585-2595.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.López-García P, López-López A, Moreira D, Rodríguez-Valera F. Diversity of free-living prokaryotes from a deep-sea site at the Antarctic Polar Front. FEMS Microbiol Ecol. 2001;36(2-3):193–202. doi: 10.1016/s0168-6496(01)00133-7. [DOI] [PubMed] [Google Scholar]
- 31.Alonso-Sáez L, Andersson A, Heinrich F, Bertilsson S. High archaeal diversity in Antarctic circumpolar deep waters. Environ Microbiol Rep. 2011;3(6):689–697. doi: 10.1111/j.1758-2229.2011.00282.x. [DOI] [PubMed] [Google Scholar]
- 32.Hugoni M, et al. Structure of the rare archaeal biosphere and seasonal dynamics of active ecotypes in surface coastal waters. Proc Natl Acad Sci USA. 2013;110(15):6004–6009. doi: 10.1073/pnas.1216863110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Belmar L, Molina V, Ulloa O. Abundance and phylogenetic identity of archaeoplankton in the permanent oxygen minimum zone of the eastern tropical South Pacific. FEMS Microbiol Ecol. 2011;78(2):314–326. doi: 10.1111/j.1574-6941.2011.01159.x. [DOI] [PubMed] [Google Scholar]
- 34.Michotey V, et al. Spatio-temporal diversity of free-living and particle-attached prokaryotes in the tropical lagoon of Ahe atoll (Tuamotu Archipelago) and its surrounding oceanic waters. Mar Pollut Bull. 2012;65(10-12):525–537. doi: 10.1016/j.marpolbul.2012.01.009. [DOI] [PubMed] [Google Scholar]
- 35.Herndl GJ, et al. Contribution of Archaea to total prokaryotic production in the deep Atlantic Ocean. Appl Environ Microbiol. 2005;71(5):2303–2309. doi: 10.1128/AEM.71.5.2303-2309.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu M, Xiao T, Wu Y, Zhou F, Zhang W. Temporal distribution of the archaeal community in the Changjiang Estuary hypoxia area and the adjacent East China Sea as determined by denaturing gradient gel electrophoresis and multivariate analysis. Can J Microbiol. 2011;57(6):504–513. doi: 10.1139/w11-037. [DOI] [PubMed] [Google Scholar]
- 37.DeLong EF. Archaeal mysteries of the deep revealed. Proc Natl Acad Sci USA. 2006;103(17):6417–6418. doi: 10.1073/pnas.0602079103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Turich C, et al. Lipids of marine Archaea: Patterns and provenance in the water-column and sediments. Geochim Cosmochim Acta. 2007;71(13):3272–3291. [Google Scholar]
- 39.Schouten S, van der Meer MTJ, Hopmans EC, Damsté JSS. Comment on “Lipids of marine Archaea: Patterns and provenance in the water column and sediments” by Turich et al. (2007) Geochim Cosmochim Acta. 2008;72(21):5342–5346. [Google Scholar]
- 40.Turich C, et al. Reply to the Comment by S. Schouten, M. van der Meer, E. Hopmans, and J.S. Sinninghe Damsté on “Lipids of marine Archaea: Patterns and provenance in the water column.”. Geochim Cosmochim Acta. 2008;72(21):5347–5349. [Google Scholar]
- 41.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frigaard N-U, Martínez A, Mincer TJ, DeLong EF. Proteorhodopsin lateral gene transfer between marine planktonic Bacteria and Archaea. Nature. 2006;439(7078):847–850. doi: 10.1038/nature04435. [DOI] [PubMed] [Google Scholar]
- 43.Iverson V, et al. Untangling genomes from metagenomes: Revealing an uncultured class of marine Euryarchaeota. Science. 2012;335(6068):587–590. doi: 10.1126/science.1212665. [DOI] [PubMed] [Google Scholar]
- 44.DeLong EF, Taylor LT, Marsh TL, Preston CM. Visualization and enumeration of marine planktonic archaea and bacteria by using polyribonucleotide probes and fluorescent in situ hybridization. Appl Environ Microbiol. 1999;65(12):5554–5563. doi: 10.1128/aem.65.12.5554-5563.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.White DC, Davis WM, Nickels JS, King JD, Bobbie RJ. Determination of the sedimentary microbial biomass by extractible lipid phosphate. Oecologia. 1979;40(1):51–62. doi: 10.1007/BF00388810. [DOI] [PubMed] [Google Scholar]
- 46.Liu X-L, Summons RE, Hinrichs K-U. Extending the known range of glycerol ether lipids in the environment: Structural assignments based on tandem mass spectral fragmentation patterns. Rapid Commun Mass Spectrom. 2012;26(19):2295–2302. doi: 10.1002/rcm.6355. [DOI] [PubMed] [Google Scholar]
- 47.Hopmans EC, Schouten S, Pancost RD, van der Meer MT, Sinninghe Damsté JS. Analysis of intact tetraether lipids in archaeal cell material and sediments by high performance liquid chromatography/atmospheric pressure chemical ionization mass spectrometry. Rapid Commun Mass Spectrom. 2000;14(7):585–589. doi: 10.1002/(SICI)1097-0231(20000415)14:7<585::AID-RCM913>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 48.Ingalls A, Huguet C. Distribution of intact and core membrane lipids of archaeal glycerol dialkyl glycerol tetraethers among size-fractionated particulate organic matter in Hood Canal, Puget Sound. Appl Environ Microbiol. 2012;78(5):1480–1490. doi: 10.1128/AEM.07016-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lipp JS, Hinrichs K-U. Structural diversity and fate of intact polar lipids in marine sediments. Geochim Cosmochim Acta. 2009;73(22):6816–6833. [Google Scholar]
- 50.Sturt HF, Summons RE, Smith K, Elvert M, Hinrichs K-U. Intact polar membrane lipids in prokaryotes and sediments deciphered by high-performance liquid chromatography/electrospray ionization multistage mass spectrometry—new biomarkers for biogeochemistry and microbial ecology. Rapid Commun Mass Spectrom. 2004;18(6):617–628. doi: 10.1002/rcm.1378. [DOI] [PubMed] [Google Scholar]
- 51.Wörmer L, Lipp JS, Schröder JM, Hinrichs K-U. Application of two new LC-ESI-MS methods for improved detection of intact polar lipids (IPLs) in environmental samples. Org Geochem. 2013;59:10–21. [Google Scholar]
- 52.Jones DR, Karl DM, Laws EA. Growth rates and production of heterotrophic bacteria and phytoplankton in the North Pacific subtropical gyre. Deep-Sea Research I. 1996;43(10):1567–1580. [Google Scholar]
- 53.Biddle JF, et al. Heterotrophic Archaea dominate sedimentary subsurface ecosystems off Peru. Proc Natl Acad Sci USA. 2006;103(10):3846–3851. doi: 10.1073/pnas.0600035103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lipp JS, Morono Y, Inagaki F, Hinrichs K-U. Significant contribution of Archaea to extant biomass in marine subsurface sediments. Nature. 2008;454(7207):991–994. doi: 10.1038/nature07174. [DOI] [PubMed] [Google Scholar]
- 55.Schubotz F, Wakeham SG, Lipp JS, Fredricks HF, Hinrichs K-U. Detection of microbial biomass by intact polar membrane lipid analysis in the water column and surface sediments of the Black Sea. Environ Microbiol. 2009;11(10):2720–2734. doi: 10.1111/j.1462-2920.2009.01999.x. [DOI] [PubMed] [Google Scholar]
- 56.Langworthy TA, Smith PF, Mayberry WR. Lipids of Thermoplasma acidophilum. J Bacteriol. 1972;112(3):1193–1200. doi: 10.1128/jb.112.3.1193-1200.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pearson A, et al. Nonmarine crenarchaeol in Nevada hot springs. Appl Environ Microbiol. 2004;70(9):5229–5237. doi: 10.1128/AEM.70.9.5229-5237.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pitcher A, Schouten S, Sinninghe Damsté JS. In situ production of crenarchaeol in two California hot springs. Appl Environ Microbiol. 2009;75(13):4443–4451. doi: 10.1128/AEM.02591-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pitcher A, et al. Crenarchaeol dominates the membrane lipids of Candidatus Nitrososphaera gargensis, a thermophilic group I.1b Archaeon. ISME J. 2010;4(4):542–552. doi: 10.1038/ismej.2009.138. [DOI] [PubMed] [Google Scholar]
- 60.Wuchter C, Schouten S, Wakeham SG, Sinninghe Damsté JS. Temporal and spatial variation in tetraether membrane lipids of marine Crenarchaeota in particulate organic matter: Implications for TEX86 paleothermometry. Paleoceanography. 2005;20(3):PA3013. doi: 10.1029/2004PA001110. [DOI] [Google Scholar]
- 61.Wuchter C, Schouten S, Wakeham SG, Sinninghe Damsté JS. Archaeal tetraether membrane lipid fluxes in the northeastern Pacific and the Arabian Sea: Implications for TEX86 paleothermometry. Paleoceanography. 2006;21(4):PA4208. doi: 10.1029/2006PA001279. [DOI] [Google Scholar]
- 62.Kim J-H, Schouten S, Hopmans EC, Donner B, Sinninghe Damsté JS. Global sediment core-top calibration of the TEX86 paleothermometer in the ocean. Geochim Cosmochim Acta. 2008;72(4):1154–1173. [Google Scholar]
- 63.Pearson A, Ingalls AE. Assessing the use of archaeal lipids as marine environmental proxies. Annu Rev Earth Planet Sci. 2013;41:359–384. [Google Scholar]
- 64.Pernthaler A, Preston CM, Pernthaler J, DeLong EF, Amann R. Comparison of fluorescently labeled oligonucleotide and polynucleotide probes for the detection of pelagic marine bacteria and archaea. Appl Environ Microbiol. 2002;68(2):661–667. doi: 10.1128/AEM.68.2.661-667.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Macalady JL, et al. Tetraether-linked membrane monolayers in Ferroplasma spp: A key to survival in acid. Extremophiles. 2004;8(5):411–419. doi: 10.1007/s00792-004-0404-5. [DOI] [PubMed] [Google Scholar]
- 66.Schoon PL, et al. Recognition of Early Eocene global carbon isotope excursions using lipids of marine Thaumarchaeota. Earth Planet Sci Lett. 2013;373:160–168. [Google Scholar]
- 67.Wuchter C, et al. Archaeal nitrification in the ocean. Proc Natl Acad Sci USA. 2006;103(33):12317–12322. doi: 10.1073/pnas.0600756103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Popendorf K, Lomas MW, Van Mooy BAS. Microbial sources of intact polar diacylglycerolipids in the western North Atlantic Ocean. Org Geochem. 2011;42(7):803–811. [Google Scholar]
- 69.Teske A, Sørensen KB. Uncultured archaea in deep marine subsurface sediments: Have we caught them all? ISME J. 2008;2(1):3–18. doi: 10.1038/ismej.2007.90. [DOI] [PubMed] [Google Scholar]
- 70.Quince C, Lanzen A, Davenport RJ, Turnbaugh PJ. Removing noise from pyrosequenced amplicons. BMC Bioinformatics. 2011;12:38. doi: 10.1186/1471-2105-12-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 72.Ludwig W, et al. ARB: A software environment for sequence data. Nucleic Acids Res. 2004;32(4):1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Letunic I, Bork P. Interactive Tree Of Life (iTOL): An online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23(1):127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- 74.Frias-Lopez J, et al. Microbial community gene expression in ocean surface waters. Proc Natl Acad Sci USA. 2008;105(10):3805–3810. doi: 10.1073/pnas.0708897105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Martínez A, Tyson GW, Delong EF. Widespread known and novel phosphonate utilization pathways in marine bacteria revealed by functional screening and metagenomic analyses. Environ Microbiol. 2010;12(1):222–238. doi: 10.1111/j.1462-2920.2009.02062.x. [DOI] [PubMed] [Google Scholar]
- 76.Lohse M, et al. RobiNA: A user-friendly, integrated software solution for RNA-Seq-based transcriptomics. Nucleic Acids Res. 2012;40(Web Server issue):W622-7. doi: 10.1093/nar/gks540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Masella AP, Bartram AK, Truszkowski JM, Brown DG, Neufeld JD. PANDAseq: Paired-end assembler for illumina sequences. BMC Bioinformatics. 2012;13:31. doi: 10.1186/1471-2105-13-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kopylova E, Noé L, Touzet H. SortMeRNA: Fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics. 2012;28(24):3211–3217. doi: 10.1093/bioinformatics/bts611. [DOI] [PubMed] [Google Scholar]
- 79.Kiełbasa SM, Wan R, Sato K, Horton P, Frith MC. Adaptive seeds tame genomic sequence comparison. Genome Res. 2011;21(3):487–493. doi: 10.1101/gr.113985.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Szymanski M, Barciszewska MZ, Barciszewski J, Erdmann VA. 5S Ribosomal RNA data bank. Nucleic Acids Res. 1999;27(1):158–160. doi: 10.1093/nar/27.1.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Huson DH, Mitra S, Ruscheweyh HJ, Weber N, Schuster SC. Integrative analysis of environmental sequences using MEGAN4. Genome Res. 2011;21(9):1552–1560. doi: 10.1101/gr.120618.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Huguet C, et al. An improved method to determine the absolute abundance of glycerol dibiphytanyl glycerol tetraether lipids. Org Geochem. 2006;37(9):1036–1041. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.