Significance
Genetic analyses require allelic markers, which are often DNA polymorphisms and can be analyzed by using short reads from high-throughput sequencing. Therefore, accuracy in genetic studies depends on correct identification of DNA polymorphic markers, but genomic structural variants increase the complexity of allelic detection and must be carefully accounted for to avoid errors. Here, we examine potential mistakes in single-nucleotide polymorphism calling caused by structural variants and their impact on detecting meiotic recombination events. Our results demonstrate that it is crucial to examine structural variants in genetic analysis with DNA marker detection by using short reads, with implications for a wide range of genetic analyses.
Keywords: structural variation, genotyping, insertions–deletions, high-throughput sequencing
Abstract
DNA polymorphisms are important markers in genetic analyses and are increasingly detected by using genome resequencing. However, the presence of repetitive sequences and structural variants can lead to false positives in the identification of polymorphic alleles. Here, we describe an analysis strategy that minimizes false positives in allelic detection and present analyses of recently published resequencing data from Arabidopsis meiotic products and individual humans. Our analysis enables the accurate detection of sequencing errors, small insertions and deletions (indels), and structural variants, including large reciprocal indels and copy number variants, from comparisons between the resequenced and reference genomes. We offer an alternative interpretation of the sequencing data of meiotic products, including the number and type of recombination events, to illustrate the potential for mistakes in single-nucleotide polymorphism calling. Using these examples, we propose that the detection of DNA polymorphisms using resequencing data needs to account for nonallelic homologous sequences.
DNA polymorphisms are ubiquitous genetic variations among individuals and include single nucleotide polymorphisms (SNPs), insertions and deletions (indels), and other larger rearrangements (1–3) (Fig. 1 A and B). They can have phenotypic consequences and also serve as molecular markers for genetic analyses, facilitating linkage and association studies of genetic diseases, and other traits in humans (4–6), animals, plants, (7–10) and other organisms. Using DNA polymorphisms for modern genetic applications requires low-error, high-throughput analytical strategies. Here, we illustrate the use of short-read next-generation sequencing (NGS) data to detect DNA polymorphisms in the context of whole-genome analysis of meiotic products.
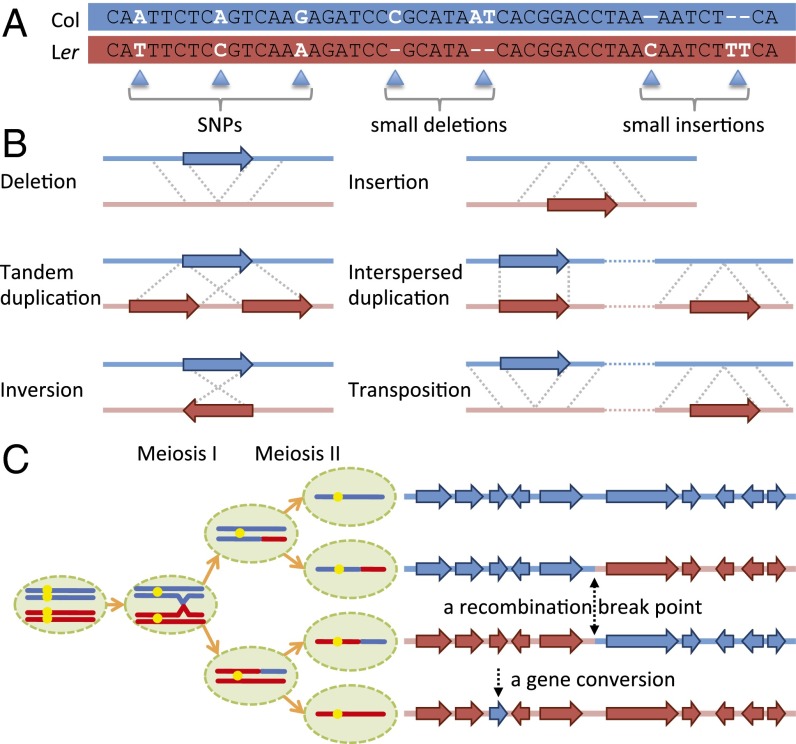
Fig. 1.
(A) SNPs and small indels between two ecotype genomes. (B) Possible types of SVs. Col genotypes are marked in blue and Ler in red. Arrows indicate DNA segments involved in SVs between the two ecotypes. (C) Meiotic recombination events including a CO and a GC (NCO). Centromeres are denoted by yellow dots.
There are many methods for detecting SNPs (11–14) and structural variants (SVs) (15–25), including NGS, which can capture nearly all DNA polymorphisms (26–28). This approach has been widely used to analyze markers in crop species such as rice (29), genes associated with diseases (6, 26), and meiotic recombination in yeast and plants (30, 31). However, accurate identification of DNA polymorphisms can be challenging, in part because short-read sequencing data have limited information for inferring chromosomal context.
Genomes usually contain repetitive sequences that can differ in copy number between individuals (26–28, 31); therefore, resequencing analyses must account for chromosomal context to avoid mistaking highly similar paralogous sequences for polymorphisms. Here, we use recently published datasets to describe several DNA sequence features that can be mistaken as allelic (32, 33) and describe a strategy for differentiating between repetitive sequences and polymorphic alleles. We illustrate the effectiveness of these analyses by examining the reported polymorphisms from the published datasets.
Meiotic recombination is initiated by DNA double-strand breaks (DSBs) catalyzed by the topoisomerase-like SPORULATION 11 (SPO11). DSBs are repaired as either crossovers (COs) between chromosomes (Fig. 1C), or noncrossovers (NCOs). Both COs and NCOs can be accompanied by gene conversion (GC) events, which are the nonreciprocal transfer of sequence information due to the repair of heteroduplex DNA during meiotic recombination. Understanding the control of frequency and distribution of CO and NCO (including GC) events has important implications for human health (including cancer and aneuploidy), crop breeding, and the potential for use in genome engineering. COs can be detected relatively easily by using polymorphic markers in the flanking sequences, but NCO products can only be detected if they are accompanied by a GC event. Because GCs associated with NCO result in allelic changes at polymorphic sites without exchange of flanking sequences, they are more difficult to detect. Recent advances in DNA sequencing have made the analysis of meiotic NCOs more feasible (30–32, 34); however, SVs present a challenge in these analyses. We recommend a set of guidelines for detection of DNA polymorphisms by using genomic resequencing short-read datasets. These measures improve the accuracy of a wide range of analyses by using genomic resequencing, including estimation of COs, NCOs, and GCs.
Results and Discussion
In many species, large-scale SVs often involve identical or highly similar sequences that differ in chromosomal contexts between individuals. Numerous genomic polymorphisms have been reported between two Arabidopsis ecotypes (geographic variants), Columbia (Col, TAIR10 assembly; ref. 35) and Landsberg erecta (Ler), including copy number variants (CNVs), large deletions, insertions, and inversions (31, 36–38). These SVs significantly influence genotyping, particularly SNP calling. Here, we focus on SV involving transposable elements (TEs) and CNVs, because their effects on false positive calling of SNPs are substantial.
Mapping Nonallelic Sequence Reads Causes Artifactual SNP Calls.
SVs between Col and Ler that include TEs (Figs. 1B and 2) create regions of high sequence similarity that map to different (nonallelic) chromosomal positions. When meiotic products from a cross between individuals with large SVs are analyzed by using unassembled short reads from resequencing, reads from the nonreference ecotype (Ler) can be misaligned to nonallelic positions on the Col reference genome, resulting in the misidentification of similar sequences as polymorphisms, including SNPs. Because these sequences are not allelic, they can assort independently if they are on different chromosomes, or be redistributed in the genomes of meiotic progeny by COs if they are on the same chromosome (Fig. 2 A and B). Mistaking these nonallelic sequences as SNPs can lead to the misidentification of gene conversion events (presumably accompanying NCOs), and recent studies have used some of the strategies described here to avoid such mistakes (31, 34).
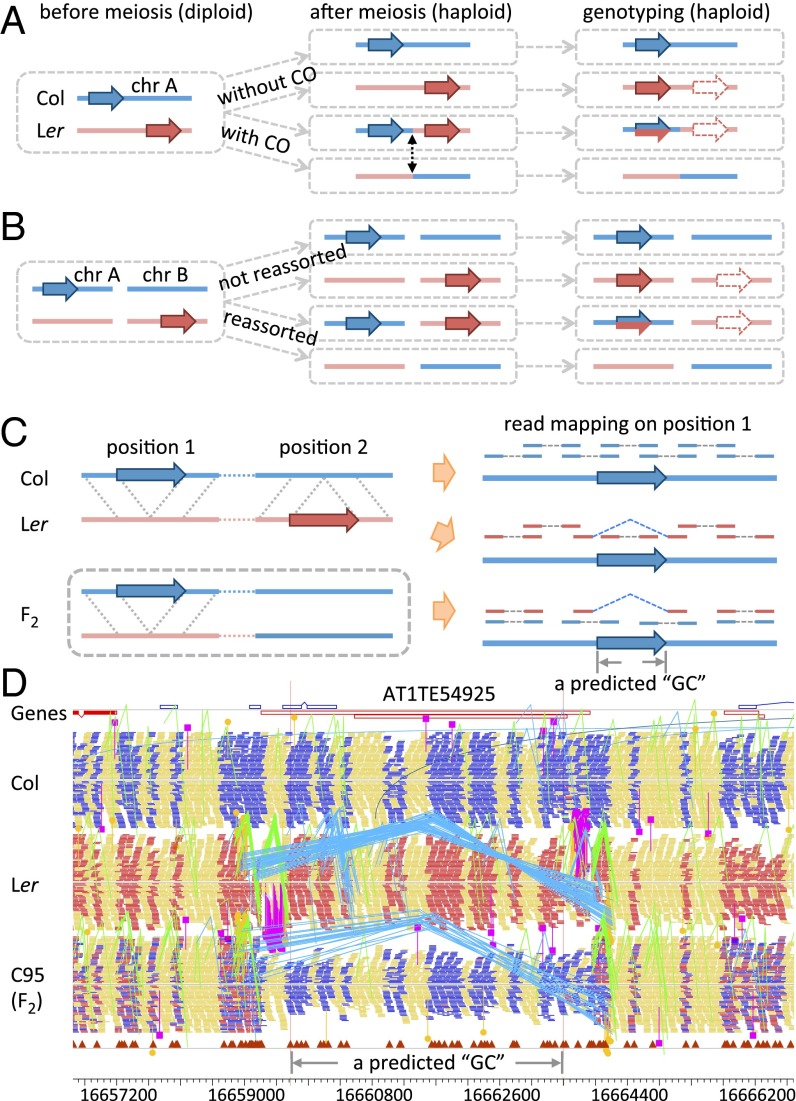
Fig. 2.
Paralogous sequences between two ecotypes and their effects on allele ratio estimation. Redistribution of SV-related paralogous DNA segments in meiotic progenies and consequent genotyping using short read mapping for paralogs on the same chromosome (A), or paralogs on different chromsomes (B). Col and Ler are marked in the same colors as in Fig. 1, whereas dashed arrows indicate actual sites of short reads which were misplaced at nonallelic sites in the reference genome (Col). (C) Read depth, mapping distance, and orientation of PE reads from Col, Ler, and the F2 on the reference genome (Col) around the SV (large blue arrows in both Left and Right). On the Right, PE reads are shown above the Col (blue) sequences; Col read pairs and most Ler read pairs are mapped normally, except those Ler read pairs flanking a deletion, shown here as distantly mapped red reads with blue linkers. Only one chromatid is shown for both Col and Ler. An F2 plant has a heterozygous (Col/Ler) genotype around position 1 and homozygous (Col/Col) around position 2. (D) PE read mapping from Col, Ler, and an F2 plant by Yang et al. (32) in a 9-kb window (16,657,200 ∼16,666,200) on chromosome 1 of Col reference genome. Col reads mapped normally; PE mapping patterns for Ler reads indicate a deletion: A TE is surrounded by a group of reads pairs (linked by blue lines) that mapped farther apart than expected, and another two group of reads pairs (marked by pink lines) mapped to different chromosomes.
Specifically, when nonallelic homologous Col and Ler sequences are redistributed by COs or independent assortment, the failure to recognize such nonallelic sequences can result in false GC events (from heterozygous to Col) in the progeny that lack Ler sequences. For example, if a DNA segment in Col (Fig. 2C, blue arrow) is nearly identical to a nonallelic sequence in Ler (Fig. 2C, red arrow), a CO between the two loci (or an independent assortment between two chromosomes if the segments are unlinked) results in an F2 plant with the second locus having the Col genotype, without the Ler allele. Consequently, short-read NGS data of the F2 plant will yield only the Col genotype at the first locus, which is different from its heterozygous flanking genotype (Col/Ler) (Fig. 2C). In this example, if the nonallelic homologous sequences are not recognized, it is not possible to distinguish between a true GC event and a CO between SVs, resulting in an overestimation of GCs.
Recently, Yang et al. reported that more than 1,000 GC events occurred in each progeny of Arabidopsis meiosis (93,696 GCs for 40 progeny of products of both male and female meiosis) (32). This result is surprising because each GC should originate from a SPO11-mediated DSB, regardless of whether the GC is associated with a CO or a NCO. Although DSBs have not been directly measured in Arabidopsis, several groups have used immunolocalization of recombination proteins such as RAD51 and DMC1 to provide indirect estimates of DSBs ranging from 120 to 220 per meiosis (39–41). Even if every DSB resulted in a GC, the reported levels appear to be much too high. Interestingly, Yang et al. reported that many of the large GC tracts (coconversion of consecutive polymorphisms) occurred at the same locations in multiple meioses (32). Our reanalysis of the sequencing data of two parental and two F2 plants (32) indicated that more than 67% of the reported large GC tracts (2 Kb ∼10 Kb; Table 1) (32) were events from different meioses and were repeatedly detected with exactly the same boundaries. The discordance between the estimated number of DSBs and the reported number of GCs, and the striking repeated occurrence of large GC tracts at the same loci, leads us to seek alternative explanations.
Table 1.
Reinterpretation of GCs by Yang et al. from sequencing data for two F2 plants (C94 and C95) and listed for various sizes
| Factors | 2 Kb ∼10 Kb | 20 bp ∼2 Kb | 2 bp ∼20 bp | 1 bp |
| Transpositions, % | 14.10 | 2.91 | 0.15 | — |
| Copy number variants, % | 30.77 | 12.56 | 0.30 | — |
| Other type of SVs, % | 7.69 | 1.79 | 0.45 | — |
| Misplacement of reads,* % | 8.97 | 16.14 | 4.69 | 2.64 |
| HDRs,† % | 19.23 | 6.95 | 1.06 | — |
| Failure of gap-opening, % | 2.56 | 32.96 | 87.44 | 32.90 |
| Incorrect SNPs,‡ % | _ | 3.36 | 2.12 | 1.98 |
| Correct SNPs but no GCs,§ % | 12.82 | 19.73 | 3.78 | 62.46 |
| Other factors,¶ % | 3.85 | 3.59 | — | — |
| Sum, % | 100 | 100 | 100 | 99.98 |
| Total reported GCs | 78 | 446 | 661 | 9,924 |
To distinguish from the type based on CNVs, this phrase refers to wrongly placing of a few reads, usually insufficient to contribute an extra coverage.
HDRs refer to highly divergent regions with insufficient identities between two ecotypes resulting in low read coverage of Ler. SNPs predicted in these regions lack enough support from sequencing data.
SNPs predicted by Yang et al. (32) are not supported by sufficient Col/Ler genotypic reads.
SNPs predicted by Yang et al. are either consistent with those from 1001 Genomes (35, 36), or supported by resequencing reads of Col/Ler. However, reads from the corresponding F2 plants do not support GCs in these SNP loci.
Refers to false positive “GCs” due to incorrect prediction of CO borders, or the absence of reads mapped to the regions of GCs from the corresponding F2 plant.
Resequencing of Arabidopsis genomes by Yang et al. produces paired-end (PE) reads from both ends of short DNA fragments of similar lengths in the sequencing library. When a genome (e.g., Ler) that has deletions compared with the reference genome (e.g., Col) is resequenced by using PE sequencing, the regions flanking the deleted DNA are adjacent and can be sequenced as PE reads from the same fragment. Such PE reads can be mapped to sites in the reference genome that span a greater distance than expected from the DNA fragment lengths of the sequencing library (Fig. 2C), providing strong support for a deletion in Ler. Furthermore, the deleted Ler sequence might be found at a different (paralogous) genomic location, possibly resulting from historic transpositions. These nonallelic Ler sequences are mapped back to the Col reference, contributing to false SNP calls in subsequent GC analysis. When an F2 plant lacks a paralogous Ler copy because of CO or chromosome reassortment, the heterozygous region would be misrepresented as having the Col genotype (Fig. 2C). The situation is illustrated by an example shown in Fig. 2D: We found a transposable element (TE; AT1TE54925) on chromosome 1 of Col (at nucleotide position 16,659,688–16,664,330 bp) that has a paralog on chromosome 2, but not on chromosome 1, in Ler (at ∼1.3 Mb; ref. 35). An F2 plant, designated C95 by Yang et al., was of the Col genotype for the entire length of chromosome 2, thus lacking the Ler copy of AT1TE54925. As a result, no reads for the Ler version of the TE were mapped to chromosome 1, producing a Col genotype at that locus. However, the regions flanking the TE on chromosome 1 were heterozygous (Col/Ler) at polymorphic markers, leading to the interpretation that the AT1TE54925 locus had experienced a GC. Accounting for limited chromosomal context information for the structural differences between Col and Ler allowed us to identify inappropriate GC calls at this site in 13 of 40 meiotic offspring, including C95 (32).
Accurate detection of meiotic GCs by using polymorphisms requires knowledge of genomic SVs between the two parental genotypes. Because of the complex nature of SVs (large reciprocal indels, CNVs), it is necessary to examine all available sequence features, including read depth, mapping distance and orientation of PE reads, and mapping boundaries revealed by split reads, to determine the types and quality of SVs (15). As described above, unexpectedly long distances between a pair of reads indicate deletions in the resequenced genome (e.g., Ler) relative to the assembled reference genome (Col) (Fig. 2C). As in the Yang et al. data, a cluster of 33 PE reads from Ler were mapped to positions at a distance of 5,100 bp on average, which was significantly longer than the average length of 474 ± 13 bp of the sequenced DNA fragments in this study (P value <<10−3 using the Kolmogorov–Smirnov test). The pattern of distantly mapped reads (Fig. 2D, short bars linked with blue dashed lines) indicates a ∼4.6-Kb deletion of a TE in the Ler genome. However, this region is fully covered by mapped Ler reads, indicating that the Ler genome has this TE, which is on chromosome 2, as supported by the two clusters of reads (marked by pink lines in Fig. 2D) adjacent to the ends of this TE.
To investigate the extent of these SVs, we analyzed the published data (32) and identified 161 sequences (Dataset S1) that mapped to different genomic positions between Col and Ler, affecting >500 Kb of the genome and leading to false positive SNP calls that relied on misplaced short reads from Ler to nonallelic Col positions. More than 14% of large GCs (2 Kb ∼10 Kb) and approximately 3% for shorter GCs (20 bp ∼2 Kb) predicted by Yang et al. (32) were associated with this type of SVs (Table 1).
Artifactual SNPs from CNVs.
Approximately 20% of the Arabidopsis genome (35) is comprised of TE-related sequences, which can vary in copy number and position among ecotypes (42–44). In addition, movement of TEs can cause CNV of nearby sequences (45–47). CNVs of both TE and non-TE sequences can also result in the misidentification of genotypes due to the mismapping of short reads (Fig. 3 A and B). We identified 1,429 CNVs between Col and Ler (Dataset S2), covering 2.7 Mb of the Col genome, and found 30% of large GCs (2 Kb ∼10 Kb) and 12% of shorter GCs (20 bp ∼2 Kb) identified by Yang et al. are located in these regions (Table 1). For example, a sequence has two copies in Ler but only one copy in Col (nucleotides 2,246,169–2,256,074 on chromosome 2; Fig. 3C). The number of reads mapped to the reference Col sequence was significantly higher (P value <<10−3) than those in its flanking regions (Fig. 3C) because reads from the additional Ler copy were mapped to the single Col copy.
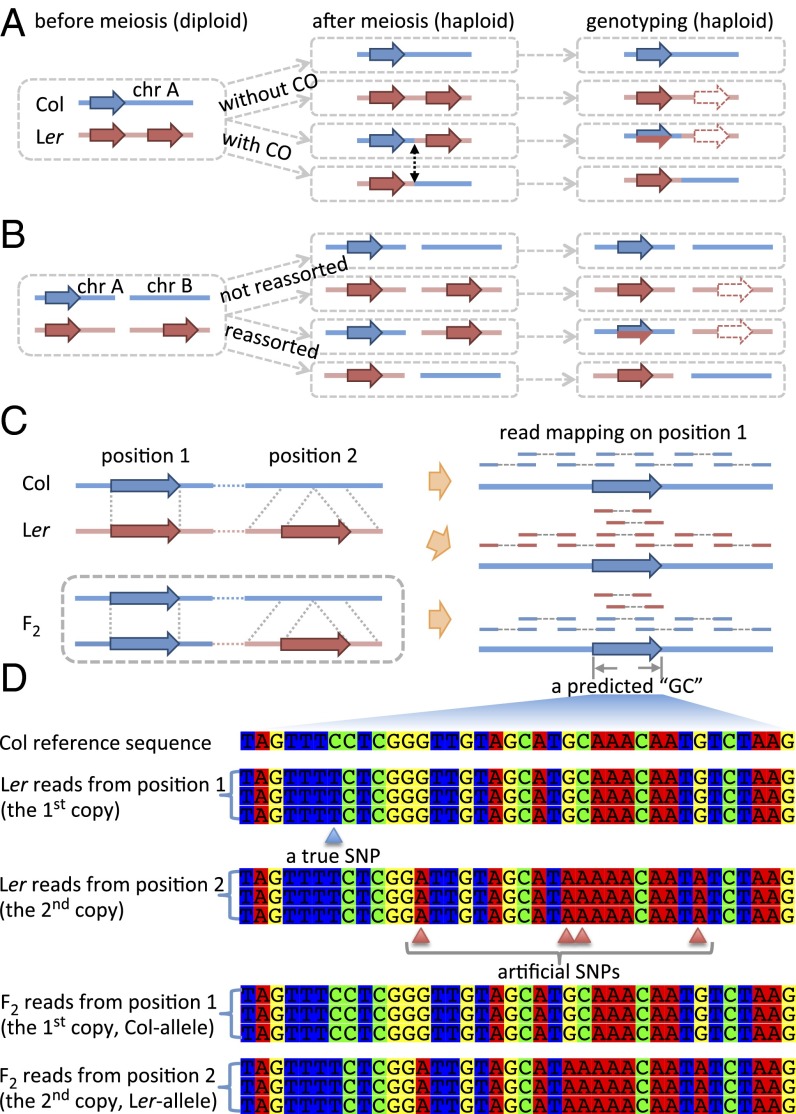
Fig. 3.
PE reads mapping patterns for CNVs and corresponding genotype predictions. Genotyping of SV-related genomic regions in meiotic progenies due to misplacement of short read in the case of CNVs on the same chromosome (A), or CNVs on different chromosomes (B). (C) Mapping pattern of reads from Col, Ler, and the F2 plant on the reference genome. The F2 plant (an example) has two Col-homologous copies and one Ler-homologous copy. Both Ler and the F2 show abnormally higher read depth within the duplicated segment. (D) Both Ler and the F2 could have two or three kinds of “genotypes,” depending on the sequence divergence of the three segments in C, where Col has one copy and Ler has two copies. Comparison of allelic sequences between Col and Ler give one true SNP (in blue), whereas misplacing the nonallelic copy of Ler creates three artifactual SNPs (in red).
When the duplicated copies have diverged slightly, their sequence differences can be mistaken for SNPs, but their true paralogous nature can be definitively recognized because they occur in the homozygous Ler parent, even when there are more than two copies. As shown in Fig. 3D, the sequence of the true Ler allele has only one substitution: from the Col “C” to “T.” However, when reads from a nonallelic duplicate copy were also mapped to the Col site, the four SNP-like differences were used inappropriately to call for GC (32) because these sequence differences were thought to be genotypes different from their flanking regions. An examination of the read depth and mapping distance/orientation of the reported large GCs (2 Kb ∼10 Kb) (32) showed that 30% are associated with loci that show evidence of copy number variation between the two parental ecotypes (Table 1). These results suggested that meiotic COs and independent assortment of chromosomes involving duplicated copies can contribute to the inflation of GC estimates from short reads.
Incomplete Coverage of Tandem Repeats Contributes to Genotyping Errors.
Tandem repeats of DNA sequences, including mono-, di-, tri-, or longer oligonucleotides, are common and may form structures (such as loops and hairpins) (48, 49) that cause DNA polymerase slippage, resulting in frequent copy number changes (small indels). In short-read NGS data, many reads may not span an entire repetitive array and are thus unable to detect small indels. At nucleotides 1,553,129–1,553,140 on chromosome 1 of the Col genome, there is an array of six “TA” dinucleotides (Fig. 4A), but the corresponding array in Ler contains only five. There are two types of reads for this region (1): The 3′ end of the reads terminates within the tandem repeats, uninformative regarding the indel (2); the reads span the array and reveal the indel. The difference in the coverage of the repeat region was interpreted as evidence for two alleles, resulting in a heterozygosity call, even when only one allele was present. These heterozygosity calls were interpreted as GCs if their flanking regions were either homozygous Col or homozygous Ler.
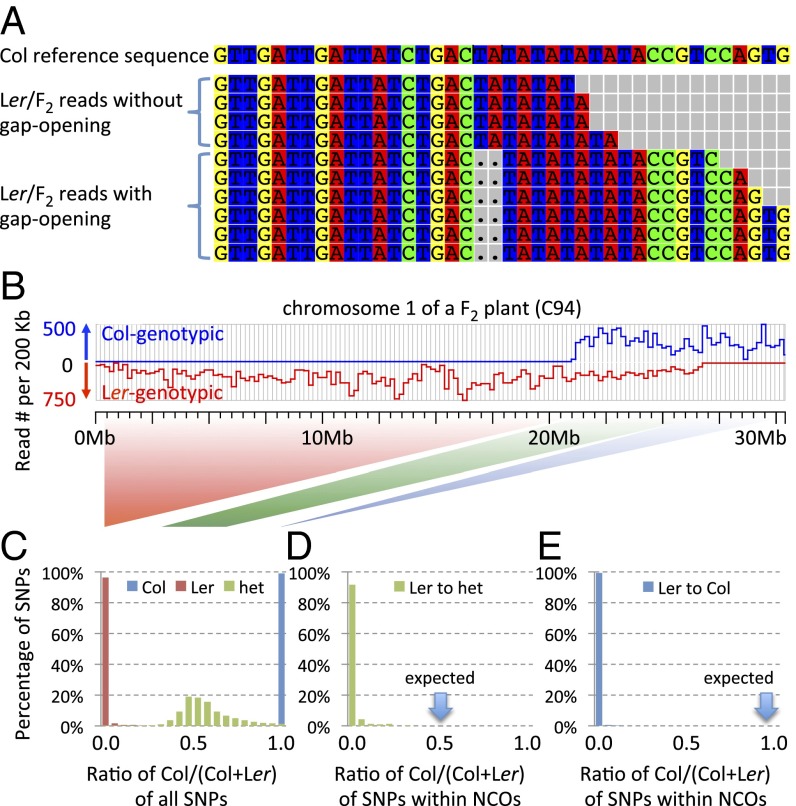
Fig. 4.
(A) Alignment of short reads with small indels in tandem repeat arrays. Gaps are opened only for Ler/F2 reads that span the entire tandem array. (B) Genotypes of chromosome 1 of an F2 plant (C94): Upper blue line indicates the number of reads genotyped as Col per 200-kb window, and the lower red line is the depth of reads genotyped as Ler. (C) Distribution of allele ratio for SNPs from left Ler homozygous region (background in red), middle heterozygous region (background in green), and right Col homozygous region (background in blue). (D). Distribution of allele ratio of SNPs within predicted GCs by Yang et al. as “heterozygous converted from Ler.” Allelic ratio of Col at these SNPs is expected to be approximately 50% (blue arrow). (E) Distribution of allele ratio of SNPs within predicted GCs predicted by Yang et al. as “Col converted from Ler”. The expected ratio (100% of Col-genotypes) is also represented by a blue arrow.
Therefore, only reads that span a repeat array and include some flanking sequences can be reliably used to detect GCs in repetitive DNA. To estimate the number of reported short GCs (2 bp ∼20 bp) (32) associated tandem repeats, we scanned the Col genome by using “Tandem Repeats Finder” (50) and found 948,905 of tandem repeat regions (8% of the genome). Strikingly, more than 87% of short GCs (2 bp ∼20 bp) from two F2 plants (32) were associated with small indels in tandem repeats, although the vast majority (91%) of the genome is covered by neither tandem repeats nor indels. Furthermore, 96% of these GCs were reported as having converted from “Col- or Ler- genotypes” to “heterozygous genotypes.” As noted above, a conversion of homozygous to heterozygous genotypes is consistent with the misinterpretation of reads of differing lengths as different alleles. Reanalysis of reads mapping to the regions of 87% of the reported GCs (2 bp ∼20 bp) indicated no support for heterozygosity or GC if reads that did not span the array were removed (Table 1). Therefore, the vast majority of the short GCs are not supported by the data when using only reads that covered the entire repeat regions and provided unambiguous genotypic evidence.
Sequencing Errors Contribute to Artifactual Alleles.
NGS technologies have error rates of ∼10−3 substitutions per nucleotide or higher (51). Sequence errors can be mistaken as evidence for polymorphism. At a given SNP site, 100× sequencing coverage will yield a 3% (10−3 × 1/3 × 102) chance of observing a read with SNP-like changes due to error. When genome-wide SNPs are examined, many such false SNP calls are expected. To evaluate the effect of sequencing errors on GC prediction, we examined the distribution of the ratio of Col and Ler reads in regions designated as Col, Ler or heterozygous, using the SNP information for chromosome 1 and the read data from an F2 plant (C94) (32). As shown in Fig. 4B, the first 21.2 Mb of the chromosome was genotyped as Ler; as expected, the ratio of Col/(Col+Ler) reads was close to zero (Fig. 4C). Similarly, the average ratio was ∼0.5 for the next 5.5 Mb heterozygous region and ∼1.0 for the last 3.6 Mb region genotyped as Col (Fig. 4C). We then performed the same analysis on SNPs from sites on chromosome 1 that were reported to have GC of 1 bp from Ler to heterozygous (32). Strikingly, the ratio of Col/(Col+Ler) reads at these SNPs was close to 0% (Fig. 4D), indicating that the number of reads called as Col was extremely small and the true genotype at these sites was likely Ler, instead of heterozygous due to GC. Fig. 4E shows an examination of SNPs from the reported “Ler to Col” type of GCs and suggests that these SNPs do not provide support for conversions. Further analysis of all 1-bp GCs from the C94 and C95 F2 plants (32) revealed that 62% lacked sufficient read support for converted genotypes (Table 1). Because the sequencing error rate of NGS is relatively low, reads of a specific SNP allele due to error are rare compared with reads for the correct genotypes. Therefore, evaluation of observed “genotype ratio” followed by a statistical test can greatly reduce false GCs calls due to sequencing errors.
Reanalysis of Reported Data for Potential GCs.
We describe a sequence analysis pipeline for detection of GCs by integrating the filters described above (SI Materials and Methods and Fig. S1). Briefly, polymorphisms between Col and Ler ecotypes including SNPs, small indels, and large SVs were either collected from 1001 Genomes (36, 37) or predicted based on alignment of Col and Ler resequencing reads (32) on the reference genome (TAIR10) (35) by using BWA (52) and inGAP-sv (14). Short reads of C94 and C95 (32) were also mapped by using the same filtering strategy and uniquely mapped reads were genotyped according to polymorphic information. COs were identified by genotyping loci along each chromosome to provide “allelic background” for the identification of GCs. Sequencing depths, allelic ratios (SI Materials and Methods) and large-scale allelic information between adjacent COs were evaluated for all three genomes (Col, Ler, and the F2 plant) for each converted SNP/indel. Candidate GCs were regarded as having insufficient support if they overlapped with SVs or CNVs.
From the data of the two reported F2 plants (32), we identified 11 COs in each of C94 and C95 (diploids resulted from one male and one female meiotic events). Consistent with prior studies, each chromosome had an average of one CO per meiosis (31, 34). Because information on other meiotic products of the same meiosis was not available, potential GC associated with these COs could not be identified. Nevertheless, after apply filtering steps to data from Col, Ler, and F2 plants (SI Materials and Methods), six potential GCs (associated with NCOs) were predicted, five in C94, and one in C95 (Table S1 and Fig. S2). Among them, only one corresponded to a small indel, consistent with the fact that there is an order of magnitude fewer small indels than SNPs in Arabidopsis (31, 33). Directions of GCs were either from “homozygous” to “heterozygous” (Fig. S3) or vice versa (Fig. S4), consistent with the allelic background of the chromosomal region. For example, all three GCs on chromosome 4 of plant C94 were from heterozygous to homozygous (one for Col/Col and two for Ler/Ler), in a background of 93% of the chromosome being Col/Ler. Two of the six GCs predicted in this study were also identified by Yang et al. (32). Optimally, predicted GCs would be validated by using PCR and conventional sequencing, but in this case, the relevant plant material was not available. The small number of GCs detected here is consistent with previous findings (31, 34, 53) and suggests relatively small sizes of the gaps repaired by NCOs, although an underestimation of GCs due to the stringent criteria here cannot be ruled out.
Variations in the Human Genome and Potential Effects on SNP Calling.
To examine the effects of SVs on SNP calling in a nonplant genome by using short reads, we examined the human genome using human chromosome 1 (hg19/GRCh37) (54) as an example. It has 432,854 repeat regions (45.7% of the chromosome), including SINE (37.4%), LINE (28.2%), and other repeats. We compared the HG00656 dataset from the 1000 Genomes Project (33) (∼5× coverage) with the human reference genome (hg19/GRCh37) (54).
As illustrated in Fig. 4A, the human genome also has small indels associated with tandem repeats, with potential problems using short reads at low coverage. Among 58,735 tandem repeats and low-complexity regions, 3,120 have small indels and were covered by reads without gap opening (see an example in Fig. 4A), making it possible for these indels to be interpreted as SNPs when coverage is not high or without proper statistical analysis. In addition, a study (33) of large deletions included 54 deletions in HG00656 (see one example in Fig. S5 A–C), 21 of which contain SNPs compared with the reference and would be considered as homozygous when they are, in fact, hemizygous. The use of these SNPs without consideration of the deletions would affect the outcome of genetic mapping, because the breakpoints of the deletions would be considered recombination points (Fig. S5D). An analysis of the distribution of the 54 deletions among 1,092 individuals from 14 populations (Dataset S3 and Fig. S5E) revealed that 16 of these deletions were detected in each population, indicative of ancient variations. Twelve other deletions were found frequently in American/East Asian/European populations, whereas the remaining 26 were less widely distributed, potentially affecting genetic studies of the relevant populations.
To investigate the influence of nonallelic similar sequences, such as those related to TEs (as illustrated in Fig. 2), we examined the HG00656 dataset by using inGAP-sv to identify complex SVs. On chromosome 1 of the HG00656 dataset, we identified 38 SVs after filtering out low quality ones: 24 of these SVs corresponded to sequences on chromosome 1 of HG00656 but in the “decoy genome” (named “hs37d5”) of the reference (54). The decoy genome contains 4,715 contigs totaling 35 Mb, including viral sequences, unassembled genomic segments, or de novo assembled sequences from other human genome projects. Thus, SVs uncovered here could be genome variations or reflect incomplete assembly of the reference. These 24 segments ranged in size from 1 to 7.4 Kb, covering 61 Kb of the decoy genome and included 133 nucleotide differences, which would be misidentified as “SNPs” when SVs are not considered. We also identified 82 duplications in HG00656, mostly tandem repeats within introns or intergenic regions. The mapping of two or more such nonallelic similar sequences to the same site would result in false “heterozygosity.” Our analyses indicate that human genomes contain a large number of variations that can potentially affect erroneous SNP calling if not accounted for properly.
Conclusions
Whole genome resequencing is now feasible for a variety of studies, many of which involve the analysis of sequence variants as genetic markers. It is important to correctly identify nonallelic sequence variants to avoid mistaking them as alleles. When the genomes being analyzed have indels, CNVs, and other types of SVs in comparison with the reference genome, short reads of nonallelic sequences can originate from a different location in the resequenced genome and be misinterpreted as polymorphisms. If such false SNPs are included, frequency measurements will be unreliable. False SNPs can be minimized by using PE reads to reveal SVs between the newly sequenced and reference genomes. In addition, reanalysis of the parental genomes of genetic crosses can uncover slightly divergent duplicates and avoid calling the variant duplicates within an individual as alleles between individuals. In outcrossing species in which individuals are heterozygous for most alleles, this analysis should reveal more than two kinds of reads for sequences with two or more similar copies, thus highlighting the need to distinguish nonallelic variants from the allelic ones.
Our reanalysis of the recently reported resequencing data for Arabidopsis meiotic recombination provides strong evidence that most of the reported GCs can be explained by the presence of highly similar but nonallelic DNA segments in the Ler genome (nonreference and unassembled) and the redistribution of such nonallelic sequence by meiotic COs or independent assortment. In addition, restricting the analysis of short GCs to stringently unambiguous genotypes drastically reduced the GC number. Therefore, there is compelling evidence that GCs are a less frequent outcome of meiotic recombination in Arabidopsis, consistent with the findings of relatively few GCs per meiosis using independent methods (31, 34, 53).
Materials and Methods
Published short read sequences from Arabidopsis (32) and human (33) were analyzed by using reported methods to detect SVs associated with TEs, CNVs, short indels relating to tandem repeats, and likely sequencing errors. The SVs and other polymorphisms in Arabidopsis were then matched with the position of reported GCs in the study (32) for evaluation. In addition, two F2 genomes were genotyped on each polymorphic locus to identify meiotic recombination events including COs and GCs. See details in SI Materials and Methods.
Supplementary Material
Acknowledgments
This research was supported by Ministry of Sciences and Technology of China 973 Program Grant 2012CB910503 (to J.Q.), National Natural Science Foundation of China Grant 91131007 (to H.M.), US National Science Foundation Grant MCB-1121563 (to G.P.C.), and the biological supercomputing server of Computing Center of Beijing Institutes of Life Science.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1321897111/-/DCSupplemental.
References
- 1.Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nat Rev Genet. 2006;7(2):85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- 2.Sharp AJ, Cheng Z, Eichler EE. Structural variation of the human genome. Annu Rev Genomics Hum Genet. 2006;7:407–442. doi: 10.1146/annurev.genom.7.080505.115618. [DOI] [PubMed] [Google Scholar]
- 3.Mitchell-Olds T, Schmitt J. Genetic mechanisms and evolutionary significance of natural variation in Arabidopsis. Nature. 2006;441(7096):947–952. doi: 10.1038/nature04878. [DOI] [PubMed] [Google Scholar]
- 4.Stankiewicz P, Lupski JR. Genome architecture, rearrangements and genomic disorders. Trends Genet. 2002;18(2):74–82. doi: 10.1016/s0168-9525(02)02592-1. [DOI] [PubMed] [Google Scholar]
- 5.Hurles ME, Dermitzakis ET, Tyler-Smith C. The functional impact of structural variation in humans. Trends Genet. 2008;24(5):238–245. doi: 10.1016/j.tig.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stankiewicz P, Lupski JR. Structural variation in the human genome and its role in disease. Annu Rev Med. 2010;61:437–455. doi: 10.1146/annurev-med-100708-204735. [DOI] [PubMed] [Google Scholar]
- 7.Johanson U, et al. Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science. 2000;290(5490):344–347. doi: 10.1126/science.290.5490.344. [DOI] [PubMed] [Google Scholar]
- 8.Michaels SD, He Y, Scortecci KC, Amasino RM. Attenuation of FLOWERING LOCUS C activity as a mechanism for the evolution of summer-annual flowering behavior in Arabidopsis. Proc Natl Acad Sci USA. 2003;100(17):10102–10107. doi: 10.1073/pnas.1531467100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koornneef M, Alonso-Blanco C, Vreugdenhil D. Naturally occurring genetic variation in Arabidopsis thaliana. Annu Rev Plant Biol. 2004;55:141–172. doi: 10.1146/annurev.arplant.55.031903.141605. [DOI] [PubMed] [Google Scholar]
- 10.Krieger U, Lippman ZB, Zamir D. The flowering gene SINGLE FLOWER TRUSS drives heterosis for yield in tomato. Nat Genet. 2010;42(5):459–463. doi: 10.1038/ng.550. [DOI] [PubMed] [Google Scholar]
- 11.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18(11):1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brockman W, et al. Quality scores and SNP detection in sequencing-by-synthesis systems. Genome Res. 2008;18(5):763–770. doi: 10.1101/gr.070227.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li R, Li Y, Kristiansen K, Wang J. SOAP: Short oligonucleotide alignment program. Bioinformatics. 2008;24(5):713–714. doi: 10.1093/bioinformatics/btn025. [DOI] [PubMed] [Google Scholar]
- 14.Qi J, Zhao F, Buboltz A, Schuster SC. inGAP: An integrated next-generation genome analysis pipeline. Bioinformatics. 2010;26(1):127–129. doi: 10.1093/bioinformatics/btp615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qi J, Zhao F. inGAP-sv: A novel scheme to identify and visualize structural variation from paired end mapping data. Nucleic Acids Res. 2011;39(Web Server issue):W567–575. doi: 10.1093/nar/gkr506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen K, et al. BreakDancer: An algorithm for high-resolution mapping of genomic structural variation. Nat Methods. 2009;6(9):677–681. doi: 10.1038/nmeth.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Korbel JO, et al. PEMer: A computational framework with simulation-based error models for inferring genomic structural variants from massive paired-end sequencing data. Genome Biol. 2009;10(2):R23. doi: 10.1186/gb-2009-10-2-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee S, Hormozdiari F, Alkan C, Brudno M. MoDIL: Detecting small indels from clone-end sequencing with mixtures of distributions. Nat Methods. 2009;6(7):473–474. doi: 10.1038/nmeth.f.256. [DOI] [PubMed] [Google Scholar]
- 19.Hormozdiari F, Alkan C, Eichler EE, Sahinalp SC. Combinatorial algorithms for structural variation detection in high-throughput sequenced genomes. Genome Res. 2009;19(7):1270–1278. doi: 10.1101/gr.088633.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hajirasouliha I, et al. Detection and characterization of novel sequence insertions using paired-end next-generation sequencing. Bioinformatics. 2010;26(10):1277–1283. doi: 10.1093/bioinformatics/btq152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeitouni B, et al. SVDetect: A tool to identify genomic structural variations from paired-end and mate-pair sequencing data. Bioinformatics. 2010;26(15):1895–1896. doi: 10.1093/bioinformatics/btq293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sindi S, Helman E, Bashir A, Raphael BJ. A geometric approach for classification and comparison of structural variants. Bioinformatics. 2009;25(12):i222–i230. doi: 10.1093/bioinformatics/btp208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. Pindel: A pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25(21):2865–2871. doi: 10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abel HJ, et al. SLOPE: A quick and accurate method for locating non-SNP structural variation from targeted next-generation sequence data. Bioinformatics. 2010;26(21):2684–2688. doi: 10.1093/bioinformatics/btq528. [DOI] [PubMed] [Google Scholar]
- 25.Wong K, Keane TM, Stalker J, Adams DJ. Enhanced structural variant and breakpoint detection using SVMerge by integration of multiple detection methods and local assembly. Genome Biol. 2010;11(12):R128. doi: 10.1186/gb-2010-11-12-r128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kidd JM, et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453(7191):56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Graubert TA, et al. A high-resolution map of segmental DNA copy number variation in the mouse genome. PLoS Genet. 2007;3(1):e3. doi: 10.1371/journal.pgen.0030003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Emerson JJ, Cardoso-Moreira M, Borevitz JO, Long M. Natural selection shapes genome-wide patterns of copy-number polymorphism in Drosophila melanogaster. Science. 2008;320(5883):1629–1631. doi: 10.1126/science.1158078. [DOI] [PubMed] [Google Scholar]
- 29.Huang X, et al. Genome-wide association studies of 14 agronomic traits in rice landraces. Nat Genet. 2010;42(11):961–967. doi: 10.1038/ng.695. [DOI] [PubMed] [Google Scholar]
- 30.Qi J, et al. Characterization of meiotic crossovers and gene conversion by whole-genome sequencing in Saccharomyces cerevisiae. BMC Genomics. 2009;10:475. doi: 10.1186/1471-2164-10-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu P, et al. Analysis of Arabidopsis genome-wide variations before and after meiosis and meiotic recombination by resequencing Landsberg erecta and all four products of a single meiosis. Genome Res. 2012;22(3):508–518. doi: 10.1101/gr.127522.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang S, et al. Great majority of recombination events in Arabidopsis are gene conversion events. Proc Natl Acad Sci USA. 2012;109(51):20992–20997. doi: 10.1073/pnas.1211827110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mills RE, et al. 1000 Genomes Project Mapping copy number variation by population-scale genome sequencing. Nature. 2011;470(7332):59–65. doi: 10.1038/nature09708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wijnker E, et al. The genomic landscape of meiotic crossovers and gene conversions in Arabidopsis thaliana. eLife. 2013;2:e01426. doi: 10.7554/eLife.01426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lamesch P, et al. The Arabidopsis Information Resource (TAIR): Improved gene annotation and new tools. Nucleic Acids Res. 2012;40(Database issue):D1202–D1210. doi: 10.1093/nar/gkr1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schneeberger K, et al. Reference-guided assembly of four diverse Arabidopsis thaliana genomes. Proc Natl Acad Sci USA. 2011;108(25):10249–10254. doi: 10.1073/pnas.1107739108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cao J, et al. Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat Genet. 2011;43(10):956–963. doi: 10.1038/ng.911. [DOI] [PubMed] [Google Scholar]
- 38.Long Q, et al. Massive genomic variation and strong selection in Arabidopsis thaliana lines from Sweden. Nat Genet. 2013;45(8):884–890. doi: 10.1038/ng.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mercier R, et al. Two meiotic crossover classes cohabit in Arabidopsis: One is dependent on MER3,whereas the other one is not. Curr Biol. 2005;15(8):692–701. doi: 10.1016/j.cub.2005.02.056. [DOI] [PubMed] [Google Scholar]
- 40.Chelysheva L, et al. AtREC8 and AtSCC3 are essential to the monopolar orientation of the kinetochores during meiosis. J Cell Sci. 2005;118(Pt 20):4621–4632. doi: 10.1242/jcs.02583. [DOI] [PubMed] [Google Scholar]
- 41.Sanchez-Moran E, Santos JL, Jones GH, Franklin FC. ASY1 mediates AtDMC1-dependent interhomolog recombination during meiosis in Arabidopsis. Genes Dev. 2007;21(17):2220–2233. doi: 10.1101/gad.439007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsukahara S, et al. Bursts of retrotransposition reproduced in Arabidopsis. Nature. 2009;461(7262):423–426. doi: 10.1038/nature08351. [DOI] [PubMed] [Google Scholar]
- 43.Initiative TAG. Arabidopsis Genome Initiative Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408(6814):796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 44.Le QH, Wright S, Yu Z, Bureau T. Transposon diversity in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2000;97(13):7376–7381. doi: 10.1073/pnas.97.13.7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiang N, Bao Z, Zhang X, Eddy SR, Wessler SR. Pack-MULE transposable elements mediate gene evolution in plants. Nature. 2004;431(7008):569–573. doi: 10.1038/nature02953. [DOI] [PubMed] [Google Scholar]
- 46.Juretic N, Hoen DR, Huynh ML, Harrison PM, Bureau TE. The evolutionary fate of MULE-mediated duplications of host gene fragments in rice. Genome Res. 2005;15(9):1292–1297. doi: 10.1101/gr.4064205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morgante M, et al. Gene duplication and exon shuffling by helitron-like transposons generate intraspecies diversity in maize. Nat Genet. 2005;37(9):997–1002. doi: 10.1038/ng1615. [DOI] [PubMed] [Google Scholar]
- 48.Djian P. Evolution of simple repeats in DNA and their relation to human disease. Cell. 1998;94(2):155–160. doi: 10.1016/s0092-8674(00)81415-4. [DOI] [PubMed] [Google Scholar]
- 49.Petruska J, Hartenstine MJ, Goodman MF. Analysis of strand slippage in DNA polymerase expansions of CAG/CTG triplet repeats associated with neurodegenerative disease. J Biol Chem. 1998;273(9):5204–5210. doi: 10.1074/jbc.273.9.5204. [DOI] [PubMed] [Google Scholar]
- 50.Benson G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999;27(2):573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Loman NJ, et al. Performance comparison of benchtop high-throughput sequencing platforms. Nat Biotechnol. 2012;30(5):434–439. doi: 10.1038/nbt.2198. [DOI] [PubMed] [Google Scholar]
- 52.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun Y, et al. Deep genome-wide measurement of meiotic gene conversion using tetrad analysis in Arabidopsis thaliana. PLoS Genet. 2012;8(10):e1002968. doi: 10.1371/journal.pgen.1002968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meyer LR, et al. The UCSC Genome Browser database: Extensions and updates 2013. Nucleic Acids Res. 2013;41(Database issue):D64–D69. doi: 10.1093/nar/gks1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.