Significance
Translation arrest regulated by nascent peptides and small cofactors controls expression of important genes, including medically relevant macrolide antibiotic resistance genes. The role of the cofactor for triggering this mechanism has remained enigmatic. Previous studies suggested that extensive interactions between the nascent chain and the antibiotic molecule juxtaposed in the ribosomal exit tunnel were critical for halting translation. However, here we show that the antibiotic induces stalling, even without significant contacts with the peptide, by allosterically altering the peptidyl transferase center. This finding unveils a previously unknown role of cofactors for translation arrest and demonstrates the existence of a functional link between the exit tunnel and the catalytic center of the ribosome.
Keywords: ketolides, solithromycin, azithromycin
Abstract
Translation arrest directed by nascent peptides and small cofactors controls expression of important bacterial and eukaryotic genes, including antibiotic resistance genes, activated by binding of macrolide drugs to the ribosome. Previous studies suggested that specific interactions between the nascent peptide and the antibiotic in the ribosomal exit tunnel play a central role in triggering ribosome stalling. However, here we show that macrolides arrest translation of the truncated ErmDL regulatory peptide when the nascent chain is only three amino acids and therefore is too short to be juxtaposed with the antibiotic. Biochemical probing and molecular dynamics simulations of erythromycin-bound ribosomes showed that the antibiotic in the tunnel allosterically alters the properties of the catalytic center, thereby predisposing the ribosome for halting translation of specific sequences. Our findings offer a new view on the role of small cofactors in the mechanism of translation arrest and reveal an allosteric link between the tunnel and the catalytic center of the ribosome.
Expression of several bacterial and eukaryotic genes is controlled by nascent peptide-dependent programmed translation arrest. In the general scenario, ribosome stalling at an upstream regulatory ORF (uORF) triggers isomerization of the mRNA structure, leading to activation of expression of downstream cistron(s). Translation arrest ensues when a distinctive amino acid sequence (the “stalling domain”) of the growing chain assembled in the ribosomal peptidyl transferase center (PTC) is placed in the nascent peptide exit tunnel (NPET). Ribosome stalling may require additional signals, thereby making this gene control mechanism sensitive to the physiological state of the cell or to the chemical composition of the environment. Often the external signal is a small molecule whose binding to the ribosome renders translation responsive to specific nascent peptides (reviewed in refs. 1, 2). In most of the examined cases of cofactor- and nascent peptide-dependent translation arrest, the binding site of the cofactor in the ribosome is unknown, which hampers understanding of the interplay among the cofactor, the nascent peptide, and the ribosome. The exception is the inducible antibiotic resistance, in which ribosome stalling and gene activation rely on binding of an antibiotic to a well-defined site in the ribosome.
Expression of macrolide resistance genes is triggered by drug-induced ribosome stalling at a defined codon of the uORF (3–5). Macrolides, from the prototype erythromycin (ERY) to the newest macrolide derivatives—ketolides, e.g., solithromycin (SOL)—bind in the NPET at a short distance from the PTC (6–9) (Fig. 1A). When a nascent peptide grows to 4–7 aa, it reaches the site of antibiotic binding and has to negotiate the drug-obstructed NPET aperture. Subsequent events depend on the properties of the nascent chain (3, 10, 11). Although for many proteins the encounter of the peptide with the antibiotic results in peptidyl–tRNA dropoff, the N-termini of certain nascent peptides can bypass the antibiotic. Translation of some of such proteins can be arrested at specific sites within the gene, resulting in formation of a stable stalled complex (11). Such translation arrest defines the role of macrolides as cofactors of programmed ribosome stalling (3, 10–12).
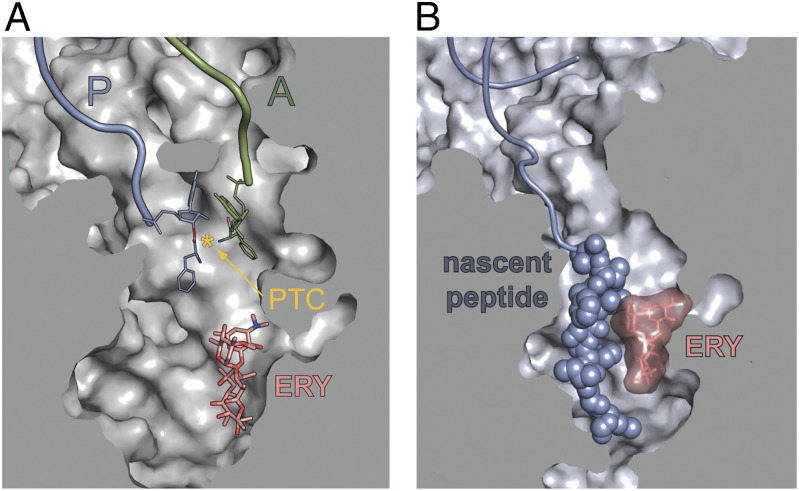
Fig. 1.
Antibiotic and nascent peptide in the ribosomal exit tunnel. (A) The relative locations of the macrolide binding site in the NPET and the PTC active site were rendered by aligning crystallographic structures of Thermus thermophilus 70S ribosome complexed with aminoacylated donor and acceptor tRNA substrates [Protein Data Bank (PDB) ID codes 2WDK, 2WDL (25)] and the vacant ribosome complexed with ERY [PDB ID codes 3OHC, 3OHJ (9)]. The PTC active site, defined as the middistance between the attacking amino group of the acceptor substrate and the carbonyl carbon atom of the donor, is marked by an asterisk. (B) The modeled position of the 9-aa–long ErmCL nascent peptide in the ribosomal tunnel obstructed by ERY (19). In the stalled complex, ErmCL is juxtaposed with the antibiotic in the tunnel.
The regulatory leader peptides of macrolide resistance genes have been classified by the structure of their known or presumed stalling domains (4, 5). Translation of ErmAL1 and ErmCL peptides is arrested after the ribosome has polymerized the 8-aa (ErmAL1) or 9-aa (ErmCL) long nascent chains that carry the C-terminal stalling domains Ile-Ala-Val-Val (IAVV) and Ile-Phe-Val-Ile (IFVI), respectively (12–14). The drug-bound ribosome stalls because it cannot catalyze transfer of the peptide from the P-site peptidyl–tRNA to the A-site aminoacyl–tRNA (12, 14). Importantly, although the N-terminal sequences of these peptides are not critical, the N-terminal segments are required for translation arrest (12). The conservation of the distance of the stalling domain from the N-terminus among peptides of these classes (4) corroborates the importance of the nascent chain length for the arrest. The 8–9-aa long ErmAL1 or ErmCL stalling peptides reach far into the NPET and must be juxtaposed with the antibiotic molecule in the NPET; such apposition has been suggested to play a key role in the mechanism of arrest (12) (Fig. 1B). This view agrees with the strict structural requirements of the macrolide cofactor in which removal or modification of the C3 cladinose abolishes stalling, possibly by disrupting drug–peptide interactions (15).
The resistance leader peptides of the third major class have been studied to a much lesser extent (16–18). These peptides were grouped together based on the presence of the Arg-Leu-Arg (RLR) motif in their sequence (4) (Table S1), although the role of this motif in programmed arrest has not been verified. Intriguingly, in striking contrast to the IAVV and IFVI classes, the placement of the RLR motif within these peptides is highly variable (4).
By analyzing translation arrest controlled by the RLR peptides, we discovered that the N-terminus is dispensable and macrolide antibiotic can block peptide bond formation and halt translation when the nascent chain is only 3-aa long and barely reaches the antibiotic in the NPET. Structural probing and molecular dynamics (MD) modeling showed the existence of an allosteric link between the NPET and the PTC, illuminating how binding of an antibiotic in the NPET predisposes the ribosome for stalling when translating specific amino acid sequences.
Results
The Position of the Conserved RLR Motif Varies in the Leader Peptides of Macrolide Resistance Genes.
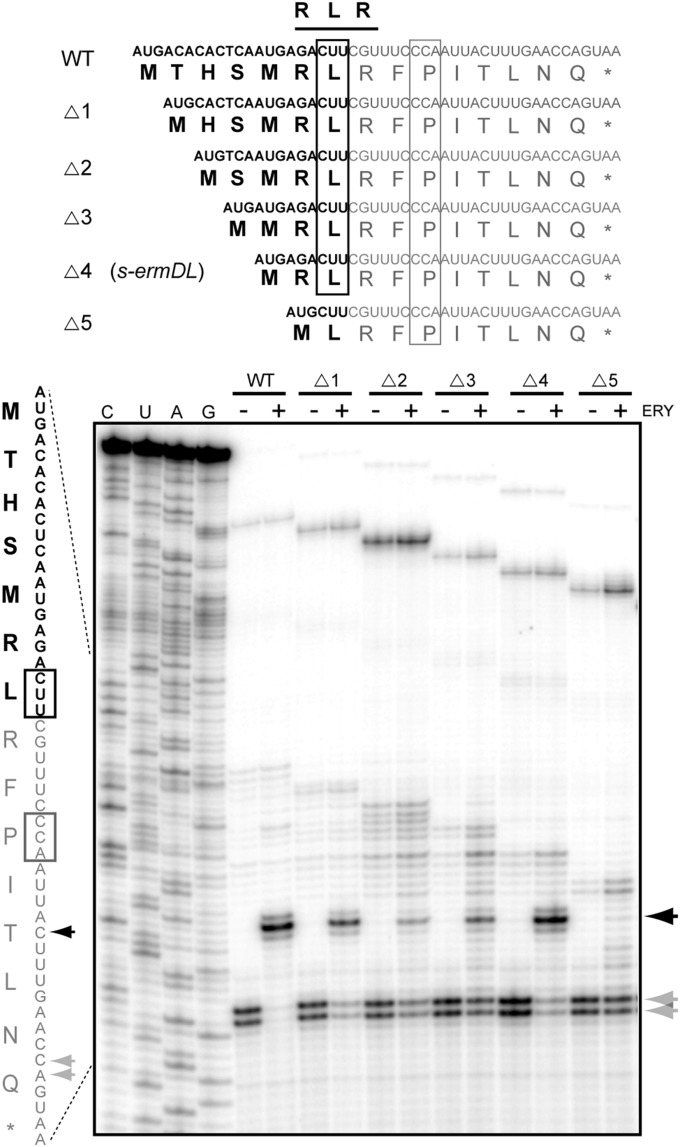
Puzzled by the variable distance of the RLR motif from the N-termini of the leader peptides of macrolide resistance genes (Table S1), we first tested whether the RLR sequence is relevant for programmed translation arrest. Several of the known or putative uORFs with varying placement of the RLR motif were generated by PCR and translated in a cell-free system, and antibiotic-dependent ribosome stalling was assayed by toeprinting. The analysis showed that irrespective of the distance of the RLR motif from the N-terminus, the drug-bound ribosome halts translation upon entrance of the Leu codon of the RLR-coding sequence into the ribosomal P site (Fig. S1). This result not only demonstrated the importance of the RLR motif for translation arrest, but also clearly indicated that for these peptides, the length of the N-terminal region preceding the stalling sequence is not as critical as it is for the IAVV and IFVI regulatory peptides (4, 5). This observation, which was strengthened further by the variability of the amino acid sequences preceding the RLR domain (Table S1), made us wonder whether the N-terminal segment is even required for antibiotic-dependent arrest. Therefore, we introduced progressive truncations in the ermDL regulatory ORF that controls expression of the resistance methyltransferase ErmD. In the presence of ERY, translation of the wild-type 14-aa–long ErmDL peptide is arrested at the Leu (L7) codon, sandwiched between two Arg codons (R6 and R8) (18, 19) (wt in Fig. 2). Remarkably, deletion of one, two, three, or even four codons preceding the ermDL stall site did not prevent drug-dependent ribosome arrest at the Leu codon of the RLR motif (Fig. 2). Although the stalling efficiency was reduced slightly when two or three codons were deleted, the four-codon deletion, resulting in a truncated ORF starting with the sequence Met-Arg-Leu-Arg (MRLR), directed arrest nearly as efficiently as the wild-type ermDL (s-ermDL in Fig. 2). However, when the truncation encompassed the first Arg codon of RLR, arrest essentially was abolished (Fig. 2, two right lanes). These results show that the N-terminal segment of the RLR peptides is dispensable for drug-dependent stalling and that binding of ERY to the NPET can trigger arrest when the nascent chain is only 4-aa (MRLR) or perhaps even 3-aa (MRL) long (see below). In contrast to ErmDL, and consistent with our previous observations (12), removal of the N-terminal segments preceding the IFVI or IAVV domains of the ErmAL1 or ErmCL peptides significantly reduced the efficiency of drug-dependent arrest (Fig. S2), suggesting that the mechanism of ribosome stalling directed by the RLR peptides significantly deviates from that proposed for the regulatory peptides of other classes.
Fig. 2.
The N-terminal segment preceding the RLR motif is dispensable for antibiotic-mediated translation arrest. (Upper) Amino acid sequences of ErmDL peptide (WT) and its N-terminally truncated mutants (∆1–∆5) (the corresponding ORFs are shown above the peptide sequences). (Lower) Toeprinting analysis of ERY-dependent ribosome stalling during cell-free translation of the wild-type and truncated ermDL ORFs. Black arrowheads and box indicate the ERY-induced toeprint signal corresponding to the arrest with the Leu codon in the P site. The nonstalled ribosomes are captured at the downstream Pro codon (gray box and arrowheads) as a result of the presence of mupirocin, an IleRS inhibitor.
Tunnel-Bound Antibiotic Inhibits Formation of Peptide Bond Between MRL Peptide and the Incoming Aminoacyl–tRNA.
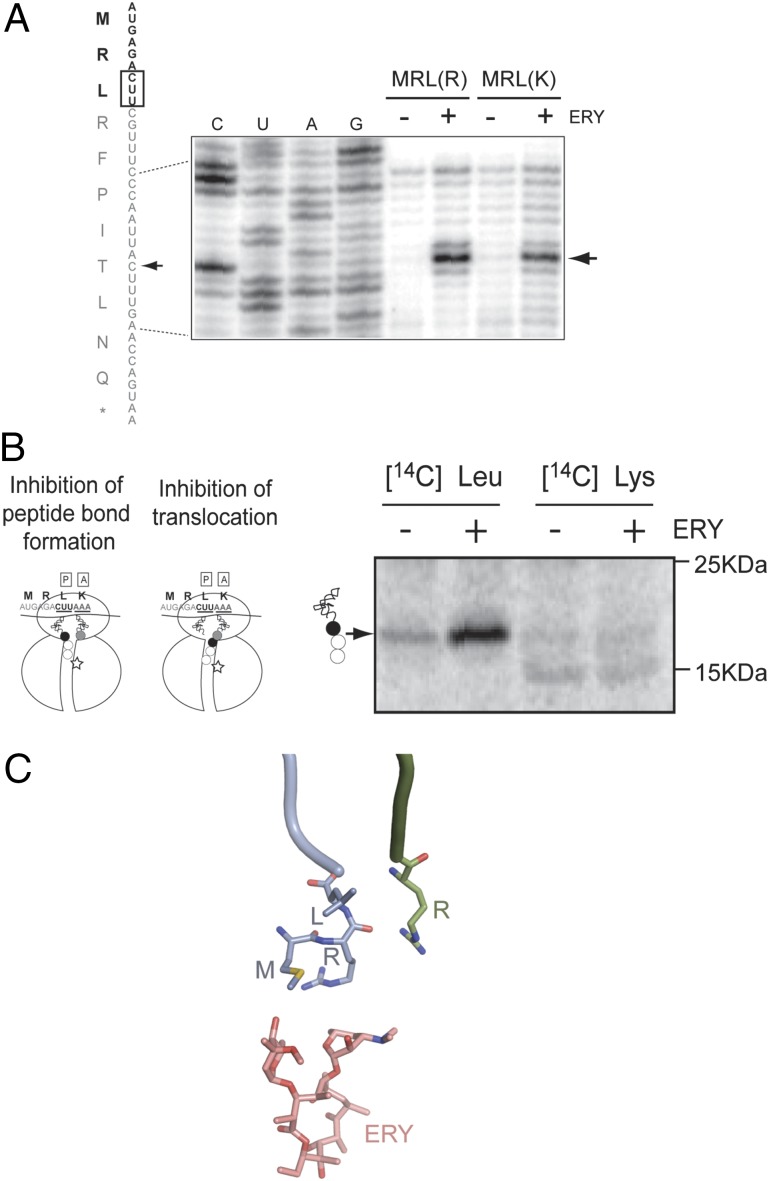
The ribosome arrested at the L3 codon of the truncated s-ermDL ORF might carry either the tripeptide MRL esterified to the P-site tRNALeu (if catalysis of peptide bond formation is impaired) or the tetrapeptide MRLR linked to the A-site tRNAArg (if translocation is inhibited) (Fig. 3B). To distinguish between these scenarios and, thus, deduce the exact length of the stalling peptide, we took advantage of the fact that the RLR peptides seem to tolerate Arg-to-Lys substitutions within the motif (Table S1). Indeed, ERY directed ribosome stalling at the L3 codon of the s-ermDL mRNA regardless of whether it was followed by an Arg (R4) or a Lys (K4) codon (Fig. 3A and Fig. S3). Thus, we used template s-ermDL(MRLK) to determine whether the fourth amino acid (K) is incorporated into the peptidyl–tRNA of the stalled ribosome. In the cell-free translation reaction, incorporation of [14C]-Leu in peptidyl–tRNA was stimulated greatly by ERY (Fig. 3B, lanes 1 and 2), whereas incorporation of [14C]-Lys remained negligible (Fig. 3B, lanes 3 and 4). Therefore, ERY arrests translation of the s-ermDL(MRLK) by blocking the transfer of the P-site tripeptide MRL to the incoming Arg or Lys aminoacyl–tRNA. This result implies that the presence of the drug in the NPET alters the catalytic properties of the PTC when the nascent chain is only three amino acid residues long (Fig. 3C).
Fig. 3.
Antibiotic inhibits the ability of the ribosome carrying the MRL nascent peptide to catalyze peptidyl transfer. (A) Toeprinting analysis of ERY-mediated stalling during translation of s-ermDL(MRLR) or s-ermDL(MRLK) ORFs. Sequencing reactions represent the s-ermDL(MRLR) template. Arrowheads show the toeprint of ribosomes stalled at the Leu codon (boxed). (B, Left) The ribosome stalled at the Leu codon of s-ermDL(MRLK) can carry either MRL tripeptide at the P-site tRNALeu or MRLK tetrapeptide at the A-site tRNALys. The Leu3 and Lys4 of the peptide are shown as black and gray circles, respectively. The codons in the P and A sites of the stalled ribosome are underlined. ERY is shown by a star. (Right) Gel electrophoresis analysis of peptidyl–tRNA accumulated during translation of the s-ermDL(MRLK) ORF in the presence of the indicated radiolabeled amino acids. Migration of markers is indicated on the right. (C) The nascent MRL tripeptide barely reaches the antibiotic in the NPET and cannot be juxtaposed with it. The P-site MRL–tRNALeu (blue) and A-site Arg–tRNAArg (green) were modeled into the structure of the E. coli ribosome–ERY complex and subjected to 2 ns equilibration to avoid immediate structural clashes.
Known Nascent Peptide Ribosomal Sensors Are Not Involved in Drug-Induced Stalling with the MRL Peptide.
Juxtaposition of 8–9-aa long IAVV and IFVI stalling peptides with the antibiotic in the tunnel brings the peptide in contact with specific rRNA sensors in the NPET that help recognize the nascent chains and relay the arrest signal to the PTC. Mutations of such 23S rRNA residues (A2062, A2503, U1782, C2610) dramatically reduce the efficiency of stalling with ErmAL1 or ErmCL (15, 19). Strikingly, neither these mutations nor changes of residues involved in recognition of other stalling peptides (G2583, U2584, U2586, A2587, U2609) (20–22) significantly affected arrest with the MRL peptide (Fig. S4). Although the search for potential MRL sensors has not been exhaustive, the available data are compatible with the possibility that it is the presence of the tunnel-bound antibiotic per se rather than the drug-imposed interaction of the nascent peptide with the tunnel sensors that is critical for the arrest of the ribosome carrying the MRL peptide.
Structurally Diverse Antibiotics Promote Ribosome Stalling with RLR Peptides.
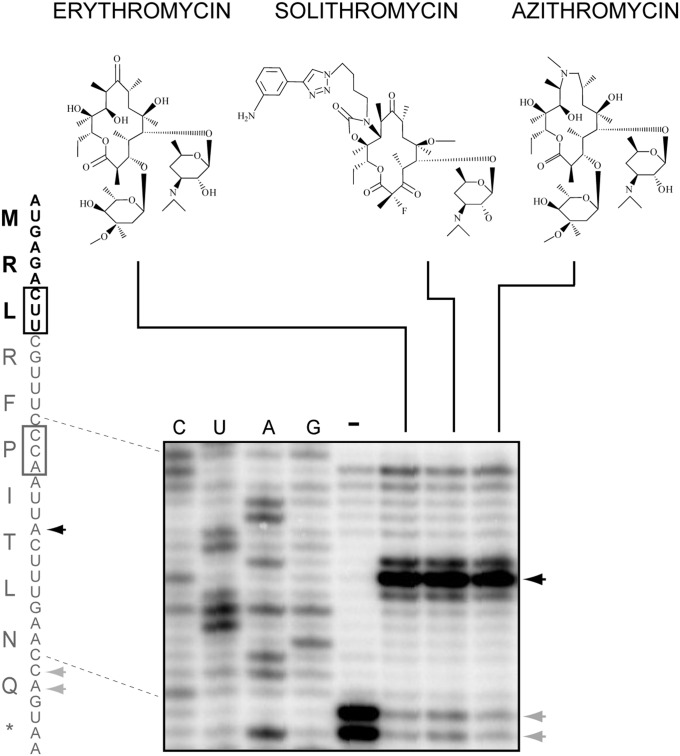
The antibiotic structure is essential for ribosome stalling directed by IAVV or IFVI peptides (12). Removal or modification of the C3 cladinose sugar of ERY abolishes arrest, possibly by disrupting specific interactions between the antibiotic and the 8–9-aa–long nascent chain (15). The MRL peptide, however, is too short to make extensive contact with the antibiotic (Fig. 3C), suggesting that the drug–peptide interface may not play any role in the arrest mechanism. Consistently, we found that stalling with MRL is triggered not only by cladinose-containing ERY, but also by azalide azithromycin (AZI) and even cladinose-lacking ketolide SOL (Fig. 4), which fails to induce arrest with IAVV or IFVI peptides. The high tolerance of MRL-dependent stalling to alterations in the antibiotic structure supports the possibility that the sole presence of the drug in the NPET, rather than its interaction with the nascent chain, is sufficient for stalling at the s-ermDL ORF.
Fig. 4.
Diverse macrolides induce ribosome stalling with the MRL nascent peptide. Toeprinting analysis of ribosomes stalled during translation of the s-ermDL ORF induced by macrolide ERY, ketolide SOL, or azalide AZI. Drug-induced toeprint representing arrest at the Leu codon (black box) is indicated by a black arrowhead. Gray arrowheads and box indicate macrolide-independent translation arrest at the Pro codon due to the presence of mupirocin, which depletes Ile–tRNA in the reaction.
Binding of Antibiotics in the NPET Allosterically Alters the Conformation of the PTC.
A distance of more than 11 Å separates the nearest atom of the tunnel-bound antibiotic (ERY, AZI, or SOL) from the PTC active site (8, 9). Hence, macrolides cannot prevent peptide bond formation by direct steric hindrance unless the peptide induces relocation of the drug to the PTC, which we view as an implausible scenario. Furthermore, the short length of the MRL peptide makes it unlikely that the drug forces it into a nonproductive conformation, as was proposed for the longer stalling peptides (12, 15). Therefore, we hypothesized that binding of the drug in the NPET may allosterically influence the structure, and hence the function, of the PTC.
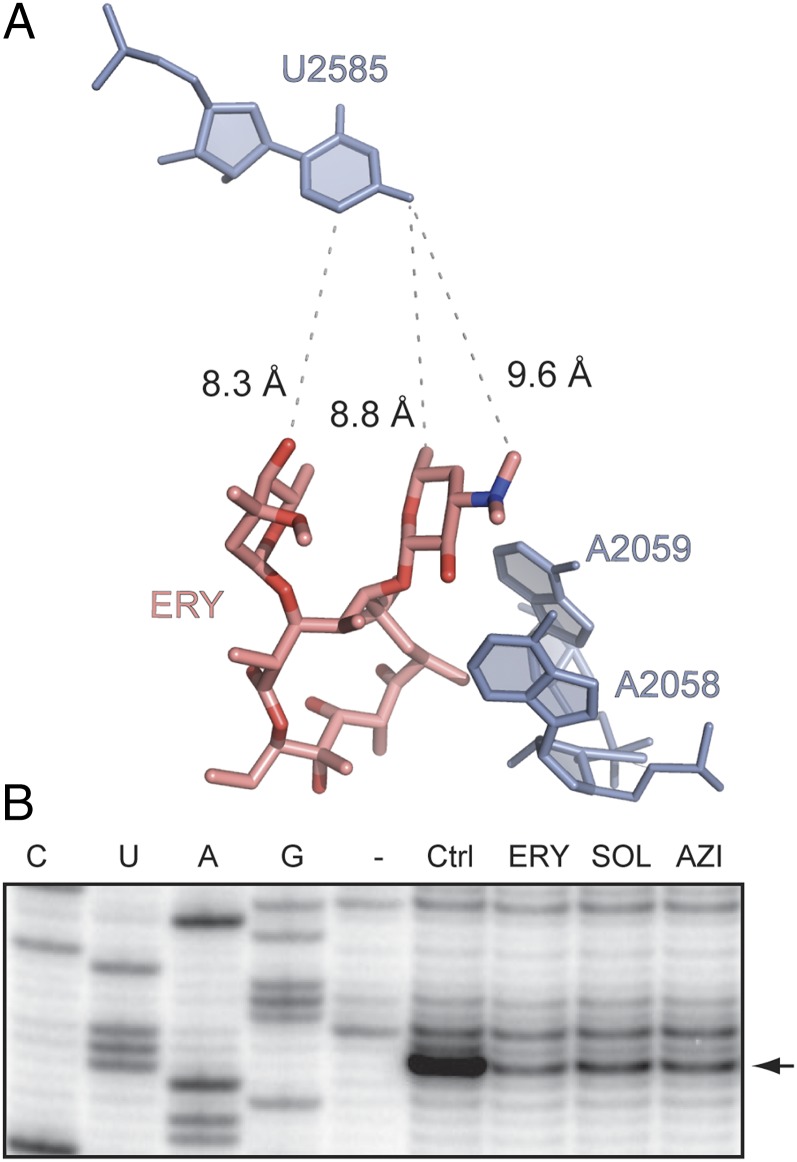
When drug-free or ERY-bound ribosomes were treated with 1-methyl-7-nitroisatoic anhydride (1M7), the reagent used for selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) (23), modification of several rRNA residues was reduced in the presence of antibiotic. Protections of some nucleotides (e.g., A2058 and A2059) were expected because these residues interact directly with the drug. Strikingly, however, modification of the distant U2585 in the PTC also was reduced significantly in response to ERY binding (Fig. 5B). Other macrolide inducers of MRL-dependent arrest (AZI and SOL) had a similar effect on the U2585 reactivity (Fig. 5B). Importantly, U2585 is located in the PTC active site and is critically involved in catalysis of peptide bond formation (24, 25). These results clearly established that the structure of the PTC and, thus, likely its catalytic properties are sensitive to the presence of the antibiotic in the NPET, thereby revealing the existence of an allosteric and functional link between the exit tunnel and the catalytic center.
Fig. 5.
Binding of antibiotics in the NPET affect the distant nucleotide U2585 in the PTC active site. (A) The relative placement of ERY in the NPET and U2585 in the PTC in the crystallographic structure of the E. coli ribosome–ERY complex (8) (PDB ID code 3OFR). Residues A2058 and A2059 in the ERY binding site are also shown. (B) Chemical probing of ribosome–antibiotic complexes with the SHAPE reagent 1M7. Ribosomes were incubated with no antibiotic (Ctrl) or with 50 µM of ERY, SOL, or AZI (see their structures in Fig. 4) and modified with 1M7. The state of modification of U2585 (arrowhead) was assessed by primer extension.
MD Simulations Substantiate the Existence of a Structural Link Between the NPET and the PTC.
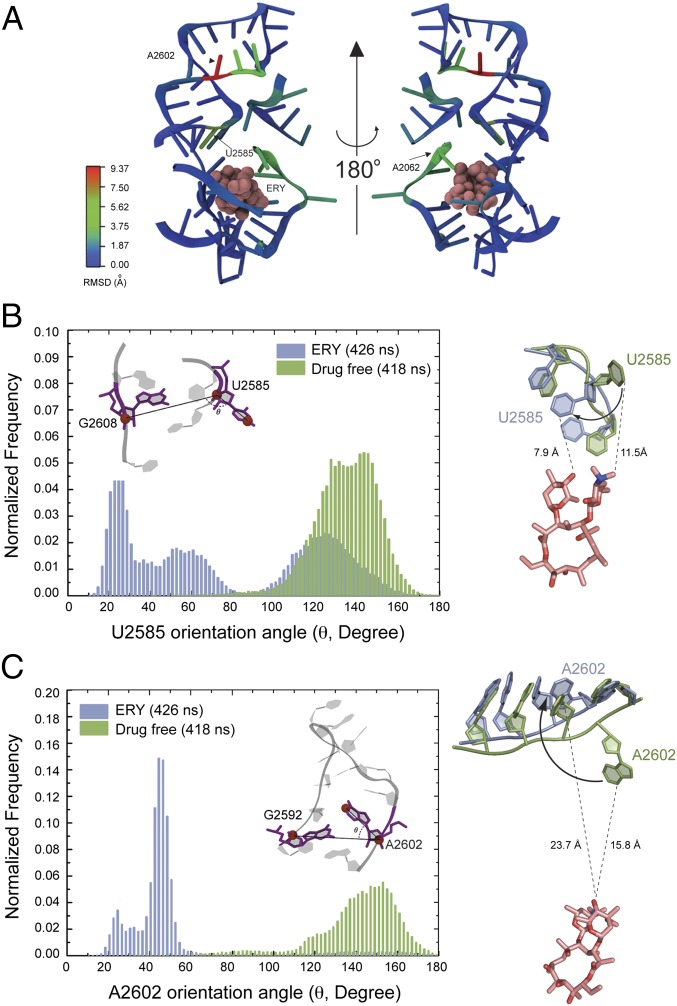
To gain independent evidence for the drug-dependent structural link between the NPET and PTC, we carried out all-atom MD simulations of the drug-free and ERY-bound Escherichia coli ribosome on the bases of the corresponding crystallographic structures (8, 26). Six independent simulations, three with the drug-free and three with the ERY-bound structure, were performed for the entire ribosome (about 3 million atoms), with production simulation time in each run ranging from 70 to 273 ns. After the preproduction equilibrations, the starting structures of both models show very similar conformations at the PTC region (Fig. S5A). Consistent with the crystal structures (9), MD simulations showed that the presence of ERY affects the placement of its immediate neighbor A2062 (Fig. S5B). Most notably, the presence of antibiotic also affects two remote sites in the PTC (Fig. 6 A, C, and D). In excellent agreement with the results of chemical probing, the drug promotes a dramatic reorientation of U2585. Although in the absence of antibiotic the nucleotide stably populates a “looped-out” configuration, ERY prompts rotation of the U2585 base by ∼100° (Fig. 6B, Figs. S5B and S6A, and Movie S1) This new “folded-in” state of U2585 is stabilized by its stacking interaction with U2584. In two of the three simulations of the drug-bound ribosome, U2585 rotation occurred within 50 ns after the start of the production simulation and remained in this orientation most of the time thereafter (Fig. S6A and Movie S1). In the third simulation, the folded-in state of U2585 was not fully achieved, but the U2585 base had rotated toward the folded-in position on average by 10° (Fig. S6 A and C) In contrast, in three simulations of the drug-free ribosome, the U2585 base barely visited the folded-in state (a total of 418 ns of the combined simulation time) and instead populated the looped-out conformation (Fig. 6B, Fig. S6 A and C, and Movie S1).
Fig. 6.
MD simulations substantiate the allosteric effect of the NPET-bound ERY on the distant PTC nucleotides U2585 and A2602. (A) Root mean square deviation (rmsd) of the preferred positions of rRNA residues in drug-free and ERY-bound ribosome. The rmsd was calculated by averaging the “last-frame” coordinates of the residues in three independent simulations of drug-free and ERY-bound ribosome. (B, Left) The frequency of visiting various conformations by U2585 in the course of MD simulations of drug-free (green) or ERY-bound (blue) ribosome [presented as angles between vectors linking atoms U2585(C3′)/U2585(C4) and U2585 C3′/G2608 C3′ (Inset)]. (Right) Placement of U2585 in drug-free (green) and drug-bound (blue) ribosomes. The averaged last-frame positions of the residues are shown. The shortest distances between the drug and U2585 base in two states are indicated. (C) Same as B but for the residue A2602.
A second, even more distant PTC residue, A2602, was also sensitive to the binding of ERY. The preferred orientations of the A2602 base in drug-free and ERY-bound states differ by ∼110° (Fig. 6C). Although in the absence of the drug A2602 is in a looped-out state away from helix 93 of 23S rRNA, the antibiotic provokes insertion of the base into the helix concomitant with its local distortion (Fig. 6C, Fig. S6 B and C, and Movie S1). Taken together, the results of the whole-ribosome MD simulations provide independent support for communication between the NPET and the PTC active site.
Discussion
The common view of the mechanism of antibiotic- and nascent peptide-controlled translation arrest presumes the key role of molecular interactions at the interface of the drug and the nascent chain. The juxtaposition of the peptide and antibiotic brings the stalling domain into contact with tunnel sensors, which relay the signal to the PTC, impairing its functions (12, 14, 19, 27). Although such a view is sufficient to rationalize the mechanism of arrest with long regulatory peptides, it fails to explain how an antibiotic can promote arrest with the only 3-aa–long MRL peptide. Furthermore, the tolerance of such arrest to alterations in antibiotic structure or to the mutations of the known nascent peptide sensors does not fit with the conventional view.
Our findings, however, may be reconciled within the framework of an alternative, potentially complementary model whose centerpiece is the allosteric link between the tunnel and the PTC. Binding of antibiotic in the tunnel alters properties of the PTC and inhibits peptidyl transfer catalysis between certain donor and acceptor substrates. Therefore, the tunnel-bound small molecule predisposes the ribosome for stalling when such combinations of substrates are encountered during translation.
The results of our biochemical testing and MD simulations clearly show that the binding of an antibiotic in the NPET alters the structural and thus likely functional features of the PTC. The data are most consistent in regard to U2585, a key residue in the PTC active site (24, 25). The reactivity of this residue to the SHAPE reagent is altered when the antibiotic is bound in the tunnel (Fig. 5). It also is one of the PTC residues that in the MD simulations reorients most dramatically in response to ERY binding and adopts a conformation rarely visited in the drug-free ribosome. The movement of U2585 seemed to be accompanied by repositioning of A2602 (Fig. S5). Although chemical probing did not provide additional evidence for rearrangements of A2602, its ERY-induced movement is supported by crystallographic structures of antibiotic-containing complexes, in which it was modeled in a conformation different from that in the drug-free ribosome (6, 9). In the absence of antibiotic, both U2585 and A2602 prefer the looped-out configuration, often stabilized by a stacking interaction between their bases (Fig. S5C and Movie S1). Drug-induced rotation of one of the residues would release the restriction and favor the repositioning of the other base as well. Because simulations of drug-free and ERY-bound ribosomes start from comparable states of the PTC, in which both residues are in looped-out conformation, their ERY-induced reorientation is compatible with either lowering the transition barrier or changing the free energy balance between the folded-in and looped-out states. Because of the limited sampling time, the available data are insufficient to distinguish between these scenarios.
Presently, we can only hypothesize how the antibiotic can promote reorientation of the distal PTC residues. Although hypothetically relocation of the antibiotic from its binding site in the NPET to the PTC is possible, neither the published crystal structures (8, 9) nor our MD simulations, in which binding of ERY in its tunnel site was extremely stable (Fig. S7A), support this scenario. Furthermore, RNA probing experiments suggest that the drug does not move from its conventional site when MRL peptide is placed in the tunnel (Fig. S7B). Therefore, we favor the model that the NPET-bound antibiotic induces changes in the PTC allosterically. One possibility is that a conformational relay could be initiated by rotation of the A2062 base located in the immediate vicinity of the drug binding site in the NPET (Fig. S5B) and connected to the PTC through its immediate neighbors, G2061 and C2063 (28, 29) (Fig. S5D). A possible alternative route might start at U2609 on the NPET wall opposite ERY. This highly flexible nucleotide is linked to the PTC via U1782 and U2586 (Fig. S5D). However, the mutations of rRNA residues in both these pathways (e.g., A2062, U2609, U2586, U1782) have either no or only a marginal effect upon ERY- and MRL peptide- dependent stalling (Fig. S4), suggesting that either the identity of these nucleotides is not critical for signal relay or other pathways are involved. In this regard, it should be noted that although biochemical and computational data clearly identified the PTC as a site sensitive to binding of antibiotic in the NPET, our MD analysis, which was performed with the tRNA-free ribosome, cannot accurately describe the placement of the PTC or NPET residues in the translating ribosome.
Although the presence of the antibiotic in the NPET is strictly required for inhibition of peptide bond formation between MRL and the incoming Arg– (or Lys–) tRNA, the requirements for drug structure are far less restrictive than those with the longer stalling peptides. Not only cladinose-containing macrolides that induce translation arrest with IAVV and IFVI peptides, but also azalides and ketolides can promote stalling after polymerization of the MRL tripeptide (Fig. 4). These results indicate that different antibiotics in the NPET can induce functionally similar changes in the PTC and reinforce our notion that specific drug–peptide interactions are inconsequential for stalling with short peptides.
The mere presence of the macrolide in the NPET does not indiscriminately inhibit catalysis of peptidyl transfer, but instead interferes with peptidyl transfer between specific substrates. The ribosome can reach the RLR motif in the full-size ErmDL and other RLR-type peptides without being arrested near the start of the leader ORFs (Table S1), clearly showing that only specific combinations of PTC donors and acceptors are problematic for the drug-bound ribosome. Thus, the antibiotic predisposes the ribosome for response to specific peptide sequences encoded in the uORFs. Importantly, the short size of the MRL peptide shows that the critical amino acid residues are those located in the PTC rather than in the tunnel. A similar trend is observed even with the longer stalling peptides in which residues critical for stalling are confined to the nascent chain C-terminus and the acceptor substrate (12, 14, 27). Even in the absence of antibiotic, the ribosomal catalytic center exhibits a considerable degree of selectivity. The rate of catalysis of peptide bond formation depends on the nature of the substrates and when peptidyl transfer becomes rate limiting, it may be manifested as context-specific ribosome stalling (30, 31). Importantly, the presence of the NPET-bound antibiotic does not seem to exacerbate the problem of the intrinsically problematic PTC substrates, but rather the macrolide-induced restrictive selectivity of the PTC makes some otherwise “normal” substrate pairs particularly difficult.
Although our findings suggest a previously unknown role of the antibiotic in the mechanism of translation arrest, they do not dismiss the importance of the drug–peptide contacts proposed previously. Conceivably, the drug may play a dual role with the longer stalling peptides: not only does it corrupt the PTC, but it may coerce the nascent chain to adopt a nonproductive conformation. Thus, the role of the cofactor in programmed translation arrest may differ depending on the nature of the peptide. With some nascent chains, the interaction between the drug and the growing protein may be absolutely critical for the arrest, which would explain why truncations of ErmCL or ErmAL1 prevent stalling. With other peptides, in which direct contacts between the antibiotic molecule and the nascent chain in the tunnel are minimal [e.g., ErmBL (27)], the allosterically altered PTC is sufficient to prevent peptide bond formation between certain donor–acceptor combinations. Such a role of the cofactor in programmed translation arrest may apply not only to antibiotics but also to other ligands that assist a broad array of peptides in halting translation (2, 32).
Experimental Procedures
Biochemical Assays.
Cell-free translation and toeprinting analyses were carried out as described (12), with additional details provided in SI Experimental Procedures and Table S2. For peptidyl–tRNA analysis, PURExpress translation reactions were supplemented with 0.5 μCi of [14C]-Leu (specific activity 306 Ci/mol) or 0.5 μCi of [14C]-Lys (specific activity 250 Ci/mol) (American Radiolabeled Chemicals). The products were analyzed in Bis-Tris polyacrylamide gels (12).
Chemical probing of rRNA (33) was performed using ribosomes prepared according to ref. 34 (see SI Experimental Procedures for details).
Molecular Modeling and MD Simulations.
The complete atomic models of the E. coli ribosome with ERY bound (the ERY model) and without the compound (drug-free model) are based on X-ray crystal structures 3OFO/3OFR and 2AVY/2AW4, respectively (8, 26). The systems were prepared as described (35). The final dimensions of both simulation systems were ∼280 × 340 × 340 Å (for details, see SI Experimental Procedures).
Supplementary Material
Acknowledgments
We thank Dr. A. Melman (Clarkson University) for the 1M7 compound, Drs. Y. Polikanov (Yale University) and B. Santarsiero (University of Illinois) for the advice on rendering molecular models, and Dr. L. Smith for proofreading the manuscript. This work was supported by National Institutes of Health Grants R01GM104370 (to A.S.M. and N.V.-L.) and 9P41GM104601 (to K.S.) and National Science Foundation Grant PHY0822613 (to K.S.). MD modeling was facilitated by the Great Lakes Consortium grant for Petascale Computation. A.B. and M.R. were supported by Estonian Ministry of Education Grant SF0180026s09 and by the European Regional Development Fund. G.C.A. was supported by Estonian Science Foundation Grant ETF9020 and European Social Fund Grant “Mobilitas” MJD99.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1403586111/-/DCSupplemental.
References
- 1.Vázquez-Laslop N, Mankin AS. Triggering peptide-dependent translation arrest by small molecules: ribosome stalling modulated by antibiotics. In: Ito K, editor. Regulatory Nascent Polypeptides. New York: Springer; 2014. [Google Scholar]
- 2.Ito K, Chiba S. Arrest peptides: Cis-acting modulators of translation. Annu Rev Biochem. 2013;82:171–202. doi: 10.1146/annurev-biochem-080211-105026. [DOI] [PubMed] [Google Scholar]
- 3.Weisblum B. Erythromycin resistance by ribosome modification. Antimicrob Agents Chemother. 1995;39(3):577–585. doi: 10.1128/AAC.39.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramu H, Mankin A, Vazquez-Laslop N. Programmed drug-dependent ribosome stalling. Mol Microbiol. 2009;71(4):811–824. doi: 10.1111/j.1365-2958.2008.06576.x. [DOI] [PubMed] [Google Scholar]
- 5.Subramanian SL, Ramu H, Mankin AS. Inducible resistance to macrolide antibiotics. In: Dougherty TJ, Pucci MJ, editors. Antibiotic Drug Discovery and Development. New York: Springer; 2011. [Google Scholar]
- 6.Schlünzen F, et al. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature. 2001;413(6858):814–821. doi: 10.1038/35101544. [DOI] [PubMed] [Google Scholar]
- 7.Tu D, Blaha G, Moore PB, Steitz TA. Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance. Cell. 2005;121(2):257–270. doi: 10.1016/j.cell.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 8.Dunkle JA, Xiong L, Mankin AS, Cate JH. Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action. Proc Natl Acad Sci USA. 2010;107(40):17152–17157. doi: 10.1073/pnas.1007988107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bulkley D, Innis CA, Blaha G, Steitz TA. Revisiting the structures of several antibiotics bound to the bacterial ribosome. Proc Natl Acad Sci USA. 2010;107(40):17158–17163. doi: 10.1073/pnas.1008685107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vazquez D. The macrolide antibiotics. In: Corcoran JW, Hahn FE, editors. Antibiotics III. Mechanism of Action of Antimicrobial and Antitumor Agents. New York: Springer; 1975. pp. 459–479. [Google Scholar]
- 11.Kannan K, Vázquez-Laslop N, Mankin AS. Selective protein synthesis by ribosomes with a drug-obstructed exit tunnel. Cell. 2012;151(3):508–520. doi: 10.1016/j.cell.2012.09.018. [DOI] [PubMed] [Google Scholar]
- 12.Vazquez-Laslop N, Thum C, Mankin AS. Molecular mechanism of drug-dependent ribosome stalling. Mol Cell. 2008;30(2):190–202. doi: 10.1016/j.molcel.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 13.Mayford M, Weisblum B. ermC leader peptide. Amino acid sequence critical for induction by translational attenuation. J Mol Biol. 1989;206(1):69–79. doi: 10.1016/0022-2836(89)90524-x. [DOI] [PubMed] [Google Scholar]
- 14.Ramu H, et al. Nascent peptide in the ribosome exit tunnel affects functional properties of the A-site of the peptidyl transferase center. Mol Cell. 2011;41(3):321–330. doi: 10.1016/j.molcel.2010.12.031. [DOI] [PubMed] [Google Scholar]
- 15.Vázquez-Laslop N, et al. Role of antibiotic ligand in nascent peptide-dependent ribosome stalling. Proc Natl Acad Sci USA. 2011;108(26):10496–10501. doi: 10.1073/pnas.1103474108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gryczan T, Israeli-Reches M, Del Bue M, Dubnau D. DNA sequence and regulation of ermD, a macrolide-lincosamide-streptogramin B resistance element from Bacillus licheniformis. Mol Gen Genet. 1984;194(3):349–356. doi: 10.1007/BF00425543. [DOI] [PubMed] [Google Scholar]
- 17.Hue KK, Bechhofer DH. Regulation of the macrolide-lincosamide-streptogramin B resistance gene ermD. J Bacteriol. 1992;174(18):5860–5868. doi: 10.1128/jb.174.18.5860-5868.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwon AR, et al. ErmK leader peptide: Amino acid sequence critical for induction by erythromycin. Arch Pharm Res. 2006;29(12):1154–1157. doi: 10.1007/BF02969307. [DOI] [PubMed] [Google Scholar]
- 19.Vázquez-Laslop N, Ramu H, Klepacki D, Kannan K, Mankin AS. The key function of a conserved and modified rRNA residue in the ribosomal response to the nascent peptide. EMBO J. 2010;29(18):3108–3117. doi: 10.1038/emboj.2010.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakatogawa H, Ito K. The ribosomal exit tunnel functions as a discriminating gate. Cell. 2002;108(5):629–636. doi: 10.1016/s0092-8674(02)00649-9. [DOI] [PubMed] [Google Scholar]
- 21.Cruz-Vera LR, Rajagopal S, Squires C, Yanofsky C. Features of ribosome-peptidyl-tRNA interactions essential for tryptophan induction of tna operon expression. Mol Cell. 2005;19(3):333–343. doi: 10.1016/j.molcel.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 22.Yang R, Cruz-Vera LR, Yanofsky C. 23S rRNA nucleotides in the peptidyl transferase center are essential for tryptophanase operon induction. J Bacteriol. 2009;191(11):3445–3450. doi: 10.1128/JB.00096-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE) J Am Chem Soc. 2005;127(12):4223–4231. doi: 10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- 24.Schmeing TM, Huang KS, Strobel SA, Steitz TA. An induced-fit mechanism to promote peptide bond formation and exclude hydrolysis of peptidyl-tRNA. Nature. 2005;438(7067):520–524. doi: 10.1038/nature04152. [DOI] [PubMed] [Google Scholar]
- 25.Voorhees RM, Weixlbaumer A, Loakes D, Kelley AC, Ramakrishnan V. Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome. Nat Struct Mol Biol. 2009;16(5):528–533. doi: 10.1038/nsmb.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schuwirth BS, et al. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310(5749):827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 27.Arenz S, et al. 2014. Molecular basis for erythromycin-dependent ribosome stalling during translation of the ErmBL leader peptide. Nat Commun 5:3501.
- 28.Seidelt B, et al. Structural insight into nascent polypeptide chain-mediated translational stalling. Science. 2009;326(5958):1412–1415. doi: 10.1126/science.1177662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gumbart J, Schreiner E, Wilson DN, Beckmann R, Schulten K. Mechanisms of SecM-mediated stalling in the ribosome. Biophys J. 2012;103(2):331–341. doi: 10.1016/j.bpj.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Woolstenhulme CJ, et al. Nascent peptides that block protein synthesis in bacteria. Proc Natl Acad Sci USA. 2013;110(10):E878–E887. doi: 10.1073/pnas.1219536110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peil L, et al. Distinct XPPX sequence motifs induce ribosome stalling, which is rescued by the translation elongation factor EF-P. Proc Natl Acad Sci USA. 2013;110(38):15265–15270. doi: 10.1073/pnas.1310642110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cruz-Vera LR, Gong M, Yanofsky C. Changes produced by bound tryptophan in the ribosome peptidyl transferase center in response to TnaC, a nascent leader peptide. Proc Natl Acad Sci USA. 2006;103(10):3598–3603. doi: 10.1073/pnas.0600082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Merryman C, Noller HF. Footprinting and modification-interference analysis of binding sites on RNA. In: Smith CWJ, editor. RNA:Protein Interactions, A Practical Approach. Oxford: Oxford Univ Press; 1998. pp. 237–253. [Google Scholar]
- 34.Onouchi H, et al. Nascent peptide-mediated translation elongation arrest coupled with mRNA degradation in the CGS1 gene of Arabidopsis. Genes Dev. 2005;19(15):1799–1810. doi: 10.1101/gad.1317105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trabuco LG, et al. The role of L1 stalk-tRNA interaction in the ribosome elongation cycle. J Mol Biol. 2010;402(4):741–760. doi: 10.1016/j.jmb.2010.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.