Significance
Previous research has investigated the interactions between linear oligonucleotides and serum nucleases but the emergence of oligonucleotides conjugated to nanoparticles opens up a new avenue of research that has yet to be fully explored. In this work, we probed the degradation of a model small interfering RNA (siRNA) immobilized on gold nanoparticles (AuNPs). We show that serum nucleases are able to recognize and degrade the immobilized siRNA at specific sites within 4 nm of the AuNP surface, which were different than the sites for siRNA free in solution. These data suggest that oligonucleotides immobilized on nanoparticle surfaces must be studied independently from siRNA free in solution due to the potential influence of the local chemical environment of the oligonucleotide–nanoparticle conjugates.
Keywords: biological recognition, nanotechnology, OliGreen, polyacrylamide gel electrophoresis
Abstract
Small interfering RNA (siRNA) is a powerful and highly effective method to regulate gene expression in vitro and in vivo. However, the susceptibility to serum nuclease-catalyzed degradation is a major challenge and it remains unclear whether the strategies developed to improve the stability of siRNA free in serum solution are ideal for siRNA conjugated to nanoparticle surfaces. Herein, we use spherical nucleic acid nanoparticle conjugates, consisting of gold nanoparticles (AuNPs) with siRNA chemisorbed to the surface, as a platform to study how a model siRNA targeting androgen receptor degrades in serum (SNA-siRNAAR). In solutions of 10% (vol/vol) FBS, we find rapid endonuclease hydrolysis at specific sites near the AuNP-facing terminus of siRNAAR, which were different from those of siRNAAR free in solution. These data indicate that the chemical environment of siRNA on a nanoparticle surface can alter the recognition of siRNA by serum nucleases and change the inherent stability of the nucleic acid. Finally, we demonstrate that incorporation of 2′-O-methyl RNA nucleotides at sites of nuclease hydrolysis on SNA-siRNAAR results in a 10-fold increase in siRNA lifetime. These data suggest that strategies for enhancing the serum stability of siRNA immobilized to nanoparticles must be developed from a dedicated analysis of the siRNA–nanoparticle conjugate, rather than a reliance on strategies developed for siRNA free in solution. We believe these findings are important for fundamentally understanding interactions between biological media and oligonucleotides conjugated to nanoparticles for the development of gene regulatory and therapeutic agents in a variety of disease models.
The challenge of short lifetimes for RNA oligonucleotides in serum-containing media is particularly important for siRNA therapeutics intended for systemic delivery, in which the therapeutic siRNA interacts with blood as it is distributed to target tissues before entry into target cells. Studies of the ability of siRNA with transfection reagents [e.g., cationic liposomes (1), polyelectrolyte micelles (2), and dendrimers (3)] to reduce the expression of genes in cell culture often take advantage of the addition of siRNA and transfection reagents to cells in media that are free of serum for a period of time [e.g., 3–24 h before replacement with media supplemented with 10% (vol/vol) FBS]. Such a procedure provides a window of time for the entry of siRNA into cells that avoids exposure of the siRNA to serum nucleases that would otherwise catalyze the hydrolysis of siRNA; such a procedure also leaves the challenge of short half-lives in serum-containing biological media unaddressed.
Spherical nucleic acid nanoparticle conjugates consisting of 13-nm gold nanoparticles (AuNPs) functionalized with complementary sense and antisense (AS) siRNA oligonucleotides (SNA-siRNA) are nanostructures under investigation and development as potential therapeutic agents for the delivery of siRNA (4, 5) (Fig. 1). Unlike linear nucleic acids, SNAs rapidly enter mammalian cells, despite their negative charge, through class A scavenger receptor-mediated endocytosis (6, 7) (>106 particles per cell in <4 h for mouse endothelial cells) (8) and have been used to regulate the expression of genes in vitro and in vivo (4, 5, 9), suggesting the promise of developing SNAs as therapeutic agents.
Fig. 1.
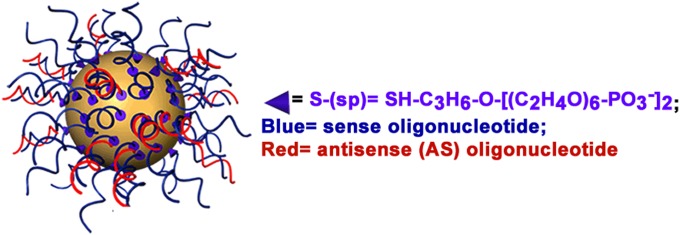
A 3D drawing of an siRNA-SNA, consisting of a gold nanoparticle with sense (blue) and AS (red) oligonucleotides. For clarity, thiol-terminated 5-kDa PEG is not drawn.
The development of SNA-siRNA with long serum lifetimes remains an outstanding challenge. An examination of an SNA-siRNA directed against the expression of luciferase (SNA-siRNAluc), and of siRNAluc free in solutions of 10% FBS, showed that SNA-siRNAluc has a half-life (τ1/2) of 816 ± 59 min, compared with 133 ± 30 min for free siRNAluc (9). This study did not however characterize the degradation products in serum or address whether the degradation was primarily the consequence of exonuclease or endonuclease activity. A strategy to systematically examine and reduce the nuclease susceptibility of siRNA chemisorbed at high density (25 pmol cm−2) and oriented at the surfaces of AuNPs has not been developed.
Chemically modified nucleotides [e.g., 2′-O-methyl (2′-OMe), 2′-deoxy-2′-fluoro (2′-F)] are commonly used to increase the stability of siRNA in serum-containing media (10–15), and research efforts to develop therapeutic siRNA often use “design rules” and patterns for the placement of chemically modified nucleotides to enhance serum lifetimes. For example, multiple reports have shown that modifying all pyrimidines in the siRNA sequence with 2′-F modifications provides high serum stability without compromising activity (16–19). The task of obtaining an siRNA with chemically modified nucleotides optimized for serum stability and gene-silencing activity, however, remains experimentally intensive. An approach that first identifies the sites of serum nuclease-catalyzed degradation of an siRNA, and then uses this information in the design of siRNA with 2′-OMe modifications (e.g., anti-MDRI siRNA) (20), offers a more focused route to improving the serum stability of a specific siRNA sequence. This observation poses the questions (i) can a similar strategy be used with siRNA-SNAs and (ii) are the sites of nuclease activity comparable to those of the free siRNA, especially given the orientation of the oligonucleotides, the potentially restricted access to nucleases due to steric effects, and the electrostatic properties of SNAs?
The study herein investigates the interaction between FBS and siRNA targeting the expression of human androgen receptor (AR; siRNAAR) immobilized on the surface of AuNPs (SNA-siRNAAR) and comparison with that of siRNAAR free in solution. We chose to analyze the interaction of serum nucleases with SNA-siRNAAR, a nanoparticle–siRNA conjugate that is under development for knocking down the expression of human AR in lymph node carcinoma of the prostate (LNCaP) cells for the treatment of prostate cancer (21). The work described herein addresses five main points: (i) the quantitative determination of the serum half-life of SNA-siRNAAR; (ii) the identification of sites of serum nuclease-catalyzed hydrolysis of SNA-siRNAAR; (iii) the differentiation between endonuclease or exonuclease-catalyzed degradation of SNA-siRNAAR; (iv) a comparison of the sites of nuclease action of SNA-siRNAAR and free siRNAAR; and (v) a demonstration that the site-selective incorporation of 2′-OMe groups in SNA-siRNAAR can provide increased resistance to nuclease-catalyzed degradation in serum-containing media. We found that the sites of nuclease-catalyzed hydrolysis for the immobilized siRNA are different from those of the free siRNA in solution. The incorporation of 2′-OMe nucleotides at the sites identified for the immobilized siRNA led to an increase in serum half-life by a factor of ∼10. Substitution at these sites and also at neighboring sites with a relatively small number (i.e., 10) of 2′-OMe or 2′-H nucleotides led to increases in serum half-life by factors >30.
Results and Discussion
Design and Synthesis of SNA-siRNAAR.
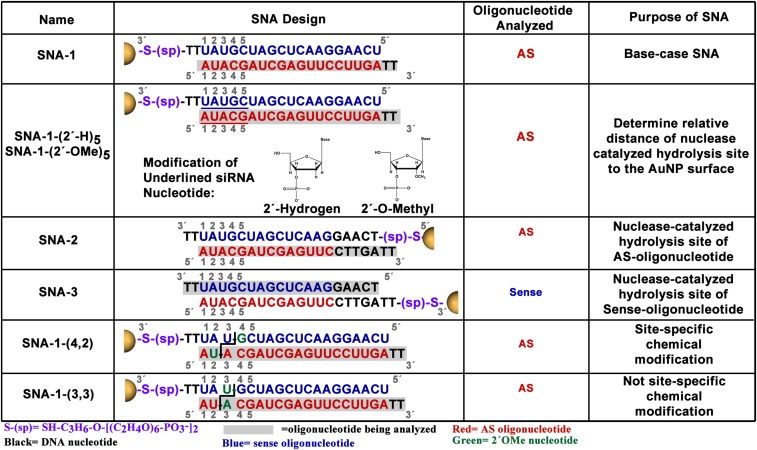
This study focused on three classes of SNAs (Fig. 2). Each SNA consists of three components: (i) 13-nm AuNP core; (ii) sense and antisense (AS) oligonucleotide, one of which is functionalized at the 3′ terminus with an oligoethylene glycol spacer and thiol group chemisorbed to the AuNP surface, and the other, which is noncovalently associated to the SNA by duplex hybridization; and (iii) thiol-terminated 5-kDa polyethylene gylcol (PEG) chemisorbed to the AuNP surface (Fig. 1). This design is one that has led to gene regulatory activity for several gene targets, including Bcl2-L12 (4, 5, 9). We used SNA-1 to establish a baseline for the stability of SNA-siRNAAR in serum solution. SNA-1-(2′-H)5 and SNA-1-(2′-OMe)5 are derivatives of SNA-1 in which the first five RNA nucleotides have been replaced with 2′-H or 2′-OMe nucleotides and were used to probe the possibility of nuclease-catalyzed hydrolysis at sites near the AuNP surface of SNA-1 (i.e., within the first five nucleotides at the terminus chemisorbed to the AuNP). To determine the specific sites of nuclease-catalyzed hydrolysis on SNA-1, we designed SNA-2 to analyze the AS oligonucleotide and SNA-3 to analyze the sense oligonucleotide, respectively. To evaluate if incorporating 2′-OMe nucleotides at sites of nuclease-catalyzed hydrolysis will enhance stability, we designed and analyzed SNA-1-(4,2). In addition, we analyzed SNA-1-(3,3), designed to have the same number of 2′-OMe nucleotides but placed at neighboring sites, to illustrate the importance of addressing the specific sites of nuclease action of a given sequence.
Fig. 2.
Design and function of SNAs with siRNAAR used in this study.
Analysis of the Noncovalently Associated Oligonucleotide.
We characterized the composition of SNA-siRNAAR studied in this work by measuring the ratio of the noncovalently associated oligonucleotide per AuNP. Dehybridization of duplex siRNA in solutions of 8 M urea heated to 45 °C, followed by centrifugation, allowed separation of the noncovalently associated oligonucleotide from the AuNP with chemisorbed oligonucleotide (Fig. 3). The noncovalently associated oligonucleotides that dissociate from the AuNPs are quantified with the fluorescence of OliGreen, a nonspecific intercalator of single-stranded oligonucleotides (22). The concentration of AuNPs was determined by measuring the absorbance at 520 nm (23). Taken together, these data allowed us to measure the ratio of noncovalently associated oligonucleotide to AuNP for all SNAs in Fig. 2 (Table S1).
Fig. 3.
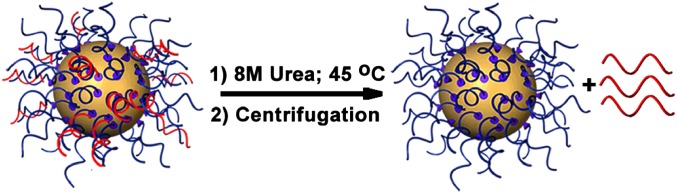
Schematic showing the removal of complementary oligonucleotide (red) from the SNA using 8 M urea and heating to 45 °C.
Degradation of SNA-1 in Solutions of FBS via Loss of the AS Oligonucleotide from SNA-1.
We performed reactions by adding SNA-1 to solutions of 10% FBS (vol/vol) in PBS at 37 °C. Following periods of incubation of 1, 2, 10, and 30 min, we stopped reactions by adding 30 mM SDS to denature the serum proteins and deactivate the enzymes from the serum solution. Separation of SNA-1 from the reaction media was accomplished by centrifugation and washing with solutions of PBS containing Tween-20 (0.01% vol/vol). We analyzed the amount of AS oligonucleotide associated with washed SNA-1 (Fig. S1). Although we have chosen to report the data as the ratio of AS oligonucleotide to AuNP, the measurement by OliGreen only reflects the total amount of noncovalently associated oligonucleotide recovered from AuNPs.
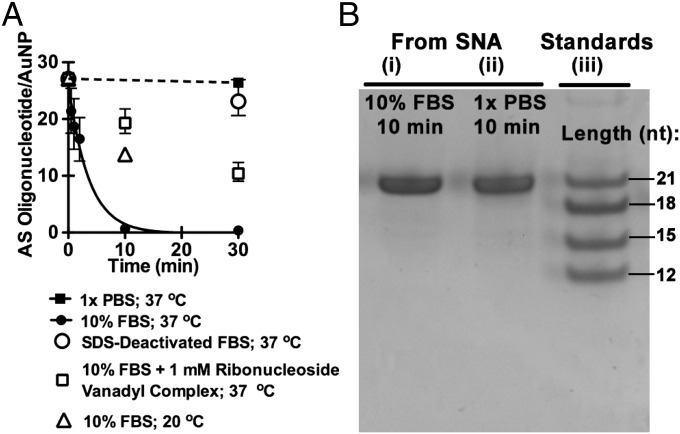
Whereas SNA-1 maintains the initial amount of AS oligonucleotides per AuNP (27 ± 1) following a 30-min incubation in PBS at 37 °C, SNA-1 rapidly loses AS oligonucleotides in solutions of 10% FBS at 37 °C; the half-life for the degradation of AS oligonucleotide is ∼2 min (Fig. 4A, black squares and circles). When the reaction medium is a solution of FBS that has been pretreated with SDS and heated to 90 °C to denature the proteins in FBS, the loss of AS oligonucleotide from SNA-1 is negligible (Fig. 4A, open circles). Comparison of the rates in Fig. 4A (PBS, FBS, SDS-denatured FBS) suggests that the cause for the rapid loss of AS oligonucleotide in solutions of FBS is serum nuclease activity. These data indicate that thermal dehybridization of siRNAAR or desorption of thiol-terminated sense oligonucleotides from Au (accompanied by the loss of AS oligonucleotide) do not contribute to the loss of AS oligonucleotide from SNA-1. The degradation of free siRNAAR oligonucleotide in solutions of 10% FBS (i.e., unconjugated to AuNPs) is also rapid (τ1/2 < 20 s; Figs. S2 and S3).
Fig. 4.
Degradation of SNA-1. (A) Plot of the ratio of AS oligonucleotide per AuNP for SNA-1 over time. The τ1/2 of the AS oligonucleotide is ∼2 min in 10% FBS at 37 °C (fit to a first order exponential decay function), whereas in PBS and deactivated FBS at 37 °C, the ratio is indistinguishable from that of the starting material. When SNA-1 is incubated in 10% FBS at 20 °C or 10% FBS with ribonucleoside vanadyl complex (an RNase A inhibitor), τ1/2 of the AS oligonucleotide is 15 min and 30 min, respectively. Error bars represent the range for n = 4 measurements. (B) Denaturing PAGE of the AS oligonucleotide from SNA-1 after 10 min in 10% FBS and 1× PBS (lanes i and ii, respectively) and analytical standards (lane iii).
We hypothesized that the loss of AS oligonucleotide in 10% FBS is serum nuclease activity likely caused by the abundance of RNase A-type enzymes in serum that catalyze the degradation of single- and double-stranded RNA (ssRNA and dsRNA) (24, 25). In solutions of FBS to which ribonucleoside vanadyl complexes (an RNase A inhibitor) have been added, the ratio of AS oligonucleotide to AuNP decreases at a slower rate (Fig. 4A, open squares). This observation implicates a contribution from RNase A as a cause for the loss of oligonucleotide in FBS. Taken together, these observations reveal that the AS oligonucleotide of SNA-1 is susceptible to rapid degradation even when conjugated to the surface of AuNPs and suggests a need for strategies to increase the lifetime in serum for some siRNA sequences formulated as SNA-siRNA.
Analysis of the AS Oligonucleotide Recovered from SNA-1 After Degradation in 10% FBS.
We hypothesized that fragments of the AS oligonucleotide that remained associated with SNA-1 following short periods of incubation in solutions of FBS would reveal evidence of exonuclease activity (by generating a “ladder” of AS oligonucleotide fragments) or endonuclease activity (by generating fragments of one or a few specific lengths). We recovered the noncovalently associated oligonucleotide remaining on SNA-1 following incubation in 10% FBS at 20 °C for 10 min; the ratio of AS oligonucleotide to AuNP following this reaction (14 ± 1) is ∼50% of the initial ratio (Fig. 4A, open triangle). Analysis of the recovered oligonucleotide by denaturing polyacrylamide gel electrophoresis (PAGE), however, revealed only the full-length [21-nucleotide (nt)] AS oligonucleotide (Fig. 4B, lane i), identical to that observed in PBS solution (Fig. 4B, lane ii). The presence of only the full-length AS oligonucleotide suggests that nuclease activity leads to very short oligonucleotide fragments (i.e., << 12 bp with Tm << 37 °C of either the sense- or AS oligonucleotide), and the dissociation of these short fragments from AuNPs following the reaction.
Nuclease-Catalyzed Hydrolysis Occurs on SNA-1 Near the AuNP Surface.
Four major classes of chemical modifications (backbone, sugar, base, and terminal) have been developed to resist nuclease-catalyzed degradation of oligonucleotides and to improve siRNA stability in both fetal bovine and human serum (10–15, 26). To test the hypothesis that enzymatic hydrolysis of SNA-1 takes places at a site near the AuNP-facing terminus of the siRNA, thus leading to oligonucleotide fragments too short to remain hybridized and associated to SNA-1, we designed and analyzed SNA-1-(2′-H)5 and SNA-1-(2′-OMe)5, where the RNA nucleotides of siRNAAR at positions 1–5 for both the sense- and AS oligonucleotide (i.e., the AuNP-facing terminus) are substituted with 2′-H or 2′-OMe nucleotides and are therefore not susceptible to RNase-catalyzed hydrolysis (Fig. 2). Analysis of SNA-1-(2′-H)5 and SNA-1-(2′-OMe)5 in solutions of FBS showed that the SNAs resist the loss of AS oligonucleotide (Fig. 5 A and B, respectively) as >80% of the initial AS oligonucleotide remains associated with AuNPs at 60 min. The stability of SNA-1-(2′-H)5 and SNA-1-(2′-OMe)5, in contrast to the rapid degradation of SNA-1, supports the hypothesis that the site of nuclease-catalyzed hydrolysis of siRNAAR of SNA-1 lies within the first five ribonucleotides of the AuNP-facing terminus of siRNAAR. These data reveal the surprising observation that serum nucleases can act upon regions of siRNA conjugated to AuNPs that would appear to be shielded by a dense layer of neighboring oligonucleotides, which is likely in part due to the curvature of the AuNP.
Fig. 5.
Chemical modifications near the AuNP surface increase the stability of the AS oligonucleotide. Plot of the ratio of AS oligonucleotide per AuNP over the course of a 60-min incubation in 10% FBS or 1× PBS at 37 °C for (A) SNA-1-(2′-H)5 and (B) SNA-1-(2′-OMe)5. For both A and B, solid black lines are first order exponential decay functions and the error bars represent the range for n = 4 measurements.
Design of SNA-2 and SNA-3 to Determine the Sites of Nuclease-Catalyzed Hydrolysis of siRNAAR Conjugated to AuNPs.
To test the hypothesis and confirm that the site of serum nuclease-catalyzed hydrolysis for siRNAAR of SNA-1 lies within positions 1–5, we designed and analyzed the reactions of SNA-2 and SNA-3 (Fig. 2) in solutions of 10% FBS. The key design features of SNA-2 and SNA-3 are that (i) the sequence of siRNAAR is conserved from SNA-1; (ii) the first five nucleotides from the point of chemisorption to the AuNP surface are DNA nucleotides; and (iii) the point of attachment to the AuNP is varied (5′ terminus of the sense oligonucleotide in SNA-2; 3′ terminus of the AS oligonucleotide in SNA-3), such that the region of siRNAAR hypothesized to contain the site of rapid hydrolysis is directed away from the AuNP surface. We designed SNA-2 and SNA-3 such that the degradation products of the noncovalently associated oligonucleotides (AS oligonucleotide for SNA-2; sense oligonucleotide for SNA-3) could be recovered from AuNPs after incubation in 10% FBS, and that characterization of these products would inform us about the sites of nuclease action on siRNAAR of SNA-1. The major hypothesis of this approach is that the site of serum nuclease activity on siRNAAR is determined by the nucleotide sequence and recognition of a motif within it, and not by the spatial relationship of siRNAAR to the AuNP surface.
Analysis of SNA-2 to Determine the Site of Nuclease-Catalyzed Hydrolysis of the AS Oligonucleotide.
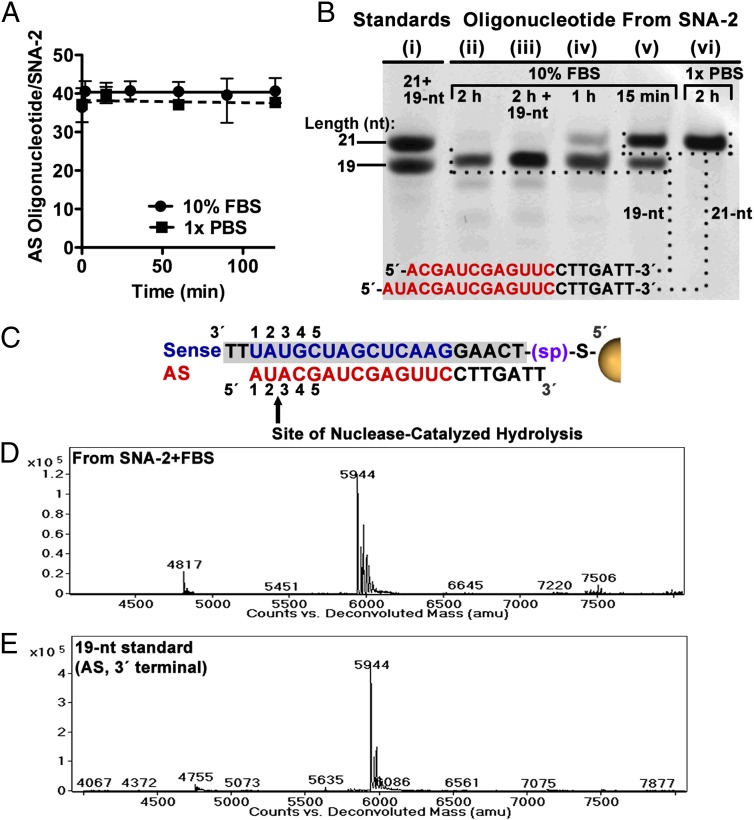
We incubated SNA-2 in solutions of 10% FBS at 37 °C and analyzed the ratio of noncovalently associated AS oligonucleotide to the AuNP. Within experimental uncertainty, the amount of AS oligonucleotide associated with SNA-2 after a 2-h incubation in 10% FBS is unchanged from the initial value, indicating negligible degradation of the AS oligonucleotide (Fig. 6A). Analysis by denaturing PAGE of the noncovalently associated oligonucleotide of SNA-2 after a 15-min and 1-h incubation in 10% FBS revealed a mixture of two bands (Fig. 6B, lanes v and iv, respectively). We interpreted the lower band as the primary product of RNase action on the AS oligonucleotide of SNA-2 (also seen after 2 h of reaction in lane ii). Analysis by PAGE of a mixture of the 19-nt analytical standard and the AS oligonucleotide recovered from SNA-2 showed the resolution of a single major band (Fig. 6B, lane iii). These results suggest that the product of nuclease action is a 19-nt fragment generated by the loss of two nucleotides from the 5′ terminus, and that the site of nuclease-catalyzed hydrolysis is the phosphodiester bond between the U and A nucleotides at positions 2 and 3 (Fig. 6C).
Fig. 6.
Site of nuclease-catalyzed hydrolysis for the AS oligonucleotide. (A) Ratio of AS oligonucleotide per AuNP remaining on SNA-2 after incubation in 10% FBS and 1× PBS at 37 °C. Error bars represent the range for n = 4 measurements. (B) Denaturing PAGE showing standards (lane i) and the noncovalently associated oligonucleotide recovered from SNA-2 after incubation in 10% FBS (lanes ii–v) and 1× PBS (lane vi) at 37 °C. For 10% FBS after 15 min, the two visible bands correspond to the full-length oligonucleotide (21 nt) and a degradation product that is shorter by two nucleotides (19 nt) (lane v). After 1 (lane iv) and 2 h (lane ii), the 19-nt product is the major species recovered, confirmed by coloading the AS oligonucleotide after 2 h with the 19-nt standard (1:1 mol ratio) and observing only a single band (lane iii). (C) The proposed site of nuclease-catalyzed hydrolysis of the AS oligonucleotide of SNA-2 is the phosphodiester bond between the U and A nucleotides at positions 2 and 3. (D) The deconvoluted ESI-MS spectra of the AS oligonucleotide recovered from SNA-2 after 2 h incubation in 10% FBS, corresponding to a loss of AU from the 5′ terminus of the AS oligonucleotide. (E) Deconvoluted mass spectra of the 19-nt analytical standard derived from the AS oligonucleotide of SNA-2.
We used electrospray ionization mass spectrometry (ESI-MS) to analyze the degradation products of the AS oligonucleotide recovered from SNA-2 after 1- and 2-h incubations in solutions of 10% FBS (Fig. 6D and Fig. S4, respectively). The observed 5,944 amu mass (Fig. 6D) agreed with the predicted and experimentally observed mass for the 19-nt standard (Fig. 6E). These observations for hydrolysis of the AS oligonucleotide at the U–A nucleotides at positions 2 and 3 support the conclusion that degradation of siRNAAR by serum nucleases occurs within the first five nucleotides at the 5′ terminus. A complete table of possible degradation products and analytical standards is provided in Fig. S5; mass spectra for the set of analytical standards for AS oligonucleotide, and those of reaction products at other time points, are in Fig. S4.
Determining the Site of Nuclease-Catalyzed Hydrolysis for the AS Oligonucleotide of siRNAAR Free in Solutions of 10% FBS.
We analyzed the degradation of siRNAAR free in solutions of 10% FBS at 37 °C by analyzing a derivative of siRNAAR consisting of a 3′-biotin-labeled AS oligonucleotide (Fig. S2A). PAGE revealed a band that is shorter than the starting material by approximately eight nucleotides (Fig. S2B), a product different from that observed for the immobilized AS oligonucleotide of SNA-2. The difference between the observed sites of nuclease action on the AS oligonucleotide, for immobilized siRNAAR and siRNAAR free in solution, shows that the immobilization of siRNA to the surface of nanoparticles influences the site preference of serum nucleases.
Analysis of SNA-3 to Determine the Site of Nuclease-Catalyzed Hydrolysis of the Sense Oligonucleotide.
Analysis of the degradation of SNA-3 in solutions of FBS allowed for identification of the site of nuclease action on the sense oligonucleotide of SNA-3, and by inference, the site of nuclease action on the sense oligonucleotide of SNA-1. We found endonuclease-catalyzed hydrolysis of the phosphodiester bond of the U–G sequence (3′–5′ at positions 3 and 4) (described in detail in SI Text and Figs. S6 and S7), which was different from the site of nuclease-catalyzed hydrolysis for the sense oligonucleotide free in solution (Fig. S3). Taken together, the analysis of the degradation of SNA-2 and SNA-3 identified sites of serum nuclease action for the sense oligonucleotide (U–G at positions 3 and 4; 3′–5′) and the AS oligonucleotide (U–A at positions 2 and 3; 5′–3′) of siRNAAR conjugated to AuNPs. Although both of these sites lie within positions 1 and 5 of siRNAAR, we were surprised to find that these sites are staggered, rather than directly opposed to each other. Previous work by Hong et al. that surveyed the degradation of 125 different siRNA sequences in serum solutions observed blunt cuts exclusively (27) although work by Volkov et al. suggests that blunt cuts are possible (20).
Site-Specific Incorporation of 2′-OMe RNA Nucleotides to Increase the Serum Stability of SNA-1.
We chose to use chemically modified nucleotides at the sites of nuclease-catalyzed hydrolysis identified herein, rather than the sites observed for siRNAAR free in solution or those predicted by design rules, to demonstrate a strategy that focuses on siRNA in the chemical context of the surface of a nanoparticle. We used 2′-OMe groups because they do not disrupt the formation of double-stranded RNA or cause cytotoxicity and can, in some cases, lead to siRNA with potent activity and extended serum lifetime (12, 18, 28–31). We therefore designed SNA-1-(4,2) and SNA-1-(3,3) (Fig. 2). SNA-1-(4,2) has the same sequence and point of chemical conjugation to the AuNP as SNA-1 and contains only two RNA nucleotides with 2′-OMe groups—one at position 4 of the sense oligonucleotide and one at position 2 of the AS oligonucleotide—each placed to block endonuclease activity at those sites. SNA-1-(3,3) contains two RNA nucleotides with 2′-OMe groups at positions that are adjacent to (one at position 3 for the sense oligonucleotide and one at position 3 for the AS oligonucleotide), but are not at the specific sites of nuclease-catalyzed hydrolysis.
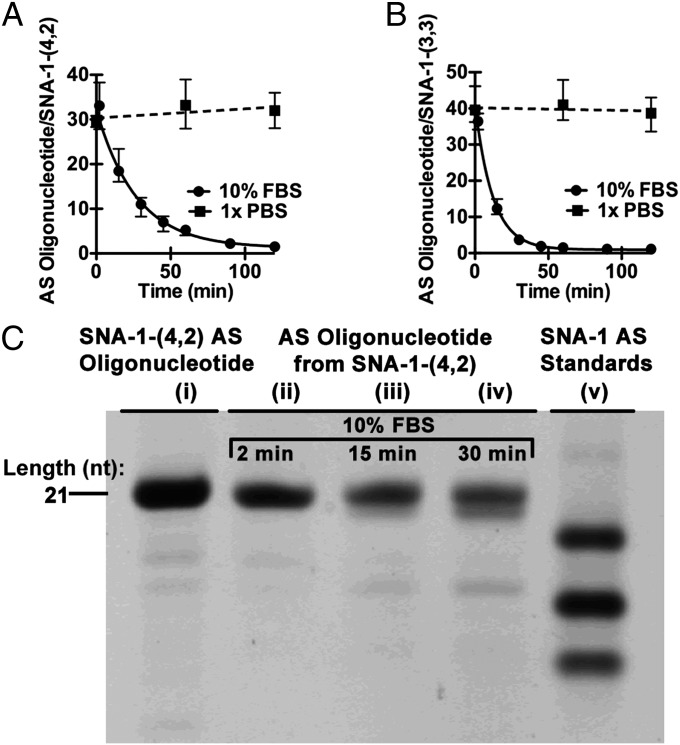
The τ1/2 of the AS oligonucleotide of SNA-1-(4,2) in solutions of 10% FBS at 37 °C was ∼20 min (Fig. 7A) and represents an approximately 10-fold increase in τ1/2 compared with SNA-1 (τ1/2 = 2 min). The value of τ1/2 for the AS oligonucleotide of SNA-1-(3,3) was ∼8 min (Fig. 7B), which illustrates that the effect of increased resistance toward nuclease activity is dependent on the sites of and not the number of chemically modified nucleotides. The contrast between the serum lifetimes of SNA-1-(4,2) and SNA-1-(3,3) illustrates the effect of placing chemically modified nucleotides at experimentally identified sites of nuclease hydrolysis.
Fig. 7.
Site-specific chemical modification increases the half-life of siRNA-SNA. (A) Plot of the ratio of AS oligonucleotide/AuNP for SNA-1-(4,2) over time. Solid line is a single-exponential fit to the points, showing a half-life of 20 min. Error bars represent the range for n = 4 measurements. (B) Plot of the ratio of AS oligonucleotide per AuNP for SNA-1-(3,3) over time. A fit to a single exponential decay function showed a half-life of 8 min. Error bars represent the range for n = 4 measurements. (C) Analysis of the AS oligonucleotide recovered from SNA-1-(4,2) by PAGE. When SNA-1-(4, 2) is incubated in 10% FBS at 37 °C for 2 (lane ii), 15 (lane iii), and 30 (lane iv) min, the full-length AS oligonucleotide remains associated with SNA-1-(4,2). Degradation of AS oligonucleotide is observed at 30 min, but the intact oligonucleotide (lane i) remains the major species associated with SNA-1-(4,2). Analytical standards for SNA-1 (those without 2′-OMe nucleotides) are analyzed in lane v, as a comparison with the lengths of degradation products.
The improvement in τ1/2 from ∼2 min for SNA-1 to ∼20 min for SNA-1-(4,2) is significant because this length of time is one over which SNA-siRNA enter mammalian cells in large quantities. In a study of the cellular uptake of SNAs by C166 cells, Choi et al. reported that ∼105 SNAs were associated per cell upon 15-min incubation with cell culture media containing SNAs (6). We wished to demonstrate that the AS oligonucleotide present on SNA-1-(4,2) following a 30-min incubation in solutions of 10% FBS at 37 °C remained full length and therefore potentially active in gene silencing. Analysis of the AS oligonucleotide by PAGE showed that the band for the full-length oligonucleotide is the major band present with a minor band of mobility close to that of a 16-nt standard (Fig. 7C). These results illustrate a potential way to improve the applicability of SNA-siRNA that is susceptible to rapid degradation in serum-containing media, but is otherwise desirable for biological application (e.g., reasons of potency in gene silencing).
Conclusion
Previous research has investigated the interactions between oligonucleotides and serum nucleases but the emergence of oligonucleotides conjugated to nanoparticles opens up a new avenue of research that has yet to be fully explored. We sought to probe how siRNA immobilized on AuNPs are recognized and degraded by serum nucleases and whether this degradation is different from that of siRNA free in serum solution. To do so, we used SNAs, which are an ideal platform for studying such interactions because of their development as agents for intracellular gene regulation and promise as therapeutic and diagnostic candidates. We found that the rates of degradation for SNA-siRNAAR are very rapid (τ1/2 ∼2 min) compared with the rates for the previously investigated SNA-siRNAluc (τ1/2 ∼103 min) (9). This contrast illustrates the wide range in serum stability of siRNA conjugated to AuNPs and therefore demonstrates the dependence of serum stability on siRNA sequence. The surface density and orientation of siRNA on SNA-siRNA suggest the possibility that electrostatic and steric effects can reduce nuclease activity at sites near the AuNP surface (9, 32). This study of SNA-siRNAAR, however, showed that rapid nuclease-catalyzed hydrolysis at such sites is possible and found that the major factor driving this activity is recognition of specific sequence motifs (U–A and G–U, 5′–3′); the distance and orientation of these sequence motifs from the surface of the AuNP core are smaller factors. Importantly, we found that the sites of nuclease-catalyzed hydrolysis of the oligonucleotides bound to the AuNP are different from those for the free oligonucleotide. This observation strongly suggests that oligonucleotides conjugated to nanoparticles should be examined independently from their linear counterparts, due to the potential influence of the local chemical environment of the oligonucleotide–nanoparticle conjugates. Development of strategies for enhancing the serum stability of oligonucleotide–nanoparticle conjugates must consider the context and influence of the nanoparticle surface and should not rely solely on strategies developed from the investigation of siRNA free in solution.
Supplementary Material
Acknowledgments
S.N.B. acknowledges Dr. Dan Sweeny and the Integrated Molecular Structure Education and Research Center at Northwestern University (IMSERC; supported by Northwestern University and the State of Illinois) for assistance with the ESI-MS experiments. C.A.M. gratefully acknowledges funding for this research through the sponsorship of the following awards: Defense Advanced Research Projects Agency HR0011-13-2-0018, Center for Cancer Nanotechnology Excellence initiative of the National Institutes of Health U54 CA151880, Alliance for Cancer Gene Therapy Agreement 4/30/2013, Prostate Cancer Foundation 20110103, and Chicago Biomedical Consortium with support from the Searle Funds at The Chicago Community Trust C2006-00997/L-003. The content of the information does not necessarily reflect the position or the policy of the Government, and no official endorsement should be inferred. S.N.B. acknowledges a National Science Foundation graduate research fellowship and A.L. acknowledges an International Institute for Nanotechnology postdoctoral fellowship.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1409431111/-/DCSupplemental.
References
- 1.Love KT, et al. Lipid-like materials for low-dose, in vivo gene silencing. Proc Natl Acad Sci USA. 2010;107(5):1864–1869. doi: 10.1073/pnas.0910603106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim SH, Jeong JH, Lee SH, Kim SW, Park TG. LHRH receptor-mediated delivery of siRNA using polyelectrolyte complex micelles self-assembled from siRNA-PEG-LHRH conjugate and PEI. Bioconjug Chem. 2008;19(11):2156–2162. doi: 10.1021/bc800249n. [DOI] [PubMed] [Google Scholar]
- 3.Tang Y, et al. Efficient in vitro siRNA delivery and intramuscular gene silencing using PEG-modified PAMAM dendrimers. Mol Pharm. 2012;9(6):1812–1821. doi: 10.1021/mp3001364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jensen SA, et al. 2013. Spherical nucleic acid nanoparticle conjugates as an RNAi-based therapy for glioblastoma. Sci Transl Med 5(209):209ra152.
- 5.Zheng D, et al. 2012. Topical delivery of siRNA-based spherical nucleic acid nanoparticle conjugates for gene regulation. Proc Natl Acad Sci USA 109(30):11975–11980.
- 6.Choi CHJ, Hao L, Narayan SP, Auyeung E, Mirkin CA. Mechanism for the endocytosis of spherical nucleic acid nanoparticle conjugates. Proc Natl Acad Sci USA. 2013;110(19):7625–7630. doi: 10.1073/pnas.1305804110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patel PC, et al. Scavenger receptors mediate cellular uptake of polyvalent oligonucleotide-functionalized gold nanoparticles. Bioconjug Chem. 2010;21(12):2250–2256. doi: 10.1021/bc1002423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giljohann DA, et al. Oligonucleotide loading determines cellular uptake of DNA-modified gold nanoparticles. Nano Lett. 2007;7(12):3818–3821. doi: 10.1021/nl072471q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giljohann DA, Seferos DS, Prigodich AE, Patel PC, Mirkin CA. Gene regulation with polyvalent siRNA-nanoparticle conjugates. J Am Chem Soc. 2009;131(6):2072–2073. doi: 10.1021/ja808719p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoerter JA, Walter NG. Chemical modification resolves the asymmetry of siRNA strand degradation in human blood serum. RNA. 2007;13(11):1887–1893. doi: 10.1261/rna.602307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Behlke MA. Chemical modification of siRNAs for in vivo use. Oligonucleotides. 2008;18(4):305–319. doi: 10.1089/oli.2008.0164. [DOI] [PubMed] [Google Scholar]
- 12.Czauderna F, et al. Structural variations and stabilising modifications of synthetic siRNAs in mammalian cells. Nucleic Acids Res. 2003;31(11):2705–2716. doi: 10.1093/nar/gkg393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shukla S, Sumaria CS, Pradeepkumar PI. Exploring chemical modifications for siRNA therapeutics: A structural and functional outlook. ChemMedChem. 2010;5(3):328–349. doi: 10.1002/cmdc.200900444. [DOI] [PubMed] [Google Scholar]
- 14.Peacock H, Kannan A, Beal PA, Burrows CJ. Chemical modification of siRNA bases to probe and enhance RNA interference. J Org Chem. 2011;76(18):7295–7300. doi: 10.1021/jo2012225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lovrinovic M, Niemeyer CM. Rapid synthesis of DNA-cysteine conjugates for expressed protein ligation. Biochem Biophys Res Commun. 2005;335(3):943–948. doi: 10.1016/j.bbrc.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 16.Birmingham A, et al. A protocol for designing siRNAs with high functionality and specificity. Nat Protoc. 2007;2(9):2068–2078. doi: 10.1038/nprot.2007.278. [DOI] [PubMed] [Google Scholar]
- 17.Layzer JM, et al. In vivo activity of nuclease-resistant siRNAs. RNA. 2004;10(5):766–771. doi: 10.1261/rna.5239604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chiu Y-L, Rana TM. siRNA function in RNAi: A chemical modification analysis. RNA. 2003;9(9):1034–1048. doi: 10.1261/rna.5103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harborth J, et al. Sequence, chemical, and structural variation of small interfering RNAs and short hairpin RNAs and the effect on mammalian gene silencing. Antisense Nucleic Acid Drug Dev. 2003;13(2):83–105. doi: 10.1089/108729003321629638. [DOI] [PubMed] [Google Scholar]
- 20.Volkov AA, et al. Selective protection of nuclease-sensitive sites in siRNA prolongs silencing effect. Oligonucleotides. 2009;19(2):191–202. doi: 10.1089/oli.2008.0162. [DOI] [PubMed] [Google Scholar]
- 21.Waltering KK, et al. Increased expression of androgen receptor sensitizes prostate cancer cells to low levels of androgens. Cancer Res. 2009;69(20):8141–8149. doi: 10.1158/0008-5472.CAN-09-0919. [DOI] [PubMed] [Google Scholar]
- 22.Zhang J, et al. Fast separation of single-stranded oligonucleotides by capillary electrophoresis using OliGreen as fluorescence inducing agent. Electrophoresis. 2005;26(23):4449–4455. doi: 10.1002/elps.200500099. [DOI] [PubMed] [Google Scholar]
- 23.Liu X, Atwater M, Wang J, Huo Q. Extinction coefficient of gold nanoparticles with different sizes and different capping ligands. Colloids Surf B Biointerfaces. 2007;58(1):3–7. doi: 10.1016/j.colsurfb.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 24.Weickmann JL, Olson EM, Glitz DG. Immunological assay of pancreatic ribonuclease in serum as an indicator of pancreatic cancer. Cancer Res. 1984;44(4):1682–1687. [PubMed] [Google Scholar]
- 25.Raines RT. Ribonuclease A. Chem Rev. 1998;98(3):1045–1066. doi: 10.1021/cr960427h. [DOI] [PubMed] [Google Scholar]
- 26.Soutschek J, et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432(7014):173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 27.Hong J, et al. Comprehensive analysis of sequence-specific stability of siRNA. FASEB J. 2010;24(12):4844–4855. doi: 10.1096/fj.09-142398. [DOI] [PubMed] [Google Scholar]
- 28.Kraynack BA, Baker BF. Small interfering RNAs containing full 2′-O-methylribonucleotide-modified sense strands display Argonaute2/eIF2C2-dependent activity. RNA. 2006;12(1):163–176. doi: 10.1261/rna.2150806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Choung S, Kim YJ, Kim S, Park HO, Choi YC. Chemical modification of siRNAs to improve serum stability without loss of efficacy. Biochem Biophys Res Commun. 2006;342(3):919–927. doi: 10.1016/j.bbrc.2006.02.049. [DOI] [PubMed] [Google Scholar]
- 30.Rettig GR, Behlke MA. Progress toward in vivo use of siRNAs-II. Mol Ther. 2012;20(3):483–512. doi: 10.1038/mt.2011.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Majlessi M, Nelson NC, Becker MM. Advantages of 2′-O-methyl oligoribonucleotide probes for detecting RNA targets. Nucleic Acids Res. 1998;26(9):2224–2229. doi: 10.1093/nar/26.9.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shack J. The influence of sodium and magnesium ions on the action of deoxyribonuclease II. J Biol Chem. 1959;234(11):3003–3006. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.