Significance
Bloom syndrome is a rare human genetic disease characterized by proportional dwarfism, immunodeficiency, and an elevated risk of many different cancer types. We used RNA expression profiling to identify networks of mRNAs and microRNAs that are differentially expressed in cells from Bloom syndrome patients and associated with cell proliferation, survival, and molecular pathways promoting cancer. Altered mRNA expression was in some cases strongly correlated with the presence of G4 motifs, which may form G-quadruplex targets that are bound by BLM. Further analysis of the genetic networks we identified may elucidate mechanisms responsible for Bloom syndrome disease pathogenesis and ways to ameliorate or prevent disease in affected individuals.
Abstract
Bloom syndrome is a rare autosomal recessive disorder characterized by genetic instability and cancer predisposition, and caused by mutations in the gene encoding the Bloom syndrome, RecQ helicase-like (BLM) protein. To determine whether altered gene expression might be responsible for pathological features of Bloom syndrome, we analyzed mRNA and microRNA (miRNA) expression in fibroblasts from individuals with Bloom syndrome and in BLM-depleted control fibroblasts. We identified mRNA and miRNA expression differences in Bloom syndrome patient and BLM-depleted cells. Differentially expressed mRNAs are connected with cell proliferation, survival, and molecular mechanisms of cancer, and differentially expressed miRNAs target genes involved in cancer and in immune function. These and additional altered functions or pathways may contribute to the proportional dwarfism, elevated cancer risk, immune dysfunction, and other features observed in Bloom syndrome individuals. BLM binds to G-quadruplex (G4) DNA, and G4 motifs were enriched at transcription start sites (TSS) and especially within first introns (false discovery rate ≤ 0.001) of differentially expressed mRNAs in Bloom syndrome compared with normal cells, suggesting that G-quadruplex structures formed at these motifs are physiologic targets for BLM. These results identify a network of mRNAs and miRNAs that may drive the pathogenesis of Bloom syndrome.
Bloom syndrome (BS; OMIM 210900), an autosomal recessive disease caused by the loss of function mutations in the BLM gene (1), is characterized by proportional dwarfism from birth; genomic instability; immune deficiency; infertility; an elevated risk of type II diabetes; a characteristic sun-sensitive “butterfly” facial rash; and a strong predisposition to a wide variety of neoplasms, especially leukemia and lymphoma (2–5). BLM encodes the BLM RECQ helicase, an ATP-dependent 3′-to-5′ DNA helicase that binds and unwinds a variety of structured DNA substrates formed during DNA replication, recombination, and repair. These substrates include G-quadruplex structures (G4 DNA), forked DNA, D loops, and Holliday junctions. BLM also interacts physically and functionally with other proteins that play key roles in DNA metabolic processes involving these DNA intermediates (6–9).
BLM is one of five members of the human RECQ helicase family, which also includes the WRN, RECQL/RECQ1, RECQL4, and RECQL5/RECQ5 helicases (10). Mutations in either WRN or RECQL4 cause heritable cancer predisposition syndromes that share some features with BS. Werner syndrome (WS), caused by WRN mutations, is characterized by a prematurely aged appearance and an increased incidence of six specific tumor types (11–13). Rothmund–Thomson syndrome (RTS), caused by RECQL4 mutations, is characterized by developmental abnormalities and an elevated risk of osteosarcoma and lymphoma (14–16). Loss of BLM or WRN function leads to genomic instability, which may drive elevated cancer risk in both syndromes. Gene expression is altered in BS, WS, and RTS cells (17), but transcriptional regulatory targets and RECQ helicase-dependent gene networks have not been identified. Thus, a better understanding of how RECQ helicases contribute to gene structure and expression will aid our understanding of the roles of these proteins in human health and disease.
G4 DNA is a stable, nonB form DNA structure that can form at sequences conforming to the consensus G≥3N1-xG≥3N1-xG≥3N1-xG≥3, known as a G4 motif (18). G4 DNA can be bound and unwound by RECQ helicases (10), and by the XPD-related helicases such as human XPD (19) and its paralogs FANCJ (20, 21), CHL1 (22), and likely RTEL1 (23). In the human genome, G4 motifs are greatly enriched near transcription start sites (TSS), at the 5′ end of first introns, and in exons and introns of many oncogenes (reviewed in refs. 18, 24, and 25). Transcription of regions containing G4 motifs promotes the formation of G-quadruplex structures (26) that may block progression of RNA polymerase II and inhibit gene expression (27, 28). G4 motif-related gene expression changes have been reported in BS and WS cells (17). We recently showed that XPD and XPB, a second helicase component of TFIIH, are enriched at G4 motifs, particularly at the TSS and near genes in pathways correlated with cancer and development (19). These results suggest that G-quadruplex structures might be in vivo regulatory targets for BLM.
Our goal here was to identify transcriptional targets of BLM on a genome scale and reveal genes and gene networks expression of which is modulated by BLM. Therefore, we compared expression of genes in cells from 16 mutation-typed BS patients with matched normal controls and in cells depleted of BLM protein. We analyzed both mRNA and also microRNA (miRNA) expression, because miRNAs had not been systematically studied and could contribute to BS by regulating development, DNA repair, stress responses, and tumorigenesis (29, 30). Differential expression characterized mRNAs transcribed from genes enriched in G4 motifs, providing support for the view that G-quadruplex structures may regulate in vivo gene expression. The partial overlap among the mRNAs, miRNAs, and pathways altered in BS and BLM-depleted cells suggest a model in which BS pathogenesis is driven by a combination of the loss of BLM function in DNA metabolism together with gene expression changes.
Results
Expression Profiling Identifies BLM-Regulated Genes.
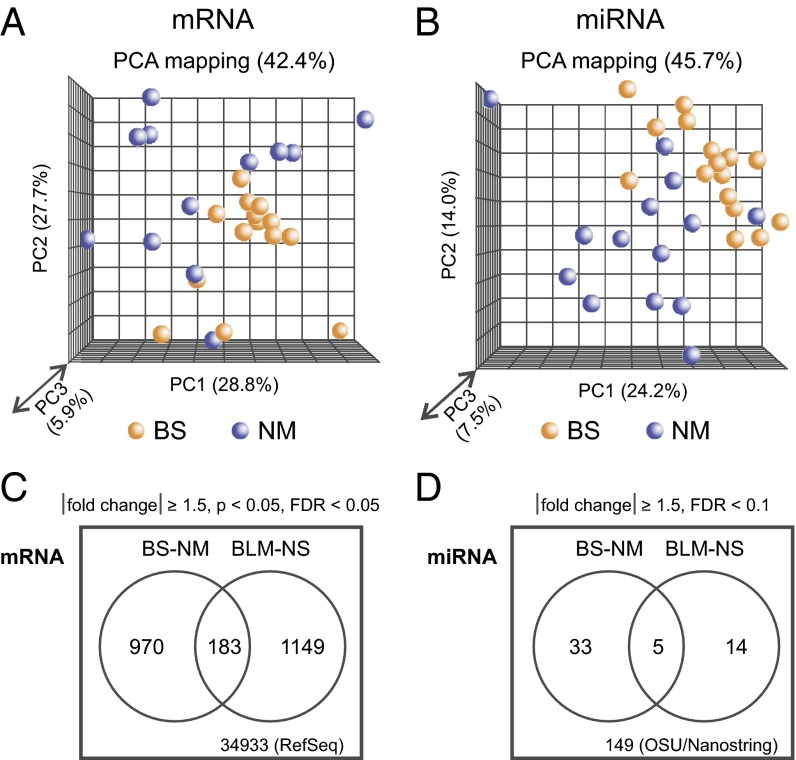
mRNA and miRNA expression was determined by array analysis of fibroblasts from 16 mutation-typed BS patients (BS) and 15 matched control donors (NM), and in BLM-depleted (BLM) or nonspecific shRNA-treated isogenic control fibroblasts (NS) (Table 1 and SI Appendix, Table S1). The experimental workflow is outlined in SI Appendix, Fig. S1. Principal component analysis (PCA) showed clustering of mRNAs and miRNAs by sample type, with no obvious clustering of BS donors by age, gender, or number of passages in culture (Fig. 1 A and B). We observed similar results for BLM-depleted cells (SI Appendix, Figs. S2 and S3). A subset of mRNA expression differences originally identified by microarray were also confirmed by quantitative PCR (qPCR) (SI Appendix, Fig. S4). Significant mRNA expression differences were identified by using an absolute expression ratio difference of ≥1.5 and a P < 0.05 with a FDR < 0.05. By these criteria, 1,153 mRNA-encoding genes were up-regulated (n = 1,012) or down-regulated (n = 141) in BS cells versus controls (Fig. 1C and SI Appendix, Table S2A). A prior analysis (17) that included cells from 3 of the 15 BS patients analyzed here identified significant expression changes in 1,111 genes (P value cutoff of 0.001) with similar proportions of up- and down-regulation (n = 867 up- and n = 247 down-regulated). BLM-depleted cells exhibited altered expression of 1,332 genes, with 580 up- and 752 down-regulated genes versus nonspecific shRNA-treated cells (Fig. 1C and SI Appendix, Table S2B). Among the 2,302 genes exhibiting altered expression, 183 (8%) were altered in both BS and BLM-depleted cells (binomial P value = 3.54 × 10−20; SI Appendix, Table S2C). Visual examination of expression data also identified other genes of interest such as cytidine deaminase (CDA), expression of which was significantly reduced in BLM-depleted cells (SI Appendix, Table S2B). CDA was previously reported to be down-regulated in BS cells (31) and is down-regulated as well in the BS patient samples analyzed here, although it did not meet our significance cutoff.
Table 1.
BS patient summary
| Cell type (n) | Age range (median), y | Sex | BLM mutations* |
| Bloom syndrome (16) | 3–43 (11.5†) | 9 M/7 F | 5 substitutions‡ |
| 1 insertion | |||
| 5 deletions | |||
| 1 indel§ | |||
| Control donors (15) | 4–45 (19) | 8 M/7 F | NA |
Mutation details are given in SI Appendix, Table S1.
Mean of middle two patients, ages 10 and 13.
Four coding/1 splice.
The BLMAsh mutation: 6-bp deletion/7-bp insertion at BLM c.2281.
Fig. 1.
mRNA and miRNA expression patterns distinguish BS and BLM-depleted cells from comparable controls. PCAs of mRNA expression (A) and miRNA expression in BS (orange) and NM (blue) cells (B) by using average distance and linkage measures. Percentages indicate the portion of expression differences that can be attributed to specific principal components. (C) Differentially expressed mRNAs identified in BS versus NM cells, and in BLM versus control fibroblasts treated with a nonspecific shRNA. (D) Differentially expressed miRNAs identified in BS versus NM cells, and in BLM versus control fibroblasts treated with a nonspecific shRNA. A total of 34,933 probes were present on mRNA arrays, whereas 149 miRNAs were represented in common on the array and Nanostring platforms used for miRNA expression profiling.
These differentially expressed gene lists were used for Ingenuity Pathway Analysis (IPA Ingenuity Systems; www.ingenuity.com) to identify functions and pathways altered in BS or BLM-depleted cells. Top-ranked molecular and cellular functions in BS cells included “cell growth/proliferation,” “cell death/survival,” “protein synthesis,” “gene expression,” and “cellular development,” with “molecular mechanisms of cancer” the top-ranked canonical pathway followed by pathways involved in protein ubiquitination and signaling (Tables 2 and 3). Top molecular and cellular functions identified in BLM-depleted cells included “cell cycle,” “cell assembly/ organization,” and “DNA replication/recombination and repair” with “cell cycle control of chromosome replication,” mitotic division and DNA damage signaling the top canonical pathways. Cellular growth and proliferation and cell death/survival were common to both BS and BLM-depleted cells (Tables 2 and 3).
Table 2.
Summary of IPA analysis driven by differentially expressed mRNA results of top molecular and cellular functions
| Bloom syndrome cells | BLM-depleted cells | ||||
| Function | P value* | Molec* | Function | P value* | Molec* |
| Cellular growth/proliferation | 3.9e−21 | 400 | Cell cycle | 4.4e−32 | 283 |
| Cell death/survival | 5.5e−20 | 391 | Cellular assembly/organization | 2.0e−30 | 296 |
| Protein synthesis | 2.8e−18 | 128 | DNA replication/recombination/repair | 2.0e−30 | 248 |
| Gene expression | 9.4e−13 | 260 | Cellular growth/proliferation | 4.2e−30 | 470 |
| Cellular development | 2.1e−12 | 339 | Cell death/survival | 5.2e−23 | 428 |
P values indicate the likelihood that the association between each set of molecules in the experiment and a given process or pathway is due to random chance. The number of molecules (molec) is the aggregate number of unique molecules in all sets within that category.
Table 3.
Summary of IPA analysis driven by differentially expressed mRNA results of top canonical pathways
| Pathway | P value* | Ratio* | Pathway | P value* | Ratio* |
| Molecular mechanisms of cancer | 1.4e−06 | 42/388 (0.11) | Mitotic roles of polo-like kinase | 4.0e−10 | 21/74 (0.28) |
| Protein ubiquitination pathway | 2.3e−06 | 34/270 (0.13) | BRCA1 role in DNA damage repair | 2.2e−09 | 19/71 (0.27) |
| Axonal guidance signaling | 4.8e−06 | 48/487 (0.10) | G2/M checkpoint regulation | 2.7e−07 | 14/49 (0.29) |
| PPARα/RXRα activation | 6.1e−06 | 26/200 (0.13) | Cell cycle control of chromosome replication | 4.6e−07 | 11/34 (0.32) |
| Glucocorticoid receptor signaling | 1.0e−05 | 32/299 (0.11) | ATM signaling | 5.9e−07 | 16/66 (0.24) |
Top canonical pathway P values provide a measure of the nonrandom association between a pathway and the dataset, whereas ratios reflect the number of molecules in a given pathway that meet expression cutoff criteria divided by the total number of pathway molecules. Ratios thus indicate fractional pathway regulation under a given experimental condition.
An expanded gene set enrichment analysis (GSEA) conditioned on BLM was used to gain a more detailed view of BLM-dependent expression changes. Among the 9,969 gene sets contained in the MSigDB C2–C7 collections, we identified highly significant enrichment for 244 gene sets in BS cells and for 101 gene sets in BLM-depleted cells. Of note, 90% (91/101) of the significant gene sets altered in BLM-depleted cells were found among the 244 gene sets significantly altered in BS cells (Table 4 and SI Appendix, Table S3).
Table 4.
Summary of GSEA analyses driven by differentially expressed mRNA results
| MSigDB collections | No. of genesets | Cutoff P value | No. of significant genesets* | ||
| BS-NM | BLM-NS | Common | |||
| C2: CGP (chemical and genetic perturbations) | 3,402 | 1.00e−08 | 39 | 10 | 9 |
| C2: CP (canonical pathways) | 1,320 | 0.05 | 31 | 8 | 7 |
| C3: MIR (microRNA targets) | 221 | 0.05 | 18 | 10 | 10 |
| C3: TFT (transcriptional factor targets) | 615 | 0.05 | 18 | 13 | 10 |
| C4: CM (cancer modules) | 431 | 0.05 | 5 | 3 | 1 |
| C4: CGN (cancer gene neighborhoods) | 427 | 0.05 | 2 | 0 | 0 |
| C5: BP (GO Biological Process Ontology) | 825 | 0.05 | 20 | 8 | 6 |
| C5: CC (GO cellular component) | 233 | 0.05 | 1 | 0 | 0 |
| C5: MF (GO molecular function) | 396 | 0.05 | 7 | 2 | 1 |
| C6: Oncogenic signatures | 189 | 0.05 | 2 | 2 | 2 |
| C7: immunologic signatures | 1,910 | 0.05 | 101 | 45 | 45 |
GSEA results are reported in greater detail in SI Appendix, Table S3.
Genesets identified in the BS vs. normal donor (BS-NM), BLM-depleted vs. nonspecific shRNA-depleted (BLM-NS) cells, and number identified in both cell types (common). See Methods for additional detail.
Canonical pathways identified by GSEA in both BS and BLM-depleted cells included Notch expression, processing, and signaling; ARF, p38MAPK, TGF-β, and NF-κB signaling; protein kinase activity; and nucleotide binding and nucleic acid metabolic processes (SI Appendix, Table S3). All 45 of the gene sets associated with diverse immunologic cell types and processes in BLM-depleted cells were present among the 101 immune function gene sets altered in BS cells (SI Appendix, Table S3). These results collectively identify expression changes in mRNA-coding genes that are common to both BS and BLM-depleted cells, and changes that are BS-specific.
Differentially Expressed miRNAs Are Often Cancer-Associated.
One-third (52/149) of the miRNAs we assayed in both BS and BLM-depleted cells exhibited significant differential expression in one or both cell types (absolute expression ratio difference of ≥1.5 and FDR < 0.1 (Fig. 1D and SI Appendix, Table S4). We verified a subset of these miRNA expression differences by qPCR (SI Appendix, Fig. S5). Of the miRNAs differentially expressed in BS cells, half (19/38) were up-regulated and half (19/30) down-regulated. The top differentially expressed miRNAs in BS cells were miR-181, miR-595, miR-155, miR-412, miR-30, miR-29, and miR-130 family members (up-regulated), and miR-143, miR-499, miR-92, miR-145, and let-7 family members (down-regulated). Of note, most of these differentially expressed miRNAs (29 of 38, or 76%, in BS cells; 14 of 19 or 74% in BLM-depleted cells) were cancer-associated in MalaCards: Human Malady Compendium (32) database (www.malacards.org). Both miR-181 family and miR-155, up-regulated in BS cells, are also up-regulated in many hematopoietic and leukemic malignancies such as B-cell lymphoma, Hodgkin lymphoma, acute myelogenous leukemia, and chronic lymphocytic leukemia (33–36). These malignancies are some of the most common cancers in BS patients (5). The miRNAs miR-143 and miR-145, down-regulated in both BS and BLM-depleted cells, may impair an MDM2-p53 feedback loop to promote cell proliferation and hinder apoptosis and, thus, could be promoting tumorigenesis in BS patients (37). Altered miRNA expression in BS might also deregulate islet cell function to promote the development of diabetes mellitus with impaired insulin signaling (38).
We used miRTarBase data (39) to determine whether altered miRNA expression might be driving BLM-dependent gene expression changes. Nineteen of the 52 miRNAs differentially expressed in BS and/or BLM-depleted cells had experimentally validated mRNA targets that were differentially expressed in the opposite direction in one or both cell types (SI Appendix, Tables S4–S6). Target genes with the potential to promote tumorigenesis that exhibited up-regulated expression in BS cells included the following: growth factor receptor/oncogene genes such as EGFR, FLI1, FZD7, IRS1, MYC, and PDGFRA; antiapoptotic genes such as BIRC6; and MDM2, which encodes a negative regulator of TP53 (SI Appendix, Table S6A). These results indicate that a subset of BLM-dependent mRNA expression changes may be driven by altered miRNA expression, where up- or down-regulation has the potential to promote cancer risk or cancer pathogenesis in BS patients.
GSEA identified 40 miRNAs that were significantly altered in BS or BLM-depleted cells, together with gene sets that share 3′-UTR microRNA-specific binding motifs (SI Appendix, Table S3). Five of these miRNAs exhibited altered expression in BS and/or BLM-depleted cells (SI Appendix, Tables S4 and S5). Gene sets that shared a transcription factor-binding site as defined in TRANSFAC (version 7.4) and were significantly altered in BS or BLM-depleted cells contained sites for MYB, FOXO1, E4BP4, HTF, and HIF1. Five other gene sets shared motifs not yet mapped to specific transcription factors (SI Appendix, Table S3). These analyses collectively identify several potential regulatory mechanisms involving miRNAs and transcription factors that could be modulating gene expression in a BLM-dependent fashion.
Enrichment of G4 Motifs Correlates with BLM-Dependent Expression.
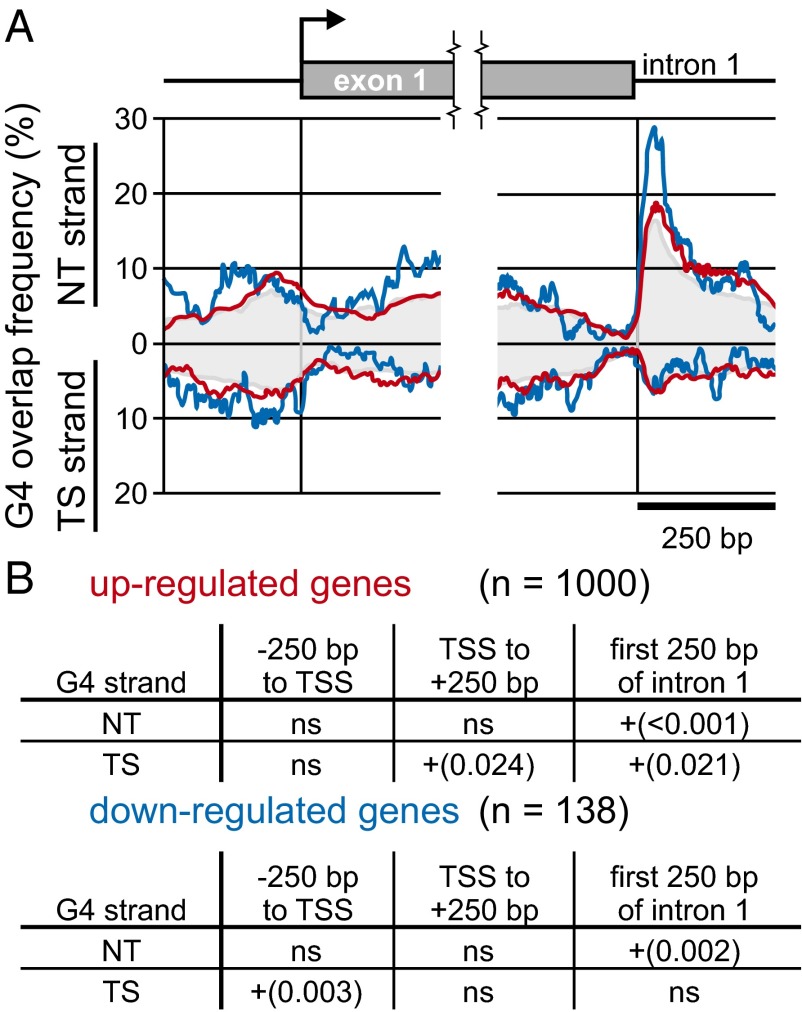
G4 DNA is bound and unwound by BLM (7). We therefore asked whether G4 motifs are enriched near TSSs and at the 5′ end of the first intron of genes encoding mRNAs differentially expressed in BS or BLM-depleted cells (SI Appendix, Tables S7 and S8). Among genes down-regulated in BS cells, we found highly significant enrichment of G4 motifs upstream of the TSS on the transcribed strand (TS; FDR = 0.003), and at the 5′ end of the first intron on the nontranscribed strand (NT; Fig. 2; FDR = 0.002). More than 30% of genes down-regulated in BS cells (40 of 131 with intron 1 annotations) carried a G4 motif overlapping the position 30 bp downstream of the exon 1/intron 1 boundary. Among genes up-regulated in BS cells, G4 motifs were enriched flanking the TSS on the transcribed strand (FDR = 0.024), and at the 5′ end of the first intron on both the nontranscribed (FDR < 0.001) and transcribed strands (FDR = 0.021) (Fig. 2). A similar analysis of BLM-depleted cells identified enrichment of G4 motifs near the 5′ end of the first intron on the NT strand of genes down-regulated upon BLM depletion (SI Appendix, Fig. S6; FDR = 0.017). This pattern of enrichment is shared with genes down-regulated in BS cells, although the position of these G4 motifs appears to be more consistently close to the exon/intron boundary. These results collectively suggest that G4 motifs in intron 1 may form G-quadruplex structures that may be targets for BLM and BLM-dependent transcription regulation.
Fig. 2.
G4 motif frequencies and enrichment near transcription start sites and intron 1 boundaries of genes with altered expression in BS cells. (A) G4 motif frequencies as a function of location in genes up- and down-regulated in BS patient cells. G4 motif counts are shown for NT and TS over the 250 bp ± gene TSSs and exon 1/intron 1 5′ boundaries for genes expression of which was up- or down-regulated (red and blue curves, respectively) versus the comparable distributions for 1,000 randomly selected gene sets of equal size drawn from the pool of all genes on expression arrays. Gray areas indicate average G4 motif frequencies among all genes surveyed on expression arrays. (B) FDR of observed G4 motif frequencies compared with 1,000 randomly selected gene sets of equal size. ns, nonsignificant (FDR > 0.05).
Some miRNA loci are also adjacent to G4 motifs. However, the relatively small number of differentially expressed miRNAs prevented us from further evaluating the relationship between miRNA G4 motifs and altered miRNA expression.
Discussion
We used RNA expression profiling of BS patient and BLM-depleted primary control fibroblasts to identify mRNAs and miRNAs exhibiting altered expression in a BLM-dependent manner. Our aim was to identify individual genes and gene networks that are regulated by BLM and may, in turn, drive BS pathogenesis. RNA expression profiling identified altered expression of 1,153 mRNAs in BS cells and 1,332 mRNAs in BLM-depleted cells, with 183 genes altered in both cell types. Thirty-eight miRNAs exhibited altered expression in BS cells and 19 in BLM-depleted cells, including 5 common to both cell types. Our results show that BLM helicase regulates gene expression and that one mechanism by which regulation occurs may involve direct binding to G-quadruplex structures.
BLM Regulation of Gene Expression.
We identified strong enrichment of G4 motifs at TSSs, and especially within the first intron of genes exhibiting altered regulation in BS patient cells. BLM binds to and unwinds G4 DNA (7, 9, 10). This enrichment of G4 motifs, also observed among targets of the transcriptional helicases XPB and XPD (19), supports the view that G-quadruplex structures are targets for regulation of gene expression genomewide. G4 motifs are enriched at the 5′ ends of first intron in many human genes (40), but the motifs associated with BLM regulation were distinguished by their proximity (within 30 bp) to the boundary between the first exon and intron. The first intron occupies a privileged genomic position, because it is close to the promoter (the average distance between the TSS +1 position and 5′ boundary of intron 1 is 98 bp), within the region where RNAP2 pauses, and it can evolve without altering protein sequence. Recent ChIP-Seq analysis of XPB/XPD showed that 40% of the sites bound by these two helicases in human cells contained G4 motifs, but significant enrichment was observed only near the TSS and not within the first intron (19). This distinction between XPB/XPD and BLM targets supports the view that different proteins recognize distinct G4 quadruplex structures in vivo to modulate gene expression (18).
BLM Mutation and Depletion Reveal Overlapping Gene Networks.
BLM mutation and BLM depletion altered the expression of different sets of mRNAs and miRNAs (Fig. 1 C and D). IPAs and GSEAs indicated a higher degree of similarity between BS and BLM-depleted cells than suggested by the modest number of mRNAs and miRNAs exhibiting altered expression in both cell types (Fig. 1 C and D). In IPAs, BLM depletion strongly perturbed molecular and cellular functions and canonical pathways that reflected the immediate loss of DNA metabolic functions of BLM (Tables 2 and 3). The top-ranked molecular and cellular functions and canonical pathways altered in BS cells, in contrast, involved cell growth, proliferation, and survival; protein synthesis; gene expression; and molecular mechanisms of cancer together with altered protein ubiquitination and cell signaling. The only prominent overlaps between BS and BLM-depleted cells identified in this analysis were molecular and cellular functions involved in cell growth, proliferation, and survival (Tables 2 and 3). GSEAs provided a more detailed view of altered gene expression that affirmed underlying similarities in gene expression in both BS and BLM-depleted cells. Of note, among the 101 gene sets altered in BLM-depleted cells, 90% (91/101) were also identified among the 244 gene sets significantly altered in BS cells (Table 4 and SI Appendix, Table S3).
Role of Transcriptional Regulation by BLM in the Pathogenesis of BS.
Our results suggest a model for the pathogenesis of BS in which loss of BLM nucleic acid metabolic functions leads to DNA damage, genomic instability and cell loss, together with altered mRNA and miRNA expression changes that are themselves modulated by the loss of BLM. The identity of the affected genes provides a useful way to begin to understand the origins of key features of BS. For example, IPAs revealed cell growth/proliferation and cell death/survival as the two top-ranked molecular and cellular functions in BS cells (Table 2). Altered cell proliferation and survival provide a ready explanation for the low birth weight and proportional dwarfism characteristic of BS (2–4, 41): Development as a process is normal, but output is scaled in response to a reduced number of available cells.
Similar quantitative and qualitative cellular defects driven by genomic instability, cell loss, and altered gene expression may explain the origin of other prominent features of BS. For example, s.c. tissue loss and the sun-sensitive rash observed in many BS patients may reflect a combination of proliferative and DNA damage repair defects, leading to cutaneous cell loss, senescence, and chronic inflammation. The immune deficiency of BS may reflect a combination of lymphoid proliferative defects together with class switch recombination defects and defective cell signaling. A surprising finding in our GSEA was the strongly perturbed expression of gene sets representing immune cell lineages, development, and function in both BS and BLM-depleted cells (Table 4 and SI Appendix, Table S3). These changes may be driving impaired immune function in BS patients. Low fertility and the elevated risk of diabetes mellitus could in similar fashion reflect lineage-specific genetic instability, proliferative defects, and cell loss together with altered gene expression, leading, respectively, to the depletion of germ cells and to dysfunction or loss of pancreatic islet β cells with altered insulin signaling (38).
A high risk of cancer, a cardinal feature of BS (4, 5), may reflect the accumulation of mutant or genetically unstable cells in many cell lineages, tissues, and organs during and after development. This reservoir of mutant cells would provide a substrate for the eventual emergence of neoplastic clones that could be further promoted by procarcinogenic, BLM-dependent gene, and miRNA expression changes. Of note, the top-ranked canonical pathway identified in IPAs of BS cells was molecular mechanisms of cancer (Table 3). The individual genes and gene networks identified as part of this analysis should be useful in guiding more detailed, mechanism-oriented analyses of the origins of BS cellular and organismal phenotypes. A deeper understanding of these disease mechanisms should provide a detailed view of the pathogenesis of BS and may suggest new opportunities to ameliorate or prevent disease in individuals affected with BS.
Materials and Methods
Detailed materials and methods are in SI Appendix, SI Materials and Methods.
Cell Sources.
Primary human skin fibroblast strains (Table 1 and SI Appendix, Table S1) from 16 mutation-typed BS patients (n = 16, donor median age 11.5 y) or from 11 control donors (n = 15, donor median age 19 y) were obtained from the Coriell Cell Repositories. NMs were age and sex-matched to BS cases. The control human primary fibroblast strain 82-6 was initiated from a foreskin fibroblast preparation as described (42). BLM protein depletion was performed by lentiviral expression of a BLM-specific shRNA, and depletions of >90% were verified by Western blot as described (43) (SI Appendix, Fig. S2) in biological triplicate samples for comparison with 82-6 fibroblasts expressing a scrambled shRNA with no known target sequence in the human genome (plasmid 1864; Addgene) or no shRNA.
mRNA and miRNA Expression Profiling.
Whole genome transcript exon profiling was performed on Affymetrix GeneChip Human Exon 1.0 ST (Affymetrix) arrays by using manufacture-recommended protocols. Samples from primary BS and NM fibroblasts were analyzed in triplicate, and 82-6 primary human fibroblasts were depleted with BLM-shRNA or treated with nonspecific shRNA, in duplicate. miRNA expression in BS and NM primary fibroblasts was analyzed by using a custom miRNA microarray chip (OSU-CCC version 4.0) that includes 898 probes to human and 704 probes to mouse mature or precursor miRNAs, spotted in duplicate (44). miRNA expression profiling of BLM-depleted 82-6 fibroblasts was performed by using the Nanostring nCounter Human miRNA Expression Assay Kit (Nanostring). A total of 149 miRNAs were shared between these two platforms.
Microarray data were preprocessed and normalized with Affymetrix Expression Console software by using RMA normalization (affymetrix.com). Differentially expressed genes were identified by using the Bioconductor limma package and a linear fixed effects model with adjustments for donor age and sex (45, 46). Preprocessing and normalization of OSU-CCC miRNA microarray data from BS and NM primary fibroblasts were done in R (version 2.6.0). nCounter RCC files generated from BLM-depleted samples were imported into nSolver (Nanostring) for quality control and normalized to the geometric mean of the top 100 expressed miRNAs. Differential expression was then analyzed by using ANOVA adjusted for age and gender for BS and control samples. Exploratory analyses and visualizations were performed by using the Partek Genomics Suite (Partek). mRNA and miRNA expression profiling data are archived in the Gene Expression Omnibus under accession no. GSE54502.
RNA Expression Analysis.
PCA (47) was used to compare global mRNA and miRNA expression in BS patients and matched NM, and in BLM and NS- or CO-treated isogenic 82-6 fibroblasts. mRNA expression data were analyzed by a combination of enhanced single gene analysis (SGA) and expanded GSEA conditioned on genotype. SGAs focused on the identification of mRNAs with significantly altered expression, as defined by an absolute fold expression difference of ≥1.5 with a P < 0.05 and a FDR of <0.05. Differential miRNA expression was defined by an absolute fold expression change of ≥1.5 with FDR < 0.1.
IPA.
We used differentially expressed gene lists to perform an Ingenuity Pathway Core Analysis (IPA Ingenuity Systems) to identify molecular and cellular functions and canonical pathways altered in BS or BLM-depleted cells. Default parameters were used to identify networks, diseases, biological functions, and canonical pathways enriched within a given sample type, and to identify both direct and indirect regulatory interactions predicted with high confidence and/or previously experimentally verified.
GSEAs.
GSEAs were performed by using the romer function in the limma package in Bioconductor (46), and the gene sets were contained in the Molecular Signature Database version 4.0 (48) (www.broad.mit.edu/gsea/msigdb/msigdb_index.html). GSEA was performed by using gene sets C2–C7. We did not analyze C1 gene sets defined by chromosomal position. The romer function allows the identification of gene sets with strong cross-correlation by boosting the signal-to-noise ratio. This function makes it possible to detect even modest changes in gene expression with confidence. The P values for the intersection of common genes with gene sets were calculated with the phyper function of R package (49), which uses a hypergeometric distribution and sampling without replacement. This test is the same statistical test used to test associations in Gene Ontology analysis (50).
miRNA Targets Analysis.
miRTarBase (39) (http://mirtarbase.mbc.nctu.edu.tw) was used to identify gene targets of miRNAs that were differentially expressed in BS and BLM-depleted cells. A hypergeometric test was then used to determine the significance of associations of a given miRNA with differentially expressed genes in BS or BLM-depleted cells versus all targets for a given miRNA.
G4 Enrichment Statistics.
G4 motifs were located in hg19 by using Quadparser (24) with settings for four or more runs of three or more consecutive Gs separated by 1- to 12-nt loops on either strand of the genome (GC 3 4 1 12) in DAS format. Quadparser results were converted to BED format by using Perl scripts to yield 722,264 G4 motif annotations. RefSeq gene annotations were retrieved from the University of California, Santa Cruz genome browser database (51). R scripts were then used to select regions flanking the RefSeq TSS that were 250 bp upstream and downstream of the transcription start site (TSS ± 250 bp) and encompassing the 250 bp at the 5′ end of the first intron (SI Appendix, Tables S7 and S8), and to calculate the number of G4 motifs that overlapped these regions on the transcribed (TS) or nontranscribed (NT) DNA strands of each gene. Genes from expression arrays were matched to RefSeq regions by using mRNA accessions. Of 17,318 genes queried by the expression arrays, 16,745 (>96%) were matched to TSSs in the RefSeq table; and 15,728 (>90%) genes were matched to intron 1 locations. Gene symbols for all differentially expressed genes with one or more G4 motifs in each region are recorded in SI Appendix, Table S7. We then calculated the number of genes in each set with one or more G4 motif, the average number of G4 motifs per gene, and the total number of G4 motifs per gene in each region (SI Appendix, Table S8). To determine significance and exclude GC richness as a potential confounder of the analysis, each set of genes with significantly altered expression was compared with 1,000 randomly selected gene sets of equal size drawn from the pool of all genes in the expression arrays (SI Appendix, Table S8). These random sets were used to empirically determine FDRs for enrichment or depletion of G4 motifs, using FDR < 0.05 (5%) as a cutoff for significance.
Supplementary Material
Acknowledgments
We thank the staffs at the Laboratory of Molecular Technology [National Cancer Institute (NCI)-Frederick], the Center for Ecogenetics and Environmental Health Functional Genomics and Proteomics Core [University of Washington (UW)], and the Microarray Core Facility at the Ohio State University for the microarray hybridization and scanning; Yizhe Tang and Scott Hudson [Laboratory of Human Carcinogenesis, NCI, National Institutes of Health (NIH)] for valuable suggestions; Alden Hackmann for graphics support; and UW RECQ Program Project colleagues for many helpful suggestions. This work was supported by the Intramural Research Program of the NCI, Center for Cancer Research, NIH; NCI of NIH Award P01CA77852 (to R.J.M. and N.M.) and by the NIH-Oxford MD/DPhil Fellowship Program support (to G.H.N.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE54502).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1404807111/-/DCSupplemental.
References
- 1.Ellis NA, et al. The Bloom’s syndrome gene product is homologous to RecQ helicases. Cell. 1995;83(4):655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- 2.Bloom D. Congenital telangiectatic erythema resembling lupus erythematosus in dwarfs; probably a syndrome entity. AMA Am J Dis Child. 1954;88(6):754–758. [PubMed] [Google Scholar]
- 3.German J. Bloom's syndrome. VIII. Review of clinical and genetic aspects. In: Goodman RM, Motulsky AG, editors. Genetic Diseases Among Askenazi Jews. Vol 1. New York: Raven; 1979. pp. 121–139. [Google Scholar]
- 4.German J. Bloom syndrome: A mendelian prototype of somatic mutational disease. Medicine (Baltimore) 1993;72(6):393–406. [PubMed] [Google Scholar]
- 5.German J. Bloom’s syndrome. XX. The first 100 cancers. Cancer Genet Cytogenet. 1997;93(1):100–106. doi: 10.1016/s0165-4608(96)00336-6. [DOI] [PubMed] [Google Scholar]
- 6.Karow JK, Chakraverty RK, Hickson ID. The Bloom’s syndrome gene product is a 3′-5′ DNA helicase. J Biol Chem. 1997;272(49):30611–30614. doi: 10.1074/jbc.272.49.30611. [DOI] [PubMed] [Google Scholar]
- 7.Sun H, Karow JK, Hickson ID, Maizels N. The Bloom’s syndrome helicase unwinds G4 DNA. J Biol Chem. 1998;273(42):27587–27592. doi: 10.1074/jbc.273.42.27587. [DOI] [PubMed] [Google Scholar]
- 8.Karow JK, Constantinou A, Li JL, West SC, Hickson ID. The Bloom’s syndrome gene product promotes branch migration of holliday junctions. Proc Natl Acad Sci USA. 2000;97(12):6504–6508. doi: 10.1073/pnas.100448097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mohaghegh P, Karow JK, Brosh RM, Jr, Bohr VA, Hickson ID. The Bloom’s and Werner’s syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 2001;29(13):2843–2849. doi: 10.1093/nar/29.13.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bachrati CZ, Hickson ID. RecQ helicases: Suppressors of tumorigenesis and premature aging. Biochem J. 2003;374(Pt 3):577–606. doi: 10.1042/BJ20030491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Epstein CJ, Martin GM, Schultz AL, Motulsky AG. Werner’s syndrome a review of its symptomatology, natural history, pathologic features, genetics and relationship to the natural aging process. Medicine (Baltimore) 1966;45(3):177–221. doi: 10.1097/00005792-196605000-00001. [DOI] [PubMed] [Google Scholar]
- 12.Yu CE, et al. Positional cloning of the Werner’s syndrome gene. Science. 1996;272(5259):258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- 13.Lauper JM, Krause A, Vaughan TL, Monnat RJ., Jr Spectrum and risk of neoplasia in Werner syndrome: A systematic review. PLoS ONE. 2013;8(4):e59709. doi: 10.1371/journal.pone.0059709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kitao S, et al. Mutations in RECQL4 cause a subset of cases of Rothmund-Thomson syndrome. Nat Genet. 1999;22(1):82–84. doi: 10.1038/8788. [DOI] [PubMed] [Google Scholar]
- 15.Wang LL, et al. Association between osteosarcoma and deleterious mutations in the RECQL4 gene in Rothmund-Thomson syndrome. J Natl Cancer Inst. 2003;95(9):669–674. doi: 10.1093/jnci/95.9.669. [DOI] [PubMed] [Google Scholar]
- 16.Siitonen HA, et al. The mutation spectrum in RECQL4 diseases. Eur J Hum Genet. 2009;17(2):151–158. doi: 10.1038/ejhg.2008.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson JE, Cao K, Ryvkin P, Wang LS, Johnson FB. Altered gene expression in the Werner and Bloom syndromes is associated with sequences having G-quadruplex forming potential. Nucleic Acids Res. 2010;38(4):1114–1122. doi: 10.1093/nar/gkp1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maizels N, Gray LT. The G4 genome. PLoS Genet. 2013;9(4):e1003468. doi: 10.1371/journal.pgen.1003468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gray LT, Vallur AC, Eddy J, Maizels N. G quadruplexes are genomewide targets of transcriptional helicases XPB and XPD. Nat Chem Biol. 2014;10(4):313–318. doi: 10.1038/nchembio.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.London TBC, et al. FANCJ is a structure-specific DNA helicase associated with the maintenance of genomic G/C tracts. J Biol Chem. 2008;283(52):36132–36139. doi: 10.1074/jbc.M808152200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu Y, Shin-ya K, Brosh RM., Jr FANCJ helicase defective in Fanconia anemia and breast cancer unwinds G-quadruplex DNA to defend genomic stability. Mol Cell Biol. 2008;28(12):4116–4128. doi: 10.1128/MCB.02210-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu Y, Sommers JA, Khan I, de Winter JP, Brosh RM., Jr Biochemical characterization of Warsaw breakage syndrome helicase. J Biol Chem. 2012;287(2):1007–1021. doi: 10.1074/jbc.M111.276022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uringa E-J, Youds JL, Lisaingo K, Lansdorp PM, Boulton SJ. RTEL1: An essential helicase for telomere maintenance and the regulation of homologous recombination. Nucleic Acids Res. 2011;39(5):1647–1655. doi: 10.1093/nar/gkq1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33(9):2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huppert JL, Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35(2):406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duquette ML, Handa P, Vincent JA, Taylor AF, Maizels N. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev. 2004;18(13):1618–1629. doi: 10.1101/gad.1200804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tornaletti S, Park-Snyder S, Hanawalt PC. G4-forming sequences in the non-transcribed DNA strand pose blocks to T7 RNA polymerase and mammalian RNA polymerase II. J Biol Chem. 2008;283(19):12756–12762. doi: 10.1074/jbc.M705003200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Belotserkovskii BP, et al. Mechanisms and implications of transcription blockage by guanine-rich DNA sequences. Proc Natl Acad Sci USA. 2010;107(29):12816–12821. doi: 10.1073/pnas.1007580107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ameres SL, Zamore PD. Diversifying microRNA sequence and function. Nat Rev Mol Cell Biol. 2013;14(8):475–488. doi: 10.1038/nrm3611. [DOI] [PubMed] [Google Scholar]
- 30.Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol. 2014;9(1):287–314. doi: 10.1146/annurev-pathol-012513-104715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chabosseau P, et al. Pyrimidine pool imbalance induced by BLM helicase deficiency contributes to genetic instability in Bloom syndrome. Nat Commun. 2011;2:368. doi: 10.1038/ncomms1363. [DOI] [PubMed] [Google Scholar]
- 32.Rappaport N, et al. MalaCards: An integrated compendium for diseases and their annotation. Database (Oxford) 2013;2013:bat018. doi: 10.1093/database/bat018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eis PS, et al. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci USA. 2005;102(10):3627–3632. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodriguez A, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316(5824):608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pekarsky Y, et al. Tcl1 expression in chronic lymphocytic leukemia is regulated by miR-29 and miR-181. Cancer Res. 2006;66(24):11590–11593. doi: 10.1158/0008-5472.CAN-06-3613. [DOI] [PubMed] [Google Scholar]
- 36.Visone R, et al. Karyotype-specific microRNA signature in chronic lymphocytic leukemia. Blood. 2009;114(18):3872–3879. doi: 10.1182/blood-2009-06-229211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang J, et al. Loss of microRNA-143/145 disturbs cellular growth and apoptosis of human epithelial cancers by impairing the MDM2-p53 feedback loop. Oncogene. 2013;32(1):61–69. doi: 10.1038/onc.2012.28. [DOI] [PubMed] [Google Scholar]
- 38.Shantikumar S, Caporali A, Emanueli C. Role of microRNAs in diabetes and its cardiovascular complications. Cardiovasc Res. 2012;93(4):583–593. doi: 10.1093/cvr/cvr300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hsu SD, et al. miRTarBase: A database curates experimentally validated microRNA-target interactions. Nucleic Acids Res. 2011;39(Database issue):D163–D169. doi: 10.1093/nar/gkq1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eddy J, Maizels N. Conserved elements with potential to form polymorphic G-quadruplex structures in the first intron of human genes. Nucleic Acids Res. 2008;36(4):1321–1333. doi: 10.1093/nar/gkm1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keller C, Keller KR, Shew SB, Plon SE. Growth deficiency and malnutrition in Bloom syndrome. J Pediatr. 1999;134(4):472–479. doi: 10.1016/s0022-3476(99)70206-4. [DOI] [PubMed] [Google Scholar]
- 42.Dhillon KK, et al. Werner syndrome protein loss leads to divergent mouse and human cell phenotypes. DNA Repair (Amst) 2008;9:11–22. doi: 10.1016/j.dnarep.2009.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mao FJ, Sidorova JM, Lauper JM, Emond MJ, Monnat RJ. The human WRN and BLM RecQ helicases differentially regulate cell proliferation and survival after chemotherapeutic DNA damage. Cancer Res. 2010;70(16):6548–6555. doi: 10.1158/0008-5472.CAN-10-0475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mathé EA, et al. MicroRNA expression in squamous cell carcinoma and adenocarcinoma of the esophagus: Associations with survival. Clin Cancer Res. 2009;15(19):6192–6200. doi: 10.1158/1078-0432.CCR-09-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smyth GK. 2004. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3(1)
- 46.Gentleman RC, et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jolliffe IT. Principle Component Analysis. 2nd Ed. New York: Springer; 2002. p. 486. [Google Scholar]
- 48.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnson NL, Kotz S, Kemp AW. 1992. Univariate Discrete Distributions (Wiley, New York), 2nd Ed.
- 50.Ashburner M, et al. The Gene Ontology Consortium Gene ontology: Tool for the unification of biology. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meyer LR, et al. The UCSC Genome Browser database: Extensions and updates 2013. Nucleic Acids Res. 2013;41(Database issue) D1:D64–D69. doi: 10.1093/nar/gks1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.