Significance
Hyperpolarization-activated cyclic nucleotide-gated (HCN) ion channels play central roles in the heart and the brain. In the heart, they are present in pacemaker cells and contribute to the regulation of the heartbeat. In the brain, they are involved in electrical signaling of neurons. HCN channels are activated by hyperpolarization of the cell membrane and are regulated by binding of cAMP to a site in an intracellular binding domain. This study shows that this binding domain undergoes major structural changes upon binding of cAMP. The results are the first step toward elucidating the molecular mechanism of gating in this important class of ion channels.
Keywords: hyperpolarization-activated ion channels, allosteric regulation
Abstract
Binding of 3′,5′-cyclic adenosine monophosphate (cAMP) to hyperpolarization-activated cyclic nucleotide-gated (HCN) ion channels regulates their gating. cAMP binds to a conserved intracellular cyclic nucleotide-binding domain (CNBD) in the channel, increasing the rate and extent of activation of the channel and shifting activation to less hyperpolarized voltages. The structural mechanism underlying this regulation, however, is unknown. We used double electron–electron resonance (DEER) spectroscopy to directly map the conformational ensembles of the CNBD in the absence and presence of cAMP. Site-directed, double-cysteine mutants in a soluble CNBD fragment were spin-labeled, and interspin label distance distributions were determined using DEER. We found motions of up to 10 Å induced by the binding of cAMP. In addition, the distributions were narrower in the presence of cAMP. Continuous-wave electron paramagnetic resonance studies revealed changes in mobility associated with cAMP binding, indicating less conformational heterogeneity in the cAMP-bound state. From the measured DEER distributions, we constructed a coarse-grained elastic-network structural model of the cAMP-induced conformational transition. We find that binding of cAMP triggers a reorientation of several helices within the CNBD, including the C-helix closest to the cAMP-binding site. These results provide a basis for understanding how the binding of cAMP is coupled to channel opening in HCN and related channels.
Ion channels are allosteric membrane proteins that open selective pores in response to various physiological stimuli, including binding of ligands and changes in transmembrane voltage (1). They are important for diverse physiological functions ranging from neurotransmission to muscle contraction. One such channel, the hyperpolarization-activated cyclic nucleotide-gated (HCN) ion channel, underlies the current (termed Ih, If, or Iq) produced in response to hyperpolarization of cardiac pacemaker cells and neurons (2). In the heart, HCN channels are responsible for pace-making activity and may have a role in the autonomic regulation of the heart rate (3–5). In the brain, HCN channels are involved in repetitive firing of neurons and dendritic integration (6–8). Despite the important physiological roles of HCN channels, the structure of the channels and molecular mechanism of their function are not completely understood.
HCN channels are part of the voltage-gated K+ channel superfamily (9). Like other members of this family, they are tetramers, with each subunit having a voltage-sensor domain of four transmembrane helices (S1–S4) and a pore-lining domain consisting of two transmembrane helices separated by a reentrant loop (S5-P-S6; Fig. 1A). However, HCN channels contain two key specializations that make them unique among the voltage-gated ion channels: (i) They are activated by membrane hyperpolarization instead of depolarization, and (ii) they are regulated by the direct binding of cyclic nucleotides, like the ubiquitous second messenger cAMP, to a cytoplasmic domain in the carboxyl-terminal region of the channel. The direct binding of the agonist cAMP to HCN channels increases the rate and extent of activation and shifts the voltage dependence of activation to more depolarizing voltages.
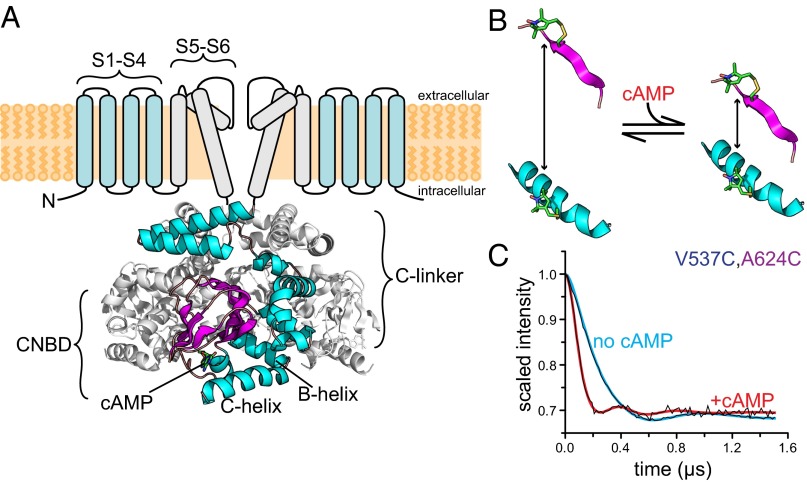
Fig. 1.
Study of conformational changes in HCN2 using DEER. (A, Upper) Putative transmembrane topology of HCN2 channels highlighting the voltage sensor domain (S1–S4) and the pore domain (S5–S6). Only two subunits are shown. (A, Lower) Crystal structure [Protein Data Bank (PDB) ID code 3ETQ] of the cysteine-free cytoplasmic carboxyl-terminal domain of HCN2. One subunit of the tetramer is shown in color. (B) Schematic diagram showing the distance change between two cysteine-attached MTSL spin labels in a protein upon cAMP binding. In this example, the two positions are closer in the presence of cAMP. (C) Raw DEER time traces for HCN2cys-free V537C,A624C labeled with MTSL are shown in black in the absence or presence of cAMP, as indicated. The colored curves are distance-distribution fits to the data. The oscillation frequency is higher in the presence of cAMP, indicating that the two positions are closer together in the ligand-bound form.
The crystal structure of the carboxyl-terminal region bound to cAMP has been solved for several HCN channels (10–14). The nearly identical structures consist of fourfold symmetrical tetramers predicted to connect directly to the S6 segments that form the ion-conducting pore (Fig. 1A). Each of the subunits contains two domains: the cyclic nucleotide-binding domain (CNBD) and the C-linker domain. The CNBD exhibits strong structural similarity to the CNBDs of other cyclic nucleotide-binding proteins, including cAMP-dependent protein kinase (PKA), the guanine nucleotide exchange factor Epac, and the Escherichia coli catabolite gene activator protein (CAP) (15–19). The CNBD consists of an eight-stranded antiparallel β-roll, followed by two α-helices (B-helix and C-helix). cAMP binds in the anticonformation between the β-roll and the C-helix. The C-linker is a unique domain found only in HCN channels and their close homologs, cyclic nucleotide-gated (CNG) channels, and KCNH family K+ channels (14, 20, 21). It is situated between the CNBD and membrane-spanning domains of the channel, and is the site of virtually all intersubunit interactions in the structure (Fig. 1A). The C-linker has been found to play a key role in coupling conformational changes in the CNBD to opening of the pore (9, 22, 23).
The ligand-induced movement of the C-helix is widely thought to initiate the conformational changes that lead to opening of the channel pore, but the structural evidence in support of this hypothesis is equivocal (10, 24–29). The crystal structure of the HCN2 carboxyl-terminal region in the absence of ligand shows little difference from the cyclic nucleotide-bound structure (12). The only significant differences between the two structures are observed in the F′-helix of the C-linker and in the C-helix. The proximal half of the C-helix is in the same position in the cAMP-bound and unbound structures, whereas the distal half is missing from the apo structure, indicating that it is disordered or can access multiple conformations. In contrast, studies on the soluble carboxyl-terminal fragment using transition metal ion FRET (tmFRET) demonstrate a relatively large movement (∼5 Å) at the proximal end of the C-helix upon binding of cAMP (12). The tmFRET studies also indicate a smaller movement at the distal end of the C-helix and increased disorder in the C-helix in the absence of cyclic nucleotides (12, 26).
In this study, we examined the cAMP-induced conformational transition in the CNBD of HCN2 using double electron–electron resonance (DEER) spectroscopy. DEER is a pulse electron paramagnetic resonance (EPR) method that can determine distances and resolve distance distributions between pairs of sites within proteins separated by about 15–80 Å (30–33). In a typical DEER experiment, two sites in a protein are mutated to cysteines and labeled with small magnetic spin labels (Fig. 1B). DEER measures the pair’s magnetic through-space coupling via excitation of one label and probing of the other with a series of short microwave pulses. This method yields an oscillating signal whose frequency falls off with the third power of the distance between the labels (Fig. 1C). Crucially, DEER measures full-distance distributions, rather than just an average distance, providing quantitative information on structural heterogeneity and variability that is not accessible from X-ray crystal structures or ensemble FRET experiments. Using DEER, we found that the binding of cAMP to the isolated C-linker/CNBD of HCN2 causes the C-helix to move substantially toward the β-roll and decreases the conformational heterogeneity of the protein. These observations are the first step in understanding the mechanisms of ligand gating of HCN channels and the activation of other CNBD-containing proteins.
Results
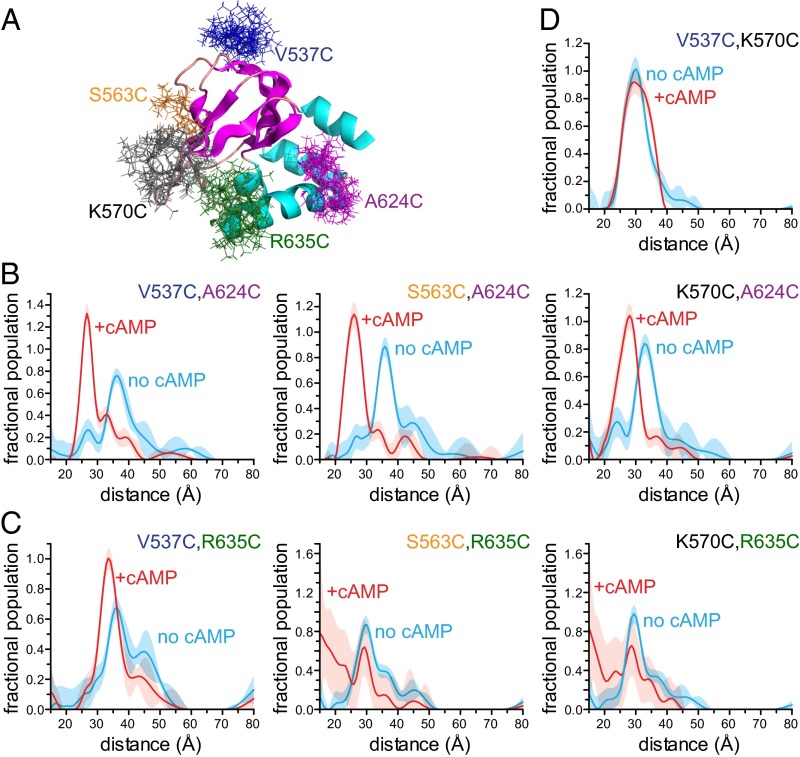
To measure the cAMP-induced conformational changes in HCN2 channels, we performed DEER experiments on an isolated carboxyl-terminal fragment containing the C-linker and CNBD. We introduced pairs of cysteines into an otherwise cysteine-free carboxyl-terminal fragment (HCN2cys-free) previously shown to have a structure nearly identical to the WT fragment (rmsd = 0.7 Å) (12). Cysteines were placed either at the proximal end of the C-helix (A624C) or at the distal end of the C-helix (R635C), as well as at each of three different positions in the β-roll (V537C, S563C, and K570C). The introduced cysteines were then modified with the nitroxide spin label S-(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl methanesulfonothioate (MTSL). Fig. 2A shows the locations of the cysteine mutations and the predicted MTSL side-chain rotamers for each residue we studied.
Fig. 2.
C-helix moves closer to the β-roll upon binding of cAMP. (A) Structure of HCN2cys-free indicating the positions and modeled rotameric distributions of MTSL spin labels used for DEER measurements. (B) Distance distributions obtained from fits to DEER traces for a spin label at the proximal end of the C-helix (A624C) relative to β-roll residues in the absence (cyan) and presence (red) of cAMP. (C) Distance distributions obtained from fits to DEER traces for a spin label at the distal end of the C-helix (R635C) relative to the β-roll residues in the absence and presence of cAMP. Poor fits were obtained for S563C,R635C and K570C,R635C in the presence of cAMP. (D) Distance distributions obtained from fits to DEER traces of MTSL-labeled V537C,K570C in the presence and absence of cAMP. In all plots, errors (2σ) are indicated by the shaded areas.
DEER was performed to measure the separations of all six possible combinations of C-helix and β-roll mutations in the absence and presence of 1 mM cAMP. At this concentration, HCN is >95% in the cAMP-bound form, based on the protein concentrations of 30–50 μM and known Kd values that range from about 0.1 μM to a few μM (11, 12, 26, 34). Fig. 1C shows the DEER time traces for HCN2cys-free spin-labeled at positions V537C and A624C. The oscillations were faster in the presence of cAMP, indicating that cAMP caused these positions to move closer together. These time traces were then converted into distance distributions, as detailed in Methods. The corresponding distance distributions demonstrate that cAMP binding induces a 9-Å movement of the peak of the distance distribution (from 36 Å to 27 Å) of the spin label at A624C at the proximal end of the C-helix toward the label at position V537C on the β-roll (Fig. 2B). Similar results were obtained for movements of the spin label at position A624C relative to other positions on the β-roll (10-Å movement toward S563C and 5-Å movement toward K570C; Fig. 2B and Fig. S1A). These data indicate that cAMP causes a large movement of the proximal end of the C-helix toward the β-roll.
When a spin label was introduced at position R635C at the distal end of the C-helix, we observed a smaller change in distance relative to the position of V537C on the β-roll (about a 3-Å shortening of the dominant distance) and a significant distribution narrowing upon cAMP binding (Fig. 2C and Fig. S1A). We obtained DEER distributions for constructs with spin labels at R635C and either S563C or K570C as well. In the absence of cAMP, we observed that the distance distributions between spin labels at S563C and R635C and at K570C and R635C had modal distances of 30 Å and 29 Å, respectively (Fig. 2C). However, in the presence of cAMP, the data analysis did not provide accurate distance distributions, as reflected by the large error bars in the distance distributions and the poor fits to the time traces for these data (Fig. 2C and Fig. S1A). A significant fraction of spin label rotamers at these positions are predicted from the crystal structure to be too close (<15 Å away) in the presence of cAMP to produce interpretable traces. Contributions from protein conformations with short separations can be underrepresented in DEER time traces relative to longer separations, complicating the quantitative analysis of such traces (35). Nevertheless, these data demonstrate that upon cAMP binding, the distal end of the C-helix also moves closer to the β-roll.
Although the distance changes we observed are consistent with movement of the C-helix, it is possible that conformational changes within the β-roll contributed to the change in distance distributions. To control for this possibility, we measured the separation of two residues on the β roll, V537C and K570C, in the absence and presence of cAMP. The distance distributions showed no change, supporting the conclusion that cAMP does not induce significant conformational changes within the β-roll but does cause the C-helix to move closer to the β-roll (Fig. 2D).
Previous analytical ultracentrifugation experiments on WT protein indicate that our carboxyl-terminal HCN2 constructs are monomeric at the concentrations used for our experiments (<50 μM) (14). A multimeric assembly of subunits would introduce intermolecular as well as intramolecular interactions between spin labels and complicate our interpretation of the results. To control for the possibility of tetramerization or nonspecific aggregation, we measured DEER traces for constructs containing only one cysteine per monomer. The DEER time traces for these mutants showed slow, quasilinear signal decays in both the absence and presence of cAMP, consistent with a monodisperse solution of HCN monomers (Fig. S1B).
For each of the DEER experiments with A624C, there is a detectable peak in the distributions in the absence of cAMP at the position of the primary peak in the presence of cAMP. These data suggest that even in the absence of cAMP, the channel samples the conformation of the cAMP-bound state. In addition, several of the distance distributions obtained from our DEER measurements were broader in the absence of cAMP, indicating increased structural heterogeneity near one or both sites to which MTSL was attached (Fig. 2 B and C). This is most obvious for the V537C,A624C pair, where the FWHM decreased from 7.5 Å in the absence of cAMP to 5.0 Å in the presence of cAMP. Similarly, the distance distribution of the V537C,R635C pair spreads out over a wider distance range in the absence than in the presence of cAMP. These results indicate that in the absence of cAMP, the position of the C-helix relative to the β-roll is heterogeneous, and the binding of cAMP restricts the conformational space of the C-helix closer to the β-roll.
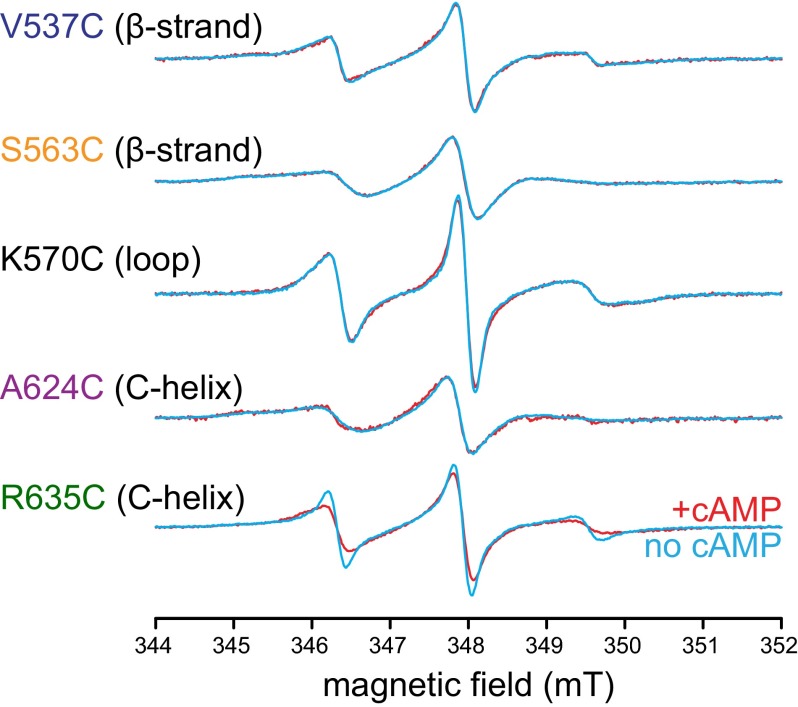
Because DEER determines the distance between the two spin labels, changes in spin-label mobility at a single position cannot be inferred from DEER data. To determine which locations undergo mobility changes, we measured the room temperature continuous wave (CW) EPR spectra for each of the single-cysteine mutants labeled with MTSL in the absence and presence of cAMP. CW spectra broaden as the rotational correlation time of the spin label attached to the protein increases and the spin label becomes less mobile. No significant agonist-induced changes in the mobility of the spin label were observed at sites other than R635C (Fig. 3). The CW spectrum of R635C broadened in the presence of cAMP, indicating a decrease in mobility upon cAMP binding. The broadening corresponded to an increase in the rotational correlation time for the spin label from 0.8 ns in the absence of cAMP to 1.3 ns in the presence of cAMP. Changes in the width of the HCN2cys-free R635C CW EPR spectrum in the presence of cAMP could reflect either a shift in the population of MTSL rotamers or a change in the mobility of the protein backbone. These results suggest that the distal end of the C-helix is more flexible in the absence than in the presence of cAMP.
Fig. 3.
cAMP binding decreased the mobility of the distal C-helix. CW EPR spectra for MTSL attached to positions V537C in the β1 strand, S563C in the β4 strand, K570C in the β4–β5 loop, A624C in the C-helix, and R635C in the C-helix in the absence and presence of cAMP. There was significant line broadening evident in the spectrum for R635C as a result of cAMP binding, indicating a reduction in mobility of the spin label.
Discussion
The cAMP-bound structure of the HCN carboxyl-terminal region has been solved for several HCN channels (10–14) and is similar to the agonist-bound structures of many other CNBD-containing proteins (15–19). However, the agonist-free structure of this protein is still controversial. Crystal structures of HCN2 in the absence of cAMP are almost identical to cAMP-bound structures, but other experiments suggest large conformational changes accompany ligand binding (12, 24–26). To resolve this controversy, we studied changes in structure associated with ligand binding using DEER and CW EPR. In our DEER experiments, we observed a large movement of the C-helix toward the β-roll and decreased heterogeneity of the C-helix upon cAMP binding. In the CW EPR spectra, we observed a decrease in the mobility of R635C at the distal end of the C-helix upon ligand binding.
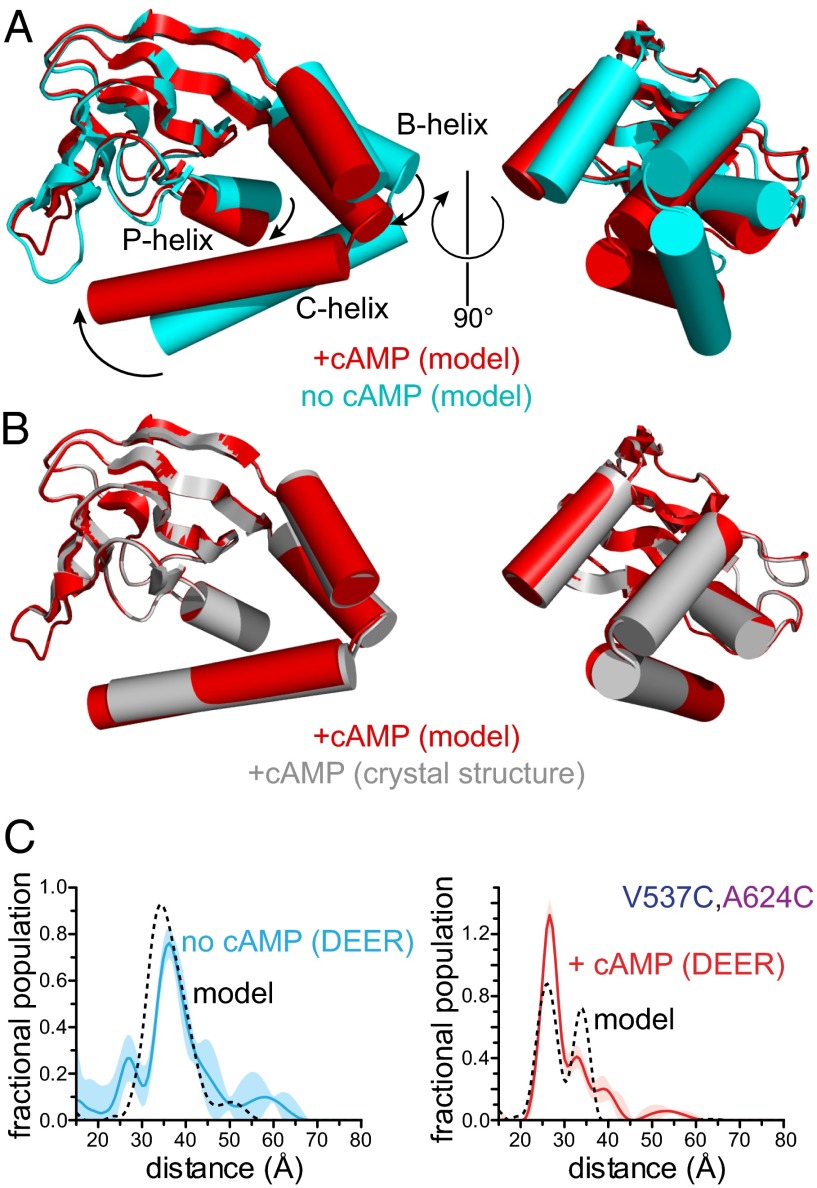
We used the distance distributions derived from our DEER data to construct a model of the conformational change in the CNBD induced by binding cAMP. The peaks of the distance distributions were used as constraints in an elastic network model to predict the structure of the HCN2 carboxyl-terminal region in the absence and presence of cAMP. The rmsd fitting errors for the two models were about 2.1 Å (apo form) and 0.9 Å (bound form). Comparison of the two structural models shows that cAMP binding induces a large movement of the C-helix toward the β-roll (Fig. 4A and Movie S1). To accommodate the large movement at the proximal end of the C-helix (5–10 Å from our DEER data), the B-helix must move as well. In the models, there is also a rotation of the C-helix about the helical axis that brings residues R632 and I636 into contact with the purine ring of the cyclic nucleotide (Movie S1). These contacts have been proposed to stabilize the ligand-bound conformation and drive the opening conformational change (36).
Fig. 4.
Conformational change in the HCN2 carboxy terminus induced by cAMP binding. (A) Elastic network models of the CNBD of HCN2 in the absence (cyan) and presence (red) of cAMP. Models were obtained using the HCN2cys-free crystal structure (PDB ID code 3ETQ) and experimental constraints from DEER. Structures were aligned at the β-rolls (residues 534–607). (B) Comparison between the modeled cAMP-bound structure (red) and the crystal structure of HCN2cys-free (gray) bound to cAMP (3ETQ). (C) Distance distributions obtained from DEER for V537C,A624C in the absence and presence of cAMP compared with predicted DEER traces (dashed lines) calculated from the modeled structures in A.
The elastic-network structural models agree very well with the experimental data. The modeled cAMP-bound structure is nearly identical (rmsd = 0.2 Å) to the HCN2cys-free crystal structure in the presence of agonist (Fig. 4B). This suggests that the DEER data in the presence of cAMP are consistent with the crystal structure. Furthermore, despite the fact that our models do not include heterogeneity in the protein backbone, the DEER distance distributions calculated from the models reasonably predict our experimental distributions (Fig. 4C and Fig. S2). Only the model prediction for the V537C,R635C DEER distribution in the absence of cAMP was significantly different from the experimental data (Fig. S2). We believe this discrepancy reflects the disorder at position R635C in the absence of cAMP, which was not directly incorporated into our model. Our coarse-grain model is also consistent with many electrophysiology studies on HCN and CNG channels (37), suggesting that the conformational change we observe in the isolated C-linker/CNBD fragment is similar in intact functional HCN channels and CNG channels as well.
We have previously used tmFRET to investigate the structural rearrangements of the C-helix induced by agonist binding (12, 26). Our tmFRET data indicated a large movement at the proximal end of the C-helix and a smaller movement at the distal end toward the β-roll subsequent to agonist binding. This is consistent with the more quantitative results from our DEER experiments, which show a 5- to 10-Å movement at the proximal end of the C-helix and a smaller movement at the distal end. In general, the distance changes reported by tmFRET were smaller in magnitude than those indicated by DEER. This discrepancy may be due to slightly different sites used for the tmFRET studies or the tendency of FRET measurements to underreport changes in distance (38).
Both DEER and CW EPR predict a decrease in the conformational heterogeneity of the C-helix upon binding to cAMP, particularly at the distal end. This observation is consistent with previous tmFRET results, which showed that agonist binding stabilized the secondary structure of the C-helix (12, 26). The structural heterogeneity predicted by EPR at the distal end of the C-helix may also explain the lack of electron density at the end of the C-helix in the apo crystal structure of HCN2 (12).
Other CNBD-containing proteins undergo similar structural changes subsequent to agonist binding. In particular, the NMR structures of the CNBD of the prokaryotic K+ channel MloK1 in the absence and presence of cAMP demonstrate a similar movement of the B- and C-helices subsequent to agonist binding (39, 40). However, the movement of the C-helix is somewhat larger than reported here. PKA and Epac also undergo similar conformational changes subsequent to agonist binding, with agonist binding accompanied by movement at a hinge between the β-roll and B-helix (18). In contrast, the solution NMR structures of CAP in the absence and presence of cAMP do not demonstrate any large translation of the B- and C-helices (41). In the NMR structure of cAMP-free CAP, the distal C terminus appears disordered. A lack of order in the C-helix was also observed in several of the cAMP-free crystal structures of the MloK1 CNBD (42). The heterogeneity that we observed in DEER studies of the C-helix in the absence of ligand could explain why the NMR and crystal structures of other CNBDs in other proteins are not well ordered.
The DEER studies reported here provide a quantitative perspective on the cAMP-induced conformational change in HCN channels. Instead of just inferring the conformational change and heterogeneity from static structures, the distance distributions provided by DEER allow us to observe the conformational space occupied in a given state of the channel directly and how it changes with ligand binding. These experiments lay the foundation for understanding how the conformational change in the C-helix is propagated to the B-helix, to the C-linker, to the voltage sensor domain, and to the gate in the ion-conducting pore. By providing information on the conformational space accessible to HCN channels, DEER will allow us to probe the relationship between conformational heterogeneity and allosteric regulation of HCN channels.
Methods
Protein Expression, Purification, and Spin Labeling.
The gene encoding residues 443–640 of a cysteine-free fragment of the mouse HCN2 ion channel (HCN2cys-free) was cloned into the pMALc2T vector (New England Biolabs). The vector contains an N-terminal maltose-binding protein (MBP) tag separated from the channel gene by a thrombin-cleavable linker. Cysteine mutations at indicated residues were introduced for EPR studies using standard PCR-based techniques.
The vector containing the HCN2cys-free gene was transfected into BL21(DE3) cells and grown at 37 °C to an OD600 of 0.6–0.8. The cells were then induced with 1 mM isopropyl β-d-1-thiogalactopyranoside and grown overnight at 18 °C. Two-liter cultures of cells were pelleted by centrifugation at 4,000 × g at 4 °C for 10 min and resuspended in 150 mL of 150 mM KCl, 30 mM Hepes, and 10% (wt/vol) glycerol (pH 7.2). DNase at a final concentration of 5 μg/mL and two tablets of protease inhibitors (cOmplete EDTA-free; Roche) were added to the buffer. The resuspended cells were lysed by an Emulsiflex-C3 homogenizer (Avestin) and clarified by centrifugation at 186,000 × g at 4 °C for 45 min. The lysate was then purified with amylose affinity chromatography, and MBP was cleaved off by thrombin.
The protein (10–50 μM) was spin-labeled with 100 μM MTSL (Toronto Research Chemicals) per cysteine mutation for 1 h at room temperature. To remove MBP and excess spin label, the sample was purified by ion exchange chromatography. The sample was diluted in buffer containing 10 mM KCl, 30 mM Hepes, and 10% (wt/vol) glycerol (pH 7.2); loaded on an SP Sepharose column (HiTrap SP FF; General Electric); and eluted with a continuous gradient between 10 mM and 1 M KCl. Fractions containing protein were pooled and concentrated to ∼50 μM using a 30,000 molecular weight cutoff (MWCO) centrifugal filter (Vivaspin; General Electric).
EPR Sample Preparation.
For both CW EPR and DEER experiments, the protein was buffer-exchanged into D2O with 150 mM KCl, 30 mM Hepes, and 10% (wt/vol) glycerol using a PD-10 column that also removes all traces of remaining unbound spin label. The protein in deuterated buffer was divided, and 1 mM cAMP was added to half. Protein in the absence and presence of cAMP was further concentrated using a 30,000 MWCO centrifugal filter. The final concentration of the protein was 30–50 µM. For CW EPR, protein was loaded into 1-mm outer diameter quartz capillaries (Q100-50–7.5; Sutter). For DEER, 200 μL of each protein sample was inserted into a 4-mm outer diameter quartz tube (707-SQ-100M; Wilmad) and flash-frozen in liquid nitrogen. The variation in modulation depths observed in our samples is likely due to different degrees of labeling.
EPR Data Acquisition.
CW EPR spectra were recorded on a Bruker EMX spectrometer with an X-band (9.78 GHz) microwave source at room temperature. A dielectric resonator with a Q-factor of 2,000–4,000 was used (ER4123D; Bruker). Spectra were recorded with incident power of 0.2 mW, modulation amplitude of 2 G, and modulation frequency of 100 kHz.
DEER data were acquired on a Bruker EleXsys E580 spectrometer at X-band (9.5 GHz) with an MD4 dielectric resonator (Bruker). Experiments were performed at 60 K using a cryostat and liquid helium cooling system (Oxford). The four-pulse, dead-time free DEER sequence [(π/2)probe − τ1 − (π)probe − τ1 + t − (π)pump − (τ2 − t) − (π)probe − τ2] was used with a 10-ns π/2 probe pulse and a 20-ns π pump pulse (30, 32). The pump frequency matched the nitroxide spectral maximum. The probe frequency was centered in the resonator dip and was 70 MHz higher than the pump frequency. Pulses were positioned using 120 ns for τ1 and 1,800 ns for τ2, and t was varied from −60 ns to 1,800 ns in increments of 10 ns. An eight-step phase-cycling protocol was used to collect data. The measurement time for each sample was 10–16 h.
Data Analysis.
DEER distance distributions were obtained using the DeerAnalysis2013 software (43). Background subtraction was performed by assuming a homogeneous 3D background. Distance distributions were generated from the time traces using Tikhonov regularization, a model-free least-squares approach. The regularization parameter was optimized separately for each dataset according to the L-curve criterion. Rotational correlation times for R635C CW EPR spectra were extracted from the experimental data using the stochastic Liouville equation solver as implemented in EasySpin (44).
Elastic Network Modeling.
Rotameric distributions of the MTSL-labeled cysteine residues were modeled using a library approach with the MMM software (45). Elastic network modeling implemented in MMM was used to create cAMP-bound and unbound models of HCN using the DEER distance constraints and the cAMP-bound HCN crystal structure (Protein Data Bank ID code 3ETQ) as a starting structure. The algorithm used for modeling was a modification of the anisotropic elastic network model developed by Zheng and Brooks (46). The model is based on harmonic oscillator potentials for the elastic interactions of all pairs of alpha-carbon atoms within a specified cutoff distance (10 Å in the implementation we used). The MMM implementation modifies the original algorithm by increasing the force constants for the nearest and next-nearest neighboring alpha-carbons by a factor of 10,000 relative to force constants between more distant atoms. It also varies the force constants according to the inverse sixth power of the separation of the alpha-carbons (33). Comparisons of experimental data with predictions based on the models were generated by modeling rotameric distributions of the MTSL-labeled cysteine residues and predicting DEER results in MMM. MMM provides noise-free predictions and does not take into account background and noise effects. We added background and Gaussian noise with a standard deviation equivalent to that observed in our experimental data to the noise-free MMM-predicted DEER time traces. Distance distributions were generated from these predicted time traces using DEERAnalysis (43).
Supplementary Material
Acknowledgments
We thank Shellee Cunnington and Stacey Camp for technical assistance, Ellen Hayes for help with the EPR and DEER experiments, and Christopher Miller for helpful comments on the manuscript. This work was supported by National Institutes of Health Grant EY10329 (to W.N.Z.) and Grant F32EY018981 (to M.C.P.), American Heart Association Grant 14IRG18770000 (to W.N.Z.), the University of Washington (S.S.), and by a fellowship from the Sackler Scholars Program in Integrative Biophysics (to H.A.D.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1405371111/-/DCSupplemental.
References
- 1.Hille B. Ion Channels of Excitable Membranes. 3rd Ed. Sunderland, MA: Sinauer; 2001. [Google Scholar]
- 2.Robinson RB, Siegelbaum SA. Hyperpolarization-activated cation currents: From molecules to physiological function. Annu Rev Physiol. 2003;65:453–480. doi: 10.1146/annurev.physiol.65.092101.142734. [DOI] [PubMed] [Google Scholar]
- 3.DiFrancesco D. Characterization of single pacemaker channels in cardiac sino-atrial node cells. Nature. 1986;324(6096):470–473. doi: 10.1038/324470a0. [DOI] [PubMed] [Google Scholar]
- 4.DiFrancesco D. Pacemaker mechanisms in cardiac tissue. Annu Rev Physiol. 1993;55:455–472. doi: 10.1146/annurev.ph.55.030193.002323. [DOI] [PubMed] [Google Scholar]
- 5.DiFrancesco D, Tortora P. Direct activation of cardiac pacemaker channels by intracellular cyclic AMP. Nature. 1991;351(6322):145–147. doi: 10.1038/351145a0. [DOI] [PubMed] [Google Scholar]
- 6.Magee JC. Dendritic Ih normalizes temporal summation in hippocampal CA1 neurons. Nat Neurosci. 1999;2(9):848. doi: 10.1038/12229. [DOI] [PubMed] [Google Scholar]
- 7.Pape HC, McCormick DA. Noradrenaline and serotonin selectively modulate thalamic burst firing by enhancing a hyperpolarization-activated cation current. Nature. 1989;340(6236):715–718. doi: 10.1038/340715a0. [DOI] [PubMed] [Google Scholar]
- 8.Williams SR, Stuart GJ. Site independence of EPSP time course is mediated by dendritic I(h) in neocortical pyramidal neurons. J Neurophysiol. 2000;83(5):3177–3182. doi: 10.1152/jn.2000.83.5.3177. [DOI] [PubMed] [Google Scholar]
- 9.Craven KB, Olivier NB, Zagotta WN. C-terminal movement during gating in cyclic nucleotide-modulated channels. J Biol Chem. 2008;283(21):14728–14738. doi: 10.1074/jbc.M710463200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flynn GE, Black KD, Islas LD, Sankaran B, Zagotta WN. Structure and rearrangements in the carboxy-terminal region of SpIH channels. Structure. 2007;15(6):671–682. doi: 10.1016/j.str.2007.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lolicato M, et al. Tetramerization dynamics of C-terminal domain underlies isoform-specific cAMP gating in hyperpolarization-activated cyclic nucleotide-gated channels. J Biol Chem. 2011;286(52):44811–44820. doi: 10.1074/jbc.M111.297606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taraska JW, Puljung MC, Olivier NB, Flynn GE, Zagotta WN. Mapping the structure and conformational movements of proteins with transition metal ion FRET. Nat Methods. 2009;6(7):532–537. doi: 10.1038/nmeth.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu X, Vysotskaya ZV, Liu Q, Zhou L. Structural basis for the cAMP-dependent gating in the human HCN4 channel. J Biol Chem. 2010;285(47):37082–37091. doi: 10.1074/jbc.M110.152033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zagotta WN, et al. Structural basis for modulation and agonist specificity of HCN pacemaker channels. Nature. 2003;425(6954):200–205. doi: 10.1038/nature01922. [DOI] [PubMed] [Google Scholar]
- 15.Clayton GM, Silverman WR, Heginbotham L, Morais-Cabral JH. Structural basis of ligand activation in a cyclic nucleotide regulated potassium channel. Cell. 2004;119(5):615–627. doi: 10.1016/j.cell.2004.10.030. [DOI] [PubMed] [Google Scholar]
- 16.Kim C, Xuong NH, Taylor SS. Crystal structure of a complex between the catalytic and regulatory (RIalpha) subunits of PKA. Science. 2005;307(5710):690–696. doi: 10.1126/science.1104607. [DOI] [PubMed] [Google Scholar]
- 17.Kim JJ, et al. Co-crystal structures of PKG Iβ (92-227) with cGMP and cAMP reveal the molecular details of cyclic-nucleotide binding. PLoS ONE. 2011;6(4):e18413. doi: 10.1371/journal.pone.0018413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rehmann H, et al. Structure and regulation of the cAMP-binding domains of Epac2. Nat Struct Biol. 2003;10(1):26–32. doi: 10.1038/nsb878. [DOI] [PubMed] [Google Scholar]
- 19.Weber IT, Gilliland GL, Harman JG, Peterkofsky A. Crystal structure of a cyclic AMP-independent mutant of catabolite gene activator protein. J Biol Chem. 1987;262(12):5630–5636. [PubMed] [Google Scholar]
- 20.Brelidze TI, Carlson AE, Sankaran B, Zagotta WN. Structure of the carboxy-terminal region of a KCNH channel. Nature. 2012;481(7382):530–533. doi: 10.1038/nature10735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brelidze TI, Gianulis EC, DiMaio F, Trudeau MC, Zagotta WN. Structure of the C-terminal region of an ERG channel and functional implications. Proc Natl Acad Sci USA. 2013;110(28):11648–11653. doi: 10.1073/pnas.1306887110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Craven KB, Zagotta WN. Salt bridges and gating in the COOH-terminal region of HCN2 and CNGA1 channels. J Gen Physiol. 2004;124(6):663–677. doi: 10.1085/jgp.200409178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paoletti P, Young EC, Siegelbaum SA. C-Linker of cyclic nucleotide-gated channels controls coupling of ligand binding to channel gating. J Gen Physiol. 1999;113(1):17–34. doi: 10.1085/jgp.113.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matulef K, Flynn GE, Zagotta WN. Molecular rearrangements in the ligand-binding domain of cyclic nucleotide-gated channels. Neuron. 1999;24(2):443–452. doi: 10.1016/s0896-6273(00)80857-0. [DOI] [PubMed] [Google Scholar]
- 25.Matulef K, Zagotta WN. Multimerization of the ligand binding domains of cyclic nucleotide-gated channels. Neuron. 2002;36(1):93–103. doi: 10.1016/s0896-6273(02)00878-4. [DOI] [PubMed] [Google Scholar]
- 26.Puljung MC, Zagotta WN. A secondary structural transition in the C-helix promotes gating of cyclic nucleotide-regulated ion channels. J Biol Chem. 2013;288(18):12944–12956. doi: 10.1074/jbc.M113.464123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tibbs GR, Liu DT, Leypold BG, Siegelbaum SA. A state-independent interaction between ligand and a conserved arginine residue in cyclic nucleotide-gated channels reveals a functional polarity of the cyclic nucleotide binding site. J Biol Chem. 1998;273(8):4497–4505. doi: 10.1074/jbc.273.8.4497. [DOI] [PubMed] [Google Scholar]
- 28.Varnum MD, Black KD, Zagotta WN. Molecular mechanism for ligand discrimination of cyclic nucleotide-gated channels. Neuron. 1995;15(3):619–625. doi: 10.1016/0896-6273(95)90150-7. [DOI] [PubMed] [Google Scholar]
- 29.Zhou L, Siegelbaum SA. Gating of HCN channels by cyclic nucleotides: Residue contacts that underlie ligand binding, selectivity, and efficacy. Structure. 2007;15(6):655–670. doi: 10.1016/j.str.2007.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pannier M, Veit S, Godt A, Jeschke G, Spiess HW. Dead-time free measurement of dipole-dipole interactions between electron spins. J Magn Reson. 2000;142(2):331–340. doi: 10.1006/jmre.1999.1944. [DOI] [PubMed] [Google Scholar]
- 31.Milov AD, Ponomarev AB, Tsvetkov YD. Electron electron double-resonance in electron-spin echo-model biradical systems and the sensitized photolysis of dDecalin. Chem Phys Lett. 1984;110(1):67–72. [Google Scholar]
- 32.Martin RE, et al. Determination of end-to-end distances in a series of TEMPO diradicals of up to 2.8 nm length with a new four-pulse double electron electron resonance experiment. Angew Chem Int Ed Engl. 1998;37(20):2834–2837. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2833::AID-ANIE2833>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 33.Jeschke G. Characterization of protein conformational changes with sparse spin-label distance constraints. J Chem Theory Comput. 2012;8(10):3854–3863. doi: 10.1021/ct300113z. [DOI] [PubMed] [Google Scholar]
- 34.Chow SS, Van Petegem F, Accili EA. Energetics of cyclic AMP binding to HCN channel C terminus reveal negative cooperativity. J Biol Chem. 2012;287(1):600–606. doi: 10.1074/jbc.M111.269563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Milov AD, Naumov BD, Tsvetkov YD. The effect of microwave pulse duration on the distance distribution function between spin labels obtained by PELDOR data analysis. Appl Magn Reson. 2004;26(4):587–599. [Google Scholar]
- 36.Zhou L, Siegelbaum SA. Pathway and endpoint free energy calculations for cyclic nucleotide binding to HCN channels. Biophys J. 2008;94(12):L90–L92. doi: 10.1529/biophysj.108.130872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Craven KB, Zagotta WN. CNG and HCN channels: Two peas, one pod. Annu Rev Physiol. 2006;68:375–401. doi: 10.1146/annurev.physiol.68.040104.134728. [DOI] [PubMed] [Google Scholar]
- 38.Taraska JW, Puljung MC, Zagotta WN. Short-distance probes for protein backbone structure based on energy transfer between bimane and transition metal ions. Proc Natl Acad Sci USA. 2009;106(38):16227–16232. doi: 10.1073/pnas.0905207106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schünke S, Stoldt M, Lecher J, Kaupp UB, Willbold D. Structural insights into conformational changes of a cyclic nucleotide-binding domain in solution from Mesorhizobium loti K1 channel. Proc Natl Acad Sci USA. 2011;108(15):6121–6126. doi: 10.1073/pnas.1015890108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schünke S, Stoldt M, Novak K, Kaupp UB, Willbold D. Solution structure of the Mesorhizobium loti K1 channel cyclic nucleotide-binding domain in complex with cAMP. EMBO Rep. 2009;10(7):729–735. doi: 10.1038/embor.2009.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Popovych N, Tzeng SR, Tonelli M, Ebright RH, Kalodimos CG. Structural basis for cAMP-mediated allosteric control of the catabolite activator protein. Proc Natl Acad Sci USA. 2009;106(17):6927–6932. doi: 10.1073/pnas.0900595106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Altieri SL, et al. Structural and energetic analysis of activation by a cyclic nucleotide binding domain. J Mol Biol. 2008;381(3):655–669. doi: 10.1016/j.jmb.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jeschke G, et al. DeerAnalysis2006—A comprehensive software package for analyzing pulsed ELDOR data. Appl Magn Reson. 2006;30(3-4):473–498. [Google Scholar]
- 44.Stoll S, Schweiger A. EasySpin, a comprehensive software package for spectral simulation and analysis in EPR. J Magn Reson. 2006;178(1):42–55. doi: 10.1016/j.jmr.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 45.Polyhach Y, Bordignon E, Jeschke G. Rotamer libraries of spin labelled cysteines for protein studies. Phys Chem Chem Phys. 2011;13(6):2356–2366. doi: 10.1039/c0cp01865a. [DOI] [PubMed] [Google Scholar]
- 46.Zheng W, Brooks BR. Modeling protein conformational changes by iterative fitting of distance constraints using reoriented normal modes. Biophys J. 2006;90(12):4327–4336. doi: 10.1529/biophysj.105.076836. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.