Significance
Histone deacetylase 1 and 2 (HDAC1/2) are sister proteins that regulate access to DNA by modulating chromatin. We have generated the first double knockout (DKO) of Hdac1/2 in embryonic stem (ES) cells and find that gene inactivation causes a loss of cell viability, which is associated with increased abnormal mitotic spindles and chromosome segregation defects. Transcriptome analysis revealed that almost 2,000 genes are deregulated in DKO cells. Significantly for the self-renewal properties of ES cells, this includes down-regulation of the core pluripotent factors, Oct4, Nanog, and Rex1. Furthermore, using the rescue of Hdac1/2-null cells as a model system to monitor HDAC1/2 activity, we have also shown that mutations that abolish inositol tetraphosphate binding reduce the activity of HDAC1 in vivo.
Abstract
Histone deacetylases 1 and 2 (HDAC1/2) form the core catalytic components of corepressor complexes that modulate gene expression. In most cell types, deletion of both Hdac1 and Hdac2 is required to generate a discernible phenotype, suggesting their activity is largely redundant. We have therefore generated an ES cell line in which Hdac1 and Hdac2 can be inactivated simultaneously. Loss of HDAC1/2 resulted in a 60% reduction in total HDAC activity and a loss of cell viability. Cell death is dependent upon cell cycle progression, because differentiated, nonproliferating cells retain their viability. Furthermore, we observe increased mitotic defects, chromatin bridges, and micronuclei, suggesting HDAC1/2 are necessary for accurate chromosome segregation. Consistent with a critical role in the regulation of gene expression, microarray analysis of Hdac1/2-deleted cells reveals 1,708 differentially expressed genes. Significantly for the maintenance of stem cell self-renewal, we detected a reduction in the expression of the pluripotent transcription factors, Oct4, Nanog, Esrrb, and Rex1. HDAC1/2 activity is regulated through binding of an inositol tetraphosphate molecule (IP4) sandwiched between the HDAC and its cognate corepressor. This raises the important question of whether IP4 regulates the activity of the complex in cells. By rescuing the viability of double-knockout cells, we demonstrate for the first time (to our knowledge) that mutations that abolish IP4 binding reduce the activity of HDAC1/2 in vivo. Our data indicate that HDAC1/2 have essential and pleiotropic roles in cellular proliferation and regulate stem cell self-renewal by maintaining expression of key pluripotent transcription factors.
Histone deacetylase 1 and 2 (HDAC1/2) are highly related enzymes (∼80% identical) that regulate chromatin structure as components of the multiprotein Sin3 (1, 2), NuRD (3), and CoREST (4) corepressor complexes. The recruitment and activity of HDAC1/2 within these multiprotein complexes was thought to be a constitutive process. However, it has recently been shown that the HDAC1/metastasis-associated 1 (MTA1) complex (of NuRD) is regulated by a molecule of inositol tetraphosphate [Ins(1,4,5,6)P4] (IP4), which regulates its enzymatic activity, presumably in response to external signaling pathways (5). Because deacetylation of histone tails results in the tightening of nucleosomal arrays (6), the physiological roles of HDAC1/2 have mostly been defined within the context of transcriptional repression. However, genome-wide mapping of HDAC1 target genes reveals a correlation between binding and transcriptional activity (7, 8), suggesting that HDAC1/2 may play a role in the cyclical acetylation of histones at active promoters (9). HDAC1/2 may thus have roles in both transcriptional activation and repression.
Although there are a burgeoning number of proteins in the acetylome in addition to histones (10), the Sin3A, NuRD, and CoREST complexes contain multiple DNA/chromatin recognition motifs (11), which, in combination with transcription factors (2, 12), target HDAC1/2 to chromatin. Their physiological roles should therefore be viewed within the framework of chromatin modulation. In the process of DNA double-strand break repair, for instance, they deacetylate histones in the vicinity of the lesion, which is a critical step in nonhomologous end joining (13). During DNA replication, new histones are deposited in an acetylated form before being rapidly deacetylated as the chromatin matures (14). A recent study that used the isolation of proteins on nascent DNA (iPOND) technique, found that HDAC1/2 are recruited close to the replication fork (15), which suggests that they play a role in the maturation of chromatin during S phase. Successful mitosis also requires appropriate patterns of histone acetylation. The Sin3A complex helps maintain pericentric heterochromatin in a hypoacetylated state to ensure correct assembly of the kinetochore and faithful chromosome segregation (16, 17). HDAC1/2 are likely to be expressed in most, if not all, mammalian cells. A generic requirement for HDAC1/2 in cell cycle progression makes them potentially useful therapeutic targets in the treatment of cancer, because blocking their activity with small-molecule inhibitors should limit cellular proliferation (11).
With a few significant exceptions [embryogenesis (18), heart (19), and brain (20)], the physiological activities of HDAC1 and HDAC2 are functionally redundant. Conditional deletion of Hdac1 or Hdac2 alone, using tissue-specific transgenic models, produced no obvious deleterious effects on the development of heart, smooth muscle, endothelial cells, neural crest cells (19), oocytes (21), epidermis (22), B cells (23), and T cells (24); whereas simultaneous deletion of Hdac1/2 in these same cell types produced a number of profound phenotypes [summarized in Kelly and Cowley (11)]. We recently described the generation and characterization of conditional knockout embryonic stem (ES) cells for Hdac1 or Hdac2 (25). Although their differentiation properties are altered, cell viability and pluripotent potential of ES cells were unaffected by loss of either HDAC1 or HDAC2 alone. To circumvent this functional redundancy, we have engineered a double conditional knockout (DKO) (HDAC1Lox/Lox; HDAC2Lox/Lox; CreER) cell line, in which we can inactivate both genes simultaneously using a tamoxifen-inducible Cre/estrogen receptor fusion expressed from the ROSA26 locus. We demonstrate that loss of HDAC1/2 causes loss of cell viability 4 days following gene inactivation, which is associated with an increase in abnormal mitotic spindles and chromosome segregation defects. Almost 2,000 genes are deregulated. Significantly for the self-renewal properties of ES cells, this includes down-regulation of the core pluripotent factors, Oct4, Nanog, and Rex1. Furthermore, using the rescue of Hdac1/2-null cells as a model system to monitor HDAC1/2 activity, we have also shown for the first time (to our knowledge) that mutations that abolish IP4 binding reduce the activity of HDAC1 in vivo.
Results
Inactivation of Hdac1/2 Causes Defective Chromosomal Segregation and a Loss of Cell Viability.
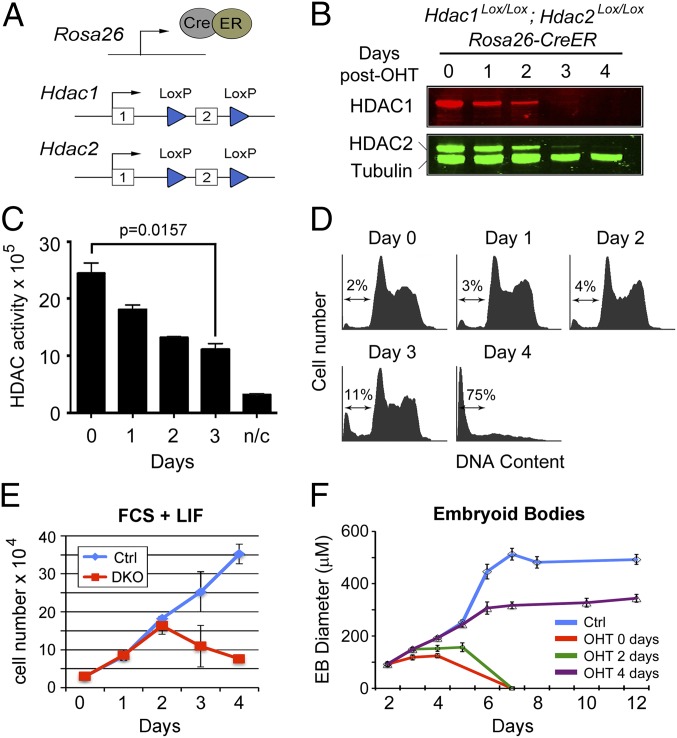
We generated a conditional Hdac1Lox/Lox; Hdac2Lox/Lox; CreER DKO ES cell line, in which exon 2 of each gene is flanked by LoxP sites (Fig. 1A), using standard gene targeting methodology. Induction of CreER, using 4-hydroxytamoxifen (OHT), causes deletion of exon 2 and disrupts the ORF such that a premature stop codon is introduced in exon 3. Consistent with our previous data (25), inactivation of the Hdac1 and Hdac2 genes resulted in loss of each protein 2–3 days after OHT treatment (Fig. 1B), indicating that both proteins have a half-life of ∼24 h (Fig. S1). The total deacetylase activity of the cell decreases by 60% over this same period (Fig. 1C), indicating that HDAC1/2 are, biochemically at least, the predominant HDAC enzymes in the cell. At 3 days following OHT treatment, Hdac1/2-deleted ES cells showed a change in morphology (Fig. S2A) and a subtle increase in cell death (2–11% sub-G1 cells; Fig. 1D). We also observed a slight reduction in the percentage of S-phase cells (60% vs. 48% 5-ethynyl-2′-deoxyuridine incorporation) in DKO cells compared with controls (Fig. S1C). However, at 4 days there was a profound loss of cell viability (2–75% sub-G1 cells). When we monitored the growth of DKO and control cells, it was observed that DKO cells fail to proliferate beyond day 2 when HDAC1/2 levels are <20% of wild type (Fig. S1). However, when we first stimulate cell cycle exit before deletion, by making embryoid bodies (Fig. 1F) or using retinoic acid treatment (Fig. S1B), the majority of cells remained viable, implying that the lethal phenotype is cell cycle dependent.
Fig. 1.
Inactivation of Hdac1/2 causes loss of cell viability. (A) Schematic diagram of the model system. An E14 ES cell line constitutively expressing a Cre/estrogen receptor (CreER) fusion from the ROSA26 locus was used to generate homozygous conditional knockout alleles for both Hdac1 and Hdac2. Both genes are inactivated by deletion of exon 2, which is flanked by LoxP sites (blue triangles). (B) Quantitative Western blot showing loss of HDAC1 and HDAC2 proteins following gene inactivation (0–4 d). Cells were cultured in the presence of 4-hydroxytamoxifen (OHT) for 24 h to induce the deletion of Hdac1/2. α-Tubulin was used to normalize protein loading, and blots were visualized and quantified using an Odyssey scanner. (C) Deacetylase activity was measured in whole-cell extracts on 4 consecutive days following OHT treatment; n/c represents the negative control. Data are representative of n > 3 independent experiments. Significance (P value) was calculated using a two-tailed t test. (D) Cell cycle distribution of Hdac1/2-deleted ES cells over a 4-d period following gene inactivation was performed using propidium iodide staining and FACS analysis. The arrow indicates the percentage of cells with a sub-G1 amount of DNA. (E) Comparative viable cell counts between control ES cells (Ctrl, untreated DKO cells) and Hdac1/2 double-knockout ES cells (DKO). All values are means (n = 3) ± SEM. (F) Embryoid bodies (EBs) were generated by plating ES cells on to bacterial dishes. OHT was add to the growth media at the indicated times following plating. The mean size of EBs size is shown (n > 30) ± SEM.
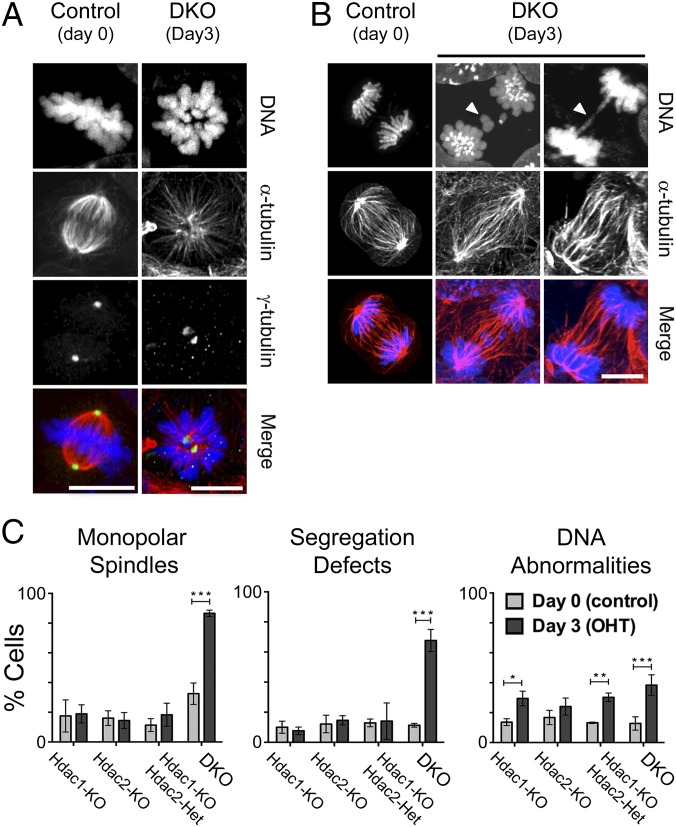
To search for potential cell cycle defects in Hdac1/2-deleted cells, we stained control (DKO cells, day 0), individual Hdac1 and Hdac2 knockout cells (25), a compound Hdac1-KO; Hdac2-Het knockout cell line (Fig. S1D), and DKO cells at day 3 after deletion, with anti–α-tubulin, anti–γ-tubulin and Hoechst 33258 to visualize chromosomes during the various stages of cell division. Initially, we focused on mitotic cells because experiments in mouse embryo fibroblasts (16) and Schizosaccharomyces pombe (17) have shown a role for the Sin3A complex in chromosome segregation. In contrast to individual and compound knockouts, a high proportion of the DKO cells in metaphase had a monopolar rather than a bipolar mitotic spindle (Fig. 2A and Fig. S2B). Consistent with this result, we also observed a sixfold increase in the number of DKO cells with segregation defects (Fig. 2B and Fig. S2C; quantified in Fig. 2C), including both lagging chromosomes and chromatin bridges, suggesting the presence of both premitotic and mitotic errors. Interphase cells also displayed a significant increase in micronucleation, multinucleate cells, and cells with lobed or highly condensed chromatin (summarized as DNA abnormalities; Fig. 2C and Fig. S2D). We conclude, then, that a double deletion of Hdac1/2 results in severe chromosome segregation defects and that this is likely a major cause of cell death in DKO cells.
Fig. 2.
Loss of HDAC1/2 causes defective chromosomal segregation. ES cells were stained with anti–α-Tubulin (red), anti–γ-Tubulin (green), and Hoechst 33258 (blue) to visualize chromosomes during various stages of cell cycle. Experiments were performed on untreated DKO (day 0, control) and DKO cells following deletion (day 3). Images show examples of mitotic cells with monopolar spindles (A) and segregation defects (B) following Hdac1/2 deletion. The white arrows indicate individual examples of lagging chromosomes (Center) and chromatin bridges (Right). The images correspond to z projections. (Scale bar, 10 µm.) (C) Quantitative analysis of chromosome segregation defects following loss of HDAC1/2. The mean (±SD) percentage of cells with abnormal DNA is indicated based on counts of at least 50 cells from n ≥ 3 experiments. Significance (P value) was calculated using a two-tailed t test (*P < 0.01; **P < 0.001; ***P < 0.0001).
Loss of HDAC1/2 Disrupts Corepressor Complex Integrity and Leads to an Increase in Global Histone Acetylation.
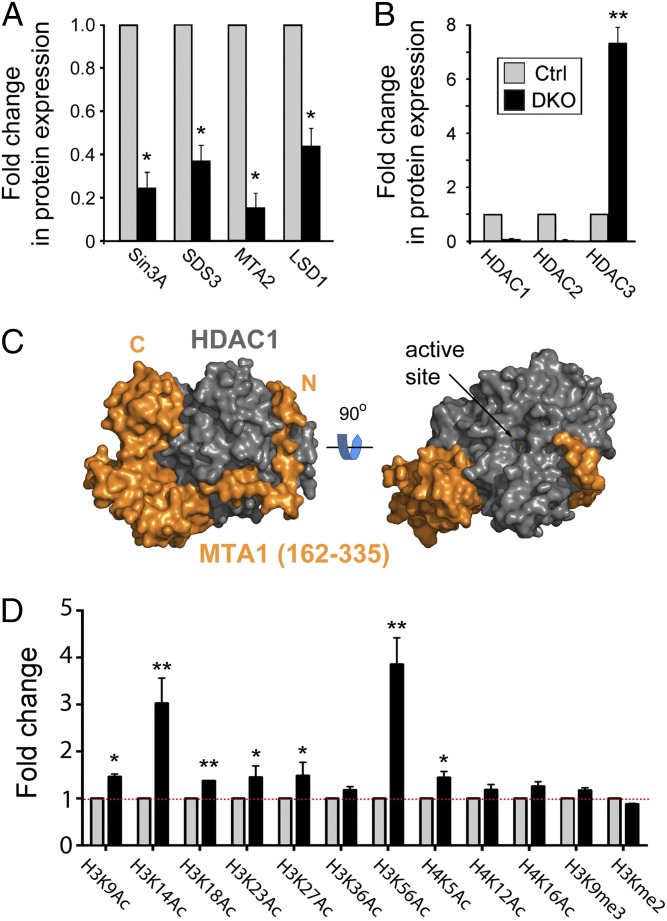
HDAC1/2 are recruited into three main transcriptional corepressor complexes: Sin3A (26), NuRD (3), and CoREST (4). Incorporation into specific complexes is fundamental to HDAC function because they do not bind DNA directly and they tend to be active only in the presence of a binding partner, often a cognate corepressor protein, such as MTA2 (27). To test the integrity of HDAC1/2-containing complexes, we performed Western blots on protein extracts from control and day 3 DKO cells (Fig. 3A). Upon simultaneous deletion of Hdac1/2, we observed a 4.2- and 7.1-fold decrease in the levels of Sin3A and MTA2, respectively, direct binding partners of HDAC1/2 (Fig. 3A). The level of SDS3, an essential gene that facilitates the interaction of Sin3A/HDAC1 (16, 28), is also reduced 2.6-fold relative to controls (Fig. 3A and Fig. S3A). However, the expression level of HDAC3, a highly related class I HDAC that forms part of the SMRT/NCoR complex, is increased sevenfold (Fig. 3B and Fig. S3B), which suggests a partial compensation for the loss of HDAC1/2. These data suggest that HDAC1/2 contribute to the structural integrity of the Sin3A and NuRD complexes. The interaction of HDAC1/2 with the NuRD complex is mediated through the ELM2-SANT domains of the MTA family, MTA1–3 (5, 29). The ELM2-SANT region of MTA1 wraps completely around the catalytic domain of HDAC1 making extensive protein–protein contacts (Fig. 3C). Importantly, the conserved N-terminal region of ELM2 (residues 162–198), which binds an extended groove on the side of HDAC1, lacks extensive secondary structure. In the absence of HDAC1/2, this region is likely to be solvent exposed and therefore lead to the increased protein turnover of MTA2 observed in DKO cells (Fig. 3A).
Fig. 3.
Loss of HDAC1/2 disrupts corepressor complex integrity and leads to increased global histone acetylation. Experiments were performed on untreated (Ctrl), or OHT-treated double-knockout (DKO) cells 3 days following gene inactivation. (A and B) Quantitative Western blot data for the indicated proteins were performed using an Odyssey scanner and normalized to the level of α-tubulin. Loss of HDAC1/2 causes increased expression of HDAC3 in DKO (day 3; black bars) cells compared with control (Ctrl; gray bars). Mean values (n > 3) ± SEM are plotted. (C) Structure of the ELM2-SANT region of MTA1 (orange) bound to the catalytic domain HDAC1 (gray). (D) Quantitative Western blotting was used to determine the levels of global histone acetylation. Acetylation levels were normalized to the total amount of H3 quantified using an Odyssey scanner. All values are means (n > 3) ± SEM. The significance (P value) of data in A, B, and D was calculated using a two-tailed t test (*P < 0.01; **P < 0.001).
Because the Sin3A and NuRD complexes appeared to be disrupted in DKO cells, we next investigated what effect this had on the levels of global histone acetylation using quantitative Western blotting. Pluripotent ES cells maintain a relatively plastic chromatin structure and consequently have a high basal level of histone acetylation (25). The loss of HDAC1/2 therefore produced a relatively modest increase in acetylation levels at most sites of histone acetylation, with the notable exceptions of H3K14Ac and H3K56Ac, which were increased threefold and fourfold, respectively (Fig. 3D). The fact that we detected an increase in the acetylation of all of the sites tested in histone H3 and H4 speaks to the pleiotropic nature of HDAC1/2 function. However, the levels H3K4me2 and H3K9me3, two methylation sites within the H3 tail that typify euchromatic and heterochromatic regions of the genome, remained largely unchanged.
HDAC1/2 Regulate the ES Cell Transcriptome and Are Required for Expression of Oct4 and Nanog.
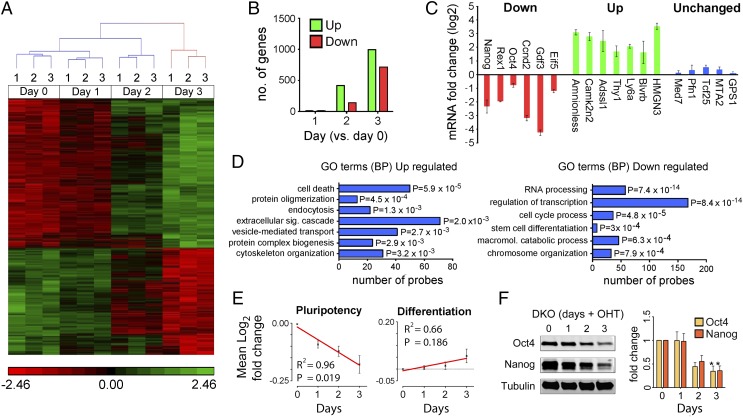
HDAC1/2 have a well-described role in the regulation of gene expression (11). Furthermore, the increase in global histone acetylation levels suggested that the pattern of gene expression may well be altered in Hdac1/2-deleted cells. We therefore performed a comparative microarray analysis on mRNA isolated from DKO cells at 0, 1, 2, and 3 days following Hdac1/2 inactivation, and transcripts deregulated ≥1.4-fold (adjusted P < 0.05) were identified from three independent experiments (Fig. 4A and Dataset S1). Interestingly, we observed a correlation between the reduction in HDAC activity and the number of deregulated genes, with very few (n = 3) aberrantly expressed transcripts observed on day 1 and an increasing number on day 2 (560 genes) and day 3 (1,708 genes), as HDAC1/2 are progressively lost (Fig. 4B). To further verify the microarray result, we quantified the levels of six down-regulated, seven up-regulated, and five unchanged transcripts by quantitative real-time PCR (qRT-PCR) (Fig. 4C). In each instance (18 of 18 transcripts), we were able to corroborate the microarray result. An analysis of functionally related gene groups among deregulated genes using Database for Annotation, Visualization, and Integrated Discovery (DAVID) (30), reveals that genes involved in cell death are up-regulated, whereas cell cycle genes are significantly down-regulated (Fig. 4D), confirming the dominant phenotype observed in the DKO cells. Down-regulated transcripts are highly enriched for genes with a role in the regulation of transcription (P = 7.4 × 10−14) and cell cycle processes (P = 4.88 × 10−5), as might be expected. However, genes involved in RNA processing are also decreased to the same level of significance (P = 8.4 × 10−14), suggesting a putative role for HDAC1/2 in the regulation of RNA splicing. Significant to the self-renewal properties of ES cells, the pluripotent factor, Nanog, was down-regulated 1.81-fold on the array and 4.6-fold by qRT-PCR (Fig. 4C). Furthermore, transcript levels of the pluripotent factors, Oct4 (Pou5f1), Rex1 (Zfp42), Esrrb,and Zfx, were also significantly reduced (between 1.45- and 1.63-fold; P < 0.005; Dataset S1). It is noteworthy that we also detected a distinct change in cell morphology of DKO cells at day 3, suggesting a loss of the stem cell phenotype (Fig. S2A). This prompted us to analyze an additional 39 genes (80 probes) relating to transcripts for factors associated with pluripotency (Fig. 4E and Table S1). We observed a progressive loss of pluripotent factor expression (R2 = 0.96; P = 0.019) over the 3-day time period in which HDAC1/2 activity is lost. The protein levels of both Oct4 and Nanog were also reduced in parallel with the decrease of HDAC1/2 activity (Fig. 4F). However, analysis of 111 genes (165 probes) associated with stem cell differentiation and lineage specification reveals only a weak positive correlation (R2 = 0.66; P = 0.186). Recent bioinformatics analyses have uncovered the “PluriNet,” a common set of characteristics shared by all pluripotent stem cell lines (31). We therefore performed gene set enrichment analysis on PluriNet target genes to determine the consequence of Hdac1/2 DKO on maintaining ES cell identity. Of the 296 PluriNet genes, 90 were significantly enriched in wild-type control ES cells (Fig. S4), demonstrating a loss of the pluripotent stem cell phenotype in DKO cells. Together, this suggests that HDAC1/2 are required for Oct4 and Nanog expression, but their loss is not sufficient to derepress genes associated with early differentiation.
Fig. 4.
HDAC1/2 regulate the ES cell transcriptome and are required for the expression of Oct4 and Nanog. (A) A heat map showing 1,708 genes that are differentially expressed in DKO ES cells over a 3-day time course following gene inactivation. The red and green labeling indicates relative gene expression levels. (B) Number of genes differentially expressed at the indicated days (compared with day 0) following deletion of Hdac1/2. (C) qRT-PCR was used to validate the change in expression of a subset of genes from the microarray. Values indicate comparative means (n = 3) ± SEM between DKO cells at day 0 and day 3 following gene inactivation. (D) Functional annotation analysis of differentially expressed genes. DAVID was used to identify biological process (BP) and gene ontology (GO) of probes up-regulated or down-regulated in DKO cells. (E) Regression analysis of pluripotency and differentiation associated genes using mean log2 fold changes of microarray data. (F) Quantitative Western blot data for Oct4 and Nanog proteins indicate a reduction in parallel with the decrease of HDAC1/2 activity. Significance (P value) was calculated using a two-tailed t test (*P < 0.01).
Rescue of DKO Cells Is Dependent upon the Integrity of the HDAC1 IP4 Binding Pocket.
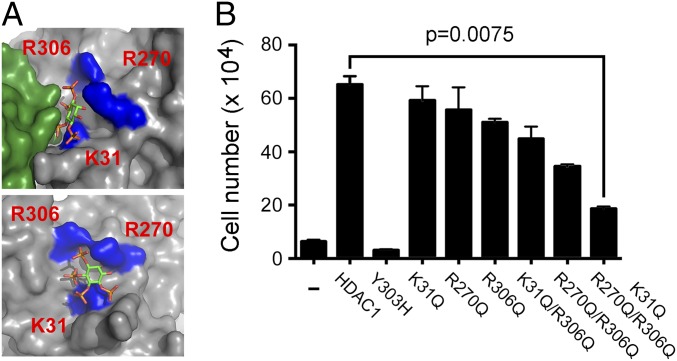
It seemed likely that the essential requirement for HDAC1/2 in cell division and their ability to influence gene expression was dependent upon deacetylase activity. To confirm this, we transfected DKO cells with cDNAs for wild type and a catalytically inactive version [HDAC1Y303H (32)] of HDAC1 to assess their ability to rescue cell viability (Fig. 5). Although wild-type HDAC1 was able to rescue DKO cells, the majority of cells transfected with HDAC1Y303H still died at day 4 following Hdac1/2 deletion. We therefore had an excellent model system in which to interrogate aspects of HDAC1 activity. It was recently shown, using in vitro assays, that HDAC1 activity is modulated by IP4 (5). The IP4 binding pocket on the surface of HDAC1 is made up of a number of positively charged residues (K31, R270, and R306), which are thought to form hydrogen bonds with the negatively charged phosphates of IP4 (Fig. 5A) and are required for IP4-dependent activation. To test the requirement for IP4 binding to the activity of HDAC1 in cells, we mutated these three residues to glutamine, a polar noncharged residue, individually (K31Q, R270Q, R306Q), as double mutants (K31Q/R270Q, R270Q/R306Q) and as a triple mutant (K31Q/R270Q/R306Q). We transfected these constructs into DKO cells to initially test their expression and relative deacetylase activity (Fig. S5). Mutation of each of the positively charged residues reduced the deacetylase activity of HDAC1, with combinations of mutations having an additive effect, such that the triple mutant (HDAC1K31Q/R270Q/R306Q) had the lowest activity of all. These same constructs were then tested for their ability to rescue the viability of Hdac1/2 DKO cells at 4 days following gene inactivation. Again, single-point mutations (K31Q, R270Q, and R306Q) produced a lower number of viable DKO cells compared with controls, whereas double and triple substitutions resulted in an additive effect, such that HDAC1K31Q/R270Q/R306Q produced the smallest number of viable cells (Fig. 5B). It is worth noting that the catalytically inactive HDAC1Y303H always produced fewer viable cells in this assay than untransfected controls, indicating a dominant-negative effect of this mutation. HDAC1K31Q/R270Q/R306Q appears to be a hypomorphic mutant, as it produces more viable DKO cells than HDAC1Y303H, but far fewer than wild-type HDAC1. This implies that IP4 binding is necessary for the full activity of HDAC1 in vivo.
Fig. 5.
Cell viability is dependent upon the integrity of the IP4 binding pocket. (A) Structure of HDAC1 (gray), with positively charged residues critical for the interaction with IP4 (K31, R270, and R306) marked in blue. Position of the MTA1 corepressor is shown in the top panel (green). (B) The number of viable cells, transfected with the indicated HDAC1 expression constructs, were counted 4 days after Hdac1/2 inactivation. All values are means (n > 3) ± SEM. Significance (P value) was calculated using a two-tailed t test.
Discussion
Essential and Pleiotropic Roles for HDAC1/2 in Cellular Proliferation.
We have generated a mouse ES cell line in which we can conditionally inactivate the highly related genes Hdac1 and Hdac2 simultaneously. Loss of HDAC1/2 results in a 60% reduction in cellular HDAC activity (Fig. 1C) despite a compensatory increase in the related class 1 HDAC, HDAC3 (Fig. 3B). The increase in HDAC3 occurs at the protein level, because Hdac3 transcripts were not significantly altered in our microarray analysis (Fig. 4A and Dataset S1). Interestingly, we observed a significant increase in DNA abnormalities with Hdac1-KO, Hdac1-KO; Hdac1-Het, and DKO cells, but not Hdac2-KO cells (Fig. 2C). A comparison of the total HDAC activity for each of these cell lines revealed a significant reduction in all but the Hdac2-KO cells (Fig. S6), suggesting that this phenotype is dependent on the dosage of HDAC1/2 activity. The predominant phenotype of Hdac1/2 DKO cells is a loss of viability 4 days after gene inactivation. The lethality is dependent on an active cell cycle, because if we first stimulate cell cycle exit, by generating embryoid bodies (Fig. 1F) or using retinoic acid (Fig. S1B), the majority of cells remained viable. A loss of cell proliferation is a common phenotype in all Hdac1/2 knockout (18, 23, 33, 34) and knockdown (35) studies. In each case, loss of HDAC1/2 is associated with up-regulation of the cyclin-dependent kinase inhibitor, p21WAF1/CIP1, which limits G1- to S-phase transition. Indeed, the proliferation of mammalian cells is largely controlled during the G1 phase, therefore implicating HDAC1/2 activity in this critical step. However, ES cells have a peculiarly short G1 phase (∼1.5 h), in which CDK2 complexes are constitutively active and Rb hyperphosphorylated (36, 37), and therefore lack the usual G1 restriction point of somatic cells. This presumably explains the difference in phenotype between DKO ES cells and other model systems, including mouse embryo fibroblasts (MEFs). Hdac1/2 KO MEFs undergo G1 arrest, associated with increased expression of the aforementioned, p21WAF1/CIP1 and p57Kip2 (23). Hdac1/2 KO ES cells lacking this G1 regulatory step are unable to arrest in G1 and therefore enter S phase, and later mitosis, where the absence of HDAC1/2 activity causes lethality. We detected a significant increase in both chromatin bridges, micronuclei (Fig. 2B and Fig. S2C), and a reduction in S-phase cells (Fig. S1C) upon Hdac1/2 deletion, indicative of DNA replication defects. This is supported by data from Sirbu et al. (15), who used the iPOND technique to show that HDAC1/2 are present at active replication forks; and Bhaskara et al. (38), who demonstrated that chemical inhibition or knockdown of HDAC1/2 reduced replication fork velocity and activates the replication stress response. In addition, we also observed that a high proportion of DKO cells in mitosis display abnormal spindles (Fig. 2), which could equally lead to chromosome segregation errors. Deletion studies of other HDAC1/2 complex components, including SDS3 (16) and Sin3A (17), suggest that HDAC1/2 maintain the hypoacetylated state of pericentric heterochromatin, a requirement for the appropriate assembly of the kinetochore. It is likely that a combination of DNA replication and mitotic defects are the major cause of death in cells lacking HDAC1/2.
HDAC1/2 Regulate the Expression of Core Pluripotency Factors in ES Cells.
Loss of HDAC1/2 activity correlates with the down-regulation of pluripotent factors (Fig. 4 E and F). Interestingly, this phenotype contrasts with the disruption of individual HDAC1/2-containing complexes in ES cells. Deletion of LSD1 perturbs the CoREST complex but does not affect Oct4 expression (39), whereas loss of MBD3 (central component of NuRD) prevents Oct4 from being repressed at all, even under differentiating conditions (40). By process of elimination, this suggests that the Sin3A complex may positively regulate Oct4 and Nanog expression; and indeed Baltus et al. (41) were able to demonstrate a direct role for the Sin3A/HDAC complex in the activation of the Nanog promoter. More recently, a genome-wide promoter analysis in ES cells revealed that HDAC1 binds close to the transcriptional start site of many pluripotent factors, including Oct4, Nanog, Sox2, and Rex1 (7), again suggesting a positive role in the maintenance of cell self-renewal. These data, and complementary genome-wide ChIP studies that reveal an enrichment of HDAC1 binding at active gene loci (8, 42), are beginning to change the view of HDAC1/2 from being exclusively repressive factors, to regulators of both gene activation and repression.
Although it was previously thought that HDAC complexes are constitutively active, it was recently shown that the deacetylase activity of HDAC1 and HDAC3 is regulated through binding of an IP4 molecule sandwiched between the HDAC and its cognate corepressor (5, 43). This raises the important question of whether IP4 actually regulates HDAC activity in vivo. We have shown here for the first time (to our knowledge) that substitution of the positively charged residues in the IP4 binding pocket of HDAC1 (K31Q, R270Q, and R306Q), essential for IP4 binding (5), reduces the activity of HDAC1 and its ability to rescue the viability of DKO cells (Fig. 5). The HDAC1 triple mutant, HDAC1K31Q/R270Q/R306Q, reduces the number of viable DKO cells by approximately threefold, which is in agreement with a previous study that showed that an HDAC1:MTA1 complex could be stimulated threefold to fourfold by addition of IP4.
In summary, we have shown that loss of HDAC1/2 activity in ES cells leads to the deregulation of almost 2,000 genes, including down-regulation of the core pluripotent factors, Oct4 and Nanog. Using these DKO cells as a model system to measure HDAC1/2 activity, we have shown for the first time (to our knowledge) that mutations that abolish IP4 binding reduce the activity of HDAC1 in vivo and, finally, that Hdac1/2 inactivation resulted in a loss of ES cell viability. Our data also suggest that inactivation of HDAC1/2, in cells that either lack (ES cells) or have a mutated G1 restriction checkpoint (cancer cells), causes cell death due to defective DNA replication and/or mitosis. This leads us to predict that specific inhibitors of HDAC1/2 should exhibit selective toxicity toward immortalized cell types, making them effective therapeutic targets in the treatment of cancer.
Methods
Generation of Hdac1 and Hdac2 DKO ES Cell Lines.
E14 ES cells expressing a CreER fusion protein from the ROSA26 locus were used to generate Hdac1Lox/Lox; Hdac2Lox/WT; CreER and Hdac1Lox/Lox; Hdac2Lox/Lox; CreER conditional knockout ES cells, using multiple rounds of gene targeting, with vectors previously described by Dovey et al. (25). Two independently derived Hdac1Lox/Lox; Hdac2Lox/Lox; CreER ES clones were used in the study.
ES Cell Culture and Differentiation.
ES cell lines were maintained on gelatinized plates in standard ES cell medium as previously described (25). To induce Hdac1/2 deletion, cells were cultured for 24 h in the presence of OHT (1 μM) to activate the CreER fusion protein. For ES cell growth curves, control and Hdac1/2 knockout ES cells were plated at 3 × 105 ES cells per well in a six-well plate, and viable cells were counted on 4 consecutive days. ES cells were differentiated by embryoid body formation in suspension culture, or using RA (1 μM) on gelatinized plates. All cells were counted using Bio-Rad TC-10 automated cell counter in triplicate.
For more details, and a complete list of antibodies and qRT-PCR primers used in this study (Tables S2 and S3), see SI Methods.
Supplementary Material
Acknowledgments
We thank Salvador Macip for critical appraisal of the manuscript. We are also grateful to the Core Biotechnology Services for providing access and support to Advanced Imaging, Genomics, and Bioinformatics facilities. A.M.F. acknowledges support from the Association for International Cancer Research (United Kingdom), the Wellcome Trust, and Kidney Research United Kingdom. J.W.R.S. is supported by Program Grant WT085408 and Senior Investigator Award WT100237 from the Wellcome Trust. S.M.C. and J.W.R.S. are jointly supported by Biotechnology and Biological Sciences Research Council Project Grant BB/J009598/1. S.M.C. is the recipient of Medical Research Council Senior Fellowship MR/J009202/1.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The microarray data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE52134).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1321330111/-/DCSupplemental.
References
- 1.Hassig CA, Fleischer TC, Billin AN, Schreiber SL, Ayer DE. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell. 1997;89(3):341–347. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- 2.Laherty CD, et al. SAP30, a component of the mSin3 corepressor complex involved in N-CoR-mediated repression by specific transcription factors. Mol Cell. 1998;2(1):33–42. doi: 10.1016/s1097-2765(00)80111-2. [DOI] [PubMed] [Google Scholar]
- 3.Xue Y, et al. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell. 1998;2(6):851–861. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- 4.You A, Tong JK, Grozinger CM, Schreiber SL. CoREST is an integral component of the CoREST- human histone deacetylase complex. Proc Natl Acad Sci USA. 2001;98(4):1454–1458. doi: 10.1073/pnas.98.4.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Millard CJ, et al. Class I HDACs share a common mechanism of regulation by inositol phosphates. Mol Cell. 2013;51(1):57–67. doi: 10.1016/j.molcel.2013.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robinson PJ, et al. 30 nm chromatin fibre decompaction requires both H4-K16 acetylation and linker histone eviction. J Mol Biol. 2008;381(4):816–825. doi: 10.1016/j.jmb.2008.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kidder BL, Palmer S. HDAC1 regulates pluripotency and lineage specific transcriptional networks in embryonic and trophoblast stem cells. Nucleic Acids Res. 2012;40(7):2925–2939. doi: 10.1093/nar/gkr1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Z, et al. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138(5):1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dovey OM, Foster CT, Cowley SM. Emphasizing the positive: A role for histone deacetylases in transcriptional activation. Cell Cycle. 2010;9(14):2700–2701. doi: 10.4161/cc.9.14.12626. [DOI] [PubMed] [Google Scholar]
- 10.Choudhary C, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325(5942):834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 11.Kelly RD, Cowley SM. The physiological roles of histone deacetylase (HDAC) 1 and 2: Complex co-stars with multiple leading parts. Biochem Soc Trans. 2013;41(3):741–749. doi: 10.1042/BST20130010. [DOI] [PubMed] [Google Scholar]
- 12.Cowley SM, et al. Functional analysis of the Mad1-mSin3A repressor-corepressor interaction reveals determinants of specificity, affinity, and transcriptional response. Mol Cell Biol. 2004;24(7):2698–2709. doi: 10.1128/MCB.24.7.2698-2709.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller KM, et al. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat Struct Mol Biol. 2010;17(9):1144–1151. doi: 10.1038/nsmb.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389(6649):349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 15.Sirbu BM, et al. Analysis of protein dynamics at active, stalled, and collapsed replication forks. Genes Dev. 2011;25(12):1320–1327. doi: 10.1101/gad.2053211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.David G, Turner GM, Yao Y, Protopopov A, DePinho RA. mSin3-associated protein, mSds3, is essential for pericentric heterochromatin formation and chromosome segregation in mammalian cells. Genes Dev. 2003;17(19):2396–2405. doi: 10.1101/gad.1109403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Silverstein RA, Richardson W, Levin H, Allshire R, Ekwall K. A new role for the transcriptional corepressor SIN3; regulation of centromeres. Curr Biol. 2003;13(1):68–72. doi: 10.1016/s0960-9822(02)01401-x. [DOI] [PubMed] [Google Scholar]
- 18.Lagger G, et al. Essential function of histone deacetylase 1 in proliferation control and CDK inhibitor repression. EMBO J. 2002;21(11):2672–2681. doi: 10.1093/emboj/21.11.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Montgomery RL, et al. Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes Dev. 2007;21(14):1790–1802. doi: 10.1101/gad.1563807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guan JS, et al. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature. 2009;459(7243):55–60. doi: 10.1038/nature07925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma P, Pan H, Montgomery RL, Olson EN, Schultz RM. Compensatory functions of histone deacetylase 1 (HDAC1) and HDAC2 regulate transcription and apoptosis during mouse oocyte development. Proc Natl Acad Sci USA. 2012;109(8):E481–E489. doi: 10.1073/pnas.1118403109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.LeBoeuf M, et al. Hdac1 and Hdac2 act redundantly to control p63 and p53 functions in epidermal progenitor cells. Dev Cell. 2010;19(6):807–818. doi: 10.1016/j.devcel.2010.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamaguchi T, et al. Histone deacetylases 1 and 2 act in concert to promote the G1-to-S progression. Genes Dev. 2010;24(5):455–469. doi: 10.1101/gad.552310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dovey OM, et al. Histone deacetylase 1 and 2 are essential for normal T-cell development and genomic stability in mice. Blood. 2013;121(8):1335–1344. doi: 10.1182/blood-2012-07-441949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dovey OM, Foster CT, Cowley SM. Histone deacetylase 1 (HDAC1), but not HDAC2, controls embryonic stem cell differentiation. Proc Natl Acad Sci USA. 2010;107(18):8242–8247. doi: 10.1073/pnas.1000478107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laherty CD, et al. Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell. 1997;89(3):349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y, et al. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 1999;13(15):1924–1935. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fleischer TC, Yun UJ, Ayer DE. Identification and characterization of three new components of the mSin3A corepressor complex. Mol Cell Biol. 2003;23(10):3456–3467. doi: 10.1128/MCB.23.10.3456-3467.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee MG, Wynder C, Schmidt DM, McCafferty DG, Shiekhattar R. Histone H3 lysine 4 demethylation is a target of nonselective antidepressive medications. Chem Biol. 2006;13(6):563–567. doi: 10.1016/j.chembiol.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 30.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 31.Müller FJ, et al. Regulatory networks define phenotypic classes of human stem cell lines. Nature. 2008;455(7211):401–405. doi: 10.1038/nature07213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fischle W, et al. Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol Cell. 2002;9(1):45–57. doi: 10.1016/s1097-2765(01)00429-4. [DOI] [PubMed] [Google Scholar]
- 33.Wilting RH, et al. Overlapping functions of Hdac1 and Hdac2 in cell cycle regulation and haematopoiesis. EMBO J. 2010;29(15):2586–2597. doi: 10.1038/emboj.2010.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zupkovitz G, et al. The cyclin-dependent kinase inhibitor p21 is a crucial target for histone deacetylase 1 as a regulator of cellular proliferation. Mol Cell Biol. 2010;30(5):1171–1181. doi: 10.1128/MCB.01500-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Senese S, et al. Role for histone deacetylase 1 in human tumor cell proliferation. Mol Cell Biol. 2007;27(13):4784–4795. doi: 10.1128/MCB.00494-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burdon T, Smith A, Savatier P. Signalling, cell cycle and pluripotency in embryonic stem cells. Trends Cell Biol. 2002;12(9):432–438. doi: 10.1016/s0962-8924(02)02352-8. [DOI] [PubMed] [Google Scholar]
- 37.Savatier P, Huang S, Szekely L, Wiman KG, Samarut J. Contrasting patterns of retinoblastoma protein expression in mouse embryonic stem cells and embryonic fibroblasts. Oncogene. 1994;9(3):809–818. [PubMed] [Google Scholar]
- 38.Bhaskara S, et al. 2013. Histone deacetylases 1 and 2 maintain S-phase chromatin and DNA replication fork progression. Epigenetics Chromatin 6(1):27.
- 39.Foster CT, et al. Lysine-specific demethylase 1 regulates the embryonic transcriptome and CoREST stability. Mol Cell Biol. 2010;30(20):4851–4863. doi: 10.1128/MCB.00521-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaji K, et al. The NuRD component Mbd3 is required for pluripotency of embryonic stem cells. Nat Cell Biol. 2006;8(3):285–292. doi: 10.1038/ncb1372. [DOI] [PubMed] [Google Scholar]
- 41.Baltus GA, Kowalski MP, Tutter AV, Kadam S. A positive regulatory role for the mSin3A-HDAC complex in pluripotency through Nanog and Sox2. J Biol Chem. 2009;284(11):6998–7006. doi: 10.1074/jbc.M807670200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kurdistani SK, Robyr D, Tavazoie S, Grunstein M. Genome-wide binding map of the histone deacetylase Rpd3 in yeast. Nat Genet. 2002;31(3):248–254. doi: 10.1038/ng907. [DOI] [PubMed] [Google Scholar]
- 43.Watson PJ, Fairall L, Santos GM, Schwabe JW. Structure of HDAC3 bound to co-repressor and inositol tetraphosphate. Nature. 2012;481(7381):335–340. doi: 10.1038/nature10728. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.