Abstract
The suppressive potentials of Bacillus and Paenibacillus strains isolated from the tomato phyllosphere were investigated to obtain new biocontrol candidates against Fusarium crown and root rot of tomato. The suppressive activities of 20 bacterial strains belonging to these genera were examined using seedlings and potted tomato plants, and two Paenibacillus strains (12HD2 and 42NP7) were selected as biocontrol candidates against the disease. These two strains suppressed the disease in the field experiment. Scanning electron microscopy revealed that the treated bacterial cells colonized the root surface, and when the roots of the seedlings were treated with strain 42NP7 cells, the cell population was maintained on the roots for at least for 4 weeks. Although the bacterial strains had no direct antifungal activity against the causal pathogen in vitro, an increase was observed in the antifungal activities of acetone extracts from tomato roots treated with the cells of both bacterial strains. Furthermore, RT-PCR analysis verified that the expression of defense-related genes was induced in both the roots and leaves of seedlings treated with the bacterial cells. Thus, the root-colonized cells of the two Paenibacillus strains were considered to induce resistance in tomato plants, which resulted in the suppression of the disease.
Keywords: Fusarium crown and root rot, Fusarium oxysporum f. sp. radicis-lycopersici, Phyllosphere bacteria, Paenibacillus, induced resistance
Fusarium crown and root rot (FCRR) of tomato is caused by Fusarium oxysporum f. sp. radicis-lycopersici (FORL), which is a disease that is commonly observed in locations worldwide, such as Europe, North America, and Japan (15, 38, 47). The symptoms of the disease consist of the browning and rotting of the crown and roots and the yellowing of leaves. Advanced symptoms are wilting and death (32), which leads to a loss in fruit yield. The fruit yield loss due to the disease in Florida, USA was reported to range between 15% and 65% (32). Although attempts to control this challenging disease in Japan have included physical and chemical approaches, such as soil solarization and the use of chemical fumigants, respectively, no biological approaches such as the use of microbes antagonistic to the causal fungi are currently available. The development of biological controls has recently received attention because of their economic and ecological advantages, as well as political demands. Several studies have examined biological control approaches against the disease (6, 28, 31, 35). In Japan, Iwamoto and Aino (20) reported that a commercial biocontrol agent (an endophytic bacterium of tomato roots, Pseudomonas fluorescens FPH9601) significantly suppressed the disease in tomato fields, although the sale of the agent is currently suspended. Another study on disease suppression indicated the successful combined use of a plant growth-promoting fungus (PGPF), Fusarium equiseti, and biodegradable pots in fields (16); however, commercial products have not yet been registered. Therefore, more novel microbial agents that are applicable to the actual practices used in tomato cultivation need to be developed.
To develop biocontrol agents based on microbial colonization and propagation principles, selection from the resident microbes of the target plant is preferable (44). Thus, microbes colonizing tomato plants are good candidates for biocontrol agents of the disease. Based on this principle, several studies on the control of the disease have been demonstrated using various microorganisms (2, 3, 6). Kamilova et al. (22) reported that a Pseudomonas fluorescens strain and Pantoea agglomerans strain selected using an enrichment method were superior root colonizers that could significantly control the tomato disease caused by FORL, which demonstrated the importance of the colonization ability of microbes for biocontrol. Although selection from resident microbes inhabiting roots appears to be more feasible for tackling the Fusarium disease in the tomato, we hypothesized that novel and unique microbial candidates may be obtained as biocontrol agents from the aboveground parts of the plants (i.e., the phyllosphere). Root-inhabiting microbes have a higher ability to colonize plants per se (46) and several of these microbes have the potential to suppress plant diseases (44, 45), whereas few attempts have been made to use phyllosphere microbes against soilborne diseases. In addition, soil is recognized to be a significant source of phyllosphere microbes (46), which suggests their potential to have suppressive functions even in soils or the rhizosphere.
The first objective of this study was to evaluate the suppressive potential of phyllosphere microbes in the tomato against FCRR, which could then be considered useful as biocontrol agents. We previously isolated and preserved culturable microbes inhabiting various plant surfaces (8, 9, 37, 49, 51). On tomato plants, Enya et al. (8, 9) reported the taxonomical grouping of 2138 culturable bacterial strains isolated from a greenhouse- and field-grown tomato phyllosphere as well as their antifungal activities against several aboveground tomato diseases. Of the microbial stocks isolated from the tomato phyllosphere, emphasis was placed on Bacillus and Paenibacillus in this study because members of these bacterial groups contribute to crop production (27) and produce endospores that are tolerant to heat and desiccation (44), leading to the easy and costless generation of the final commercial products. Several commercial products originating from these bacterial groups have already been developed (11). Once two Paenibacillus strains from the tomato phyllosphere were verified to have suppressive potentials, we determined the mode of action for the suppressive abilities of the Paenibacillus strains.
Materials and Methods
Tomato plants
The tomato plant (Solanum lycopersicum Mill.) cv. House-momotaro (Takii Seed, Kyoto, Japan) was used in this experiment. The seeds were sown in each cell (3×3×4.5 cm) of a plastic 128-cell tray (Takii Seed) containing commercial soil (0.1 g N, 1.25 g P, 0.1 g K, and 0.1 g Mg kg−1; Ryousaibaido Pp; Nihon Hiryo, Tokyo, Japan), grown in a greenhouse at 23°C for 3 weeks, and then used for seedling or potted plant experiments as described below.
Bacterial strains
The 13 Bacillus and 7 Paenibacillus strains summarized in Table 1 were used in this study. They were isolated from the healthy leaves of field or greenhouse-grown tomatoes in Tsukuba, Japan in 2003 (8) and preserved in the National Institute for Agro-Environmental Sciences, Tsukuba. These strains were chosen as representatives belonging to each cluster in the phylogenetic tree comprising 190 Bacillus and Paenibacillus strains, as discussed in a previous study (9).
Table 1.
Bacterial strains (Bacillus and Paenibacillus) isolated from the tomato phyllosphere and used for seedling experiments
| Straina | Accession No.b | Closely related speciesc | Homology (%)c | Disease severity (DS)d | Protective value (PV)e |
|---|---|---|---|---|---|
| 12HD2 | AB242753 | Paenibacillus alginolyticus DSM 5050 | 98.6 | 27.5 bc | 40.6 |
| 12HD4 | AB242667 | Bacillus niacini IFO15566 | 98.8 | 33.4 abc | 27.9 |
| 22HD1 | AB242662 | Bacillus solisalsi YC1 | 98.4 | 37.5 abc | 18.9 |
| 22HD4 | AB242763 | Paenibacillus xylanilyticus XIL14 | 99.3 | 40.0 abc | 13.6 |
| 31ND2 | AB242757 | Paenibacillus kobensis DSM 10249 | 95.8 | 34.5 abc | 25.5 |
| 31NP3 | AB242674 | Bacillus parviboronicapiensf BAM-582 | 97.7 | 39.2 abc | 15.3 |
| 42ND16 | AB242755 | Paenibacillus alginolyticus DSM 5050 | 98.4 | 34.2 abc | 26.1 |
| 42ND17 | AB242663 | Bacillus weihenstephanensis DSM11821 | 98.4 | 44.5 ab | 3.9 |
| 42ND20 | AB242668 | Bacillus foraminis CV53 | 95.1 | 37.8 ab | 18.3 |
| 42NP7 | AB242671 | Paenibacillus favisporus GMP01 | 99.7 | 22.5 c | 51.4 |
| 42NP15 | AB242664 | Bacillus megaterium IAM 13418 | 99.6 | 36.7 abc | 20.6 |
| 42NP17 | AB242672 | Bacillus simplex DSM 1321 | 99.8 | 42.2 abc | 8.8 |
| 52HD3 | AB242659 | Bacillus stratosphericus 41KF2a | 100 | 39.2 abc | 15.3 |
| 62HD17 | AB242675 | Bacillus mojavensis IFO15718 | 99.7 | 35.9 abc | 22.5 |
| 62ND1 | AB181686 | Bacillus safensis FO-036b | 98.7 | 34.2 abc | 26.1 |
| 62NP15 | AB242758 | Paenibacillus lautus JCM 9073 | 96.2 | 37.5 abc | 19.0 |
| 62NP21 | AB242756 | Paenibacillus glycanilyticus DS-1 | 98.3 | 31.7 abc | 31.6 |
| 83ND30 | AB242669 | Bacillus panaciterrae Gsoil 1517 | 99.5 | 36.7 abc | 20.7 |
| 104NP2 | AB242644 | Bacillus solisalsi YC1 | 97.8 | 35.9 abc | 22.5 |
| 104NP13 | AB242645 | Bacillus benzoevorans NCIMB 12555 | 97.4 | 30.0 abc | 35.2 |
| Controlg | 46.3 a |
Each strain was obtained in a previous study (9).
The accession number of the 16S rRNA gene sequence of each strain was deposited in the GenBank data library.
Closely related species and its homology (%) in the library based on the gene sequence.
Different letters indicate significant differences using Fisher’s LSD test (P<0.05).
Calculation was performed based on the mean DS value.
Present name is Lysinibacillus parviboronicapiens.
Seedlings were treated with SDW as a control.
Preparation of the F. oxysporum f. sp. radicis-lycopersici (FORL) inoculum
FORL strain For6-3 was isolated from greenhouse-grown tomato roots (20) and preserved in the Hyogo Prefectural Technology Center for Agriculture, Forestry and Fisheries (Kasai, Japan) for use in the present study. For6-3 was shaken (120 rpm) in potato dextrose liquid medium (Difco Laboratories Inc., Detroit, MI, USA) for 5 d at 28°C in the dark, and the culture was filtered through cheesecloth to remove mycelial fragments. The resulting budcells were then harvested by centrifugation at 4,000×g for 10 min, washed twice with sterilized distilled water (SDW), suspended in SDW to achieve a concentration of 1.0×107 budcells ml−1, and then used in the seedling experiment. In pot experiments, 40 mL of the prepared budcell suspension was poured into 600 g of an autoclaved soil– wheat bran medium (a mixture of clay soil and wheat bran at a ratio of 4:1) in a plastic case (299×224×62 mm) and incubated for 2 weeks at 25°C in the dark. After incubation, fungal propagules grown in the medium were ground using a mortar and stored at 4°C until use. The fungal density in the medium was checked by a dilution plating method before use and confirmed to range between 3 and 8×107 CFU g−1 of medium.
Seedling experiment
To screen bacterial strains having a suppressive effect on FCRR, each Bacillus and Paenibacillus strain was shaken (120 rpm) in R2A liquid medium (Wako Pure Chemical Industries, Osaka, Japan) for 48 h at 28°C in the dark, collected by centrifugation at 6000×g for 10 min, washed twice, and suspended in SDW to achieve an OD600 =0.5. Five milliliters of the bacterial suspension of each strain was drenched into the soil of each seedling with a pipette, and 5 mL of SDW was similarly applied to another eight seedlings as a control. Two d after the bacterial treatment, 15 mL of the budcell suspension of FORL For6-3 prepared was inoculated into the soil of each seedling with a pipette. After incubation for 4 weeks in a greenhouse at 23°C, the soil was carefully dug away from the roots of the seedlings and the roots were thoroughly washed with tap water. The degree of root rot was then evaluated based on a lesion index value of 0, 1, 2, 3, 4, or 5 (0, no lesion; 1, lesion areas on 1/10 to 1/4 of the root area; 2, lesion areas on >1/4 to 1/3 of the root area; 3, lesion areas on >1/3 to 2/3 of the root area; 4, lesion areas on >2/3 of the root area; 5, dying). The disease severity (DS) and protective value (PV) were calculated based on the following formulas: DS = (0n0 + 1n1 + 2n2 + 3n3 + 4n4 + 5n5)/5(n0 + n1 + n2 + n3 + n4 + n5) × 100, where n0–5 is the number of seedlings with each of the 1–5 lesion index values; PV = 100 − ([DS of the sample/DS of the control] × 100). The calculation was performed using eight replicated seedlings and the experiment was repeated four times. Results were analyzed by ANOVA followed by Fisher’s LSD test using Kaleida-Graph (Synergy Software, PA, USA)
Potted plant experiment
Bacterial suspensions of the strains 12HD2 and 42NP7, which displayed suppressive effects in the seedling experiment, were again applied to each of the eight seedlings using the same procedure, except for bacterial suspensions achieving an OD600=0.3. SDW-treated seedlings were also prepared as a control. Two d after the bacterial treatment, the seedlings were individually transplanted to vinyl pots 9 cm in diameter that contained FORL-contaminated soil, which was prepared by mixing sieved and air-dried clay soil and a commercial soil (Engeibaido-1-gou; Nihon Hiryo, Tokyo, Japan) at a ratio of 3:1, followed by mixing with an appropriate amount of FORL propagules in a soil–wheat bran medium, to reach a final concentration of 1.0×105 CFU g−1 soil. After transplanting, the pots were placed in a greenhouse for 4 weeks at 23°C, and the degree of root rot on each potted tomato plant was then evaluated. The DS was calculated using the same procedures as those in the seedling experiment. The calculation was performed using eight replicated seedlings and the experiment was repeated three times. Results were subjected to ANOVA followed by Fisher’s LSD test as described above. To estimate the FORL density in the root tissues of the potted plants, 1 g (fresh weight) of the roots of a representative potted plant, with an average severity of root rot among the eight plants treated with each bacterial strain, was excised from the middle of the roots using sterilized scissors. The roots were surface-sterilized in 70% ethanol for 1 min and then rinsed twice with 10 mM potassium phosphate buffer (pH 7.4). Roots obtained from uninoculated healthy plants were prepared in the same way to act as negative controls. The roots were subsequently ground in a mortar with 10 mL of the potassium phosphate buffer, diluted appropriately, and spread on agar plates of Komada’s selective medium (23). This medium specifically detects nonpathogenic and pathogenic F. oxysporum, including FORL and other formae specialists. After 10 d of incubation at 25°C, the number of presumable FORL colonies on each plate was counted and the FORL density (CFU g−1 roots) was calculated based on these counts. The FORL recovery experiment was repeated three times. Results were analyzed by ANOVA followed by Fisher’s LSD test as described above.
Field experiment
A field experiment on biological control for FCRR was carried out in a plastic greenhouse at the experimental farm of Hyogo Prefectural Technology Center for Agriculture, Forestry and Fisheries, Japan in 2013. A FORL-contaminated field was prepared by mixing the soil and FORL propagules grown on barley grain medium at a ratio of 24 kg dry weight are−1 (20), and a chemical fertilizer S604 (160 g N, 100 g P, 140 g K kg−1; JCAM AGRI, Tokyo, Japan) was also applied to the field soil at the ratio of 4 kg are−1. The tomato seedlings were individually grown in vinyl pots 9 cm in diameter that contained Sumi soil N-100 (0.1 g N, 0.25 g P, 0.1 g K l−1; Sumika Agrotech, Osaka, Japan) and grown for 8 weeks. Thirty milliliters of the bacterial suspension of the strains 12HD2 and 42NP7, prepared as described above, were poured into the soil of each plant. SDW-treated plants were also prepared as a control. Five d after the treatments, the plants were transplanted in mid-February 2013 in two rows with 30 cm between plants in a row and with 120 cm of a ridge width. Each plot (2 m2) included 9 plants and 6 replicated plots per treatment were systematically distributed in the field. After transplanting, the plants were cultivated in the field for 18 weeks, and the degree of root rot on each placed tomato plant was then evaluated. The DS in each plot was calculated using the same procedures as in the seedling experiment. Results were subjected to ANOVA followed by Fisher’s LSD test as described above.
The suppressive effect of bacterial cells and culture filtrate of strains 12HD2 and 42NP7
Bacterial strains 12HD2 and 42NP7 were shaken in R2A, a cell suspension of each strain was prepared by the centrifugation method described above, and a culture filtrate of each strain following centrifugation was obtained. The cell suspension and culture filtrate of the strains were individually applied to the soils of eight seedlings in a cell tray. Five milliliters of SDW and R2A were also applied to the soils of another eight seedlings, as controls for the cell suspension and culture filtrate, respectively. FORL inoculation and the subsequent DS calculations were based on the degree of root rot and performed using the same procedures as those used in the seedling experiment. The experiment was repeated three times, and results were analyzed by ANOVA followed by Fisher’s LSD test.
In vitro antifungal activity
The direct antifungal activities of the 12HD2 and 42NP7 strains were evaluated using two nutrient media, PDA or 1.5% agar-supplied R2A (AR2A) plates containing FORL budcells. One ml of the FORL For6-3 budcell suspension (1×107 mL−1) was mixed with 10 mL of PDA or AR2A at 55°C, and the mixture was immediately poured into a petri dish with a diameter of 9 cm. After solidification, paper discs (8 mm diameter) immersed with each cell suspension of the 12HD2 and 42NP7 strains were placed onto the plates. Paper discs immersed with a cell suspension of Escherichia coli DH5α grown on Luria–Bertani (LB) medium and iturin-producing Bacillus amyloliquefaciens RC-2 (48) grown on R2A were also placed onto the plates as negative and positive controls, respectively. Antifungal activity was determined by the appearance of growth inhibition zones around the paper disks after incubation for 3 d (PDA plate) or 10 d (AR2A plate) at 28°C in the dark.
Scanning electron microscopy
To obtain semi-aseptic tomato seedlings, seeds of cv. House-momotaro were surface-sterilized by immersion in 70% ethanol for 1 min and 5% antiformin for 5 min, followed by rinsing twice with SDW. After eliminating the extra water on the sterilized filter paper, the sterilized seeds were sown in a cell tray containing commercial soil (Ryousaibaido Pp) that had been autoclaved twice for 20 min at 121°C and placed in a growth chamber for 2 weeks at 23°C under fluorescent light (340 μmol m−2 s−1) with a photoperiod of 14 h (light)/10 h (dark) to grow seedlings. A cell suspension (5 mL) of 12HD2 and 42NP7 prepared as described above and 5 mL SDW (as a control) were each applied to each soil of the seedlings. After incubation in the growth chamber for 4 d, the soil was carefully dug away from the roots of the seedlings and the roots were washed by shaking thoroughly in SDW. The roots were immediately cut out, divided into small segments, and fixed in 2% (v/v) glutaraldehyde in 0.1 M phosphate buffer (pH 7.4) for 1 d at 4°C. The fixed samples were then passed through increasing concentrations of ethanol (30%, 50%, 70%, 90%, 95%, and 100% [v/v]). The dehydrated samples were subsequently dried, spattered with gold–palladium, and observed with a scanning electron microscope (SEM) (JSM- 5610LV; JEOL Tokyo, Japan) using a previously reported procedure (50).
Enumeration of strain 42NP7 on roots of tomato seedlings
Strain 42NP7rk, a spontaneous mutant of strain 42NP7, which is resistant to both rifampicin and kanamycin, was obtained on AR2A containing both 50 μg mL−1 antibiotics and used for the experiment. No significant differences were observed in the growth rates or nutrient availability between the mutant strain and wild type (data not shown). Although attempts were made to generate mutants of strain 12HD2 using several similar procedures, no mutated strains were obtained. Strain 42NP7rk was cultured on AR2A plates containing 50 μg mL−1 each of rifampicin and kanamycin for 4 d at 28°C in the dark, then cells on the plates were harvested and suspended in SDW after washing by centrifugation at 6000×g for 10 min. After the concentration of the cell suspension was adjusted to 6.7×107 CFU mL−1, aliquots (5 mL) of the cell suspension were inoculated into the soil of each tomato seedling grown in the cell tray, using the same procedures for the above seedling experiment as described above. Seedlings that were not inoculated were also prepared as controls. The inoculated seedlings were incubated for 0 (i.e. immediately after the inoculation), 3, 7, 14, 21, or 28 d in the greenhouse at 23°C, were then carefully plucked from the cell tray, and the surface of the roots were thoroughly washed in tapped distilled water using a sterilized artist brush to remove the attached small soil particles. After extra water was absorbed using a paper towel, the washed roots (fresh weight: 0.14–0.52 g) of each seedling were cut into small segments using a scissors and ground in a mortar with 10 mM phosphate buffer (pH 7.0) (5 mL per g of root segments). Appropriate dilutions of the obtained macerates were spread onto AR2A plates containing 50 μg mL−1 of both antibiotics. Colony counts were carried out 7 d after the incubation of plates at 28°C in the dark. Two seedlings were used for the bacterial enumeration at each recovery.
Antifungal activities of the extracts from roots treated with the bacterial cells of strains 12HD2 and 42NP7
Potted tomato plants treated with each bacterial suspension and transplanted into the 9-cm pot containing FORL-infested soil were prepared, as described above in the potted plant experiment, to ascertain antifungal activity in the tomato roots following the treatment with the bacterial strains 12HD2 and 42NP7. Similarly, potted tomato plants treated with SDW in the infested soils (SDW-treated roots) or those without the treatment and grown in uninfested soils (untreated roots) were also prepared as controls. Four weeks after transplanting, the DS values in the treated and untreated roots were evaluated as described above, the roots were then cut out using sterilized scissors (approximately 5.03–10.12 g fresh weight), transferred into glass petri dishes, and immersed in 50 mL acetone for 30 min at room temperature. After passing through filter paper to remove debris, each acetone extract was air-dried and dissolved with 50% methanol at a concentration of 60 mg mL−1 (dry weight). Aliquots (10 μL) of the concentrated extracts were each placed onto a small mycelial block (approximately 1 mm3) of FORL For6-3 cultured on 1.5% agar plates following the procedures used in a previous study (48). Ten microliters of 50% methanol was similarly placed onto another mycelial block as a relative control. To ascertain the activity against a non-tomato pathogen, Colletotrichum dematium S9733, the causal fungus of mulberry anthracnose, was also examined on PDA (Difco Laboratories Inc.) plates. This fungal species was chosen because it was assumed to be more susceptible to antibiotics based on the findings of a previous study (48). After incubation for 1 d with For6-3 and 2 d with S9733 at 25°C, the diameter of the mycelial colony that developed in each mycelial block was measured and four replicate measurements were averaged. The antifungal activity of each extract was evaluated based on the relative averaged diameter of the mycelial colony treated with the relative control on each fungal strain, and the results were analyzed by ANOVA followed by Fisher’s LSD test.
RT-PCR analysis
To isolate total RNA from tomato seedlings, the seeds of cv. House-momotaro were sown in plastic pots (7 cm in diameter) containing 110 g quartz sand, placed in the growth chamber for 18 d under the same conditions as described above, and fertilized with 2000-fold diluted Hyponex solution (Hyponex Japan, Osaka, Japan) at 1 d intervals. Ten milliliters of each cell suspension (OD600=0.3) of the 12HD2 and 42NP7 strains prepared according to the above procedures was then poured into the sand of each tomato seedling. Cell suspensions of E. coli DH5α prepared from the LB culture were also applied to other seedlings as a negative control. The tomato seedlings were thoroughly pulled out 2 d after the bacterial treatment, and total RNA was immediately isolated from a 0.1 g segment from the roots at 15–20 mm under the surface of the sands and a second leaf of the individual seedlings using an ISOGEN kit (Nippon gene, Tokyo, Japan) according to the instruction manual. First-strand cDNA was synthesized from total RNA using a PrimeScript RT-PCR Kit (Takara, Ohtsu, Japan) according to the instruction manual. First-strand cDNA (the equivalent of 20 ng reverse-transcribed RNA) was added to 20 μL of the PCR reaction mixture (Ex Taq; Takara) with 0.2 μM of each primer plus a negative control (non-reverse-transcripted RNA). The reaction was run with the following program: 23–30 cycles at 95°C for 20 s, 60°C for 30 s, and 72°C for 30 s. The expression levels of the following defense-related genes were monitored: PR-1 (42) and PR-5 (33) (encoding an acidic antifungal protein and an acidic thaumatin-like protein, respectively) are salicylic acid (SA)-responsive marker genes, and PR-3 (5) and PR-6 (12) (encoding a basic chitinase and basic proteinase-inhibitor, respectively) are jasmonic acid/ethylene (JA/ET)-responsive maker genes. The internal standard actin gene Act was also monitored. The gene-specific primers used in this study are listed in Table 2.
Table 2.
Primers used for RT-PCR
| Name | Sequence (5′→3′) | Target gene |
|---|---|---|
| LePRP6f | TGTCCGAGAGGCCAAGCTAT | PR-1 |
| LePRP6r | AGGACGTTGTCCGATCCAGT | PR-1 |
| LeChi9f | GACCATACGCATGGGGTTAC | PR-3 |
| LeChi9r | CTCCAATGGCTCTTCCACAT | PR-3 |
| LePRPA5f | AGGTCCATGTGGCCCTACTG | PR-5 |
| LePRPA5r | TCACTTGAGGGCATCTCCAA( | PR-5 |
| LeCEVI57Gf | TGTACGACGTGTTGCACTGG | PR-6 |
| LeCEVI57Gr | TGCAACCCTCTCCTGCACTA | PR-6 |
| LeActinF | GCCCCACCTGAGAGGAAGTA | Act |
| LeActinR | AGGGAGCTGCTCTGGAAATG | Act |
Results
Seedling experiments
A screening was performed for suppressive strains against FCRR. Although the reductions observed in the disease severity were slightly more with all strains than with the control, only two strains, 12HD2 and 42NP7, significantly (P<0.05) suppressed the disease. Their disease severity (DS) values were 27.5 and 22.5, respectively, while that of the control was 46.3 (Table 1). The protective values (PVs) of 12HD2 and 42NP7 corresponded to 40.6 and 51.4, respectively. Based on similarities in the 16S rRNA gene sequences, 12HD2 and 42NP7 were found to be the most closely related to Paenibacillus alginolyticus DSM5050 (98.6% homology) and Paenibacillus favisporus GMP01 (99.7% homology), respectively.
Potted plant experiment
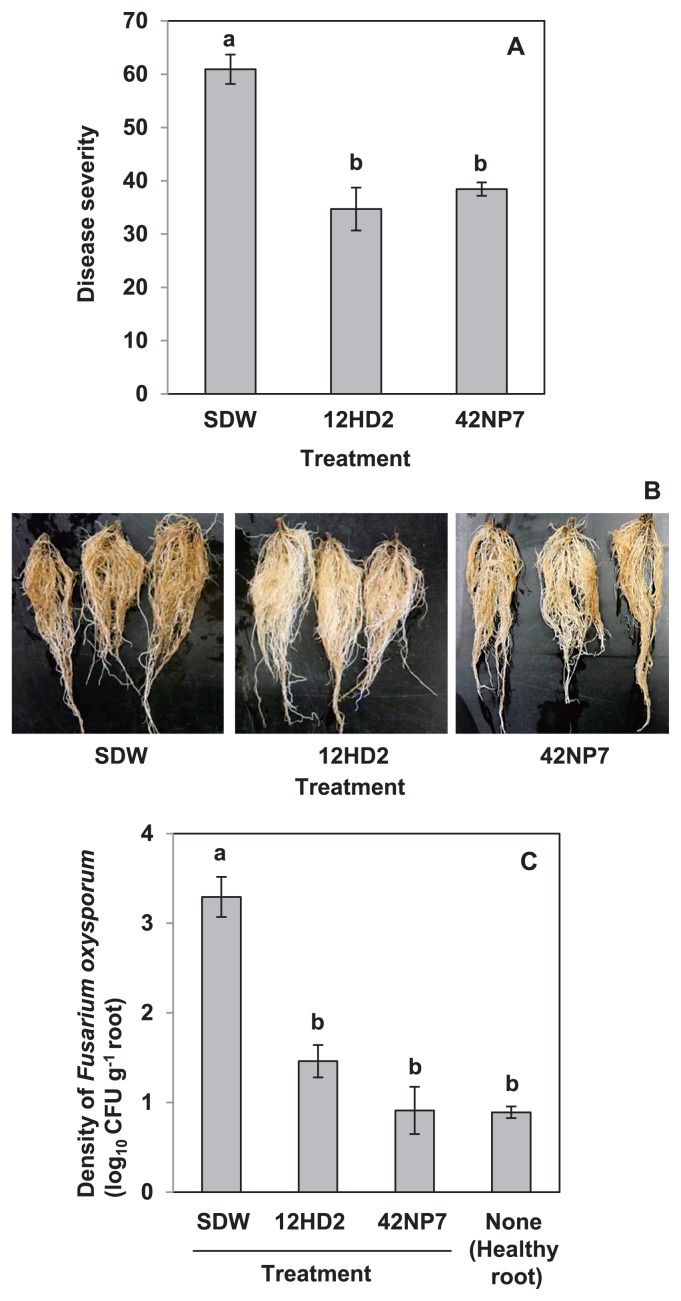
Because the 12HD2 and 42NP7 strains were selected as positive for the suppression of FCRR in the seedling experiment, they were examined further in the potted plant experiment. Consequently, the DS values of roots treated with 12HD2 and 42NP7 were 34.7 and 38.4, respectively, and were significantly (P<0.05) lower than that of the sterilized distilled water (SDW)-treated control roots (DS = 60.9; Fig. 1A), with a reduction being observed in the typical browning symptoms (Fig. 1B). When the density of total F. oxysporum in these roots was calculated using Komada’s selective medium, the densities of roots treated with both 12HD2 and 42NP7 were significantly (P<0.05) lower (less than one 80th) than those of SDW-treated roots (Fig. 1C). Although the medium used could detect not only pathogenic, but also nonpathogenic strains, the densities of the fungal species isolated from uninoculated healthy roots (negative control), most of which were assumed to be nonpathogenic or species other than FORL, were approximately 10 CFU g−1 roots (Fig. 1C). This indicated that the majority of the colonies isolated from each bacterial strain- or SDW-treated root were FORL.
Fig. 1.
Suppressive effects against Fusarium crown and root rot by Paenibacillus strains 12HD2 and 42NP7 in a potted plant experiment. (A) Disease severity caused by Fusarium oxysporum f. sp. radicis-lycopersici in the roots of tomato seedlings treated with each strain. Each value indicates the mean of three experiments and the bar denotes the standard error of the mean. Different letters within each column indicate significant differences (P<0.05) according to Fisher’s LSD test. (B) Representatives of the washed roots of seedlings treated with SDW (left) and a cell suspension of 12HD2 (right). Note that rot symptoms were suppressed by the treatment with a cell suspension of 12HD2. (C) Density (CFU g−1 of root) of Fusarium oxysporum isolated from the roots of potted tomato seedlings treated with each strain. The fungal species were also isolated from the healthy roots of uninoculated seedlings as a negative control (Healthy roots). Each value indicates the mean of three experiments and the bar denotes the standard error of the mean. Different letters within each column indicate significant differences (P<0.05) according to Fisher’s LSD test.
Field experiment
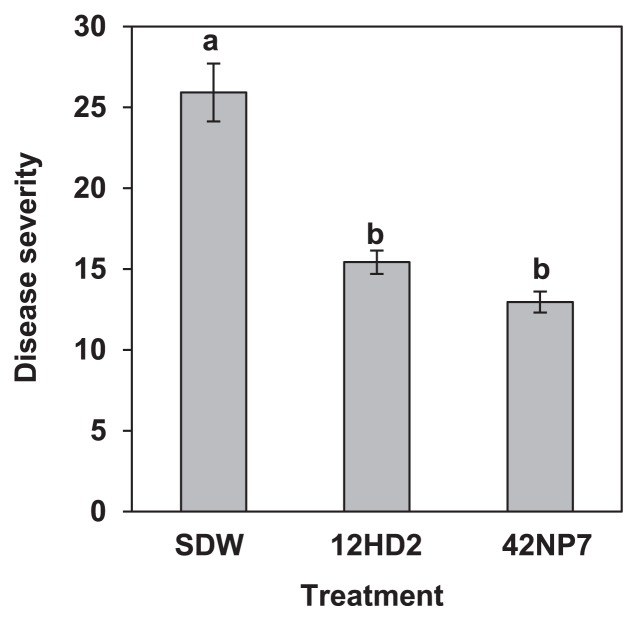
The suppressive strains against FCRR in the potted plant experiment, 12HD2 and 42NP7, were further examined in the field experiment. Consequently, the DS values on the roots treated with 12HD2 and 42NP7 were 15.4 and 12.9, respectively, and were significantly (P<0.05) lower than that of the SDW-treated control roots (DS=25.9; Fig. 2). PVs by the treatments with 12HD2 and 42NP7 were 40.4 and 50.0, respectively.
Fig. 2.
Suppressive effects against Fusarium crown and root rot by Paenibacillus strains 12HD2 and 42NP7 in a field experiment. Each value indicates the mean of six experiments and the bar denotes the standard error of the mean. Different letters within each column indicate significant differences (P<0.05) according to Fisher’s LSD test.
Disease suppressive effects by the bacterial cells and culture filtrate of strains 12HD2 and 42NP7
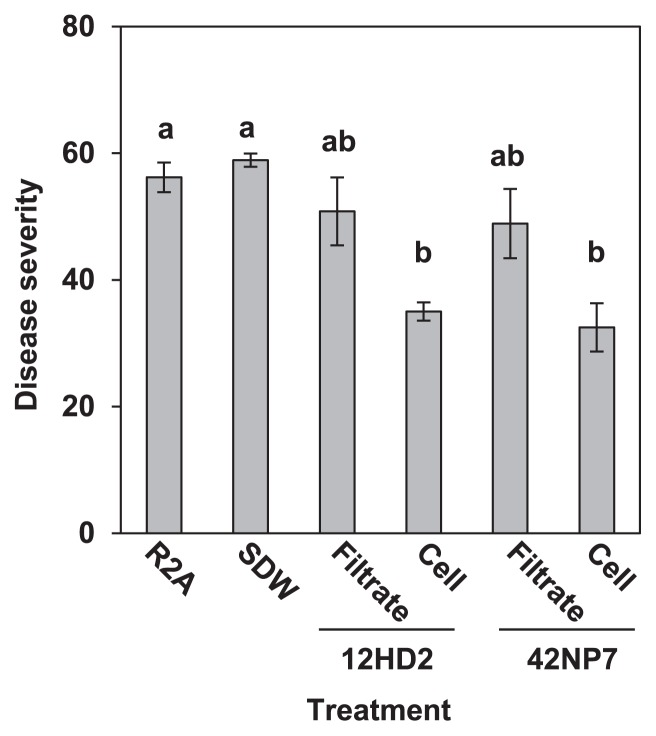
Fig. 3 shows that the DS values on the roots treated with either cell suspension of 12HD2 or 42NP7 were significantly (P<0.05) lower than those with the control treatment with SDW, whereas DS values on the roots treated with the culture filtrates of either strain were not significantly different from the control value (R2A treatment).
Fig. 3.
Disease severity caused by Fusarium oxysporum f. sp. radicis-lycopersici in the roots of potted tomato seedlings treated with bacterial cells and a culture filtrate of Paenibacillus strains 12HD2 and 42NP7. Each value indicates the mean of three experiments and bars denote the standard error of the mean. Different letters within each column indicate significant differences (P<0.05) according to Fisher’s LSD test.
In vitro antifungal activity
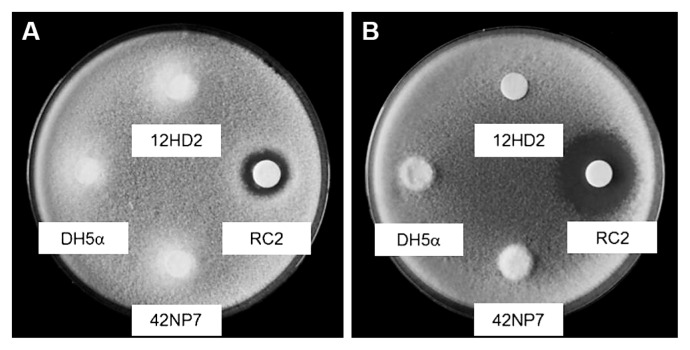
Filter disks containing a cell suspension of each Paenibacillus strain, 12HD2 and 42NP7, formed no clear inhibitory zones around the disks on PDA or AR2A plates containing FORL (Fig. 4). Only suspensions of B. amyloliquefaciens RC-2, as a positive control, formed clear zones on both plates.
Fig. 4.
In vitro antifungal activities of the Paenibacillus strains 12HD2 and 42NP7 against Fusarium oxysporum f. sp. radicis-lycopersici on PDA (A) and 1.5% agar-supplied R2A (B) plates. Paper discs immersed with each cell suspension of strains 12HD2, 42NP7, Escherichia coli DH5α (negative control), and iturin-producing Bacillus amyloliquefaciens RC-2 (positive control) were placed on each agar plate containing budcells of Fusarium oxysporum f. sp. radicis-lycopersici. Observations were made 3 (A) and 10 (B) d after the incubation at 28°C. Note only the positive control formed clear zones, which indicated antifungal activity in both plates.
Scanning electron microscopy
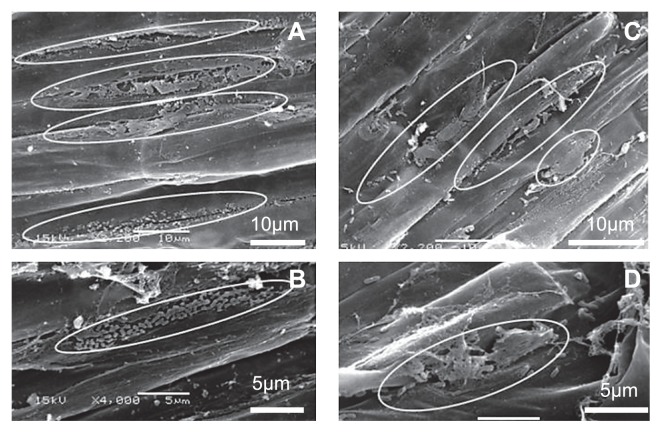
Fig. 5 shows scanning electron microscopic images of tomato roots treated with a cell suspension of Paenibacillus strains 12HD2 (Fig. 5A, B) and 42NP7 (Fig. 5C, D). Cells of both strains colonized the surfaces, particularly at the interspaces between epidermal cells. Cells were often viewed as assemblages or clusters at the interspaces in 12HD2 cell-inoculated roots, and appeared to be embedded in an outer layer of the roots (the circles in Fig. 5A, B). 42NP7 cells also formed clusters at the interspaces (Fig. 5C). They were also observed at other locations on the root surface, presumably utilizing extracellular compounds (Fig. 5D). Similar bacterial cells and structures were not observed on the surface of the control roots.
Fig. 5.
Scanning electron microscopic images of the roots of semi-aseptic tomato seedlings treated with the cells of Paenibacillus strains 12HD2 (A, B) and 42NP7 (C, D). The circles in A–D indicate assemblages or clusters of cells at the interspaces between epidermal cells.
Enumeration of colonized strain 42NP7 on roots of tomato seedlings
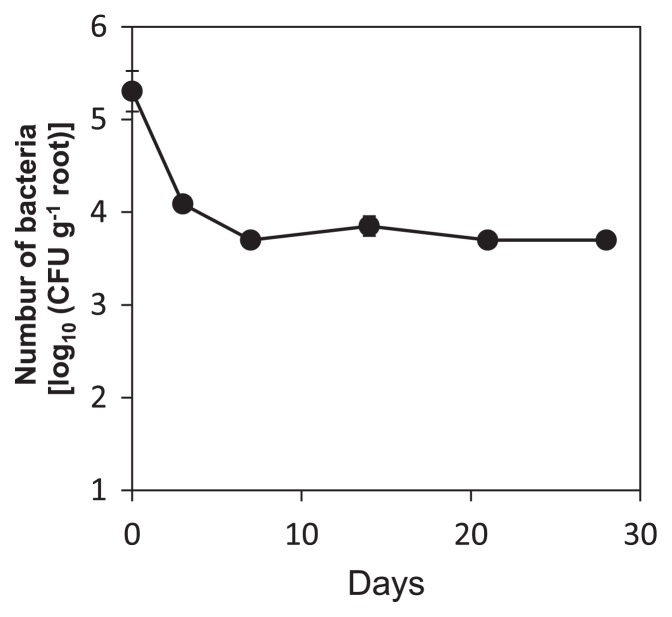
Fig. 6 shows the dynamics of the bacterial density of strain 42NP7rk, an antibiotic-resistant mutant of strain 42NP7, inoculated to the roots of tomato seedlings. The initial density of the inoculated bacterial strain was 2.5×105 CFU g−1 roots. Although the density in the root macerates rapidly decreased by approximately 1.2×104 CFU g−1 roots until 3 d after the inoculation, the bacterial density afterwards was nearly flat, at least until 28 d after the inoculation. No bacterial colonies appeared from the macerates of non-inoculated control roots on the plates.
Fig. 6.
Population dynamics of 42NP7rk in the roots of the tomato after the inoculation. The cell suspension of strain 42NP7rk, the antibiotic-resistant strain of 42NP7, was inoculated into the roots of tomato seedlings and the CFU was determined. Each value indicates the mean of three experiments and bars denote the standard error of the mean.
Antifungal activities of extracts from roots treated with bacterial cells of strains 12HD2 and 42NP7
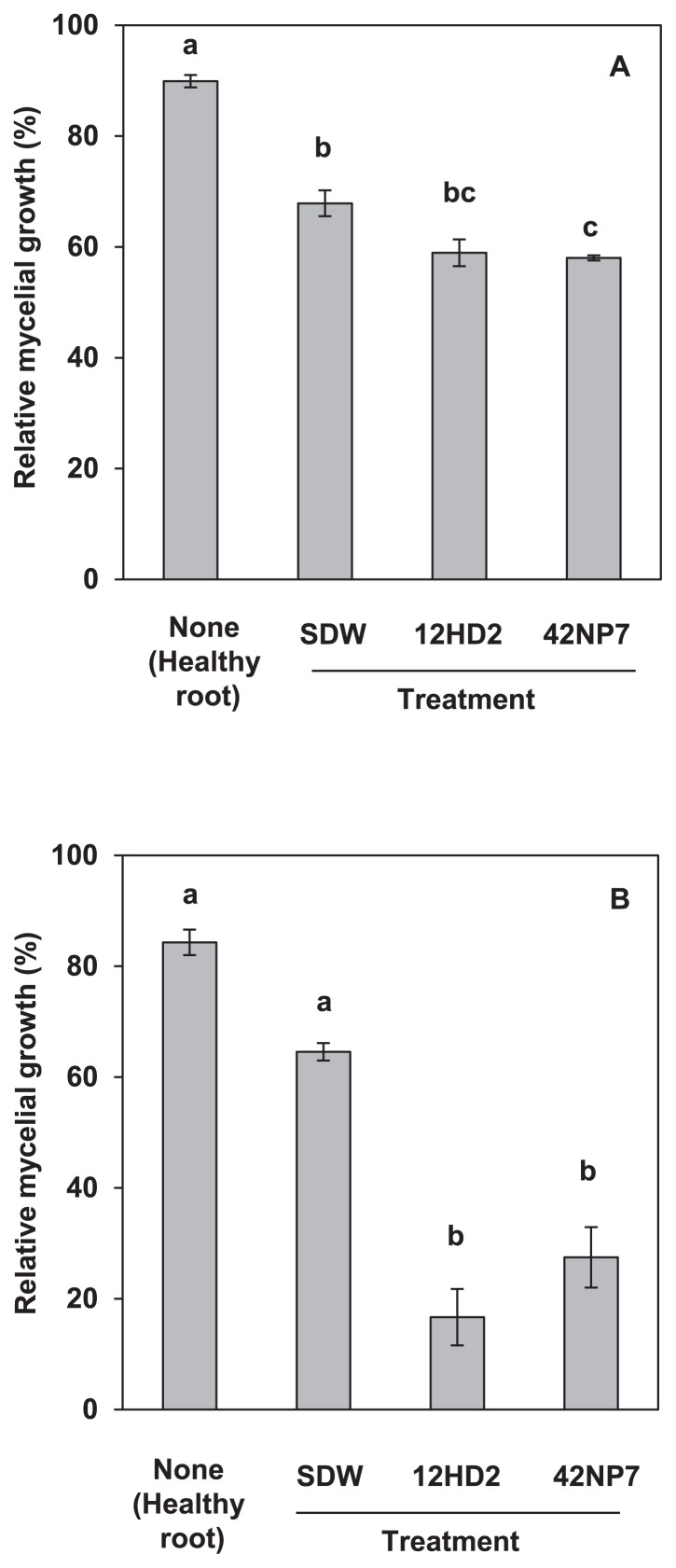
Fig. 7 shows the relative mycelial growth of FORL and C. dematium treated with acetone extracts from tomato roots with or without the bacterial treatment. DS values on the plant roots treated with Paenibacillus strains 12HD2 and 42NP7 were 48.6 and 37.8, respectively, while that of the SDW-treated roots was 68.3. Under such conditions, inhibition of the mycelial growth of FORL and C. dematium, based on the dropping assay, was slightly stronger in extracts obtained from roots treated with both Paenibacillus strains. Against FORL, the colony diameters of the extracts from untreated roots and SDW-treated roots were 89.9 and 67.9% that of the control (50% methanol), respectively, and the extract obtained from 42NP7-treated roots significantly (P<0.05) more reduced the diameter comparing to them (Fig. 7A), although the relative mycelial growth by that of 12HD2- treated roots was not significantly different. Similarly, the inhibitory effects of the extracts from both Paenibacillus strain-treated roots against C. dematium were 16.7 and 26.5% in the colony diameter of the relative control and the inhibition of growth was significantly stronger than that from untreated roots and SDW-treated roots (Fig. 7B).
Fig. 7.
Antifungal activity in acetone extracts from the roots of tomato seedlings treated with bacterial cells of Paenibacillus strains 12HD2 and 42NP7 against FORL (A) and an indicator phytopathogenic fungus, Colletotrichum dematium (B). Antifungal activity was evaluated based on the relative diameter of mycelial growth from a small mycelial block of the fungus relative to growth following a treatment with a control solution (50% methanol). Each value indicates the mean of three experiments and bars denote the standard error of the mean. Different letters within each column indicate significant differences according to Fisher’s LSD test.
RT-PCR analysis
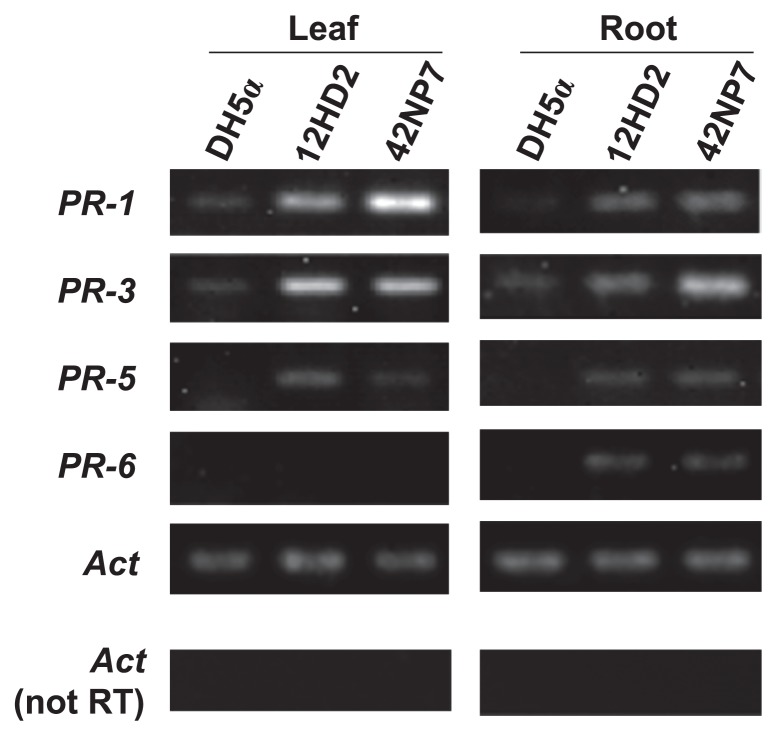
The expression of defense-related genes in tomato plants treated with each cell suspension of the Paenibacillus strains 12HD2 and 42NP7 is shown in Fig. 8. The expression levels of all the targeted genes were stronger on the roots following the treatment with both strains than on roots treated with DH5α as a control, and a clearer signal band on PR-3 appeared following the treatment with 42NP7. The expression of PR-1, PR-3, and PR-5, but not PR-6, was also observed in leaf samples following the treatment with both strains.
Fig. 8.
Expression of defense-related genes in the leaves and roots of tomato seedlings treated with the cells of Paenibacillus strains 12HD2 and 42NP7. SA-responsive (PR-1, PR-5) and JA/ET-responsive (PR-3, PR-6) genes were used as representative markers in RT-PCR. The expression of genes in the seedlings treated with Escherichia coli DH5α are displayed as a control. The actin gene, Act, was used as an internal standard for expression. Act (not RT) represents the amplification of Act using nonreverse-transcripted RNA as a negative control.
Discussion
Of 20 strains obtained from the tomato phyllosphere in the seedling experiment, two Paenibacillus strains, 12HD2 and 42NP7, were confirmed to significantly suppress FCRR. This result indicated that the phyllosphere harbors bacterial strains that have the potential to suppress soilborne FCRR. The suppressive activities of the selected strains were ascertained in potted and field experiments, and the population density of F. oxysporum (mostly FORL) was reduced to less than 80% of the control; therefore, these strains can be considered to have potential as biological control agents (BCAs) against the disease. In previous studies on biological control using Paenibacillus bacteria, P. polymyxa was shown to suppress Ralstonia wilt of tomato (1), crown rot of peanut (13), phytophthora blight of pepper plant (21), and the oomycete plant pathogens Phytophthora palmivora and Pythium aphanidermatum in an Arabidopsis thaliana model system (39). In addition, P. alvae was found to protect the eggplant, tomato, and potato against Verticillium dahliae (40). To the best of our knowledge, this is the first study to demonstrate the suppressive potential of bacterial strains belonging to Paenibacillus against FCRR. Additionally, these two candidate strains, 12HD2 and 42NP7, were taxonomically different from P. polymyxa and P. alvae, but were closely related to P. alginolyticus and P. favisporus, respectively, based on similarities in their 16S rRNA gene sequences. P. alginolyticus was originally obtained from soil (29, 36). Although P. favisporus has been obtained from the rhizosphere (14), this species was originally isolated from aged cow dung (43). These findings imply that both bacterial species have the potential to colonize both the phyllosphere and soil.
The suppressive activities of the selected Paenibacillus strains against the disease were found to occur following the application of the washed cell suspension, but not the culture filtrate (Fig. 3). In addition, neither strain displayed direct antifungal activity against FORL on both PDA and AR2A plates, which consisted of different ingredients (Fig. 4). These results suggested that both disease suppression and reductions in the FORL population in the root tissue following the treatment with these two strains were not based on the secretion of antifungal compounds. Although Saidi et al. (35) reported two Bacillus strains as BCA candidates against FCRR and showed antifungal activity against FORL, the causal factor of the suppressive activities of the Paenibacillus strains in this study appeared to differ between the Bacillus strains.
SEM observations revealed that the colonization of bacterial cells occurred on the surface of roots treated with cells of each Paenibacillus strain, with assemblages or clusters being formed particularly in the interspaces of epidermal root cells (Fig. 5). In addition, 42NP7 cells were detected in root tissue inoculated with the strain for 28 d (Fig. 6). These results suggested that suppressive activity may occur from the root colonization of these strains. The importance of the colonization of roots for the biological control of soilborne diseases has been well documented, particularly for several Pseudomonas strains (45). Regarding Paenibacillus, Haggag and Timmusk (13) reported that P. polymyxa colonized and formed biofilms on peanut roots, and suppressed crown root rot disease caused by Aspergillus niger. Similar associations between bacterial colonization and potential biocontrol activity were demonstrated in the present study.
Acetone extracts from both Paenibacillus strain-treated tomato roots exhibited stronger antifungal activities against FORL and the indicator fungus, C. dematium than those obtained from healthy roots (Fig. 7), although the activity of the extract obtained from 12HD2 treated-roots against FORL was not significantly different from that of the SDW-treated roots. Since antibiotic activity by the bacteria was absent in vitro, colonized bacterial cells may have induced the accumulation of antifungal compounds in the roots of the plant, leading to the suppression of the disease. We speculated that the main antifungal compound of the extracts may be a-tomatine, a tomato phytoanticipin, based on our preliminary experiments. Although antifungal activity was weaker against FORL than C. dematium, this may have been due to the secretion of tomatinase, an enzyme for the degradation of a-tomatine by FORL (19), while the production of enzymes by C. dematium has not yet been reported. Further investigations are needed to identify the antifungal compounds accumulated in the roots.
Together with the significant increase observed in the antifungal activity in the extracts obtained from Paenibacillus-treated roots, the colonization of these bacterial cells on roots may trigger the accumulation of antifungal compounds in plants as a defense response mechanism in the roots, leading to the suppression of FCRR. Furthermore, we analyzed the expression of defense-related genes in tomato plants by root treatments with each cell suspension of the two strains (Fig. 8), and found that the expression of SA-responsive genes (PR-1 and PR-5) and JA/ET-responsive genes (PR-3 and PR-6) were induced in the root tissue. Thus, both strains have the potential to accumulate defense-related proteins in the treated-root tissues, in addition to the accumulation of antifungal compounds, as above described, and may additively or synergistically play a suppressive role against the disease. The SA defense pathway, which plays a major role in the activation of defenses against biotrophic pathogens, and the JA/ET defense pathway, which is more commonly associated with defense against necrotrophic pathogen attacks, have generally been considered to work antagonistically (7, 26). Previous studies on the induction of plant defense pathways by Paenibacillus and Bacillus spp. (4, 18, 30, 34, 41) revealed that P. alvae strain K165 (41) and B. subtilis strain GB03 (34) activated the SA and ET pathways in Arabidopsis thaliana, respectively. On the other hand, Niu et al. (30) reported that B. cereus AR156 activated both of these pathways simultaneously. Similarly, the co-activation of both pathways following the treatment of the two selected Paenibacillus strains on the tomato roots was suggested in this study. Various compounds, such as volatile organic compounds produced by Paenibacillus and Bacillus spp., have been shown to elicit plant defense pathways (4, 10, 17, 24, 25, 34). In the present study, we determined no compound eliciting defense pathways in the tomato plants. We showed that the cellular fraction, but not the culture filtrate exhibited disease suppression activity, which indicated that a cellular component of the strains or a compound produced by the strains after the colonization on the roots activated the defense pathway. In addition to the enhanced expression of defense-related genes in the root, bacterial treatments induced the expression of SA-responsive genes (PR-1 and PR-5) and the JA/ET-responsive gene (PR-3) in the second leaf tissues of tomato seedlings, which suggested that tomato plants developed systemic resistance due to the action of the Paenibacillus strains. Hence, these strains may suppress not only FCRR, but also several airborne diseases of the tomato, and experiments are ongoing to elucidate this possibility.
Acknowledgements
This work was financially supported by the Ministry of Agriculture, Forestry and Fisheries, Japan, through a research project titled “Development of technologies for mitigation and adaptation to climate change in agriculture, forestry and fisheries.”
We thank M. Imai and S. Takahashi for technical assistance.
References
- 1.Algam SAE, Xie G, Li B, Yu S, Su T, Larsen J. Effects of Paenibacillus strains and chitosan on plant growth promotion and control of Ralstonia wilt in tomato. J Plant Pathol. 2010;92:593–600. [Google Scholar]
- 2.Bolwerk A, Lagopodi AL, Lugtenberg BJJ, Bloemberg GV. Visualization of interactions between a pathogenic and beneficial Fusarium strain during biocontrol of tomato foot and root rot. Mol Plant Microbe Interact. 2005;18:710–721. doi: 10.1094/MPMI-18-0710. [DOI] [PubMed] [Google Scholar]
- 3.Chin-A-Woeng TFC, Bloemberg GV, Mulders IHM, Dekkers LC, Lugtenberg BJJ. Root colonization by phenazine-1- carboxamide-producing bacterium Pseudomonas chlororaphis PCL1391 is essential for biocontrol of tomato foot and root rot. Mol Plant Microbe Interact. 2000;13:1340–1345. doi: 10.1094/MPMI.2000.13.12.1340. [DOI] [PubMed] [Google Scholar]
- 4.Choudhary DK, Johri BN. Interactions of Bacillus spp. and plants—with special reference to induced systemic resistance (ISR) Microbiol Res. 2009;164:493–513. doi: 10.1016/j.micres.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 5.Danhash N, Wagemakers CA, Van Kan JAL, de Wit PJ. Molecular characterization of four chitinase cDNAs obtained from Cladosporium fulvum-infected tomato. Plant Mol Biol. 1993;22:1017–1029. doi: 10.1007/BF00028974. [DOI] [PubMed] [Google Scholar]
- 6.Datnoff LE, Nemec S, Pernezny K. Biological control of Fusarium crown and root rot of tomato in Florida using Trichoderma harzianum and Glomus intraradices. Biol Control. 1995;5:427–431. [Google Scholar]
- 7.Durrant WE, Dong X. Systemic acquired resistance. Annu Rev Phytopathol. 2004;42:185–209. doi: 10.1146/annurev.phyto.42.040803.140421. [DOI] [PubMed] [Google Scholar]
- 8.Enya J, Shinohara H, Yoshida S, Tsukiboshi T, Negishi H, Suyama K, Tsushima S. Culturable leaf-associated bacteria on tomato plants and their potential as biological control agents. Microb Ecol. 2007;53:524–536. doi: 10.1007/s00248-006-9085-1. [DOI] [PubMed] [Google Scholar]
- 9.Enya J, Koitabashi M, Shinohara H, Yoshida S, Tsukiboshi T, Negishi H, Suyama K, Tsushima S. Phylogenetic diversities of dominant culturable Bacillus, Pseudomonas and Pantoea species on tomato leaves and their possibility as biological control agents. J Phytopathol. 2007;155:446–453. [Google Scholar]
- 10.Farag MA, Zhang H, Ryu CM. Dynamic chemical communication between plants and bacteria through airborne signals: induced resistance by bacterial volatiles. J Chem Ecol. 2013;39:1007–1018. doi: 10.1007/s10886-013-0317-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fravel DR. Commercialization and implementation of biocontrol. Annu Rev Phytopathol. 2005;43:337–359. doi: 10.1146/annurev.phyto.43.032904.092924. [DOI] [PubMed] [Google Scholar]
- 12.Gadea J, Mayda ME, Conejero V, Vera P. Characterization of defense-related genes ectopically expressed in viroid-infected tomato plants. Mol Plant Microbe Interact. 1996;9:409–415. doi: 10.1094/mpmi-9-0409. [DOI] [PubMed] [Google Scholar]
- 13.Haggag WM, Timmusk S. Colonization of peanut roots by biofilm-forming Paenibacillus polymyxa initiates biocontrol against crown rot disease. J Appl Microbiol. 2008;104:961–969. doi: 10.1111/j.1365-2672.2007.03611.x. [DOI] [PubMed] [Google Scholar]
- 14.Han J, Xia D, Sun L, Yang K, Zhang L. Diversity of culturable bacteria isolated from root domains of moso bamboo (Phyllostachys edulis) Microb Ecol. 2009;58:363–373. doi: 10.1007/s00248-009-9491-2. [DOI] [PubMed] [Google Scholar]
- 15.Hartman JR, Fletcher JT. Fusarium crown and root rot of tomatoes in the UK. Plant Pathol. 1991;40:85–92. [Google Scholar]
- 16.Horinouchi H, Katsuyama N, Taguchi Y, Hyakumachi M. Control of Fusarium crown and root rot of tomato in a soil system by combination of a plant growth-promoting fungus, Fusarium equiseti, and biodegradable pots. Crop Prot. 2008;27:859–864. [Google Scholar]
- 17.Huang C-J, Tsay JF, Chang SY, Yang HP, Wu WS, Chen CY. Dimethyl disulfide is an induced systemic resistance elicitor produced by Bacillus cereus C1L. Pest Manag Sci. 2012;68:1306–1310. doi: 10.1002/ps.3301. [DOI] [PubMed] [Google Scholar]
- 18.Hyakumachi M, Nishimura M, Arakawa T, Asano S, Yoshida S, Tsushima S, Takahashi H. Bacillus thuringiensis suppresses bacterial wilt disease caused by Ralstonia solanacearum with systemic induction of defense-related gene expression in tomato. Microb Environ. 2013;28:128–134. doi: 10.1264/jsme2.ME12162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ito S, Kawaguchi T, Nagata A, et al. Distribution of the FoToml gene encoding tomatinase in formae speciales of Fusarium oxysporum and identification of a novel tomatinase from F. oxysporum f. sp. radicis-lycopersici, the causal agent of Fusarium crown and root rot of tomato. J Gen Plant Pathol. 2004;70:195–201. [Google Scholar]
- 20.Iwamoto Y, Aino M. Suppressive effect of Pseudomonas fluorescens FPH9601 on crown and root rot disease of tomato (Lycopersicon esculentum) caused by Fusarium oxysporum f. sp. radicis-lycopersici. Soil Microorg. 2008;62:3–8. [Google Scholar]
- 21.Jung WJ, Jin YL, Kim KY, Park RD, Kim TH. Changes in pathogenesis-related proteins in pepper plants with regard to biological control of phytophthora blight with Paenibacillus illinoisensis. BioControl. 2005;50:165–178. [Google Scholar]
- 22.Kamilova F, Validov S, Azarova T, Mulders I, Lugtenberg B. Enrichment for enhanced competitive plant root tip colonizers selects for a new class of biocontrol bacteria. Environ Microbiol. 2005;7:1809–1817. doi: 10.1111/j.1462-2920.2005.00889.x. [DOI] [PubMed] [Google Scholar]
- 23.Komada H. Development of a selective medium for quantitative isolation of Fusarium oxysporum from natural soil. Rev Plant Prot Res. 1975;8:114–125. [Google Scholar]
- 24.Lee B, Farag MA, Park HB, Kloepper JW, Lee SH, Ryu CM. Induced resistance by a long-chain bacterial volatile: elicitation of plant systemic defense by a C13 volatile produced by Paenibacillus polymyxa. PLoS One. 2012;7:e48744. doi: 10.1371/journal.pone.0048744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee SH, Cho YE, Park SH, Balaraju K, Park JW, Lee SW, Park K. An antibiotic fusaricidin: a cyclic depsipeptide from Paenibacillus polymyxa E681 induces systemic resistance against Phytophthora blight of red-pepper. Phytoparasitica. 2013;41:49–58. [Google Scholar]
- 26.Lorenzo O, Solano R. Molecular players regulating the jasmonate signaling network. Curr Opin Plant Biol. 2005;8:532–540. doi: 10.1016/j.pbi.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 27.McSpadden Gardener BB. Ecology of Bacillus and Paenibacillus spp. in agricultural systems. Phytopathology. 2004;94:1252– 1258. doi: 10.1094/PHYTO.2004.94.11.1252. [DOI] [PubMed] [Google Scholar]
- 28.Muslim A, Horinouchi H, Hyakumachi M. Control of Fusarium crown and root rot of tomato with hypovirulent binucleate Rhizoctonia in soil and rock wool systems. Plant Dis. 2003;87:739–747. doi: 10.1094/PDIS.2003.87.6.739. [DOI] [PubMed] [Google Scholar]
- 29.Nakamura LK. Bacillus alginolyticus sp. nov. and Bacillus chondroitinus sp. nov., two alginate degrading species. Int J Syst Bacteriol. 1987;37:284–286. [Google Scholar]
- 30.Niu DD, Liu HX, Jiang CH, Wang YP, Wang QY, Jin HL, Guo JH. The plant growth–promoting rhizobacterium Bacillus cereus AR156 induces systemic resistance in Arabidopsis thaliana by simultaneously activating salicylate- and jasmonate/ethylene-dependent signaling pathways. Mol Plant Microbe Interact. 2011;24:533–542. doi: 10.1094/MPMI-09-10-0213. [DOI] [PubMed] [Google Scholar]
- 31.Omar I, O’Neill TM, Rossall S. Biological control of Fusarium crown and root rot of tomato with antagonistic bacteria and integrated control when combined with the fungicide carbendazim. Plant Pathol. 2006;55:92–99. [Google Scholar]
- 32.Ozbay N, Newman SE. Fusarium crown and root rot of tomato and control methods. Plant Pathol J. 2004;3:9–18. [Google Scholar]
- 33.Rodrigo I, Vera P, Tornero P, Hernandez-Yago J, Conejero V. cDNA cloning of viroid-induced tomato pathogenesis related protein P23 (characterization as a vacuolar antifungal factor) Plant Physiol. 1993;102:939–954. doi: 10.1104/pp.102.3.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ryu CM, Farag MA, Hu CH, Reddy MS, Kloepper JW, Paré PW. Bacterial volatiles induce systemic resistance in Arabidopsis. Plant Physiol. 2004;134:1017–1026. doi: 10.1104/pp.103.026583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saidi N, Kouki S, M’Hiri F, Hajlaoui MR, Mahrouk M, Ouzari H, Jedidi N, Hassen A. Characterization and selection of Bacillus sp. strains, effective biocontrol agents against Fusarium oxysporum f. sp. radicis-lycopersici, the causal agent of Fusarium crown and root rot in tomato. Ann Microbiol. 2009;59:191–198. [Google Scholar]
- 36.Shida O, Takagi H, Kadowaki K, Nakamura LK, Komagata K. Transfer of Bacillus alginolyticus, Bacillus chondroitinus, Bacillus curdlanolyticus, Bacillus glucanolyticus, Bacillus kobensis, and Bacillus thiaminolyticus to the genus Paenibacillus and emended description of the genus Paenibacillus. Int J Syst Bacteriol. 1997;47:289–298. doi: 10.1099/00207713-47-2-289. [DOI] [PubMed] [Google Scholar]
- 37.Shinohara H, Yoshida S, Enya J, Watanabe Y, Tsukiboshi T, Negishi H, Tsushima S. Culturable bacterial communities on leaf sheaths and panicles of rice plants in Japan. Folia Microbiol. 2011;56:505–517. doi: 10.1007/s12223-011-0084-3. [DOI] [PubMed] [Google Scholar]
- 38.Sonoda RM. The occurrence of Fusarium root rot of tomatoes in south Florida. Plant Dis Reptr. 1976;60:271–274. [Google Scholar]
- 39.Timmusk S, Van West P, Gow NAR, Huffstutler RP. Paenibacillus polymyxa antagonizes oomycete plant pathogens Phytophthora palmivora and Pythium aphanidermatum. J Appl Microbiol. 2009;106:1473–1481. doi: 10.1111/j.1365-2672.2009.04123.x. [DOI] [PubMed] [Google Scholar]
- 40.Tjamos EC, Tsitsigiannis DI, Tjamos SE, Antoniou PP, Katinakis P. Selection and screening of endorhizosphere bacteria from solarized soils as biocontrol agents against Verticillium dahliae of solanaceous hosts. Eur J Plant Pathol. 2004;110:35–44. [Google Scholar]
- 41.Tjamos SE, Flemetakis E, Paplomatas EJ, Katinakis P. Induction of resistance to Verticillium dahliae in Arabidopsis thaliana by the biocontrol agent K-165 and pathogenesis-related proteins gene expression. Mol Plant Microbe Interact. 2005;18:555–561. doi: 10.1094/MPMI-18-0555. [DOI] [PubMed] [Google Scholar]
- 42.Van Kan JAL, Joosten MHAJ, Wagemakers CAM, Van Den Berg-Velthuis GCM, De Wit PJGM. Differential accumulation of mRNAs encoding extracellular and intracellular PR proteins in tomato induced by virulent and avirulent races of Cladosporium fulvum. Plant Mol Biol. 1992;20:513–527. doi: 10.1007/BF00040610. [DOI] [PubMed] [Google Scholar]
- 43.Velázquez E, de Miguel T, Poza M, Rivas R, Rosselló-Mora R, Villa TG. Paenibacillus favisporus sp. nov., a xylanolytic bacterium isolated from cow faeces. Int J Syst Evol Microbiol. 2004;54:59–64. doi: 10.1099/ijs.0.02709-0. [DOI] [PubMed] [Google Scholar]
- 44.Weller DM. Biological control of soilborne plant pathogens in the rhizosphere with bacteria. Annu Rev Phytopathol. 1988;26:379–407. [Google Scholar]
- 45.Weller DM. Pseudomonas biocontrol agents of soilborne pathogens: Looking back over 30 years. Phytopathology. 2007;97:250–256. doi: 10.1094/PHYTO-97-2-0250. [DOI] [PubMed] [Google Scholar]
- 46.Whipps JM, Hand P, Pink D, Bending GD. Phyllosphere microbiology with special reference to diversity and plant genotype. J App Microbiol. 2008;105:1744–1755. doi: 10.1111/j.1365-2672.2008.03906.x. [DOI] [PubMed] [Google Scholar]
- 47.Yamamoto I, Komada H, Kuniyasu K, Saito M, Ezuka A. A new race of Fusarium oxysporum f. sp. lycopersici inducing root rot of tomato. Proc Kansai Plant Protect Soc. 1974;16:17–29. [Google Scholar]
- 48.Yoshida S, Hiradate S, Tsukamoto T, Hatakeda K, Shirata A. Antimicrobial activity of culture filtrate of Bacillus amyloliquefaciens RC-2 isolated from mulberry leaves. Phytopathology. 2001;91:181– 187. doi: 10.1094/PHYTO.2001.91.2.181. [DOI] [PubMed] [Google Scholar]
- 49.Yoshida S, Tsukiboshi T, Arie T, Shinohara H, Koitabashi M, Tsushima S. Inhabitancy and colonization on healthy rice plants by Glomerella cingulata. J Phytopathol. 2007;155:38–44. [Google Scholar]
- 50.Yoshida S, Ogawa N, Fujii T, Tsushima S. Enhanced biofilm formation and 3-chlorobenzoate degrading activity by the bacterial consortium of Burkholderia sp. NK8 and Pseudomonas aeruginosa PAO1. J Appl Microbiol. 2009;106:790–800. doi: 10.1111/j.1365-2672.2008.04027.x. [DOI] [PubMed] [Google Scholar]
- 51.Yoshida S, Ohba A, Liang YM, Koitabashi M, Tsushima S. Specificity of Pseudomonas isolates on healthy and Fusarium head blight-infected spikelets of wheat heads. Microb Ecol. 2012;64:214–225. doi: 10.1007/s00248-012-0009-y. [DOI] [PubMed] [Google Scholar]