Abstract
Recent advances in understanding the genetics of enterotoxigenic Clostridium perfringens, including whole genome sequencing of a chromosomal cpe strain and sequencing of several cpe-carrying large plasmids, have led to the development of molecular approaches to more precisely investigate isolates involved in human gastrointestinal diseases and isolates present in the environment. Sequence-based PCR genotyping of the cpe locus (cpe genotyping PCR assays) has provided new information about cpe-positive type A C. perfringens including: 1) Foodborne C. perfringens outbreaks can be caused not only by chromosomal cpe type A strains with extremely heat-resistant spores, but also less commonly by less heat-resistant spore-forming plasmid cpe type A strains; 2) Both chromosomal cpe and plasmid cpe C. perfringens type A strains can be found in retail foods, healthy human feces and the environment, such as in sewage; 3) Most environmental cpe-positive C. perfringens type A strains carry their cpe gene on plasmids. Moreover, recent studies indicated that the cpe loci of type C, D, and E strains differ from the cpe loci of type A strains and from the cpe loci of each other, indicating that the cpe loci of C. perfringens have remarkable diversity. Multi-locus sequence typing (MLST) indicated that the chromosomal cpe strains responsible for most food poisoning cases have distinct genetic characteristics that provide unique biological properties, such as the formation of highly heat-resistant spores. These and future advances should help elucidate the epidemiology of enterotoxigenic C. perfringens and also contribute to the prevention of C. perfringens food poisoning outbreaks and other CPE-associated human diseases.
Keywords: molecular assays, cpe-genotyping assay, MLST
Introduction
Clostridium perfringens is a Gram-positive, rod-shaped, spore-forming, anaerobic bacterium that causes a broad spectrum of human and veterinary diseases (35, 36). The virulence of C. perfringens largely results from its prolific toxin-producing ability (36). Based on the production of four major toxins (alpha, beta, epsilon, and iota), this organism is commonly classified into one of five types (A to E) (35). Some C. perfringens strains produce another important toxin named Clostridium perfringens enterotoxin (CPE), which is responsible for several human gastrointestinal (GI) diseases, including C. perfringens type A food poisoning and many cases of antibiotic-associated diarrhea (AAD), sporadic diarrhea (SD), and nosocomial diarrheal disease (1, 2, 23, 25, 52, 56). Therefore, detection of CPE produced by C. perfringens in feces specimens of ill individuals is a criterion for clinical diagnosis.
CPE production, which is responsible for the diarrhea symptoms of diseases caused by cpe-positive type A strains, is sporulation-associated. Intact cpe genes can also be found in some type C, D and E strains. CPE expression is also sporulation-associated in those type C and D strains and, probably, also in those type E isolates, based upon sequence data indicating the presence of sigE- and sigK-dependent promoters upstream of the cpe gene in those type E strains (10, 16, 18, 32, 43, 50, 59).
Despite the medical importance of enterotoxigenic C. perfringens, the ecology of these bacteria remains poorly understood. In part this is because, while C. perfringens has widespread distribution in the environment, only a small fraction (~1 to 5%) of the global C. perfringens population carries the enterotoxin (cpe) gene (8, 14, 24, 29, 34, 38, 44, 55, 57). However, enterotoxigenic C. perfringens are a suitable target bacterium for microbial source tracking (MST) for identifying contamination processes (21). Recently, the accumulation of genetic information about chromosomal and plasmid cpe type A strains has facilitated the development of molecular methods using MST tools for detecting and identifying enterotoxigenic C. perfringens (41, 42, 45, 48, 58). These molecular methods to detect the cpe gene and to identify the cpe locus represent a useful alternative approach for MST (41, 58). Using recently developed molecular assays, several new findings about enterotoxigenic C. perfringens ecology have been reported (29, 44); therefore, new strategies for preventing human and animal GI diseases caused by enterotoxigenic C. perfringens may be developed in the near future.
Molecular assays for detecting the cpe gene
C. perfringens type A food poisoning usually develops after the ingestion of foods contaminated with large numbers (>106 bacteria g−1) of CPE-positive vegetative cells (36). Those bacteria then sporulate in the intestines and produce CPE. The stool from diseased persons typically contains large numbers (>106 bacteria g−1) of CPE-positive C. perfringens spores (36). To prove C. perfringens as the etiologic agent of an outbreak, serotyping or molecular genotyping assays, such as pulsed-field gel electrophoresis (PFGE) have been developed (23, 33).
However, in some outbreaks, enterotoxigenic C. perfringens can only be isolated from feces of sick individuals and not from any food source, and only low numbers of viable bacteria remain in those feces (this is a particular problem if fecal samples are not collected soon after the onset of diarrhea). To identify the contaminated food in these cases, molecular methods such as conventional PCR, nested PCR, real-time PCR, and other recently developed assays, such as loop-mediated isothermal amplification (LAMP assay) can be useful tools (21). In these assays, ~103 cpe-positive bacterial cells are necessary for detection, while these assays can detect 0.1 to 10 pg of purified bacterial DNA; however, combined with enrichment culture, these assays can detect less than three viable cpe-positive C. perfringens strains (21); therefore, these assays are also helpful to identify how and when enterotoxigenic C. perfringens isolates enter the food supply. The results of future surveys using molecular assays will be useful to fully understand and prevent C. perfringens type A food poisoning outbreaks.
These assays have been applied to isolates from food poisoning outbreaks, diarrheic patients, and other sources. It is often easier to detect the cpe gene with a molecular assay than to detect in vitro CPE production by strains, because the production of CPE is sporulation-associated, i.e., demonstrating in vitro CPE production requires the in vitro sporulation of isolates (3, 18, 59). Unfortunately, sporulation is often difficult to achieve in vitro using sporulation media such as Duncan-Strong medium (36).
Despite the advantages of molecular methods for detecting the presence of the cpe gene when screening many samples, an epidemiological protocol for detecting cpe-positive strains has not been fully established. The reasons are: 1) The presence of cpe-positive C. perfringens in non-outbreak foods and the environment is often at a low frequency of 1 to 5% or less and it is expensive and time consuming to identify cpe-positive C. perfringens from large numbers of isolates; 2) In non-outbreak retail foods, the number of contaminated C. perfringens is usually very low, less than 3 MPN/gram in many samples (38, 57); and 3) To detect cpe-positive C. perfringens with molecular assays, a large number of bacteria, (approximately 103 g−1 sample) are necessary to prepare template DNA using commercial DNA purification kits (21). Because of these difficulties, studies using molecular methods for the detection of cpe-positive C. perfringens in food samples have rarely been published (38, 39).
To overcome these issues, an additional enrichment culture step is usually used before the preparation of DNA template for PCR analysis (21, 38, 46, 57). Adding an enrichment culture step allowed the isolation and detection of enterotoxigenic C. perfringens from retail uncooked food samples (21, 38, 57). In a recent study using molecular methods for detecting cpe-positive C. perfringens, the addition of an enrichment culture step markedly improved the detection efficiency of the cpe gene in retail raw meat samples (21). Moreover, an enrichment culture can reduce known (collagen molecule) and unknown inhibitors present in food samples (21).
Although molecular methods can detect non-viable cpe-positive C. perfringens in tested samples, significant numbers of these bacteria are needed to obtain a PCR-positive sample, i.e., cpe-positive C. perfringens should be propagated in samples before testing. Even using an enrichment culture step, not all cpe-positive C. perfringens might be detected in all samples if many more cpe-negative C. perfringens are present in the sample.
Collectively, many issues remain for detecting cpe-positive C. perfringens in foods and the environment for epidemiological purposes, although repressing food contamination by cpe-positive C. perfringens is important to reduce the occurrence of C. perfringens food poisoning outbreaks.
Genetic diversity of cpe loci amongst type A, C, D and E cpe-carrying C. perfringens
In type A isolates, the cpe gene can reside on the C. perfringens chromosome or on large plasmids (12). Some type C, D, and E, isolates also carry functional cpe genes on large plasmids (32, 43). In most or all CPE-positive type A, C, and D isolates, the cpe gene encodes a 957 bp ORF whose sequence is identical; however, some type E isolates encode a variant of the functional cpe gene, while other type E isolates carry a silent cpe gene (10, 12, 13, 30, 43).
Early studies indicated that classical type A food poisoning C. perfringens isolates carry a chromosomal cpe gene (11, 12). It was also reported that CPE-positive C. perfringens type A isolates causing non-foodborne human diarrhea disease such as AAD and SD carry their cpe gene on a large plasmid (5, 6, 11, 12).
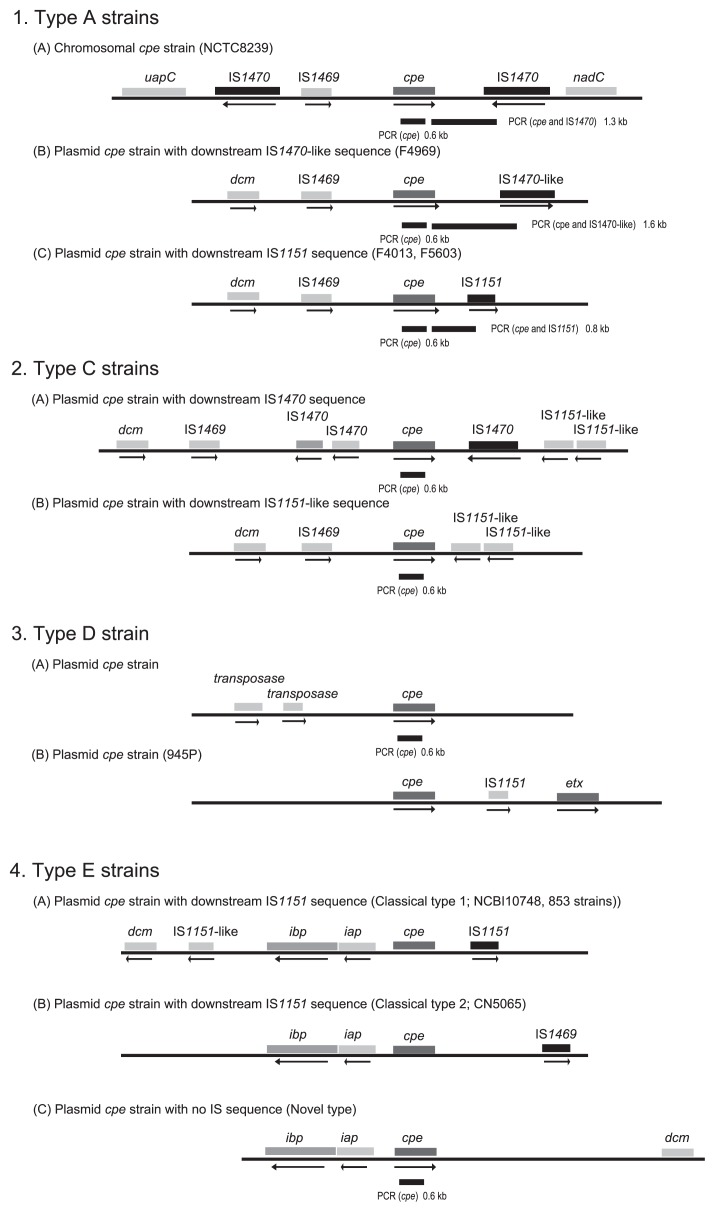
Comparing the organization of the chromosomal cpe locus versus plasmid cpe loci in type A isolates revealed an identical ~3 kb region, which contains an upstream IS1469 element, the cpe gene, and downstream sequences, present in both the plasmid and chromosomal cpe loci (Fig. 1) (7, 40). Beyond this conserved region immediately surrounding the cpe gene, substantial differences were identified between these cpe loci amongst type A isolates. When chromosomally-located, the cpe gene appears to be associated with mobile genetic elements, i.e., the chromosomal cpe gene is present on a putative 6.3 kb transposon, named Tn5565 (7). This putative transposon is flanked by upstream and downstream copies of IS1470 and is apparently inserted between the purine permease (uapC) and quinolinate phosphoribosyltransferase (nadC) genes on the chromosome (Fig. 1) (7).
Fig. 1.
The organization of cpe loci in cpe-positive type A, C, D, and E C. perfringens. In type A strains, comparison of the genetic organization of chromosomal cpe locus (A), and plasmid cpe loci (B, C) revealed three downstream sequences. The bar below the open reading frames depicts the cpe-IS1470 cpe genotyping PCR assay product from chromosomal cpe strains, from cpe-IS1470-like plasmid strains, and from cpe-IS1151 plasmid strains. In type C strains, two types of genetic organization of plasmid cpe loci have been found, i.e., (A) cpe locus with downstream IS1470 and two IS1151-like sequences and (B) the cpe locus with two IS1151-like sequences. In type D strains, the genetic organization of the cpe locus lacks any apparent IS sequence. In type E strains, the genetic organization of (A) iota toxin locus with the disrupted cpe gene and IS1151, (B) iota toxin locus with the disrupted cpe gene and IS1469, and (C) iota toxin locus with functional cpe gene and no apparent IS element. The bar below the open reading frames of the cpe gene depicts the cpe genotyping PCR assay product by internal primers of the cpe gene.
The type A plasmid cpe locus lacks the IS1470 element that is present upstream of the chromosomal cpe gene, instead carrying an upstream cytosine methyltransferase gene (dcm) (Fig. 1) (40, 42). Additionally, the IS1470 element located downstream of the chromosomal cpe locus has been replaced in type A plasmid loci by either a defective IS1470-like element, or by an IS1151 element (40, 42). Moreover, a study determining complete sequence and diversity analysis determined that the two kinds of plasmid cpe genotypes (downstream IS1151 or downstream IS1470-like sequence) share a conserved region, including a replication region and a plasmid conjugative transfer region, but have different variable regions that can encode bacteriocins, metabolic genes or toxin genes, such as the cpb2 gene (42).
Some strains of type C, D, and E C. perfringens also carry the cpe gene (4, 32, 43). The organization of the cpe loci of type C, and D isolates is different from that of type A plasmid cpe strains, while the cpe ORF sequence is identical amongst type A, C, and D isolates (32). Flanking sequences of the cpe gene in type C strains can be divided into two groups with one group carrying an upstream IS1470 sequence and downstream IS1470 and IS1151-like sequences, and the other group sharing a resemblance to the plasmid-borne cpe locus of pCPF5603 carried by type A isolate F5603 (Fig. 1). Both types of the type C cpe locus are located downstream of the cytosine methyltransferase gene (dcm), which is almost always located upstream of the cpe gene in type A strains (25, 32, 40). Overall, the cpe gene has been localized near dcm in those cpe loci where the cpe gene is flanked by various combinations of IS1469, IS1470, IS1470-like, IS1151 or IS1151-like sequences in type A and C C. perfringens isolates. Investigated type D strains carry a unique cpe locus, which is different from that in any other characterized cpe-positive C. perfringens. In these type D isolates, upstream of the cpe gene, there are two copies of the putative transposase gene in Tn154; however, no IS element is found downstream of the cpe gene (Fig. 1) (32).
Amongst type E isolates, two groups of cpe loci have been identified (4, 43). In classical type E isolates, a silent cpe sequence associated with IS1151 and IS1469 is located on the same plasmid adjacent to the iota toxin genes (Fig. 1) (4, 47); however, in some recently-identified type E isolates, an intact cpe gene is next to the iota genes (Fig. 1) (43). Interestingly, the newly identified cpe gene in those the novel type E isolates has several nucleotide differences from the classical cpe gene of type A, C, and D isolates (32, 43). The iota toxin genes in these novel type E isolates also exhibit nucleotide differences from the classical iota toxin genes (43); therefore, this newly identified type E isolate was initially classified as type A by the current PCR-based toxin genotyping assay as the iap primer in this assay could not amplify the variant iap gene present in these type E isolates (17, 37, 51). Moreover, complete sequencing of this toxin plasmid showed that no intact transposase gene and a disrupted dcm gene are present on this putative conjugatively transferable toxin plasmid (43). Collectively, these studies have revealed greater diversity in the cpe gene and the cpe loci in type C, D, and E isolates than those in type A strains.
Usefulness of cpe-genotyping assays
Using the sequence information generated for the cpe locus on the chromosome and plasmids in type A isolates, simple and rapid PCR cpe genotyping assays were developed (41, 58). Because these PCR assays can distinguish between the cpe locus on the chromosome and the two well characterized cpe loci present on the plasmids in type A isolates, these cpe genotyping assays are a useful diagnostic and epidemiological tools for investigating CPE-associated GI disease cases, including food poisoning, AAD, and SD caused by cpe-positive type A strains.
Applying these PCR based cpe-genotype assays to chromosomal or plasmid cpe-positive type A isolates from various sources, several new insights have been reported (34). First, ~30% of C. perfringens food poisoning outbreaks in Japan and Europe appear to be caused by type A plasmid cpe strains (19, 25, 53), with the remainder of C. perfringens food poisoning outbreaks caused by chromosomal cpe strains. Before sequencing analysis and development of sequence-based cpe-genotyping PCR assays, it had been thought that all food poisoning isolates carry cpe on their chromosome, while isolates from AAD and SD cases bear cpe on the plasmids (11, 12). This conclusion was based upon strains isolated from a limited number of food poisoning outbreaks that were investigated using RFLP Southern blotting analysis, an approach that can only be performed in research laboratories with a limited number of isolates. Second, the cpe genotyping assays, which can be easily performed with large numbers of isolates by clinical labs, identified chromosomal cpe-strains in ~1.4% of retail meats in USA (57). In contrast, ~4% of retail meat products in Japan were contaminated with type A plasmid cpe-positive strains, as determined using a PCR assay that can distinguish chromosomal or plasmid cpe strains (38). Also, three meat plasmid-cpe isolates were identified as being the IS1470-like cpe genotype and these isolates also formed relatively heat-labile spores, i.e., D100 value of spores was less than 3 min in an isolate from sporadic diarrhea (38). These results indicated that both chromosomal cpe and plasmid cpe strains can contaminate food and potentially induce food poisoning, although the relative importance of these cpe genotypes for causing food poisoning may vary, perhaps due to cultural differences in food preparation. Third, chromosomal cpe-positive strains are rarely isolated from feces of healthy humans, although those feces do contain type A plasmid cpe strains (8, 19). Therefore, feces of healthy humans might be a reservoir for plasmid cpe-positive isolates and, perhaps less commonly, for chromosomal cpe-positive isolates. Because of their rarity, chromosomal cpe isolates might be found only transiently in healthy human feces. Fourth, at least two unusual variants of the cpe locus have been found amongst type A isolates recovered from foods, human feces and the environment, including sewage (19, 29, 34, 44). At least, some of these isolates with unusual cpe loci, obtained from feces of healthy humans, can produce CPE (19). These findings suggest that there might be several other cpe genotypes in type A isolates but the clinical significance of the variant cpe loci-carrying isolates has not yet been investigated; however, many of these cpe isolates that are untypeable by current genotype assays originated from the environment, such as sewage, and these isolates are rarely found in human feces and foods. These findings suggest a need for further investigation of how and when foods become contaminated with cpe-positive isolates. Use of molecular methods, including the cpe detection PCR method and/or cpe-genotyping PCR assay combined with prior enrichment culture, should facilitate these epidemiological studies (21, 38, 57).
Type C cpe-positive strains have been isolated from feces of patients suffering from enteritis necroticans (Pigbel) (26); however, the involvement, if any, of CPE in this disease is not fully understood because of the rarity of Pigbel and the lack of an established clinical diagnosis protocol. The involvement of type D cpe-positive strains in disease has also not yet been established (35). Use of recently developed cpe-genotyping assays should make it easier to distinguish these cpe-positive type C, D and/or E strains from type A cpe-positive isolates at the clinical stage, which might help the diagnosis and the investigation of cpe-positive type C, D, and E strains.
Multi-locus sequence typing (MLST) of C. perfringens
To epidemiologically link isolates obtained from patients with isolates found in suspected food vehicles, classical serotype assays have been used; however, many cpe-positive food poisoning strains cannot be serotyped using existing antisera. As an alternative, pulsed-field gel electrophoresis (PFGE) has more recently been applied for the epidemiological study of isolates from C. perfringens food poisoning or from nosocomial outbreaks (23, 33). The PFGE method has advantages over serotyping for demonstrating a link between various isolates associated with an outbreak (23, 33). These advantages include high reproducibility, high typeability with substantial discrimination ability (more than 30 types are distinguishable), and applicability to many kinds of bacterial species.
While the PFGE approach can demonstrate a clonal lineage of outbreak strains, it does not reflect the properties based on gene sequence diversity; as a result, data interpretation might be subjective in some cases. In addition, in some C. perfringens strains, bacterial DNA is rapidly degraded partly by internal DNase of the bacterial cell; as a result, DNA fingerprinting shows smearing (22). On the other hand, sequence-based molecular analysis, an approach known as multi-locus sequence typing (MLST), makes it possible to investigate more precise relationships among isolates with respect to disease presentation and/or host preference; the major origin and route of pathogenic strain spread (15, 20, 48). Moreover, MLST is a very good candidate for bacterial genotyping because MLST generates unambiguous nucleotide sequence data and does not have the potential for subjectivity in data interpretation. In addition, this technique has high reproducibility, high typeability and applicability to many bacterial species, similar to PFGE (20, 54); therefore, MLST has been applied to a number of bacterial pathogens, with the subsequent creation of databases, to which new MLST data can be added as it is generated (http://www.mlst.net) (20, 54).
The genes used in MLST analysis for cpe-positive C. perfringens isolates included genes for toxin genes (plc, colA), stress response (groEL, sod), putative metabolic genes (pgk, nadA), genes in DNA replication (gyrB) and genes for a sigma factor involved in sporulation (sigK), which is essential for cpe expression (15). These MLST studies indicated that human food poisoning isolates carrying the cpe gene on the chromosome are clearly related among strains isolated from several food poisoning outbreaks in Europe, United States, and Japan (Fig. 2) (15, 48). Plasmid-cpe isolates from human gastrointestinal diseases such as food-borne diarrhea and nosocomial diarrhea, were also clustered in the MLST assay, indicating that there also appeared to be a genetic relationship among plasmid-cpe isolates in feces of sickened individuals; however, cpe-negative isolates from healthy human feces exhibited considerable genetic diversity in the same MLST analysis (Fig. 2) (15).
Fig. 2.
Phylogenetic relationships among cpe-positive and cpe-negative strains. The phylogenetic tree was constructed by Clustal W analysis based on the concatenated nucleotide sequence of 8 house-keeping genes. # indicates type A chromosomal cpe isolates from foods and food poisoning outbreaks. $ indicates type A plasmid cpe isolates from foods, sporadic diarrhea patients, food-borne outbreak, nosocomial outbreaks, and healthy individual. Isolates with a gray background are cpe-positive. * indicates type B to E animal diseases isolates. @ indicates novel type E isolate in retail meat sample. + indicates cpe-negative healthy human feces isolates.
By definition, toxin types B to E express one or more of beta, epsilon and iota toxins; as a result, these isolates are highly associated with specific diseases and animal hosts (9, 20). MLST approaches can identify host-species relationships with respect to the animal origin of isolates, even for isolates that are not clonal by PFGE profile (20, 48). Consistent with these findings, MLST indicated that type B to E strains from animal diseases was clustered with some diversity, while novel type E isolates from retail meat have a different genetic background from animal isolates (Fig. 2) (15, 43); however, there is no evidence that these type E meat isolates can cause food poisoning.
For C. perfringens isolates from various origins, the superoxide dismutase (sod) gene has been identified as a useful and sensitive PCR target gene for distinguishing chromosomal cpe strains from type A plasmid cpe-positive and other cpe-negative strains. This is because only a single locus sequence of the sod gene has been identified for chromosomal cpe strains (38).
Other common properties of cpe-positive C. perfringens
Type A C. perfringens strains carrying the cpe gene on the chromosome, whether as vegetative cells or spores, usually possess much higher resistance properties against heat, cold, pH, and nitrites than type A strains carrying cpe on a plasmid (6, 27, 28, 31, 49). In addition, chromosomal cpe strains typically grow faster at optimal temperature and have a broader growth temperature range (27, 28). These complex differences in biological properties may reflect broad genetic variations between type A chromosomal cpe isolates and other C. perfringens strains. In fact, a newly identified product (Ssp4) of the ssp4 gene (one of four C. perfringens genes encoding a small acid-soluble protein), was recently reported to be the most important protein for heat and nitrite resistance of spores made by chromosomal cpe strains (31). It was found that type A chromosomal cpe isolates producing resistant spores have a single amino acid substitution in their Ssp4 protein that mediates, in large part, their resistance phenotype (31).
Interestingly, most, if not all, of type A chromosomal cpe strains do not carry the θ toxin gene (pfoA), which indicates that chromosomal cpe strains produce non-hemolytic colonies on a sheep blood agar plate (15). This property is common, but not specific to chromosomal cpe strains, because some strains of type A plasmid cpe-positive and/or cpe-negative strains also do not carry the pfoA gene (15). In some cases, this property might help in the detection and identification of type A chromosomal cpe C. perfringens isolates.
Conclusion
Recent advances in understanding the genetics of type A cpe-positive C. perfringens have facilitated the detection and identification of enterotoxigenic C. perfringens isolates in food-borne outbreaks and outbreaks of nosocomial diarrhea; using molecular assays to identify three types of the cpe loci in type A isolates, several novel insights into diseases caused by enterotoxigenic C. perfringens have been published; however, newly identified issues were also recognized in type A cpe-positive C. perfringens identification. In most isolates obtained from patient specimens, the current cpe-genotyping assays are useful tools; however, a small population of cpe-positive C. perfringens, including variant(s) of the cpe locus, has been found in the environment and these strains do not react appropriately in the current cpe genotyping assays (29, 43). The organization of the cpe locus in C. perfringens type A, C, D, and E has various arrangements (4, 32, 43); therefore, based on the sequence differences of the cpe locus of all types, molecular assays, including the cpe-genotyping assays for type A strains, should distinguish and identify these other cpe locus-carrying isolates. When developed, applying these assays to clinical isolates, human and veterinary GI diseases caused by enterotoxigenic C. perfringens should provide accurate and rapid diagnoses, allowing improved appreciation for the clinical significance of these strains.
References
- 1.Abrahao C, Carman RJ, Hahn H, Liesenfeld O. Similar frequency of detection of Clostridium perfringens enterotoxin and Clostridium difficile toxins in patients with antibiotic-associated diarrhea. Eur J Clin Microbiol Infect Dis. 2001;20:676–677. doi: 10.1007/s100960100571. [DOI] [PubMed] [Google Scholar]
- 2.Asha NJ, Wilcox MH. Laboratory diagnosis of Clostridium perfringens antibiotic-associated diarrhea. J Med Microbiol. 2002;51:891–894. doi: 10.1099/0022-1317-51-10-891. [DOI] [PubMed] [Google Scholar]
- 3.Augustynowicz E, Gzyl A, Slusarczyk J. Detection of enterotoxigenic Clostridium perfringens with a duplex PCR. J Med Microbiol. 2002;51:169–172. doi: 10.1099/0022-1317-51-2-169. [DOI] [PubMed] [Google Scholar]
- 4.Billington SJ, Wieckowski EU, Sarker MR, Bueschel D, Songer JG, McClane BA. Clostridium perfringens type E animal enteritis isolates with highly conserved, silent enterotoxin sequences. Infect Immun. 1998;66:4531–4536. doi: 10.1128/iai.66.9.4531-4536.1998. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 5.Borriello SP, Barclay FE, Welch AR, Stringer MF, Watson GN, Williams RK, Seal DV, Sullens K. Epidemiology of diarrhea caused by enterotoxigenic Clostridium perfringens. J Med Microbiol. 1985;20:363–372. doi: 10.1099/00222615-20-3-363. [DOI] [PubMed] [Google Scholar]
- 6.Borriello SP, Larson HE, Welch AR, Barclay F, Stringer MF, Bartholomew BA. Enterotoxigenic Clostridium perfringens: a possible cause of antibiotic-assocaited diarrhea. Lancet. 1984;1:305–307. doi: 10.1016/s0140-6736(84)90359-3. [DOI] [PubMed] [Google Scholar]
- 7.Brynestad S, Synstad B, Granum PE. The Clostridium perfringens enterotoxin gene is on a transposable element in type A human food poisoning strains. Microbiology. 1997;143:2109–2115. doi: 10.1099/00221287-143-7-2109. [DOI] [PubMed] [Google Scholar]
- 8.Carman RJ, Sayeed S, Li J, Genheimer CW, Hiltonsmith MF, Wilkins TD, McClane BA. Clostridium perfringens toxin genotypes in the feces of healthy North Americans. Anaerobe. 2008;14:102–108. doi: 10.1016/j.anaerobe.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chalmers G, Bruce HL, Hunter DB, Parreira VR, Kulkarni RR, Jiang YF, Prescott JF, Boerlin P. Multilocus sequence typing analysis of Clostridium perfringens isolates from necrotic enteritis outbreaks in broiler chicken populations. J Clin Microbiol. 2008;46:3957–3964. doi: 10.1128/JCM.01548-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collie RE, Kokai-Kun JF, McClane BA. Phenotypic characterization of enterotoxigenic Clostridium perfringens isolates from non-foofborne human gastrointestinal diseases. Anaerobe. 1998;4:69– 79. doi: 10.1006/anae.1998.0152. [DOI] [PubMed] [Google Scholar]
- 11.Collie RE, McClane BA. Evidence that the enterotoxin gene can be episomal in Clostridium perfringens isolates associated with nonfoodborne human gastrointestinal diseases. J Clin Microbiol. 1998;36:30–36. doi: 10.1128/jcm.36.1.30-36.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cornillot E, Saint-Joanis B, Daube G, Katayama S, Granum PE, Canard B, Cole ST. The enterotoxin gene (cpe) of Clostridium perfringens can be chromosomal or plasmid-borne. Mol Microbiol. 1995;15:639–647. doi: 10.1111/j.1365-2958.1995.tb02373.x. [DOI] [PubMed] [Google Scholar]
- 13.Czeczulin JR, Hanna PC, McClane BA. Cloning, nucleotide sequencing, and expression of the Clostridium perfringens enterotoxin gene in Escherichia coli. Infect Immun. 1993;61:3429–3439. doi: 10.1128/iai.61.8.3429-3439.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daube G, Simon P, Limbourg B, Manteca C, Mainil J, Kaeckenbeeck A. Hybridization of 2,659 Clostridium perfringens isolates with gene probes for seven toxins (α, β, ε, ι, theta, μ and enterotoxin) and for sialidase. Am J Vet Res. 1996;57:496–501. [PubMed] [Google Scholar]
- 15.Deguchi A, Miyamoto K, Kuwahara T, Miki Y, Kaneko I, Li J, McClane BA, Akimoto S. Genetic characterization of type A enterotoxigenic Clostridium perfringens strains. PLoS One. 2009;4:e5598. doi: 10.1371/journal.pone.0005598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fisher DJ, Fernandez-Miyakawa ME, Sayeed S, Poon R, Adams V, Rood JI, Uzal FA, McClane BA. Dissecting the contributions of Clostridium perfringens type C toxins to lethality in the mouse intravenous injection model. Infect Immun. 2006;74:5200–5210. doi: 10.1128/IAI.00534-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garmony HS, Chanter N, French NP, Bueschel D, Songer JG, Titball RW. Occurrence of Clostridum perfringens b2-toxin amongst animals, determined using genotyping and subtyping PCR assays. Epidemiol Infect. 2000;124:61–67. doi: 10.1017/s0950268899003295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harry KH, Zhou R, Kroos L, Melville SB. Sporulation and enterotoxin (CPE) synthesis are controlled by the sporulation-specific sigma factors SigE and SigK in Clostridium perfringens. J Bacteriol. 2009;191:2728–2742. doi: 10.1128/JB.01839-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heikinheimo A, Lindström M, Granum PE, Korkeala H. Humans as reservoir for enterotoxin gene-carrying Clostridium perfringens type A. Emerg Infect Dis. 2006;12:1724–1729. doi: 10.3201/eid1211.060478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jost BH, Trinh HT, Songer JG. Clonal relationship among Clostridium perfringens of porcine origin as determined by multilocus sequence typing. Vet Microbiol. 2006;116:158–165. doi: 10.1016/j.vetmic.2006.03.025. [DOI] [PubMed] [Google Scholar]
- 21.Kaneko I, Miyamoto K, Mimura K, Yumine N, Utsunomiya H, Akimoto S, McClane BA. Detection of enterotoxigenic C. perfringens in meat samples by using molecular methods. Appl Environ Microbiol. 2011;77:7526–7532. doi: 10.1128/AEM.06216-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kato H, Kato N, Watanabe K, Ueno K, Ushijima H, Hashira S, Abe T. Application of typing by pulsed-field gel electrophoresis to the study of Clostridium difficle in a neonatal intensive care unit. J Clin Microbiol. 1994;32:2067–2070. doi: 10.1128/jcm.32.9.2067-2070.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kobayashi S, Wada A, Shibasaki S, et al. Spread of a large plasmid carrying the cpe gene and the tcp locus amongst Clostridium perfringens isolates from nosocomial outbreaks and sporadic cases of gastrointestiritis in a geriatric hospital. Epidemiol Infect. 2008;137:108–113. doi: 10.1017/S0950268808000794. [DOI] [PubMed] [Google Scholar]
- 24.Kokai-Kun JF, Songer JG, Czeczulin JR, Chen F, McClane BA. Comparison of Western Immunoblots and gene detection assays for identification of potentially enterotoxigenic isolates of Clostridium perfringens. J Clin Microbiol. 1994;32:2533–2539. doi: 10.1128/jcm.32.10.2533-2539.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lahti P, Heikinheimo A, Johansson T, Korkeala H. Clostridium perfringens type A isolates carrying the plasmid-borne enterotoxin gene (genotype IS1151-cpe or IS1470-like-cpe) are a common cause of food poisoning. J Clin Microbiol. 2008;46:371–373. doi: 10.1128/JCM.01650-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lawrence G, Walker PD. Pathogenesis of enteritis necroticans in Papua, New Guinea. Lancet. 1976;1:125–128. doi: 10.1016/s0140-6736(76)93160-3. [DOI] [PubMed] [Google Scholar]
- 27.Li J, McClane BA. Comparative effects of osmoptic, sodium nitrate-induced, and pH-induced stress on growth and survival of Clostridium perfringens type A isolates carrying chromosomal or plasmid-borne enterotoxin genes. Appl Environ Microbiol. 2006;72:7620–7625. doi: 10.1128/AEM.01911-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li J, McClane BA. Further comparison of temperature effects on growth and survival of Clostridium perfringens type A isolates carrying a chromosomal or plasmid-borne enterotoxin gene. Appl Environ Microbiol. 2006;72:4561–4568. doi: 10.1128/AEM.00177-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li J, Sayeed S, McClane BA. Prevalence of enterotoxigenic Clostridium perfringens isolates in Pittsburgh (Pennsylvania) area soil and home kitchens. Appl Environ Microbiol. 2007;73:7218–7224. doi: 10.1128/AEM.01075-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li J, Miyamoto K, McClane BA. Comparison of virulence plasmids among Clostridium perfringens type E isolates. Infect Immun. 2007;75:1811–1819. doi: 10.1128/IAI.01981-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li J, McClane BA. A novel small acid soluble protein variant is important for spore resistance of most Clostridium perfringens food poisoning isolates. PLoS Pathog. 2008;4:e1000056. doi: 10.1371/journal.ppat.1000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li J, Miyamoto K, Sayeed S, McClane BA. Organization of the cpe locus in CPE-positive Clostridium perfringens type C and D isolates. PLoS One. 2010;5:e10932. doi: 10.1371/journal.pone.0010932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin YT, Labbe R. Enterotoxigenicity and genetic relatedness of Clostridium perfringens isolates from retail foods in the United States. Appl Environ Microbiol. 2003;69:1642–1646. doi: 10.1128/AEM.69.3.1642-1646.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lindström M, Heikinheimo A, Lathi P, Korkeara H. Novel insight into the epidemiology of Clostridium perfringens type A food poisoning. Food Microbiol. 2011;28:192–198. doi: 10.1016/j.fm.2010.03.020. [DOI] [PubMed] [Google Scholar]
- 35.McClane BA. Clostridial enterotoxins. In: Druse P, editor. Handbook on Clostridia. CRC Press; Boca Raton: 2005. pp. 385–406. [Google Scholar]
- 36.McClane BA. Clostridium perfringens. In: Doyle MP, Beuchat LR, Montville TJ, editors. Food Microbiology: Fundamentals and Frontier. ASM Press; Washington DC: 2001. pp. 351–372. [Google Scholar]
- 37.Meer RR, Songer JG. Multiplex polymerase chain reaction assay for genotyping Clostridium perfringens. Am J Vet Res. 1997;58:702–705. [PubMed] [Google Scholar]
- 38.Miki Y, Miyamoto K, Kaneko-Hirano I, Fujiuchi K, Akimoto S. Prevalence and characterization of enterotoxin gene-carrying Clostridium perfringens isolates from retail meat products in Japan. Appl Environ Microbiol. 2008;74:5366–5372. doi: 10.1128/AEM.00783-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miwa N, Nishina T, Kubo S, Atsumi M, Honda H. Amount of enterotoxigenic Clostridium perfringens in meat detected by nested PCR. Int J Food Microbiol. 1998;42:195–200. doi: 10.1016/s0168-1605(98)00082-8. [DOI] [PubMed] [Google Scholar]
- 40.Miyamoto K, Chakrabarti G, Morino Y, McClane BA. Organization of the plasmid cpe locus in Clostridium perfringens type A isolates. Infect Immun. 2002;70:4261–4272. doi: 10.1128/IAI.70.8.4261-4272.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miyamoto K, Wen Q, McClane BA. Multiplex PCR genotyping assay that distinguishes between isolates of Clostridium perfringens type A carrying a chromosomal enterotoxin gene (cpe) locus, a plasmid cpe locus with an IS1470-like sequence, or a plasmid cpe locus with an IS1151 sequence. J Clin Microbiol. 2004;42:1552–1558. doi: 10.1128/JCM.42.4.1552-1558.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miyamoto K, Fisher DJ, Li J, Sayeed S, Akimoto S, McClane BA. Complete sequencing and diversity analysis of the enterotoxin-encoding plasmids in Clostridium perfringens type A non-food-borne human gastrointestinal disease isolates. J Bacteriol. 2006;188:1585–1598. doi: 10.1128/JB.188.4.1585-1598.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miyamoto K, Yumine N, Mimura K, Nagahama M, Li J, McClane BA, Akimoto S. Identification of novel Clostridium perfringens type E strains that carry an iota toxin plasmid with a functional enterotoxin gene. PLoS One. 2011;6:e20376. doi: 10.1371/journal.pone.0020376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mueller-Spitz SR, Stewart LB, Klump JV, McLellan SL. Freshwater suspended sediments and sewage are reservoirs for enterotoxin-positive Clostridium perfringens. Appl Environ Microbiol. 2010;76:5556–5562. doi: 10.1128/AEM.01702-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Myers GS, Rasko DA, Cheung JK, et al. Skewed genomic variability in strains of the toxinogenic bacterial pathogen Clostridium perfringens. Genome Res. 2006;16:1031–1040. doi: 10.1101/gr.5238106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pillai SD, Vega E. Molecular detection and characterization tools. In: Santo Domingo JW, Sadowsky MJ, editors. Microbial Source Tracking. ASM Press; Washington DC: 2007. pp. 65–91. [Google Scholar]
- 47.Popoff MR, Stiles BG. Clostridial toxins vs. other bacterial toxins. In: Druse P, editor. Handbook on Clostridia. CRC Press; Boca Raton: 2005. pp. 324–383. [Google Scholar]
- 48.Rooney AP, Swezey JL, Friedman R, Hecht DW, Maddox CW. Analysis of core housekeeping and virulence genes reveals cryptic lineages of Clostridium perfringens that are associated with distinct disease presentation. Genetics. 2006;172:2081–2092. doi: 10.1534/genetics.105.054601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sarker MR, Shivers RP, Sparks SG, Juneja VK, McClane BA. Comparative experiments to examine the effects of heating on vegetative cells and spores of Clostridium perfringens isolates carrying plasmid versus chromosomal enterotoxin genes. Appl Environ Microbiol. 2000;66:3234–3240. doi: 10.1128/aem.66.8.3234-3240.2000. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 50.Sayeed S, Fernandez-Miyakawa ME, Fisher DJ, Adams V, Poon R, Rood JI, Uzal FA, McClane BA. Epsilon-toxin is required for most Clostridium perfringens type D vegetative culture supernatants to cause lethality in the mouse intravenous injection model. Infect Immun. 2005;73:7413–7421. doi: 10.1128/IAI.73.11.7413-7421.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Songer JG, Meer RR. Genotyping of Clostridium perfringens by polymerase chain reaction is a useful adjunct to diagnosis of clostridial enteric disease in animals. Anaerobe. 1996;2:197–203. [Google Scholar]
- 52.Tanaka D, Isobe J, Hosorogi S, et al. An outbreak of food-borne gastroenteritis caused Clostridium perfringens carrying the cpe gene on a plasmid. Jpn J Infect Dis. 2003;56:137–139. [PubMed] [Google Scholar]
- 53.Tanaka D, Kimata K, Shimizu M, et al. Genotyping of Clostridium perfringens isolates collected from food poisoning outbreaks and healthy individuals in Japan based on the cpe locus. Jpn J Infect Dis. 2007;60:68–69. [PubMed] [Google Scholar]
- 54.Urwin R, Maiden M. Multi-locus sequence typing: a tool for global epidemiology. Trends Microbiol. 2003;11:479–487. doi: 10.1016/j.tim.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 55.Van Damme-Jongsten M, Rodhouse J, Gilbert RJ, Notermans S. Synthetic DNA probes for detection of enterotoxigenic Clostridium perfringens strains isolated from outbreaks of food poisoning. J Clin Microbiol. 1990;28:131–133. doi: 10.1128/jcm.28.1.131-133.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Watanabe M, Hitomi S, Sawahara T. Nosocomial diarrhea caused by Clostridium perfringens in the Tsukuba-Tsuchiura district, Japan. J Infect Chemother. 2008;14:228–231. doi: 10.1007/s10156-008-0605-4. [DOI] [PubMed] [Google Scholar]
- 57.Wen Q, McClane BA. Detection of enterotoxigenic Clostridium perfringens type A isolates in American retail foods. Appl Environ Microbiol. 2004;70:2685–2691. doi: 10.1128/AEM.70.5.2685-2691.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wen Q, Miyamoto K, McClane BA. Development of a duplex PCR genotyping assay for distinguishing Clostridium perfringens type A isolates carrying chromosomal enterotoxin (cpe) genes from those carrying plasmid-borne enterotoxin (cpe) genes. J Clin Microbiol. 2003;41:1494–1498. doi: 10.1128/JCM.41.4.1494-1498.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao Y, Melville SB. Identification and characterization of sporulation-dependent promoters upstream of the enterotoxin gene (cpe) of Clostridium perfringens. J Bacteriol. 1998;180:136–142. doi: 10.1128/jb.180.1.136-142.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]