Abstract
Eight Aeromonas hydrophila-like arabinose-negative isolates from diverse sources (i.e., river freshwater, cooling-system water pond, diseased wild European eels, and human stools) sampled in Valencia (Spain) during 2004–2005, were characterized by 16S rRNA gene sequencing and extensive biochemical testing along with reference strains of most Aeromonas species. These isolates and all reference strains of A. hydrophila subsp. dhakensis and A. aquariorum showed a 16S rRNA sequence similarity of 99.8–100%, and they all shared an identical phenotype. This matched exactly with that of A. hydrophila subsp. dhakensis since all strains displayed positive responses to the Voges-Prokauer test and to the use of dl-lactate. This is the first report of A. hydrophila subsp. dhakensis recovered from environmental samples, and further, from its original isolation in India during 1993–1994. This was accurately identified and segregated from other clinical aeromonads (A. hydrophila subsp. hydrophila, A. caviae, A. veronii biovars veronii and sobria, A. trota, A. schubertii and A. jandaei) by using biochemical key tests. The API 20 E profile for all strains included in A. hydrophila subsp. dhakensis was 7047125. The prevalence of this species in Spanish sources was higher for water (9.4%) than for feces (6%) or eels (1.3%). Isolates recovered as pure cultures from diseased eels were moderately virulent (LD50 of 3.3×106 CFU fish−1) to challenged eels in experimental trials. They were all resistant to ticarcillin, amoxicillin-clavuranic acid, cefoxitin, and imipenem, regardless of its source. Our data point to A. hydrophila subsp. dhakensis as an emerging pathogen for humans and fish in temperate countries.
Keywords: A. hydrophila subsp. dhakensis, phenotypic profile, 16S rRNA gene sequencing, emerging pathogen
The genus Aeromonas (family Aeromonadaceae) is ubiquitous in aquatic ecosystems that include chlorinated drinking water, raw sewage, and natural waters (i.e., fresh and brackish water) and free-living fish in such habitats (27). Aeromonas species are most commonly implicated as the causative agents of gastroenteritis in tropical countries, with sepsis a fatal complication of Aeromonas infectious diseases, particularly in immunocompromised patients (22). In addition, some Aeromonas species are one of the major causative agents of diseases in reared and wild fish (2, 13).
Aeromonas hydrophila subsp. dhakensis was originally isolated from patients with diarrhea in Dhaka, Bangladesh (India) during 1993–1994 (20, 23). Since then A. hydrophila subsp. dhakensis has not been isolated in any region of the world, but many reports have focused on its relationship with other A. hydrophila subspecies. Thus, depending on the housekeeping genes used (i.e., 16S rRNA, rpoB, or dnaJ), A. hydrophila subsp. dhakensis exhibits high divergence from A. hydrophila subsp. hydrophila and A. hydrophila subsp. ranae (31, 32) or it does not (24, 35). In addition, it has been recently reported that type strains of the species A. hydrophila subsp. dhakensis and A. aquariorum clustered together in the phylogenetic tree derived from concatenated gyrB-rpoD sequences (31). However, the phenotypic profile of these two arabinose-negative Aeromonas species mostly differed (20, 30), as well as their respective genomic homology (DNA-DNA relatedness) with the type strain of the species A. hydrophila, which was 46% in the case of A. aquariorum (30) but 78 to 84% in the case of A. hydrophila subsp. dhakensis (20).
Identification of aeromonads by using biochemical schemes is difficult (1) while numerical studies have obtained quite high success showing a good correlation with genotypic identification (42). Most clinical microbiology laboratories still routinely rely on easy-to-use phenotypic methods (39); therefore, key traits for discriminating at least those Aeromonas species relevant in clinical sources should be well defined (25, 39). In a previous study, 215 Aeromonas isolates were recovered from river water and cooling systems (8), from wild European eels (13), and from human feces, collected during a 1-year period at locations in the river Xúquer floodplain, a highly urbanized and industrialized metropolitan area (753,552 inhabitants) around the city of Valencia (800,469 inhabitants). Eight out of these 215 aeromonads were presumptively identified as A. hydrophila, despite their negative production of acid from l-arabinose. The aim of the present study was to fully identify these A. hydrophila-like arabinose-negative strains, and to assess their clinical and veterinary relevance. For this purpose we used 16S rRNA gene sequencing and extensive biochemical testing, and also other assays to check for its susceptibility to antimicrobials and its virulence in fish.
Materials and Methods
Bacterial isolates
Strains ABF132, ABF144, and ABF145 were isolated from a wild European eel caught in the Albufera lake in El Palmar (Valencia, Spain) in November 2004 (13); strains MA17 and MA26 were isolated from two freshwater samples collected in the River Júcar in Alberique (Valencia, Spain), in October 2004 and February 2005, respectively (8); and strain MA131 was isolated from the water of a cooling-system pond in Catarroja (Valencia, Spain) in November 2005 (8). Strains 133.341 and 133.343 were isolated from stools on xylose-galactosidase plates (16) at the “Servicio de Microbiología, del Hospital Universitario La Fe” (Valencia, Spain), in September 2005. Strain CECT 4588 was originally recovered from feces of a patient with diarrhea in the Netherlands as isolate AH290 (12). This was identified as “A. hydrophila-like arabinose-negative” by others (42).
PCR amplification, 16S rRNA sequencing, and phylogenetic analysis
The almost complete 16S rRNA gene sequence of strains ABF132, ABF144, ABF145, MA17, MA26, MA131, 133.341, 133.343, CECT4588, A. hydrophila subsp. dhakensis CECT5743, A. hydrophila subsp. dhakensis CECT5745, A. aquariorum MDC310, and A. aquariorum MDC317 was obtained by the Colección Española de Cultivos Tipo (CECT) service (Valencia, Spain). Bacterial genomic DNAs were extracted according to a method described previously (37). Universal primers (Invitrogen, Life Technologies) used were 616V (forward) and 699R (reverse) for a stretch around 1,000 nt close to the 5′ end (targeting positions for these primers are 8–25 and 1,099–1,113, respectively [Escherichia coli numbering]) (5), and P609D (forward) and P1525R (reverse) targeting positions 785–802 and 1,525–1,541, respectively (Escherichia coli numbering) (26). PCR mixtures were composed of 5.0 μL PCR buffer (10×), 0.75 μL MgCl2 (100 mM), 1.0 μL dNTPs (10 mM each), 1.0 μL each forward and reverse primers (50 μM), 0.5 μL Taq polymerase (6 U μL−1; New England Biolabs) and 5.0 μL template DNA (50 ng μL−1) in a total volume of 50 μL. PCR amplifications of the DNA templates were performed using a PTC-100 ThermoCycler (MJ Research). The conditions for 16S rRNA gene amplification were (i) 4 min at 94°C; (ii) 30 cycles of 1 min at 94°C, 1 min 30 s at 52°C and 2 min at 72°C; and (iii) a final elongation step of 10 min at 72°C. Amplified products were examined by agarose gel electrophoresis (1.2%) and ethidium bromide staining. Purified amplicons (Mo Bio Laboratories) were sequenced by the dideoxy method using the BigDye Terminator v. 3.0 Ready Reaction cycle sequencing kit and analyzed in an ABI PRISM 3730 sequencer (Applied Biosystems). Sequencing primers were the same as those used in the amplification reaction but diluted tenfold (5 pmol).
The sequences obtained were aligned by CLUSTAL W program, version 1.83 (41) with the sequences of the type and reference strains of the members of the genus Aeromonas (Fig. 1) that were available in GenBank. Genetic distances and clustering were obtained using Kimura’s 2-parameter method. Phylogenetic trees were constructed by the neighbor-joining and maximum-parsimony methods using MEGA4 program (40). Stability of the relationships was assessed by bootstrapping (1,000 replicates).
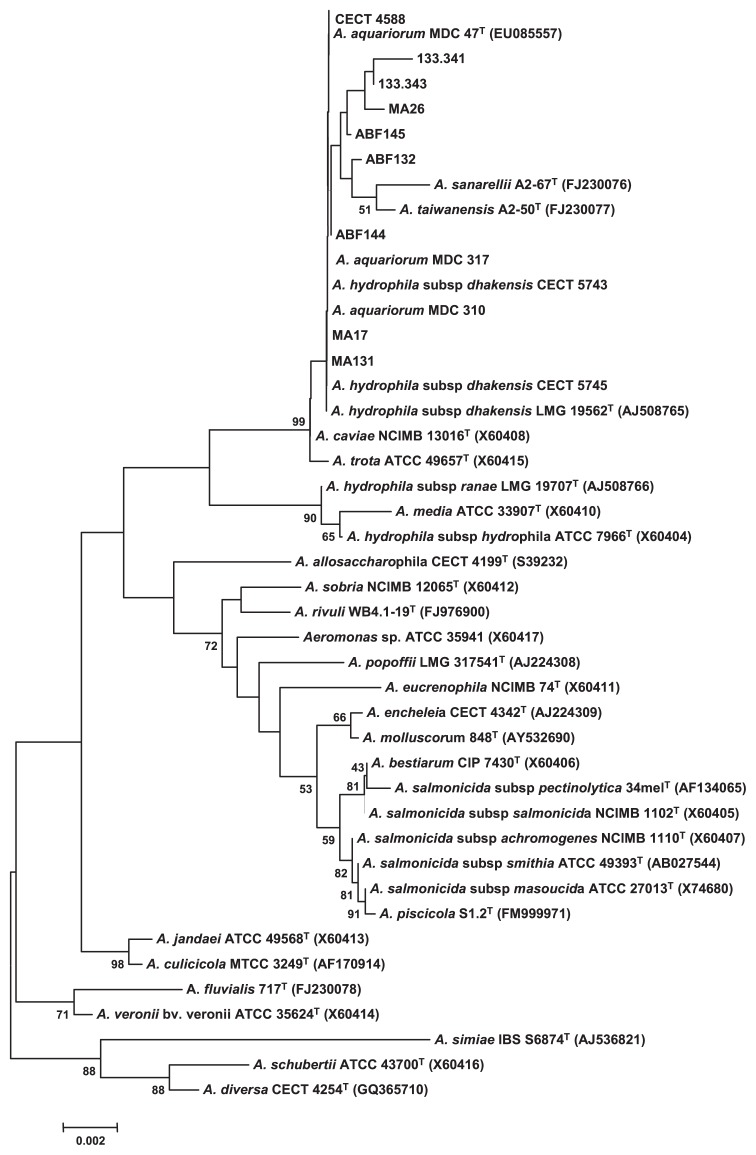
Fig. 1.
Unrooted neighbor-joining phylogenetic tree derived from the 16S rRNA gene sequences of the Aeromonas strains used. GenBank accession numbers for 16S rRNA gene sequences obtained in the present study are JQ034588 to JQ034600.
Phenotypic characterization
Spanish isolates, strain A. hydrophila-like CECT4588, and reference strains of A. hydrophila subsp. dhakensis (CECT5744T, CECT5743, and CECT5745), A. aquariorum (MDC47T, MDC310, and MDC317), A. hydrophila subsp. hydrophila (CECT839T), A. veronii bv. veronii (CECT4257T), A. veronii bv. sobria (CECT4835), A. sobria (CECT4245T), A. jandaei (CECT4228T), A. popoffii (CECT4995), A. bestiarum (CECT4227T), A. allosaccharophila (CECT4199T), A. eucrenophila (CECT4224T), A. encheleia (CECT4342T), A. trota (CECT4255T), A. enteropelogenes (CECT4487T), A. caviae (CECT838T), A. media (CECT4232T), A. schubertii (CECT4240T), and A. diversa (CECT4254T) were examined in 45 tests described by us as valuable traits for identifying Aeromonas (42). In addition, testing of the use of urocanic acid and β-hemolytic activity against sheep blood was performed as reported by others (18, 23). All tests were incubated at 28°C. These data for A. hydrophila subsp. ranae LMG19707T, A. fluvialis CECT 7401T, A. simiae CIP 107798T, A. tecta CECT 7082T, A. molluscorum CECT 5864T, A. bivalvium CECT 7113T, A. piscicola CECT 7443T, A. taiwanensis CECT7403T, A. sanarellii CECT7402T, and A. rivuli CECT 7518T were searched in published reports (3, 4, 7, 11, 15, 19, 21, 32, 33). Phenotypes were compared using the Simple Matching and Jacard’s similarity coefficients, and clustering was achieved by the unweighted pair group mathematical averaging method (UPGMA). All analyses were performed using NTSYSpc version 2.0.
In addition, API 20 E Strips (BioMérieux) were used in all isolates in order to know their API 20 E code.
Antimicrobial susceptibility assays
Minimal inhibitory concentrations (MICs) of kanamycin (K), tetracycline (TET), nalidixic (NA), oxolinic (OA) acid, flumequine (UB), erythromycin (ERY), rifampicin (RD) and chloramphenicol (CHL) were determined by the microbroth dilution method (9). In addition, MICs of amoxicillin/clavulanic (AMC), cefotaxime (CTX), cefoxitin (FOX), ceftazidime (CAZ), imipenem (IPM), piperacillin (PRL), ticarcillin (TIC), aztreonan (ATM), cefepime (FEP), ciprofloxacin (CIP), netilmicin (NET), norfloxacin (NOR) and levofloxacin (LEF) were determined using E-strips (MICE; Oxoid, Madrid, Spain). Finally, susceptibility to the antibiotics tested was evaluated on the basis of MIC values obtained, in accordance with the breakpoints recommended by the Clinical and Laboratory Standards Institute (9).
Virulence for European eel
For the challenge experiment, six young eels of around 10–20 g were challenged by intraperitoneal injection with each of the bacterial doses (108, 107, 106, 105, 104, 103, 102, 101 CFU mL−1), or with 0.1 mL PBS (controls). Each set of six eels was kept in a 20 L aquarium under the following conditions: i) dechlorinated tap water, ii) water temperature around 20°C, iii) oxygen concentration in water was above 90% saturation, and iv) fish were not fed. Mortality was recorded daily for 10 days and was only considered if the challenged bacterium was recovered as pure culture from the internal organs.
The bacterial strains used were ABF132, ABF144, and ABF145, which had been isolated by Esteve and Alcaide (13) from a specimen of wild European eel that suffered from hemorrhagic disease. The primary culture obtained originally from the kidney and liver of this specimen had been pure (i.e., strains ABF132 and ABF144, respectively), but that recovered from the ulcer was not (13). In the latter case we found three types of colonies and, among them, the colony matching ABF154 was the most abundant (personal communication). Bacterial virulence (LD50 expressed as inoculated colony forming units, CFU fish−1) was calculated (38).
Accession numbers for the 16S rRNA sequence data obtained from GenBank
EU085557 (A. aquariorum MDC 47T), FJ230076 (A. sanarellii A2-67T), FJ230077 (A. taiwanensis A2-50T), AJ508765 (A. hydrophila subsp. dhakensis LMG 19562T), X60408 (A. caviae NCIMB 13016T), X60415 (A. trota ATCC 49657T), AJ508766 (A. hydrophila subsp. ranae LMG 19707T), X60410 (A. media ATCC 33907T), X60404 (A. hydrophila subsp. hydrophila ATCC 7966T), S39232 (A. allosaccharophila CECT 4199T), X60412 (A. sobria NCIMB 12065T), FJ976900 (A. rivuli WB4.1-19T), X60417 (Aeromonas sp. ATCC 35941T), AJ224308 (A. popoffii LMG 317541T), X60411 (A. eucrenophila NCIMB 74T), AJ224309 (A. encheleia CECT 4342T), AY532690 (A. molluscorum 848T), X60406 (A. bestiarum CIP 7430T), AF134065 (A. salmonicida subsp. peptinolytica 34melT), X60405 (A. salmonicida subsp. salmonicida NCIMB 1102T), X60407 (A. salmonicida subsp. achromogenes NCIMB 1110T), AB027544 (A. salmonicida subsp. smithia ATCC 49393T), X74680 (A. salmonicida subsp. masoucida ATCC 27013T), FM999971 (A. piscicola S1.2T), X60413 (A. jandaei ATCC 49568T), AF170914 (A. culicicola MTCC 3248T), FJ230078 (A. fluvialis 717T), X60414 (A. veronii bv. veronii ATCC 35624T), AJ536821 (A. simiae IBS S6874T), X60416 (A. schubertii ATCC 43700T), GQ365710 (A. diversa CECT 4254T).
Nucleotide sequence accession numbers
GenBank accession numbers for 16S rRNA gene sequences obtained in the present study are JQ034588 to JQ034600.
Results and Discussion
Phylogenetic analysis
The 16S rRNA gene sequence was obtained from eight A. hydrophila-like arabinose-negative isolates, from strain CECT4588, and from A. hydrophila subsp. dhakensis CECT 5743 and CECT 5745, and A. aquariorum MDC310 and MDC317 (GenBank accession numbers: JQ034588 to JQ034600). These were compared with those of A. hydrophila subsp. dhakensis LMG 19562T and A. aquariorum MDC 47T and with those from other Aeromonas-type strains (Fig. 1). The 16S rRNA gene sequence similarity among the A. hydrophila-like arabinose-negative Spanish isolates, strain CECT 4588, and the A. hydrophila subsp. dhakensis and A. aquariorum reference strains was 100–99.8% (0 to 3 bp differences), in accordance with that reported solely for the type strain of these species (31). These sequence similarity values of the 16S rRNA gene were in line with those reported, among strains, for other Aeromonas species (7, 10, 15, 19, 28, 29, 32, 33). Thus, our phylogenetic results indicated that A. hydrophila subsp. dhakensis and A. aquariorum strains are extremely similar, suggesting that their genomic homology should be checked, especially as experiments of DNA-DNA hybridization between them fail to be performed.
On the other hand, A. hydrophila-like arabinose-negative Spanish isolates, strain CECT 4588, and the A. hydrophila subsp. dhakensis and A. aquariorum reference strains belonged to the “A. caviae-A. trota” branch (Fig. 1) of the Aeromonas phylogenetic tree, which also includes the species A. sanarelli and A. taiwanensis (4). At present, 16S rRNA gene sequencing is widely available in reference laboratories, while biochemical testing is still appropriate for separating those Aeromonas species which are phylogenetically close (17). Regarding this finding, the fifteen strains tested by us were clearly segregated from the species A. caviae and A. trota (Table 1; Fig. 2), as well as from A. sanarelli and A. taiwanensis (Fig. 2)
Table 1.
Key phenotypic profile of the A. hydrophila subsp. dhakensis–A. aquariorum clustera compared with Aeromonas type strains characterized in the study
| Biochemical test | A. hydrophila subsp. dhakensis–A. aquariorum cluster | A. hydrophila subsp. hydrophila | A. caviae | A. veronii bv. veronii | A. bestiarum | A. encheleia | A. eucrenophila | A. media | A. veronii bv. sobria | A. trota | A. popoffii | A. sobria | A. jandaei | A. allosaccharophila | A. schubertii | A. diversa |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hydrolysis of arbutin | + | + | + | + | + | + | + | + | − | − | − | − | − | − | − | − |
| aesculin | + | + | + | + | + | + | + | + | − | − | − | − | − | + | − | − |
| SDS | + | − | − | + | − | − | − | − | − | + | + | + | + | − | + | + |
| Gas from d-glucose | + | + | − | + | + | + | + | − | + | + | + | + | + | + | − | − |
| Voges-Proskauer | + | + | − | + | − | − | − | − | − | − | + | + | + | − | + | + |
| ADH (Moellers’) | + | + | − | − | + | + | + | − | + | + | + | + | + | + | + | + |
| LDC (Moellers’) | + | + | − | + | + | − | − | − | − | + | − | + | + | + | + | − |
| ODC (Moellers’) | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − |
| Acid from l-arabinose | − | + | + | − | + | − | + | + | − | − | − | − | − | + | − | − |
| salicin | + | + | + | + | + | + | + | + | − | − | − | − | − | − | − | − |
| d-cellobiose | − | − | + | + | + | − | + | + | + | + | − | + | − | + | − | − |
| l-fucose | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| l-rhamnose | − | − | − | − | − | + | − | − | − | − | − | − | − | + | − | − |
| Use of dl-Lactate | + | + | + | − | − | − | − | + | − | + | − | − | − | − | + | − |
| Urocanic acid | + | − | + | − | + | + | − | + | − | + | + | − | − | − | − | − |
+, all strains are positive; −, all strains are negative
Strains included in the cluster are: A. hydrophila subsp. dhakensis CECT 5744T, CECT 5743 and CECT 5745; isolates ABF132, ABF144, ABF145, MA17, MA26, MA131, 133.341, and 133.343; strain CECT4588; and A. aquariorum MDC47T, MDC310, and MDC317.
Fig. 2.
Phenogram obtained from numerical analysis of 48 phenotypic test results using the simple matching coefficient (SSM) and unweighted pair group method with arithmetic averages (UPGMA).
Phenotypic analysis
The A. hydrophila-like arabinose-negative isolates, strain A. hydrophila-like CECT 4588, and type and reference strains of the species A. hydrophila subsp. dhakensis and A. aquariorum were grouped by numerical analysis at 100% phenotypic similarity (S), using both simple matching (SSM) (Fig. 2) and Jaccard’s (SJ) coefficients. Moreover, they were all clearly segregated from the other type strains of the Aeromonas species whose phenotypic profile was included in the numerical analysis (Fig. 2). All strains included in the “A. hydrophila subsp. dhakensis-A. aquariorum” cluster (Fig. 2) showed the key responses detailed in Table 1. In addition, they all were positive for: motility; cytochrome-oxidase; catalase; gluconate oxidation; O/F metabolism; growth with 0–3% (w/v) NaCl; production of H2S from cysteine; production of indole from tryptophan; hydrolysis of arbutin, casein, starch, elastin, and gelatin (liquefaction); β-haemolysis of sheep red blood cells; and acid production from d-fructose, d-galactose, maltose, d-mannitol, d-mannose, sucrose, and d-trehalose; however, they all were negative for: Gram staining; production of brown diffusible pigment; susceptibility to O/129 (150 μg) (Oxoid discs); growth with 6% (w/v) NaCl; hydrolysis of urea; and acid production from d-amygdalin, dulcitol, d-fucose, meso-inositol, melibiose, d-raffinose, d-sorbitol, and d-xylose.
Phenotypic profile of the “A. hydrophila subsp. dhakensis-A. aquariorum” cluster exactly matched that described for A. hydrophila subsp. dhakensis (20) but differed from that described for A. aquariorum (30). Previous data on the response of A. aquariorum strains to the VP test are confusing; the species was originally described as VP-negative (i.e., in text) and -positive (i.e., in the diagnostic table) simultaneously (30) and hence were reported as VP-positive (7, 34) or VP-negative (4, 15) in subsequent reports. Once again, species A. aquariorum was originally described as both negative (i.e., in the diagnostic table) and positive (i.e., in text) for the utilization of dl-lactate (30), but was later reported as dl-lactate-negative by taxonomic reports (7, 15, 34), mostly based on the diagnostic table (30). We have found that the three A. aquariorum strains assayed by us displayed a clear positive response in the VP semisolid medium (42) and in the Lactate utilization medium (18) respectively, similarly to that shown by Spanish isolates as well as by the reference strains of A. hydrophila subsp. dhakensis (Table 1). It should be noted that our results came from the first extensive phenotypic characterization that compares strains of A. aquariorum and of A. hydrophila subsp. dhakensis, along with other Aeromonas species. Accordingly, other authors have recently reported the positive responses to both, VP test and dl-lactate utilization, for A. aquariorum strains isolated from diverse sources in Western Australia (6).
In line with the present results, A. aquariorum cannot be differentiated from A. hydrophila subsp. hydrophila by the use of dl-lactate, contrary to other reports (14). In fact, the pair A. hydrophila subsp. dhakensis-A. aquariorum, which shared an identical phenotype, was separated in our study from A. hydrophila subsp. hydrophila by its inability to produce acid from l-arabinose and l-fucose, and its use of urocanic acid, and further by its ability to hydrolyze sodium dodecyl sulfate (SDS) (i.e., alkyl sulfatase activity) (Table 1). Finally, we have identified the eight A. hydrophila-like arabinose-negative Spanish isolates, and strain CECT4588, as belonging to the species A. hydrophila subsp. dhakensis, on the basis of the overall results (Table 1; Figs. 1 and 2). The API 20 E profile for all strains included in the “A. hydrophila subsp. dhakensis-A. aquariorum” cluster was 7047125, which previously had shown very little prevalence (i.e., 9%; 8 out of 86), which was obtained from Aeromonas strains (13). In addition, we found that A. hydrophila subsp. dhakensis can be accurately identified and segregated from other clinical aeromonads, such as A. hydrophila subsp. hydrophila, A. caviae, A. veronii biovars veronii and sobria, A. trota, A. schubertii and A. jandaei (25, 39), using biochemical key tests (Table 1).
Incidence and clinical and veterinary relevance of Aeromonas hydrophila subsp. dhakensis in Mediterranean Spain
Aeromonas hydrophila subsp. dhakensis was previously isolated in India (23), and was encountered in Valencia (Mediterranean Spain) during a 1-year study (2004–2005) in which 32 water samples (8), 75 wild European eels (13), and an unknown number of feces from humans suffering acute gastroenteritis were analyzed. Among them, the number of Aeromonas-positive samples was 17 (53.1%) and 20 (26.7%), respectively, with 32 patients suffering from Aeromonas gastroenteritis. The overall prevalence of A. hydrophila subsp. dhakensis in these Aeromonas-positive specimens was of 8.7% (6/69), although it was higher for water (17.7%) than for feces (6.25%) or eels (5%). Thus, we have found a wider distribution of A. hydrophila subsp. dhakensis in Spain in comparison with its unique previous finding in association with patients but not with water or fish (23). It is important to note that such water sources were agricultural ponds linked to the Xúquer river basin, which are used for the irrigation of agricultural products; the counts of Aeromonas in these waters ranged from (102 to 104 CFU mL−1) in winter to (104 to 107 CFU mL−1) in summer (8). In addition, wild European eels are caught in Albufera Lake for commercial purposes and are used for human consumption (13). Hence, these findings could have public health implications because Aeromonas infections might be transmitted through the ingestion of contaminated water or food (i.e., vegetables, fish and shellfish, etc.), or by contact with them (22).
Our clinical A. hydrophila subsp. dhakensis isolates (i.e., 133.341 and 133.343) were recovered from two siblings, a 1-year-old boy and a 6-year-old girl, who had acute gastroenteritis accompanied by bloody stools and high-grade (≥39°C) fever and required antibiotic treatment. The fact that these clinical isolates were rather multi-resistant to antibiotics (Table 2) complicated the management of this Aeromonas-mediated diarrhea, similarly to in other geographical areas (39). Thus, strains 133.341 and 133.343 showed the highest MICs for ticarcillin, piperacillin, amoxicillin/clavuranic, cefoxitin, imipenem, flumequine, nalidix acid, oxolinic acid, and erythromycin (Table 2); however, resistance to ticarcillin, amoxicillin-clavuranic acid, cefoxitin, and imipenem were common in the Spanish isolates of A. hydrophila subsp. dhakensis from any source (i.e., stool, water, fish) (Table 2). Fortunately, the latest generation cephalosporins and the fluoroquinolones had the best inhibitory activity in vitro against A. hydrophila subsp. dhakensis isolates (Table 2), as also was described for other clinical Aeromonas (43).
Table 2.
Antimicrobial resistance in the Spanish A. hydrophila subsp. dhakensis isolates
| MIC (μg mL−1) | |||||
|---|---|---|---|---|---|
| Antibiotic | Range | For the 50% tested strains | For the 90% tested strains | Break-points (μg mL−1) | Resistant strains (%) |
| Amoxicillin/Clavulanic (AMC) | 128–512 | 128 | 256 | 32a | 100b |
| Cefoxitin (FOX) | 32–128 | 64 | 128 | 32a | 100b |
| Imipenem (IPM) | 0.1–128 | 32 | 64 | 16a | 75b |
| Ticarcillin (TIC) | 128–512 | 128 | 256 | 128a | 100b |
| Piperacillin (PRL) | 0.1–256 | 0.5 | 256 | 128a | 50b |
| Flumequine (UB) | 0.1–64 | 0.3 | 64 | 8a | 50b |
| Nalidixic acid (NA) | 0.1–512 | 0.5 | 512 | 32a | 37.5b |
| Oxolinic acid (OA) | 0.2–64 | 0.2 | 32 | 4a | 37.5b |
| Erythromycin (ERY) | 0.1–64 | 16 | 64 | 8a | 62.5b |
| Rifampicin (RD) | 0.1–4 | 0.1 | 2 | 4a | 12.5 |
For cefotaxime (CTX), ceftazidime (CAZ), cefepime (CPM), aztreonan (ATM), kanamycin (K), tetracyline (TET), ciprofloxacin (CIP), levofloxacin (LEF), norfloxacin (NOR), choramphenicol (CHL) and netilmicin (NET), the MIC at which 90% of isolates were inhibited was inferior to 0.5 μg mL−1, so all strains were sensitive to these drugs.
This value of MIC is the cut-off to consider resistance to the antimicrobial (9).
The two clinical isolates (i.e., 133.341 and 133.343) were resistant.
The three isolates of A. hydrophila subsp. dhakensis (i.e., ABF132, ABF144, and ABF145) which have been recovered from a wild European eel suffering from hemorrhagic septicemia (13), displayed an LD50 dose of 2.6×105 to 3.3×106 CFU fish−1 in experimental challenges using healthy eels. These data are the first report on the active role of A. hydrophila subsp. dhakensis as fish pathogen, although others have published that a clinical isolate of this species was moderately virulent for challenged trout (36). Interestingly, A. aquariorum, its closest species, was originally recovered from imported ornamental fish that showed symptoms of weakness (30).
In summary, species A. hydrophila subsp. dhakensis can be recovered from natural waters, fish, and clinical specimens in Mediterranean Spain. This result suggests its potential role as a waterborne pathogen for humans and fish in temperate countries. In fact, strain CECT4588 was recovered from feces of a patient with diarrhea in the Netherlands in the eighties. Aeromonas hydrophila subsp. dhakensis can be identified by biochemical key tests. In the present study the phenotypic profile of A. hydrophila subsp. dhakensis was also demonstrated by the type and reference strains of A. aquariorum. Up to now, species A. hydrophila subsp. dhakensis as well as its closest species A. aquariorum have been reported to be distributed throughout warm countries (6, 14. 23, 30). The present results constitute the first report of A. hydrophila subsp. dhakensis from a temperate country, suggesting the worldwide distribution of this species.
Acknowledgements
This study has been supported by project CGL2007-60565 from the Ministerio de Ciencia e Innovación (Spain). M.D. Blasco is the recipient of a PhD fellowship from the Spanish Government (Ministerio de Educación y Ciencia). We thank Antonio J. Martínez-Murcia for kindly supplying the MDC strains of A. aquariorum. We thank Francisco Javier Pascual Martínez for his technical support with phylogenetic analysis.
References
- 1.Abbot SL, Cheung WKW, Janda JM. The genus Aeromonas: biochemical characteristics, atypical reactions, and phenotypic identification schemes. J Clin Microbiol. 2003;41:2348–2357. doi: 10.1128/JCM.41.6.2348-2357.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Austin B. Taxonomy of bacterial fish pathogens. Vet Res. 2011;42:20. doi: 10.1186/1297-9716-42-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alperi A, Martínez-Murcia AJ, Monera A, Saavedra MJ, Figueras MJ. Aeromonas fluvialis sp. nov., isolated from Spanish river. Int J Syst Evol Microbiol. 2010;60:72–77. doi: 10.1099/ijs.0.011643-0. [DOI] [PubMed] [Google Scholar]
- 4.Alperi A, Martínez-Murcia AJ, Ko WC, Monera A, Saavedra MJ, Figueras MJ. Aeromonas taiwanensis sp. nov. and Aeromonas sanarelli sp. nov., two new clinical species from Taiwan. Int J Syst Evol Microbiol. 2010;60:2048–2055. doi: 10.1099/ijs.0.014621-0. [DOI] [PubMed] [Google Scholar]
- 5.Arahal DR, Sánchez E, Macián MC, Garay E. Value of recN sequences for species identification and as a phylogenetic marker within the family “Leuconostocaceae”. Int Microbiol. 2008;11:33–39. [PubMed] [Google Scholar]
- 6.Aravena-Román M, Harnett GB, Riley TV, Inglis TJ, Chang BJ. Aeromonas aquariorum is widely distributed in clinical and environmental specimens and can be misidentified as Aeromonas hydrophila. J Clin Microbiol. 2011;49:3006–3008. doi: 10.1128/JCM.00472-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beaz-Hidalgo R, Alperi A, Figueras MJ, Romalde JL. Aeromonas piscicola sp. nov., isolated from diseased fish. Syst Appl Microbiol. 2009;32:471–479. doi: 10.1016/j.syapm.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 8.Blasco MD, Esteve C, Alcaide E. Multiresistant waterborne pathogens isolated from water reservoirs and cooling systems. J Appl Microbiol. 2008;105:469–475. doi: 10.1111/j.1365-2672.2008.03765.x. [DOI] [PubMed] [Google Scholar]
- 9.Clinical and Laboratory Standards Institute (CLSI) 16th informational supplement document M2-A9 and M7-A7. Wayne: 2006. Performance Standards for antimicrobial susceptibilty testing. [Google Scholar]
- 10.Demarta A, Huys G, Tonolla M, Swings J, Peduzzi R. Polyphasic taxonomic study of “Aeromonas eucrenophila-like” isolates from clinical and environmental sources. Syst Appl Microbiol. 2004;27:343–349. doi: 10.1078/0723-2020-00276. [DOI] [PubMed] [Google Scholar]
- 11.Demarta A, Küpfer M, Riegel P, et al. Aeromonas tecta sp. nov., isolated from clinical and environmental sources. Syst Appl Microbiol. 2009;31:278–286. doi: 10.1016/j.syapm.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 12.Esteve C, Alcaide E, Canals R, Merino S, Blasco D, Figueras MJ, Tomás JM. Pathogenic Aeromonas hydrophila Serogroup O:14 and O:81 Strains with an S Layer. Appl Environ Microbiol. 2004;70:5898–5904. doi: 10.1128/AEM.70.10.5898-5904.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esteve C, Alcaide E. Influence of diseases on the wild eel stock: the case of Albufera lake. Aquaculture. 2009;289:143–149. [Google Scholar]
- 14.Figueras MJ, Alperi A, Saavedra MJ, Ko WC, Gonzalo N, Navarro M, Martínez-Murcia AJ. Clinical relevance of the recently described species Aeromonas aquariorum. J Clin Microbiol. 2009;47:3742–3746. doi: 10.1128/JCM.02216-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Figueras MJ, Alperi A, Beaz-Hidalgo R, Stackebrandt E, Brambilla E, Monera A, Martínez-Murcia AJ. Aeromonas rivuli sp. nov., isolated from the upstream region of a karst water rivulet. Int J Syst Evol Microbiol. 2011;61:242–248. doi: 10.1099/ijs.0.016139-0. [DOI] [PubMed] [Google Scholar]
- 16.García-Aguayo JM, Úbeda P, Gobernado M. Evaluation of xylose-galactosidase médium, a new plate for the isolation of Salmonella, Shigella, Yersinia and Aeromonas species. Eur J Clin Microbiol Infect Dis. 1999;18:77–78. doi: 10.1007/s100960050234. [DOI] [PubMed] [Google Scholar]
- 17.Graf J. Diverse restriction fragment length polymorphism patterns of the PCR-amplified 16S rRNA genes in Aeromonas veronii strains and possible misidentification of Aeromonas species. J Clin Microbiol. 1999;37:3194–3197. doi: 10.1128/jcm.37.10.3194-3197.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hänninen ML. Phenotypic characteristics of the three hybridization groups of Aeromonas hydrophila complex isolated from different sources. J Appl Bacteriol. 1994;76:455–462. [Google Scholar]
- 19.Harf-Monteil C, Le Flèche A, Riegel P, Prèvost G, Bermond D, Grimont PAD, Monteil H. Aeromonas simiae sp. nov., isolated from monkey faeces. Int J Syst Evol Microbiol. 2004;54:481–485. doi: 10.1099/ijs.0.02786-0. [DOI] [PubMed] [Google Scholar]
- 20.Huys G, Kämpfer P, Albert MJ, Kühn I, Denys R, Swings J. Aeromonas hydrophila subsp. dhakensis subsp. nov., isolated from children with diarrhoea in Bangladesh, and extended description of Aeromonas hydrophila subsp hydrophila (Chester 1901) Stanier 1943 (Approved Lists 1980) Int J Syst Evol Microbiol. 2002;52:705–712. doi: 10.1099/00207713-52-3-705. [DOI] [PubMed] [Google Scholar]
- 21.Huys G, Pearson M, Kämpfer P, Denys R, Cnockaert M, Inglis V, Swings J. Aeromonas hydrophila subsp. ranae subsp. nov., isolated from septicaemic facmed frogs in Thailand. Int J Syst Evol Microbiol. 2003;53:885–891. doi: 10.1099/ijs.0.02357-0. [DOI] [PubMed] [Google Scholar]
- 22.Janda JM, Abbot SL. Evolving concepts regarding the genus Aeromonas: an expanding panorama of species, disease presentation, and unanswere questions. Clin. Infect Dis. 1998;27:332–344. doi: 10.1086/514652. [DOI] [PubMed] [Google Scholar]
- 23.Kühn I, Albert MJ, Ansaruzzaman M, et al. Characterization of Aeromonas spp. isolated from humans with diarrea, from healthy controls, and from surface water in Bangladesh. J Clin Microbiol. 1997;35:369–373. doi: 10.1128/jcm.35.2.369-373.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Küpfer M, Kuhnert P, Korczak BM, Peduzzi R, Demarta A. Genetic relationships of Aermonas strains inferred from 16S rRNA, gyrB and rpoB gene sequences. Int J Syst Evol Microbiol. 2006;56:2743–2751. doi: 10.1099/ijs.0.63650-0. [DOI] [PubMed] [Google Scholar]
- 25.Lamy B, Kodjo A the colBVH Study Group. Laurent F. Prospective nationwide study of Aeromonas infections in France. J Clin Microbiol. 2009;47:1234–1237. doi: 10.1128/JCM.00155-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lucena T, Pascual J, Garay E, Arahal DR, Macián MC, Pujalte MJ. Haliea mediterranea sp. nov., a new marine gammaproteobacterium. Int J Syst Evol Microbiol. 2010;60:1844–1848. doi: 10.1099/ijs.0.017061-0. [DOI] [PubMed] [Google Scholar]
- 27.Martin-Carnahan A, Joseph SW. Genus I Aeromonas Stanier 1943, 213AL. In: Garrity GM, Brenner DJ, Krieg NR, Staly JT, editors. Bergey’s Manual of Systematic Bacteriology, vol. 2, part B. Springer-Verlag; New York: 2005. pp. 557–578. [Google Scholar]
- 28.Martínez-Murcia AJ, Benlloch S, Collins MD. Phylogenetic interrelationships of members of the genera Aeromonas and Plesiomonas as determined by 16S ribosomal DNA sequencing: lack of congruence with results of DNA-DNA hybridizations. Int J Syst Bacteriol. 1992;42:412–421. doi: 10.1099/00207713-42-3-412. [DOI] [PubMed] [Google Scholar]
- 29.Martínez-Murcia AJ, Esteve C, Garay E, Collins MD. Aeromonas allosaccharophila sp. nov., a new mesophilic member of the genus Aeromonas. FEMS Microbiol Lett. 1992;70:199–205. doi: 10.1016/0378-1097(92)90698-n. [DOI] [PubMed] [Google Scholar]
- 30.Martínez-Murcia AJ, Saavedra MJ, Mota VR, Maier T, Stackebrandt E, Cousin S. Aeromonas aquariorum sp. nov., isolated from aquaria of ornamental fish. Int J Syst Evol Microbiol. 2008;58:1169–1175. doi: 10.1099/ijs.0.65352-0. [DOI] [PubMed] [Google Scholar]
- 31.Martínez-Murcia AJ, Monera A, Alperi A, Figueras MJ, Saavedra MJ. Phylogenetic evidence suggests that strains of Aeromonas hydrophila subsp. dhakensis belong to the species Aeromonas aquariorum sp. nov. Curr Microbiol. 2009;58:76–80. doi: 10.1007/s00284-008-9278-6. [DOI] [PubMed] [Google Scholar]
- 32.Miñana-Galbis D, Farfán M, Fusté MC. Aeromonas molluscorum sp. nov., isolated from bivalve molluscs. Int J Syst Evol Microbiol. 2004;54:2073–2078. doi: 10.1099/ijs.0.63202-0. [DOI] [PubMed] [Google Scholar]
- 33.Miñana-Galbis D, Farfán M, Fusté MC, Lorén JG. Aeromonas bivalvium sp. nov., isolated from bivalve molluscs. Int J Syst Evol Microbiol. 2007;57:582–587. doi: 10.1099/ijs.0.64497-0. [DOI] [PubMed] [Google Scholar]
- 34.Miñana-Galbis D, Farfán M, Lorén JG, Fusté MC. Proposal to assign Aeromonas diversa sp. nov. as a novel species designation for Aeromonas group 501. Syst Appl Microbiol. 2010;33:15–19. doi: 10.1016/j.syapm.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 35.Nhung PH, Hata H, Ohkusu K, Noda M, Shah MM, Goto K, Ezaki T. Use of the novel phylogenetic marker dnaJ and DNA-DNA hybridization to clarify interrelationships within the genus Aeromonas. Int J Syst Evol Microbiol. 2007;57:1232–1237. doi: 10.1099/ijs.0.64957-0. [DOI] [PubMed] [Google Scholar]
- 36.Orozova P, Barker M, Austin DA, Austin B. Identification and pathogenicity to rainbow trout, Oncorhynchus mykiss ( Walbaum), of some aeromonads. J Fish Dis. 2009;32:865–871. doi: 10.1111/j.1365-2761.2009.01065.x. [DOI] [PubMed] [Google Scholar]
- 37.Pitcher DG, Saunders NA, Owen RJ. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett Appl Microbiol. 1989;8:151–156. [Google Scholar]
- 38.Reed MJ, Müench M. A simple method for estimating fifty percent endopoints. Am J Hyg. 1938;27:493–497. [Google Scholar]
- 39.Sinha S, Shimada T, Ramamurthy T, Bhattacharya SK, Yamasaki S, Takeda Y, Nair GB. Prevalence, serotype distribution, antibiotic susceptibility and genetic profiles of mesophilic Aeromonas species isolated from hospitalized diarrhoeal cases in Kolkata, India. J Med Microbiol. 2004;53:527–534. doi: 10.1099/jmm.0.05269-0. [DOI] [PubMed] [Google Scholar]
- 40.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 41.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Valera L, Esteve C. Phenotypic study by numerical taxonomy of strains belonging to the genus Aeromonasr. J Appl Microbiol. 2002;93:77–95. doi: 10.1046/j.1365-2672.2002.01665.x. [DOI] [PubMed] [Google Scholar]
- 43.Vila J, Marco F, Soler L, Chacón M, Figueras MJ. In vitro antimicrobial susceptibility of clinical isolates of Aeromonas caviae, Aeromonas hydrophila and Aeromonas veronii biotype sobria. J Antimicrob Chemother. 2002;49:701–702. doi: 10.1093/jac/49.4.701. [DOI] [PubMed] [Google Scholar]