Abstract
The current nitrogen fertilization for sugarcane production in Guangxi, the major sugarcane-producing area in China, is very high. We aim to reduce nitrogen fertilization and improve sugarcane production in Guangxi with the help of indigenous sugarcane-associated nitrogen-fixing bacteria. We initially obtained 196 fast-growing bacterial isolates associated with the main sugarcane cultivar ROC22 plants in fields using a nitrogen-deficient minimal medium and screened out 43 nitrogen-fixing isolates. Analysis of 16S rRNA gene sequences revealed that 42 of the 43 nitrogen-fixing isolates were affiliated with the genera Enterobacter and Klebsiella. Most of the nitrogen-fixing enterobacteria possessed two other plant growth-promoting activities of IAA production, siderophore production and phosphate solubilization. Two Enterobacter spp. strains of NN145S and NN143E isolated from rhizosphere soil and surface-sterilized roots, respectively, of the same ROC22 plant were used to inoculate micropropagated sugarcane plantlets. Both strains increased the biomass and nitrogen content of the sugarcane seedlings grown with nitrogen fertilization equivalent to 180 kg urea ha−1, the recommended nitrogen fertilization for ROC22 cane crops at the seedling stage. 15N isotope dilution assays demonstrated that biological nitrogen fixation contributed to plant growth promotion. These results suggested that indigenous nitrogen-fixing enterobacteria have the potential to fix N2 associated with sugarcane plants grown in fields in Guangxi and to improve sugarcane production.
Keywords: biological nitrogen fixation, enterobacteria, nifH, plant growth-promoting bacteria, sugarcane
Guangxi is the major sugarcane- and sugar-producing area in China and produces about 60% of China’s sugarcane and sugar. The present sugarcane mean yields are between 70 and 80 Mg ha−1. The cost of sugarcane production in Guangxi is much higher than in Brazil. One of the major factors of the high cost is high N-fertilization. Over 60% of the sugarcane fields are applied with urea at over 600 kg ha−1 yr−1 (32). In Brazil, the present sugarcane mean yields are also between 70 and 80 Mg ha−1, but N-fertilizer mean applications are between 60 and 70 kg N ha−1 yr−1 (50). 15N isotope assays have demonstrated that some Brazilian sugarcane varieties are able to obtain considerable nitrogen from biological nitrogen fixation (BNF; 25, 49, 50). A number of nitrogen-fixing bacteria have been isolated from the rhizosphere and interior of sugarcane plants, and have shown potential to fix N2 associated with sugarcane plants (5, 13, 35, 43).
BNF may help farmers to maintain sugarcane yields under reduced N-fertilization and develop environmentally benign sugarcane production in Guangxi. At present, little is known about the diversity and predominant population of nitrogen-fixing bacteria associated with the sugarcane plants growing in Guangxi. Recently, some nitrogen-fixing bacteria have been isolated from sugarcane plants grown in Guangxi (26, 28, 44, 52, 53) using NFb, JNFb and LGI-P media that were respectively used to isolate Azospirillum (10), Herbaspirillum (6) and Gluconacetobacter diazotrophicus (8); however, nitrogen-fixing bacteria belonging to the genera Azospirillum, Herbaspirillum, and Gluconacetobacter, which are predominantly associated with sugarcane plants in Brazil, have not been isolated. The diversity of nitrogen-fixing bacteria associated with sugarcane plants grown with high N-fertilization in Guangxi may be different from in Brazil.
The ROC22 cultivar is the main sugarcane cultivar, growing in over 60% of sugarcane-planting areas in Guangxi. It is sensitive to low nitrogen stress and requires at least 150 kg ha−1 urea fertilization at the seedling stage for tillering and elongation of the plant cane crops (23). The recommended dose of urea fertilization for plant cane crops at the seedling stage is 180 kg ha−1, 30% of urea fertilization for a season (46). Recent studies have shown that nitrogen-fixing bacterial strains isolated from other sugarcane cultivars are able to provide nitrogen to micropropagated ROC22 sugarcane seedlings via BNF and promote sugarcane growth (26, 29); however, neither indigenous nitrogen-fixing bacteria associated with ROC22 sugarcane plants nor their associative BNF under recommended N-fertilization have been investigated.
Here, we attempted to isolate a large number of nitrogen-fixing bacteria associated with ROC22 sugarcane plants, investigate their diversity and predominant affiliation, and evaluate their potential for plant-growth promotion and associative BNF under the recommended N-fertilization. We initially obtained 196 fast-growing isolates from rhizosphere soil and roots of ROC22 sugarcane plants grown in 14 production areas. Nitrogen-fixing isolates were screened using the acetylene reduction assay (ARA) and PCR amplification of the nifH gene encoding the iron protein of nitrogenase (55). We found that enterobacteria were predominant among the obtained nitrogen-fixing bacteria by analyzing their 16S rRNA gene (rrs) sequences. We further screened their plant growth-promoting activities, including the production of indole acetic acids (IAA) and siderophores, phosphate solubilization and ACC (1-aminocyclopropane-1-carboxylic acid) deamination. Finally, we chose two Enterobacter spp. strains isolated from the same ROC22 plant to inoculate micropropagated ROC22 sugarcane seedlings and investigate their plant growth-promoting and associative BNF activities under the recommended N-fertilization for ROC22 crops using the 15N isotope dilution technique.
Materials and Methods
Bacterial isolation
Root samples were taken from ROC22 sugarcane plants grown for five to eight months in the fields in 14 production areas (Table 1). Root systems of six sugarcane plants in each production area were dug out; bulk soil loosely adhering to the roots was shaken off; rhizosphere soil tightly adhering to the roots was suspended in autoclaved distilled water; roots were washed in sequence once by autoclaved distilled water, 70% (v/v) ethanol for 30 s, 0.1% (w/v) HgCl2 for 1 min and 70% (v/v) ethanol for 30 s, five times by autoclaved distilled water, and ground with autoclaved quartz sand and phosphate-buffered saline (40) with a mortar and pestle. Soil suspensions and root homogenates were 10-fold serially diluted using autoclaved distilled water. One hundred microliters of each suspension were spread on modified nitrogen-deficient Ashby’s agar medium (per liter contains 10 g sucrose, 0.2 g NaCl, 0.2 g KH2PO4, 0.2 g MgSO4·7H2O, 0.1 g CaSO4·2H2O, 5 g CaCO3, 15 g agar, pH 7.0) (3). After incubation at 30°C for 3–5 d, colonies with distinguished morphology were purified three times by streaking on Ashby’s agar. Purified isolates were maintained on Luria-Bertani (LB) agar (40). Their liquid LB cultures were stored with 15% (v/v) glycerol at −70°C.
Table 1.
Nitrogen-fixing bacterial isolates associated with sugarcane cultivar ROC22 grown in Guangxi
| Isolates | Isolation site | Isolation source | Genus affiliation | IAA production | Siderophore production | Phosphate solubilization | ACC deaminase |
|---|---|---|---|---|---|---|---|
| CZ150S | Chongzuo | soil | Klebsiella | + | + | + | − |
| CZ152S | Chongzuo | soil | Burkholderia | − | + | + | + |
| CZ186S | Chongzuo | soil | Klebsiella | + | + | + | − |
| GG164S | Guigang | soil | Klebsiella | + | + | + | − |
| HX148S | Hengxian | soil | Enterobacter | − | + | + | − |
| HX149S | Hengxian | soil | Klebsiella | + | + | + | − |
| LA16S | Longan | soil | Klebsiella | + | + | + | − |
| LC55S | Liuchen | soil | Klebsiella | + | + | + | − |
| NN145S | Nanning | soil | Enterobacter | − | + | + | − |
| NN169S | Nanning | soil | Klebsiella | + | + | + | − |
| PG132S | Pingguo | soil | Enterobacter | + | + | + | − |
| QZ25S | Qinzhou | soil | Enterobacter | − | + | + | − |
| QZ33S | Qingzhou | soil | Klebsiella | − | + | + | − |
| TD153S | Tiandong | soil | Klebsiella | + | + | + | − |
| YL34S | Yulin | soil | Enterobacter | − | + | + | − |
| YX115S | Yangxi | soil | Klebsiella | + | + | + | − |
| YX116S | Yangxi | soil | Klebsiella | + | + | + | − |
| YX117S | Yangxi | soil | Klebsiella | + | + | + | − |
| YX118S | Yangxi | soil | Klebsiella | + | + | + | − |
| DX120E | Daxin | root | Klebsiella | + | + | + | − |
| DX194E | Daxin | root | Klebsiella | + | − | + | − |
| GG41E | Guigang | root | Klebsiella | − | + | + | − |
| GG42E | Guigang | root | Klebsiella | − | + | + | − |
| GG49E | Guigang | root | Enterobacter | + | + | + | − |
| GG50E | Guigang | root | Klebsiella | − | + | − | − |
| GG53E | Guigang | root | Klebsiella | − | + | + | − |
| GG160E | Guigang | root | Klebsiella | + | + | + | − |
| GG165E | Guigang | root | Enterobacter | + | + | − | − |
| LC89E | Liucheng | root | Klebsiella | + | + | − | − |
| LZ83E | Liuzhou | root | Klebsiella | + | + | − | − |
| LZ84E | Liuzhou | root | Enterobacter | + | + | + | − |
| LZ87E | Liuzhou | root | Klebsiella | + | + | − | − |
| LA3E | Longan | root | Klebsiella | + | + | + | − |
| LA4E | Longan | root | Enterobacter | + | + | − | − |
| LA11E | Longan | root | Klebsiella | − | + | + | − |
| LA14E | Longan | root | Enterobacter | − | + | − | − |
| NN143E | Nanning | root | Enterobacter | − | + | + | − |
| NN144E | Nanning | root | Enterobacter | − | + | + | − |
| NN208E | Nanning | root | Enterobacter | − | + | + | − |
| PG122E | Pingguo | root | Klebsiella | + | + | + | − |
| QZ80E | Qinzhou | root | Enterobacter | + | + | − | − |
| SS107E | Shangsi | root | Klebsiella | + | + | + | − |
| SS82E | Shangsi | root | Klebsiella | − | + | − | − |
“+” presents positive, “−” presents negative
ARA
One milliliter of each isolate grown overnight in liquid LB medium was harvested, washed twice, and suspended in 10 mL of liquid Ashby’s medium in a 60-mL Erlenmeyer flask. After static incubation at 28°C for 24 h, the flask was sealed with a rubber stopper and then 5 mL (10%) gas volume in the flask was replaced with acetylene. After incubation for another 24 h, ethylene was detected with a Shimadzu GC-9A gas chromatograph (Shimadzu, Kyoto, Japan) equipped with a flame ionization detector and a column filled with GDX-502 (Borui Jianhe Chromatography Technology, Tianjin, China) (53). To ARA-negative in initial ARA screening but nifH-positive isolates of LA16S, DX194E and LC89E, each isolate was suspended in modified liquid Ashby’s medium supplemented with 0.05% (w/v) yeast extract in flasks at 28°C; ARA was performed every 2 h on bacterial cultures during a 24 h growth period.
Colony PCR
A colony approximately 1 mm in diameter grown on LB agar was picked up with an autoclaved 10-μL pipette tip and transferred to 10 μL sterilized Millipore water in a PCR tube. The bacterial suspension was heated in a P7021TP-6 microwave oven (Galanz, Foshan, China) at full power for 3 min. After centrifugation, 1 μL of the bacterial lysate was used as the template for PCR. The G. diazotrophicus strain PAL5 isolated from sugarcane plants (13) and the Escherichia coli strain DH5α were respectively used as positive and negative controls for nifH amplification.
Amplification of partial nifH sequences was performed with the degenerate Z-primers (55). Amplification of near full-length rrs sequences was performed with universal 27F/1492R primers (22). The primers were synthesized by Sangon (Sangon Biotech, Shanghai, China) and Takara (Takara Biotechnology, Dalian, China). The primers synthesized by Sangon were purified by ULTRAPAGE and determined by mass spectrometry. The primers synthesized by Takara were purified by HPLC. The Z-primers were used at 2 μM. The 27F/1492R primers were used at 0.25 μM. Ready-to-use 2×concentrated PCR masters (0.1 U μL−1 Taq DNA polymerase, 0.2 mM dNTP, 3 mM MgCl2, 2×PCR buffer) produced by Sangon and Tiangen (Tiangen Biotech, Beijing, China) were used for reactions. PCR amplification was performed in a PTC-200 DNA Engine thermal cycler and an S1000 thermal cycler (Bio-Rad Laboratories, Hercules, CA, USA). A touchdown PCR strategy was used for nifH amplification to improve amplification specificity. In the 20 touchdown cycles, the annealing temperature was decreased by 0.5°C every cycle from 67 to 57°C. Fifteen additional cycles were performed at the annealing temperature of 57°C. PCR products were electrophoresed on agarose gels, stained with ethidium bromide, visualized and recorded with a JS-680B Gel Documentation and Analysis System (Shanghai Peiqing Science and Technology, Shanghai, China), and compared with molecular weight markers (Takara).
Cloning and sequencing of nifH and rrs fragments
The expected amplicons were excised from agarose gels, purified by a TIANgel Midi Purification Kit (Tiangen), cloned into the pMD18-T vector (Takara) and screened as described by the manufacturers. Three to six clones containing inserts of the correct size for each amplicon were chosen for sequencing (Invitrogen, Carlsbad, CA, USA).
Sequence analysis
The sequence segments corresponding to Z-primers were removed from the amplified nifH sequences. The partial nifH sequences were translated using the MEGA 4.0 program (45). The deduced amino acid sequences were aligned using the MUSCLE program (11) implemented in the MEGA 4.0 program. The rrs sequences were screened with the Mallard program (4) and BLASTed (1). The rrs sequences of enterobacteria were aligned with those from the type strains of the species belonged to the genera Enterobacter and Klebsiella in the List of Prokaryotic names with Standing in Nomenclature (http://www.bacterio.cict.fr/m/microbacterium.html) using the MUSCLE program. Phylogenetic analysis was performed using the PhyML 3.0 program based on the maximum-likelihood principle (16). The best-fit TIM1+G model of nucleotide substitution was estimated using the jModelTest program based on the Akaike information criterion (38). The branch support measurement was assessed by a nonparametric, Shimodaira-Hasegawa-like test implemented in the PhyML 3.0 program.
Determination of IAA production, siderophore production, phosphate solubilization, and ACC deaminase activity
IAA production from a 3-d culture in DF medium (36) supplemented with 0.02% (w/v) L-tryptophan was determined by a microplate colorimetric assay (41). Siderophore production was determined by the Chrome Azurol S plate assay (42). Mineral phosphate solubilization activity was determined using Pikovskaya’s agar medium containing 0.5% (w/v) Ca3(PO4)2 (37). Bacterial utilization of ACC was screened using a colorimetric assay of ACC based on the ninhydrin reaction (24); bacterial ACC deaminase activity was measured as described by Penrose and Glick (2003) (36).
Inoculation and acclimatization of micropropagated sugarcane plantlets
Sugarcane micropropagated plantlets were developed from stem apical meristems of cultivar ROC22 (51). Two nitrogen-fixing Enterobacter spp. strains of NN145S and NN143E isolated from rhizosphere soil and surface-sterilized roots, respectively, of the same ROC22 plant were used to inoculate micropropagated plantlets. Both strains were also able to produce siderophores and solubilize Ca3(PO4)2. Inoculation and acclimatization of micropropagated plantlets were performed as described by Lin et al. (26). Rooted plantlets were co-cultured with bacteria in liquid one-tenth MS medium (sucrose and basal salt mixture) (39). Initial density of the bacteria was approximately 2.0×105 cells per milliliter medium. Plantlets without inoculation were prepared as the control. Seven days after inoculation, plantlets were transferred to autoclaved sands and acclimatized for 14 d (26).
Counting of NN145S and NN143E cells colonizing sugarcane plantlets
Seven days after inoculation, plantlets were briefly washed with autoclaved distilled water, dried with autoclaved tissue paper, weighed, and ground with autoclaved quartz sand and phosphate-buffered saline in a mortar and pestle. The homogenates were serially diluted and spread on Ashby’s agar. Bacterial colonies were counted after incubation at 30°C for 3 d.
15N isotope dilution assay
Sand and perlite were mixed 1:1 (v:v), autoclaved twice at 121°C for 2 h, and cooled for 1 d at room temperature after each autoclave. The soil mixtures were added to 10 mg (NH4)2SO4 (10.11 atom % 15N excess) per kilogram wet weight of soil mixture (kg−1 soil) and mixed thoroughly twice daily for 2 weeks prior to planting to ensure uniform distribution of 15N isotope (19). Two and a half kilograms of 15N-labeled soil mixtures filled each autoclaved 2.5-L plastic pot. Each pot was then planted with three acclimatized micropropagated sugarcane seedlings. Six seedlings for each treatment were fertilized by adding 200 mL of one-tenth MS basal salt mixture to each pot 5 d after transplant. As a result, 8.82 mg N kg−1 soil including 15N isotope was added. Oliveira et al. (35) estimated that 10 mg (NH4)2SO4 (2.1 mg N) kg−1 soil is equivalent to 20 kg N ha−1. The N-fertilization of 8.82 mg N kg−1 soil is thus equivalent to 84 kg N ha−1 or 180 kg urea ha−1. The seedlings were grown under a 14-h light period with approximately 100 μmol m−2 s−1 photon flux density at 26±2°C, and watered as needed with autoclaved distilled water. Fifty-five days after transplant, plant roots were washed by distilled water to remove the attached soil mixtures. Roots and shoots were separated, freeze-dried, weighed, and ground. Ten milligrams of ground root and shoot materials were respectively analyzed for 15N isotope content by a Delta Plus AD EA-IRMS Mass Spectrometer (Thermo Fisher Scientific, Waltham, MA, USA). The contribution of nitrogen derived from air (Ndfa) was estimated by the equation: %Ndfa=100×(1−[atom % 15N excess inoculated plant/atom % 15N excess control plant]) (19). Data plotting and statistical T test were carried out using Microsoft Excel 2007. The confidence intervals were 95% (α=0.05).
Nucleotide sequence accession numbers
The partial nifH, anfH (encoding the iron protein of Fe-only alternative nitrogenase; 20) and rrs sequences were deposited in GenBank under accession numbers HQ204222 to HQ204264, HQ204265 to HQ204276, and HQ204277 to HQ204319, respectively (Table S1).
Results
Bacterial isolation and ARA screening
Bacterial colonies grown well on nitrogen-deficient Ashby’s agar generally appeared from 10-fold diluted soil suspensions and root homogenates and were selected for purification. One hundred ninety-six fast-growing isolates were obtained. Acetylene reduction activity in liquid nitrogen-deficient Ashby’s medium was repeatedly detected from 18 isolates obtained from rhizosphere soil and 22 isolates obtained from surface-sterilized roots.
PCR amplification and sequencing confirmation of partial nifH genes
PCR amplification of nifH (nifH PCR) based on Z-primers amplified approximately 360 bp fragments from all 40 ARA-positive isolates (Fig. S1A). Cloning and sequencing of the amplicons showed that all 40 ARA-positive isolates contained sequences that were the closest match to nifH (Table S1). In addition, sequences matching anfH encoding the iron protein of Fe-only alternative nitrogenase (20) were obtained from 12 isolates (Table S1). The conserved residues of C86, C98, R101, D126, D130, and C133 (Azotobacter vinelandii OP NifH numbering; GenBank accession number AAA64709) for nitrogenase iron protein were present in all the obtained partial NifH and AnfH sequences (data not shown).
Application of Z-primer-based colony nifH PCR
In the initial screening of the 196 bacterial isolates, ARA did not detect nitrogenase activity from 156 isolates; however, ARA may miss true nitrogen-fixing bacteria whose nitrogenase activities would be induced under more favorable growth conditions or at more active growth stages. To verify this assumption, we used Z-primer-based colony nifH PCR for the 156 ARA-negative isolates. Positive amplification was obtained from three isolates of LA16S, DX194E, and LC89E (Fig. S1B). Subsequent cloning and sequencing confirmed that the three isolates contained nifH (Table S1). We further performed ARA on the three isolates grown in liquid Ashby’s medium supplemented with 0.05% (w/v) yeast extract. Bacterial growth was promoted by yeast extract and reached the exponential phase after 4 h. Weak acetylene reduction activity was detected from the cultures of three isolates grown for 8–16 h (data not shown); therefore, Z-primer-based colony nifH PCR is able to rapidly screen nifH-positive isolates from a large number of bacterial isolates and largely reduce the number of isolates for further ARA determination.
IAA production, siderophore production, phosphate solubilization and ACC deaminase activity
Among the 43 nitrogen-fixing bacterial isolates, 42 isolates produced siderophores; 34 isolates including 19 isolates obtained from rhizosphere soil and 15 isolates obtained from roots were able to dissolve Ca3(PO4)2; 27 isolates including 13 isolates obtained from rhizosphere soil and 14 isolates obtained from roots produced IAA; one isolate CZ152S displayed ACC deaminase activity (24); 19 isolates showed two of the four tested plant growth-promoting activities; 21 isolates showed three of the four activities (Table 1).
Analysis of rrs sequences
Approximately 1,500 bp of rrs sequences for all 43 nitrogen-fixing isolates were obtained by PCR amplification and confirmed by subsequent cloning and sequencing. Sequence BLASTing suggested that 28 isolates belonged to the genus Klebsiella, 14 isolates belonged to the genus Enterobacter, and isolate CZ152S belonged to the genus Burkholderia (Table 1 and S1). The rrs sequence of CZ152S showed the highest sequence similarity to those of B. cepacia strain LMG 12614 (99.8%) and B. cenocepacia strain J2315 (99.8%). The phylogenetic tree of the rrs sequences of the 28 Klebsiella spp. isolates, 14 Enterobacter isolates, and the type strains of the Klebsiella and Enterobacter species are shown in Fig. 1.
Fig. 1.
Phylogenetic tree based on 16S rRNA gene sequences for nitrogen-fixing enterobacteria associated with sugarcane cultivar ROC22 and the type strains of the species belonging to the genera Enterobacter and Klebsiella. Accession numbers of the 16S rRNA genes in the GenBank database are given in parentheses. The tree was generated by the maximum-likelihood principle using the PhyML 3.0 program. The TIM1+G model was used. Branch support measure was assessed by a Shimodaira-Hasegawa-like test and values >50 are indicated at the nodes. Bar, 0.01 nucleotide substitutions per site.
Inoculation effects on micropropagated sugarcane seedlings
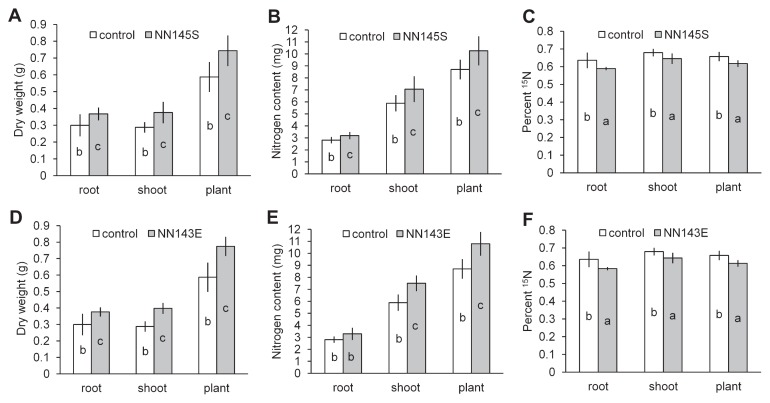
Seven days after inoculation, approximately 6.5×107 NN145S cells and 8.0×107 NN143E cells per gram of the micropropagated sugarcane plantlets were counted, whereas no bacterial colonies were formed from uninoculated controls. After 14 d acclimatization without N-fertilization and 55 d growth with 8.82 mg N kg−1 soil including 15N isotope, inoculated sugarcane seedlings showed higher dry weights and nitrogen contents than those of uninoculated controls (Fig. 2). Inoculation of strain NN145S increased the dry weights of roots, shoots, and whole seedlings at 22.7%, 30.6%, and 26.7%, respectively (Fig. 2A), and increased nitrogen contents of roots, shoots, and whole seedlings at 13.7%, 20.0%, and 17.9%, respectively (Fig. 2B); all the increases were statistically significant at the 95% confidence level (Fig. 2A and B). Inoculation of strain NN143E increased dry weights of roots, shoots, and whole seedlings at 25.6%, 38.4%, and 31.9%, respectively (Fig. 2D), and increased nitrogen contents of roots, shoots, and whole seedlings at 17.2%, 27.4%, and 24.0%, respectively (Fig. 2E); the increases were statistically significant at the 95% confidence level, except that the increase of the root nitrogen content was significant at the 90% confidence level (Fig. 2D and E). Moreover, the 15N isotope concentrations of the inoculated plants were significantly lower than those of the uninoculated controls (Fig. 2C and F) as a result of dilution by N2 fixation. Seedlings inoculated with NN145S received 7.3%, 5.0%, and 6.1% nitrogen in roots, shoot, and whole plants from N2, respectively; seedlings inoculated with NN143E received 8.3%, 5.3%, and 6.7% nitrogen in roots, shoot, and whole plants from N2, respectively. The dry weight (0.77 g plant−1), nitrogen content (10.79 mg plant−1), and %Ndfa (6.7) of sugarcane seedlings inoculated with NN143E were respectively higher than those (0.74 g plant−1, 10.26 mg plant−1, and 6.1) of seedlings inoculated with NN145S but were not statistically significant.
Fig. 2.
Inoculation effects of Enterobacter sp. NN145S (A, B, C) and NN143E (D, E, F) on micropropagated sugarcane ROC22 seedlings. (A, D) dry weight, (B, E) nitrogen content, and (C, F) percent 15N of roots, shoots, and whole seedlings are presented for comparison of inoculated seedlings with uninoculated controls. The columns represent the mean of the data for each treatment. Bars represent the standard error. Different letters within the columns indicate significant differences between the treatments at the 95% confidence level.
Discussion
This initial large-scale isolation of nitrogen-fixing bacteria from root samples of the main sugarcane cultivar ROC22 grown in Guangxi obtained a limited number of nitrogen-fixing isolates that showed limited diversity and predominantly belonged to the genera Klebsiella and Enterobacter. Previous studies showed that high levels of N-fertilization reduced the bacterial cell number (12, 34) and genetic diversity (7) of G. diazotrophicus in sugarcane plants. The high levels of N-fertilization in the sugarcane fields in Guangxi may act as a selective factor to reduce the population and diversity of nitrogen-fixing bacteria.
Enterobacteria are seemingly predominant in the fast-growing culturable nitrogen-fixing bacteria associated with ROC22 sugarcane plants growing in Guangxi. On the one hand, the selective nitrogen-deficient Ashby’s medium used in the isolation procedure may lead to the predominant isolation of enterobacteria. On the other hand, nitrogen-fixing enterobacteria have been isolated from sugarcane plants cultivated in Guangxi using NFb and JNFb media (28, 44, 53) and also isolated from sugarcane plants cultivated in other countries (15, 27, 31, 33, 48). Mehnaz et al. (31) found that enterobacteria were predominant among the nitrogen-fixing bacteria isolated from sugarcane plants in Pakistan. Taulé et al. (48) found that enterobacteria formed a major group among the endophytic nitrogen-fixing bacteria isolated from sugarcane plants in Uruguay. Magnani et al. (30) showed that enterobacteria, including three nitrogen-fixing isolates, comprised a major group of endophytic bacteria isolated from Brazilian sugarcane using potato agar medium. Moreover, the detection of nifH sequences in sugarcane plants grown in Japan without culture showed that nifH sequences homologous to those of enterobacteria were predominant (2). The frequent association between nitrogen-fixing enterobacteria and sugarcane crops may develop from the long use of organic manures in agriculture (21).
Recent studies have shown that nitrogen-fixing enterobacteria can fix N2 associated with sugarcane plants and promote sugarcane growth. Luo et al. (29) showed that a Klebsiella sp. strain fixed N2 and increased nitrogen content of the ROC22 seedlings under gnotobiotic conditions. Wu et al. (52) showed that a nitrogen-fixing Pantoea sp. strain promoted the growth of sugarcane plants grown in non-sterile sand. Govindarajan et al. (15) showed that a nitrogen-fixing Klebsiella sp. strain increased the biomass and nitrogen content of sugarcane plants grown in non-sterile soil. Mirza et al. (33) showed that a Klebsiella sp. isolate SC20 (identified on the basis of its rrs sequence [GenBank accession number AJ278447]) fixed N2 and increased the nitrogen content of sugarcane plants under gnotobiotic conditions. Here, we showed that two Enterobacter spp. strains provided nitrogen to sugarcane plants via BNF and promoted sugarcane growth under gnotobiotic conditions and relatively high N-fertilization (equivalent to 180 kg urea ha−1) recommended for sugarcane plant crops at the seedling stage in Guangxi. To our knowledge, this is the first report that indigenous nitrogen-fixing Enterobacter spp. strains can fix N2 associated with the hosts of sugarcane plants and promote their growth.
Using the method of introducing endophytic bacteria into the rooted micropropagated sugarcane plantlets established by Reis et al. (39), both the rhizosphere soil isolate NN145S and the root isolate NN143E obtained from the same ROC22 sugarcane plant were able to heavily colonize the ROC22 seedlings at respective densities of 6.5×107 and 8.0×107 cells per gram fresh weight. It would be interesting to study their colonization patterns, interactions and coordinated BNF contributions after coinoculation into the host ROC22 plant.
ARA is a sensitive method to detect nitrogenase activity of microbial cultures and has been widely used to identify nitrogen-fixing isolates (17). It is also known that bacterial nitrogenase activities vary with the media, culture conditions, and growth stages; therefore, using ARA to screen diazotrophs from a large number of isolates may miss true diazotrophs in the silent state of N2 fixation and is rather time-consuming. Here, we have shown that colony nifH PCR can circumvent the disadvantages of ARA and rapidly screen nitrogen-fixing bacteria from a large number of isolates. Firstly, nifH PCR amplification based on Z-primers is sensitive and specific for the identification of ARA-positive isolates belonging to broad taxonomic classes (9, 18, 47). Demba Diallo et al. (9) have revealed that Z-primers have the highest rate of match to nifH sequences in the databases among the widely used universal nifH primers. Moreover, Z-primer-based PCR can amplify the corresponding fragments of nifH, vnfH, and anfH (9, this study). Secondly, colony nifH PCR skipping DNA extraction is easy to perform, rapid, reproducible, and not related to bacterial growth stages. Commercially available reagents, such as the ready-to-use 2×concentrated PCR masters, can facilitate PCR manipulation. In a modern microbiology laboratory, a thermal cycler for PCR is usually more available than a gas chromatograph for ARA. Using colony nifH PCR for the initial screening can largely reduce the number of isolates for further ARA determination; however, great care should be taken regarding the contamination of primers and other PCR reagents by trace levels of nifH (14, 54).
In conclusion, colony nifH PCR followed by ARA determination of nifH-positive isolates enables highly efficient workflow to screen and identify diazotrophs. The initial large-scale isolation of nitrogen-fixing bacteria from ROC22 sugarcane root samples obtained predominantly nitrogen-fixing enterobacteria that possessed multiple plant growth-promoting activities of IAA production, siderophore production, or phosphate solubilization. The initial inoculation of two Enterobacter spp. strains showed the potential for nitrogen-fixing enterobacteria to provide nitrogen via BNF to sugarcane cultivars requiring a high level of N-fertilization and increase sugarcane production in fields with relative high N-fertilization in Guangxi.
Supplementary Material
Acknowledgements
This work was supported by the Natural Science Foundation of China (Grant No. 30660085), Zhejiang Provincial Natural Science Foundation of China (Grant No. Y307143), Qianjiang Talents Project of the Science and Technology Department of Zhejiang Province (Grant No. 2007R10037), and the Science Foundation of Chinese University (Grant No. 2009QNA6030).
References
- 1.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ando S, Goto M, Meunchang S, Thongra-ar P, Fujiwara T, Hayashi H, Yoneyama T. Detection of nifH sequences in sugarcane (Saccharurn officinarum L.) and pineapple (Ananas comosus[L.] Merr.) Soil Sci Plant Nutr. 2005;51:303–308. [Google Scholar]
- 3.Ashby SF. Some observations on the assimilation of atmospheric nitrogen by a free living soil organism—Azotobacter chroococcum of Beijerinck. J Agric Sci. 1907;2:35–51. [Google Scholar]
- 4.Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ. New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Appl Environ Microbiol. 2006;72:5734–5741. doi: 10.1128/AEM.00556-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baldani JI, Baldani VLD. History on the biological nitrogen fixation research in graminaceous plants: special emphasis on the Brazilian experience. An Acad Bras Cienc. 2005;77:549–579. doi: 10.1590/s0001-37652005000300014. [DOI] [PubMed] [Google Scholar]
- 6.Baldani VLD, Baldani JI, Olivares FL, Döbereiner J. Identification and ecology of Herbaspirillum seropedicae and closely related Pseudomonas rubrisubalbicans. Symbiosis. 1992;13:65–73. [Google Scholar]
- 7.Caballero-Mellado J, Fuentes-Ramirez LE, Reis VM, Martinez-Romero E. Genetic structure of Acetobacter diazotrophicus populations and identification of a new genetically distant group. Appl Environ Microbiol. 1995;61:3008–3013. doi: 10.1128/aem.61.8.3008-3013.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cavalcante VA, Döbereiner J. A new acid-tolerant nitrogen-fixing bacterium associated with sugarcane. Plant Soil. 1988;108:23–31. [Google Scholar]
- 9.Demba Diallo M, Reinhold-Hurek B, Hurek T. Evaluation of PCR primers for universal nifH gene targeting and for assessment of transcribed nifH pools in roots of Oryza longistaminata with and without low nitrogen input. FEMS Microbiol Ecol. 2008;65:220–228. doi: 10.1111/j.1574-6941.2008.00545.x. [DOI] [PubMed] [Google Scholar]
- 10.Döereiner J, Marriel IE, Nery M. Ecological distribution of Spirillum lipoferum Beijerinck. Can J Microbiol. 1976;22:1464–1473. doi: 10.1139/m76-217. [DOI] [PubMed] [Google Scholar]
- 11.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuentes-Ramírez LE, Caballero-Mellado J, Sepúlveda J, Martínez-Romero E. Colonization of sugarcane by Acetobacter diazotrophicus is inhibited by high N fertilization. FEMS Microbiol Ecol. 1999;29:117–128. [Google Scholar]
- 13.Gillis M, Kersters K, Hoste B, et al. Acetobacter diazotrophicus sp. nov., a nitrogen-fixing acetic acid bacterium associated with sugarcane. Int J Syst Bacteriol. 1989;39:361–364. [Google Scholar]
- 14.Goto M, Ando S, Hachisuka Y, Yoneyama T. Contamination of diverse nifH and nifH-like DNA into commercial PCR primers. FEMS Microbiol Lett. 2005;246:33–38. doi: 10.1016/j.femsle.2005.03.042. [DOI] [PubMed] [Google Scholar]
- 15.Govindarajan M, Kwon S-W, Weon H-Y. Isolation, molecular characterization and growth-promoting activities of endophytic sugarcane diazotroph Klebsiella sp. GR9. World J Microbiol Biotechnol. 2007;23:997–1006. [Google Scholar]
- 16.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 17.Hardy RWF, Burns RC, Holsten RD. Applications of the acetylene-ethylene assay for measurement of nitrogen fixation. Soil Biol Biochem. 1973;5:47–81. [Google Scholar]
- 18.Hou W, Peng G-X, Xu Z-J, Chen S-X, Tan Z-Y. Diversity of endophytic diazotrophs isolated from Bambusa blumeana in Guangdong Province. J Agric Biotechnol. 2007;15:290–294. (in Chinese, English abstract available) [Google Scholar]
- 19.Iniguez AL, Dong Y, Triplett EW. Nitrogen fixation in wheat provided by Klebsiella pneumonia 342. Mol Plant Microbe Interact. 2004;17:1078–1085. doi: 10.1094/MPMI.2004.17.10.1078. [DOI] [PubMed] [Google Scholar]
- 20.Joerger RD, Jacobson MR, Premakumar R, Wolfinger ED, Bishop PE. Nucleotide sequence and mutational analysis of the structural genes (anfHDGK) for the second alternative nitrogenase from Azotobacter vinelandii. J Bacteriol. 1989;171:1075–1086. doi: 10.1128/jb.171.2.1075-1086.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kennedy IR, Choudhury ATMA, Kecskés ML. Nonsymbiotic bacterial diazotrophs in crop-farming systems: can their potential for plant growth promotion be better exploited? Soil Biol Biochem. 2004;36:1229–1244. [Google Scholar]
- 22.Lane DJ. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic Acid Techniques in Bacterial Systematics. John Wiley & Sons; New York: 1991. pp. 115–175. [Google Scholar]
- 23.Li X, Wang L, Fang F, et al. The effects of low nitrogen stress on different sugarcane genotypes. Chin Agric Sci Bull. 2011;27:208–211. (in Chinese, English abstract available) [Google Scholar]
- 24.Li Z, Chang S, Lin L, Li Y, An Q. A colorimetric assay of 1-aminocyclopropane-1-carboxylate (ACC) based on ninhydrin reaction for rapid screening of bacteria containing ACC deaminase. Lett Appl Microbiol. 2011;53:178–185. doi: 10.1111/j.1472-765X.2011.03088.x. [DOI] [PubMed] [Google Scholar]
- 25.Lima E, Boddey RM, Döbereiner J. Quantification of biological nitrogen fixation associated with sugar cane using a 15N-aided nitrogen balance. Soil Biol Biochem. 1987;19:165–170. [Google Scholar]
- 26.Lin L, Guo W, Xing Y, et al. The actinobacterium Microbacterium sp. 16SH accepts pBBR1-based pPROBE vectors, forms biofilms, invades roots, and fixes N2associated with micropropagated sugarcane plants. Appl Microbiol Biotechnol. 2012;93:1185–1195. doi: 10.1007/s00253-011-3618-3. [DOI] [PubMed] [Google Scholar]
- 27.Loiret FG, Ortega E, Kleiner D, Ortega-Rodés P, Rodés R, Dong Z. A putative new endophytic nitrogen-fixing bacterium Pantoea sp. from sugarcane. J Appl Microbiol. 2004;97:504–511. doi: 10.1111/j.1365-2672.2004.02329.x. [DOI] [PubMed] [Google Scholar]
- 28.Luo T, Ouyang X, Yang L, Li Y. An endophytic nitrogen-fixing bacterium Klebsiella sp. strain L03 from sugarcane: isolation, identification and characterization. Chin. J. Trop Crops. 2010;31:972–978. (in Chinese, English abstract available). [Google Scholar]
- 29.Luo T, Ouyang X, Yang L, Li Y. Effect of nitrogen-fixing bacteria inoculation on biological nitrogen fixation in sugarcane by 15N isotope dilution technique. J Nucl Agric Sci. 2010;24:1026–1031. (in Chinese, English abstract available) [Google Scholar]
- 30.Magnani GS, Didonet CM, Cruz LM, Picheth CF, Pedrosa FO, Souza EM. Diversity of endophytic bacteria in Brazilian sugarcane. Genet Mol Res. 2010;9:250–258. doi: 10.4238/vol9-1gmr703. [DOI] [PubMed] [Google Scholar]
- 31.Mehnaz S, Baig DN, Lazarovits G. Genetic and phenotypic diversity of plant growth promoting rhizobacteria isolated from sugarcane plants growing in Pakistan. J Microbiol Biotechnol. 2010;20:1614–1623. doi: 10.4014/jmb.1005.05014. [DOI] [PubMed] [Google Scholar]
- 32.Meng S. Status quo, problems and countermeasures of sugarcane fertilization in Guangxi. J Guangxi Agric. 2007;22(5):37–39. (in Chinese, English abstract available) [Google Scholar]
- 33.Mirza MS, Ahmad W, Latif F, Haurat J, Bally R, Normand P, Malik KA. Isolation, partial characterization, and the effect of plant growth-promoting bacteria (PGPB) on micropropagated sugarcane in vitro. Plant Soil. 2001;237:47–54. [Google Scholar]
- 34.Muthukumarasamy R, Revathi G, Lakshminarasimhan C. Influence of N fertilisation on the isolation of Acetobacter diazotrophicus and Herbaspirillum spp. from Indian sugarcane varieties. Biol. Fertil Soils. 1999;29:157–164. [Google Scholar]
- 35.Oliveira ALM, Urquiaga S, Döbereiner J, Baldani JI. The effect of inoculating endophytic N2-fixing bacteria on micropropagated sugarcane plants. Plant Soil. 2002;242:205–215. [Google Scholar]
- 36.Penrose DM, Glick BR. Methods for isolating and characterizing ACC deaminase-containing plant growth-promoting rhizobacteria. Physiol Plant. 2003;118:10–15. doi: 10.1034/j.1399-3054.2003.00086.x. [DOI] [PubMed] [Google Scholar]
- 37.Pikovskaya RI. Mobilization of phosphorus in soil in connection with vital activity of some microbial species. Mikrobiologiya. 1948;17:362–370. [Google Scholar]
- 38.Posada D. jModelTest: phylogenetic model averaging. Mol Biol Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 39.Reis VM, Olivares FL, Oliveira ALM, Reis FB, Jr, Baldani JI, Döbereiner J. Technical approaches to inoculate micropropagated sugarcane plants with Acetobacter diazotrophicus. Plant Soil. 1999;206:205–211. [Google Scholar]
- 40.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3rd ed. Cold Spring Harbor Laboratory Press; New York: 2001. [Google Scholar]
- 41.Sarwar M, Kremer RJ. Determination of bacterially derived auxins using a microplate method. Lett Appl Microbiol. 1995;20:282–285. [Google Scholar]
- 42.Schwyn B, Neilands JB. Universal chemical assay for the detection and determination of siderophores. Anal Biochem. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- 43.Sevilla M, Burris RH, Gunapala N, Kennedy C. Comparison of benefit to sugarcane plant growth and 15N2incorporation following inoculation of sterile plants with Acetobacter diazotrophicus wild-type and Nif−mutant strains. Mol Plant Microbe Interact. 2001;14:358–366. doi: 10.1094/MPMI.2001.14.3.358. [DOI] [PubMed] [Google Scholar]
- 44.Su J-B, Yang R-Z, Gui Y-Y, Li Y-R, Lei X-T, Zhang H-L. Isolation, identification and characteristics of endophytic bacteria with nitrogenase from sugarcane. Southwest China J Agric Sci. 2007;20:1055–1059. (in Chinese, English abstract available) [Google Scholar]
- 45.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 46.Tan H, Zhou L, Xie R, Huang M, Huang J. Sugarcane nutrition and fertilization. In: Li Y, editor. Current Knowledge of Sugarcane. Chinese Agriculture Publishing; Beijing: 2010. pp. 218–263. (in Chinese) [Google Scholar]
- 47.Tan Z, Peng G, Xu P, Ai S, Tang S, Zhang G, Zeng F. Diversity and high nitrogenase activity of endophytic diazotrophs isolated from Oryza rufipogon Griff. Chin Sci Bull. 2009;54:2839–2848. [Google Scholar]
- 48.Taulé C, Mareque C, Barlocco C, Hackembruch F, Reis VM, Sicardi M, Battistoni F. The contribution of nitrogen fixation to sugarcane (Saccharum officinarum L.), and the identification and characterization of part of the associated diazotrophic bacterial community. Plant Soil. 2012;356:35–49. [Google Scholar]
- 49.Urquiaga S, Cruz KHS, Boddey RM. Contribution of nitrogen fixation to sugar cane: Nitrogen-15 and nitrogen-balance estimates. Soil Sci Soc Am J. 1992;56:105–114. [Google Scholar]
- 50.Urquiaga S, Xavier RP, de Morais RF, et al. Evidence from field nitrogen balance and 15N natural abundance data for the contribution of biological N2 fixation to Brazilian sugarcane varieties. Plant Soil. 2012;356:5–21. [Google Scholar]
- 51.Wang L-W, Li Y-R, He W-Z, Xian W, Liang J, Tan Y-M. The detection of activity for endophytic nitrogen fixing bacteria in sugarcane (Saccharrum officinarum L.) by stem apical culture seedlings. Plant Physiol Commun. 2007;43:65–68. (in Chinese, English abstract available) [Google Scholar]
- 52.Wu K-C, Liang J, Luo T, Xing Y-X, Yang L-T, Li Y-R. Effects of inoculating nitrogen fixing bacteria on agronomic characters in different sugarcane genotypes. Guangxi Agric Sci. 2010;41:528–530. (in Chinese, English abstract available) [Google Scholar]
- 53.Xing Y-X, Yang L-T, Huang S-L, Li Y-R. The 16S rRNA gene sequences and growth characteristics of two nitrogen fixing endobacteria strains from sugarcane stalk. Microbiol China. 2008;35:1732–1737. (in Chinese, English abstract available). [Google Scholar]
- 54.Zehr JP, Crumbliss LL, Church MJ, Omoregie EO, Jenkins BD. Nitrogenase genes in PCR and RT-PCR reagents: implications for studies of diversity of functional genes. Biotechniques. 2003;35:996–1002. 1004–1005. doi: 10.2144/03355st08. [DOI] [PubMed] [Google Scholar]
- 55.Zehr JP, McReynolds LA. Use of degenerate oligonucleotides for amplification of the nifH gene from the marine cyanobacterium Trichodesmium thiebautii. Appl Environ Microbiol. 1989;55:2522–2526. doi: 10.1128/aem.55.10.2522-2526.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.