Abstract
Rapidly growing mycobacteria (RGM) inhabit soil and water but certain strains represent a health risk for human and animals. Both clinical and soil RGM may be under selection pressure for resistance to tetracycline (TET) antibiotics, since tetracyclines are administrated to humans and farm animals, and TET residues enter soil through manuring; however, resistance to TET and the presence of TET-resistance genes have been assessed only in clinical isolates. We were therefore interested in comparing soil and clinical RGM in terms of TET resistance and the presence of TET-resistance genes. We used 44 RGM from grasslands with different exposure to animal manure, and 38 clinical RGM from Czech hospitals. There was no difference between the clinical and soil isolates in TET resistance, with >50% resistant isolates in both groups. otr(A), otr(B), tet(K), tet(L) or tet(M) were not detected in any soil or clinical isolate. In contrast, most isolates harbored tet(V) and tap, both encoding mycobacterial efflux pumps, including species where these genes have never been evidenced before. The phylogeny of tet(V) correlated with isolates’ BOX-PCR profiles, suggesting that this gene evolved along with mycobacterial genomes as a part of the intrinsic resistome. In certain cases, tet(V) and/or tap were found in TET-sensitive isolates, or inversely, were not found in resistant strains. Concluding, intrinsic efflux pumps may be more important for TET resistance than horizontally transferred genes in both soil and clinical RGM. Their simple presence, however, does not attest to resistance, and therefore their diversity, function and expression merit further research.
Keywords: efflux pump, rapidly growing Mycobacterium, tetracycline resistance, tap, tet(V)
Rapidly growing mycobacteria (RGM) are common inhabitants of soil and water. In past decades, they have been increasingly recognized as a cause of human and animal diseases (15, 25). These include respiratory infections, a spectrum of hard and soft tissue infections or bacteremia in immunocompromised patients, as well as infections related to injuries in healthy individuals (8, 29, 48). The natural and man-transformed environment is an important reservoir of RGM, from which transmission to humans occurs (50). Although the direct relationship between an environmental source and clinical disease is difficult to evidence, several RGM outbreaks have been associated with the exposure to soil and water (2, 16), including drinking water (49).
The prevention and treatment of infections due to RGM is not trivial because of their high resistance to disinfectants and antibiotics. One group of antibiotics used in the therapy of RGM infections is the tetracyclines (TET) such as doxycycline and minocycline (15). Mycobacteria in general have intrinsic resistance to many antibiotics ensured by the composition of their cell wall and the presence of several multidrug efflux pumps (33, 36). Tetracycline/multidrug efflux pumps Tet(V) and Tap may belong to this intrinsic resistome as they have been so far found only in certain RGM species (17, 22).
Acquired resistance is not often seen in Mycobacterium; however, Pang et al. (38) reported four genes conferring TET resistance in clinical RGM that have been most probably horizontally transferred from other bacteria. These included otr(A) and otr(B), self-protection genes from the oxytetracycline producer Streptomyces rimosus (9), and tet(K) and tet(L), low-G+C-content genes encoding tetracycline efflux pumps, which are typically found in Firmicutes (Streptococcus, Staphylococcus, Enterococcus) but also in some Gram-negative bacteria (40). More recently, tet(M), a widely distributed gene with a low G+C content encoding a ribosomal protection protein, was found in a human-associated Mycobacterium sp. (41).
Resistance to TET and the presence of TET-resistance genes in RGM have been studied in clinical isolates only; however, soil is an important reservoir of TET resistance genes, both indigenous (14) and introduced by manuring (10). In addition, tetracycline residues are detectable in manured soil (10), which may help select resistant strains. Soildwelling RGM may therefore represent an important pool of TET resistance genes.
The main objective of this study was to compare soil and clinical isolates of RGM (both from the Czech Republic) in terms of the resistance to tetracycline and presence of seven TET-resistance determinants. The genome relatedness of isolates was assessed with BOX-PCR as a high correlation exists between BOX-PCR fingerprints and DNA-DNA homology data (31, 39, 51). 16S rRNA gene sequencing was performed to identify the isolates. Resistance to TET was assessed with the agar disk diffusion method, and the presence of TET resistance genes that were previously described in clinical RGM, i.e., otr(A), otr(B), tet(K), tet(L), tet(M), tet(V) and the multi-drug efflux pump-encoding gene tap, was checked with PCR and sequencing. The phylogeny of tet(V) was compared to the BOX-PCR profiles of isolates.
Materials and Methods
Soil isolates of rapidly-growing mycobacteria
Soil RGM were isolated from four sites of three farms located in South Bohemia, Czech Republic in 2007–2010 (Table S1). The distance between farms was up to 10 km. Farm 1 is a conventional farm engaged in intensive pig fattening, where animals (about 2,000) are commonly treated with antibiotics including chlortetracycline and doxycycline. At Farm 1, we sampled a permanent grassland, which had been periodically manured (2–3 times per year) with pig slurry for the previous 30 years (designated Site 1). At Farm 2, which is a small family farm in a neighborhood community, we sampled permanent grassland that had not been manured for the previous 20 years (Site 2). Samples were taken from Site 1 and Site 2 in June 2007 and 2009. Farm 3 has performed outdoor cattle husbandry since 1993 and is an organic farm without the application of antibiotics. Two sites (Site 3 and Site 4) were sampled at Farm 3 in May 2010. Site 3 is part of the pasture where cattle stay from October until May. It is highly impacted by the cattle, i.e., the soil is highly enriched with excrement and vegetation cover is damaged (28). Site 4 is a pasture with low impact by the cattle and preserved vegetation. At each site, soil from a depth of about 10–30 cm (under the plant roots) was sampled with a sterile spade from three points 5–20 m apart. The soil from the three points was mixed and sieved. Soils were kept at 4°C during transport to the laboratory and prior to the accompanying physicochemical and microbial analyses (Table S1). RGM were isolated from soil using the NaOH/malachite green/cycloheximide decontamination method of Iivanainen (27) or the olive oil/SDS decontamination method (with 10 mg SDS per plate) of Yamamura and Harayama (53) or directly on Tryptic-Soy agar plates with 25 mg L−1 chlortetracycline (2 isolates from 2007) (Table S2).
Clinical isolates of rapidly-growing mycobacteria
Clinical RGM were obtained from the National Reference Laboratory for Pathogenic Actinomycetes, Regional Hospital in Trutnov, and from the Institute of Public Health, Ostrava, Czech Republic. They were isolated in 2006–2011 from various samples such as abscess, urine, hemoculture, corneal ulcer and sputum from 18 hospitals in the Czech Republic (Table S3). Their role in the etiology of infection was confirmed in isolates from abscesses, hemocultures and corneal ulcers. Incidental isolates from sputa of patients screened for Mycobacterium tuberculosis without clinical and imaging correlates were usually colonizers, with the exception of Mycobacterium chelonae OS10 associated with a pulmonary disease in a 47-year-old patient.
Susceptibility to tetracycline
Susceptibility to tetracycline was assessed with a disc diffusion test (26). Pure isolates were first grown on M2 (42) or Šula’s medium (43) with the addition of 1.5% agar, at 28°C for 5 to 7 d. A homogenous bacterial suspension was prepared by vortexing (Vortex-Genie2; Mo Bio Laboratories, Carlsbad, CA, USA) and ultrasonication (Ultrasonic Compact Cleaner UC 006DM1, Tesla, Czech Republic) of several colonies in 4 mL of sterile 0.9% NaCl. The turbidity of the suspension was adjusted with sterile 0.9% NaCl to match the McFarland standard 0.5 (densitometer DEN-1; Biosan, Latvia) (52). The suspension was spread onto Mueller-Hinton agar medium (Bio-Rad Laboratories, Hercules, CA, USA) supplied with TET disks (30 μg; Bio-Rad). Inhibition zone diameters were recorded after 5 d of incubation at 28°C. The ranked zone sizes of environmental and clinical strains were statistically compared with Wilcoxon rank sum test in R (http://www.r-project.org). Strains OS18, OS2/1, OS2/2 and OS2/4 did not grow on Mueller-Hinton agar and disc diffusion analysis was therefore performed on Šula’s medium. These strains were not included in the statistical analysis.
BOX-PCR genomic DNA fingerprints
Prior to PCR amplification, cell lysates were prepared as follows. One bacteriological loop of mycobacterial biomass grown on an agar plate was resuspended in 100 μL ultra-pure water. The suspensions were then boiled three times (water bath, 100°C) for 5 min and frozen at −20°C for 1 h. The lysates were stored at −20°C and 1 μL of the lysates was used as a template for PCR.
DNA amplification followed the procedure of Lanoot et al. (31) using the BOXA1R primer (5′-CTACGGCAAGGCGACGCT GACG-3′) (51). PCR products (20 μL) were separated on 20×20 cm gels using 130 V, 400 mA for 240 min in 1×TBE buffer (Tris base 53 g, boric acid 27.5 g, 0.5 M EDTA 20 ml, pH 8.0). The gels were stained for 30 min in a 1×TBE bath supplemented with ethidium bromide (1 mg L−1). A photograph of the gel was stored as a TIFF file through a CCD coupled camera using Photo-Doc software (Vilber-Lourmat, Marne-la-Vallée, France). Gels were imported into the software package GelCompar II (Applied Maths, Sint-Martens-Latem, Belgium) and similarity matrices of densitometric curves of the gel tracks were calculated using the Pearson correlation coefficient followed by dendogram construction using the UPGMA algorithm. We used the limit of 70% similarity to define distinct BOX-PCR groups (12).
Isolate identification
Clinical strains TR-1378, OS1, OS8, OS9, OS10, OS11, OS13, OS21, OS24, OS25, OS28, OS30, OS2/7 and OS2/8 were identified with the GenoType Mycobacterium CM (Common Mycobacteria) Test based on DNA Strip technology (Hain Lifescience, Nehren, Germany). The 16S rRNA gene of the clinical strain TR-1380 was sequenced by the commercial system MicroSeq 500 16S rDNA Bacterial Identification Kit (Applied Biosystems, Foster City, CA, USA). Both analyses were performed at the Institute of Public Health Ostrava (Czech Republic).
The remaining clinical isolates and at least one soil isolate from each BOX-PCR group were identified with 16S rRNA gene amplification using universal bacterial primers (21) pA (5′-AGAGTTTGATCCTGGCTCAG-3′) and pH (5′-AAGGAGGT GATCCAGCCGCA-3′), and sequencing. The total volume of PCR reactions was 50 μL. The final reaction mixtures contained (final concentrations) Expand Long Template PCR System Buffer #1 (Roche Applied Science, Mannheim, Germany; 1×), dNTPs (Fermentas, Thermo Fisher Scientific, Waltham, MA, USA; 0.3 mM each), primers (500 nM each) and Expand Long Template polymerase (Roche; 0.05 U μL−1). Mycobacterial lysate (1 μL, see above) served as a template. Thermal cycling was performed as follows: Initial denaturation at 94°C for 2 minutes; followed by 35 cycles of denaturation (94°C/15s), annealing (61°C/30s) and extension (68°C/45s; the duration of extension was prolonged to 90s after the first ten cycles); and final extension at 68°C for 7 min. Amplified 16S rRNA genes were cleaned-up with the GenElute PCR Clean-Up Kit (Sigma-Aldrich, St. Louis, MO, USA) and sequenced using the primers pA, pH and 519r (21, 30).
The obtained 16S rRNA gene sequences were edited by Bioedit 7.0.4.1 software (23) and assembled using SeqMAN (DNAStar, Madison, WI, USA) (45). The edited sequences were compared against the database of type strains Ez-Taxon Database (http://www.eztaxon.org) (11) to retrieve the most relative species.
Detection of tetracycline resistance genes
The presence of tetracycline resistance genes otr(A), otr(B), tet(K), tet(L), tet(M), tet(V) and tap was assessed with PCR using gene-specific primers (Table 1). The total volume of PCR reactions was 25 μL and 1 μL of mycobacterial lysates (see above) or ~20–50 ng of chromozomal DNA from positive control strains or plasmid DNA were used as a template. The cycling conditions for each gene are shown in Table 1.
Table 1.
Primers and PCR conditions used for tetracycline resistance gene amplification
| Gene | Primers | Primer sequences 5′-3′ (Reference) | PCR cycles | Amplicon (bp) | Positive control |
|---|---|---|---|---|---|
| otr(A) | otr(A) (F) otr(A) (R) |
GAACACGTACTGACCGAGAAG CAGAAGTAGTTGTGCGTCCG (37) |
5 min/95°C; 35×(1 min/94°C, 30 s/60°C, 30 s/72°C); 5 min/72°C | 778 | Streptomyces rimosus subsp. rimosus DSMZ 40260 (ATTC 10970) |
| otr(B) | otr(B) (F) otr(B) (R) |
CCGACATCTACGGGCGCAAGC GGTGATGACGGTCTGGGACAG (37) |
5 min/95°C; 35×(1 min/94°C, 1 min/68°C, 1 min/72°C); 7 min/72°C | 947 | Streptomyces rimosus subsp. rimosus DSMZ 40260 (ATTC 10970) |
| tap | Tap1 Tap2 |
GTCGCGTTCCCGTGGCTGGT CGATACCGGGGCCGACGATG (22) |
10 min/94°C; 35×(1 min/94°C, 30 s/68°C, 30 s/72°C); 3 min/72°C | 400 | Mycobacterium fortuitum TR-1242 (this study) |
| tet(K) and tet(L) | tetKL-FW tetKL-RV |
TTACCTGATATTGCAA GACCAATGAATATAAT (this study) |
5 min/95°C; 35×(30 s/94°C, 30 s/40°C, 30 s/72°C); 3 min/72°C | 397 | Staphylococcus haemolyticus CB-N (tet(K); this study) and Staphylococcus aureus pSTS9-like (tet(L); 1) |
| tet(M) | TetM-FW TetM-RV |
ACAGAAAGCTTATTATATAAC TGGCGTGTCTATGATGTTCAC (6) |
4 min/94°C; 35×(20 s/94°C, 30 s/52.3°C, 1 min/72°C); 7 min/68°C | 171 | Plasmid pAT101 that carries tet(M) gene from Streptococcus transposon Tn1545 (34) |
| tet(V) | tetV-FW tetV-RV |
GCCTACGGTTTCATCCTGGC CGAGACCACCTTCGACAGCG (this study) |
7 min/95°C; 35×(1 min/94°C, 15 s/65°C, 30 s/72°C); 5 min/ 72°C | 351 | Mycobacterium sp. Site2-2C (this study) |
The genes otr(A), otr(B) and tap were amplified with the help of Qiagen Taq polymerase and Q-solution (Qiagen, Hilden, Germany). The final reaction mixtures contained (final concentrations) Qiagen Taq buffer (1×), Qiagen Q-solution (1×), dNTPs (Fermentas, Thermo Fisher Scientific; 0.2 mM each), primers [500 nM in the case of otr(A) and tap, and 100 nM in the case of otr(B)] and Qiagen Taq polymerase (0.05 U μL−1). The genes tet(K) and (L) (co-amplified), tet(V) and tet(M) were amplified using Dream Taq polymerase (Fermentas, Thermo Fisher Scientific) and the final reaction mixtures contained Dream Taq buffer [with (NH4)2SO4, without MgCl2; 1×], MgCl2 (1.5 mM), dNTPs (0.2 mM each), primers (500 nM each), dimethyl sulfoxide (Sigma-Aldrich; 5%), bovine serum albumin (Fermentas, 1.2 mg mL−1) and Dream Taq polymerase (0.05 U μL−1). Five microliters of PCR products were analyzed in 1–2% agarose gel (Top Vision agarose, Fermentas, Thermo Fisher Scientific) stained with ethidium bromide (1 mg L−1), 30 min, for the presence of bands of the expected size.
The specificity of primer pairs designed in this study, i.e., tetKL-FW/tetKL-RV, and tetV-FW/tetV-RV was tested by PCR with other tetracycline efflux pump genes as a template (negative controls). The tetKL-FW/tetKL-RV were tested against plasmids containing genes tet(A), tet(B), tet(C), tet(D), tet(E), tet(G), tet(H), tet(J), tet(V), tet(Y), tet(Z) and agaist Streptomyces rimosus subsp. rimosus DSMZ 40260 chromosomal DNA containing otr(B). Primers tetV-FW/tetV-RV were tested against tet(A), tet(B), tet(C), tet(D), tet(E), tet(H), tet(J), tet(K), tet(L), tet(Y), tet(Z) and otr(B) (6, 7).
Amplified tet(V) or tap genes were cleaned-up with the GenElute PCR Clean-Up Kit (Sigma-Aldrich) and sequenced from both ends in the case of tet(V) and from the forward primer in the case of tap, as described above. The sequences were edited and assembled with Bioedit 7.0.4.1 software (23) and compared to the GenBank database (www.ncbi.nml.nih.gov) using blastn and blastp algorithms. Altogether, 46 PCR products of tet(V) and 14 PCR products of tap were sequenced.
Phylogeny analyses of tet(V)
Phylogeny analyses were conducted in MEGA software version 4.0 (http://www.megasoftware.net/) (46). The partial sequences of tet(V) (311 nucleotides) were aligned together with the published sequence of M. smegmatis tet(V) (GenBank, gb|CP000480.1|: 5286339–5287598) and M. vanbaalenii H+ antiporter gene (Gen-Bank, CP000511.1). The neighbor-joining phylogenetic tree was constructed based on the nucleotide sequences using Kimura-2 parameter. The correlation between the distance matrices of tet(V) sequences of 43 isolates and corresponding BOX-PCR profiles was assessed with the Mantel test in R (http://www.r-project.org/), ADE-4 package (47), using 1,000 repetitions.
Accession numbers
The tet(V) sequences were deposited in GenBank under accession numbers JF290326–JF290351 and JQ348076–JQ348095, and the tap sequences under accession numbers JF290352–JF290365. The 16S rRNA gene sequences are available in GenBank under accession numbers JF304573–JF304610 and JQ348096–JQ348111.
Results
Isolate identification and genome relatedness
Forty-four isolates from grasslands and 38 clinical isolates were included in this study (Table S2, S3 and S4). The environmental isolates were divided into 14 distinct groups and the clinical isolates into 28 distinct groups according to their BOX-PCR profile similarity, using the Pearson correlation coefficient threshold of 0.7 (Table 2, 3 and Fig. S1). Clinical and environmental strains always belonged to separate BOX-PCR groups.
Table 2.
Tetracycline resistance and presence of resistance genes in the environmental isolates
| Isolate | BOX group | Identificationa | TET resistance (zone in mm)b | Presence of genec | |
|---|---|---|---|---|---|
|
| |||||
| tap | tet(V) | ||||
| Site3-B14 | A | M. litorale (99.46%) | 12 | + | + |
| Site3-B33 | A | M. septicum (100%) | 35 | + | + |
| Site4-B4 | C | M. alvei (98.99%) | 9 | + | + |
| Site4-B30 | C | M. septicum (100%) | 12 | + | + |
| Site4-B31 | D | M. septicum (99.06%) | 11 | + | + |
| Site1-IIA/46 | E | M. septicum (99.84%) | 12 | + | + |
| Site4-B5 | F | M. septicum (99.58%) | 20 | + | + |
| Site4-B18 | F | ND | 25 | + | + |
| Site4-B19 | F | ND | 18 | + | + |
| Site1-8A | F | M. septicum (99.93%) | 14 | + | + |
| Site1-10A | F | M. septicum (100%) | 13 | + | + |
| Site4-B2 | G | M. septicum (100%) | 11 | + | − |
| Site4-B7 | G | ND | 15 | + | − |
| Site4-B8 | G | ND | 9 | − | − |
| Site4-B29 | G | M. septicum (99.77%) | 11 | + | + |
| Site2-2C | H | M. septicum (100%) | 14 | + | + |
| Site2-3C | H | M. septicum (100%) | 14 | + | + |
| Site2-5C | H | M. septicum (99.78%) | 17 | + | + |
| Site2-7C | H | M. septicum (99.76%) | 10 | + | + |
| Site4-B1 | I | M. septicum (99.55%) | 11 | + | + |
| Site4-B3 | I | M. septicum (98.93%) | 37 | + | + |
| Site4-B9 | I | M. septicum (99.25%) | 11 | + | + |
| Site4-B23 | I | ND | 10 | + | + |
| Site4-B24 | I | ND | 39 | + | + |
| Site4-B26 | I | M. fortuitum subsp. acetamidolyticum (99.46%) | 35 | + | + |
| Site4-B27 | I | ND | 35 | + | + |
| Site4-B28 | I | ND | 10 | + | + |
| Site2-4C | J | M. fortuitum subsp. acetamidolyticum (99.42%) | 31 | + | + |
| Site4-B6 | M | M. septicum (99.17%) | 10 | − | − |
| Site4-B16 | M | ND | 9 | − | − |
| Site4-B17 | M | ND | 8 | − | − |
| Site4-B21 | M | ND | 9 | − | − |
| Site4-B39 | M | ND | 20 | − | − |
| Site4-B15 | P | M. septicum (99.71%) | 50 | + | + |
| Site4-B25 | P | M. septicum (99.56%) | 55 | + | + |
| Site4-B38 | Q | M. septicum (100%) | 14 | + | + |
| Site1-2A | Q | M. septicum (99.71%) | 53 | + | + |
| Site1-3A | Q | M. septicum (99.71%) | 56 | + | + |
| Site1-9A | Q | M. septicum (99.71%) | 56 | + | + |
| Site1-11A | Q | M. septicum (99.71%) | 52 | + | + |
| Site3-B10 | U | M. septicum (100%) | 15 | + | − |
| Site3-B34 | U | ND | 10 | + | − |
| Site4-B36 | U | M. septicum (99.67%) | 10 | + | − |
| Site2-IIIC/14 | δ | M. aubagnense (99.04%) | 12 | − | − |
In parentheses, % pairwise sequence similarity with the closest type strain on EzTaxon is shown. ND, not done.
Resistance in bold.
The genes otr(A), otr(B), tet(K)(L) and tet(M) were detected in none of the isolates.
Table 3.
Tetracycline resistance and presence of resistance genes in the clinical isolates
| Isolate | BOX group | Identification | TET resistance (zone in mm) | Presence of geneb | |
|---|---|---|---|---|---|
|
| |||||
| tap | tet(V) | ||||
| OS6 | B | M. novacastrense (99.56%) | 60* | + | − |
| OS2/8 | K | M. peregrinuma | 6.5 | + | + |
| TR-1378 | L | M. fortuituma | 17 | + | − |
| OS2/2 | N | M. arupense (100%) | 6.5* | − | − |
| TR-1536 | O | M. franklinii (100%) | 6.5 | − | − |
| OS19 | R | M. septicum (100%) | 6.5 | + | − |
| OS2/7 | R | M. fortuituma | 11 | + | − |
| OS14 | S | M. septicum (100%) | 11 | + | + |
| OS16 | T | M. septicum (100%) | 6.5 | + | − |
| TR-1358 | V | M. goodii (99.77%) | 41 | + | + |
| OS2 | V | M. neoaurum (99.37%) | 58 | + | − |
| TR-1344 | W | M. llatzerense (98.40%) | 11 | − | − |
| TR-1380 | X | M. arupense (98.95%) | 55 | − | + |
| OS22 | Y | M. neoaurum (100%) | 48 | + | + |
| OS29 | Y | M. neoaurum (100%) | 54 | + | − |
| OS2/1 | Z | M. nonchromogenicum (98.91%) | 6.5* | − | − |
| TR-1294 | α | M. neoaurum (99.76%) | 47 | + | − |
| OS3 | α | M. neoaurum (100%) | 48 | + | − |
| OS8 | β | M. fortuituma | 9 | + | + |
| OS26 | γ | M. neoaurum (100%) | 57 | + | − |
| OS27 | ɛ | M. obuense (99.78%) | 48 | + | − |
| OS4 | ζ | M. rufum (100%) | 52 | + | − |
| TR-1359 | η | M. rufum (99.76%) | 61 | + | − |
| OS13 | θ | M. abscessusa | 9 | − | − |
| TR-1242 | ι | M. fortuitum subsp. fortuitum (99.88%) | 52 | + | + |
| TR-1266 | ι | M. fortuitum subsp. fortuitum (100%) | 6.5 | + | + |
| OS24 | ι | M. fortuituma | 6.5 | + | + |
| OS25 | ι | M. fortuituma | 6.5 | + | + |
| OS9 | κ | M. fortuituma | 6.5 | + | + |
| OS28 | κ | M. fortuituma | 55 | + | + |
| OS10 | λ | M. abscessusa | 6.5 | − | − |
| OS30 | λ | M. fortuituma | 6.5 | + | + |
| OS21 | μ | M. fortuituma | 31 | + | + |
| OS18 | ν | M. hiberniae (99.78%) | 6.5* | − | − |
| OS2/4 | ν | M. hiberniae (99.78%) | 6.5* | − | − |
| OS11 | ξ | M. mucogenicuma | 49 | + | − |
| OS7 | π | M. frederiksbergense (99.56%) | 55 | + | − |
| OS1 | ρ | M. smegmatisa | 38 | − | − |
Identified with the GenoType Mycobacterium CM (Common Mycobacteria) Test based on DNA Strip technology (Hain Lifescience)
The genes otr(A), otr(B), tet(K)(L) and tet(M) were detected in none of the isolates.
Measured on Šula’s medium (43).
A minimum of one isolate from each environmental BOX-PCR group was identified by sequencing the 16S rRNA gene and comparing to the type strain database using EzTaxon v 2.1 software (Tables 2 and S2). Most of the soil isolates (i.e., 26) had sequences identical or almost identical (98–100% pairwise sequence similarity) to the species Mycobacterium septicum, 2 isolates to Mycobacterium fortuitum subsp. acetamidolyticum, 1 isolate to Mycobacterium alvei, 1 isolate to Mycobacterium litorale and 1 isolate to Mycobacterium aubagnense. The isolates from one BOX-PCR group were usually assigned to the same species, with the exception of group A, where isolates were attributed either to M. septicum or to M. litorale, group C where one isolate was attributed to M. alvei and another to M. septicum, and group I (3 isolates attributed to M. septicum and one to M. fortuitum). The set of clinical isolates was more diverse and comprised 17 species, of which M. fortuitum was the most prevalent (29%), followed by M. neoaurum (16%) (Table 3 and S3). As in the case of the soil isolates, different species sometimes occurred in the same BOX-PCR group, e.g., M. septicum and M. fortuitum in group R, M. goodie and M. neoaurum in group V, and M. abscessus and M. fortuitum in group λ.
Resistance to tetracycline
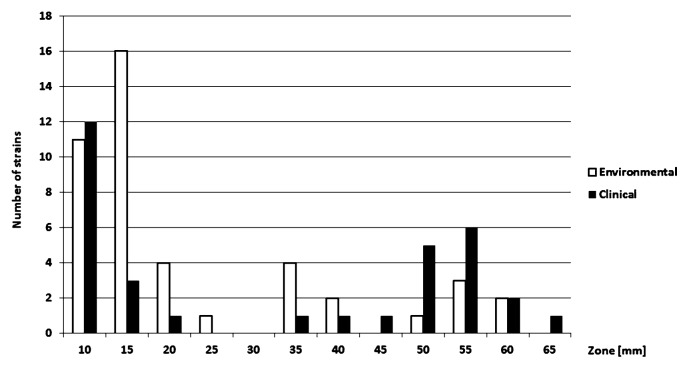
The distribution of TET resistance was bimodal in both clinical and environmental strains (Fig. 1). Since standard breakpoints of the disc diffusion method are not available for RGM, we arbitrarily set them according to the zone size distributions. An isolate was considered to be resistant if the zone was up to 25 mm. Based on the arbitrary breakpoint, thirty-two environmental (70%) and sixteen clinical isolates (53%) were resistant to TET (Table 2 and 3). There was no significant difference in TET resistance (in terms of zone sizes, assessed with Wilcoxon rank sum test) between the environmental and clinical isolates. Tetracycline resistance was not a specific characteristic of individual BOX-PCR groups, i.e., both resistant and sensitive isolates could be found within the same BOX-PCR group.
Fig. 1.
Tetracycline resistance in the clinical and the environmental isolates of rapidly growing mycobacteria. Bars represent the number of isolates with the corresponding inhibition zone size around 30 μg tetracycline disks.
Detection of tetracycline resistance genes
The two primer pairs designed in this study, i.e., tetV-FW/tetV-RV [tet(V) detection], and tetKL-FW/tetKL-RV [simultaneous detection of tet(K) and tet(L)], were tested for their specificity in PCR using different tetracycline efflux pump genes as templates. Both primer pairs were specific, i.e., they amplified only tet(V) in the case of tetV-FW/tetV-RV and tet(K) and tet(L) in the case of tetKL-FW/tetKL-RV (data not shown). The specificity of the tetV-FW/tetV-RV primer pair was further corroborated by sequencing the PCR products (see below).
All isolates were tested for the presence of otr(A), otr(B), tet(K)/tet(L), tet(M), tet(V) and tap. The gene tet(V) was detected in 32 of the total 44 environmental strains (73%) and in fourteen clinical isolates (37%) (Table 2 and 3). The environmental isolates harboring tet(V) were assigned to the species M. septicum, M. litorale, M. alvei and M. fortuitum. The clinical isolates with tet(V) belonged to the species M. fortuitum, M. peregrinum, M. septicum, M. goodii, M. arupense and M. neoaurum. Surprisingly, tet(V) was found also in isolates that were sensitive to tetracycline (even in those with zones over 50 mm) (Table 2 and 3). The gene tap was detected in 37 environmental isolates (84%) and in 28 clinical isolates (74%). The environmental isolates with tap belonged to the species M. fortuitum, M. alvei, M. septicum, and the clinical isolates to the species M. fortuitum, M. novacastrense, M. peregrinum, M. septicum, M. goodi, M. neoaurum, M. rufum, M. obuense, M. frederiksbergense and M. mucogenicum. Similarly to tet(V), tap was detected also in strains that were sensitive to tetracycline. In contrast, the genes otr(A), otr(B), tet(K)/tet(L) and tet(M) were not detected in any isolate, and eight clinical and seven environmental isolates were resistant to TET but possessed none of the tested genes.
Analyses of tet(V) and tap sequences
The gene tet(V) was sequenced in all isolates (i.e., 46) where we obtained positive signals from PCR. Sequences of isolates Site2-3C, Site2-5C and Site2-7C (all from the BOX-PCR group H) were not used for further analyses since they contained several ambiguous peaks that could not be resolved (although PCR and sequencing were attempted twice). It is possible that these isolates had two copies of tet(V) that differed slightly in their sequences.
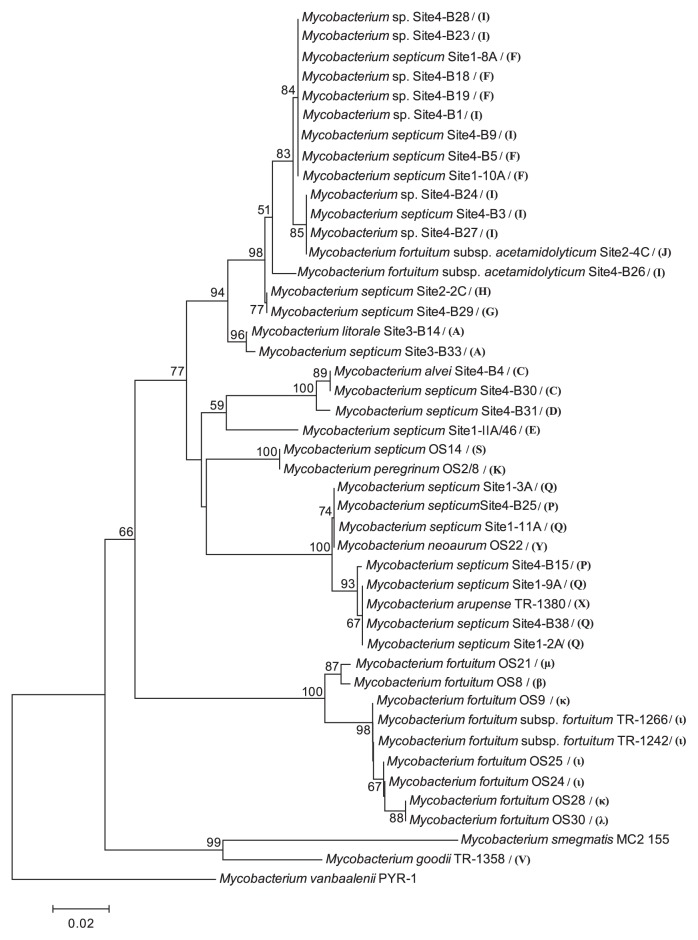
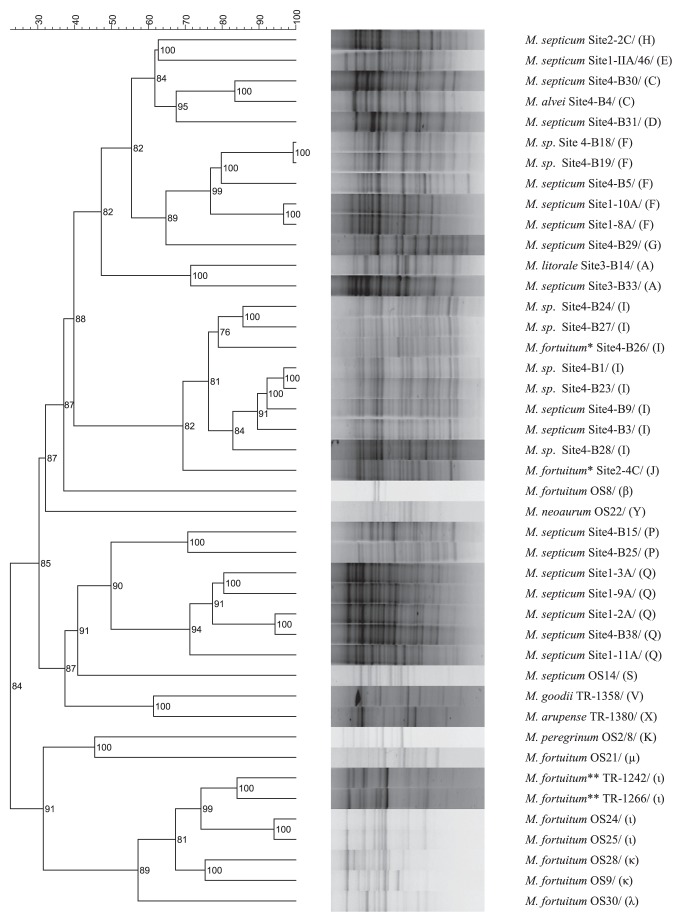
The 43 analyzed partial sequences of the gene tet(V) [corresponding to positions 823–1,132 of M. smegmatis MC2-155 tet(V)] differed substantially among the isolates, with 17% differences between two most distant isolates (Fig. 2). The isolates shared 82–88% and 90–95% identity, respectively, of nucleic and inferred amino acid sequences with the published tet(V) sequence of M. smegmatis MC2-155. Since all the recovered sequences had amino acid identity above 80% with the published tet(V), they can be attributed to the tet(V) gene class (32). Interestingly, M. neoarum OS22 had a one-nucleotide deletion at position 915. The phylogeny of tet(V) (Fig. 2) shared overall similarity with the genome relatedness of the strains (Fig. 3), and the soil isolates usually clustered separately from clinical RGM. Indeed, there was a significant correlation (r=0.64, P=0.001) between the BOX-PCR profile- and tet(V) sequence-based distant matrices.
Fig. 2.
Phylogenetic tree based on the nucleotide sequences of tet(V), constructed by the neighbor-joining method using Kimura-2 parameter. Bootstrap values are indicated at the nodes as a percentage of 1,000 replications, if they were higher than 50%. Letters following the isolate names indicate the BOX-PCR groups the isolates belonged to.
Fig. 3.
BOX-PCR profiles of 43 RGM isolates with sequenced tet(V). Left, UPGMA-clustering of the isolates based on the similarity matrix of their BOX-PCR profiles. Right, isolate names preceded by a letter indicating the BOX-PCR groups (based on the ≥70% similarity threshold). See Fig. S1 for BOX-PCR profiles of all isolates from this study.
Fourteen PCR products of tap (corresponding to positions 764–1,080 of tap M. fortuitum) (4) were chosen for sequencing in order to verify the specificity of primers and check for tap gene diversity. Similarly to tet(V), differences in the partial sequences of tap between our isolates and the published sequence were found (data not shown). Out of the sequenced isolates, M. fortuitum subsp. fortuitum TR-1242 had the tap sequence most similar to the published sequence (99% identity), while the M. rufum TR-1359 tap sequence was the most dissimilar (only 79% identity). The environmental isolates had 82–89% sequence identity with the published tap sequence, and there was quite a high diversity of the sequences among the environmental isolates (up to 15% substitutions). The number of differences was lower at the amino acid level (96–100% amino acid identities).
Discussion
In general, the distribution of TET resistance did not differ between the soil and the clinical RGM and the same TET-resistance genes were found in both groups. The gene tet(V) encoding a tetracycline efflux pump (19) was found in 73% soil isolates and 37% clinical isolates, including species where it has not been reported previously, i.e., M. septicum, M. litorale, M. alvei, M. peregrinum, M. goodii, M. arupense and M. neoaurum. This study also showed the high diversity of tet(V) (up to 17% difference between two sequences), which is unusual among other tet gene classes. For example, the published sequences of tet(B), a horizontally transferred TET efflux pump with at least 20 reported host genera (40), did not differ from each other in more than 1% nucleotides (analysis not shown). This finding, together with the positive correlation between the BOX-PCR profiles and tet(V) phylogeny, suggest that tet(V) evolved together with the mycobacterial genomes rather than being acquired horizontally. These findings therefore support the hypothesis that tet(V) belongs to the mycobacterium intrinsic resistome; however, in certain cases, different RGM species (e.g., M. arupense and M. septicum) shared the same partial tet(V) sequence, so the horizontal exchange of tet(V) among mycobacteria cannot be completely excluded. It is also conceivable that this gene underwent a long evolution within a certain group of RGM species from which it was horizontally transferred to other RGM species later. Comparative studies of the tet(V) gene and its sourroundings in a broader set of RGM species might be the next step to learn more about antibiotic resistance evolution in mycobacteria.
BLAST search revealed the presence of tet(V) homologues in the sequenced genomes of seven mycobacterial strains and in seven other actinomycetes; for example, there was a gene for H+ antiporter of M. vanbaalenii PYR-1 (86% identity), Mycobacterium sp. JLS (CP000580.1, 82% identity), Mycobacterium sp. KMS (CP000518.1, 82% identity) or major facilitator superfamily protein of Geodermatophilus obscurus DSM 43160 (CP001867.1, 73% identity). These genes may potentially code for drug resistance and therefore merit further attention. It is also possible that tet(V) is present in more mycobacterial species but has not been detected with PCR because of its high diversity and thus possible lack of complementarity with the primers used here.
The gene tap, which confers low-level resistance to tetracycline and certain aminoglycosides (4), seems to be very common among RGM. In our study, we found it in 84% environmental and 74% clinical isolates. Likewise, Esteban et al. (22) found tap in most (66%) of the clinical RGM tested. Besides the species in which tap has already been reported (22), it was detected also in M. litorale, M. novacastrense, M. septicum, M. goodi, M. neoaurum, M. rufum, M. obuense, M. frederiksbergense and M. mucogenicum. Based on the number of identities with the published tap sequence, it seems that we recovered more diverse tap sequences (83–99% identity) than Esteban et al. (22) (92–95% identity), using the same primers. Analysis of more sequences would be necessary to show whether there is a correlation between BOX-PCR profiles and tap phylogeny, as in the case of tet(V).
Surprisingly, tet(V) and/or tap were found also in the isolates that were highly susceptible to TET. Although our sequences of Tet(V) differed from that for which functionality was previously shown (19), there was no correlation between the tet(V) alleles and TET inhibition zone size; for example, two clinical M. fortuitum had the same sequence (at least in the part analyzed) but markedly differed in resistance. In general, sense mutations prevailed over the mutations changing the amino acid sequence, indicating that the function was rather to be maintained; however, M. neoaurum OS22 had one nucleotide deletion in the analyzed part of tet(V) that would change the reading frame, and this isolate was sensitive to tetracycline. It is possible that point deletions in the unanalyzed parts of the tet(V) sequence occurred also in other isolates. In addition, the observed discrepancy between genotype and phenotype may be due to mutations in distant regulation regions or to gene expression regulation; for example, Nash et al. (35) noticed that high clarithromycin resistance was inducible by overnight incubation of certain sensitive mycobacteria with a low concentration of the antibiotic. The discrepancy between the presence of a resistance gene and the resistance to TET in RGM or other bacteria was reported previously (5, 22). TET-resistant isolates in which tet(V) and tap were not detected could either differ in their tet(V) and tap sequences from the used primers or possess other TET resistance genes that were not tested. The search for horizontally transferred TET resistance genes so far described in RGM was unsuccessful, though it included the genes tet(M) and tet(L), which are commonly found in Gram-positive bacteria in manure (1, 3). The number of studies reporting horizontally transferred TET resistance genes in RGM is low (38, 41) and it could be that there are marked local differences in the distribution of these genes in RGM.
The majority of the soil as well as clinical isolates from this study belonged to the Mycobacterium fortuitum group. Most soil isolates had the highest 16S rRNA sequence similarity to the species M. peregrinum, M. septicum or M. fortuitum, which all include potential human pathogens (29, 48). Some of the isolates close to M. septicum indeed grew at 37°C (data not shown), indicating they may be able to colonize the human body; however, the soil and clinical isolates of M. septicum from this study were genetically different, as shown by analyses of their BOX-PCR profiles (and the same applies also to M. fortuitum isolates). The species in the M. fortuitum group are frequently recovered from soil (25), including agricultural soils (20, 53), as well as from clinical samples (24, 50), but so far a direct relationship between a clinical manifestation and exposure to soil has not been clearly shown in this group. The group M. chelonae-M. abscessus was reported as being even more frequent than the M. fortuitum group in clinical samples (24, 50), but not in our case. The discrepancy between BOX-PCR groups and the assignment of isolates to species (e.g., BOX-PCR groups A, C or I) can be because the BOX-PCR groups were defined quite broadly (70% similarity) (12), but it can also indicate the need for more comprehensive taxonomical evaluation of RGM, as already shown in the case of streptomycetes (31).
In this study, we tested resistance to tetracycline and not to clinically used doxycycline, which was relevant in the context of soil since tetracycline residues are found in soil and manure (10). Ultimately, the zone sizes with tetracycline and doxycycline disks were correlated (tested with 32 isolates, Pearson correlation 0.88, P<0.01, data not shown). It was previously shown with the disk dilution method that RGM susceptible to tetracycline were also susceptible to doxycline, although the MIC values were 1–2 dilutions lower for doxycycline (52). Interestingly, there was no difference in the zone size distribution between clinical and environmental RGM and more than half of the isolates from both groups were resistant to tetracycline. The bimodal distribution of resistance to tetracycline antibiotics was reported previously (44, 52). Both resistant and sensitive phenotypes occurred within one species (e.g., within M. fortuitum or within M. septicum), and even within one BOX-PCR group (Table 2 and 3). We thus did not observe any consistency between BOX-PCR fingerprints and antibiotic resistance phenotypes, in contrast to Davelos Baines et al. (13), who found significant correlations between antibiotic phenotypes and BOX-PCR fingerprints in soil streptomycetes. This could be because the selection pressure for antibiotic resistance in soil acts on a much smaller scale than we sampled (13) and because antibiotic resistance in individual mycobacterial isolates can be affected by point mutations that would not significantly affect their BOX-PCR profiles. The variable resistance to tetracycline antibiotics within the M. fortuitum group is consistent with previous studies, usually reporting 40–60% sensitive isolates (24, 44, 52).
Conclusion
In conclusion, the studied soil and the clinical RGM from the Czech Republic did not differ in the distribution of TET resistance and occurrence of TET resistance genes, most possessing efflux-pump encoding genes tet(V) and/or tap. This study shows for the first time the presence of tet(V) and tap in soil mycobacteria. The correlation between the tet(V) phylogeny and isolate genomic profiles indicates that tet(V) belongs to the mycobacterial intrinsic resistom. The intrinsic efflux pumps may therefore play an important role in the antibiotic resistance of RGM, as in the case of M. tuberculosis (18, 33). The simple presence of efflux-pump encoding genes, however, did not always match TET resistance. Further research should therefore be performed on gene diversity, distribution and expression in order to better understand the mycobacterial intrinsic resistom.
Supplementary Material
Acknowledgements
This work was funded by the Grant Academy of the Czech Republic, project P504/10/2077, Ministry of Education, Youth and Sport of the Czech Republic, project LC06066. We are grateful to F. M. Aarestrup, National Food Institute, Technical University of Denmark, Copenhagen, and H. Schmitt, Institute for Risk Assessment Sciences (IRAS), Utrecht University (with the permission of R. Aminov, Rowett Institute of Nutrition and Health, University of Aberdeen) for the gift of strains. We thank Z. Stehlíková, (Biology Center AS CR, v.v.i., Institute of Soil Biology, Czech Republic) for technical help. GENERI BIOTECH, s.r.o., and the Laboratory of Genomics (Biology Centre AS CR, v.v.i., Institute of Plant Molecular Biology) are acknowledged for sequencing.
References
- 1.Aarestrup FM, Agerso Y, Gerner-Smidt P, Madsen M, Jensen LB. Comparison of antimicrobial resistance phenotypes and resistance genes in Enterococcus faecalis and Enterococcus faecium from humans in the community, broilers, and pigs in Denmark. Diagn Microbiol Infect Dis. 2000;37:127–137. doi: 10.1016/s0732-8893(00)00130-9. [DOI] [PubMed] [Google Scholar]
- 2.Adékambi T, Stein A, Carvajal J, Raoult D, Drancourt M. Description of Mycobacterium conceptionense sp. nov., a Mycobacterium fortuitum group organism isolated from a posttraumatic osteitis inflammation. J Clin Microbiol. 2006;44:1268–1273. doi: 10.1128/JCM.44.4.1268-1273.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agersø Y, Jensen LB, Givskov M, Roberts MC. The identification of a tetracycline resistance gene tet M., on a Tn916-like transposon, in the Bacillus cereus group. FEMS Microbiol Lett. 2002;214:251–256. doi: 10.1111/j.1574-6968.2002.tb11355.x. [DOI] [PubMed] [Google Scholar]
- 4.Aínsa JA, Blokpoel MC, Otal I, Young DB, De Smet KA, Martí C. Molecular cloning and characterization of Tap, a putative multidrug efflux pump present in Mycobacterium fortuitum and Mycobacterium tuberculosis. J Bacteriol. 180(1998):5836–5843. doi: 10.1128/jb.180.22.5836-5843.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alexander TW, Reuter T, Sharma R, Yanke LJ, Topp E, McAllister TA. Longitudinal characterization of resistant Escherichia coli in fecal deposits from cattle fed subtherapeutic levels of antimicrobials. Appl Environ Microbiol. 2009;75:7125–7134. doi: 10.1128/AEM.00944-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aminov RI, Garrigues-Jeanjean N, Mackie RI. Molecular ecology of tetracycline resistance: development and validation of primers for detection of tetracycline resistance genes encoding ribosomal protection proteins. Appl Environ Microbiol. 2001;67:22–32. doi: 10.1128/AEM.67.1.22-32.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aminov RI, Chee-Sanford JC, Garrigues N, Teferedegne B, Krapac IJ, White BA, Mackie RI. Development, validation, and application of PCR primers for detection of tetracycline efflux genes of Gram-negative bacteria. Appl Environ Microbiol. 2002;68:1786–1793. doi: 10.1128/AEM.68.4.1786-1793.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Appelgren P, Farnebo F, Dotevall L, Studahl M, Jönsson B, Petrini B. Late-onset posttraumatic skin and soft-tissue infections caused by rapid-growing mycobacteria in tsunami survivors. Clin. Infect. Dis. 2008;47:e11–6. doi: 10.1086/589300. [DOI] [PubMed] [Google Scholar]
- 9.Butler MJ, Friend EJ, Hunter IS, Kaczmarek FS, Sugden DA, Warren M. Molecular cloning of resistance genes and architecture of a linked gene cluster involved in biosynthesis of oxytetracycline by Streptomyces rimosus. Mol Gen Genet. 1989;215:231–238. doi: 10.1007/BF00339722. [DOI] [PubMed] [Google Scholar]
- 10.Chee-Sanford JC, Mackie RI, Koike S, Krapac IG, Lin Y-F, Yannarell AC, Maxwell S, Aminov RI. Fate and transport of antibiotic residues and antibiotic resistance genes following land application of manure waste. J Environ Qual. 2009;38:1086–1108. doi: 10.2134/jeq2008.0128. [DOI] [PubMed] [Google Scholar]
- 11.Chun J, Lee J-H, Jung Y, Kim M, Kim S, Kim BK, Lim YW. EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int J Syst Evol Microbiol. 2007;57:2259–2261. doi: 10.1099/ijs.0.64915-0. [DOI] [PubMed] [Google Scholar]
- 12.Coenye T, Spilker T, Martin A, LiPuma JJ. Comparative assessment of genotyping methods for epidemiologic study of Burkholderia cepacia genomovar III. J Clin Microbiol. 2002;40:3300–3307. doi: 10.1128/JCM.40.9.3300-3307.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davelos Baines AL, Xiao K, Kinkel LL. Lack of correspondence between genetic and phenotypic groups amongst soil-borne streptomycetes. FEMS Microbiol Ecol. 2007;59:564–575. doi: 10.1111/j.1574-6941.2006.00231.x. [DOI] [PubMed] [Google Scholar]
- 14.D’Costa VM, McGrann KM, Hughes DW, Wright GD. Sampling the antibiotic resistome. Science. 2006;311:374–377. doi: 10.1126/science.1120800. [DOI] [PubMed] [Google Scholar]
- 15.De Groote MA, Huitt G. Infections due to rapidly growing mycobacteria. Clin Infect Dis. 2006;42:1756–1763. doi: 10.1086/504381. [DOI] [PubMed] [Google Scholar]
- 16.De Groote MA, Pace NR, Fulton K, Falkinham JO., III Relationships between Mycobacterium isolates from patients with pulmonary mycobacterial infection and potting soils. Appl Environ Microbiol. 2006;72:7602–7606. doi: 10.1128/AEM.00930-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Rossi E, Aínsa JA, Riccardi G. Role of mycobacterial efflux transporters in drug resistance: an unresolved question. FEMS Microbiol Rev. 2005;30:36–52. doi: 10.1111/j.1574-6976.2005.00002.x. [DOI] [PubMed] [Google Scholar]
- 18.De Rossi E, Arrigo P, Bellinzoni M, Silva PEA, Martín C, Aínsa JA, Guglierame P, Riccardi G. The multidrug transporters belonging to major facilitator superfamily (MFS) in Mycobacterium tuberculosis. Mol Med. 2002;8:714–724. [PMC free article] [PubMed] [Google Scholar]
- 19.De Rossi E, Blokpoel MCJ, Cantoni R, Branzoni M, Riccardi G, Young DB, De Smet KAL, Ciferri O. Molecular cloning and functional analysis of a novel tetracycline resistance determinant, tet(V), from Mycobacterium smegmatis. Antimicrob Agents Chemother. 1998;42:1931–1937. doi: 10.1128/aac.42.8.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Donoghue HD, Overend E, Stanford JL. A longitudinal study of environmental mycobacteria on a farm in south-west England. J Appl Microbiol. 1997;82:57–67. doi: 10.1111/j.1365-2672.1997.tb03297.x. [DOI] [PubMed] [Google Scholar]
- 21.Edwards U, Rogall T, Blöcker H, Emde M, Böttger EC. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 1989;17:7843–7853. doi: 10.1093/nar/17.19.7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Esteban J, Martín-de-Hijas NZ, Ortiz A, Kinnari TJ, Bodas Sánchez A, Gadea I, Fernández-Roblas R. Detection of lfrA and tap efflux pump genes among clinical isolates of non-pigmented rapidly growing mycobacteria. Int. J. Antimicrob Agents. 2009;34:454–456. doi: 10.1016/j.ijantimicag.2009.06.026. [DOI] [PubMed] [Google Scholar]
- 23.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 24.Han XY, Dé I, Jacobson KL. Rapidly growing mycobacteria: clinical and microbiologic studies of 115 cases. Am J Clin Pathol. 2007;128:612–621. doi: 10.1309/1KB2GKYT1BUEYLB5. [DOI] [PubMed] [Google Scholar]
- 25.Hartmans S, De Bont JAM, Stackebrandt E. The genus Mycobacterium—nonmedical. In: Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E, Dworkin M, editors. The Prokaryotes. Vol. 3. Springer-Verlag; New York: 2006. pp. 889–918. [Google Scholar]
- 26.Hindler JF, Munro S. Disk diffusion test. In: Garcia LS, Isenberg HD, editors. Clinical Microbiology Procedures Handbook. 3rd ed. Vol. 2. ASM Press; Washington, DC: 2010. pp. 5.1.1–5.1.13. [Google Scholar]
- 27.Iivanainen E. Isolation of mycobacteria from acidic forest soil samples: comparison of culture methods. J Appl Bacteriol. 1995;78:663–668. doi: 10.1111/j.1365-2672.1995.tb03113.x. [DOI] [PubMed] [Google Scholar]
- 28.Jirout J, Tříska J, Růžičková K, Elhottová D. Disturbing impact of outdoor cattle husbandry on community of arbuscular mycorrhizal fungi in upland pasture soil. Commun Soil Sci Plant Anal. 2009;40:736–745. [Google Scholar]
- 29.Lamy B, Marchandin H, Hamitouche K, Laurent F. Mycobacterium setense sp. nov., a Mycobacterium fortuitum-group organism isolated from a patient with soft tissue infection and osteitis. Int J Syst Evol Microbiol. 2008;58:486–490. doi: 10.1099/ijs.0.65222-0. [DOI] [PubMed] [Google Scholar]
- 30.Lane DJ, Pace B, Olsen GJ, Stahl DA, Pace NR. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Nat Acad Sci USA. 1985;82:6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lanoot B, Vancanneyt M, Dawyndt P, Cnockaert MC, Zhang J, Huang Y, Liu Z, Swings J. BOX-PCR fingerprinting as a powerful tool to reveal synonymous names in the genus Streptomyces. Emended descriptions are proposed for the species Streptomyces cinereorectus, S. fradiae, S. tricolor, S. colombiensis, S. filamentous, S. vinaceus and S. phaeopurpureus. Syst Appl Microbiol. 2004;27:84–92. doi: 10.1078/0723-2020-00257. [DOI] [PubMed] [Google Scholar]
- 32.Levy SB, McMurry LM, Barbosa TM, Burdett V, Courvalin P, Hillen W, Roberts MC, Rood JI, Taylor DE. Nomenclature for new tetracycline resistance determinants. Antimicrob Agents Chemother. 1999;43:1523–1524. doi: 10.1128/aac.43.6.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Louw GE, Warren RM, Gey van Pittius NC, McEvoy CRE, Van Helden PD, Victor TC. A balancing act: efflux/ influx in mycobacterial drug resistance. Antimicrob Agents Chemother. 2009;53:3181–3189. doi: 10.1128/AAC.01577-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin P, Trieu-Cuot P, Courvalin P. Nucleotide sequence of the tetM tetracycline resistance determinant of the streptococcal conjugative shuttle transposon Tn1545. Nucleic Acids Res. 1986;14:7047–7058. doi: 10.1093/nar/14.17.7047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nash KA, Andini N, Zhang Y, Brown-Elliott BA, Wallace RJ., Jr Intrinsic macrolide resistance in rapidly growing mycobacteria. Antimicrob Agents Chemother. 2006;50:3476–3478. doi: 10.1128/AAC.00402-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nguyen L, Thompson CJ. Foundations of antibiotic resistance in bacterial physiology: the mycobacterial paradigm. Trends Microbiol. 2006;14:304–312. doi: 10.1016/j.tim.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 37.Nikolakopoulou TL, Egan S, van Overbeek LS, et al. PCR detection of oxytetracycline resistance genes otr(A) and otr(B) in tetracycline-resistant streptomycete isolates from diverse habitats. Curr Microbiol. 2005;51:211–216. doi: 10.1007/s00284-004-4430-4. [DOI] [PubMed] [Google Scholar]
- 38.Pang Y, Brown BA, Steingrube VA, Wallace RJ, Jr, Roberts MC. Tetracycline resistance determinants in Mycobacterium and Streptomyces species. Antimicrob Agents Chemother. 1994;38:1408–1412. doi: 10.1128/aac.38.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Redemaker JLW, Hoste B, Louws FJ, Kersters K, Swings J, Vauterin L, Vauterin P, van Bruijn JF. Comparison of AFLP and rep-PCR genomic fingerprinting with DNA-DNA homology studies: Xanthomonas as a model system. Int J Syst Evol Microbiol. 2000;50:665–677. doi: 10.1099/00207713-50-2-665. [DOI] [PubMed] [Google Scholar]
- 40.Roberts MC. Update on acquired tetracycline resistance genes. FEMS Microbiol Lett. 2005;245:195–203. doi: 10.1016/j.femsle.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 41.Rossi-Fedele G, Scott W, Spratt D, Gulabivala K, Roberts AP. Incidence and behaviour of Tn916-like elements within tetracycline-resistant bacteria isolated from root canals. Oral Microbiol Immunol. 2006;21:218–222. doi: 10.1111/j.1399-302X.2006.00279.x. [DOI] [PubMed] [Google Scholar]
- 42.Shirling EB, Gottlieb D. Methods for characterization of Streptomyces species. Int J Syst Bacteriol. 1966;16:313–340. [Google Scholar]
- 43.Šula L. WHO co-operative studies on a simple culture technique for the isolation of mycobacteria. 1. Preparation, lyophilization and reconstitution of a simple semi-synthetic concentrated liquid medium; culture technique, growth pattern of different mycobacteria. Bull World Health Organ. 1963;29:589–606. [PMC free article] [PubMed] [Google Scholar]
- 44.Swenson JM, Wallace RJ, Jr, Silcox VA, Thornsberry C. Antimicrobial susceptibility of five subgroups of Mycobacterium fortuitum and Mycobacterium chelonae. Antimicrob Agents Chemother. 1985;28:807–811. doi: 10.1128/aac.28.6.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Swindell SR, Plasterer TN. SEQMAN. Contig assembly. Methods Mol Biol. 1997;70:75–89. [PubMed] [Google Scholar]
- 46.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 47.Thioulouse J, Chessel D, Dolédec S, Olivier JM. ADE-4: a multivariate analysis and graphical display software. Stat Comput. 1997;7:75–83. [Google Scholar]
- 48.Tortoli E. Impact of genotypic studies on mycobacterial taxonomy: the new mycobacteria of the 1990s. Clin Microbiol Rev. 2003;16:319–354. doi: 10.1128/CMR.16.2.319-354.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vaerewijck MJM, Huys G, Palomino JC, Swings J, Portaels F. Mycobacteria in drinking water distribution systems: ecology and significance for human health. FEMS Microbiol Rev. 2005;29:911–934. doi: 10.1016/j.femsre.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 50.van Ingen J, Boeree MJ, Dekhuijzen PNR, van Soolingen D. Environmental sources of rapid growing nontuberculous mycobacteria causing disease in humans. Clin Microbiol Infect. 2009;15:888–893. doi: 10.1111/j.1469-0691.2009.03013.x. [DOI] [PubMed] [Google Scholar]
- 51.Versalovic J, Schneider M, de Bruijn FJ, Lupski JR. Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods Mol Cell Biol. 1994;5:25–40. [Google Scholar]
- 52.Wallace RJ, Dalovisio JR, Pankey GA. Disk diffusion testing of susceptibility of Mycobacterium fortuitum and Mycobacterium chelonei to antibacterial agents. Antimicrob Agents Chemoter. 1979;16:611–614. doi: 10.1128/aac.16.5.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamamura H, Harayma S. Method for selective isolation of mycobacteria using olive oil emulsified with SDS. Biosci Biotechnol Biochem. 2007;71:1553–1556. doi: 10.1271/bbb.60687. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.