Abstract
Roseobacter litoralis OCh149 is a type strain of aerobic anoxygenic phototrophic bacteria in marine Roseobacter clade. Its full genome has been sequenced; however, proteomic research, which will give deeper insights into the environmental stimuli on gene expression networks, has yet to be performed. In the present study, a proteomic approach was employed to analyze the status of R. litoralis OCh149 in carbon starvation during the stationary phase and its responses to a dark/light regimen (12 h:12 h) in both exponential and stationary phases. LC-MS/MS-based analysis of highly abundant proteins under carbon starvation revealed that proteins involved in transport, the transcription/translation process and carbohydrate metabolism were the major functional categories, while poly-β-hydroxyalkanoate (PHA), previously accumulated in cells, was remobilized after stress. Glucose, as the sole carbon source in the defined medium, was broken down by Entner-Doudoroff and reductive pentose phosphate (PP) pathways. Carbohydrate catabolism-related proteins were down-regulated under light regardless of the growth phase, probably due to inhibition of respiration by light. In contrast, responses of amino acid metabolisms to light regimen varied among different proteins during growth phases depending on cellular requirements for proliferation, growth or survival. Fluorescence induction and relaxation measurements suggested that functional absorption cross-sections of the photosynthetic complexes decreased during the dark period and always recovered to about the previous level during the light period. Although the photosynthetic genes in R. litoralis OCh149 are located on the plasmid, these data indicate the regulatory mechanism of photoheterotroph metabolism by both carbon and light availability.
Keywords: Roseobacter litoralis, proteomic analysis, carbohydrate metabolism, transporter, light regimen
On the ocean surface, 15%–25% of microbial communities consist of the Roseobacter clade, occupying diverse marine habitats (6, 7, 55). Some members of this clade are aerobic anoxygenic phototrophic bacteria (AAPB), which possess bacteriochlorophyll a (BChl a) and utilize sunlight as additional energy for their heterotrophic growth (2, 25). In addition to their ecological significance and biogeographic properties, a wide range of physiological features and capabilities have been revealed, such as symbiosis with algae (54), degradation of dimethylsulfoniopropionate (20), CO oxidation (32), and denitrification (16). Including generalists as well as specialists, the clade plays a significant role in marine biogeochemical cycles (35). Over the past two decades, from the first strain identified to the first genome sequenced, a great deal of knowledge has been gained for a better understanding of their enormous success in marine environments (32, 46). So far, 39 genomes of Roseobacter strains have been completely sequenced and annotated (up to 7 March 2012). Furthermore, some functional post-genomic studies have been carried out to examine predictive frameworks and explore metabolic pathways, such as the proteomic and fluxomic responses of Phaeobacter gallaeciensis and Dinoroseobacter shibae to different growth phases and the availability of glucose (53, 60). In contrast to the well-studied heterotrophic properties, especially the heterotrophic metabolic pathways of the Roseobacter clade, little is known about the proteomic response of photoheterotrophic growth in this clade (4).
Among the AAPB members in the marine Roseobacter clade, Roseobacter litoralis OCh149 is of special interest for photoheterotrophic studies. Unlike its closest phylogenetic species R. denitrificans OCh114, R. litoralis OCh149 features pufLM genes located on a linear plasmid and a much lower cellular content of BChl a (40, 46). The photosynthetic genes in R. litoralis OCh149 are obtained presumably through horizontal gene transfer (41). This special genomic organization of R. litoralis OCh149, distinct from all other BChl a containing members of the Rosobacter clade, makes it a specific model for photoheterotrophic metabolism. Considering the unusual location of the photosynthetic genes, we carried out proteomic analysis of R. litoralis OCh149 in response to carbon starvation in the stationary growth phase and light regimen in different growth phases, and demonstrated not only its photoheterotrophic property, but also the mechanism involved in starvation- and light-regulated proteins.
Materials and Methods
Cultivation of experimental bacterium
The R. litoralis OCh149 strain was obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ, Braunschweig, Germany). It was maintained and precultured aerobically at 25°C in rich organic medium (58), and stirred at a rate of 120 rpm in the dark. To simplify metabolic process analysis, a reported defined marine minimal medium (MMM) (27, 36) was used in the present study. Cells on entering the exponential phase (1/2 ODmax) were used to inoculate the main cultures, which were sampled for subsequent analysis. The main cultures were grown in 250 mL Erlenmeyer flasks containing 100 mL MMM. The final concentration of the carbon source in 100 mL MMM was provided by 0.2 g glucose.
Two groups of cells were incubated in a growth chamber (Conviron, Winnipeg, Canada) with four incandescent lamps (low light intensity 30 μE m−2 s−1): (1) light regimen incubation (light/dark cycle of 12 h:12 h), providing the dark period necessary for the synthesis of BChl a to retain the availability of the photosynthetic apparatus (PA); and (2) dark incubation (bottles wrapped tightly with aluminum foil), abolishing photosynthetic activity, as the control. The growth curves (Fig. S1C) of R. litoralis OCh149 in both experimental and control groups were determined by measuring optical density at 600 nm. Cells were harvested in either the exponential (after incubation for 118 h) or stationary phase (after incubation for 190 h) by centrifugation at 10,000×g for 10 min at 4°C. The resulting pellets were stored at −80°C until proteomic analysis was performed.
2D-GE
The cell pellets were suspended in 1 mL lysis buffer (7 M urea, 2 M thiourea, 2% CHAPS, 1% DTT, 4% TRITON-X 100, 2% carrier ampholytes, 5% protease inhibitor cocktail) (17). After incubation at room temperature for 30 min with period vortexing, the suspensions were sonicated on ice and centrifuged. The supernatants were directly taken for measurement of protein concentration with the RC DC kit (Bio-rad Laboratories, Hercules, CA, USA). The 2D-GE analysis was performed as follows: In brief, for isoelectric focusing (IEF), approximately 100 μg protein was loaded onto each precast 18 cm immobilized non-linear pH 4–7 strip (Bio-Rad Laboratories) and the strips were rehydrated for 12 h. IEF was carried out in an Ettan IPG-phor3 System (GE Healthcare, Buckinghamshire, UK). SDS-PAGE was carried out on 11.5% polyacrylamide gels in an Ettan DALT apparatus (GE Healthcare). To analyze the reproducibility of 2D gels, we repeated the experiments three times for each sample. Proteins were visualized using silver staining and scanned with an Image Scanner II system (GE Healthcare). Images were analyzed and quantitatively compared using PDQuest 8.0 Software (Bio-Rad Laboratories). After loading the gel images of each growth phase into PDQuest, the same set of triplicate gels was grouped together to determine the average quantities of the protein spots. There were two groups for each growth phases, the dark group and the light regimen group. The “dark” group was chosen as the control group. Spot intensities were normalized according to the mode “total quantity of valid spots” and analyses were performed in quantitative and qualitative modes. The confidence threshold for up- and down-regulation of protein spots was set at two-fold above or below the spot intensity seen in the control.
MALDI-TOF/TOF-MS/MS analysis
The in-gel digestion of proteins was performed according to a previous protocol with modifications (45). Protein spots of interest were cut from the silver-stained gels and were destained with a solution of 15 mM potassium ferricyanide and 50 mM sodium thiosulfate (1:1) for 20 min at room temperature. The gel pieces were then washed twice with deionized water, shrunk by dehydration in acetonitrile (ACN), and then swollen in digestion buffer containing 10 mM ammonium bicarbonate and 4 ng μL−1 trypsin (Promega, Madison, WI, USA) at 4°C. After 30 min incubation, the pieces were digested for more than 12 h at 37°C. Peptides were extracted twice using 5% trifluoroacetic acid (TFA) in 50% ACN. The extracts were dried under the protection of N2. MALDI-TOF/TOF analysis was carried out on an AB Sciex TOF/TOF 5800 (Applied Biosystems/Life Technologies, Carlsbad, CA, USA). Peptides were eluted onto the target with 0.7 μL matrix solution (a-cyano-4-hydroxy-cinnamic acid in 0.1% TFA, 50% ACN). Samples were allowed to air dry before loading them into the mass spectrometer.
LC-MS/MS analysis
Cellular proteins without fractionation from each treatment were obtained using a gel-free approach and digested according to filter-aided sample preparation as outlined by Jacek et al. (57) before being loaded onto nano-LC using an Ettan MDLC capillary LC system (GE Healthcare). Peptides were trapped and captured using a 100 μm i.d.×15 mm long pre-column packed with 200 Å, 5 μm MAGIC C18AQ particles. Peptides were separated on a 75 μm i.d.×150 mm long analytical column packed with 100 Å, 5 μm MAGIC C18AQ particles. Peptides were eluted using an acidified (formic acid, 0.1% [v/v]) water–acetonitrile gradient (5–35% acetonitrile for 60 min, 35%–95% acetonitrile for 35 min) at 250 nL min−1. Mass spectrometry was performed on a LTQ-Orbitrap mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA) with NSI. Data-dependent MS/MS spectra were obtained simultaneously. The heated capillary temperature and spray voltage were 200°C and 2.2 kV, respectively. The MS scan range was 300–2,000 m/z with a resolution R = 60,000 at m/z 400. For each cycle, the five most abundant ions from MS analysis were selected for MS/MS analysis using a collision energy setting of 35%. The dynamic exclusion settings were as follows: repeat count 1, repeat duration 30 s, exclusion duration 180 s. The experiments were repeated twice and these results were combined into the final result.
MS/MS data analysis
For MALDI-MS/MS spectra, all data searches were performed using MASCOT version 2.1 (Matrix Science, London, UK). The following settings were used: NCBInr database (release 20070627; 4182491 sequences; 1439956234 residues), bacteria, trypsin digestion with one missing cleavage, peptide tolerance of ±0.2 Da, fragment mass tolerance of ±0.3 Da, and possible oxidation of methionine.
For LC-MS/MS spectra, each data set was matched using Bioworks 3.2 software (Thermo Fisher Scientific, Waltham, MA, USA) against the R. litoralis OCh149 protein file containing 4,746 protein entries compiled from the annotation of the R. litoralis OCh149 genome (downloaded from NCBI on 24 March 2010, http://www.ncbi.nlm.nih.gov/). To estimate the false positive rate (FPR), reverse sequences of the R. litoralis OCh149 protein file were added to the database. The SEQUEST parameters were set to allow 0.5 Da fragment ion tolerance and 2 missed internal cleavage sites by trypsin. The results of the SEQUEST search were filtered using the Trans-Proteomic Pipeline 4.2.1 (downloaded from http://tools.proteomecenter.org/TPP.ph) with the minimum Peptide-Prophet and ProteinProphet thresholds set at 0.05 and 0.9, respectively. The criterion for protein identification was that at least one of its unique peptides was identified in three or more spectra or that at least two of its unique peptides were identified in one or more spectra. We applied reverse-database searching to determine the FPR. No sequence from the reversed database was detected in the results.
Morphological and physiological analysis
Cells of R. litoralis OCh149 for ultrastructural analysis were collected by centrifugation at 5,000×g for 15 min, and fixed in 0.1 M phosphate buffer (2.5% glutaraldehyde) for 12 h at 4°C. Cells were dehydrated with ethanol and treated for embedment in Spurr resin (Sigma-Aldrich, St. Louis, MO, USA). After ultrathin sectioning, the samples were observed under a transmission electron microscope.
The in vivo photophysiological parameter of the functional absorption cross section (σ) was measured using the FIRe fluorometer system (Satlantic, Halifax, Canada) at 12 h intervals of sampling, as described previously (50). Fluorescence signals were separated using the infrared filter (880 nm, 50 nm bandwidth) for BChl a fluorescence, and the total duration of signal photosynthetic turnover was 400 μs, which gradually closed the reaction centers, and average signals were obtained from 30 iterations of one sample.
We analyzed the C:N ratio of cells retained on a precombusted glass-fiber filter (Whatman GF/F, Whatman/GE Healthcare, Buckinghamshire, UK) with a Vario EL III elemental analyzer (Elementar Analysensysteme, Hanau, Germany). Triplicate samples were run, and the results were converted and calculated as atoms.
Results and Discussion
Abundant proteomic profiles of R. litoralis OCh149 in carbon starvation
LC-MS/MS proteomic analysis of the cells harvested in the stationary phase showed that light incubation yielded 710 unique peptides corresponding to 206 proteins, while dark incubation yielded 844 unique peptides corresponding to 219 proteins. Taken together, 271 abundant proteins (154 common proteins shared by both incubations) were identified from 1,119 unique peptides that collectively matched 3,315 tandem spectra.
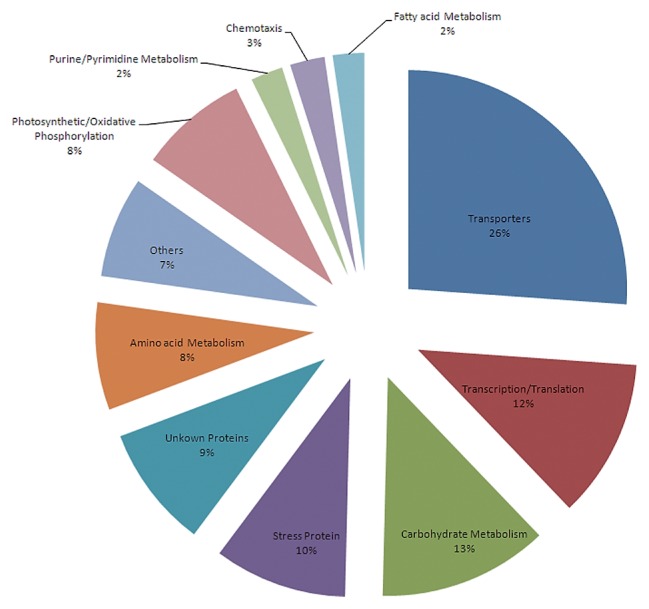
All detected proteins were divided into 11 groups by functional category (Fig. 1). Based on the number of tandem spectra, the transporters registered the largest proportion, accounting for 26% of the total. The percentages of housekeeping proteins, including proteins involved in carbohydrate metabolism, transcription/translation process, stress response, amino acid metabolism and photosynthetic/oxidative phosphorylation, ranged from 8% to 13% of the total number of tandem spectra. Other proteins, which were not associated with definite metabolic processes, accounted for 7%. Unknown proteins, expressed by genes encoding hypothetical or conserved hypothetical proteins, amounted to 9% of the detected spectra. The results concerning the proteome in the stationary phase using LC-MS/MS revealed abundant proteins and represented the dominant metabolism of R. litoralis OCh149 under carbon-limited conditions.
Fig. 1.
Percentages of the 11 protein groups identified by LC-MS/MS in R. litoralis OCh149 in the stationary phase. The functional categories were determined according to the database of the Kyoto Encyclopedia of Genes and Genomes (KEGG). The percentage of each protein group was calculated as follows: the number of tandem spectra of each functional category divided by the number of tandem spectra of total proteins.
Central metabolic pathways of R. litoralis OCh149
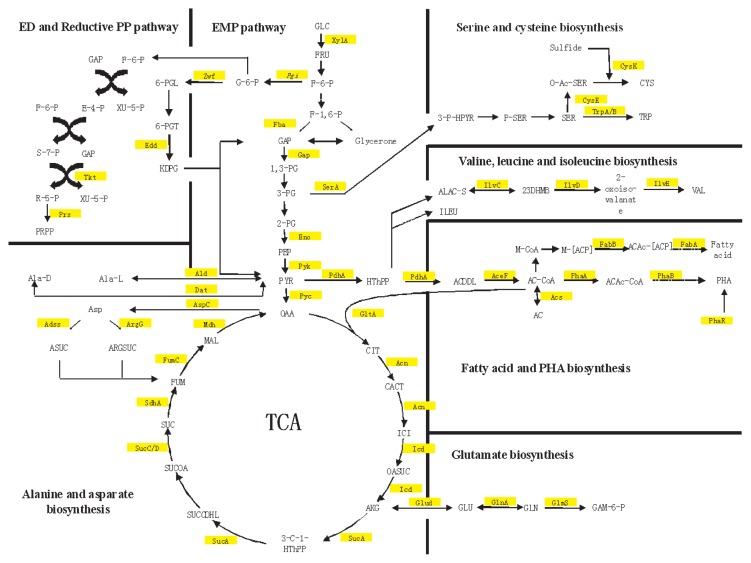
Based on the abundant proteomic profiles, the central metabolic pathways in R. litoralis OCh149 were characterized. This not only allowed improved understanding of the carbon metabolism initiated by glucose, but provided clues to the exploited nutrient metabolism as glucose became gradually unavailable. The deduced central pathways included carbohydrate, amino acid, fatty acid and purine/pyrimidine metabolisms (Fig. 2).
Fig. 2.
Central metabolic pathways of R. litoralis OCh149 in the stationary phase. Enzymes identified using LC-MS/MS are highlighted. Abbreviations (metabolites): GLC, glucose; FRU, fructose; F-6-P, fructose-6P; F-1,6-P, fructose-1,6P; GAP, glyceraldehyde-3P; 1,3-PG, glycerate-1,3P; 3-PG, glycerate-3P; 2-PG, glycerate-2P; PEP, phosphoenol-pyruvate; PYR, pyruvate; OAA, oxaloacetate; CIT, citrate; CACT, cis-aconitate; ICI, isocitrate; OASUC, oxalosuccinate; AKG, 2-oxo-glutarate; 3-C-1-HThPP, 3-carboxy-1-hydroxypropyl-ThPP; SUCCDHL, S-succinyldihydrolipoamide-E; SUCOA, succinyl-CoA; SUC, succinate; FUM, fumarate; MAL, malate; G-6-P, glucose-6P; 6-PGL, 6-phosphogluconolactone; 6-PGT, 6-phosphogluconate; KDPG, 2-dehydro-3-deoxygluconate-6P; E-4-P, erythrose-4P; XU-5-P, xylulose-5P; S-7-P, sedoheptulose-7P; R-5-P, ribose-5P; GLU, glutamate; GLN, glutamine; GAM-6-P, glucosamine-6P; Ala-L, L-alanine; Ala-D, D-alanine; Asp, asparatate; ARGSUC, arginino-succinate; ASUC, adenylo-succinate; 3-P-HPYR, 3P-hydroxy-pyruvate; P-SER, phosphoserine; SER, serine; O-Ac-SER, O-acetyl-serine; CYS, cysteine; TRP, L-tryptophan; ALAC-S, (s)-2-acetolactate; 23DHMB, (R)-2,3-dihydroxy-3-methylbutanoate; ILEU, isoleusine; VAL, valine; HTHPP, 2-hydroxyethyl-ThPP; ACDDL, S-acetyl-dihydrolipoamide-E; AC-CoA, acetyl-CoA; ACAc-CoA, acetoacetyl-CoA; M-CoA, malonyl-CoA; M-[ACP], malonyl-[acp]; ACAc-[ACP], acetoacetyl-[acp]; AC, acetate. Abbreviations (enzymes): XylA, xylose isomerase; Pgi, glucose-6-phosphate isomerase; Zwf, glucose-6-phosphate-1-dehydrogenase; Fba, fructose-bisphosphate aldolase; Gap, glyceraldehyde-3-phosphate dehydrogenase; Eno, enolase; Pyk, pyruvate kinase; Pyc, pyruvate carboxylase; Edd, phosphogluconate dehydratase; Tkt, transketolase; Prs, ribose-phosphate pyrophosphokinase putative; GltA, citrate synthase; Acn, aconitate hydratase; Icd, isocitrate dehydrogenase (NADP-dependent); SucA, alpha-ketoglutarate dehydrogenase; SucC, succinyl-CoA synthetase, beta subunit; SucD, succinyl-CoA synthetase, alpha subunit; SdhA, succinatedehydrogenase flavoprotein subunit; FumC, fumarate hydratase; Mdh, malate dehydrogenase; Ald, alanine dehydrogenase; Dat, D-alanine aminotransferase; AspC, aspartate aminotransferase; Adss, adenylosuccinate synthetase; ArgG, argininosuccinate synthase; CysE, serine acetyltransferase; CysK, O-acetylserine sulfhydrylase; TrpA, tryptophan synthase, alpha subunit; TrpB, tryptophan synthase, beta subunit; IlvC, ketol-acidreductoisomerase; IlvD, dihydroxy-acid dehydratase; IlvE, branched-chain amino acid aminotransferase; Glud, glutamate dehydrogenase; GlnA, glutamine synthetase, type I; GlmS, D-fructose-6-phosphate amidotransferase; PdhA, pyruvate dehydrogenase complex, E1 component, alpha subunit; AceF, dihydrolipoamide acetyltransferase; PhaA, acetyl-CoA acetyltransferase; PhaB, acetoacetyl-CoA reductase; PhaR, polyhydroxyalkanoate synthesis repressor; FabA, 3-hydroxydecanoyl-ACP dehydratase; FabB, 3-oxoacyl-(acyl-carrier-protein) synthase; Acs, acetyl-coenzyme A synthetase. Detailed information about the proteins is provided in supplemental material Table S1.
Carbohydrate metabolism is fundamental for bacterial heterotrophic metabolism, providing carbon skeletons, energy and intermediate products. In general, there are three metabolic pathways for the utilization of glucose as a carbon source: the Embden-Meyerhof-Parnas (EMP) pathway, the Entner-Doudoroff (ED) pathway and the pentose phosphate (PP) pathway (oxidative and non-oxidative). In the genome of R. litoralis OCh149, all the genes required for the ED pathway are present, while pfk (6-phosphofrucokinase) and pgd (6-phosphogluconate dehydrogenase) genes, which encode the key enzymes of the EMP and the oxidative PP pathway, respectively, are missing. In this study, Edd, Eda (2-keto-3-deoxy-phosphogluconate aldolase, see the 2-DE results below) and Tkt proteins were identified. The combination of genomic and proteomic data implied that R. litoralis OCh149 used the ED pathway for glucose breakdown and the non-oxidative PP pathway for nucleic acid, ATP and coenzymes synthesis.
For comparison of the glucose metabolic pathways in R. litoralis OCh149 and other members of the marine Roseobacter clade, all 39 available genomes were investigated (Table 1). The non-oxidative PP pathway exists in all 39 known Roseobacter genomes and almost all have the eda gene, except Roseovarius nubinhibens ISM and Roseovarius sp. 217. The EMP pathway only exists in Citreicella sp. SE45, Oceanicola granulosus HTCC2516, Pelagibaca bermudensis HTCC2601, Dinoroseobacter shibae DFL12, Maritimibacter alkaliphilus HTCC2654 and Sagittula stellata E-37, with the first two strains possessing all five key genes of the three metabolic pathways, while the EMP pathway has not been reported to be active in AAPB, such as D. shibae DFL12, which has been shown to catabolize glucose exclusively via the ED pathway (53). In contrast, some anaerobic anoxygenic phototrophic bacteria (AnAPB) do not possess the related genes in the ED pathway (52). This indicates that the ED pathway is mainly restricted to aerobic prokaryotes (both bacteria and archaea) (8, 15, 51) and members of the Roseobacter clade belong to AAPB, which depend on molecular oxygen to synthesize BChl a, suggesting that the ED pathway may have a significant role in glucose metabolism in AAPB (53, 60).
Table 1.
Survey of key genes in glucose metabolic pathways from 39 Roseobacter clade genomesa
| Genome | EMP | ED | Oxidative PP | Non-oxidative PP | |
|---|---|---|---|---|---|
|
| |||||
| pfk | eda | pgd | tkt | tal | |
| Citreicella sp. SE45 | + | + | + | + | + |
| Oceanicola granulosus HTCC2516 | + | + | + | + | + |
| Pelagibaca bermudensis HTCC2601 | + | + | + | + | |
| Dinoroseobacter shibae DFL12 | + | + | + | + | |
| Maritimibacter alkaliphilus HTCC2654 | + | + | + | + | |
| Sagittula stellata E-37 | + | + | + | + | |
|
| |||||
| Roseibium sp TrichSKD4 | + | + | + | + | |
| Jannaschia sp. CCS1 | + | + | + | + | |
| Oceanibulbus indolifex HEL-45 | + | + | + | + | |
| Octadecabacter antarcticus 238 | + | + | + | + | |
| Octadecabacter antarcticus 307 | + | + | + | + | |
| Ketogulonicigenium vulgare Y25 | + | + | + | + | |
| Rhodobacterales bacterium HTCC2150 | + | + | + | + | |
| Roseobacter sp. CCS2 | + | + | + | + | |
| Roseobacter sp. GAI101 | + | + | + | + | |
| Roseovarius sp. TM1035 | + | + | + | + | |
| Sulfitobacter sp. EE-36 | + | + | + | + | |
| Sulfitobacter sp. NAS-14.1 | + | + | + | + | |
| Thalassiobium sp. R2A62 | + | + | + | + | |
|
| |||||
| Rhodobacterales bacterium Y4I | + | + | + | ||
| Ruegeria pomeroyi DSS-3 | + | + | + | ||
| Ruegeria sp. TM1040 | + | + | + | ||
| Roseobacter denitrificans OCh 114 | + | + | + | ||
| Roseobacter litoralis OCh149 | + | + | + | ||
| Loktanella vestfoldensis SKA53 | + | + | + | ||
| Oceanicola batsensis HTCC2597 | + | + | + | ||
| Phaeobacter gallaeciensis BS107 | + | + | + | ||
| Phaeobacter gallaeciensis 2.10 | + | + | + | ||
| Rhodobacterales bacterium HTCC2083 | + | + | + | ||
| Roseobacter sp. AzwK-3b | + | + | + | ||
| Roseobacter sp. MED193 | + | + | + | ||
| Roseobacter sp. SK209-2-6 | + | + | + | ||
| Ruegeria lacuscaerulensis ITI-1157 | + | + | + | ||
| Ruegeria sp. TrichCH4B | + | + | + | ||
| Ruegeria sp. R11 | + | + | + | ||
| Rhodobacterales bacterium KLH11 | + | + | + | ||
|
| |||||
| Rhodobacterales bacterium HTCC2255 | + | + | |||
|
| |||||
| Roseovarius nubinhibens ISM | + | + | + | ||
| Roseovarius sp. 217 | + | + | + | ||
+ indicates the presence of a homolog with an E value ≤10−40 and amino acid percent sequence identity ≥40%.
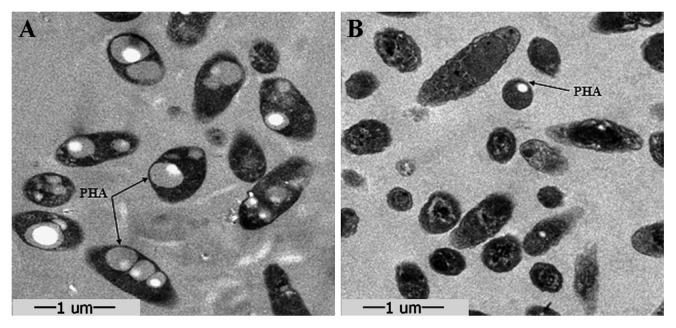
In order to further examine the central metabolism, the TCA cycle and anaplerotic pathways were reconstructed from the proteomic data. The capability for synthesis of amino acids (alanine, asparate, cysteine, serine and glutamate etc.), fatty acids and poly-β-hydroxyalkanoates (PHAs) was verified (Fig. 2). These substances were converted from glucose and contributed to the dissolved organic carbon pool in the cells; some of the substances could even be transformed to inclusion bodies for carbon storage in the cells (29). For example, PHAs are universal prokaryotic storage material with the proposed designation “carbonosome” (22). In heterotrophic bacteria they are accumulated under unbalanced growth conditions (N or P starvation) when carbon sources are still available, and provide the cells with carbon and energy when the external sources are deficient (60). R. litoralis OCh149 proteomic data provided evidence of the enzymes for the synthesis of PHAs. Six genes are involved in PHA biosynthesis and are organized into two distant clusters (phaAB and phaZCPR) in the R. litoralis OCh149 genome. The proteins encoded by three of these appeared in our proteomic data: PhaA, PhaB and PhaR. The existence of PHAs in R. litoralis OCh149 was also observed using electron microscopy of ultrathin sections and the identity of PHAs was also confirmed with Nile Blue staining (data not shown). In the cytoplasm of cells from pre-cultivation (rich organic medium), PHA granules were clearly accumulated (Fig. 3A), while in research about phototrophic bacteria, growing photoautotrophically under balanced or unbalanced conditions, glycogen was much more heavily accumulated than PHAs (14). The accumulation of PHAs in R. litoralis OCh149 therefore highlighted its heterotrophic characteristics. When R. litoralis OCh149 suffered from nutrient limitation in the stationary phase, the previously accumulated PHA granules were found to be smaller or even disappeared from the cells (Fig. 3B). This indicated that PHAs were degraded as carbon and energy sources during starvation; however, PhaZ (polyhydroxyalkanoate depolymerase), responsible for catalyzing the depolymerization of PHAs, was not detected. This could be due to the effects of other molecular mechanisms for PHA degradation. First, PhaR plays a negative role in the regulation of PHA synthesis (12, 28). Excess PhaR can repress the expression of PhaP (Phasin) by binding to upstream DNA regions of phaP and phaR (39). This could also explain why PhaR was detected, but PhaP was not detected in the present study. Second, PHA-associated proteins may also contribute to PHA degradation. PHAs are complexly organized subcellular structures with many proteins on the granule surface, some of which are essential for PHA metabolism (22). Further work is necessary to reveal the in vivo functions of these proteins.
Fig. 3.
Electron micrographs of ultrathin sections of R. litoralis OCh149 and PHA granules. (A) Cells grown in the preliminary medium (rich organic medium). (B) Cells in the late stationary phase in glucose (marine minimal medium).
Transporter proteins in R. litoralis OCh149
The large proportion of transporter proteins, as revealed by LC-MS/MS, was a noteworthy finding (Fig. 1). As Gram-negative bacteria, R. litoralis OCh149 exchanges substances between the periplasm and cytoplasm by specific substrate-binding transport systems (such as ATP-binding cassette [ABC] transporters, tripartite ATP-independent periplasmic [TRAP] transporters and TonB-dependent [TBD] transporters) and uses porins as molecular sieves in the outer membrane to allow hydrophilic molecules into the periplasmic space (3). In our experiment, three transport systems were detected: ABC transporters, TRAP transporters and porins. Spectra of ABC and TRAP transporters in particular accounted for 20% of the total spectra overall. This number situates R. litoralis OCh149 next to a mostly oligotrophic-adapted ubiquitous heterotrophic clade SAR11 (28%–35%) but above the phototrophic representative Rhodobacter sphaeroides (11%) (9, 47). This means that the ability of nutrient scavenging in R. litoralis OCh149 is more effective than in photoautotrophic bacteria, but less than in typical heterotrophic bacteria. From the viewpoint of physiology, this may be because ATP converted from light energy can substitute for the energy generated from organic substrate oxidation. A transcriptomic study of coastal bacterioplankton showed that Roseobacter is the taxon with the most abundant dissolved organic carbon-related transporter sequences (37). Roseobacter and SAR11, two clades of ubiquitous and abundant bacterioplankton in the surface ocean (18, 19, 34), possess a high proportion of transporters, which can be interpreted as an advantageous adaptation to diverse marine habitats and an ecologic strategy in low-nutrient environments.
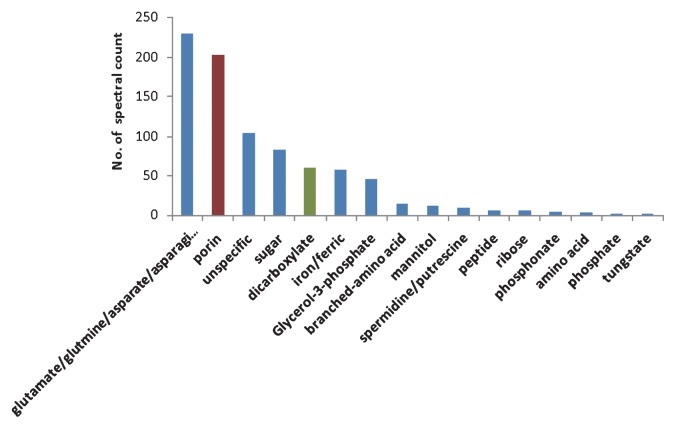
In addition to abundant transporters, further details on dissolved organic matter (DOM) utilization could be obtained. Among the ABC transporters, extracytoplasmic solute receptors (ESR) were identified, which were involved in the binding of amino acid, branched amino acid, peptide, sugar, mannitol, ribose, phosphate, phosphonate, ferric ion, and tungstate, and among the TARP transporters which were distributed across DctP and TAXI families, only C4-dicarboxylate-binding proteins were detected as ESR (Fig. 4, Table S1). In particular, glutamate/glutamine/aspartate/asparagine-binding protein was the most frequently detected, accounting for 26% of the total transporter spectra. The pathway for the synthesis of glutamate and aspartate from glucose could also be inferred (Fig. 2). This indicated that glutamate or aspartate is an important carbon and nitrogen source for R. litoralis OCh149. The dominant uptake of amino acid by R. litoralis OCh149 might explain the decrease of the bacterial carbon: nitrogen (C:N) ratio during its growth process (from 5.47±0.33 at the beginning to 3.86±0.02 at the end of culture). Such selective utilization of substrates may also influence the ambient DOM composition, which supports the proposition of the microbial carbon pump in the ocean (23). Transport systems for spermidine/putrescine and dicarboxylate were also detected and were considered to contribute to the uptake of algal osmolytes and the subsequent photo-oxidative products of DOM released by algae (10, 32, 33). Silicibacter pomeroyi DSS-3 and R. litoralis OCh149 genomes analyses, as genetic proofs, revealed the significant feature of the transport systems for algal osmolytes (13, 32). Roseobacter clade members commonly attach to, or live symbiotically with algae in the field (42, 54, 56), such as R. denitrificans with Enteromorpha linza (46), and Dinoroseobacter shibae DFL12 with a dinoflagellate (5). This indicates that Roseobacter closely associates with algae by substrate transporters, which are necessary for the transformation and uptake of DOM released from photosynthetically fixed organic carbon by phytoplankton (11).
Fig. 4.
Distribution of spectra from transporters according to the binding substances. Blue column, ABC transporters; green column, TRAP transporters; red column, outer membrane porin proteins.
Light-induced differential protein expression in different growth phases
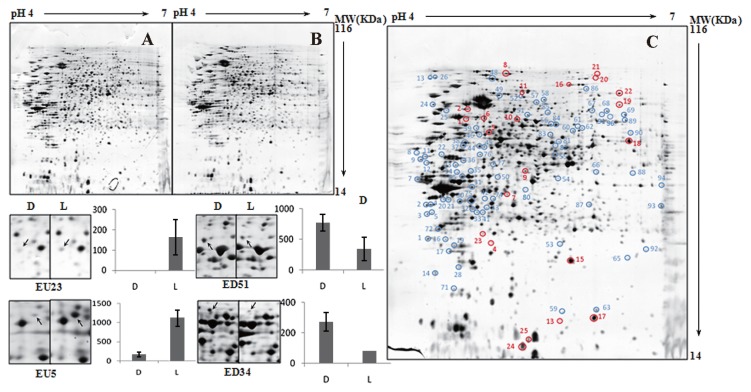
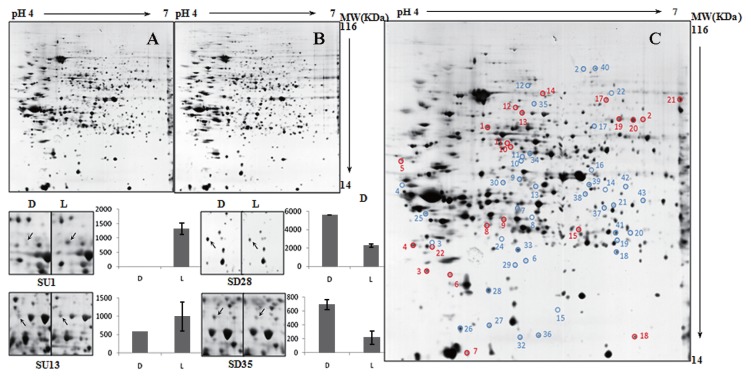
The results of comparative by 2D-GE proteomic analysis of cells harvested in both exponential and stationary phases provided insights into the differences in proteomic responses to light and nutrient stresses (Fig. 5 and 6).
Fig. 5.
2D-GE image analysis of the R. litoralis OCh149 proteome in exponential phase under dark and light regimen treatments. (A, B) A set of two gels from samples treated with dark (A) and light (B) regimens. (C) A representative gel showing the identified differentially expressed proteins. Red, proteins up-regulated in light regimen. Blue, proteins down-regulated in light regimen. (D) Typical examples of spots showing different expression profiles. D, under dark condition. L, under light regimen.
Fig. 6.
2D-GE image analysis of the R. litoralis OCh149 proteome in stationary phase under dark and light regimen treatments. (A, B) A set of two gels from samples treated with dark (A) and light (B) regimen. (C) A representative gel showing the identified differentially expressed proteins. Red, proteins up-regulated in light regimen. Blue, proteins down-regulated in light regimen. (D) Typical examples of spots showing different expression profiles. D, under dark condition. L, under light regimen.
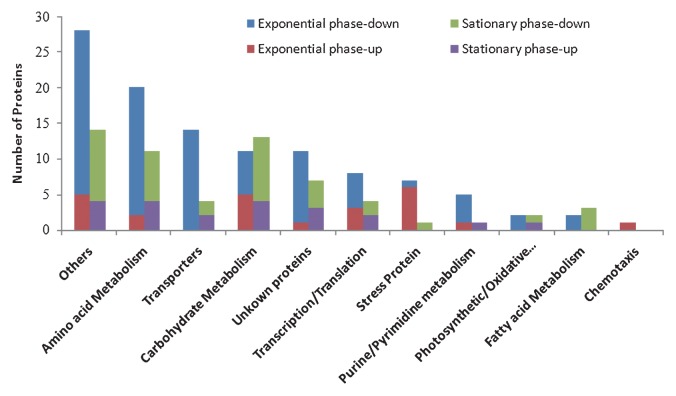
From cells in the exponential phase, 109 proteins were reported to exhibit differential expressions (Table 2), with 24 proteins being up-regulated and 85 proteins down-regulated under the light regimen. In particular, three protein groups were exclusively down-regulated, including transporters (14 proteins), those involved in the photosynthetic/oxidative phosphorylation process (two proteins) and fatty acid metabolism (two proteins), and one protein group involved in chemotaxis (one protein) was exclusively up-regulated. From cells in the stationary phase, 60 proteins were shown to be differentially expressed (Table 3), with 21 proteins being up-regulated and 39 down-regulated under the light regimen. Two protein groups, namely those involved in the stress response (one protein) and fatty acid metabolism (three proteins), were exclusively down-regulated, and one protein group associated with purine/pyrimidine metabolism (one protein) was exclusively up-regulated.
Table 2.
Proteins up- or down-regulated in exponential phase
| No.a | Accession No. | Gene | Protein Name | MW kDa | PI | Pep.Hits | Protein Score | Fold Changeb |
|---|---|---|---|---|---|---|---|---|
| Carbohydrate Metabolism | ||||||||
| EU9 | gi|163732692 | alcohol dehydrogenase, zinc-containing, putative | 35.40 | 5.05 | 13 | 804 | −2.33 | |
| EU16 | gi|163732764 | acsA | acetyl-coenzyme A synthetase | 69.53 | 5.37 | 25 | 682 | 1.58 |
| EU21 | gi|163732974 | tme | malic enzyme | 80.96 | 5.58 | 26 | 469 | L |
| ED17 | gi|163731504 | eda | KHG/KDPG aldolase | 21.62 | 4.55 | 6 | 131 | −5 |
| ED34 | gi|163731505 | frk | fructokinase | 32.66 | 4.62 | 8 | 540 | −6.66 |
| ED43 | gi|163734895 | tpiA | triosephosphate isomerase | 28.06 | 4.86 | 14 | 360 | −4.55 |
| ED60 | gi|163731508 | glgC | glucose-1-phosphate adenylyltransferase | 47.01 | 5.46 | 30 | 983 | −2.44 |
| ED63 | gi|163734455 | coxG | carbon monoxide dehydrogenase G protein, putative | 15.85 | 5.50 | 12 | 549 | −1.53 |
| ED79 | gi|163732070 | mvaB | hydroxymethylglutaryl-CoA lyase | 30.53 | 4.96 | 16 | 644 | −33.3 |
| ED61 | gi|163735981 | UDP-glucose/GDP-mannose dehydrogenase, putative | 48.88 | 5.44 | 7 | 169 | −5.88 | |
| ED36 | gi|163732615 | PfkB family kinase, putative | 35.03 | 4.66 | 15 | 531 | −3.45 | |
| Photosynthesis and Oxidative Phosphorylation | ||||||||
| ED54 | gi|163736059 | cobT | nicotinate-nucleotide--dimethylbenzimidazole phosphoribosyltransferase | 20.54 | 4.59 | 3 | 216 | −2.44 |
| ED72 | gi|110677947 | petA | ubiquinol-cytochrome c reductase, iron-sulfur subunit | 20.10 | 4.55 | 8 | 529 | −1.89 |
| Fatty acid Metabolism | ||||||||
| ED28 | gi|163731533 | accB | acetyl-CoA carboxylase, biotin carboxyl carrier protein, putative | 16.94 | 4.70 | 4 | 124 | −7.15 |
| Amino acid Metabolism | ||||||||
| EU10 | gi|163734687 | hisD | histidinol dehydrogenase | 51.40 | 5.46 | 14 | 411 | 2.20 |
| ED39 | gi|163732719 | hisD | histidinol dehydrogenase | 45.93 | 4.78 | 17 | 604 | −2.08 |
| EU18 | gi|163732684 | ald | alanine dehydrogenase | 38.74 | 5.71 | 19 | 722 | 2.23 |
| ED16 | gi|163732180 | hisA | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methyli deneamino] imidazole-4-carboxamide isomerase | 24.80 | 4.55 | 14 | 822 | −2.63 |
| ED35 | gi|163735980 | fmdA | formamidase, putative | 33.38 | 4.76 | 6 | 168 | −3.45 |
| ED32 | gi|163731988 | trpC | indole-3-glycerol phosphate synthase | 29.42 | 4.71 | 17 | 503 | −2.22 |
| ED41 | gi|163733282 | leuB | 3-isopropylmalate dehydrogenase | 39.96 | 4.63 | 10 | 263 | −2.78 |
| ED42 | gi|163733060 | arcB | ornithine cyclodeaminase, putative | 33.21 | 4.84 | 17 | 782 | −5 |
| ED45 | gi|163734587 | aspB | aspartate aminotransferase | 43.35 | 4.86 | 17 | 494 | −3.03 |
| ED76 | gi|163731559 | aspB | aspartate aminotransferase, putative | 39.31 | 4.67 | 16 | 773 | −1.72 |
| ED50 | gi|163733195 | serB | phosphoserine phosphatase, putative | 30.72 | 4.90 | 7 | 128 | −2.04 |
| ED51 | gi|163731381 | mmsA | methylmalonate-semialdehyde dehydrogenase | 53.98 | 5.15 | 24 | 684 | −2.22 |
| ED56 | gi|163735924 | paaK | phenylacetate-CoA ligase, putative | 44.68 | 5.29 | 13 | 349 | −2.04 |
| ED65 | gi|163731568 | ilvH | acetolactate synthase 3 small subunit | 20.13 | 5.77 | 8 | 277 | −2.78 |
| ED67 | gi|163733251 | serA | D-3-phosphoglycerate dehydrogenase | 56.27 | 5.67 | 11 | 268 | −2.63 |
| ED68 | gi|163733251 | serA | D-3-phosphoglycerate dehydrogenase | 56.27 | 5.67 | 23 | 834 | −2.86 |
| ED85 | gi|163733589 | ipuC | gamma-glutamylisopropylamide synthetase, putative | 51.04 | 5.28 | 20 | 346 | −1.92 |
| ED89 | gi|163731868 | aroG | phospho-2-dehydro-3-deoxyheptonate aldolase | 51.19 | 5.82 | 27 | 1,080 | −1.82 |
| ED90 | gi|163733560 | thrC | threonine synthase | 50.56 | 5.84 | 22 | 852 | −2.38 |
| ED93 | gi|163731356 | cysE | serine acetyltransferase | 29.35 | 6.10 | 22 | 965 | −1.96 |
| Purine/Pyrimidine Metabolism | ||||||||
| ED48 | gi|163734274 | polyribonucleotide nucleotidyltransferase | 76.75 | 4.88 | 30 | 1,090 | −2.33 | |
| ED77 | gi|163732267 | thioredoxin-disulfide reductase | 35.16 | 4.90 | 8 | 174 | −1.75 | |
| ED91 | gi|163731781 | guaB | inosine-5′-monophosphate dehydrogenase | 50.75 | 5.83 | 24 | 736 | −1.85 |
| ED69 | gi|163731781 | guaB | inosine-5′-monophosphate dehydrogenase | 50.75 | 5.83 | 26 | 984 | −2.08 |
| Transcription and Translation | ||||||||
| EU4 | gi|163734097 | npdA | NAD-dependent deacetylase | 26.53 | 5.04 | 7 | 367 | 2.07 |
| EU5 | gi|163732886 | tufA | elongation factor Tu | 42.84 | 4.90 | 19 | 802 | 5.41 |
| EU7 | gi|110679879 | rpsB | 30S ribosomal protein S2 | 28.19 | 5.02 | 10 | 448 | 2.05 |
| EU19 | gi|163733470 | proS | prolyl-tRNA synthetase | 51.03 | 5.71 | 27 | 696 | L |
| ED44 | gi|163734600 | endonuclease, putative | 39.54 | 4.76 | 18 | 898 | −2.13 | |
| ED81 | gi|163735131 | rnd | ribonuclease D | 43.88 | 5.39 | 16 | 298 | −1.79 |
| ED86 | gi|163735041 | thrS | threonyl-tRNA synthetase | 73.82 | 5.53 | 35 | 977 | −1.92 |
| ED87 | gi|163732970 | rph | ribonuclease PH | 25.55 | 5.55 | 9 | 514 | −5.26 |
| ED96 | gi|163735421 | ppsR | transcriptional regulator PpsR | 51.50 | 5.68 | 27 | 850 | D |
| Transporters | ||||||||
| ED2 | gi|163735254 | amino-acid ABC transporter, periplasmic binding protein | 30.06 | 4.37 | 11 | 355 | −3.13 | |
| ED4 | gi|163735254 | amino-acid ABC transporter, periplasmic binding protein | 30.06 | 4.37 | 16 | 558 | −3.12 | |
| ED22 | gi|163731684 | branched-chain amino acid ABC transporter, amino acid-binding protein | 42.35 | 4.55 | 16 | 715 | −2 | |
| ED3 | gi|163735168 | putative extracellular solute-binding protein | 29.86 | 4.48 | 8 | 365 | −2.5 | |
| ED25 | gi|163733639 | bacterial extracellular solute-binding protein, family 5 | 57.44 | 4.56 | 19 | 752 | −3.23 | |
| ED7 | gi|163735575 | ferric iron ABC transporter, periplasmic ferric iron-binding protein | 36.51 | 4.31 | 8 | 356 | −2.00 | |
| ED9 | gi|163731673 | ABC transporter, binding protein | 39.77 | 4.40 | 11 | 644 | −2.28 | |
| ED11 | gi|163731860 | pstS | phosphate ABC transporter, periplasmic phosphate-binding protein | 35.56 | 4.19 | 9 | 801 | −2.78 |
| ED12 | gi|163733174 | potF | putrescine ABC transporter, periplasmic putrescine-binding protein | 39.57 | 4.30 | 8 | 362 | −3.45 |
| ED20 | gi|163735933 | C4-dicarboxylate binding protein, putative | 37.17 | 4.65 | 15 | 680 | −4.76 | |
| ED21 | gi|163735933 | C4-dicarboxylate binding protein, putative | 37.17 | 4.65 | 16 | 660 | −2.44 | |
| ED23 | gi|163735737 | TRAP dicarboxylate transporter, DctP subunit, putative | 36.53 | 4.57 | 8 | 141 | −5 | |
| ED38 | gi|163734761 | sugar ABC transporter, periplasmic binding protein, putative | 47.91 | 4.72 | 17 | 885 | −2.22 | |
| ED82 | gi|163735383 | spermidine/putrescine ABC transporter, ATP-binding protein | 40.37 | 5.30 | 14 | 175 | −33.3 | |
| Stress Proteins | ||||||||
| EU8 | gi|163735915 | clpB | ATP-dependent Clp protease, ATP-binding subunit ClpB | 95.34 | 5.03 | 43 | 937 | 2.49 |
| EU15 | gi|163731525 | clpP | Clp protease, putative | 23.06 | 5.27 | 10 | 343 | 2.00 |
| EU11 | gi|163734797 | htpG | heat shock protein 90 | 73.10 | 5.26 | 11 | 76 | L |
| EU17 | gi|163732530 | heat shock protein, Hsp20 family | 17.52 | 5.66 | 15 | 580 | 2.14 | |
| EU13 | gi|163732530 | heat shock protein, Hsp20 family | 17.52 | 5.66 | 10 | 299 | L | |
| EU23 | gi|163734278 | antioxidant, AhpC/Tsa family, putative | 23.58 | 4.95 | 11 | 209 | 50 | |
| ED24 | gi|163733305 | serine protease DO-like precursor, putative | 51.46 | 4.50 | 25 | 749 | −4.17 | |
| ED84 | gi|163735794 | pepA | leucyl aminopeptidase | 51.59 | 5.33 | 21 | 1,090 | −1.79 |
| Chemotaxis | ||||||||
| EU24 | gi|110680249 | cheY | chemotaxis protein CheY, putative | 13.11 | 5.07 | 9 | 351 | L |
| Others | ||||||||
| ED71 | gi|163732956 | protein-export protein SecB, putative | 18.92 | 4.70 | 11 | 436 | −2.16 | |
| ED37 | gi|163732221 | dacC | D-alanyl-D-alanine carboxypeptidase | 41.66 | 4.71 | 17 | 685 | −2.33 |
| EU1 | gi|163734456 | arylsulfatase | 58.27 | 4.73 | 11 | 184 | 2.95 | |
| EU6 | gi|163734456 | arylsulfatase | 58.27 | 4.73 | 20 | 936 | 2.01 | |
| EU2 | gi|163735358 | sulfatase, putative | 60.10 | 4.78 | 21 | 720 | 2.38 | |
| EU20 | gi|163734500 | pta | phosphate acetyltransferase | 82.30 | 5.55 | 29 | 554 | 3.42 |
| EU22 | gi|115345666 | bifunctional sulfate adenylyltransferase subunit 1/adenylylsulfate kinase protein | 64.39 | 5.67 | 23 | 691 | 2.48 | |
| ED1 | gi|163731557 | outer membrane protein, putative | 27.77 | 4.44 | 14 | 803 | −2.33 | |
| ED5 | gi|163732591 | VacJ-like lipoprotein, putative | 26.01 | 4.44 | 16 | 679 | −3.45 | |
| ED8 | gi|163732359 | ggt | gamma-glutamyltranspeptidase | 61.18 | 4.52 | 8 | 202 | −8.33 |
| ED14 | gi|163735712 | YceI-like family protein | 20.76 | 4.58 | 7 | 305 | −3.23 | |
| ED26 | gi|115345661 | TPR repeat-containing protein | 88.21 | 4.37 | 14 | 300 | −2.13 | |
| ED30 | gi|163733017 | cysQ | 3′(2′),5′-bisphosphate nucleotidase | 28.71 | 4.62 | 17 | 850 | −2.13 |
| ED49 | gi|163734774 | monooxygenase protein, putative | 67.94 | 4.95 | 21 | 508 | −3.57 | |
| ED52 | gi|163732119 | metallopeptidase, family M24, putative | 64.79 | 5.12 | 22 | 454 | −5.56 | |
| ED53 | gi|163731615 | putative acetyltransferase | 24.09 | 5.33 | 11 | 179 | −2.33 | |
| ED57 | gi|163731748 | acy | penicillin acylase, putative | 91.03 | 5.13 | 18 | 459 | −2.27 |
| ED58 | gi|163734508 | AMP-binding domain protein | 58.70 | 5.23 | 19 | 702 | −2.13 | |
| ED62 | gi|163733176 | putative aminotransferase | 50.48 | 5.46 | 9 | 121 | −2.38 | |
| ED66 | gi|163733218 | zinc-binding dehydrogenase | 36.77 | 5.59 | 13 | 383 | −2.00 | |
| ED75 | gi|163732812 | immunogenic protein, putative | 34.13 | 4.87 | 13 | 515 | −2.86 | |
| ED83 | gi|163735942 | amidohydrolase family protein | 41.94 | 5.27 | 18 | 528 | −4.17 | |
| ED88 | gi|163735799 | pdxA | 4-hydroxythreonine-4-phosphate dehydrogenase | 34.31 | 5.77 | 8 | 285 | −2.84 |
| ED92 | gi|163732023 | oxidoreductase, putative | 22.12 | 5.93 | 10 | 356 | −2.07 | |
| ED94 | gi|163735137 | metallo-beta-lactamase family protein, putative | 33.56 | 6.06 | 9 | 252 | −3.13 | |
| Hypothetical Proteins | ||||||||
| EU25 | gi|163735582 | unkown protein RLO149_07769 | 17.02 | 5.16 | 4 | 117 | 7.93 | |
| ED13 | gi|163735835 | unkown protein RLO149_00770 | 43.35 | 4.27 | 10 | 369 | −2.44 | |
| ED19 | gi|163731610 | unkown protein RLO149_19939 | 21.62 | 4.59 | 8 | 313 | −4.76 | |
| ED31 | gi|163732009 | unkown protein RLO149_01122 | 33.60 | 4.69 | 12 | 336 | −2.70 | |
| ED33 | gi|163732240 | unkown protein RLO149_02277 | 36.23 | 5.59 | 13 | 316 | −3.45 | |
| ED46 | gi|163734706 | unkown protein RLO149_14803 | 45.06 | 4.85 | 11 | 198 | −3.33 | |
| ED55 | gi|163731979 | unkown protein RLO149_00972 | 42.28 | 5.46 | 11 | 316 | −2.33 | |
| ED59 | gi|163732720 | unkown protein RLO149_12800 | 18.17 | 5.37 | 17 | 592 | −2.56 | |
| ED74 | gi|163732284 | unkown protein RLO149_02497 | 31.69 | 4.88 | 11 | 251 | −1.89 | |
| ED80 | gi|163735585 | unkown protein RLO149_07784 | 34.70 | 5.20 | 11 | 426 | −33.3 | |
| ED95 | gi|163732428 | unkown protein RLO149_03217 | 28.68 | 4.86 | 13 | 500 | −1.72 | |
EU, up-regulated in light regimen in exponential phase; ED, down-regulated in light regimen in exponential phase
L, only expressed under light regimen conditions; D, only expressed under dark conditions
Table 3.
Proteins up- or down-regulated in stationary phase
| No.a | Accession No. | Gene | Protein Name | MWkDa | PI | Pep. Hits | Protein Score | Fold Changeb |
|---|---|---|---|---|---|---|---|---|
| Carbohydrate Metabolism | ||||||||
| SU10 | gi|163731361 | pgk | phosphoglycerate kinase | 41.43 | 5.09 | 15 | 466 | 2.34 |
| SU11 | gi|163734512 | phaA | acetyl-CoA acetyltransferase | 43.23 | 5.05 | 15 | 477 | 4.60 |
| SU12 | gi|115345672 | xanB | mannose-1-phosphate guanylyltransferase | 50.85 | 5.20 | 19 | 954 | 5.24 |
| SU21 | gi|163734174 | IpdA | dihydrolipoamide dehydrogenase | 49.37 | 6.15 | 14 | 357 | 2.33 |
| SD8 | gi|115345653 | rfbA | glucose-1-phosphate thymidylyltransferase | 31.67 | 4.90 | 4 | 99 | D |
| SD13 | gi|163735839 | nagZ | beta-hexosaminidase, putative | 36.13 | 5.19 | 21 | 917 | −11.11 |
| SD14 | gi|163733474 | malate/L-lactate dehydrogenase, putative | 37.29 | 6.08 | 3 | 223 | −7.69 | |
| SD24 | gi|163731409 | tpiA | triosephosphate isomerase | 25.60 | 4.92 | 7 | 359 | D |
| SD27 | gi|163734747 | fucU | fucose operon FucU protein, putative | 16.28 | 4.86 | 13 | 1,010 | −3.85 |
| SD33 | gi|163731776 | pgl | 6-phosphogluconolactonase | 23.75 | 5.06 | 8 | 503 | −2.08 |
| SD38 | gi|163733019 | gtaB | UTP--glucose-1-phosphate uridylyltransferase | 33.21 | 5.22 | 18 | 525 | −4.17 |
| SD42 | gi|163733025 | kdsD | arabinose 5-phosphate isomerase | 33.94 | 5.74 | 11 | 533 | −2.63 |
| Photosynthesis and Oxidative Phosphorylation | ||||||||
| SU13 | gi|163732297 | atpA | F0F1 ATP synthase subunit alpha | 54.68 | 5.26 | 23 | 633 | 4.03 |
| SD22 | gi|163732324 | electron transfer flavoprotein-ubiquinone oxidoreductase, putative | 60.01 | 5.59 | 26 | 1,120 | D | |
| SD29 | gi|163734669 | cobH | precorrin-8X methylmutase | 22.45 | 5.08 | 5 | 254 | −3.70 |
| Fatty acid Metabolism | ||||||||
| SD16 | gi|163731882 | lpxD | UDP-3-O-3-hydroxymyristoyl glucosamine N-acyltransferase | 38.27 | 5.52 | 19 | 727 | −6.67 |
| SD2 | gi|163734510 | fadJ | fatty acid oxidation complex alpha subunit, putative | 78.48 | 5.61 | 32 | 815 | −2.21 |
| SD40 | gi|163734510 | fadJ | fatty acid oxidation complex alpha subunit, putative | 78.48 | 5.61 | 33 | 1,080 | −2.33 |
| Amino acid Metabolism | ||||||||
| SU2 | gi|163731868 | aroG | phospho-2-dehydro-3-deoxyheptonate aldolase | 51.19 | 5.82 | 16 | 464 | L |
| SU19 | gi|163733991 | gill | glutamate dehydrogenase | 51.48 | 5.91 | 26 | 569 | 2.64 |
| SU20 | gi|163733991 | gill | glutamate dehydrogenase | 51.48 | 5.91 | 15 | 445 | 6.09 |
| SU22 | gi|163732180 | hisA | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methyli deneamino] imidazole-4-carboxamide isomerase | 24.80 | 4.55 | 6 | 157 | L |
| SD6 | gi|163734513 | gst | glutathione S-transferase, putative | 23.79 | 5.14 | 16 | 659 | −25 |
| SD20 | gi|110678669 | fahd | fumarylacetoacetate hydrolase, putative | 24.61 | 5.66 | 5 | 115 | −8.33 |
| SD21 | gi|163734433 | ilvE | branched-chain amino acid aminotransferase | 33.73 | 5.84 | 18 | 816 | −2.08 |
| SD30 | gi|163732015 | alr | alanine racemase | 36.75 | 4.99 | 17 | 1,040 | −2.00 |
| SD34 | gi|163732077 | ivd | isovaleryl-CoA dehydrogenase | 41.89 | 5.09 | 16 | 471 | −2.56 |
| SD35 | gi|163731381 | mmsA | methylmalonate-semialdehyde dehydrogenase | 53.98 | 5.15 | 19 | 1,020 | −3.03 |
| SD39 | gi|163734103 | argF | ornithine carbamoyltransferase | 34.26 | 5.41 | 15 | 747 | −2.44 |
| Purine/Pyrimidine Metabolism | ||||||||
| SU17 | gi|163735199 | hydA | dihydropyrimidinase | 53.12 | 5.51 | 20 | 391 | 9.68 |
| Transcription and Translation | ||||||||
| SU1 | gi|163732886 | tufA | elongation factor Tu | 42.84 | 4.90 | 17 | 1,020 | L |
| SU9 | gi|163733850 | chvI | DNA-binding response regulator ChvI, putative | 26.68 | 4.96 | 17 | 897 | 2.05 |
| SD18 | gi|163733026 | 3′-5′ exonuclease family protein, putative | 22.98 | 5.70 | 13 | 669 | −50 | |
| SD32 | gi|163732327 | greA | transcription elongation factor GreA | 17.14 | 5.03 | 5 | 133 | −2.18 |
| Transporters | ||||||||
| SU5 | gi|163731673 | ABC transporter, binding protein | 39.77 | 4.40 | 15 | 769 | 2.64 | |
| SU6 | gi|163732488 | ABC transporter, ATP-binding protein | 21.94 | 4.65 | 10 | 528 | 2.57 | |
| SD4 | gi|163735372 | sugar ABC transporter, substrate-binding protein, putative | 36.49 | 4.48 | 9 | 674 | D | |
| SD10 | gi|163733177 | polyamine ABC transporter, ATP-binding protein, putative | 41.39 | 5.08 | 20 | 694 | −12.5 | |
| Stress Proteins | ||||||||
| SD11 | gi|163734560 | phoH | phosphate starvation inducible protein, putative | 36.90 | 5.09 | 14 | 396 | −10.72 |
| Others | ||||||||
| SU4 | gi|163731557 | outer membrane protein, putative | 27.77 | 4.44 | 14 | 524 | 7.33 | |
| SU14 | gi|163731748 | acy | penicillin acylase, putative | 91.03 | 5.13 | 23 | 604 | 9.58 |
| SU15 | gi|163733001 | ubiG | 3-demethylubiquinone-9 3-methyltransferase | 27.74 | 5.41 | 11 | 387 | 1.05 |
| SU18 | gi|163732359 | ggt | gamma-glutamyltranspeptidase | 61.18 | 4.52 | 10 | 739 | 2.30 |
| SD3 | gi|163735850 | phospholipase/carboxylesterase family protein | 23.59 | 4.38 | 5 | 153 | D | |
| SD7 | gi|163736040 | oxidoreductase, putative | 29.79 | 5.07 | 15 | 683 | −33.3 | |
| SD9 | gi|163734732 | oxidoreductase, putative | 40.46 | 5.28 | 25 | 914 | −33.3 | |
| SD12 | gi|163732119 | metallopeptidase, family M24, putative | 64.79 | 5.12 | 21 | 494 | −5.88 | |
| SD17 | gi|163733176 | putative aminotransferase | 50.48 | 5.46 | 10 | 329 | −11.11 | |
| SD19 | gi|163735045 | hydrolase, putative | 26.05 | 5.73 | 11 | 394 | −20 | |
| SD26 | gi|163731362 | peptidyl-prolyl cis-trans isomerase, cyclophilin-type, putative | 18.17 | 4.74 | 7 | 566 | −2.08 | |
| SD28 | gi|163734126 | NifU-like domain protein | 20.21 | 4.92 | 7 | 356 | −2.22 | |
| SD36 | gi|163734683 | decarboxylase, putative | 18.74 | 5.16 | 4 | 237 | −2.33 | |
| SD43 | gi|163732693 | qor | quinone oxidoreductase | 34.36 | 5.88 | 10 | 534 | −2.33 |
| Unkown Proteins | ||||||||
| SU3 | gi|163732454 | unkown protein RLO149_03347 | 19.88 | 4.66 | 10 | 732 | 3.02 | |
| SU7 | gi|163731750 | unkown protein RLO149_20639 | 22.84 | 6.29 | 3 | 193 | 2.54 | |
| SU8 | gi|163734708 | unkown protein RLO149_14813 | 27.79 | 4.85 | 5 | 99 | 4.12 | |
| SD15 | gi|163732606 | unkown protein RLO149_12230 | 21.07 | 5.27 | 11 | 382 | −7.69 | |
| SD25 | gi|163731436 | unkown protein RLO149_19069 | 32.66 | 4.45 | 11 | 652 | −3.45 | |
| SD37 | gi|163734540 | unkown protein RLO149_13973 | 35.81 | 5.54 | 6 | 179 | −2.63 | |
| SD41 | gi|163735269 | unkown protein RLO149_06937 | 25.45 | 5.66 | 23 | 983 | −2.27 | |
SU, up-regulated in light regimen in stationary phase; SD, down-regulated in light regimen in stationary phase
L, only expressed under light regimen conditions; D, only expressed under dark conditions
In five functional categories of metabolism, down-regulated proteins outnumbered up-regulated proteins under the light regimen in both exponential and stationary phases, including amino acid metabolism, carbohydrate metabolism, and fatty acid metabolism, as well as other proteins and unknown proteins (Fig. 7). In some metabolic processes, such as transporters, transcription/translation, photosynthetic/oxidative phosphorylation, down-regulated proteins dominated exclusively in the exponential phase, while stress proteins and chemotaxis proteins were markedly up-regulated. Purine/pyrimidine metabolism-related proteins were mainly down-regulated in the exponential phase but up-regulated in the stationary phase (Fig. 7).
Fig. 7.
Proteins with differential expression were grouped by functional category. Up-regulated in the light regimen in exponential phase (red), down-regulated in the light regimen in exponential phase (blue), up-regulated in the light regimen in stationary phase (purple), down-regulated in the light regimen in stationary phase (green). Bar heights represent the numbers of proteins.
Glycolysis and TCA cycle-related proteins were enhanced in both exponential and stationary phases under dark conditions, and Eda protein, essential for the ED pathway, was also up-regulated in the exponential phase in the dark (Table 2). In addition, the abundance of fructose 1,6-bisphosphate aldolase (Fba), as determined by the above LC-MS/MS results, was higher in the dark (10 spectra) than in the light regimen condition (5 spectra) (Fig. 2, Table S1). These indicated that glucose catabolism was inhibited by light in R. litoralis OCh149 due to the common electron carriers shared by photosynthetic and respiratory electron transfer systems (21, 59); however, Synechocystis sp. PCC 6803, an oxygenic photoautotroph, exhibits light-activated heterotrophic growth (LAHG) with the induced expression of Fba (49), and LAHG is common to other photoautrophic organisms (61). The differential responses of heterotrophic growth to light between the two phototrophic clades (anoxygenic and oxygenic) were also verified in field research, which demonstrated that light enhanced leucine incorporation by Cyanobacteria, while there was no such reaction in AAPB (30). Furthermore, in the present experiment, the synthesis of peptidoglycan (nagZ) and lipopolysaccharide (rfbA, xanB, kdsD), the metabolism of gluconeogenesis and other carbohydrates (tpiA, glgC, mvaB, pgl, gtaB) were also activated in the dark. CO oxidation by members of the widespread Roseobacter clade, which is a non-ignorable process in the global carbon cycle, was examined previously through their genome sequences (7, 31, 32, 48). Here, carbon-monoxide dehydrogenase G protein (coxG), which can oxidize CO, was detected and repressed by light. Further, by carbonic anhydrase, the production of CO2 was attached to PEP, or PYR to form OAA, as a CO2 assimilation process.
The response of amino acid metabolism to light varied with the growth phase. In the exponential growth phase, the majority of differentially expressed proteins were down-regulated under light, which participated in the synthesis of serine (serA/B), cysteine (cysE), tryptophane (trpC), leucine (leuB), glutamine (ipuC), aspartate (aspB), threonine (thrC), proline (arcB), histidine (hisA), aromatic (aroG) and other branched-chain amino acids (ilvH). In the stationary growth phase, the up-regulated proteins were involved in the synthesis of glutamate (gill), histidine (hisA) and aromatic amino acid (aroG), while the down-regulated proteins were pertinent to the synthesis of alanine (alr), arginine (argF) and branched-chain amino acid (ilvE). The above data implied that eugonic cells enhanced amino acid synthesis in dark conditions so as to support cell growth and proliferation with enough carbon and nitrogen sources. The metatranscriptomic results for the microbial community also suggest that nighttime accumulation of amino acid is an important pathway of cellular nitrogen storage (38). Furthermore, in the stationary phase, cells under the light regimen and dark conditions may require different amino acids as a result of the light-stimulated cellular DOM availability to heterotrophic metabolism (1), and light-enhanced synthesis of glutamate in the stationary phase may be involved in the production of glutathione that protects cells against light stress. Considering leucine, in the exponential phase it was synthesized and converted into components of general metabolism, such as acetoacetate and acetyl-CoA, to participate in active carbohydrate metabolism, and the initiation of the stationary phase led to its degradation in the dark by the detection of down-regulated isovaleryl-CoA dehydrogenase (ivd).
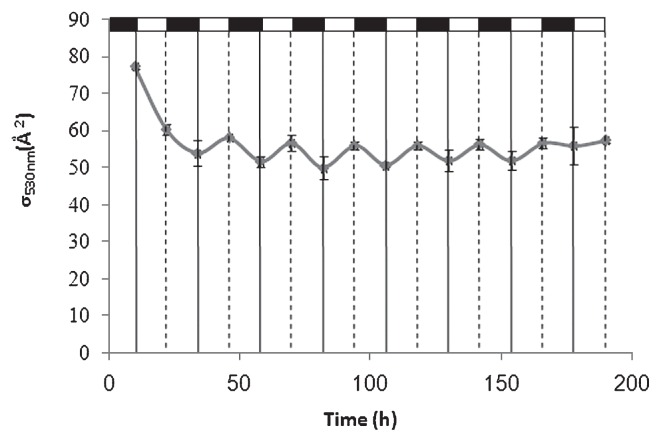
The response of photosynthetic units in R. litoralis OCh149 to the light regimen was directly followed by fluorescence induction and relaxation (FIRe) measurements that generated variable BChl a fluorescence transients (26). Analysis of the induction kinetics provided information on the functional absorption cross-section (σ) size of the photosynthetic unit (reaction center and light-harvesting systems) (24). The time-series detection of the value at 12 h intervals showed that it displayed a mild increase under light conditions, but under dark incubation it decreased reversibly at the end of the dark period and reached an elevated level after 12 hours of light (Fig. 8). This indicated that low light intensity stimulated the synthesis of the light-harvesting complexes. In some purple photosynthetic bacteria, such as Rhodopseudomonas palustris and Rhodospirillum photometricum, more antenna rings developed under low-light conditions (43, 44). In addition, proteins related to stress and chemotaxis were up-regulated in the exponential phase. This phenomenon may be considered as the response of active cells to light stimulation by the functioning of PA proteins.
Fig. 8.
Functional absorption cross-sections (σ) determined by FIRe from whole cell samples after excitation at 530 nm. The top zebra stripe indicates the dark:light (12 h:12 h) cycles of the light regimen. The experiment was performed three times, and the means ± standard errors of the mean are shown.
In summary, light can regulate R. litoralis OCh149 photoheterotrophic metabolism by inhibiting its carbohydrate metabolism and stimulating the formation of a functional absorption cross-section and cellular chemotaxis. In addition, in different growth phases, light can enhance different types of amino acid biosynthesis.
Conclusions
Proteomic analysis of R. litoralis OCh149 in the present study provided evidence for an adaptive mechanism in AAPB to stressful environments and the role of light in regulating photoheterotrophic metabolism. The significant proportion of transporters in R. litoralis OCh149 demonstrated its effective strategy for rapid carbon/nutrient acquisition; the observed PHA storage and re-utilization showed an intracellular carbon regulation mechanism of R. litoralis OCh149 in accordance with extracellular carbon availability. The recorded high proportion of periplasmic glutamate/glutamine/asparate/asparagines-binding proteins in the total transporter spectra along with their corresponding metabolism pathways confirmed the preference for glutamate by R. litoralis OCh149, suggesting bacterial selective use of carbon sources. Light-induced variations in carbohydrate and amino acid metabolism indicated that light plays a role in the regulation of heterotrophic metabolism in AAPB. These findings indicate that proteomics allow the analysis of comprehensive mechanisms such as metabolic adaptations to environmental conditions. Further proteomic study of more AAPB type strains will provide insights into photoheterotrophic processes mediating carbon and energy cycles in the ocean.
Supplementary Material
Acknowledgements
This work was supported by NSFC projects 91028001 and 41076063; SOA project 201105021. We thank Dr. Meinhard Simon, the editor and two anonymous reviewers for the valuable comments and helpful suggestions. Professor John Hodgkiss of The University of Hong Kong is thanked for his assistance with polishing the English.
References
- 1.Alonso-Sáez L, Gasol JM, Lefort T, Hofer J, Sommaruga R. Effect of natural sunlight on bacterial activity and differential sensitivity of natural bacterioplankton groups in northwestern Mediterranean coastal waters. Appl Environ Microbiol. 2006;72:5806–5813. doi: 10.1128/AEM.00597-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Béjà O, Suzuki MT, Heidelberg JF, Nelson WC, Preston CM, Hamada T, Eisen JA, Fraser CM, DeLong EF. Unsuspected diversity among marine aerobic anoxygenic phototrophs. Nature. 2002;415:630–633. doi: 10.1038/415630a. [DOI] [PubMed] [Google Scholar]
- 3.Benz R, Bauer K. Permeation of hydrophilic molecules through the outer membrane of gram-negativ bacteria. Eur J Biochem. 1988;176:1–19. doi: 10.1111/j.1432-1033.1988.tb14245.x. [DOI] [PubMed] [Google Scholar]
- 4.Berghoff BA, Glaeser J, Nuss AM, Zobawa M, Lottspeich F, Klug G. Anoxygenic photosynthesis and photooxidative stress: a particular challenge for Roseobacter. Environ Microbiol. 2011;13:775–791. doi: 10.1111/j.1462-2920.2010.02381.x. [DOI] [PubMed] [Google Scholar]
- 5.Biebl H, Allgaier M, Tindall BJ, Koblizek M, Lünsdorf H, Pukall R, Wagner-Döbler I. Dinoroseobacter shibae gen. nov., sp nov., a new aerobic phototrophic bacterium isolated from dinoflagellates. Int J Syst Evol Microbiol. 2005;55:1089–1096. doi: 10.1099/ijs.0.63511-0. [DOI] [PubMed] [Google Scholar]
- 6.Brinkhoff T, Giebel HA, Simon M. Diversity, ecology, and genomics of the Roseobacter clade: a short overview. Arch Microbiol. 2008;189:531–539. doi: 10.1007/s00203-008-0353-y. [DOI] [PubMed] [Google Scholar]
- 7.Buchan A, González JM, Moran MA. Overview of the marine Roseobacter lineage. Appl Environ Microbiol. 2005;71:5665–5677. doi: 10.1128/AEM.71.10.5665-5677.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Budgen N, Danson M. Metabolism of glucose via a modified Entner-Doudoroff pathway in the thermoacidophilic archaebacterium Thermoplasma acidophilum. FEBS Lett. 1986;196:207–210. [Google Scholar]
- 9.Callister SJ, Dominguez MA, Nicora CD, Zeng X, Tavano CL, Kaplan S, Donohue TJ, Smith RD, Lipton MS. Application of the accurate mass and time tag approach to the proteome analysis of sub-cellular fractions obtained from Rhodobacter sphaeroides 2.4.1. aerobic and photosynthetic cell cultures. J Proteome Res. 2006;5:1940–1947. doi: 10.1021/pr060050o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan KL, New D, Ghandhi S, Wong F, Lam CMC, Wong JTY. Transcript levels of the eukaryotic translation initiation factor 5A gene peak at early G1phase of the cell cycle in the dinoflagellate Crypthecodinium cohnii. Appl Environ Microbiol. 2002;68:2278–2284. doi: 10.1128/AEM.68.5.2278-2284.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen W, Wangersky PJ. Rates of microbial degradation of dissolved organic carbon from phytoplankton cultures. J Plankton Res. 1996;18:1521–1533. [Google Scholar]
- 12.Chou ME, Chang WT, Chang YC, Yang MK. Expression of four pha genes involved in poly-β-hydroxybutyrate production and accumulation in Rhodobacter sphaeroides FJ1. Mol. Genet Genomics. 2009;282:97–106. doi: 10.1007/s00438-009-0448-4. [DOI] [PubMed] [Google Scholar]
- 13.Kalhoefer D, Thole S, Voget S, Lehmann R, Liesegang H, Wollher A, Daniel R, Simon M, Brinkhoff T. Comparative genome analysis and genome-guided physiological analysis of Roseobacter litoralis. BMC Genomics. 2011;12:324–342. doi: 10.1186/1471-2164-12-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Philippis R, Ena A, Guastiini M, Sili C, Vincenzini M. Factors affecting poly-β-hydroxybutyrate accumulation in cyanobacteria and in purple non-sulfur bacteria. FEMS Microbiol Lett. 1992;103:187–194. [Google Scholar]
- 15.De Rosa M, Gambacorta A, Nicolaus B, Giardina P, Poerio E, Buonocore V. Glucose metabolism in the extreme thermoacidophilic archaebacterium Sulfolobus solfataricus. Biochem J. 1984;224:407–414. doi: 10.1042/bj2240407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doi M, Shioi Y. Enhancement of denitrifying activity in cells of Roseobacter denitrificans grown aerobically in the light. Plant Cell Physiol. 1991;32:365–370. [Google Scholar]
- 17.Gade D, Theiss D, Lange D, et al. Towards the proteome of the marine bacterium Rhodopirellula baltica: Mapping the soluble proteins. Proteomics. 2005;5:3654–3671. doi: 10.1002/pmic.200401201. [DOI] [PubMed] [Google Scholar]
- 18.Giebel HA, Kalhoefer D, Lemke A, Thole S, Gahl-Janssen R, Simon M, Brinkhoff T. Distribution of Roseobacter RCA and SAR11 lineages in the North Sea and characteristics of an abundant RCA isolate. ISME J. 2010;5:8–19. doi: 10.1038/ismej.2010.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giebel HA, Brinkhoff T, Zwisler W, Selje N, Simon M. Distribution of Roseobacter RCA and SAR11 lineages and distinct bacterial communities from the subtropics to the Southern Ocean. Environ Microbiol. 2009;11:2164–2178. doi: 10.1111/j.1462-2920.2009.01942.x. [DOI] [PubMed] [Google Scholar]
- 20.González JM, Simó R, Massana R, Covert JS, Casamayor EO, Pedrós-Alió C, Moran MA. Bacterial community structure associated with a dimethylsulfoniopropionate-producing North Atlantic algal bloom. Appl Environ Microbiol. 2000;66:4237–4246. doi: 10.1128/aem.66.10.4237-4246.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gray K, Daldal F. Mutational studies of the cytochrome bc1 complexes. Anoxygenic Photosynthetic Bacteria. 2004;2:747–774. [Google Scholar]
- 22.Jendrossek D. Polyhydroxyalkanoate granules are complex subcellular organelles (carbonosomes) J Bacteriol. 2009;191:3195–3202. doi: 10.1128/JB.01723-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiao N, Herndl GJ, Hansell DA, et al. Microbial production of recalcitrant dissolved organic matter: long-term carbon storage in the global ocean. Nat Rev Microbiol. 2010;8:593–599. doi: 10.1038/nrmicro2386. [DOI] [PubMed] [Google Scholar]
- 24.Koblizek M, Shih JD, Breitbart SI, Ratcliffe EC, Kolber ZS, Hunter CN, Niederman RA. Sequential assembly of photosynthetic units in Rhodobacter sphaeroides as revealed by fast repetition rate analysis of variable bacteriochlorophyll a fluorescence. Biochim Biophys Acta—Bioenergetics. 2005;1706:220–231. doi: 10.1016/j.bbabio.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 25.Kolber ZS, Plumley FG, Lang AS, et al. Contribution of aerobic photoheterotrophic bacteria to the carbon cycle in the ocean. Science. 2001;292:2492–2495. doi: 10.1126/science.1059707. [DOI] [PubMed] [Google Scholar]
- 26.Kolber ZS, Prášil O, Falkowski PG. Measurements of variable chlorophyll fluorescence using fast repetition rate techniques: defining methodology and experimental protocols. Biochim Biophys Acta—Bioenergetics. 1998;1367:88–106. doi: 10.1016/s0005-2728(98)00135-2. [DOI] [PubMed] [Google Scholar]
- 27.Mårdén P, Tunlid A, Malmcrona-Friberg K, Odham G, Kjelleberg S. Physiological and morphological changes during short term starvation of marine bacterial isolates. Arch Microbiol. 1985;142:326–332. [Google Scholar]
- 28.Maehara A, Taguchi S, Nishiyama T, Yamane T, Doi Y. A repressor protein, PhaR, regulates polyhydroxyalkanoate (PHA) synthesis via its direct interaction with PHA. J Bacteriol. 2002;184:3992–4002. doi: 10.1128/JB.184.14.3992-4002.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mas J, Gemerden HV. Storage products in purple and green sulfur bacteria. Anoxygenic Photosynthetic Bacteria. 2004;2:973–990. [Google Scholar]
- 30.Michelou VK, Cottrell MT, Kirchman DL. Lightstimulated bacterial production and amino acid assimilation by Cyanobacteria and other microbes in the North Atlantic Ocean. Appl Environ Microbiol. 2007;73:5539–5546. doi: 10.1128/AEM.00212-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moran MA, Belas R, Schell MA, et al. Ecological genomics of marine roseobacters. Appl Environ Microbiol. 2007;73:4559. doi: 10.1128/AEM.02580-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moran MA, Buchan A, Gonzalez JM, et al. Genome sequence of Silicibacter pomeroyi reveals adaptations to the marine environment. Nature. 2004;432:910–913. doi: 10.1038/nature03170. [DOI] [PubMed] [Google Scholar]
- 33.Moran MA, Zepp RG. Role of photoreactions in the formation of biologically labile compounds from dissolved organic matter. Limnol Oceanogr. 1997;42:1307–1316. [Google Scholar]
- 34.Morris RM, Rappe MS, Connon SA, Vergin KL, Siebold WA, Carlson CA, Giovannoni SJ. SAR11 clade dominates ocean surface bacterioplankton communities. Nature. 2002;420:806–810. doi: 10.1038/nature01240. [DOI] [PubMed] [Google Scholar]
- 35.Newton RJ, Griffin LE, Bowles KM, et al. Genome characteristics of a generalist marine bacterial lineage. ISME J. 2010;4:784–798. doi: 10.1038/ismej.2009.150. [DOI] [PubMed] [Google Scholar]
- 36.Östling J, Goodman A, Kjelleberg S. Behaviour of IncP-1 plasmids and a miniMu transposon in a marine Vibrio sp.: isolation of starvation inducible lac operon fusions. FEMS Microbiol Lett. 1991;86:83–94. [Google Scholar]
- 37.Poretsky R, Sun S, Mou X, Moran M. Transporter genes expressed by coastal bacterioplankton in response to dissolved organic carbon. Environ Microbiol. 2010;12:616–627. doi: 10.1111/j.1462-2920.2009.02102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poretsky RS, Hewson I, Sun SL, Allen AE, Zehr JP, Moran MA. Comparative day/night metatranscriptomic analysis of microbial communities in the North Pacific subtropical gyre. Environ Microbiol. 2009;11:1358–1375. doi: 10.1111/j.1462-2920.2008.01863.x. [DOI] [PubMed] [Google Scholar]
- 39.Potter M, Madkour MH, Mayer F, Steinbuchel A. Regulation of phasin expression and polyhydroxyalkanoate (PHA) granule formation in Ralstonia eutropha H16. Microbiology. 2002;148:2413–2426. doi: 10.1099/00221287-148-8-2413. [DOI] [PubMed] [Google Scholar]
- 40.Pradella S, Allgaier M, Hoch C, Pauker O, Stackebrandt E, Wagner-Dobler I. Genome organization and localization of the pufLM genes of the photosynthesis reaction center in phylogenetically diverse marine Alphaproteobacteria. Appl Environ Microbiol. 2004;70:3360–3369. doi: 10.1128/AEM.70.6.3360-3369.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raymond J, Zhaxybayeva O, Gogarten JP, Gerdes SY, Blankenship RE. Whole-genome analysis of photosynthetic prokaryotes. Science. 2002;298:1616–1620. doi: 10.1126/science.1075558. [DOI] [PubMed] [Google Scholar]
- 42.Riemann L, Steward GF, Azam F. Dynamics of bacterial community composition and activity during a mesocosm diatom bloom. Appl Environ Microbiol. 2000;66:578–587. doi: 10.1128/aem.66.2.578-587.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scheuring S, Goncalves RP, Prima V, Sturgis JN. The photosynthetic apparatus of Rhodopseudomonas palustris: structures and organization. J Mol Biol. 2006;358:83–96. doi: 10.1016/j.jmb.2006.01.085. [DOI] [PubMed] [Google Scholar]
- 44.Scheuring S, Sturgis JN. Chromatic adaptation of photosynthetic membranes. Science. 2005;309:484–487. doi: 10.1126/science.1110879. [DOI] [PubMed] [Google Scholar]
- 45.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc. 2006;1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 46.Shiba T. Roseobacter litoralis gen. nov., sp. nov., and Roseobacter denitrificans sp. nov., aerobic pink-pigmented bacteria which contain bacteriochlorophyll a. Syst Appl Microbiol. 1991;14:140–145. [Google Scholar]
- 47.Sowell S, Wilhelm L, Norbeck A, Lipton M, Nicora C, Barofsky D, Carlson C, Smith R, Giovanonni S. Transport functions dominate the SAR11 metaproteome at low-nutrient extremes in the Sargasso Sea. ISME J. 2008;3:93–105. doi: 10.1038/ismej.2008.83. [DOI] [PubMed] [Google Scholar]
- 48.Swingley WD, Sadekar S, Mastrian SD, et al. The complete genome sequence of Roseobacter denitrificans reveals a mixotrophic rather than photosynthetic metabolism. J Bacteriol. 2007;189:683. doi: 10.1128/JB.01390-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tabei Y, Okada K, Makita N, Tsuzuki M. Light-induced gene expression of fructose 1,6-bisphosphate aldolase during heterotrophic growth in a cyanobacterium, Synechocystis sp. PCC 6803. FEBS J. 2009;276:187–198. doi: 10.1111/j.1742-4658.2008.06772.x. [DOI] [PubMed] [Google Scholar]
- 50.Tang K, Zong R, Zhang F, Xiao N, Jiao NZ. Characterization of the photosynthetic apparatus and proteome of Roseobacter denitrificans. Curr Microbiol. 2010;60:124–133. doi: 10.1007/s00284-009-9515-7. [DOI] [PubMed] [Google Scholar]
- 51.Tang KH, Feng XY, Tang YJJ, Blankenship RE. Carbohydrate metabolism and carbon fixation in Roseobacter denitrificans OCh114. PLoS ONE. 2009;4:e7233. doi: 10.1371/journal.pone.0007233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tang KH, Tang YJ, Blankenship RE. Carbon metabolic pathways in phototrophic bacteria and their broader evolutionary implications. Fron Microbiol. 2011;2:165. doi: 10.3389/fmicb.2011.00165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fürch T, Preusse M, Tomasch J, Zech H, Wagner-Döbler I, Rabus R, Wittmann C. Metabolic fluxes in the central carbon metabolism of Dinoroseobacter shibae and Phaeobacter gallaeciensis, two members of the marine Roseobacter clade. BMC Microbiol. 2009;9:209–219. doi: 10.1186/1471-2180-9-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wagner-Döbler I, Ballhausen B, Berger M, et al. The complete genome sequence of the algal symbiont Dinoroseobacter shibae: a hitchhiker’s guide to life in the sea. ISME J. 2010;4:61–77. doi: 10.1038/ismej.2009.94. [DOI] [PubMed] [Google Scholar]
- 55.Wagner-Döbler I, Biebl H. Environmental biology of the marine Roseobacter lineage. Annu Rev Microbiol. 2006;60:255–280. doi: 10.1146/annurev.micro.60.080805.142115. [DOI] [PubMed] [Google Scholar]
- 56.Wagner-Döbler I, Thiel V, Eberl L, et al. Discovery of complex mixtures of novel long-chain quorum sensing signals in free-living and host-associated marine alphaproteobacteria. ChemBioChem. 2005;6:2195–2206. doi: 10.1002/cbic.200500189. [DOI] [PubMed] [Google Scholar]
- 57.Wiśniewski J, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 58.Yurkov V, Krieger S, Stackebrandt E, Beatty JT. Citromicrobium bathyomarinum, a novel aerobic bacterium isolated from deep-sea hydrothermal vent plume waters that contains photosynthetic pigment-protein complexes. J Bacteriol. 1999;181:4517–4525. doi: 10.1128/jb.181.15.4517-4525.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yurkov VV, Beatty JT. Aerobic anoxygenic phototrophic bacteria. Microbiol Mol Biol Rev. 1998;62:695–724. doi: 10.1128/mmbr.62.3.695-724.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zech H, Thole S, Schreiber K, Kalhöfer D, Voget S, Brinkhoff T, Simon M, Schomburg D, Rabus R. Growth phase-dependent global protein and metabolite profiles of Phaeobacter gallaeciensis strain DSM 17395, a member of the marine Roseobacter-clade. Proteomics. 2009;9:3677–3697. doi: 10.1002/pmic.200900120. [DOI] [PubMed] [Google Scholar]
- 61.Zotina T, Köster O, Jüttner F. Photoheterotrophy and light-dependent uptake of organic and organic nitrogenous compounds by Planktothrix rubescens under low irradiance. Freshwat Biol. 2003;48:1859–1872. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.