Abstract
Denitrification occurs markedly in rice paddy fields; however, few microbes that are actively involved in denitrification in these environments have been identified. In this study, we used a laboratory soil microcosm system in which denitrification activity was enhanced. DNA and RNA were extracted from soil at six time points after enhancing denitrification activity, and quantitative PCR and clone library analyses were performed targeting the 16S rRNA gene and denitrification functional genes (nirS, nirK and nosZ) to clarify which microbes are actively involved in denitrification in rice paddy soil. Based on the quantitative PCR results, transcription levels of the functional genes agreed with the denitrification activity, although gene abundance did not change at the DNA level. Diverse denitrifiers were detected in clone library analysis, but comparative analysis suggested that only some of the putative denitrifiers, especially those belonging to the orders Neisseriales, Rhodocyclales and Burkholderiales, were actively involved in denitrification in rice paddy soil.
Keywords: denitrification, nirS, nirK, nosZ
Denitrification is a microbial reaction in which NO3− and NO2− are successively reduced to NO, N2O and N2 (16, 47). It is an important part of the nitrogen cycle in the biosphere (8, 28). In upland agricultural fields, denitrification can cause the loss of fertilizer nitrogen and the emission of N2O, a greenhouse gas. Although denitrification commonly occurs in rice paddy soil, the emission of N2O gas is much lower than in upland crop fields (1, 16, 25). This indicates that microbes are highly active in N2O reduction in rice paddy field soils.
Because many types of bacteria (3, 6, 20), archaea (8, 27) and fungi (8, 35) are known to have the ability to denitrify, it is difficult to detect denitrifiers by a culture-independent approach using the 16S rRNA gene alone (28). Instead, primers targeting the cytochrome cd1-containing nitrite reductase gene (nirS) and the copper-containing nitrite reductase gene (nirK) are frequently used as marker genes to examine denitrifier communities in various environmental samples (4, 17, 18, 29, 31, 38, 44). In addition, the nitrous oxide reductase gene (nosZ) can be used to detect N2O-reducing denitrifiers (10, 15, 22, 30).
Previously, we analyzed the community structure of the denitrifiers in rice paddy soil by DNA-based clone library analysis targeting nirS and nirK (45, 46). In these studies, clones related to the nirS of Burkholderiales and the nirK of Rhizobiales were frequently detected in soil with strong denitrification activity; however, clones distantly related to the known denitrifiers were also detected. Although DNA-based analysis is useful for examination of the structure of denitrifier communities, it is not clear whether these microbes actually perform denitrification. RNA-based analysis can detect the transcription of functional genes; therefore, it can be used to detect active denitrifiers in the environment (9, 21, 26, 34, 36, 43).
Consequently, the objectives of this study were (1) to analyze the amount of bacterial 16S rRNA and the transcription levels of nirS and nirK, (2) to analyze microbial community structures before and after inducing denitrification, (3) to examine the diversity of nirS, nirK and nosZ and their transcripts and (4) to compare results obtained by DNA- and RNA-based analyses.
Materials and Methods
On 23 April 2009, soil was collected from a rice paddy field at Niigata Crop Research Center, Niigata Agricultural Research Institute, Nagaoka, Niigata, Japan (37°44′N, 138°87′E). The soil samples were sieved with 2-mm mesh to remove gravel and plant roots, and the samples were stored at 4°C. The soil type was gley soil. Physicochemical characteristics of the soil have been described elsewhere (12).
A previously established laboratory soil microcosm system (32) was used with modifications in this study. In brief, 5 g moist soil (corresponding to ca. 3 g air-dried soil) was pre-incubated in a serum vial at 30°C for 1 week with 5.4 mL sterilized distilled water to decrease soil redox potential. After pre-incubation, excess water was removed, and 0.3 mg N NO3− and 1.5 mg C succinate were added as an electron acceptor and electron donor, respectively, for denitrification. The vials were then anaerobically incubated with Ar gas at 30°C. Soil samples were collected from replicated vials (n=3) after 0-, 6-, 12-, 16-, 20- and 24-h incubation. Another set of vials was incubated with Ar-C2H2 (90:10, v/v) to measure denitrifying activities in the soil microcosm by the C2H2 block method (32).
DNA was extracted and purified from 0.5 g soil using an UltraClean Soil DNA Isolation kit (MoBio Laboratories, Carlsbad, CA, USA). Purified DNA was diluted 50- and 10-fold for PCR and quantitative PCR (qPCR), respectively, to adjust the DNA concentration to the proper condition for PCR, and to reduce the influence of the PCR inhibitors (e.g., humic substances).
RNA extraction was performed as described previously (41). RNA was extracted from 3.2 g soil using a RNA PowerSoil Total RNA Isolation kit (MoBio Laboratories). Extracted RNA was purified using Illustra MicroSpin Columns S-400 HR (GE Healthcare, Buckinghamshire, UK) to remove humic substances, and the remaining DNA was removed using a Turbo DNA-free kit (Applied Biosystems, Foster City, CA, USA). RNA was further purified and concentrated using an RNA Clean & Concentrator-5 kit (Zymo Research, Orange, CA, USA). The amount of purified RNA was measured using a NanoDrop 1000 (NanoDrop Products, Wilmington, DE, USA). The absence of DNA carryovers in the RNA samples was verified by PCR targeting the 16S rRNA gene without reverse transcription. Purified RNA was reverse transcribed using random hexamers and PrimeScript Reverse Transcriptase (Takara Bio, Otsu, Shiga, Japan) with 20 U of SUPERase-In RNase Inhibitor (Applied Biosystems). Synthesized cDNA was diluted 10-fold for PCR and qPCR, and 1,000-fold for qPCR targeting 16S rRNA gene. Replicate DNA and cDNA samples were pooled and used for clone library analyses.
qPCR was performed using a StepOne Real-Time PCR System (Applied Biosystems) with Power SYBR Green PCR Master Mix (Applied Biosystems). Primers 357F and 520R (19, 24), modified cd3aF and R3cd (17), and nirK876F and nirK1040R (17) were used for quantification of the bacterial 16S rRNA gene, nirS, and nirK, respectively. PCR was performed under the conditions described elsewhere (5, 17, 23). Amplification of correctly sized products was verified by dissociation curve analysis and agarose gel electrophoresis. Statistical significance among the amount of genes and gene transcripts was examined by analysis of variance at α=0.05 using R program version 2.8.1, as previously described (13).
For clone library analysis, DNA and cDNA samples were amplified with the modified primers 27F and 1492R (39), Cd3aF and R3cd (38), F1aCu and R3Cu (38) and nosZ-F-1181 and nosZ-R-1880 (30) for the bacterial 16S rRNA gene, nirS, nirK and nosZ, respectively. These primers, different from those used for qPCR, were employed because longer PCR products were suitable for clone library analysis. The PCR reaction mixture (20 μL) contained 10 mM Tris-HCl (pH 7.5), 10 mM NaCl, 0.01 mM EDTA, 0.2 mM dithiothreitol, 5% (w v−1) glycerol, 1.5 mM MgCl2, 0.2 mM each dNTP, 1 μM each of the forward and reverse primers, 10 μg bovine serum albumin, 2 units of BioTaq HS DNA polymerase (Bioline, London, UK) and 1 μL soil DNA or cDNA. After incubation for 10 min at 95°C, PCR was performed in a thermal cycler, the GeneAmp PCR System 9700 (Applied Biosystems), under the conditions described elsewhere (30, 38, 39). For samples with low concentrations of PCR template (nirK and nosZ amplicons from RNA samples and nirS amplicons from RNA samples extracted from the soil before incubation), a second PCR reaction was performed using a 10-fold diluted product of the first PCR reaction as the template. The PCR products were ligated into the pGEM-T vector system (Promega, Madison, WI, USA) and transferred to Escherichia coli JM109 high-efficiency competent cells (Promega) according to the manufacturer’s instructions. Colonies were randomly selected, and the inserted fragments were sequenced as described previously (45).
The 16S rRNA clones sharing nucleotide sequence homology at more than 99% were grouped into one operational taxonomic unit (OTU) using DOTUR program ver. 1.53 (33). The 16S rRNA clones were classified into bacterial taxa using the Ribosomal Database Project classifier program (40). Sequences from nirS, nirK and nosZ clones were translated, and OTU values and diversity indices were calculated as described previously (46). Phylogenetic trees were constructed using ClustalX ver. 1.83 based on the nucleotide sequences of 16S rRNA or 16S rRNA gene, and the deduced amino acid sequences from nirS, nirK and nosZ.
The nucleotide sequences obtained in this study have been deposited in the DDBJ, EMBL and GenBank databases under the following accession numbers: AB672106–AB672500.
Results
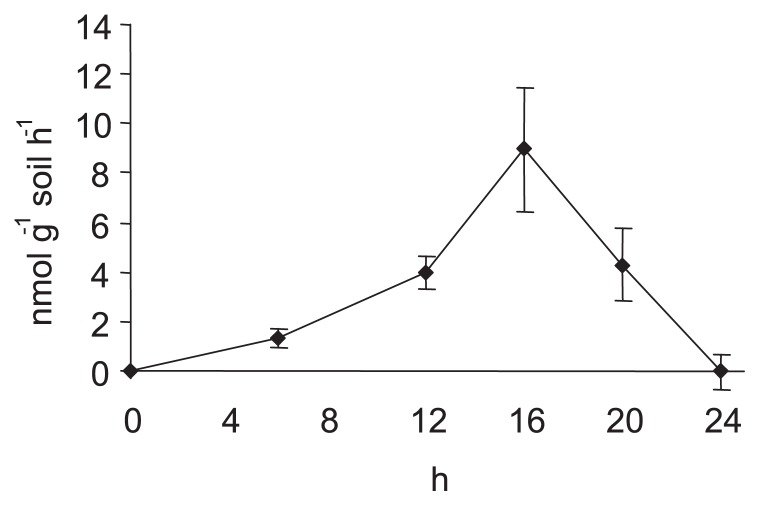
The denitrification activity based on N2O production increased with incubation time and peaked 16 h after the start of incubation (Fig. 1).
Fig. 1.
Denitrifying activities in the soil microcosm system as measured by the C2H2 block method.
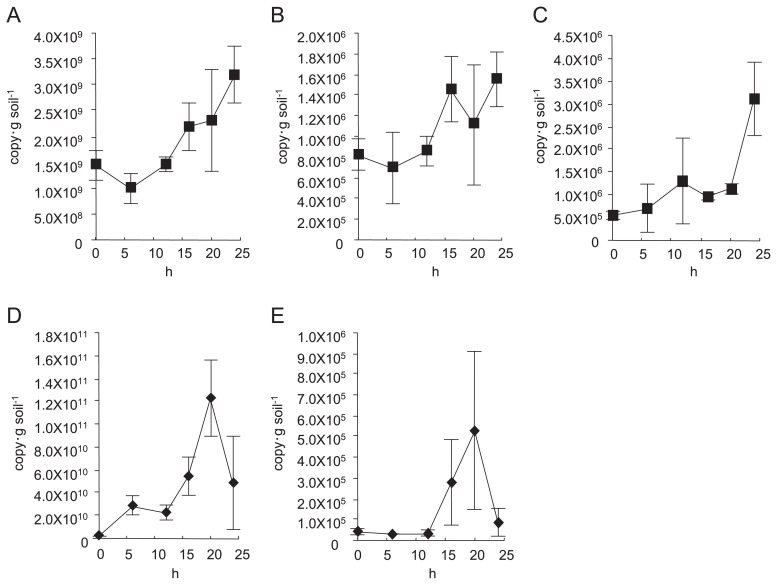
Based on DNA-based analysis, copy numbers of the 16S rRNA gene and nirK increased as the incubation proceeded (p=0.014 and p=0.006) (Fig. 2A and C), whereas the amount of nirS did not show a significant change (Fig. 2B). Conversely, based on RNA-based analysis, the amount of 16S rRNA significantly increased and peaked 20 h after the start of incubation (p=0.002). The amount of nirS showed a similar trend to that of 16S rRNA, but the tendency was not significant (p=0.13) (Fig. 2E). The amount of nirK product was below the detection limit.
Fig. 2.
Changes in the amounts of (A) 16S rRNA gene, (B) nirS, (C) nirK from DNA samples and (D) 16S rRNA, (E) nirS transcripts from cDNA samples. X axes show incubation time. Y axes show numbers of gene copies or gene transcripts.
Clone library analysis was performed with DNA and RNA samples obtained from soil before incubation and after 20-h incubation, the time-point at which the amount of 16S rRNA transcripts peaked (Fig. 2D). The numbers of obtained clones are shown in Table 1. We could not obtain nosZ amplicons from the RNA samples extracted from the soil before incubation. All of the nirK clones obtained from the RNA samples extracted from the soil before incubation and half of the nirK clones obtained from the soil after incubation were pseudogenes, and therefore, they were removed from subsequent analysis.
Table 1.
Diversity indices of the clone libraries obtained in this study
| Name of soil sample | clones | OTUs | ChaoI | Shannon (H′) | Simpson (1/D) | |
|---|---|---|---|---|---|---|
| D0 | 16S rRNA | 93 | 82 | 637.0 | 4.342 | 267.375 |
| NirS | 84 | 43 | 64.2 | 3.499 | 34.515 | |
| NirK | 31 | 7 | 8.0 | 1.410 | 3.370 | |
| NosZ | 26 | 12 | 19.5 | 2.302 | 12.500 | |
| D20 | 16S rRNA | 63 | 54 | 348.0 | 3.901 | 130.200 |
| NirS | 129 | 51 | 76.3 | 3.676 | 42.557 | |
| NirK | 45 | 7 | 8.0 | 1.648 | 4.853 | |
| NosZ | 82 | 17 | 24.0 | 2.199 | 5.796 | |
| R0 | 16S rRNA | 35 | 19 | 26.5 | 2.781 | 22.037 |
| NirS | 89 | 9 | 12.3 | 0.811 | 1.514 | |
| R20 | 16S rRNA | 148 | 63 | 149.0 | 3.455 | 16.383 |
| NirS | 99 | 21 | 26.3 | 2.527 | 8.601 | |
| NirK | 23 | 2 | 2.0 | 0.669 | 1.992 | |
| NosZ | 88 | 8 | 8.5 | 1.496 | 3.455 |
D0, DNA extracted from the soil before incubation; D20, DNA extracted from the soil after incubation; R0, cDNA extracted from the soil before incubation; R20, cDNA extracted from the soil after incubation.
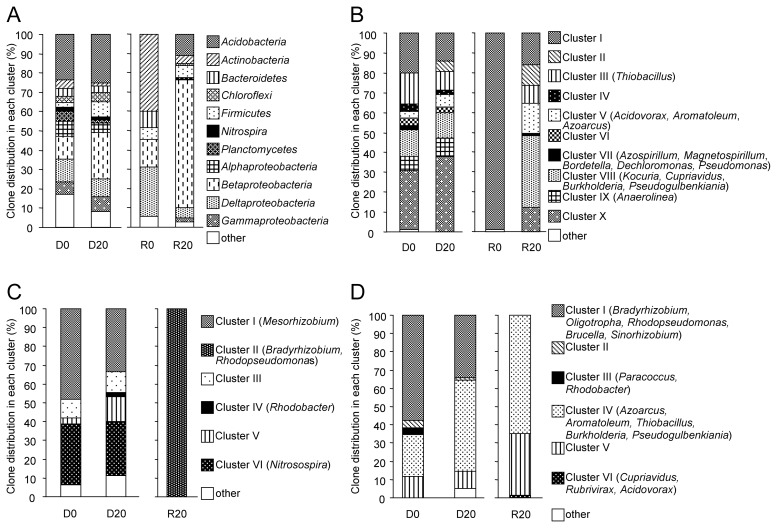
Clones of the 16S rRNA gene were widespread in the phylogenetic tree (Fig. S1). Based on DNA-based analysis, community structures were similar between soil samples before and 20 h after incubation (Fig. 3A); however, RNA-based analysis showed a marked population change between these samples (Fig. 3A). In addition, the diversity in the 16S rRNA gene was larger in the DNA samples than in the RNA samples (Table 1). In the RNA samples, proportions of the clones related to the class Betaproteobacteria and the phylum Acidobacteria increased by 20-h incubation from 14.3% to 66.2% and from 0.0% to 10.8%, respectively. Within Betaproteobacteria, clones related to the orders Neisseriales and Rhodocyclales accounted for a large proportion. In contrast, the proportions of clones related to the phyla Actinobacteria and Bacteroidetes and the class Deltaproteobacteria were decreased by incubation from 40.0% to 4.1%, 8.6% to 0% and 25.7% to 5.4%, respectively.
Fig. 3.
Changes in the distribution of (A) 16S rRNA, (B) nirS, (C) nirK and (D) nosZ clones in response to denitrification-inducing conditions. Clones were grouped into clusters based on the phylogenetic trees shown in Supplementary Materials (Fig. S1, 2, 3, and 4).
Clones of nirS were also widespread in the phylogenetic tree (Fig. S2). Similar to the results based on the 16S rRNA gene, the composition of clones did not change significantly in the DNA samples, whereas that of clones in the RNA samples changed greatly. In addition, the diversity of nirS was larger in the DNA samples than in the RNA samples (Table 1). In the RNA samples, almost all nirS clones obtained from soil before incubation were grouped in Cluster I, whereas clones from soil after incubation were spread throughout the phylogenetic tree. Clones obtained from both DNA and RNA samples were found in Clusters I, II, III, V, VII, VIII, and X. Although clones in Clusters I, II, and X were distantly related to the nirS of known denitrifiers, those in Cluster III were related to the nirS of Hydrogenophilales bacteria (Thiobacillus spp.), those in Cluster V to the nirS of Rhodocyclales bacteria (Acidovorax spp. and Aromatoleum spp.), those in Cluster VII to the nirS of Rhodospirillales bacteria (Azospirillum spp. and Magnetospirillum spp.), Burkholderiales bacteria (Bordetella spp.), Rhodocyclales bacteria (Dechloromonas spp.) and Pseudomonadales bacteria (Pseudomonas spp.), and those in Cluster VIII to the nirS of Burkholderiales bacteria (Burkholderia spp. and Cupriavidus spp.) and Actinobacteria (Kocuria spp.). Clones in Cluster VIII accounted for 36.4% of clones obtained from the RNA samples after incubation (Fig. 3B). All clones in Clusters IV, VI and IX were from DNA samples, not from RNA samples.
The nirK clones were also widespread in the phylogenetic tree (Fig. S3); however, the sequence compositions were different between DNA and RNA samples. The diversity indices of nirK were smaller than those of the other functional genes (Table 1). All clones found in Clusters I, III, IV, V, and VI were obtained only from DNA samples. On the other hand, clones in Clusters II were obtained only from RNA samples (Fig. 3C).
Clones of nosZ were also widespread in the phylogenetic tree (Fig. S4). Clones in Cluster I were related to the nosZ of Rhizobiales (Bradyrhizobium spp., Brucella spp. and Sinorhizobium spp.) and were only observed in the DNA samples. The proportion of the clones in Cluster I decreased after 20-h incubation (Fig. 3D), suggesting that these clones were not actively involved in N2O reduction. In contrast, the proportion of clones in Cluster IV that increased after incubation and were dominant in the RNA samples (related to Azoarcus spp., Aromatoleum spp., Thiobacillus spp., Burkholderia spp. and, Pseudogulbenkiania spp.) and Cluster V (distantly related to known bacteria) were also dominant in cDNA.
Discussion
In this study, denitrifier communities were investigated both by DNA- and RNA-based analyses targeting 16S rRNA and denitrification functional genes, and their transcripts. DNA-based analysis revealed that the denitrifier community did not change much by incubation, whereas RNA-based analysis showed dynamic changes in the abundance and composition of transcripts as a result of incubation (Fig. 3). The results of this study were compared with those obtained previously by culture-dependent analyses (14, 37) and DNA-based culture-independent analyses (45, 46) with rice paddy field soils. In the microcosm setup used in this study, the amount of 16S rRNA and nirS transcripts increased along with denitrification activity. In contrast to the RNA-based study, the amount of each gene from DNA samples did not change significantly within 24-h incubation. In addition, the amounts of the functional gene transcripts (mRNA) were markedly smaller than those of the copy numbers (DNA). These results indicated that only a few microbes may have transcribed their denitrification functional genes, and their growth was not significant enough to contribute to the overall increase in the amount of the functional genes within 24-h incubation; however, degradation of mRNA may also have caused the relatively small number of gene transcripts. Härtig reported the half-life of nirS transcripts to be 12.6 min (7), indicating that mRNA may be degraded during extraction.
Active microbes under denitrification-inducing conditions were identified by comparative analysis of the clone libraries. Although diverse bacteria were present in the rice paddy soil, as observed in a previous study (11), only a few were active under denitrification-inducing conditions. Among these, many clones related to the class Betaproteobacteria, especially those of the order Neisseriales, were the most abundant. Previous culture-based analysis also identified many Neisseriales-related bacteria (e.g. Pseudogulbenkiania sp. NH8B) in the same rice paddy soil (37). In addition to Betaproteobacteria, the proportion of clones related to the phylum Acidobacteria also increased after 20-h incubation. These microbes may also be involved in denitrification or N2O reduction. Based on genome analysis, some Acidobacteria may have the ability to reduce nitrate and nitrite (42). On the other hand, the proportion of 16S rRNA related to the phyla Actinobacteria and Bacteroidetes and the class Deltaproteobacteria decreased. These microbes may be present in soil, but they did not respond to denitrification-inducing conditions.
Similar to comparative clone library analysis based on the 16S rRNA or the 16S rRNA gene, analysis of the denitrification functional gene or the gene transcripts suggested that only a small number of denitrifiers were active under denitrification-inducing conditions. By comparative analysis of the clone libraries, we were able to identify the nirS and nosZ clones that increased in response to denitrification-inducing conditions. Some of these were distantly related to the nirS of known denitrifiers, suggesting that denitrifiers carrying these functional gene sequences have not been isolated yet. In Cluster X, the nirS clone sequences obtained in this study from the rice paddy soil in Niigata were similar to the nirS from Bradyrhizobium sp. TSA1, which was isolated previously from rice paddy soil in Tokyo (14). Similar nirS sequences were also obtained from clone library analyses based on rice paddy soil in Tokyo (45, 46), indicating that these denitrifiers may be present in various types of rice paddy soils. Many clones related to the nirS of Actinobacteria and Betaproteobacteria (Cluster VIII in Fig. S2) and the nosZ of Betaproteobacteria (Cluster IV in Fig. S4) were frequently obtained from the soil when denitrification was actively performed, indicating that these bacteria may also play important roles in denitrification and N2O reduction.
In contrast to nirS transcripts, the number of nirK transcripts was not detected. This indicated that most of the active denitrifiers in this soil may use NirS, not NirK, for nitrite reduction. A previous culture-based study also reported that most denitrifiers isolated from rice paddy soil carried nirS (14); however, bias caused by the primers used in this study may also have caused the constant level of nirK expression. Although the contribution of nirK-carrying denitrifiers may not be large, we identified nirK clone sequences that were related to clone sequences of the order Rhizobiales (Cluster II) in RNA samples obtained from the soil after incubation. These microbes may have been involved in denitrification in the soil used in this study. Similar to the present study, these denitrifiers were previously isolated from rice paddy soils (2, 14, 37).
In conclusion, denitrifiers belonging to the class Betaproteobacteria (e.g. the orders Neisseriales, Rhodocyclales and Burkholderiales) may be involved in denitrification in rice paddy soils under the conditions used. In addition, denitrifiers harboring previously uncharacterized nirS may be involved in denitrification. Isolation and characterization of the denitrifiers carrying these genes (e.g., 14) are necessary in the future. Furthermore, the soil was incubated with succinate in this study, however, it was reported that the denitrifier community responded differently to the carbon source (9). Further studies using diverse carbon sources may be needed for greater understanding of the active dentrifying community. Our study also clearly showed that only some of the denitrifiers actively performed denitrification in the environment. Although DNA-based analysis is useful to assess the diversity of denitrifier populations, an RNA-based approach is important for the identification of active denitrifiers.
Supplementary Material
Acknowledgements
This work was supported by the Program for the Promotion of Basic and Applied Researches for Innovations in Bio-oriented Institution, Tokyo, Japan. Additional financial support was provided by a Grant-in-Aid for Scientific Research (B) (No. 19380041) from the Japan Society for the Promotion of Science (JSPS) and a Grant-in-Aid (Soil eDNA) from the Ministry of Agriculture, Forestry and Fisheries of Japan. M.Y. was supported by a Grant-in-Aid for JSPS Fellows (No. 20.4259). We thank Yutaka Shiratori for his help in collecting soil samples.
References
- 1.Akiyama H, Yan X, Yagi K. Estimations of emission factors for fertilizer-induced direct N2O emissions from agricultural soils in Japan: Summary of available data. Soil Sci Plant Nutr. 2006;52:774–787. [Google Scholar]
- 2.Ashida N, Ishii S, Hayano S, Tago K, Tsuji T, Yoshimura Y, Otsuka S, Senoo K. Isolation of functional single cells from environments using a micromanipulator: application to study denitrifying bacteria. Appl Microbiol Biotechnol. 2009;85:1211–1217. doi: 10.1007/s00253-009-2330-z. [DOI] [PubMed] [Google Scholar]
- 3.Bothe H, Jost G, Schloter M, Ward BB, Witzel KP. Molecular analysis of ammonia oxidation and denitrification in natural environments. FEMS Microbiol Rev. 2000;24:673–690. doi: 10.1111/j.1574-6976.2000.tb00566.x. [DOI] [PubMed] [Google Scholar]
- 4.Bremer C, Braker G, Matthies D, Reuter A, Engels C, Conrad R. Impact of plant functional group, plant species, and sampling time on the composition of nirK-type denitrifier communities in soil. Appl Environ Microbiol. 2007;73:6876–6884. doi: 10.1128/AEM.01536-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fierer N, Jackson JA, Vilgalys R, Jackson RB. Assessment of soil microbial community structure by use of taxon-specific quantitative PCR assays. Appl Environ Microbiol. 2005;71:4117–4120. doi: 10.1128/AEM.71.7.4117-4120.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gamble TN, Betlach MR, Tiedje JM. Numerically dominant denitrifying bacteria from world soils. Appl Environ Microbiol. 1977;33:926–939. doi: 10.1128/aem.33.4.926-939.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Härtig E, Zumft WG. Kinetics of nirS expression (cytochrome cd1 nitrite reductase) in Pseudomonas stutzeri during the transition from aerobic respiration to denitrification: evidence for a denitrification-specific nitrate- and nitrite-responsive regulatory system. J Bacteriol. 1999;181:161–166. doi: 10.1128/jb.181.1.161-166.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hayatsu M, Tago K, Saito M. Various players in the nitrogen cycle: diversity and functions of the microorganisms involved in nitrification and denitrification. Soil Sci Plant Nutr. 2008;54:33–45. [Google Scholar]
- 9.Henderson SL, Dandie CD, Patten CL, Zebarth BJ, Burton DL, Trevors JT, Goyer C. Changes in denitrifier abundance, denitrification gene mRNA levels, nitrous oxide emissions, and denitrification in anoxic soil microcosms amended with glucose and plant residues. Appl Environ Microbiol. 2010;76:2155–2164. doi: 10.1128/AEM.02993-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henry S. Quantitative detection of the nosZ gene, encoding nitrous oxide reductase, and comparison of the abundances of 16S rRNA, narG, nirK, and nosZ genes in soils. Appl Environ Microbiol. 2006;72:5181–5189. doi: 10.1128/AEM.00231-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ishii S, Yamamoto M, Kikuchi M, Oshima K, Hattori M, Otsuka S, Senoo K. Microbial populations responsive to denitrification-inducing conditions in rice paddy soil, as revealed by comparative 16S rRNA gene analysis. Appl Environ Microbiol. 2009;75:7070–7078. doi: 10.1128/AEM.01481-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishii S, Yamamoto M, Tago K, Otsuka S, Senoo K. Microbial populations in various paddy soils respond differently to denitrification-inducing conditions, albeit background bacterial populations are similar. Soil Sci Plant Nutr. 2010a;56:220–224. [Google Scholar]
- 13.Ishii S, Yan T, Vu H, Hansen DL, Hicks RE, Sadowsky MJ. Factors controlling long-term survival and growth of naturalized Escherichia coli populations in temperate field soils. Microbes Environ. 2010b;25:8–14. doi: 10.1264/jsme2.me09172. [DOI] [PubMed] [Google Scholar]
- 14.Ishii S, Ashida N, Otsuka S, Senoo K. Isolation of oligotrophic denitrifiers carrying previously uncharacterized functional gene sequences. Appl Environ Microbiol. 2011a;77:338–342. doi: 10.1128/AEM.02189-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishii S, Ohno H, Tsuboi M, Otsuka S, Senoo K. Identification and isolation of active N2O reducers in rice paddy soil. ISME J. 2011b;5:1936–1945. doi: 10.1038/ismej.2011.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishii S, Ikeda S, Minamisawa K, Senoo K. Nitrogen cycling in rice paddy environments: past achievements and future challenges. Microbes Environ. 2011;26:282–292. doi: 10.1264/jsme2.me11293. [DOI] [PubMed] [Google Scholar]
- 17.Kandeler E, Deiglmayr K, Tscherko D, Bru D, Philippot L. Abundance of narG, nirS, nirK, and nosZ genes of denitrifying bacteria during primary successions of a glacier foreland. Appl Environ Microbiol. 2006;72:5957–5962. doi: 10.1128/AEM.00439-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katsuyama C, Kondo N, Suwa Y, Yamagishi T, Itoh M, Ohte N, Kimura H, Nagaosa K, Kato K. Denitrification activity and relevant bacteria revealed by nitrite reductase gene fragments in soil of temperate mixed forest. Microbes Environ. 2008;23:337–345. doi: 10.1264/jsme2.me08541. [DOI] [PubMed] [Google Scholar]
- 19.Lane DJ. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. New York, NY: John Wiley and Sons, New York; 1991. pp. 115–175. [Google Scholar]
- 20.Liesack W, Schnell S, Revsbech NP. Microbiology of flooded rice paddies. FEMS Microbiol Rev. 2000;24:625–645. doi: 10.1111/j.1574-6976.2000.tb00563.x. [DOI] [PubMed] [Google Scholar]
- 21.McGrath KC, Mondav R, Sintrajaya R, Slattery B, Schmidt S, Schenk PM. Development of an environmental functional gene microarray for soil microbial communities. Appl Environ Microbiol. 2010;76:7161–7170. doi: 10.1128/AEM.03108-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morales SE, Cosart T, Holben WE. Bacterial gene abundances as indicators of greenhouse gas emission in soils. ISME J. 2010;4:799–808. doi: 10.1038/ismej.2010.8. [DOI] [PubMed] [Google Scholar]
- 23.Murakami Y, Otsuka S, Senoo K. Abundance and community structure of Sphingomonads in leaf residues and nearby bulk soil. Microbes Environ. 2010;25:183–189. doi: 10.1264/jsme2.me10114. [DOI] [PubMed] [Google Scholar]
- 24.Muyzer G, de Waal EC, Uitterlinden AG. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nishimura S, Sawamoto T, Akiyama H, Sudo S, Cheng W, Yagi K. Continuous, automated nitrous oxide measurements from paddy soils converted to upland crops. Soil Sci Soc Am J. 2005;69:1977–1986. [Google Scholar]
- 26.Nogales B, Timmis KN, Nedwell DB, Osborn AM. Detection and diversity of expressed denitrification genes in estuarine sediments after reverse transcription-PCR amplification from mRNA. Appl Environ Microbiol. 2002;68:5017–5025. doi: 10.1128/AEM.68.10.5017-5025.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Philippot L. Denitrifying genes in bacterial and Archaeal genomes. Biochim Biophys Acta. 2002;1557:355–376. doi: 10.1016/s0167-4781(02)00420-7. [DOI] [PubMed] [Google Scholar]
- 28.Philippot L, Hallin S, Schloter M. Ecology of denitrifying prokaryotes in agricultural soils. Adv Agron. 2007;96:249–305. [Google Scholar]
- 29.Priemé A, Braker G, Tiedje JM. Diversity of nitrite reductase (nirK and nirS) gene fragments in forested upland and wetland soils. Appl Environ Microbiol. 2002;68:1893–1900. doi: 10.1128/AEM.68.4.1893-1900.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rich JJ, Heichen RS, Bottomley PJ, Cromack K, Jr, Myrold DD. Community composition and functioning of denitrifying bacteria from Adjacent meadow and forest soils. Appl Environ Microbiol. 2003;69:5974–5982. doi: 10.1128/AEM.69.10.5974-5982.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ryuda N, Hashimoto T, Ueno D, Inoue K, Someya T. Visualization and drect counting of individual denitrifying bacterial cells in soil by nirK-targeted direct in situ PCR. Microbes Environ. 2011;26:74–80. doi: 10.1264/jsme2.me10180. [DOI] [PubMed] [Google Scholar]
- 32.Saito T, Ishii S, Otsuka S, Nishiyama M, Senoo K. Identification of novel Betaproteobacteria in a succinate-assimilating population in denitrifying rice paddy soil by using stable isotope probing. Microbes Environ. 2008;23:192–200. doi: 10.1264/jsme2.23.192. [DOI] [PubMed] [Google Scholar]
- 33.Schloss PD, Handelsman J. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol. 2005;71:1501–1506. doi: 10.1128/AEM.71.3.1501-1506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sharma S, Aneja MK, Mayer J, Munch JC, Schloter M. Diversity of transcripts of nitrite reductase genes (nirK and nirS) in rhizospheres of grain legumes. Appl Environ Microbiol. 2005;71:2001–2007. doi: 10.1128/AEM.71.4.2001-2007.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shoun H, Kim DH, Uchiyama H, Sugiyama J. Denitrification by fungi. FEMS Microbiol Lett. 1992;94:277–282. doi: 10.1016/0378-1097(92)90643-3. [DOI] [PubMed] [Google Scholar]
- 36.Smith CJ, Nedwell DB, Dong LF, Osborn AM. Diversity and abundance of nitrate reductase genes (narG and napA), nitrite reductase genes (nirS and nrfA), and their transcripts in estuarine sediments. Appl Environ Microbiol. 2007;73:3612–3622. doi: 10.1128/AEM.02894-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tago K, Ishii S, Nishizawa T, Otsuka S, Senoo K. Phylogenetic and functional diversity of denitrifying bacteria isolated from various rice paddy and rice-soybean rotation fields. Microbes Environ. 2011;26:30–35. doi: 10.1264/jsme2.me10167. [DOI] [PubMed] [Google Scholar]
- 38.Throbäck IN, Enwall K, Jarvis A, Hallin S. Reassessing PCR primers targeting nirS, nirK and nosZ genes for community surveys of denitrifying bacteria with DGGE. FEMS Microbiol Ecol. 2004;49:401–417. doi: 10.1016/j.femsec.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 39.Tyson GW, Chapman J, Hugenholtz P, Allen EE, Ram RJ, Richardson PM, Solovyev VV, Rubin EM, Rokhsar DS, Banfield JF. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature. 2004;428:37–43. doi: 10.1038/nature02340. [DOI] [PubMed] [Google Scholar]
- 40.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Y, Morimoto S, Ogawa N, Fujii T. A survey of the cellular responses in Pseudomonas putida KT2440 growing in sterilized soil by microarray analysis. FEMS Microbiol Ecol. 2011;78:220–232. doi: 10.1111/j.1574-6941.2011.01146.x. [DOI] [PubMed] [Google Scholar]
- 42.Ward NL, Challacombe JF, Janssen PH, et al. Three genomes from the phylum Acidobacteria provide insight into the lifestyles of these microorganisms in soils. Appl Environ Microbiol. 2009;75:2046–2056. doi: 10.1128/AEM.02294-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wertz S, Dandie CE, Goyer C, Trevors JT, Patten CL. Diversity of nirK denitrifying genes and transcripts in an agricultural soil. Appl Environ Microbiol. 2009;75:7365–7377. doi: 10.1128/AEM.01588-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wolsing M, Priemé A. Observation of high seasonal variation in community structure of denitrifying bacteria in arable soil receiving artificial fertilizer and cattle manure by determining T-RFLP of nir gene fragments. FEMS Microbiol Ecol. 2004;48:261–271. doi: 10.1016/j.femsec.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 45.Yoshida M, Ishii S, Otsuka S, Senoo K. Temporal shifts in diversity and quantity of nirS and nirK in a rice paddy field soil. Soil Biol Biochem. 2009;41:2044–2051. [Google Scholar]
- 46.Yoshida M, Ishii S, Otsuka S, Senoo K. nirK-harboring denitrifiers are more responsive to denitrification-inducing conditions in rice paddy soil than nirS-harboring bacteria. Microbes Environ. 2010;25:45–48. doi: 10.1264/jsme2.me09160. [DOI] [PubMed] [Google Scholar]
- 47.Zumft WG. Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev. 1997;61:533–616. doi: 10.1128/mmbr.61.4.533-616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.