Abstract
Motivation: In recent years, metabolomics has emerged as an approach to perform large-scale characterization of small molecules in biological systems. Metabolomics posed a number of bioinformatics challenges associated in data analysis and interpretation. Genome-based metabolic reconstructions have established a powerful framework for connecting metabolites to genes through metabolic reactions and enzymes that catalyze them. Pathway databases and bioinformatics tools that use this framework have proven to be useful for annotating experimental metabolomics data. This framework can be used to infer connections between metabolites and diseases through annotated disease genes. However, only about half of experimentally detected metabolites can be mapped to canonical metabolic pathways. We present a new Cytoscape 3 plug-in, MetDisease, which uses an alternative approach to link metabolites to disease information. MetDisease uses Medical Subject Headings (MeSH) disease terms mapped to PubChem compounds through literature to annotate compound networks.
Availability and implementation: MetDisease can be downloaded from http://apps.cytoscape.org/apps/metdisease or installed via the Cytoscape app manager. Further information about MetDisease can be found at http://metdisease.ncibi.org
Contact: akarnovs@med.umich.edu
Supplementary information: Supplementary Data are available at Bioinformatics online
1 INTRODUCTION
A wide range of pathway mapping and visualization tools have been developed for the analysis of transcriptomics, proteomics and metabolomics data. They provide a quick and intuitive way to place experimental data into the context of prior biological knowledge. Most of the existing tools for the analysis of metabolomics data rely on genome-based metabolic reconstruction databases (Frolkis et al., 2010; Garcia-Alcalde et al., 2011; Karnovsky et al., 2012; Xia and Wishart 2011). Metabolic reconstruction databases such as KEGG (Kanehisa, 2006), BioCyc (Caspi et al., 2012; Romero et al., 2005) and their subsequent refinements (Caspi et al., 2012; Duarte et al., 2007; Hao et al., 2010; Kanehisa, 2006; Ma et al., 2007; Sigurdsson et al., 2010; Thiele et al., 2013) provide carefully curated information about metabolic pathways, metabolites, metabolic reactions, enzymes and the genes that encode them.
Recent progress in the field of metabolomics now allows rapid and quantitative measurement of hundreds of named metabolites and thousands of chromatographic features representing additional metabolites. As the experimental data analysis methods improve, metabolomics has increasing potential to provide informative readouts of metabolic changes in a variety of diseases (Sreekumar et al., 2009; Urayama et al., 2010; Wang et al., 2011; Wisloff et al., 2005; Yap et al., 2010). However, biological interpretation of metabolomic experiments is hindered by relatively low coverage of experimentally identified metabolites in pathway databases. In fact, most metabolic reconstructions cover the majority of primary metabolites, whereas the coverage of lipids, secondary metabolites and volatile metabolites is significantly lower (Barupal et al., 2012). Additional reasons for low metabolite coverage include the presence of metabolites from different organisms (e.g. presence of bacterial metabolites in human samples originating from microbiome), drug metabolites and compounds of environmental origin, few of which are contained in most pathway databases.
To explore the possibility of expanding metabolite annotation through biomedical literature, we have developed Metab2MeSH (http://metab2mesh.ncibi.org), which links PubChem compounds to MeSH terms via substances that are annotated to PubMed articles (Sartor et al., 2012). National Library of Medicine (NLM) provides a list of chemical substances that contain a wide range of synonyms used in biomedical publications that can be linked to the compound synonyms used in PubChem. Using the compounds and their occurrences in PubMed literature, statistical tests were performed to estimate the significance of the associations between compound–MeSH descriptor pairs. The resulting associations, supporting data and literature are stored in a relational database and can be accessed via Metab2MeSH web interface and web services. However, incorporating these annotations into data analysis workflow remains a challenge.
To further demonstrate the utility of Metab2MeSH data set and provide a quick and easy way to annotate metabolic networks, we developed MetDisease, a plug-in for the open-source tool Cytoscape [29] that uses disease-related MeSH terms. Users can import and annotate any network where metabolites (compounds) are represented as nodes, referenced by KEGG or PubChem IDs. The edges can be arbitrarily defined by the users. MetDisease allows users to highlight and explore parts of metabolic networks related to one or more MeSH disease terms and provides links to relevant PubMed literature. Users have an option to import their own metabolic networks or to use MetDisease to annotate metabolic networks generated with the Cytoscape plug-in Metscape (Karnovsky et al., 2012). Both use cases are illustrated below.
2 METHODS
MetDisease uses the Metab2MeSH database (Sartor et al., 2012). This dataset is created twice a year by downloading the PubChem Compound and Substance databases and the NLM PubMed database, parsing them, and loading them into an in-house relational database. Associations between compounds and MeSH terms are calculated using 2-sided Fisher’s exact tests, and any results with P < 0.0001 are retained in the database. MetDisease then uses an internal service to access this database via SQL queries to determine relevant MeSH disease terms for the compounds in a given metabolic network.
3 RESULTS
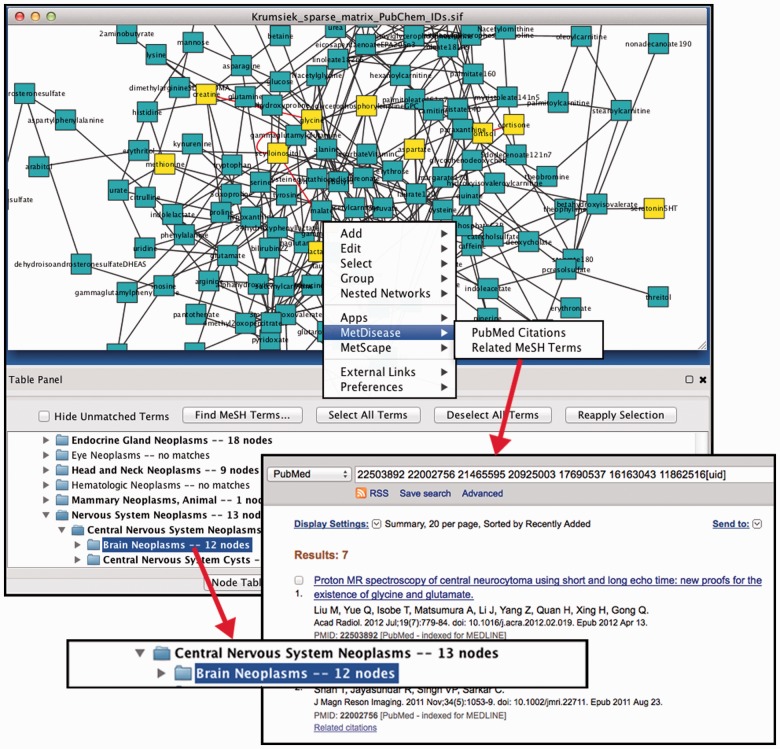
To illustrate the use and features of MetDisease, we created a network using a publicly available metabolomics dataset (Krumsiek et al., 2012). A subset of known metabolites was downloaded from Supplementary Data (GGM.xlsx). Partial correlation coefficient values <5e-4 were used to draw the edges in the resulting network (See Supplementary Data File). Once the network was loaded to Cytoscape, we used MetDisease to identify MeSH disease terms associated with compound nodes as shown in Figure 1.
Fig. 1.
The network was constructed using metabolites reported by Krumsiek et al. as nodes and partial correlation coefficients as edges (see network file in Supplementary Data). MetDisease plug-in was used to annotate the metabolites with MeSH disease terms using PubChem IDs. When a MeSH term is selected in the lower panel (e.g. Brain Neoplasms highlighted in blue), the metabolites that were mapped to it are selected in the network [in this case 12 (yellow) compounds are selected]. Numbers to the right of the term indicate the number of mapped metabolites in the active network. By right clicking on the node of interest (e.g. glycine), user can access the publications in PubMed that substantiate that connection
Once the network has been loaded, users can select MetDisease from the Cytoscape Apps menu. When the Filter Options dialog box appears, users are prompted to select an appropriate identifier (KEGG or PubChem ID) and the column in their input file that contains that identifier. After the mapping has been completed, the Disease branch of the MeSH tree is displayed. The MeSH terms that have mapped compounds in an active network are shown in bold. Users have an option to hide unmatched terms. By right clicking on the node of interest, users can access PubMed publications as shown in Figure 1, or identify other MeSH terms related to the compound of interest by going to the Metab2MeSH web application.
Another characteristic feature of MetDisease is the ability to annotate metabolic networks generated by Metscape (Karnovsky et al., 2012). A use case where a glycine metabolic network generated by Metscape was annotated using MetDisease is shown in Supplementary Figure S1, where sarcosine has been appropriately linked to prostatic neoplasms.
In conclusion, MetDisease provides a quick convenient way to identify compounds associated with a disease term, and identify publications that link them together. Just like the underlying Metab2MeSH data set, because of its dependence on what is in the literature, MetDisease is not meant to predict novel associations. However, as the examples illustrate, it is a useful tool for identifying disease associations that would otherwise be difficult to find.
Importantly, because the associations are not derived from genome-based metabolic reconstructions, MetDisease can be used to annotate a broader range of compounds, including drugs, nutritional compounds and environmental toxins.
Funding: Support was provided by the NIH U54 DA021519 01A1, P30 DK089503, P30 ES017885 and DK097153.
Conflict of Interest: none declared.
Supplementary Material
REFERENCES
- Barupal DK, et al. MetaMapp: mapping and visualizing metabolomic data by integrating information from biochemical pathways and chemical and mass spectral similarity. BMC Bioinformatics. 2012;13:99. doi: 10.1186/1471-2105-13-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi R, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2012;40:D742–D753. doi: 10.1093/nar/gkr1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte NC, et al. Global reconstruction of the human metabolic network based on genomic and bibliomic data. Proc. Natl Acad. Sci. USA. 2007;104:1777–1782. doi: 10.1073/pnas.0610772104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolkis A, et al. SMPDB: the small molecule pathway database. Nucleic Acids Res. 2010;38:D480–D487. doi: 10.1093/nar/gkp1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Alcalde F, et al. Paintomics: a web based tool for the joint visualization of transcriptomics and metabolomics data. Bioinformatics. 2011;27:137–139. doi: 10.1093/bioinformatics/btq594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao T, et al. Compartmentalization of the Edinburgh human metabolic network. BMC Bioinformatics. 2010;11:393. doi: 10.1186/1471-2105-11-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, et al. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:354–357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnovsky A, et al. Metscape 2 bioinformatics tool for the analysis and visualization of metabolomics and gene expression data. Bioinformatics. 2012;28:373–380. doi: 10.1093/bioinformatics/btr661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumsiek J, et al. Mining the unknown: a systems approach to metabolite identification combining genetic and metabolic information. PLoS Genet. 2012;8:e1003005. doi: 10.1371/journal.pgen.1003005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, et al. The Edinburgh human metabolic network reconstruction and its functional analysis. Mol. Syst. Biol. 2007;3:135. doi: 10.1038/msb4100177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero P, et al. Computational prediction of human metabolic pathways from the complete human genome. Genome Biol. 2005;6:R2. doi: 10.1186/gb-2004-6-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartor MA, et al. Metab2MeSH: annotating compounds with medical subject headings. Bioinformatics. 2012;28:1408–1410. doi: 10.1093/bioinformatics/bts156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigurdsson MI, et al. A detailed genome-wide reconstruction of mouse metabolism based on human Recon 1. BMC Syst. Biol. 2010;4:140. doi: 10.1186/1752-0509-4-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreekumar A, et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009;457:910–914. doi: 10.1038/nature07762. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Thiele I, et al. A community-driven global reconstruction of human metabolism. Nat. Biotechnol. 2013;31:419–425. doi: 10.1038/nbt.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urayama S, et al. Comprehensive mass spectrometry based metabolic profiling of blood plasma reveals potent discriminatory classifiers of pancreatic cancer. Rapid Commun. Mass Spectrom. 2010;24:613–620. doi: 10.1002/rcm.4420. [DOI] [PubMed] [Google Scholar]
- Wang TJ, et al. Metabolite profiles and the risk of developing diabetes. Nat. Med. 2011;17:448–453. doi: 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisloff U, et al. Cardiovascular risk factors emerge after artificial selection for low aerobic capacity. Science. 2005;307:418–420. doi: 10.1126/science.1108177. [DOI] [PubMed] [Google Scholar]
- Xia J, Wishart DS. Web-based inference of biological patterns, functions and pathways from metabolomic data using MetaboAnalyst. Nat. Protoc. 2011;6:743–760. doi: 10.1038/nprot.2011.319. [DOI] [PubMed] [Google Scholar]
- Yap IK, et al. Metabolome-wide association study identifies multiple biomarkers that discriminate north and south Chinese populations at differing risks of cardiovascular disease: INTERMAP study. J Proteome Res. 2010;9:6647–6654. doi: 10.1021/pr100798r. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.