Abstract
Summary: Model Quality Assessment Programs (MQAPs) are used to predict the quality of modeled protein structures. These usually use two approaches: methods using consensus of many alternative models and methods requiring only a single model to do its prediction. The consensus methods are useful to improve overall accuracy; however, they frequently fail to pick out the best possible model and cannot be used to generate and score new structures. Single-model methods, on the other hand, do not have these inherent shortcomings and can be used to both sample new structures and improve existing consensus methods. Here, we present ProQM-resample, a membrane protein-specific single-model MQAP, that couples side-chain resampling with MQAP rescoring by ProQM to improve model selection. The side-chain resampling is able to improve side-chain packing for 96% of all models, and improve model selection by 24% as measured by the sum of the Z-score for the first-ranked model (from 25.0 to 31.1), even better than the state-of-the-art consensus method Pcons. The improved model selection can be attributed to the improved side-chain quality, which enables the MQAP to rescue good backbone models with poor side-chain packing.
Availability and implementation: http://proqm.wallnerlab.org/download/.
Contact: bjornw@ifm.liu.se
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Protein structure modeling represents a fundamental challenge in structural bioinformatics and is crucial for a detailed understanding of the structure and biological function of molecules. It can be used to guide and explain experiments, as well as for prediction of proteins whose structure, in particular for membrane proteins, for the most part is unknown (∼0.5% membrane protein in the Protein Data Bank ∼25% in most genomes). A common technique in structure modeling is to generate many alternative models and use a Model Quality Assessment Program (MQAP) to select the best model. Alternatively, an MQAP can also be used to assess the absolute quality of a single model, i.e. a measure that is related to similarity to true native structure (Wallner and Elofsson, 2003; Wang et al., 2009).
ProQM (Ray et al., 2010) is an MQAP that uses a support vector machine to predict the quality of a membrane protein model by combining structural and sequence-based features calculated from the model. In its original implementation, external programs were used to calculate features, e.g. PSI-BLAST (Altschul et al., 1997), PSIPRED (McGuffin et al., 2000), Naccess (Hubbard and Thornton, 1993), Stride (Frishman and Argos, 1995), ProQres (Wallner and Elofsson, 2006), Zpred (Granseth et al., 2006), Topcons (Bernsel et al., 2009), MPRAP (Illergård et al., 2010) and SVM-light (Joachims, 2002). These dependencies made it difficult to distribute the program, run large batches and use it in conformational sampling. Therefore, ProQM has only been available as a webserver for small-scale use. To overcome these issues, ProQM was incorporated as scoring function in the Rosetta modeling framework. This gives in one hand full access to the modeling machinery within Rosetta and allows for easy integration with any Rosetta protocol. In particular, ProQM-resample uses the repack protocol to sample side-chain conformations followed by rescoring using ProQM to improve model selection.
2 METHOD DEVELOPMENT
ProQM (Ray et al., 2010) was implemented as a scoring function in Rosetta (Das and Baker, 2008). ProQM uses two sets of features, one that only depends on the model sequence and one that is calculated from the structural model. The sequence-based features only need to be calculated once for a given sequence and are used as input to Rosetta. While all structural features such as atom–atom contacts, residue–residue contacts, surface areas and secondary structures, as well as the SVM prediction are calculated by Rosetta during scoring, there is still a dependency on external programs for the sequence-based features. The programs and the scripts to prepare input files are provided on the download page. There is also a server available at: http://ProQSeq.wallnerlab.org/.
For the structural-based features, we adapted an already existing implementation of Naccess (Hubbard and Thornton, 1993) and DSSP (Kabsch and Sander, 1983) to calculate exposed residue surface area and for assigning secondary structure, respectively. The atom–atom and residue–residue contact matrices, previously calculated by ProQres, were implemented directly in Rosetta as well as the functionality to read and predict SVM models. To account for changes in implementation details, the SVM model weights were retrained using the original 5-fold cross-validated ProQM training set (Ray et al., 2010).
2.1 ProQM resampling protocol
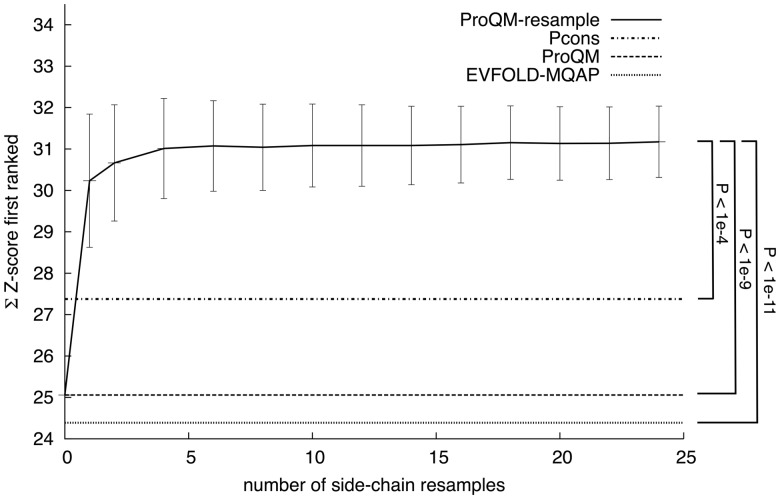
An advantage of implementing ProQM as a scoring function in the Rosetta modeling framework is that it enables conformational sampling using the MQAP as part of the scoring function. However, because an MQAP should measure the quality of any input model it cannot make large change to the model and claim it is assessing the quality of the input model. Therefore, we decided to sample only the side-chain rotamers while keeping the backbone fixed, effectively keeping the quality measures based on Cα coordinates such as TMscore (Zhang and Skolnick, 2004) constant. This was achieved by rebuilding side-chains with a backbone-dependent rotamer library implemented in the repack protocol using the score_membrane scoring function (Barth et al., 2007), followed by rescore using ProQM. Based on Figure 1 sampling and rescoring, 10 different model decoys for each initial model seem to be a good choice.
Fig. 1.
Overall target selection for ProQM-resample measured by the sum of the Z-score for first-ranked models against the number of generated resampled models; Pcons, ProQM and EVFOLD-MQAP are included as reference; error bars correspond to SD resulting from 100 replicas; these were also used to calculate the P-values
2.2 Benchmark
ProQM-resample was benchmarked on the independent EVfold_membrane data set (Hopf et al., 2012) consisting of 15 340 models for 25 targets generated with CNS (Brunger et al., 1998) using distance constraints from evolutionary couplings extracted from large multiple sequence alignments (the set is actually larger, but only 25 targets had a known structure). Results were compared with Pcons (Larsson et al., 2009), a state-of-the-art consensus method, and EVFOLD-MQAP (Hopf et al., 2012), a predicted ranking based on satisfaction of unused constraints, predicted secondary structure and predicted lipid exposure agreement. In the cases where the EVfold_membrane set contained homologous proteins (BLAST E < 0.01) to the original ProQM training set, ProQM was retrained with non-homologous proteins. Model selection accuracy was measured by Z-scores calculated from TMscore.
3 RESULTS
First, the ProQM implementation in Rosetta was compared with the original ProQM-webserver version. They should be similar, but because of different implementations of Naccess and DSSP versus STRIDE, and the fixing of some minor bugs, the results will not be identical. Still, there is a clear correlation, R = 0.98, between ProQM-Rosetta and ProQM-webserver and the prediction performance is also maintained, R = 0.62 to true answer (data not shown).
The benchmark on the EVfold_membrane set showed that model selection is significantly improved by resampling the side-chains (Fig 1). Already without any sampling ProQM selects slightly better models than EVFOLD-MQAP (Z = 25.0 versus 24.4). Side-chain sampling increases the performance significantly to Z = 31.1 and levels out at around 10 resamples per model, surpassing even the state-of-the-art consensus Pcons. This demonstrates the usefulness of single-model MQAPs in model selection. It also highlights the need to include side-chain sampling into the MQAP procedure to avoid losing good backbone models suffering from poor side-chain packing. A possible reason for the improved selection is the fact that almost all models (96%) improved the side-chain packing after resampling (Supplementary Fig. S1). Before resampling, the side-chain quality is roughly the same for all targets (Table 1). But after resampling the side-chain improvement is larger for the set of targets that also show backbone improvement after resampling compared with targets that show no improvement in backbone, indicating that improved side-chains help the MQAP to select better backbone models.
Table 1.
Side-chain quality before and after resampling
| Set | Before resampling (%) | After resampling (%) | Number of targets | Number of models |
|---|---|---|---|---|
| All | 13.5 ± 0.1 | 20.5 ± 0.2 | 25 | 15 340 |
| No improvement | 13.3 ± 0.2 | 19.0 ± 0.3 | 11 | 6538 |
| Improvement | 13.6 ± 0.2 | 22.0 ± 0.3 | 8 | 4377 |
Note: Side-chain quality measured by fraction chi1 and chi2 within 40 from correct. Error estimates represent 99.999% confidence intervals. The sets correspond to all models, models from targets without and with backbone improvement. See Supplementary Information for exact definition of the sets.
Funding: Swedish Research Council (Dnr 2012-5270) and Carl Tryggers Stiftelse (Dnr 12:516).
Conflict of Interest: none declared.
Supplementary Material
REFERENCES
- Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth P, et al. Toward high-resolution prediction and design of transmembrane helical protein structures. Proc. Natl Acad. Sci. USA. 2007;104:15682–15687. doi: 10.1073/pnas.0702515104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernsel A, et al. TOPCONS: consensus prediction of membrane protein topology. Nucleic Acids Res. 2009;37:W465–W468. doi: 10.1093/nar/gkp363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Das R, Baker D. Macromolecular modeling with rosetta. Annu. Rev. Biochem. 2008;77:363–382. doi: 10.1146/annurev.biochem.77.062906.171838. [DOI] [PubMed] [Google Scholar]
- Frishman D, Argos P. Knowledge-based protein secondary structure assignment. Proteins. 1995;23:566–579. doi: 10.1002/prot.340230412. [DOI] [PubMed] [Google Scholar]
- Granseth E, et al. ZPRED: predicting the distance to the membrane center for residues in alpha-helical membrane proteins. Bioinformatics. 2006;22:e191–e196. doi: 10.1093/bioinformatics/btl206. [DOI] [PubMed] [Google Scholar]
- Hopf TA, et al. Three-dimensional structures of membrane proteins from genomic sequencing. Cell. 2012;149:1607–1621. doi: 10.1016/j.cell.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard SJ, Thornton JM. 1993. NACCESS [computer program], Department of Biochemistry and Molecular Biology, University College London. [Google Scholar]
- Illergård K, et al. MPRAP: an accessibility predictor for a-helical transmembrane proteins that performs well inside and outside the membrane. BMC Bioinformatics. 2010;11:333. doi: 10.1186/1471-2105-11-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joachims T. Learning to Classify Text Using Support Vector Machines. Kluwer, Waltham, MA: 2002. [Google Scholar]
- Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- Larsson P, et al. Assessment of global and local model quality in CASP8 using Pcons and ProQ. Proteins. 2009;77:167–172. doi: 10.1002/prot.22476. [DOI] [PubMed] [Google Scholar]
- McGuffin LJ, et al. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- Ray A, et al. Model quality assessment for membrane proteins. Bioinformatics. 2010;26:3067–3074. doi: 10.1093/bioinformatics/btq581. [DOI] [PubMed] [Google Scholar]
- Wallner B, Elofsson A. Can correct protein models be identified? Protein Sci. 2003;12:1073–1086. doi: 10.1110/ps.0236803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallner B, Elofsson A. Identification of correct regions in protein models using structural, alignment, and consensus information. Protein Sci. 2006;15:900–913. doi: 10.1110/ps.051799606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, et al. Evaluating the absolute quality of a single protein model using structural features and support vector machines. Proteins. 2009;75:638–647. doi: 10.1002/prot.22275. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Skolnick J. Scoring function for automated assessment of protein structure template quality. Proteins. 2004;57:702–710. doi: 10.1002/prot.20264. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.