Abstract
Summary: We present methylC track, an efficient mechanism for visualizing single-base resolution DNA methylation data on a genome browser. The methylC track dynamically integrates the level of methylation, the position and context of the methylated cytosine (i.e. CG, CHG and CHH), strand and confidence level (e.g. read coverage depth in the case of whole-genome bisulfite sequencing data). Investigators can access and integrate these information visually at specific locus or at the genome-wide level on the WashU EpiGenome Browser in the context of other rich epigenomic datasets.
Availability and implementation: The methylC track is part of the WashU EpiGenome Browser, which is open source and freely available at http://epigenomegateway.wustl.edu/browser/. The most up-to-date instructions and tools for preparing methylC track are available at http://epigenomegateway.wustl.edu/+/cmtk.
Contact: twang@genetics.wustl.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
DNA methylation is a well-studied epigenetic mark. It plays crucial roles in development, gene regulation, chromatin structure, imprinting and many diseases (Esteller, 2007). DNA methylation primarily occurs as 5-methylcytosine in the CpG context. In specific cell types, such as embryonic stem cells (ESCs), 5-methylcytosine sometimes occurs in the context of CHG and CHH (H is A, T or C) (Lister et al., 2009). Usually CpG methylation is symmetrical between the two DNA strands, but hemi-methylation, where only one strand is methylated, can be observed. Several modern techniques measure DNA methylation at single-base resolution, including whole-genome bisulfite sequencing (WGBS or methylC-seq; Lister et al., 2009), reduced representation bisulfite sequencing (RRBS; Meissener et al., 2005) and Illumina Infinium arrays. Bisulfite sequencing-based methods can provide information including non-CpG menthylation, strand-specific methylation and confidence level (read depth; Ziller et al., 2013) and are considered the gold standard in the field.
Typically, DNA methylation data can be visualized on a genome browser as a track of bar plots, where the position of each bar marks a measured cytosine, and the height represents the methylation level. An example of a WGBS track and Infinium 450 K track on the UCSC Genome Browser is shown in Supplementary Figure S1. However, several parameters important for data interpretation are missing in this visual representation. First, non-CG methylation is omitted; second, strand-specific information provided by WGBS is removed; third, sequencing coverage, a key measurement of the data confidence, cannot be assessed visually; fourth, a cytosine with ‘zero percent’ methylation (or completely unmethylated) is not visually distinguishable from ‘no value’. The Anno-J Browser (http://www.annoj.org) can display strand-specific and context-specific methylation as composite bar charts (Supplementary Fig. S2). However, it does not distinguish unmethylated cytosines from cytosines with no data and can not combine the two strands. Anno-J requires a special data storage format that is not compatible with most popular genomics tools.
To address these issues, we extended the WashU EpiGenome Browser (Zhou et al., 2011, 2013) by inventing the methylC track for genome-wide single-base DNA methylation data represented by WGBS. Cytosine methylation levels are displayed as conventional bar plots, but investigators can display and distinguish cytosines in different context and choose to display strand-specific DNA methylation or to dynamically combine both strands. An overlayed curve displays read-depth data at each cytosine, allowing investigators to assess quality of the data. By using a different background color, investigators can easily focus on unmethylated cytosines without confusing those cytosines with no measurement. Importantly, investigators do not need to reformat their data. Our data format is fully compatible with the UCSC Genome Browser. Investigators can mimic some features of our visualization with the UCSC Genome Browser (Supplementary Fig. S3), but the WashU EpiGenome Browser provides more flexibility in combining data and toggling between views.
2 IMPLEMENTATION
We store all relevant data in bedGraph format for compatibility. Depending on the experiment type, up to eight bedGraph files are generated for one experiment (e.g. WGBS) to store CG, CHG and CHH methylation level data and read-depth data for each DNA strand. Most files are optional. For example, for Infinium arrays only CG methylation level is necessary. Data files are compressed, indexed by Tabix (Li, 2009) and hosted on Web servers. The bedGraph file URLs are organized into a single methylC track using JSON (Fig. 1a) and displayed via the datahub mechanism (http://epigenomegateway.wustl.edu/+/hub).
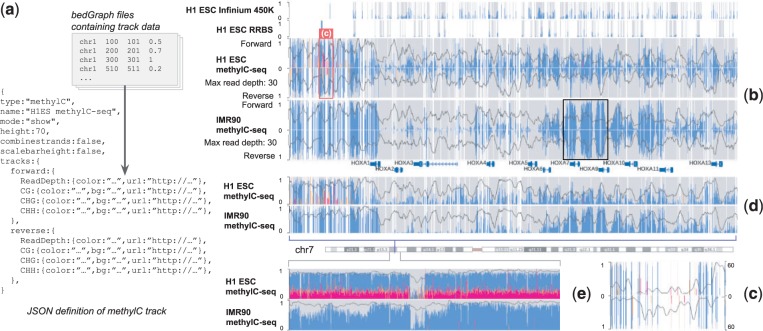
Fig. 1.
Format and display of the methylC track. (a) Track definition in JSON. (b) methylC track showing various DNA methylation assays over the HOXA gene cluster in the human genome. Four tracks are displayed: Infinium 450 K assay for H1 ESC, RRBS for H1 ESC, WGBS for both H1 ESC and IMR90. WGBS data are displayed in strand-specific mode. Cytosine methylation levels and contexts are identified using combinations of foreground/background bar colors (blue/gray for CG, orange/light orange for CHG and magenta/light magenta for CHH). Read depth (smoothed over 4 kb windows) is displayed as a black curve. (c) A zoomed-in view of H1 ESC WGBS data [marked by light red box in (b)]. The left axis marks methylation level. The axis on the right marks read depth. (d) WGBS data as in (b) with two strands combined. (e) A zoomed-out view of WGBS data with two strands combined
To illustrate the different visualization options, we displayed Infinium, RRBS and WGBS of ESC H1 (H1 ESC) and WGBS of IMR90 cells in Figure 1b. WGBS data are from Lister et al. (2009) and are displayed as context-specific and strand-specific values. Bar plots in the foreground indicate methylation levels with context-specific colors. Completely unmethylated cytosines are distinguished from cytosines with no data by a different user-configurable background color. The read-depth data are displayed as a line plot. In Figure 1b, the sparseness of Infinium and RRBS is visually obvious, and unmethylated CpGs are visually distinct from CpGs with no value. Compared with Infinium and RRBS data, WGBS clearly reveals more dynamics of DNA methylation across the regions and between the samples. Both H1 ESC and IMR90 are lowly methylated over the HOXA gene cluster. A region with elevated methylation in IMR90 is marked by a black box. It lies at the boundary of two chromatin domains (Supplementary Fig. S4). The methylation increase correlates with the bipartite histone modification pattern over HOXA cluster in IMR90 (Zhou et al., 2013). Figure 1d shows the WGBS data with two DNA strands combined. Figure 1e displays a zoomed-out view after combining data of the two strands, where the prevalence of non-CpG methylation in H1 ESC can be easily appreciated.
3 CONCLUSION
The methylC track on WashU EpiGenome Browser contributes simple but powerful innovations for visually exploring genome-wide single-base resolution DNA methylation data. The track is easy to prepare using standard bedGraph format files and is compatible with most common genomics tools. The track design is highly flexible and can be easily extended to incorporate new aspects of DNA methylation including allelic methylation, hydroxymethylation, and other complex genomics datasets.
Supplementary Material
ACKNOWLEDGEMENT
The authors thank an anonymous reviewer for identifying bugs in our code.
Funding: March of Dimes Foundation (grant number 5-FY10-491) and National Institutes of Health (grant numbers 5U01ES017154, R01HG007354, DA027995).
Conflict of Interest: none declared.
REFERENCES
- Esteller M. Cancer epigenomics: DNA methylomes and histone-modifications maps. Nat. Rev. Genet. 2007;8:286–298. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- Li H. Tabix: fast retrieval of sequence features from generic TAB-delimited files. Bioinformatics. 2009;27:718–719. doi: 10.1093/bioinformatics/btq671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–332. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissener A, et al. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 2005;33:5868–5877. doi: 10.1093/nar/gki901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, et al. Human epigenome browser at Washington University. Nat. Methods. 2011;8:989–990. doi: 10.1038/nmeth.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, et al. Exploring long-range genome interactions using the WashU EpiGenome Browser. Nat. Methods. 2013;10:375–376. doi: 10.1038/nmeth.2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziller MJ, et al. Charting a dynamic DNA methylation landscape of the human genome. Nature. 2013;500:477–481. doi: 10.1038/nature12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.