Abstract
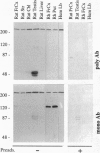
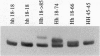
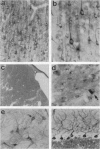
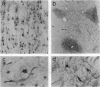
The Huntington disease (HD) phenotype is associated with expansion of a trinucleotide repeat in the IT15 gene, which is predicted to encode a 348-kDa protein named huntington. We used polyclonal and monoclonal anti-fusion protein antibodies to identify native huntingtin in rat, monkey, and human. Western blots revealed a protein with the expected molecular weight which is present in the soluble fraction of rat and monkey brain tissues and lymphoblastoid cells from control cases. In lymphoblastoid cell lines from juvenile-onset heterozygote HD cases, both normal and mutant huntingtin are expressed, and increasing repeat expansion leads to lower levels of the mutant protein. Immunocytochemistry indicates that huntingtin is located in neurons throughout the brain, with the highest levels evident in larger neurons. In the human striatum, huntingtin is enriched in a patch-like distribution, potentially corresponding to the first areas affected in HD. Subcellular localization of huntingtin is consistent with a cytosolic protein primarily found in somatodendritic regions. Huntingtin appears to particularly associate with microtubules, although some is also associated with synaptic vesicles. On the basis of the localization of huntingtin in association with microtubules, we speculate that the mutation impairs the cytoskeletal anchoring or transport of mitochondria, vesicles, or other organelles or molecules.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albin R. L., Greenamyre J. T. Alternative excitotoxic hypotheses. Neurology. 1992 Apr;42(4):733–738. doi: 10.1212/wnl.42.4.733. [DOI] [PubMed] [Google Scholar]
- Ambrose C. M., Duyao M. P., Barnes G., Bates G. P., Lin C. S., Srinidhi J., Baxendale S., Hummerich H., Lehrach H., Altherr M. Structure and expression of the Huntington's disease gene: evidence against simple inactivation due to an expanded CAG repeat. Somat Cell Mol Genet. 1994 Jan;20(1):27–38. doi: 10.1007/BF02257483. [DOI] [PubMed] [Google Scholar]
- Andrew S. E., Goldberg Y. P., Kremer B., Telenius H., Theilmann J., Adam S., Starr E., Squitieri F., Lin B., Kalchman M. A. The relationship between trinucleotide (CAG) repeat length and clinical features of Huntington's disease. Nat Genet. 1993 Aug;4(4):398–403. doi: 10.1038/ng0893-398. [DOI] [PubMed] [Google Scholar]
- Beal M. F. Does impairment of energy metabolism result in excitotoxic neuronal death in neurodegenerative illnesses? Ann Neurol. 1992 Feb;31(2):119–130. doi: 10.1002/ana.410310202. [DOI] [PubMed] [Google Scholar]
- Blackstone C. D., Moss S. J., Martin L. J., Levey A. I., Price D. L., Huganir R. L. Biochemical characterization and localization of a non-N-methyl-D-aspartate glutamate receptor in rat brain. J Neurochem. 1992 Mar;58(3):1118–1126. doi: 10.1111/j.1471-4159.1992.tb09370.x. [DOI] [PubMed] [Google Scholar]
- Burry R. W., Vandré D. D., Hayes D. M. Silver enhancement of gold antibody probes in pre-embedding electron microscopic immunocytochemistry. J Histochem Cytochem. 1992 Dec;40(12):1849–1856. doi: 10.1177/40.12.1453003. [DOI] [PubMed] [Google Scholar]
- Ferrante R. J., Kowall N. W., Beal M. F., Richardson E. P., Jr, Bird E. D., Martin J. B. Selective sparing of a class of striatal neurons in Huntington's disease. Science. 1985 Nov 1;230(4725):561–563. doi: 10.1126/science.2931802. [DOI] [PubMed] [Google Scholar]
- Gerfen C. R. The neostriatal mosaic: multiple levels of compartmental organization. Trends Neurosci. 1992 Apr;15(4):133–139. doi: 10.1016/0166-2236(92)90355-c. [DOI] [PubMed] [Google Scholar]
- Hedreen J. C., Folstein S. E. Early loss of neostriatal striosome neurons in Huntington's disease. J Neuropathol Exp Neurol. 1995 Jan;54(1):105–120. doi: 10.1097/00005072-199501000-00013. [DOI] [PubMed] [Google Scholar]
- Hersch S. M., Gutekunst C. A., Rees H. D., Heilman C. J., Levey A. I. Distribution of m1-m4 muscarinic receptor proteins in the rat striatum: light and electron microscopic immunocytochemistry using subtype-specific antibodies. J Neurosci. 1994 May;14(5 Pt 2):3351–3363. doi: 10.1523/JNEUROSCI.14-05-03351.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeste D. V., Barban L., Parisi J. Reduced Purkinje cell density in Huntington's disease. Exp Neurol. 1984 Jul;85(1):78–86. doi: 10.1016/0014-4886(84)90162-6. [DOI] [PubMed] [Google Scholar]
- Kremer B., Goldberg P., Andrew S. E., Theilmann J., Telenius H., Zeisler J., Squitieri F., Lin B., Bassett A., Almqvist E. A worldwide study of the Huntington's disease mutation. The sensitivity and specificity of measuring CAG repeats. N Engl J Med. 1994 May 19;330(20):1401–1406. doi: 10.1056/NEJM199405193302001. [DOI] [PubMed] [Google Scholar]
- Levey A. I., Edmunds S. M., Hersch S. M., Wiley R. G., Heilman C. J. Light and electron microscopic study of m2 muscarinic acetylcholine receptor in the basal forebrain of the rat. J Comp Neurol. 1995 Jan 16;351(3):339–356. doi: 10.1002/cne.903510303. [DOI] [PubMed] [Google Scholar]
- Levey A. I., Kitt C. A., Simonds W. F., Price D. L., Brann M. R. Identification and localization of muscarinic acetylcholine receptor proteins in brain with subtype-specific antibodies. J Neurosci. 1991 Oct;11(10):3218–3226. doi: 10.1523/JNEUROSCI.11-10-03218.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S. H., Schilling G., Young W. S., 3rd, Li X. J., Margolis R. L., Stine O. C., Wagster M. V., Abbott M. H., Franz M. L., Ranen N. G. Huntington's disease gene (IT15) is widely expressed in human and rat tissues. Neuron. 1993 Nov;11(5):985–993. doi: 10.1016/0896-6273(93)90127-d. [DOI] [PubMed] [Google Scholar]
- Mhatre A. N., Trifiro M. A., Kaufman M., Kazemi-Esfarjani P., Figlewicz D., Rouleau G., Pinsky L. Reduced transcriptional regulatory competence of the androgen receptor in X-linked spinal and bulbar muscular atrophy. Nat Genet. 1993 Oct;5(2):184–188. doi: 10.1038/ng1093-184. [DOI] [PubMed] [Google Scholar]
- Penney L., Kilshaw P. J., MacDonald T. T. Regional variation in the proliferative rate and lifespan of alpha beta TCR+ and gamma delta TCR+ intraepithelial lymphocytes in the murine small intestine. Immunology. 1995 Oct;86(2):212–218. [PMC free article] [PubMed] [Google Scholar]
- Perutz M. F., Johnson T., Suzuki M., Finch J. T. Glutamine repeats as polar zippers: their possible role in inherited neurodegenerative diseases. Proc Natl Acad Sci U S A. 1994 Jun 7;91(12):5355–5358. doi: 10.1073/pnas.91.12.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotrel A., Paskevich P. A., Kiely D. K., Bird E. D., Williams R. S., Myers R. H. Morphometric analysis of the prefrontal cortex in Huntington's disease. Neurology. 1991 Jul;41(7):1117–1123. doi: 10.1212/wnl.41.7.1117. [DOI] [PubMed] [Google Scholar]
- Strong T. V., Tagle D. A., Valdes J. M., Elmer L. W., Boehm K., Swaroop M., Kaatz K. W., Collins F. S., Albin R. L. Widespread expression of the human and rat Huntington's disease gene in brain and nonneural tissues. Nat Genet. 1993 Nov;5(3):259–265. doi: 10.1038/ng1193-259. [DOI] [PubMed] [Google Scholar]
- Telenius H., Kremer B., Goldberg Y. P., Theilmann J., Andrew S. E., Zeisler J., Adam S., Greenberg C., Ives E. J., Clarke L. A. Somatic and gonadal mosaicism of the Huntington disease gene CAG repeat in brain and sperm. Nat Genet. 1994 Apr;6(4):409–414. doi: 10.1038/ng0494-409. [DOI] [PubMed] [Google Scholar]