Abstract
• Premise of the study: The Compositae (Asteraceae) are a large and diverse family of plants, and the most comprehensive phylogeny to date is a meta-tree based on 10 chloroplast loci that has several major unresolved nodes. We describe the development of an approach that enables the rapid sequencing of large numbers of orthologous nuclear loci to facilitate efficient phylogenomic analyses.
• Methods and Results: We designed a set of sequence capture probes that target conserved orthologous sequences in the Compositae. We also developed a bioinformatic and phylogenetic workflow for processing and analyzing the resulting data. Application of our approach to 15 species from across the Compositae resulted in the production of phylogenetically informative sequence data from 763 loci and the successful reconstruction of known phylogenetic relationships across the family.
• Conclusions: These methods should be of great use to members of the broader Compositae community, and the general approach should also be of use to researchers studying other families.
Keywords: base tree, conserved, exon capture, next-generation sequencing, orthologs, phylogenomics
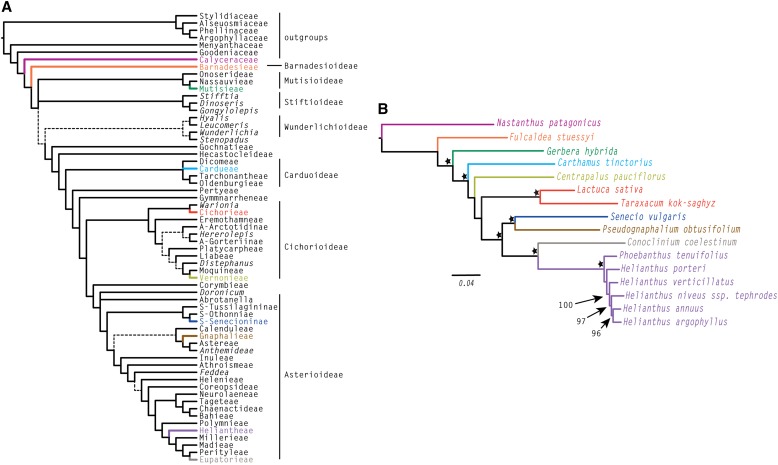
Ten to twelve percent of all flowering plants (25,000–33,000 species) belong to the Compositae family (Asteraceae). They occur throughout the world, but are most abundant in open areas with seasonal climates such as Mediterranean climates, deserts, prairies and steppes, and mountains. Some family members are widespread and a few are aggressive weeds; most, however, have restricted ranges, and many are in danger of extinction. The Compositae are monophyletic based on morphology as well as molecular genetic data. The most comprehensive phylogeny to date is a meta-tree (Funk and Specht, 2007) that was constructed using a base tree (i.e., the tree used to build the larger phylogeny/meta-tree) of 10 chloroplast loci (Funk et al., 2009; based on Panero and Funk, 2008; Funk and Chan, 2009; Pelser and Watson, 2009; Baldwin, 2009); this meta-tree included ∼900 of the 1700 genera found in the family. Several areas of the tree remain poorly resolved (Fig. 1A). These areas are important because the unresolved taxa vary in key morphological traits; thus, well-supported hypotheses of character evolution cannot be developed (e.g., Ortiz et al., 2009).
Fig. 1.
Compositae base tree and COS loci tree. (A) Summary of 10-locus chloroplast tree for ca. 900 genera redrawn from Funk et al. (2009). (B) Maximum likelihood tree based on 763 COS loci. Species sampled in our study are colored the same as their tribe in (A). Bootstrap values greater than 75% are indicated with a star. Bootstraps for the Heliantheae-only data set are given in numeric values with arrows. Note that within the Heliantheae-only data set, Phoebanthus was used as the outgroup so no support value is given for the node distinguishing it and the Helianthus species.
The combination of next-generation sequencing and large-scale phylogenomics is a promising avenue for efficiently assaying hundreds of loci across multiple taxa to resolve species relationships. One potential approach to identify and sequence loci for phylogenetic analysis is transcriptome sequencing (e.g., RNA-seq; e.g., McKain et al., 2012); however, in the many cases where obtaining fresh RNA is difficult or not feasible, a method based on genomic DNA would be preferable. This would also enable the use of museum specimens. Recent work in vertebrates has used DNA sequence capture of mostly noncoding nuclear regions flanking and including so-called “ultraconserved elements” (UCEs; Faircloth et al., 2012). The use of UCEs for phylogenomics is promising, but their detection in plant genomes may be more limited than in vertebrates (or absent altogether; see Reneker et al., 2012), partly due to the abundance of highly similar repetitive elements in the noncoding portions of plant genomes (Kritsas et al., 2012; Hupalo and Kern, 2013). Previous studies using target capture in plants have been more limited in scope: focusing primarily upon the plastome (Parks et al., 2012; Stull et al., 2013), or a single species (Populus trichocarpa, Zhou and Holliday, 2012; Fragaria vesca, Tennessen et al., 2013), or genus (Pinus, Neves et al., 2013; Gossypium, Salmon et al., 2012). A number of recent reviews have also been published promoting target capture and next-generation sequencing as an efficient and effective method for evolutionary and phylogenetic studies in plants (Cronn et al., 2012; Egan et al., 2012; Grover et al., 2012). We have thus designed sequence capture probes targeting a conserved ortholog set identified from expressed sequence tags (ESTs) from across the Compositae. The goal here is to improve both the family base tree and the level of resolution among related taxa. This project seeks to facilitate research in the Compositae by providing a robust framework for microevolutionary and systematic studies, and clearly delineating areas for which molecular and morphological studies are especially desirable. Our work also serves as a model for the development of similar tools in other taxonomic groups. Here we describe the design and workflow of our method including the wet laboratory protocol and downstream bioinformatic and phylogenetic analyses.
METHODS AND RESULTS
Taxon selection
Fifteen taxa were selected for this study to serve two purposes. First, 10 species were selected to span the entire family and its sister group, the Calyceraceae (species list in Table 1). These taxa allowed us to investigate the utility of our sequence capture probes across the family. Second, we added four additional representatives of the genus Helianthus (H. annuus L. was one of the original 10) and a taxon from its sister genus, Phoebanthus, so that we could also investigate the ability of our COS loci to resolve relationships among more closely related species.
Table 1.
Species sampled, fold-enrichment, number of COS loci per species, and collection information.a
| Tribe | Genus | Species | Authority | Enr. | No. of locib | Location | Collection date | Collector(s) | Collection no. |
| Calyceraceae | Nastanthus | patagonicus | Spreg. | 17 | 306 | Argentina: Santa Cruz, Rio Chico | 14-Dec-2009 | Bonifacino & Funk | 4016* |
| Barnadesieae | Fulcaldea | stuessyi | Roque & V. A. Funk | 4 | 174 | Brazil: Bahia | 09-Aug-2010 | Abreu, I. S. | 123* |
| Mutisieae | Gerbera | hybrida | n/a | 19 | 279 | Greenhouse grown cutting, Terra Nigra, USA | 22-Oct-2013 | Mandel, J. R. | 105ǂ |
| Cardueae | Carthamus | tinctorius | L. | 21 | 396 | Voucher n/a, USDA, PI 592391 | n/a | n/a | n/a |
| Cichorieae | Taraxacum | kok-saghyz | L. E. Rodin | 25 | 407 | Greenhouse grown seed≠ USDA, W6 35156 | 27-Aug-13 | Mandel, J. R. | 102ǂ |
| Vernonieae | Centrapalus | pauciflorus | (Willd.) H. Rob. | 28 | 408 | Greenhouse grown seed USDA, PI 312852 | 22-Oct-2013 | Mandel, J. R. | 104ǂ |
| Senecioneae | Senecio | vulgaris | L. | 28 | 316 | Washington, D.C.: NMNH | 07-Nov-2011 | Funk, V. A. | 12774* |
| Gnaphalieae | Pseudognaphalium | obtusifolium | (L.) Hilliard & B. L. Burtt | 15 | 208 | USA: Fairfax Co., Falls Church, Virginia | 12-Sep-2011 | Funk, V. A. | 12773* |
| Eupatorieae | Conoclinium | coelestinum | (L.) DC. | 30 | 416 | USA: Fairfax Co., Falls Church, Virginia | 11-Sep-2001 | Funk, V. A. | 12769* |
| Heliantheae | Phoebanthus | tenuifolius | S. F. Blake | 3ˆ | 372 | Greenhouse grown seed collected USA: Liberty Co., Florida | 10-Sep-2010 | Mason, C. M. | 101ǂ |
| Heliantheae | Helianthus | porteri | (A. Gray) Pruski | 46 | 325 | Greenhouse grown seed collected USA: DeKalb Co., Georgia≠ | 22-Oct-2013 | Mandel, J. R. | 103ǂ |
| Heliantheae | Helianthus | verticillatus | Small | 33 | 379 | Greenhouse grown seed collected USA: Madison Co., Tennesee | 01-Sep-2004 | Mandel, J. R. | 101ǂ |
| Heliantheae | Helianthus | niveus subsp. tephrodes | (A. Gray) Heiser | 62 | 395 | Voucher n/a, USDA, PI 613758 | n/a | n/a | n/a |
| Heliantheae | Helianthus | argophyllus | Torr. & A. Gray | 34 | 339 | Voucher n/a, USDA, PI 435623 | n/a | n/a | n/a |
| Heliantheae | Helianthus | annuus | L. | 38 | 385 | Voucher n/a, USDA, PI 603989 | n/a | n/a | n/a |
Note: Enr. = fold enrichment; n/a = not available; NMNH = National Museum of Natural History; USDA = U.S. Department of Agriculture.
Five hundred ninety-one COS loci were extracted from the lettuce genome. DNA from some taxa studied had been extracted for other projects and the plants were not vouchered; they are listed as n/a.
Number of COS loci identified by PHYLUCE.
Deposited at the United States National Herbarium, Smithsonian Institution (US), Washington, D.C., USA.
Deposited at the University of Georgia Herbarium (GA), Athens, Georgia, USA.
Voucher specimen is an individual from the same population.
The Phoebanthus WGS sample resulted in far fewer reads than other taxa (∼2% of the average for other WGS samples) and could bias the enrichment calculation downward.
Identification of conserved orthologous sequences
A set of ∼1300 conserved genes including approximately 300 single- or low-copy genes for the Compositae was previously identified via BLAST (version 2.2.6) searches of H. annuus (sunflower; Asteroideae) and Lactuca sativa L. (lettuce; Cichorioideae) ESTs against a set of Arabidopsis single-copy genes (the spliced gene models only; see putative intron position determination below) (Kozik et al., unpublished; see http://www.cgpdb.ucdavis.edu/COS_Arabidopsis/ for a description of the pipeline and sequence files). To broaden the representation of Compositae sequences in our analysis, we subsequently used ca. 19,000 Carthamus tinctorius L. (safflower; Carduoideae) unigenes derived from ca. 41,000 ESTs (data available at http://www.cgpdb.ucdavis.edu/asteraceae_assembly/) in a BLAST (version 2.2.26) search against the set of ∼1300 genes (hereafter simply referred to as the conserved ortholog set loci, or COS loci). The best safflower hits with an E-value ≤E-40 and spanning ≥150 bp were added to the COS alignments using MUSCLE (version 3.8; Edgar, 2004). We were able to generate safflower alignments to 624 out of the ∼1300 COS loci. These sequences and the alignments are deposited in the Dryad Digital Repository: http://doi.org/10.5061/dryad.gr93t (Mandel et al., 2014).
Probe design, sequence capture, and sequencing
Custom biotinylated RNA bait/probe libraries were designed using the MYbaits target enrichment system (MYcroarray, Ann Arbor, Michigan, USA) to enrich genomic DNA libraries from 15 species from across the family for the COS loci (species list in Table 1). The biotinylated 120-mer baits were tiled across each locus with a 60-base overlap between baits. The putative intron positions of each locus were taken into account during the design process such that probes were positioned so as not to span putative splice sites. Putative intron positions for these loci were determined following the methods of Chapman et al. (2007); briefly, the Arabidopsis sequence from each alignment was used in a BLAST search against the full Arabidopsis genome database (available from http://www.arabidopsis.org/). The BLAST output was mapped onto the global alignments (i.e., the multispecies alignments) via custom scripts (available from Chapman et al., 2007), thus allowing the identification of putative intron positions for these loci. For each COS locus, we designed three probes when possible, i.e., matching the lettuce, sunflower, and safflower sequences. Ultimately, the probe set included 9678 baits targeting 1061 orthologous genes (note that some genes were not able to be covered by baits during the MYcroarray design process due to short putative exon lengths). The sequences for the baits and source ESTs can be found in Appendix S1 (960.3KB, xlsx) .
Genomic DNA of each species was extracted using the DNeasy Plant Mini Kit (QIAGEN, Valencia, California, USA), and a barcoded sequencing library was constructed using the TruSeq DNA Sample Preparation kit (Illumina, San Diego, California, USA). Sequence capture was performed following the manufacturer’s protocol (MYcroarray) with two additional steps to improve the hybridization efficiency of the baits to the DNA: (1) the amount of “Hyb #1” used in Step III of the manufacturer’s protocol was reduced from 20 μL to 15 μL per sample, and (2) the magnetic beads were incubated for 30 min at room temperature with 5 μg of denatured salmon sperm DNA prior to transferring the hybridization/capture solution to the beads. We also produced libraries with unique TruSeq barcodes of noncaptured DNA (i.e., an unenriched library) for each species for whole genome shotgun (WGS) sequencing. The enriched samples (hereafter referred to as COS-DNA) and WGS-DNA libraries were quantified using a Qubit 2.0 Fluorometer (Life Technologies, Grand Island, New York, USA), pooled, and sequenced on two lanes of an Illumina HiSeq 2000 sequencer (paired end, 100 base reads). The number of reads for each taxon is located in Appendix S1 (960.3KB, xlsx) . Overall, we obtained a high number of reads for each sample (COS-DNA and WGS-DNA). The only exception was for the Phoebanthus WGS-DNA sample, which had poor sequencing results. Fold-enrichment was calculated for each species by using BLAST similarity (blastn, “-e 1e-5”) of the COS-DNA reads and WGS-DNA reads (both quality trimmed; see below for details) to the COS loci and taking the percentage of reads that had a significant BLAST hit out of the total number of reads for each species (COS-DNA vs. WGS-DNA). Fold enrichment is indicated for each species in Table 1. There was no overwhelming trend with respect to the fold enrichment for each species and the phylogenetic distance from the species used in bait design. However, species in the Heliantheae tended to have higher enrichment when compared to the other taxa, and the outgroup and most basal taxon had lower values of enrichment.
Bioinformatic and phylogenetic analyses
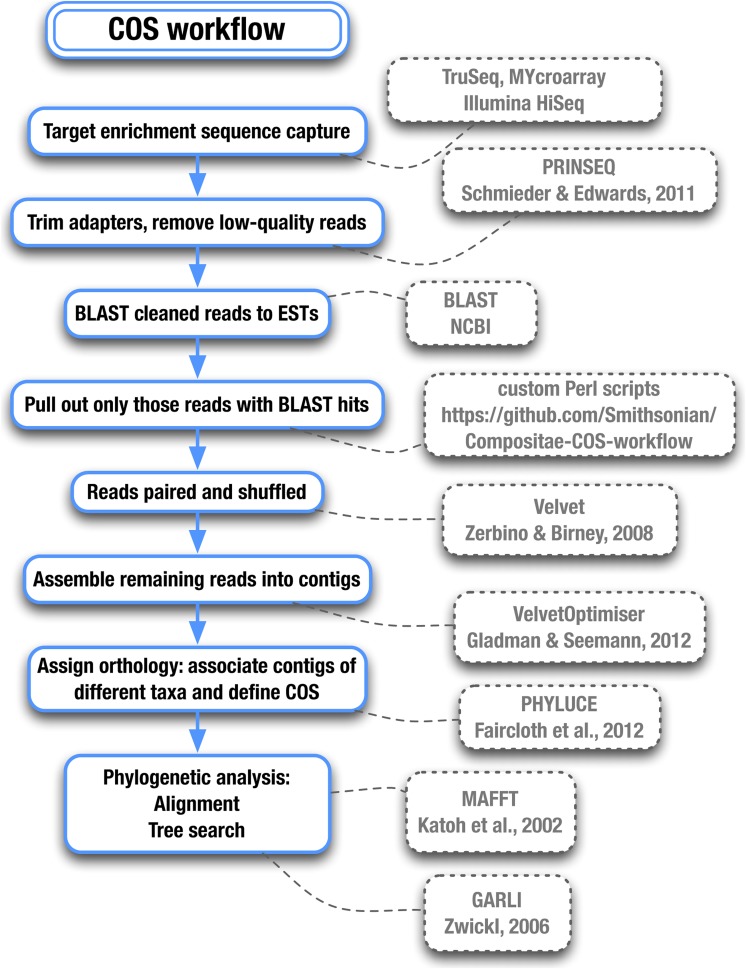
All custom scripts, FASTA files, and links to publicly available code have been placed on GitHub (https://github.com/Smithsonian/Compositae-COS-workflow). The workflow is also illustrated in Fig. 2. Note that we also included two optional Perl wrapper scripts on GitHub that can be used to streamline several of the steps performed here; see the README files on the GitHub repository for details. The bioinformatic and phylogenetic workflow was as follows: for each sample, the COS-DNA paired-end reads (the following steps were performed on the two pairs from each species separately: R1, R2) were quality trimmed and reformatted from FASTQ to FASTA using PRINSEQ (version 0.18.2; Schmieder and Edwards, 2011) with the parameters: ‘-min_len 40 –noniupac –min_qual_mean 15 –lc_method entropy –lc_threshold 60 –trim_ns_right 10 –ns_max_p 20’ to remove low-quality and short sequences. While we enriched for the targeted loci by performing the sequence capture, the abundance of remaining off-target reads, combined with the sheer amount of data, made de novo assemblies (see below) computationally demanding. Therefore, we performed a prefiltering step to screen for reads that contained DNA of targeted loci. To do this, the cleaned reads in FASTA format were subjected to BLAST for a similarity search to the EST sequences used for probe design (COS_sunf_lett_saff_all.fasta), and a custom Perl script (Top_Hits.pl) was used to take only the best read hit from the BLAST output and create a new file with only those read sequences from the original FASTA reads files. The BLAST-filtered Illumina reads have been deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) as BioProject PRJNA236448. The R1 and R2 reads were paired with Pairfq (https://github.com/sestaton/Pairfq) and then shuffled (using shuffleSequences_fasta.pl from the Velvet de novo sequence assembler package; Zerbino and Birney, 2008), and the singles (from R1 or R2) were concatenated for assembly via Velvet, with hash lengths k = 51–99 using VelvetOptimiser (version 2.2.0; http://bioinformatics.net.au/software.velvetoptimiser.shtml) to find the optimal k-mer length, expected coverage, and coverage cutoffs for each assembly. The number of contigs assembled for each species is located in Appendix S1 (960.3KB, xlsx) . To get a first look at how many COS loci were recovered via de novo assembly, we performed BLAST searches of each species’ contigs to the COS loci using the same parameters as before and noted the number of COS loci obtained as a best hit for each species (see Appendix S1 (960.3KB, xlsx) ). The most basal taxon, Fulcaldea stuessyi Roque & V. A. Funk, which had the lowest number of BLAST filtered reads, also had the lowest number of Velvet contigs and COS BLAST hits—even lower than the outgroup. At the present time, we do not know if this is related to DNA/library quality or whether this taxon is extremely divergent in the family. Future studies will include more taxon sampling from this area of the Compositae and probes designed from additional basal taxa. After Fulcaldea, the number of COS BLAST loci generally followed a trend of recovering more loci the closer the query was in relatedness to the species used for probe design, though overall a good number of loci were recovered for the majority of these species.
Fig. 2.
COS workflow. Schematic of the laboratory method, bioinformatics, and phylogenetic analyses for this project.
Following assembly of the captured reads, contigs from each of the 15 species were analyzed in the PHYLUCE pipeline (version 0.1.0; Faircloth et al., 2012) specifically using the programs: match_contigs_to_probes.py, get_match_counts.py, get_fastas_from_match_counts.py, and seqcap_align_2.py. Briefly, the program uses LASTZ (version 1.02.00; Harris, 2007) to align the baits/probes to the assembled contigs from each taxon to determine which contigs match the COS loci. PHYLUCE then associates the contigs across species that are putative orthologs, and is quite conservative in that it rejects putative orthologs with more than one match assuming possible paralogy (i.e., ensuring that only one contig matches probes from one COS locus and that only probes from one COS locus match one contig). In addition to the 15 species for which sequence capture was performed, we also included lettuce sequences derived from the publicly available whole genome assembly (version 4; https://lgr.genomecenter.ucdavis.edu). This was done by creating a BLAST database from the lettuce genome scaffolds and performing BLAST searches of each captured COS (from each species) to the lettuce genome. Nucleotide alignments were performed in MAFFT (version 7.029b; Katoh et al., 2002; as implemented in PHYLUCE) for the 763 COS loci with sufficient coverage in a minimum of three species. The number of COS loci analyzed for each species is listed in Table 1. A comparison of the COS loci recovered via BLAST for each species (Appendix S1 (960.3KB, xlsx) ) and the COS loci retained following PHYLUCE revealed that, in general, the hybridization and sequencing captured a large portion of the 1061 targeted loci for each taxon, and the stage where the majority of loci were lost (i.e., not recovered for phylogenetic analyses out of the 1061) occurred at the orthology assignment step in PHYLUCE. Notably, the one species that is a polyploid, Senecio vulgaris L., did not suffer from fewer loci recovered, despite the conservative process of rejecting potential paralogs in PHYLUCE. To assess the utility of the probe set within closely related species, and to ascertain whether PHYLUCE would homologize additional data, we also ran the PHYLUCE pipeline using only the taxa from the Heliantheae tribe, and this resulted in 415 COS loci aligned with MAFFT. These 415 loci had a mean length of 404 bp (range: 200–1101 bp), while the original loci across all taxa had a mean length of 353 bp (range: 27–1545 bp). Alignments have been deposited in the Dryad Data Repository (http://doi.org/10.5061/dryad.gr93t; Mandel et al., 2014).
Phylogenetic analyses of concatenated data sets were completed for all 763 COS loci (269,585 bp; 59% missing data), all COS loci for which 10/16 (lettuce included) species were represented (186 loci; 49,918 bp; 37% missing data), and all COS loci for which 8/16 species were represented (347 loci; 96,649 bp; 45% missing data) under the maximum likelihood optimality criterion in GARLI (version 2.0; Zwickl, 2006; GTR-gamma model of nucleotide substitution; 100 search replicates; 1000 bootstrap replicates). The significant fraction of missing data is indicative of the stringency of PHYLUCE. In future work, we are planning to implement strategies that will allow us to explore the effects of different methods of orthology detection. The 763-locus tree, along with bootstrap information, is presented in Fig. 1B. This 763-locus alignment contained a total of 28,324 parsimony informative characters. Comparison with Fig. 1A shows that the relationships found in the 763-locus tree mirror what is expected based on the base tree (Funk et al., 2009; Fig. 1A), and bootstrap support was greater than 75% on almost all nodes. The 186-locus tree (not shown) was congruent with the 763-locus tree, and the 347-locus tree (also not shown) differed only in the placement of species within Heliantheae, suggesting there may be a tradeoff between taxon representation and total data/number of genes when compared to the 186- and 763-locus trees. The Heliantheae-only tree (415 loci; 167,650 bp) also reconstructed the expected relationships (and the same as the 763-locus tree) based on current phylogenetic information for the taxa studied here with high bootstrap support (Timme et al., 2007; Schilling and Panero, 2011). Bootstrap values for this data set are displayed on Fig. 1B with arrows. As noted previously, restricting the PHYLUCE pipeline to only taxa within the Heliantheae often produced longer alignments for the 415 loci when compared to these same loci in the whole data set alignments (see MAFFT alignments in the Dryad Data Repository [http://doi.org/10.5061/dryad.gr93t; Mandel et al., 2014]). This may, in part, be due to difficulty in aligning across introns, or other highly divergent regions of a locus, when including taxa from across the entire family. These findings suggest that orthology detection is dependent on taxon sampling and the development of new strategies that take into account the hierarchical nature of homology, and how to include as many data as possible in the initial orthology assessment, will be explored in the future. A broader study of the phylogenetic relationships within the Compositae using this method and their relationship with chloroplast loci is underway (Mandel et al., in prep). Data matrices and phylogenetic trees have been deposited in the Dryad Data Repository (http://doi.org/10.5061/dryad.gr93t; Mandel et al., 2014).
CONCLUSIONS
Targeted sequence capture of COS loci facilitated phylogenetic analyses based on a large number of genes across the Compositae. To date, phylogenetic reconstruction in the Compositae has mostly relied on a small number of chloroplast DNA loci, and many relationships among family members remain poorly understood. The motivation for choosing the taxa sequenced here was that the relationships among them are well-established (based on both morphological and molecular data); thus, these known relationships provide a benchmark against which the COS-DNA phylogenies can be compared. We were able to generate usable sequence information for a total of 763 loci and to recover a phylogeny consistent with known relationships within the family with high bootstrap support at most of the nodes within the tree. Moreover, our workflow also proved successful in reconstructing relationships within the Heliantheae tribe, demonstrating that this method is useful for both broader- and finer-scaled phylogenetic reconstruction. We are continuing to add taxa to this base tree using the methods described herein with an ultimate goal of generating a phylogeny that includes at least 200 species selected to represent key nodes within the family. These methods should be of great use to members of the broader Compositae community, and the general approach outlined herein should also be of use to researchers studying other families.
Supplementary Material
LITERATURE CITED
- Baldwin B. 2009. Heliantheae alliance. In V. A. Funk, A. Susanna, T. F. Stuessy, and R. J. Bayer [eds.], Systematics, evolution, and biogeography of Compositae, 689–711. International Association for Plant Taxonomy, Vienna, Austria. [Google Scholar]
- Chapman M. A., Chang J.-C., Weisman D., Kesseli R. V., Burke J. M. 2007. Universal markers for comparative mapping and phylogenetic analysis in the Asteraceae (Compositae). Theoretical and Applied Genetics 115: 747–755 [DOI] [PubMed] [Google Scholar]
- Cronn R., Knaus B., Liston A., Maughan P. J., Parks M., Syring J., Udall J. 2012. Targeted enrichment strategies for next generation plant biology. American Journal of Botany 99: 291–311 [DOI] [PubMed] [Google Scholar]
- Edgar R. C. 2004. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan A. N., Schlueter J., Spooner D. M. 2012. Applications of next-generation sequencing in plant biology. American Journal of Botany 99: 175–185 [DOI] [PubMed] [Google Scholar]
- Faircloth B. C., McCormack J. E., Crawford N. G., Harvey M. G., Brumfield R. T., Glenn T. C. 2012. Ultraconserved elements anchor thousands of genetic markers spanning multiple evolutionary timescales. Systematic Biology 61: 717–726 [DOI] [PubMed] [Google Scholar]
- Funk V. A., Chan R. 2009. Introduction to Cichorioideae. In V. A. Funk, A. Susanna, T. F. Stuessy, and R. J. Bayer [eds.], Systematics, evolution, and biogeography of Compositae, 336–342. International Association for Plant Taxonomy, Vienna, Austria. [Google Scholar]
- Funk V. A., Specht C. 2007. Meta-trees: Grafting for a global perspective. Proceedings of the Biological Society of Washington 120: 232–240 [Google Scholar]
- Funk V. A., Susanna A., Stuessy T. F., Bayer R. J. 2009. Systematics, evolution, and biogeography of the Compositae. International Association for Plant Taxonomy, Vienna, Austria [Google Scholar]
- Grover C. E., Salmon A., Wendel J. F. 2012. Targeted sequence capture as a powerful tool for evolutionary analysis. American Journal of Botany 99: 312–319 [DOI] [PubMed] [Google Scholar]
- Harris R. S. 2007. Improved pairwise alignment of genomic DNA. Ph.D. Thesis, The Pennsylvania State University, University Park, Pennsylvania, USA.
- Hupalo D., Kern A. D. 2013. Conservation and functional element discovery in 20 angiosperm plant genomes. Molecular Biology and Evolution 30: 1729–1744 [DOI] [PubMed] [Google Scholar]
- Katoh K., Misawa K., Kuma K., Miyata T. 2002. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Research 30: 3059–3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kritsas K., Wuest S. E., Hupalo D., Kern A. D., Wicker T., Grossniklaus U. 2012. Computational analysis and characterization of UCE-like elements (ULEs) in plant genomes. Genome Research 22: 2455–2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel J. R., Dikow R. B., Funk V. A., Masalia R. R., Staton S. E., Kozik A., Michelmore R. W., et al. 2014. Data from: A target enrichment method for gathering phylogenetic information from hundreds of loci: An example from the Compositae. Dryad Digital Repository. http://doi.org/10.5061/dryad.gr93t [DOI] [PMC free article] [PubMed]
- McKain M. R., Wickett N., Zhang Y., Ayyampalayam S., McCombie W. R., Chase M. W., Pires J. C., et al. 2012. Phylogenomic analysis of transcriptome data elucidates co-occurrence of a paleopolyploid event and the origin of bimodal karyotypes in Agavoideae (Asparagaceae). American Journal of Botany 99: 397–406 [DOI] [PubMed] [Google Scholar]
- Neves L. G., Davis J. M., Barbazuk W. B., Kirst M. 2013. Whole-exome targeted sequencing of the uncharacterized pine genome. Plant Journal 75: 146–156 [DOI] [PubMed] [Google Scholar]
- Ortiz S., Bonifacino J. M., Crisci J. V., Funk V. A., Hansen H., Hind D. J. N., Katinas L., et al. 2009. The basal grade of Compositae: Mutisieae (sensu Cabrerra). In V. A. Funk, A. Susanna, T. F. Stuessy, and R. J. Bayer [eds.], Systematics, evolution, and biogeography of Compositae, 193–213. International Association for Plant Taxonomy, Vienna, Austria. [Google Scholar]
- Panero J. L., Funk V. A. 2008. The value of sampling anomalous taxa in phylogenetic studies: Major clades of the Asteraceae revealed. Molecular Phylogenetics and Evolution 47: 757–782 [DOI] [PubMed] [Google Scholar]
- Parks M., Cronn R., Liston A. 2012. Separating the wheat from the chaff: Mitigating the effects of noise in a plastome phylogenomic data set from Pinus L. (Pinaceae). BMC Evolutionary Biology 12: 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelser P. B., Watson L. 2009. Introduction to Asteroideae. In V. A. Funk, A. Susanna, T. F. Stuessy, and R. J. Bayer [eds.], Systematics, evolution, and biogeography of Compositae, 495–502. International Association for Plant Taxonomy, Vienna, Austria. [Google Scholar]
- Reneker J., Lyons E., Conant G. C., Pires J. C., Freeling M., Shyu C.-R., Korkin D. 2012. Long identical multispecies elements in plant and animal genomes. Proceedings of the National Academy of Sciences, USA 109: E1183–E1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon A., Udall J. A., Jeddeloh J. A., Wendel J. 2012. Targeted capture of homoeologous coding and noncoding sequence in polyploid cotton. G3: Genes|Genomes|Genetics 2: 921–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling E. E., Panero J. L. 2011. A revised classification of subtribe Helianthinae (Asteraceae: Heliantheae). II. Derived lineages. Botanical Journal of the Linnean Society 167: 311–331 [Google Scholar]
- Schmieder R., Edwards R. 2011. Quality control and preprocessing of metagenomic datasets. Bioinformatics (Oxford, England) 27: 863–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stull G. W., Moore M. J., Mandala V. S., Douglas N. A., Kates H.-R., Qi X., Brockington S. F., et al. 2013. A targeted enrichment strategy for massively parallel sequencing of angiosperm plastid genomes. Applications in Plant Sciences 1: 1200497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennessen J. A., Govindarajulu R., Liston A. 2013. Targeted sequence capture provides insight into genome structure and genetics of male sterility in a gynodioecious diploid strawberry, Fragaria vesca ssp. bracteata (Rosaceae). G3: Genes|Genomes|Genetics 3: 1341–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timme R. E., Simpson B. B., Linder C. R. 2007. High-resolution phylogeny for Helianthus (Asteraceae) using the 18S-26S ribosomal DNA external transcribed spacer. American Journal of Botany 94: 1837–1852 [DOI] [PubMed] [Google Scholar]
- Zerbino D. R., Birney E. 2008. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Research 18: 821–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Holliday J. A. 2012. Targeted enrichment of the black cottonwood (Populus trichocarpa) gene space using sequence capture. BMC Genomics 13: 703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwickl D. J. 2006. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. PhD thesis, University of Texas at Austin, Austin, Texas, USA.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.