SUMMARY
Cells of the inner cell mass (ICM) of the mouse blastocyst differentiate into the pluripotent epiblast (EPI) or the primitive endoderm (PrE), marked by the transcription factors NANOG and GATA6, respectively. To investigate the mechanistic regulation of this process, we applied an unbiased, quantitative, single-cell resolution image analysis pipeline, to analyze embryos lacking or exhibiting reduced levels of GATA6. We find that Gata6 mutants exhibit a complete absence of PrE, and demonstrate that GATA6 levels regulate the timing and speed of lineage commitment within the ICM. Furthermore, we show that GATA6 is necessary for PrE specification by FGF signaling, and propose a model where interactions between NANOG, GATA6 and the FGF/ERK pathway determine ICM cell fate. This study provides a framework for quantitative analyses of mammalian embryos, and establishes GATA6 as a nodal point in the gene regulatory network driving ICM lineage specification.
Keywords: mouse, blastocyst, inner cell mass, cell lineage commitment, primitive endoderm, epiblast, GATA6, NANOG, FGF, live imaging, quantitative immunofluorescence
INTRODUCTION
The first cell differentiation events during mammalian development result in the segregation of two extra-embryonic lineages, the trophectoderm (TE) and the primitive endoderm (PrE), from the pluripotent epiblast (EPI), the founder tissue of most of the embryo-proper (Saiz and Plusa, 2013; Schrode et al., 2013). These three cell types, found in the late blastocyst, are thought to arise through two sequential rounds of binary cell fate decisions. In the mouse, the first cell fate decision begins at the 8-16-cell stage, when the morula undergoes compaction and cells on the surface acquire apicobasal polarity, eventually becoming TE (Johnson and Ziomek, 1981). The second decision involves scattered cell differentiation within the inner group of cells, which are referred to as the inner cell mass (ICM), followed by cell sorting, and results in the differentiation of PrE and EPI lineages.
GATA6 and NANOG are the earliest markers of the PrE and EPI lineages, respectively; however, they are co-expressed in all ICM cells at the early blastocyst (32–64 cells) stage (Plusa et al., 2008). As embryos develop, individual ICM cells acquire exclusive GATA6 or NANOG expression, in an apparently stochastic manner, which is thought to reflect the specification of PrE and EPI fates (Chazaud et al., 2006; Plusa et al., 2008). This process, proposed to be mediated both by stimulation of the FGF/ERK pathway (Chazaud et al., 2006; Kang et al., 2013a; Krawchuk et al., 2013; Nichols et al., 2009; Yamanaka et al., 2010) and by reciprocal repression between GATA6 and NANOG (Singh et al., 2007), results in a salt-and-pepper distribution of PrE and EPI precursors in the ICM of mid blastocysts (64–100 cells) (Chazaud et al., 2006). This scattered lineage specification is followed by the sorting of PrE precursors to the surface of the ICM in the late blastocyst (>100 cells), where they come to form an epithelium separating the blastocyst cavity and the EPI (Meilhac et al., 2009; Plusa et al., 2008; Saiz et al., 2013). However, the molecular mechanisms and gene regulatory networks governing the specification of PrE and pluripotent EPI within the ICM of the early blastocyst are still poorly understood.
In the mouse, FGF signaling is critical for PrE specification (Chazaud et al., 2006; Goldin and Papaioannou, 2003; Kang et al., 2013a; Krawchuk et al., 2013; Nichols et al., 2009; Yamanaka et al., 2010). FGF4 and FGFR2 are reciprocally expressed in EPI- and PrE-biased cells, respectively. FGF4, produced by EPI-biased cells (Guo et al., 2010; Ohnishi et al., 2014b), has been proposed to activate the FGF/ERK pathway in neighboring cells, leading to the down-regulation of NANOG and induction of the PrE program. However, the mechanism has not yet been experimentally addressed. Importantly, single-cell gene expression profiling studies coupled with the analysis of mutants (Kang et al., 2013a; Ohnishi et al., 2014a), have suggested that FGF signaling is not required for the initial establishment of the gene regulatory network (GRN) in ICM cells, but is essential for cells to exit this immature multi-lineage priming state and differentiate into EPI or PrE.
GATA6, a member of the GATA family of zinc-finger transcription factors, is the earliest PrE marker expressed in the early mouse embryo (Morrisey et al., 1996). It is first detected at around the 8-cell stage in all blastomeres, and by the mid blastocyst (~64-cell stage) it is restricted to PrE progenitors (Chazaud et al., 2006; Plusa et al., 2008). Ectopic expression of GATA6 in mouse embryonic stem (ES) cells is sufficient to direct them to a PrE-like state (Artus et al., 2010; Fujikura et al., 2002; Shimosato et al., 2007). GATA6 therefore likely acts near the top of the hierarchy regulating PrE development. However, the position of GATA6 relative to NANOG and the FGF/ERK pathway in the GRN driving ICM cell fate specification in vivo remains to be established.
In this study we have undertaken a quantitative, single-cell resolution analysis to understand the process of PrE segregation from the pluripotent EPI, and begin to mechanistically decipher the networks in which GATA6 engages to regulate this event. To investigate the role of GATA6 in ICM development, we have analyzed a wild-type, heterozygote and null mutant Gata6 allelic series (Sodhi et al., 2006) using automated nuclear segmentation (Lou et al., 2014) followed by single-cell resolution quantitative three-dimensional (3D) image analyses.
Our results demonstrate that the early spatial pattern of differentiation of PrE versus EPI precursors is stochastic, and that spatial order emerges gradually at later stages. GATA6 is required for PrE cell fate specification, and for the execution of the PrE program. Gata6 null mutant embryos lack a PrE entirely, and exhibit pan-ICM expression of the pluripotency-associated factors NANOG, OCT4 and SOX2. In Gata6 heterozygotes the proportion of ICM cells adopting a PrE fate is reduced, and their commitment decelerated, such that the period of time over which ICM cells make a PrE fate choice is extended. Exposure to exogenous FGF4 failed to restore PrE precursors within Gata6 null mutant embryos, indicating that GATA6 is required for activation of the PrE program, and the concomitant down-regulation of Nanog induced by FGF4. Collectively, our findings place GATA6 at the top of the hierarchy regulating PrE specification.
RESULTS
Cell fate choice is, in large part, determined by the action of key lineage-specific transcription factors. PrE and EPI lineage specification within the ICM of the mouse blastocyst appears to be undertaken in a stochastic manner. A sequence of events involving lineage specification and subsequent positional segregation has been defined. It involves the initial co-expression of factors within all ICM cells, progressive restriction of gene expression to lineage precursors, followed by a combination of cell sorting and cell death to refine their position (Artus et al., 2013; Chazaud et al., 2006; Gerbe et al., 2008; Meilhac et al., 2009; Plusa et al., 2008). Within this emergent mechanistic framework, GATA6 is the earliest expressed PrE-specific transcription factor, while NANOG is the earliest expressed EPI-specific transcription factor. However, these factors are initially co-expressed within the ICM, and so are only markers once they become mutually-exclusive, thus this initiation and transition in marker localization is likely to be key to understanding the establishment of respective PrE and EPI fates.
A pipeline for single-cell resolution quantitative analyses of expression and position: progressive distribution of GATA6 and NANOG
A rigorous mechanistic understanding of how single cells can operate coordinately to produce global effects relies on methods to resolve single-cell resolution information in the context of a population. Thus far, attempts at single-cell analyses of cell fate decisions in pre-implantation mammalian embryos have been hindered by time consuming, manual data processing at a small scale. To decipher the details of the GRN operating within the ICM, we assembled a novel unbiased single-cell resolution analysis pipeline. This pipeline comprised software specifically developed for automated nuclear segmentation of 3D image data of mouse pre-implantation stage embryos (Lou et al., 2014), followed by quantitative fluorescent and spatial data analyses. The highly accurate segmentation afforded by our pipeline facilitates single-cell resolution, large-scale comparisons of protein concentrations, represented by fluorescence intensities after immunostaining and confocal imaging (Figure 1A). In this way, an analysis could be undertaken at the level of the entire ICM, taking into account all cells within every embryo analyzed.
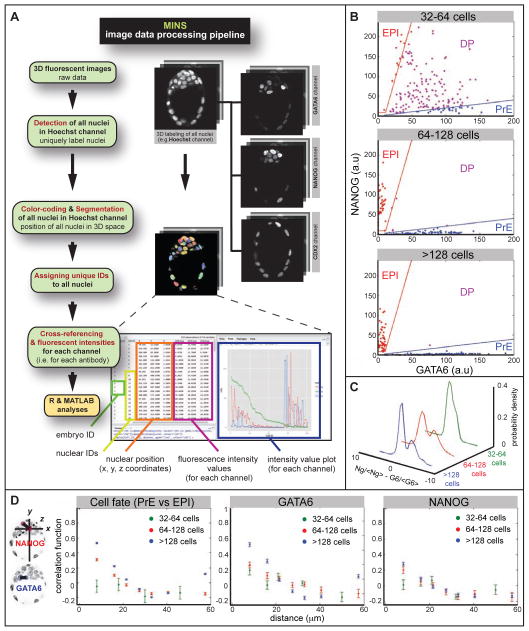
Figure 1. Quantitative analysis of GATA6 and NANOG expression.
A Image data processing pipeline incorporating MINS software. B Nuclear concentration of GATA6 and NANOG at different stages. Blue and red lines show the threshold function used to define cells as PrE and EPI precursors respectively. Cells that do not fall in either group are co-expressing NANOG and GATA6, and are classified as DP (double positive, uncommitted ICM) cells. C Probability density of normalized GATA6 concentration minus normalized NANOG concentration (<G6> and <Ng> indicate the average concentrations of GATA6 and NANOG, respectively). Earliest stage (defined by total cell number N) cells express both GATA6 and NANOG (unimodal distribution), but at the “salt-and-pepper” stage cells are either GATA6-positive or NANOG-positive, resulting in a bimodal distribution. D Correlation coefficient of cell fates and of levels of GATA6 and NANOG as a function of cell-cell distance for three different stages. Correlation = 0, cells are making their fate choice independent of other cells, no fate prediction can be made; correlation =1, certain prediction that cells with close distance are alike; Correlation = −1, certain prediction that cells with close distance are unalike.
A method for unbiased assignment of cell fate
We performed immunohistochemistry using antibodies directed against GATA6 and NANOG proteins on wild-type embryos at several successive stages. As expected, wild-type morulae and early blastocysts exhibited co-expression of GATA6 and NANOG, which resolved in a mutually-exclusive expression pattern at the mid blastocyst, and in sorted lineages by the late blastocyst (Figure 1B, 2A). Applying our segmentation pipeline, we extracted fluorescence intensities of GATA6 and NANOG in each nucleus, estimated nuclear concentration by subtracting background fluorescence, and corrected for attenuation of fluorescence signals by tissue depth. Analysis of nuclear concentrations as a function of stage (32–64 cells, 64–128 cells, >128 cells) revealed that at the early stage most cells expressed high levels of both GATA6 and NANOG. At the 64–128 cells stage, however, most cells expressed either high-GATA6 and low-NANOG or vice versa. At the last stage analyzed, all cells were either GATA6-positive and NANOG-negative or NANOG-positive and GATA6-negative (Figure 1B, C). We used the nuclear concentrations at this stage to empirically deduce a range over which cells could be defined as either EPI or PrE. This procedure provided an unbiased, reproducible and quantitative method to assign fates to each cell of any given embryo. It allowed us to assign cell fate (Figure 1D), based on the automatic threshold procedure we developed (Figure 1B), in the absence of arbitrary factors, such as human user error.
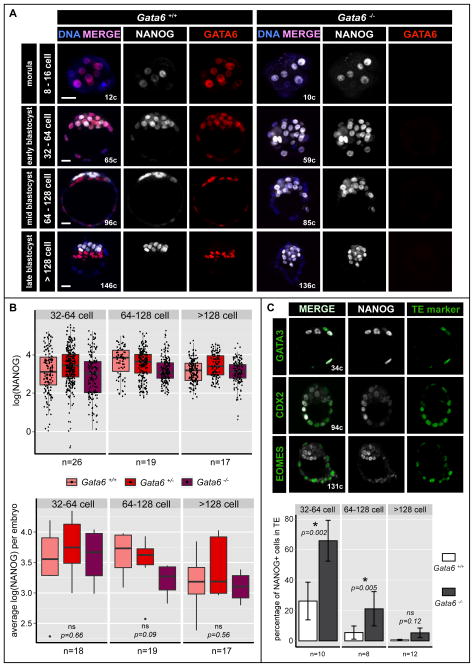
Figure 2. NANOG is expressed ectopically in Gata6 mutants.
A Localization and distribution of NANOG (white) and GATA6 (red) in wild-type (Gata6+/+) and Gata6 mutant mouse embryos from the morula to late/implanting blastocyst stage. Blue in merge: Hoechst. Scale bars: 20 μm. B Distribution of NANOG protein levels as measures of logarithm transformed fluorescence intensity of each EPI cell (upper panel) and averaged for each embryo (lower panel) in NANOG-positive ICM cells of Gata6+/+, Gata6+/− and Gata6−/− embryos of the stages indicated. ns: not significant. C Localization and distribution of NANOG (white) and TE markers GATA3, CDX2 and EOMES (green) in Gata6 mutant embryos at early, mid and late blastocyst stages (upper panel). Percentage of NANOG-positive cells in the TE of Gata6+/+ and Gata6−/− embryos at early, mid and late blastocyst stages (lower panel). Asterisks indicate statistical significance. ns: not significant.
The early spatial pattern of differentiation of PrE versus EPI precursors is stochastic, while spatial order emerges gradually at later stages
We analyzed the pattern of emergence of cell differentiation to determine the importance of stochastic effects versus cell-cell communication or positional effects. We sought to determine if there are spatial correlations between the fates of cells and/or the expression of NANOG and GATA6. Such correlations would suggest that cell-cell interaction/communication or position within the embryo play a prominent role in early differentiation. Alternatively, a lack of correlation would suggest that stochastic, cell autonomous processes are the main initial determinants of fate choice. To distinguish between these two scenarios, we computed the Pearson’s correlation coefficient as a function of cell-cell distance. The coefficient measures the strength of the relationship between two variables (for example, cell fates). The coefficient can assume values between −1 (a perfect negative relationship) and 1 (a perfect positive relationship). A correlation coefficient of zero implies that the two variables are independent, so that the status of one variable does not inform the status of the other. To compute the correlation coefficient, we binned data based on cell-cell distance choosing bin sizes of about 10 microns, an estimate of cell diameter, ensuring several data points present in each bin.
Cell fates (EPI, PrE, or undifferentiated ICM) were assigned in an automatic unbiased fashion (Figure 1B). We computed the correlation coefficient as a function of distance for differentiated cells (i.e. EPI and PrE), with undifferentiated cells excluded from the calculation. We found that at the earliest stage (~32–64 cells) there was no correlation in the spatial pattern of cell differentiation (Figure 1D), as expected for a random cell autonomous decision (Figure S1A). Consistently, the position of cells that differentiated early was independent of the embryo’s coordinates, for example distance from the blastocyst cavity (Figure S1B). This random pattern of early differentiation likely reflected a lack of spatial correlation in the levels of NANOG and GATA6 at this stage (measured across all cells), and suggested that the earliest stages of cell specification are dominated by cell autonomous stochastic factors. At the salt-and-pepper stage (~64–128 cells), a pattern of correlation began to emerge in both cell fate and NANOG, GATA6 levels, so that adjacent cells had a slightly positive chance of exhibiting the same fate (Figure 1D). This could, in principle, originate from the previous uncorrelated pattern, if cells retained similar expression levels and/or fate upon division. At the last stage analyzed (>128 cells), after cells had sorted, a clear spatial correlation was present between cell fates and the levels of both GATA6 and NANOG (Figure 1D). Importantly, the dependency of the correlation coefficient on cell-cell distance at this stage, was similar to that expected given the geometry of the embryo and sorted cell fates (see the comparison between a “virtual” embryo and our data (Figure S1)). Collectively, these results are consistent with a model in which the earliest steps of cell differentiation are dominated by stochastic fluctuations of NANOG and GATA6, which later resolve into a clear spatial pattern, as a result of cell sorting. Finally, we observed that the correlation in NANOG was slightly lower than for GATA6 (many EPI cells at the >128 cells stage were already down-regulating NANOG expression), probably as a result of its highly transient expression, and its extinguishment coincident with embryo implantation.
Gata6 mutants exhibit pan-ICM NANOG expression
GATA6, like NANOG, is initially expressed by all ICM cells, but thereafter becomes exclusive to the PrE (Plusa et al., 2008). An antagonistic interaction of GATA6 with NANOG has been proposed to be at the core of the GRN regulating PrE versus EPI specification (Frankenberg et al., 2011). Thus, we hypothesized that if GATA6 acts near the top of the hierarchy regulating PrE cell fate specification, its elimination must cause a profound defect in lineage specification within the ICM. We therefore determined the phenotype arising from the complete loss, as well as reduction, of GATA6 in Gata6−/− mutants and Gata6+/− heterozygotes, respectively. By contrast to wild-type stage-matched embryos, Gata6−/− embryos expressed NANOG starting at the 8-cell stage in all cells, and thereafter throughout the ICM (Figure 2A). Notably, GATA6 was not detected at any stage analyzed, indicating that the allele is a protein null, the zygotic ablation of Gata6 produces no protein, and suggesting no maternal mRNA is transcribed and/or translated into detectable protein (Figure 2A). To compound this observation, we analyzed Gata6 maternal zygotic (mz) mutant embryos. These exhibited an equivalent phenotype to Gata6 zygotic mutant embryos, referred to as Gata6−/− throughout the text (Figure S2A).
Since Gata6 mutant embryos have been reported as exhibiting a defect at post-implantation stages, affecting the cardiac mesoderm or visceral endoderm (Koutsourakis et al., 1999; Morrisey et al., 1996), we collected embryos from Gata6+/− intercrosses at E5.5. We failed to recover any Gata6−/− embryos among a total of 42 embryos obtained from 7 litters at E5.5 (Table S1). Immunofluorescence revealed that both wild-type and heterozygous embryos exhibited a normal morphology with a GATA4-positive visceral endoderm layer (Figure S2B). The apparent discrepancy between our observation of a pre-implantation defect, which never gave rise to egg cylinder stage embryos, and previous studies reporting early post-implantation defects could be attributed to allele or strain-specific differences (Koutsourakis et al., 1999; Morrisey et al., 1996).
Cells in Gata6 mutant ICMs do not up-regulate NANOG expression
Previous studies have proposed that GATA6 and NANOG function, at least in part, through mutual-repression (Frankenberg et al., 2011; Singh et al., 2007). Such a mutual antagonism could explain the mutually-exclusive expression of markers, and the salt-and-pepper distribution of cells committed to PrE versus EPI lineages. In accordance with such a model of mutual-repression, the absence of GATA6 could relieve Nanog from GATA6-mediated repression, and result in elevated levels of NANOG. To determine if this is the case in vivo, we quantified protein levels in all cells of wild-type, Gata6+/− and Gata6−/− embryos by measuring fluorescence intensities after immunostaining and confocal imaging using the analysis pipeline we constructed (Figure 1A).
Levels of NANOG protein were not elevated in the ICM cells of Gata6−/− embryos (n=364cells /12embryos), compared to NANOG-positive ICM cells in Gata6+/− or wild-type embryos (n=476cells /25embryos, and n=323cells /17embryos, respectively) (Figure 2B), suggesting that some distinct or additional mechanism must control NANOG levels within the ICM in vivo. Interestingly, we noted that in wild-type embryos, the levels of NANOG exhibited a broad dynamic range in the early blastocyst, but became more homogeneous as development proceeded, accompanying the maturation of the EPI lineage (Figure 2B). By contrast, Gata6−/− embryos exhibited a decreased dynamic range of NANOG levels earlier, which was comparable to that observed in mature EPI cells in wild-type late blastocysts. These data therefore suggest that in the absence of GATA6, all ICM cells prematurely commit to an EPI fate.
GATA6 is involved in NANOG repression in TE cells
In addition to pan-ICM expression of NANOG, we also noted a significantly increased number of NANOG-positive cells in the TE of Gata6−/− embryos compared to wild-type embryos at early and mid blastocyst stages, which appeared to recover by the late blastocyst stage (Figure 2C). These findings suggest that GATA6 functions, at least in part, to actively repress NANOG in TE cells. TE cells ectopically expressing NANOG also co-expressed TE markers such as CDX2, GATA3 and EOMES (Figure 2C). These TE cells appeared morphologically normal and blastocyst outgrowths of Gata6−/− embryos showed expansion of the TE and differentiation of trophoblast giant cells (Figure S2C). Additionally, while post-implantation Gata6−/− embryos could not be recovered, we noted Mendelian ratios of empty implantation sites in uteri dissected after E5.5. Collectively these data suggest that, despite the ectopic expression of NANOG, the TE is functional in Gata6−/− embryos.
The PrE lineage is not specified in the absence of GATA6
PrE formation is accompanied by the sequential activation of a number of factors regulating cell fate specification, proliferation and polarity, such as PDGFRA, SOX17, and GATA4, among others (Artus et al., 2011; Gerbe et al., 2008; Kurimoto et al., 2006; Morris et al., 2010; Niakan et al., 2010; Plusa et al., 2008; Saiz et al., 2013). Having determined that Gata6 mutants exhibit pan-ICM NANOG expression, we wanted to investigate whether the PrE program is activated in the absence of GATA6. To this end we determined the expression of these secondary PrE markers in Gata6+/+, Gata6+/− and Gata6−/− embryos. Embryos lacking GATA6 failed to activate Pdgfra expression, as assessed in Gata6−/−; PdgfraH2B-GFP/+ embryos (Video S1), and exhibited no SOX17 (n=13 embryos) or GATA4 (n= 4 embryos) expression at either mid (64–100 cells) or late blastocyst (>128 cells) stages (Figure 3A).
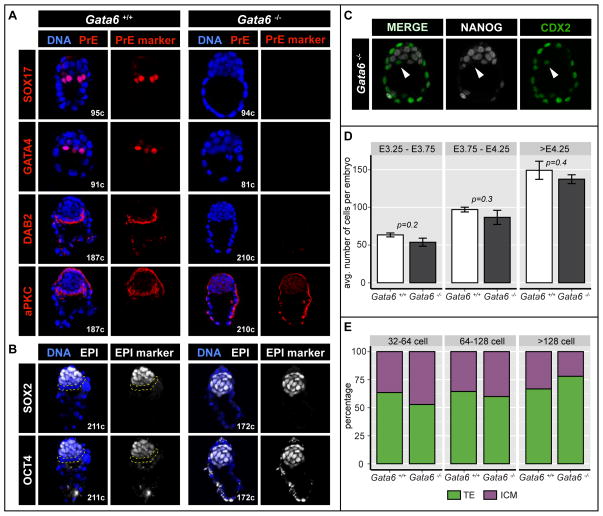
Figure 3. PrE cannot be specified in the absence of GATA6.
A Localization and distribution of PrE markers SOX17, GATA4 and epithelial markers DAB2 and aPKC (red) in wild-type (Gata6+/+) and Gata6−/− embryos at the 64-128-cell and the >128-cell stage respectively. Blue in merge: Hoechst. B Localization and distribution of EPI markers OCT4 and SOX2 (white) in wild-type (Gata6+/+) and Gata6−/− embryos at the >128-cell stage. OCT4 staining in TE cells is cytoplasmic. Dashed, yellow lines indicate PrE layer. Blue in merge: Hoechst. C Localization and distribution of NANOG (white) and CDX2 (green) in Gata6 mutant mouse embryos at the 64-128-cell stage. Arrowheads indicate CDX2 expressing cells in the ICM. D Average number of total cells in Gata6+/+ and Gata6−/− embryos at early, mid and late blastocyst stages. Error bars: standard error. E Cell composition in Gata6+/+, Gata6+/− and Gata6−/− embryos at stages indicated. Cell numbers are shown as percentages of the total number of cells per embryo. TE: trophectoderm; ICM: inner cell mass.
At the late blastocyst stage PrE cells polarize and form an epithelium (Smyth et al., 1999). Markers of apicobasal polarity, including aPKC and DAB2, become localized to the apical surface of PrE cells, revealing their progressive epithelial maturation (Gerbe et al., 2008; Saiz et al., 2013). By contrast to wild-type embryos, we found that Gata6 mutants failed to apically express both aPKC and DAB2 (Figure 3A), indicating that their ICM cells not only failed to up-regulate PrE markers, but also were unable to form an epithelium, one of the defining features of the PrE (Saiz et al., 2013).
Since all ICM cells in Gata6−/− embryos expressed NANOG, we assessed whether ICM cells in these embryos adopted an EPI identity in the absence of GATA6. We found the pluripotency-associated factors SOX2 and OCT4 were expressed in all ICM cells in Gata6−/− embryos at all blastocyst stages, whereas they were restricted to the EPI in control Gata6+/+ or Gata6+/− late blastocysts (Figure 3B), thus suggesting all ICM cells had acquired EPI identity in the absence of GATA6.
Of note, in 9 out of 20 Gata6−/− blastocysts analyzed, we identified between 1 and 3 cells on the surface of the ICM, out of a total of between 15 and 40 cells, that did not express NANOG, and thus were GATA6/NANOG double-negative. Surprisingly, these cells expressed the TE marker CDX2 (Figure 3C), which could suggest that in the absence of both NANOG and GATA6, ICM cells might acquire an alternative cell fate resulting in their up-regulation of TE markers such as CDX2, if positioned adjacent to the blastocyst cavity. However when embryos were allowed to develop in vitro, this cell population did not expand over time, suggesting that CDX2-positive GATA6/NANOG-negative cells were either transient, or the result of delayed CDX2 down-regulation in some ICM cells, however the latter might also be expected to be CDX2/GATA6 and/or NANOG-positive.
One mechanism to ensure balanced lineage representation and proper allocation of progenitors within the ICM of the blastocyst is apoptosis (Plusa, 2008; Artus 2013). To determine whether the lack of PrE cells was due to selective apoptosis at an early stage or to a cell fate switch, we calculated the average total cell number per embryo, and the relative distribution of cells to TE and ICM compartments. We noted that neither embryo size nor relative ICM size differed significantly between wild-type (n=17) and Gata6−/− embryos (n=12) at any stage (Figure 3 D, E), suggesting there was no increase in cell death in the absence of GATA6.
Collectively, our data demonstrate that, in the absence of GATA6, ICM cells failed to adopt a PrE fate, and a PrE epithelial layer was altogether absent. Absence of GATA6 results in absence of the PrE lineage coupled to acquisition of EPI fate.
GATA6 levels influence both the timing and frequency of PrE cell specification
Since GATA6 is necessary for PrE specification, we wanted to explore whether the amount of GATA6 protein present in individual ICM cells has any influence on their fate choice. To address this question, we quantified PrE and EPI cell numbers as well as GATA6 expression levels in wild-type and Gata6+/− embryos. Quantification of fluorescence intensity levels revealed a significant reduction in GATA6 protein in Gata6+/− PrE cells (n=444cells /25embryos) compared to wild-type cells (n=470cells /17embryos) (Figure 4A).
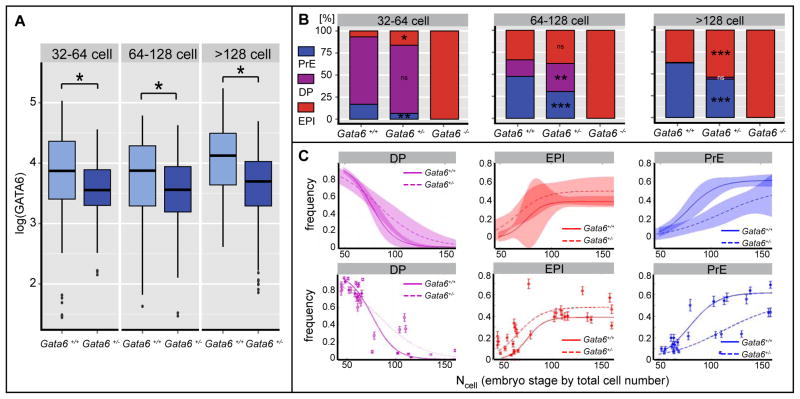
Figure 4. Protein level analysis reveals dosage effect in Gata6+/− embryos.
A GATA6 protein levels as measures of logarithmically transformed fluorescence intensity in GATA6-positive ICM cells of Gata6+/+ and Gata6+/− embryos at indicated stages. B ICM composition in Gata6+/+, Gata6+/− and Gata6−/− embryos. Cell numbers are shown as percentages of total ICM number. PrE, primitive endoderm (GATA6); DP, double positive (NANOG, GATA6) EPI, epiblast (NANOG). Asterisks indicate statistical significance. C Frequency of cell fate specification as a function of cell number for undifferentiated ICM (DP) cells, EPI precursors and PrE precusors. Probability functions were estimated by logistic regression. Shaded areas show 95% confidence intervals (upper panel). Data points were fitted using logistic regression. Error bars represent standard deviation estimated using binomial distribution. p-values were computed using χ2 test. PrE induction is slower (p-value<10-5), loss of ICM is also slower (p-value<10-4), while induction of EPI is faster (p-value=0.01).
Interestingly, Gata6+/− embryos displayed co-expression of GATA6 and NANOG within a subpopulation of ICM cells until later stages than wild-type embryos (Figure 4B), suggesting that cell fate specification is delayed or less abrupt when GATA6 levels are reduced. To distinguish between these two scenarios, and to further characterize the effects of reduced levels of GATA6 on the dynamics of cell fate specification, we quantified the frequency of PrE and EPI cells as a function of cell number. This analysis revealed that in wild-type embryos the specification of both EPI and PrE cells occurs over a short period of time (until ~100 cell stage).. In Gata6+/− embryos, specification of PrE cells was significantly retarded, indicating that wild-type GATA6 levels are required for rapid commitment to PrE cell fate (Figure 4C). Interestingly, specification of the EPI exhibited a similar abruptness, but it occurred at a slightly earlier time (Figure 4C). As a consequence of the slow commitment of PrE cells, undifferentiated ICM cells were observed at later stages than in wild-type embryos (Figures 4B, 4C). Furthermore, the number and proportion of PrE cells in Gata6+/− embryos was significantly reduced (n=25) compared to wild-type embryos (n=17), while the number of EPI cells was increased (Figure 4B). Collectively, these data indicate that GATA6 protein concentration is important for PrE fate specification, and that a reduction leads to a deceleration in the rate of PrE fate specification, as well as a reduction in the number of cells adopting a PrE fate within the ICM. Since Gata6+/− animals were viable, this reduction in the number of PrE cells did not appear to affect the function of the PrE lineage. It is possible that PrE cell numbers either recovered at post-implantation stages, or that the PrE might tolerate cell number variability.
FGF signaling is not sufficient to direct PrE specification in the absence of GATA6
There is increasing evidence that differential activation of the FGF/MAPK pathway in individual ICM cells determines EPI or PrE cell fate choice in mice (Chazaud et al., 2006; Nichols et al., 2009; Yamanaka et al., 2010). Our experiments have revealed a failure in PrE cell specification in Gata6−/− embryos, a phenotype which resembles that observed in mutants in which FGF signaling components have been perturbed (Chazaud et al., 2006; Kang et al., 2013a; Krawchuk et al., 2013), or in wild-type embryos treated with inhibitors of FGF signaling (Nichols et al., 2009; Yamanaka et al., 2010). In our previous analysis of Fgf4 mutant embryos, we noted that GATA6 expression was initiated at the early blastocyst stage, but failed to be maintained in the absence of FGF4, resulting in a lack of PrE cells (Kang et al., 2013a).
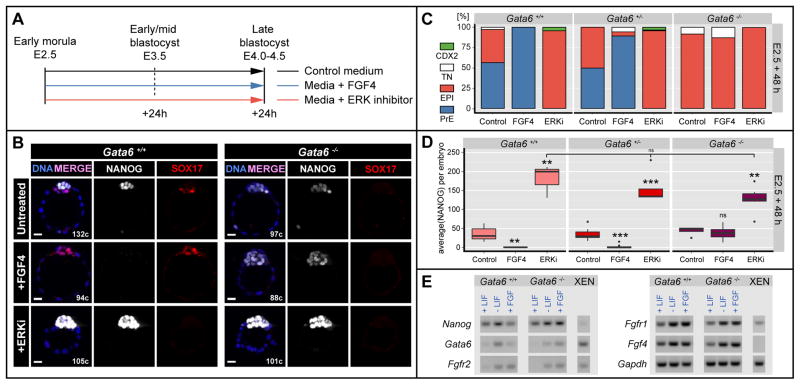
To determine the relative positions of FGF signaling and GATA6 in the hierarchy governing PrE specification, we treated Gata6−/− embryos with saturating doses of FGF, which have been previously shown to induce a PrE identity throughout the ICM in wild-type embryos (Yamanaka et al., 2010), and Fgf4 mutants (Kang et al., 2013a). Since timed treatments have revealed variations in the sensitivity to FGF signaling through pre-implantation development (Frankenberg et al., 2011; Grabarek et al., 2012; Yamanaka et al., 2010), we used two regimes in a parallel set of experiments (Figures 5A and 6A). First, we applied exogenous FGF for 48h starting at E2.5 (8–16 cell stage), prior to the initiation of any differentiation, when NANOG and GATA6 are co-expressed (Figure 5A). Gata6−/− embryos failed to respond to doses of between 500 and 1000 ng/ml of either FGF4 (Figure 5A, B) or FGF2 (Figure S3). They maintained NANOG expression throughout the ICM, with levels comparable to those of untreated embryos (Figure 5B, D). By contrast, wild-type embryos displayed pan-ICM GATA6 or SOX17 expression, and complete absence of NANOG expression (Figure 5B–D), as previously described (Saiz et al., 2013; Yamanaka et al., 2010). Gata6+/− embryos exhibited a comparable response to wild-type embryos, although some cells expressing NANOG could be identified (Figure 5C), suggesting that the kinetics of the effect produced upon FGF treatment were altered by a reduction in the dose of GATA6. FGF treatment also failed to induce expression of later PrE markers, such as SOX17, in Gata6−/− embryos (Figure 5B).
Figure 5. FGF4 is not sufficient to direct PrE specification in the absence of GATA6, while ERK inhibition results in NANOG upregulation.
A Experimental timeline. B Representative immunofluorescence images of Gata6+/+ and Gata6−/− embryos cultured from the morula (E2.5) stage for 48h in culture medium alone (Untreated), medium + 1μg/ml FGF4 + 1μg/ml Heparin (+FGF4) or medium + 1μM PD0325901 (ERKi). Blue: Hoechst; white: NANOG; red: SOX17. C ICM composition in Gata6+/+, Gata6+/− and Gata6−/− embryos after culture as in (A). Cell numbers are shown as percentages of the total ICM number. TN: Triple negative for NANOG, PrE marker and CDX2; PrE: primitive endoderm; EPI: epiblast. D Fluorescence levels of NANOG as measures of average per embryo in Gata6+/+, Gata6+/− and Gata6−/− ICMs after culture as in (A). Asterisks indicate statistical significance. ns: not significant. E RT-PCR on Gata6+/+ and Gata6−/− ES cells after 48 h culture in standard LIF containing medium (+LIF), after LIF withdrawal and in the presence of 250 ng/mL FGF.
Figure 6. Changes in FGF signaling cannot induce cell fate switch after PrE and EPI specification.
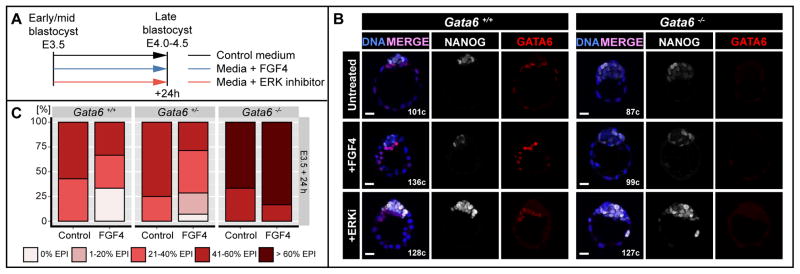
A Experimental timeline. B Representative immunofluorescence images of Gata6+/+ and Gata6−/− embryos cultured from the salt-and-pepper (E3.5) stage for 24h in medium without (untreated) or with 500ng/ml FGF4 + 1μg/ml Heparin (+FGF4) or medium + 1μM PD0325901 (ERKi). Blue: Hoechst; white: NANOG; red: GATA6. C Percentages of embryos falling into 1 of five FGF-response categories: 0% EPI, 1–20% EPI, 21–40% EPI, 41–60% EPI and >60% EPI in ICMs of embryos treated as in (A). EPI: Epiblast.
Collectively these data lead us to hypothesize that while FGF4 acts as an instructive signal to activate the PrE program, GATA6 is necessary to execute it. Thus, in the absence of GATA6, FGF treatment of embryos fails to activate the PrE program. Alternatively, Gata6−/− embryos may exhibit aberrant expression of FGF receptors on their ICM cells, and fail to mount a response to an FGF signal.
ERK inhibition promotes NANOG upregulation in the absence of GATA6
Upon FGF/ERK stimulation of wild-type E2.5 embryos NANOG is repressed, and GATA6 is activated in all ICM cells (Yamanaka et al., 2010). If GATA6 were the factor directly mediating NANOG repression upon FGF/ERK stimulation, we would hypothesize that inhibition of the pathway in Gata6−/− embryos would have no effect on NANOG expression compared to untreated, Gata6−/− embryos. Culture in the presence of 1μM of the ERK1/2 inhibitor PD0325901 (Nichols et al., 2009) resulted in a marked up-regulation of NANOG throughout the ICM in all embryos, regardless of their genotype (Figure 5D). These data indicate that, whereas GATA6 is necessary for NANOG down-regulation, an additional ERK1/2-mediated mechanism operates to maintain NANOG levels within EPI cells.
Collectively, these results demonstrate that GATA6 is necessary to cell-autonomously mediate FGF/ERK signaling upstream of ERK1/2, which in turn mediates NANOG repression. Furthermore, the effects of ERK inhibition reveal a distinct role for FGF signaling in EPI cells, potentially acting to inhibit levels of NANOG so as to maintain them within a physiological range.
Modulation of FGF signaling cannot induce an ICM fate switch after PrE and EPI specification
Next, we cultured embryos in the presence of FGF4 for 24 hours from E3.5 (early-to-mid blastocyst stage), when PrE and EPI specification has already begun (Figure 6A). By contrast to FGF4 treatment experiments from E2.5, exposure to 500ng/ml of FGF4 from E3.5 did not result in pan-ICM expression of GATA6 in either wild-type or Gata6+/− embryos (Figure 6B, C). Although we did observe a moderate expansion of the PrE compartment, most wild-type embryos retained a NANOG-positive population (Figure 6C). Similarly, Gata6−/− embryos exhibited no reduction in the size of the NANOG-positive compartment upon FGF4 treatment (Figure 6B, C). We observed a proportion of ICM cells that expressed neither NANOG, nor PrE markers (GATA6 or SOX17) regardless of their genotype (TN). This population, albeit larger in Gata6+/− and Gata6−/− embryos, did not change significantly upon FGF treatment, suggesting it resulted from developmental down-regulation of NANOG as the EPI matures, rather than a consequence of FGF treatment.
In agreement with these results, we observed that inhibition of ERK1/2 at E3.5 with 1μM of PD0325901 did not result in down-regulation of GATA6 in wild-type or Gata6+/− embryos, as previously described (Nichols et al., 2009). On the other hand, consistent with our previous observations from incubations initiated at E2.5, ERK1/2 inhibition resulted in up-regulation of NANOG throughout the EPI regardless of embryo genotype (Figure 6B).
These results suggest that de-sensitizing cells to FGF may be a mechanism operating to prevent changes in the ICM lineage composition upon sustained FGF production. Such a model is supported by the fact that XEN stem cells, which represent the primitive endoderm lineage, can be derived from Fgf4 mutants, demonstrating a temporally restricted requirement for FGF/ERK signaling within the PrE lineage (Kang et al., 2013b).
The induction of Gata6 expression is independent of NANOG repression
It has been proposed that NANOG directly represses Gata6 (Mitsui et al., 2003; Singh et al., 2007) and that down-regulation of NANOG through FGF signaling alleviates Gata6 suppression, leading to its up-regulation and differentiation of cells towards a PrE fate. It has been shown in ES cells that activation of FGF/Erk signaling directly represses Nanog transcription (Santostefano et al., 2012). To test if this down-regulation of NANOG leads to an up-regulation of Gata6 or if FGF signaling can directly influence Gata6 expression, we derived Gata6+/+ and Gata6−/− ES cells (Czechanski et al., 2014) and analyzed gene expression after culture in the presence of FGF. While undifferentiated Gata6+/+ and Gata6−/− ES cells cultured in LIF exhibited little or no expression of Gata6, Gata6 transcripts were detected in Gata6+/+ and Gata6−/− ES cells maintained in the presence of FGF (Figure 5E). As a control, Gata6+/+ and Gata6−/− ES cells were also allowed to differentiate in the absence of LIF and without addition of FGF. These cells exhibited an up-regulation of Gata6 comparably to cells treated with FGF. These data suggest that Gata6 is activated upon differentiation of ES cells in a GATA6-independent manner. Additionally, the absence of a down-regulation of Nanog in the absence of GATA6 (Figure 5D–E) suggests that, if FGF signaling is involved in the up-regulation of Gata6, it does so directly and independently of NANOG repression. However, since Gata6 was detected both in the presence and absence of FGF, we were unable to exclude the possibility that this repression occurred independently of FGF signaling.
DISCUSSION
With the aim of obtaining unbiased quantitative information on individual cells to understand how they direct the behavior of a population within early mammalian embryos, we developed a pipeline involving single-cell resolution, quantitative immunofluorescence image analysis, coupled with cartesian coordinate information of cellular position. This allowed us to account for how the salt-and-pepper distribution of emergent PrE and EPI lineage progenitors arises within the ICM of the mouse blastocyst. The method we developed extends previous findings using single-cell microarray-based expression analyses (Ohnishi et al., 2014b), as it also incorporates positional information for individual cells. Our results suggest that the early pattern of cell differentiation is purely stochastic, and might be dominated by cell autonomous noisy expression of NANOG and GATA6. Following this initial phase, order starts to emerge, most likely as the result of cell division, cell motility and the acquisition of apicobasal polarity (Gerbe et al., 2008; Meilhac et al., 2009; Saiz et al., 2013), as well as apoptosis of misplaced cells (Artus et al., 2013; Plusa et al., 2008). In future studies, it will be important to determine if the early stochastic differentiation is propagated to later stages, or if corrective feedback mechanisms operate to ensure that the embryo ends up with the correct number of EPI and PrE progenitors, despite an early phase of stochastic differentiation.
The distribution of lineage progenitors is reflected by the expression of the key lineage-specific transcription factors GATA6 and NANOG. Since it is the earliest expressed PrE factor we reasoned that GATA6 might be responsible for directing ICM cell fate choice. To gain insight into the mechanisms driving cell fate decision away from pluripotency, we analyzed embryos exhibiting a reduction (heterozygotes) or absence (null mutants) of Gata6. Our studies reveal a strict GATA6 dose-dependence in both the rate and probability of PrE specification within the ICM, which is manifest as a reduction in PrE cells in Gata6 heterozygotes, and an absence of a PrE lineage in Gata6 mutants. The phenotype observed in Gata6 mutants appears more specific, and more severe, with respect to the ICM than mutants in FGF signaling components, such as Fgf4, which also exhibit a lack of PrE (Kang et al., 2013a). By contrast to Gata6 mutants, which fail to activate any PrE markers, Fgf4 mutants initially express, but fail to maintain GATA6, as well as later PrE-specific factors, such as PDGFRA (Artus et al., 2010; Kang et al., 2013a; Plusa et al., 2008). Furthermore, and unlike Gata6 mutants, Fgf4 mutant embryos exhibit a reduction in embryo cell number, as well as defects in the TE, due to a mitogenic role of FGF/ERK signaling in this lineage (Kang et al., 2013a). While Gata6 mutants display ectopic expression of NANOG in TE cells, this did not appear to impair TE function. Rather, it suggests that GATA6 may be involved in the repression of NANOG in these cells. Interestingly, the mechanism by which this happens appears to be distinct from that in ICM cells, since Fgf4 mutant embryos exhibit expression of NANOG in the TE (Kang et al., 2013a), suggesting this process is independent of FGF signaling.
All cells within Gata6 mutant ICMs express the pluripotency-associated factors NANOG, SOX2 and OCT4 at all blastocyst stages. Indeed, NANOG has been proposed to interact with GATA6 in a mutually repressive manner (Frankenberg et al., 2013). In the absence of GATA6-mediated repression, one might predict an increase in NANOG levels. However, NANOG levels were not observed to increase in Gata6 mutant embryos, suggesting that GATA6 itself may be regulating the on/off switch of NANOG, rather than fine-tuning its expression. On the other hand, NANOG expression was decreased and its range narrowed earlier in Gata6−/− than in Gata6+/+ ICM cells (64–128 cell stage, compared to >128 cell stage). NANOG levels therefore appeared more comparable to mature EPI cells observed in late blastocyst stage wild-type embryos. We speculate that the absence of GATA6 does not necessarily result in a novel ICM cell type; rather, cells maintain NANOG levels comparable to those of mature EPI cells of wild-type embryos, where GATA6 is repressed without a concomitant increase in NANOG levels. One could argue that cells in Gata6−/− early ICMs are less plastic than cells in stage-matched wild-type ICMs, and so may already be committed to an EPI fate. Relative, rather than absolute, protein concentrations in any given cell might ensure this, and one might envisage a scenario in which GATA6 and NANOG, acting through mutual-repression, maintain an equilibrium that restrains cells from committing to either EPI or PrE fate until an appropriate time. At some point the balance is stochastically disturbed, disequilibrium ensues, and one of these two factors predominates. This event drives the onset of cell lineage choice and commitment to PrE or EPI fates. In Gata6−/− embryos this initial balance of transcription factors is never established, and consequently all ICM cells prematurely differentiate to a mature EPI state.
In heterozygous embryos GATA6 protein levels were reduced compared to stage-matched wild-type embryos. Indeed, Gata6+/− embryos exhibited a significantly lower number of PrE cells and a slower commitment so that at later stages more GATA6/NANOG double positive (DP) ICM cells were observed in an uncommitted (NANOG+; GATA6+) state. The commitment to an EPI fate appeared slightly faster, although more data are required to confirm this hypothesis.
Given these data, the GRN driving ICM lineage choice can be subject to geometric reasoning (Corson and Siggia, 2012). Our observations are compatible with a simple geometric model of cell differentiation (Waddington landscape). We propose that GATA6 and NANOG mutual-repression generates a system with two stable states (valleys): NANOG-positive; GATA6-negative and NANOG-negative; GATA6-positive. Stochastic fluctuations in the levels of GATA6 and NANOG push the cells toward one or the other fate. The time required to reach a stable state is controlled by how much the relative levels of the two factors deviate from the unstable region in which cells remain DP (double positive, Figure 1B) ICM (the peak in the landscape). In the absence of GATA6, the landscape changes, so that the only stable state is the NANOG-positive; GATA6-negative state, which should, therefore, display similar levels of NANOG to wild-type EPI cells. In Gata6+/− embryos, levels of GATA6 are reduced and, therefore, a higher number of cells end up in the EPI state, as a result of the relatively higher levels of NANOG compared to GATA6. In cells in which levels of GATA6 are still sufficient to drive the PrE fate, the GATA6:NANOG ratio should be reduced, and, therefore, result in a slower transition toward the PrE fate. The model predicts that the opposite would be observed in Nanog+/− embryos. Additional experiments are required to test this model, and determine the importance of relative versus absolute levels of GATA6 and NANOG for ICM cell differentiation.
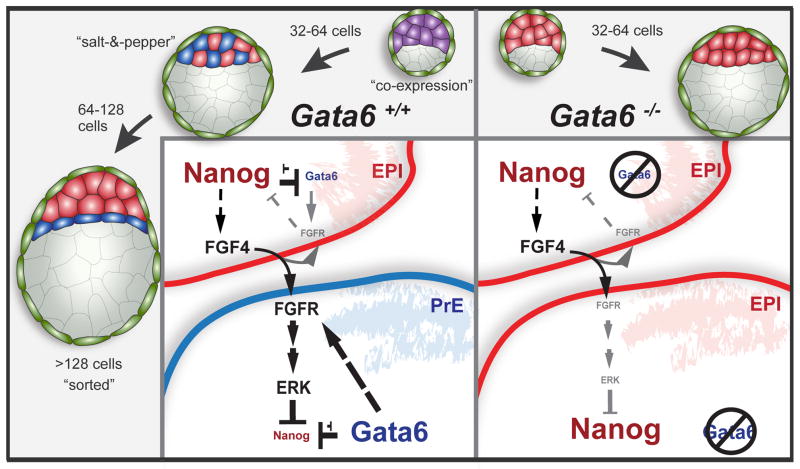
How the transcriptional network involving this interaction of GATA6 and NANOG is integrated with signaling cues to regulate ICM cell fate specification remains an open question. Modulation of FGF signaling, in mutants or using inhibitors or growth factors, affects NANOG and GATA6 expression in wild-type blastocysts (Frankenberg et al., 2011; Kang et al., 2013a; Nichols et al., 2009; Yamanaka et al., 2010). However, treatment of Gata6 mutants with exogenous FGF4 failed to direct cells towards a PrE fate. These results show that GATA6 is required for FGF mediated down-regulation of NANOG. However, inhibition of ERK in Gata6 mutants resulted in robust up-regulation of NANOG, suggesting that FGF/ERK signaling can directly regulate NANOG levels. These results reveal that FGF signaling is necessary but not, as previously assumed, sufficient to repress NANOG and induce PrE differentiation. Instead this process is GATA6-dependent, with GATA6 acting upstream of ERK. We suggest a tunable system with three nodes, consisting of GATA6, FGFR and NANOG, in which GATA6 is up-regulating and possibly mediating stabilization of FGFR (or another upstream element in the cascade), thereby cell-autonomously allowing for elevated stimulation or attenuation of the FGF/ERK pathway by FGF4. Increased ERK signaling would then be able to repress NANOG, which in turn would lead to decreased GATA6 repression by NANOG (Figure 7). Through this extended feedback loop a PrE fate would be reinforced. In EPI cells, very low levels of GATA6 would be insufficient to up-regulate FGFR, and baseline levels of the receptor would only allow for marginal induction of FGF/ERK signaling, which would not be able to repress, but instead merely moderate NANOG levels, as observed in wild-type EPI cells (Figure 7). Single-cell gene expression profiling demonstrated expression of Fgfr1 in all ICM cells, and notably a correlation of Gata6 and Fgfr2 mRNA levels in blastocysts (Ohnishi et al., 2014a). Further studies will be needed to determine if there is a direct link between GATA6 and FGFR expression, but such a mechanism could produce a general effect on RTKs, as our results also demonstrate the absence of the related RTK PDGFRA in the absence of GATA6.
Figure 7. Working model.
GATA6 potentiates the upregulation of FGFR2, thereby cell-autonomously stimulating the FGF/ERK pathway, activated by FGF4. Increased ERK signaling leads to repression of NANOG, which in turn releases GATA6 repression. This feedback loop reinforces a PrE fate. In EPI cells, very low levels of GATA6 maintain but do not up-regulate FGFR2. Baseline levels of the receptor allows for marginal induction of FGF/ERK signaling, which in turn moderate NANOG levels.
Unexpectedly, we also noted that, independently of genotype, stimulation of the FGF/ERK pathway at blastocyst stages failed to induce a PrE fate in cells that had presumably already acquired an EPI state. These results suggest that a subpopulation of EPI cells may be lineage committed soon after the establishment of a salt-and-pepper distribution of cells within the ICM, and less responsive to FGF signaling cues, in agreement with previous reports (Grabarek et al., 2012). Since FGF signaling fails to repress NANOG in the absence of GATA6 (Figure 5), this phenomenon can be explained if in mature EPI cells GATA6 is repressed and they concomitantly cease to respond to FGF signaling. Such a mechanism could operate to prevent committed EPI cells from switching their fate in the FGF-rich environment of the sorted EPI compartment of the late blastocyst. However, additional experiments will be required to elucidate the mechanistic details behind these observations.
Importantly, recent studies in other mammals (Kuijk et al., 2012; Niakan and Eggan, 2013; Roode et al., 2012; Van der Jeught et al., 2013), have revealed that GATA6 and NANOG are expressed early within the ICM and exhibit a sequence of expression that is comparable to that of the mouse (Chazaud et al., 2006; Dietrich and Hiiragi, 2007; Plusa et al., 2008), suggesting that some aspects of the GRN driving ICM lineage choice are evolutionarily conserved. Surprisingly, it was recently noted that by contrast to rodents (Kang et al., 2013a; Nichols et al., 2009; Roode et al., 2012; Yamanaka et al., 2010), modulation of FGF signaling does not elicit an effect on ICM lineage commitment in human and bovine blastocysts (Kuijk et al., 2012; Roode et al., 2012; Van der Jeught et al., 2013). If the role of FGF signaling in ICM lineage specification is a rodent species-specific adaptation, our studies lead us to speculate that GATA6 might be a key evolutionarily conserved transcription factor driving PrE lineage specification across eutherian mammals.
EXPERIMENTAL PROCEDURES
Mouse husbandry
Mouse strains used were: Gata6cKO/cKO (Sodhi et al., 2006), CAG:Cre (Sakai and Miyazaki, 1997), Zp3:Cre (Lewandoski et al., 1997), PdgfraH2B–GFP/+ (Hamilton et al., 2003) and wild-type CD1 (Taconic). Gata6KO/+ animals were of a mixed (CD1/B6/129) strain background and were generated by crossing of Gata6cKO/cKO to a CAG:Cre strain. Embryos with maternal and zygotic ablation of Gata6 were obtained by breeding Gata6cKO/cKO; Zp3:CreTg/+ males with Gata6cKO/cKO females.
Embryo collection and handling
Mice were maintained under a 12 hour light cycle. Pre-implantation embryos were flushed from uteri or oviducts in FHM (Millipore) as described (Nagy et al., 2003). The zona pellucida was removed from blastocysts by brief incubation in acid Tyrode’s solution (Sigma). Embryos were fixed in 4% paraformaldehyde (PFA) in PBS for 10 min at room temperature and stored in PBS at 4°C.
Embryo in vitro culture
Embryos were cultured in drops of KSOM-AA (Millipore) under mineral oil (Sigma) at 37°C in a 5% CO2 atmosphere. For live imaging, embryos were cultured in glass-bottom dishes (MatTek) in an environmental chamber (Solent Scientific) as described previously (Piliszek et al., 2011). Embryos at post-cavitation stages were cultured without zona pellucidae. For FGF treatment experiments, FGF4 or FGF2 (R&D Systems) were added to culture medium at 500–2000 ng/ml, in the presence of 1 μg/ml heparin (Sigma). For ERK inhibition experiments 1μM of the ERK1/2 inhibitor PD0325901 (Stemgen) was added to the culture. Embryos were cultured from E2.5 or E3.5 various periods of time.
Immunostaining
Immunofluorescent staining was performed as previously described (Frankenberg et al., 2013). Details of antibodies used are provided in the Supplementary Experimental Procedures. The following primary antibodies were used: goat anti-SOX2 (R&D Systems) at a dilution of 1/50; mouse anti-OCT4 (Santa Cruz), goat anti-GATA4 (Santa Cruz), goat anti-GATA6 (R&D Systems) and goat anti-SOX17 (R&D Systems) at 1/100; rabbit anti-PKCζ (Santa Cruz), mouse anti-DAB2 (BD Transduction Laboratories) at 1/300 and mouse anti-CDX2 (BioGenex) at 1/200; rabbit anti-NANOG (CosmoBio), Mouse anti-GATA3 (Biolegend) and rabbit anti-EOMES (Abcam) at 1/500. Secondary Alexa Fluor-conjugated antibodies (Invitrogen) were used at a dilution of 1/500. DNA was visualized using 5 μg/ml Hoechst 33342 (Invitrogen) in PBS.
Image data acquisition and processing
Laser scanning confocal images were acquired on a Zeiss LSM 510 METALSM510META. Embryos were mounted in PBS on glass-bottom dishes. Fluorescence was excited with a 405-nm laser diode (Hoechst), a 488-nm Argon laser (GFP, Alexa Fluor 488), a 543-nm HeNe laser (Alexa Fluor 543, 555) and a 633-nm HeNe laser (Alexa Fluor 633 and 647). Images were acquired using a Plan-Neofluar 40×/1.3 Oil DIC objective, with an optical section thickness of 1–1.2 μm. Raw data were processed using ZEN (Carl Zeiss Microsystems), ImageJ (NIH) and MINS (Lou et al., 2014) software. 3D time-lapse imaging was performed on a Zeiss LSM 510 META. Embryos were placed in drops of KSOM under mineral oil on glass-bottom dishes (MatTek Corp). Time intervals between z-stacks were 15 minutes, for 15–20 hours total. Raw data were processed using Zeiss ZEN software (Carl Zeiss Microsystems).
Image segmentation and quantitative fluorescence intensity analysis pipeline
For cell counting and quantification MINS software (http://katlab-tools.org/) was used (Lou et al., 2014). Details of the analyses are provided in the Supplementary Experimental Procedures.
Supplementary Material
HIGHLIGHTS.
Early fate selection within the inner cell mass appears random and noisy
The GATA6 transcription factor is essential for primitive endoderm (PrE) formation
GATA6 levels control the speed and proportion of PrE (vs. epiblast) fate commitment
GATA6 governs PrE cell fate choice by mediating the response to FGF signaling
Acknowledgments
We thank S. Duncan and P. Soriano for Gata6cKO and PdgfraH2B-GFP mouse lines respectively; V. Seshan of the MSKCC Biostatistics Core Facility for advice on fluorescence intensity data transformations; M. Kang, J. Nichols, S. Nowotschin, B. Plusa, C. Schroeter and P. Xenopoulos for discussions and comments on the manuscript. Our work is supported by NIH-RO1-HD052115, NIH-RO1-DK084391 and NYSTEM (AKH), and NIH-K99-HD074670 (SDT).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Artus J, Kang M, Cohen-Tannoudji M, Hadjantonakis AK. PDGF signaling is required for primitive endoderm cell survival in the inner cell mass of the mouse blastocyst. Stem Cells. 2013;31:1932–1941. doi: 10.1002/stem.1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artus J, Panthier JJ, Hadjantonakis AK. A role for PDGF signaling in expansion of the extra-embryonic endoderm lineage of the mouse blastocyst. Development. 2010;137:3361–3372. doi: 10.1242/dev.050864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artus J, Piliszek A, Hadjantonakis AK. The primitive endoderm lineage of the mouse blastocyst: sequential transcription factor activation and regulation of differentiation by Sox17. Developmental biology. 2011;350:393–404. doi: 10.1016/j.ydbio.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chazaud C, Yamanaka Y, Pawson T, Rossant J. Early lineage segregation between epiblast and primitive endoderm in mouse blastocysts through the Grb2-MAPK pathway. Developmental cell. 2006;10:615–624. doi: 10.1016/j.devcel.2006.02.020. [DOI] [PubMed] [Google Scholar]
- Corson F, Siggia ED. Geometry, epistasis, and developmental patterning. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:5568–5575. doi: 10.1073/pnas.1201505109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechanski A, Byers C, Greenstein I, Schrode N, Donahue LR, Hadjantonakis AK, Reinholdt LG. Derivation and characterization of mouse embryonic stem cells from permissive and nonpermissive strains. Nature protocols. 2014;9:559–574. doi: 10.1038/nprot.2014.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich JE, Hiiragi T. Stochastic patterning in the mouse pre-implantation embryo. Development. 2007;134:4219–4231. doi: 10.1242/dev.003798. [DOI] [PubMed] [Google Scholar]
- Frankenberg S, Gerbe F, Bessonnard S, Belville C, Pouchin P, Bardot O, Chazaud C. Primitive endoderm differentiates via a three-step mechanism involving Nanog and RTK signaling. Developmental cell. 2011;21:1005–1013. doi: 10.1016/j.devcel.2011.10.019. [DOI] [PubMed] [Google Scholar]
- Frankenberg S, Shaw G, Freyer C, Pask AJ, Renfree MB. Early cell lineage specification in a marsupial: a case for diverse mechanisms among mammals. Development. 2013;140:965–975. doi: 10.1242/dev.091629. [DOI] [PubMed] [Google Scholar]
- Fujikura J, Yamato E, Yonemura S, Hosoda K, Masui S, Nakao K, Miyazaki Ji J, Niwa H. Differentiation of embryonic stem cells is induced by GATA factors. Genes & development. 2002;16:784–789. doi: 10.1101/gad.968802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbe F, Cox B, Rossant J, Chazaud C. Dynamic expression of Lrp2 pathway members reveals progressive epithelial differentiation of primitive endoderm in mouse blastocyst. Developmental biology. 2008;313:594–602. doi: 10.1016/j.ydbio.2007.10.048. [DOI] [PubMed] [Google Scholar]
- Goldin SN, Papaioannou VE. Paracrine action of FGF4 during periimplantation development maintains trophectoderm and primitive endoderm. Genesis. 2003;36:40–47. doi: 10.1002/gene.10192. [DOI] [PubMed] [Google Scholar]
- Grabarek JB, Zyzynska K, Saiz N, Piliszek A, Frankenberg S, Nichols J, Hadjantonakis AK, Plusa B. Differential plasticity of epiblast and primitive endoderm precursors within the ICM of the early mouse embryo. Development. 2012;139:129–139. doi: 10.1242/dev.067702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G, Huss M, Tong GQ, Wang C, Li Sun L, Clarke ND, Robson P. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Developmental cell. 2010;18:675–685. doi: 10.1016/j.devcel.2010.02.012. [DOI] [PubMed] [Google Scholar]
- Hamilton TG, Klinghoffer RA, Corrin PD, Soriano P. Evolutionary divergence of platelet-derived growth factor alpha receptor signaling mechanisms. Molecular and cellular biology. 2003;23:4013–4025. doi: 10.1128/MCB.23.11.4013-4025.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MH, Ziomek CA. Induction of polarity in mouse 8-cell blastomeres: specificity, geometry, and stability. The Journal of cell biology. 1981;91:303–308. doi: 10.1083/jcb.91.1.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang M, Piliszek A, Artus J, Hadjantonakis AK. FGF4 is required for lineage restriction and salt-and-pepper distribution of primitive endoderm factors but not their initial expression in the mouse. Development. 2013a;140:267–279. doi: 10.1242/dev.084996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang M, Xenopoulos P, Munoz-Descalzo S, Lou X, Hadjantonakis AK. Live imaging, identifying, and tracking single cells in complex populations in vivo and ex vivo. Methods Mol Biol. 2013b;1052:109–123. doi: 10.1007/7651_2013_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutsourakis M, Langeveld A, Patient R, Beddington R, Grosveld F. The transcription factor GATA6 is essential for early extraembryonic development. Development. 1999;126:723–732. [PubMed] [Google Scholar]
- Krawchuk D, Honma-Yamanaka N, Anani S, Yamanaka Y. FGF4 is a limiting factor controlling the proportions of primitive endoderm and epiblast in the ICM of the mouse blastocyst. Developmental biology. 2013;384:65–71. doi: 10.1016/j.ydbio.2013.09.023. [DOI] [PubMed] [Google Scholar]
- Kuijk EW, van Tol LT, Van de Velde H, Wubbolts R, Welling M, Geijsen N, Roelen BA. The roles of FGF and MAP kinase signaling in the segregation of the epiblast and hypoblast cell lineages in bovine and human embryos. Development. 2012;139:871–882. doi: 10.1242/dev.071688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurimoto K, Yabuta Y, Ohinata Y, Ono Y, Uno KD, Yamada RG, Ueda HR, Saitou M. An improved single-cell cDNA amplification method for efficient high-density oligonucleotide microarray analysis. Nucleic acids research. 2006;34:e42. doi: 10.1093/nar/gkl050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewandoski M, Wassarman KM, Martin GR. Zp3-cre, a transgenic mouse line for the activation or inactivation of loxP-flanked target genes specifically in the female germ line. Current biology: CB. 1997;7:148–151. doi: 10.1016/s0960-9822(06)00059-5. [DOI] [PubMed] [Google Scholar]
- Lou X, Kang M, Xenopoulos P, Munoz-Descalzo S, Hadjantonakis AK. A Rapid and Efficient 2D/3D Nuclear Segmentation Method for Analysis of Early Mouse Embryo and Stem Cell Image Data. Stem Cell Reports. 2014;2:382–397. doi: 10.1016/j.stemcr.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meilhac SM, Adams RJ, Morris SA, Danckaert A, Le Garrec JF, Zernicka-Goetz M. Active cell movements coupled to positional induction are involved in lineage segregation in the mouse blastocyst. Developmental biology. 2009;331:210–221. doi: 10.1016/j.ydbio.2009.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- Morris SA, Teo RT, Li H, Robson P, Glover DM, Zernicka-Goetz M. Origin and formation of the first two distinct cell types of the inner cell mass in the mouse embryo. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:6364–6369. doi: 10.1073/pnas.0915063107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrisey EE, Ip HS, Lu MM, Parmacek MS. GATA-6: a zinc finger transcription factor that is expressed in multiple cell lineages derived from lateral mesoderm. Developmental biology. 1996;177:309–322. doi: 10.1006/dbio.1996.0165. [DOI] [PubMed] [Google Scholar]
- Nagy A, Gertsenstein M, Vinterstein K, Behringer R. Manipulating the Mouse Embryo. Cold Spring Harbor Press; 2003. [Google Scholar]
- Niakan KK, Eggan K. Analysis of human embryos from zygote to blastocyst reveals distinct gene expression patterns relative to the mouse. Developmental biology. 2013;375:54–64. doi: 10.1016/j.ydbio.2012.12.008. [DOI] [PubMed] [Google Scholar]
- Niakan KK, Ji H, Maehr R, Vokes SA, Rodolfa KT, Sherwood RI, Yamaki M, Dimos JT, Chen AE, Melton DA, et al. Sox17 promotes differentiation in mouse embryonic stem cells by directly regulating extraembryonic gene expression and indirectly antagonizing self-renewal. Genes & development. 2010;24:312–326. doi: 10.1101/gad.1833510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J, Silva J, Roode M, Smith A. Suppression of Erk signalling promotes ground state pluripotency in the mouse embryo. Development. 2009;136:3215–3222. doi: 10.1242/dev.038893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi Y, Huber W, Tsumura A, Kang M, Xenopoulos P, Kurimoto K, Oles AK, Arauzo-Bravo MJ, Saitou M, Hadjantonakis AK, et al. Cell-to-cell expression variability followed by signal reinforcement progressively segregates early mouse lineages. Nature cell biology. 2014a;16:27–37. doi: 10.1038/ncb2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi Y, Huber W, Tsumura Y, Kang M, Xenopoulos P, Kurimoto K, Oles AK, Arauzo-Bravo MJ, Saitou M, Hadjantonakis AK, et al. Cell-to-cell expression variability follwed by signal reinforcement progressively segregates early mouse lineages. Nature Cell Biology. 2014b doi: 10.1038/ncb2881. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piliszek A, Kwon GS, Hadjantonakis AK. Ex utero culture and live imaging of mouse embryos. Methods Mol Biol. 2011;770:243–257. doi: 10.1007/978-1-61779-210-6_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plusa B, Piliszek A, Frankenberg S, Artus J, Hadjantonakis AK. Distinct sequential cell behaviours direct primitive endoderm formation in the mouse blastocyst. Development. 2008;135:3081–3091. doi: 10.1242/dev.021519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roode M, Blair K, Snell P, Elder K, Marchant S, Smith A, Nichols J. Human hypoblast formation is not dependent on FGF signalling. Developmental biology. 2012;361:358–363. doi: 10.1016/j.ydbio.2011.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiz N, Grabarek JB, Sabherwal N, Papalopulu N, Plusa B. Atypical protein kinase C couples cell sorting with primitive endoderm maturation in the mouse blastocyst. Development. 2013;140:4311–4322. doi: 10.1242/dev.093922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiz N, Plusa B. Early cell fate decisions in the mouse embryo. Reproduction. 2013;145:R65–80. doi: 10.1530/REP-12-0381. [DOI] [PubMed] [Google Scholar]
- Sakai K, Miyazaki J. A transgenic mouse line that retains Cre recombinase activity in mature oocytes irrespective of the cre transgene transmission. Biochemical and biophysical research communications. 1997;237:318–324. doi: 10.1006/bbrc.1997.7111. [DOI] [PubMed] [Google Scholar]
- Santostefano KE, Hamazaki T, Pardo CE, Kladde MP, Terada N. Fibroblast growth factor receptor 2 homodimerization rapidly reduces transcription of the pluripotency gene Nanog without dissociation of activating transcription factors. The Journal of biological chemistry. 2012;287:30507–30517. doi: 10.1074/jbc.M112.388181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrode N, Xenopoulos P, Piliszek A, Frankenberg S, Plusa B, Hadjantonakis AK. Anatomy of a blastocyst: cell behaviors driving cell fate choice and morphogenesis in the early mouse embryo. Genesis. 2013;51:219–233. doi: 10.1002/dvg.22368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimosato D, Shiki M, Niwa H. Extra-embryonic endoderm cells derived from ES cells induced by GATA factors acquire the character of XEN cells. BMC developmental biology. 2007;7:80. doi: 10.1186/1471-213X-7-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AM, Hamazaki T, Hankowski KE, Terada N. A heterogeneous expression pattern for Nanog in embryonic stem cells. Stem Cells. 2007;25:2534–2542. doi: 10.1634/stemcells.2007-0126. [DOI] [PubMed] [Google Scholar]
- Smyth N, Vatansever HS, Murray P, Meyer M, Frie C, Paulsson M, Edgar D. Absence of basement membranes after targeting the LAMC1 gene results in embryonic lethality due to failure of endoderm differentiation. The Journal of cell biology. 1999;144:151–160. doi: 10.1083/jcb.144.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodhi CP, Li J, Duncan SA. Generation of mice harbouring a conditional loss-of-function allele of Gata6. BMC developmental biology. 2006;6:19. doi: 10.1186/1471-213X-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Jeught M, O’Leary T, Ghimire S, Lierman S, Duggal G, Versieren K, Deforce D, Chuva de Sousa Lopes S, Heindryckx B, De Sutter P. The combination of inhibitors of FGF/MEK/Erk and GSK3beta signaling increases the number of OCT3/4- and NANOG-positive cells in the human inner cell mass, but does not improve stem cell derivation. Stem cells and development. 2013;22:296–306. doi: 10.1089/scd.2012.0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka Y, Lanner F, Rossant J. FGF signal-dependent segregation of primitive endoderm and epiblast in the mouse blastocyst. Development. 2010;137:715–724. doi: 10.1242/dev.043471. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.