Abstract
Multiplexed surface-enhanced Raman scattering (SERS) nanoparticles (NPs) offer the potential for rapid molecular phenotyping of tissues, thereby enabling accurate disease detection as well as patient stratification to guide personalized therapies or to monitor treatment outcomes. The clinical success of molecular diagnostics based on SERS NPs would be facilitated by the ability to accurately identify tissue biomarkers under time-constrained staining and detection conditions with a portable device. In vitro, ex vivo and in vivo experiments were performed to optimize the technology and protocols for the rapid detection (0.1-s integration time) of multiple cell-surface biomarkers with a miniature fiber-optic spectral-detection probe following a brief (5 min) topical application of SERS NPs on tissues. Furthermore, we demonstrate that the simultaneous detection and ratiometric quantification of targeted and nontargeted NPs allows for an unambiguous assessment of molecular expression that is insensitive to nonspecific variations in NP concentrations.
INNOVATION
Multidisciplinary advances have been made to develop a technology for the multiplexed molecular phenotyping of fresh ex vivo and in vivo tissues under time-constrained conditions that are relevant for point-of-care clinical applications. By developing high-affinity targeted SERS NPs, a sensitive portable spectral-detection device, and an optimized topical-delivery protocol, we demonstrate for the first time a ratiometric method to rapidly quantify the specific binding of a panel of biomarker-targeted NPs on fresh tissues, thereby eliminating the ambiguities that often arise due to nonspecific sources of contrast.
INTRODUCTION
A major focus of biomedical optics has been to develop technologies for the detection of some of the most prevalent diseases worldwide such as epithelial cancers of the colon, esophagus, oral cavity, cervix and skin, as well as to image surgical margins to guide tumor-resection procedures1–6. The general approach of optical diagnostics is to deduce tissue status through the measurement of optical signals generated either intrinsically by cell and tissue constituents7,8 or extrinsically by targeted contrast agents with known signatures9–11. While the simplicity and regulatory ease of imaging intrinsic signatures is compelling, the use of exogenous contrast agents allows for the assessment of highly informative biomarkers such as cell-surface receptors. A challenging issue is that molecular biomarkers of disease vary greatly among subjects, between disease subtypes, and even within a single subject over time12. Thus, exogenous probes should ideally be capable of being multiplexed to simultaneously detect multiple biomarkers. A technology for the rapid molecular phenotyping of fresh tissues at the point of care could enable accurate disease diagnosis, the monitoring of treatment response, and patient stratification to guide personalized therapies.
Although numerous molecular probes are being developed to label disease biomarkers13–15, their utility for cancer detection is often limited by various factors. For example, fluorescent dyes are easily photobleached, have a wide emission spectrum, and must often be excited at disparate wavelengths when combined, thus limiting their multiplexing capability. Although quantum dots (QD) offer a narrower emission bandwidth, higher sensitivity and higher photostability than fluorescent dyes16, their potential toxicity has thus far precluded their in vivo use in humans17. Surface-enhanced Raman-scattering (SERS) nanoparticles (NPs), here-after referred to as “SERS NPs” or “NPs”, have attracted interest due to their brightness, low toxicity, and potential for sensitive and multiplexed biomarker detection18. The SERS NPs utilized in this study are available in multiple “flavors,” each of which emits a characteristic Raman fingerprint spectrum that allows for the identification and quantification of large multiplexed mixtures of different NP flavors when illuminated at a single wavelength19–21. It is important to emphasize that these SERS NPs are engineered to emit a stable and unique Raman spectra that is insensitive to the environment19. This is accomplished by encapsulating the SERS NPs within a protective silica shell, such that their gold core and Raman-active layer are shielded from other NP cores as well as from their surroundings. The gold cores at the center of these SERS NPs provide an electromagnetic enhancement that dramatically increases the Raman fingerprint signal (which uniquely identifies each NP flavor) compared with non-enhanced Raman signals19,22. Since the Raman signals emitted by these SERS NPs are much brighter than background Raman signals from tissue components or buffers, the background Raman signals are negligible at the measurement conditions (laser power, detector integration times, and optical set up) utilized in this study. Here we design each flavor of NP to target a unique protein biomarker by conjugating the NPs to monoclonal antibodies. The SERS NP spectra can be easily identified by their distinct and narrow spectral peaks whereby the quantity of each NP flavor in a mixture may be determined through a spectral-demultiplexing software algorithm21,23–25.
In recent years, a number of groups have begun to investigate the advantages of various types of SERS NPs for the detection of cancer biomarkers18,26. For example, a few studies have explored the feasibility of using SERS NPs to specifically label cell-surface protein biomarkers under in vitro conditions9,27–31. One study has demonstrated the basic feasibility of targeting a single biomarker using SERS-NPs on ex vivo tissues topically stained for 1 hour and detected with a large Raman microscope system9. In addition, a few groups have demonstrated the feasibility of the multiplexed detection of large panels of nontargeted SERS NPs21,22,32–36. Finally, studies have utilized large Raman microscope systems to explore the behavior of targeted and nontargeted SERS NPs after intravenous injection in tumor-bearing mice26,37.
Although the basic sensitivity, targeting ability and multiplexing capabilities of SERS NPs have been investigated, there is a need to demonstrate that multiplexed molecular detection is possible under time-limited tissue-staining and detection conditions, as encountered in point-of-care clinical settings, using a miniaturized spectral-detection probe. In particular, in this study we focus on establishing the feasibility of topical application as a means of rapidly delivering targeted SERS NPs to exposed tissue surfaces (e.g. epithelial surfaces and surgical margins), resulting in sufficient contrast to enable in vivo molecular detection at short integration times (0.1 s). Topical application potentially offers an expedited route for the regulatory approval of SERS NPs, especially since recent studies have shown that SERS NPs applied topically in the gastrointestinal tract result in negligible systemic uptake38,39. An example application for this technology would be to screen the oral cavity, esophagus, or colon for dysplasia and/or cancer by irrigating the tissue with, or having the patient drink, a cocktail of targeted SERS NPs prior to performing multiplexed molecular imaging with a spectral-imaging endoscope. In the case of oral cancer detection, a major clinical challenge is the fact that many lesions appear suspicious upon visual examination but are actually noncancerous. Physicians (and dentists) are often hesitant to obtain physical biopsy specimens due to the discomfort and potential complications from such biopsies that are often of benign lesions40. A noninvasive imaging method to screen for molecular alterations indicative of cancer or precancer could play a valuable role in guiding biopsies (i.e. image-guided biopsy). Another example would be the intraoperative molecular phenotyping/imaging of in vivo or freshly excised surgical specimens to identify residual tumor at the surgical margins and to improve the completeness of resection15.
In the specific case of breast-conserving surgery (lumpectomy), numerous reports reveal that 20% to 60% of lumpectomy patients must undergo re-excision surgeries after post-operative pathology reveals residual tumor at the surgical margins41,42. With our technology, tissues could potentially be topically stained with a panel of SERS NPs (in vivo or ex vivo), and imaged after one or more rinse steps to rapidly identify/quantify the presence of tumor-related biomarkers during surgery. In this study, we demonstrate the general feasibility to address both ex vivo and in vivo imaging applications. In this study, we also demonstrate the advantages that SERS NPs confer for the ratiometric quantification of molecularly specific vs. nonspecific binding. The importance of distinguishing between specific and nonspecific probe accumulation is critical. For example, in this study we observe that nontargeted NPs that are topically applied on tissues show higher retention in normal tissues versus xenograft tumors, i.e. inverse contrast. This same phenomenon has also been seen with intravenously injected contrast agents (e.g. fluorescent dyes) in implanted tumors43,44. By contrast, in many animal models and clinical samples, the opposite effect of enhanced penetration and retention (EPR) in tumors versus normal tissues is often observed10,45,46. Regardless of the mechanisms for the nonspecific behavior of contrast agents, there is increasing consensus that utilizing a dual-reporter ratiometric detection strategy, in which one contrast agent serves as a negative control for a simultaneously delivered targeted probe, is valuable to accurately identify and quantify molecularly specific binding43–45. Here we show that multiplexed SERS NPs are well suited for this purpose, owing to the fact that one nontargeted NP may serve as a negative control to quantify the specific vs. nonspecific binding of several other targeted NPs. In particular, our SERS NPs are excited under identical illumination conditions from a single laser (785 nm), and emit Raman spectra within the same narrow band of wavelengths (approximately 850 – 900 nm). Therefore, their ratios are insensitive to variables such as tissue optical properties [see Supplementary Information (SI) for additional information]. In short, we stain tissues with an equimolar mixture of targeted NPs and one untargeted NP that controls for nonspecific effects. By first obtaining a calibration measurement of an equimolar mixture of NP flavors, it is possible to accurately quantify the relative concentration ratio of the NP flavors when the mixture is used to stain tissues (see SI), and consequently to determine the specific vs. nonspecific binding ratio of the NPs. This assumes that the nontargeted NP flavor accurately mimics the nonspecific behavior of the targeted NP flavors in the mixture (see Supplementary Fig. 3 for information on the optimal design of a nontargeted control NP). We demonstrate the robustness of this ratiometric measurement approach via in vitro and ex vivo, as well as in vivo experiments.
RESULTS
Specific-binding ability of mAb-conjugated NPs
Flow cytometry and confocal microscopy were first performed to investigate the specific-binding abilities of targeted vs. nontargeted SERS NPs. As described in the Methods section, SERS NPs were conjugated with fluorophores (for flow cytometry) as well as with one of the following three monoclonal antibodies (mAbs): an anti-EGFR mAb (to NP flavor S440), an anti-HER2 mAb (to NP flavor S420), or an isotype-control (nontargeted) mAb (to NP flavor S421). Conjugation parameters (e.g. dosage of antibody) and staining conditions (e.g. NP concentration and staining duration) were all optimized to maximize the ratio of specific vs. nonspecific binding of the targeted NPs (Supplementary Table 1). In addition, the design of a nontargeted NP to control for nonspecific binding was also optimized (Supplementary Fig. 3). For cell labelling, a 200-pM staining concentration and a 15-min staining duration resulted in high levels of specific vs. nonspecific binding. This study utilized three cell lines: (1) A431, which highly overexpresses the epidermal growth factor receptor (EGFR) with ~2 × 106 receptors/cell and moderately expresses the human epidermal growth factor receptor 2 (HER2) with ~2 × 104 receptors/cell47,48; (2) SkBr3, which highly overexpresses HER2 with ~1 × 106 receptors/cell and moderately expresses EGFR with 9 × 104 receptors/cell47,48; and (3) 3T3, a normal mouse fibroblast cell line that expresses negligible amounts of EGFR and HER249. The density of overexpressed cell-surface receptors in the A431 and SkBr3 cell lines is representative of the receptor densities measured in many naturally occurring tumors50.
Figure 1 shows the flow cytometry data in the form of fluorescence histograms of a 10,000-cell analysis. The geometric mean of the fluorescent intensity (MFI) of the cell samples is used to compare binding levels. For the negative-control cell line 3T3, the MFI of cells stained with all three NP conjugates is similar. For the A431 cell line, which highly overexpresses EGFR, the MFI of cells stained with EGFR-NPs is 65 times higher than cells stained with isotype-NPs. Since A431 cells express moderate levels of the HER2 receptor47,51, cells stained with HER2-NPs also show a slightly elevated MFI compared to cells stained with isotype-NPs (~5 times). SkBr3 cells have been reported to highly express HER2 as well as EGFR at a slightly lower level47,51. Our data in Fig. 1c support these reports, showing that cells stained with HER2-NPs have a MFI that is 92 times higher than cells stained with isotype-NPs, and that cells stained with EGFR-NPs have a MFI that is 15 times higher than cells stained with isotype-NPs. When competitively inhibited with an excess of mAb, A431 cells (inhibited with anti-EGFR mAb and then labeled with EGFR-NPs) show a 26-fold reduction in MFI, while SkBr3 cells (inhibited with anti-HER2 mAb and then labeled with HER2-NPs) show a 17-fold reduction, suggesting that the EGFR- and HER2-conjugated NPs bind specifically to cell-surface EGFR and HER2 receptors, respectively.
Figure 1.
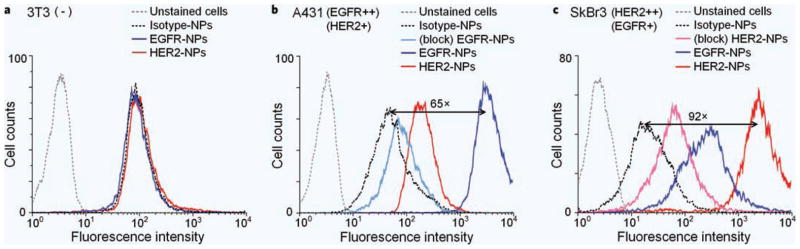
Flow cytometry validation of conjugated NPs with cultured cells. NPs were conjugated with either anti-EGFR, anti-HER2 or isotype control monoclonal antibodies. These NPs were individually used to stain (a) 3T3 (−), (b) A431 (EGFR++, HER2+) and (c) SkBr3 (HER2++, EGFR+) cell lines for 15 min, followed by flow cytometry analysis. Fluorescence histograms from unlabeled cells as well as cells stained with isotype-NPs, EGFR-NPs and HER2-NPs are shown. In addition, the results of competitive-binding experiments are displayed, in which excess mAb is used to block cell-surface receptors prior to NP staining (see text for details).
In order to further confirm the specific-binding ability of the NP conjugates, cells from flow-cytometry experiments were transferred to glass slides and observed under a confocal microscope (Leica TCS SP5). By setting a tight detection pinhole to produce a thin optical section, it was observed that the fluorescence signal was primarily localized to the periphery of the cells (Supplementary Fig. 4), indicating that the NPs were bound to the cell surface rather than internalized. To compare the total amount of NPs bound to each cell (Supplementary Fig. 5), the pinhole was enlarged. The same objective (20×; dry), laser power, and detector gain settings were used to image all samples. Supplementary Fig. 5 shows the bright-field and fluorescence images of 9 stained samples, including all combinations of 3 cell lines and 3 types of conjugated NPs.
Ratiometric analysis of multiplexed NPs on cell monolayers
In vitro experiments were performed to demonstrate the ability to rapidly assess molecular expression on cell monolayers via multiplexed detection of SERS NPs with a miniature spectral-detection probe (Fig. 2). To simulate the topical application of NPs on tissue surfaces, confluent monolayers of cells were cultured in 96-well plates (see Methods and Supplementary Fig. 7–9). Equimolar mixtures of positive and negative NPs were simultaneously applied to the monolayers for 15 min. Unbound NPs were then removed via multiple rounds of rinsing with phosphate-buffered saline (PBS). SERS spectra were collected with a miniature Raman detection probe, in which the NP mixtures were illuminated with a single low-power (10 mW) diode laser (see Fig. 2c, Methods, and design optimization in Supplementary Fig. 1). Several approaches have been developed to demultiplex (unmix) a spectrum generated by two or more flavors of SERS NPs in the presence of a competing background21,24. An analysis and comparison between various demultiplexing approaches is beyond the scope of this paper but has been explored by others52. Here, a simple and commonly used direct classical least squares (DCLS) demultiplexing method was employed23, along with a calibrated ratiometric strategy to quantify specific vs. nonspecific binding (see SI). We have shown through validation studies (Supplementary Fig. 6) that the DCLS demultiplexing approach provides accurate quantification of NP concentrations and ratios across our measurement conditions.
Figure 2.
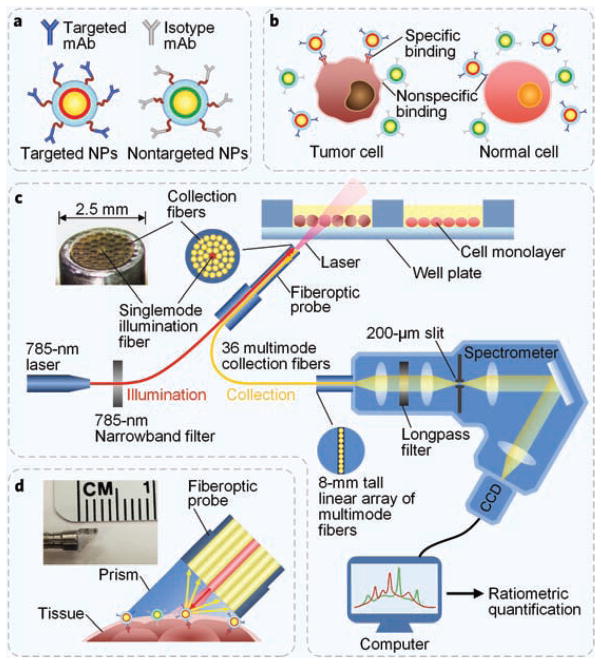
Schematic illustrations of a customized spectral-imaging device to detect bioconjugated NPs on cells and tissues. (a) NPs conjugated with targeted-and isotype-control (nontargeted) monoclonal antibodies (mAb), where a unique flavor of NP is used for each of the targeted and nontargeted contrast agents; (b) A depiction (not to scale) of the specific binding of multiplexed NPs to cell-surface receptors (in the case of targeted NPs) or nonspecific locations (in the case of all NP flavors); (c) Experimental platform to detect multiplexed NPs on cell monolayers; (d) Angled contact probe for ex vivo and in vivo tissue measurements.
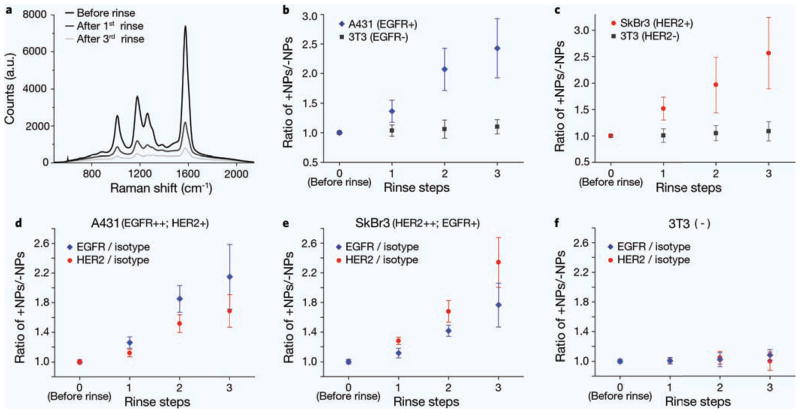
In initial experiments, a dual-flavor mixture (150 pM per flavor) consisting of a targeted (positive) NP flavor and a nontargeted (negative) NP flavor, was applied to each well to stain cell monolayers for 15 min, followed by 3 rounds of rinsing to remove unbound NPs (each rinse replaced ~90% of the liquid in each well). Raman spectra from an A431 monolayer stained with NPs, before and after rinsing, are shown in Fig. 3a. The Raman peaks are distinguishable in spite of a competing broadband spectral background, even after multiple stages of rinsing. For each cell line, raw spectra were analyzed (3 wells of each cell line, 3 different positions per well, and 2 acquisitions at each position for a total n = 18) to calculate the weight of each flavor, the weight ratio of positive and negative NPs, and the actual concentration ratio of the NPs (+NPs/−NPs). The weight ratios measured before rinsing were used to calibrate subsequent ratiometric measurements, assuming that a 1:1 equimolar ratio of NPs was used in the staining mixtures. The calibrated NP concentration ratios are plotted in Fig. 3b,c (additional details are provided in the SI).
Figure 3.
Demultiplexing analysis of the concentration ratio of multiplexed NPs on cell monolayers. Each cell monolayer was simultaneously stained with two (a–c) or three (d–f) flavors of NPs and then rinsed with PBS. (a) Raw spectra before and after rinses (from an A431 cell monolayer); (b) Concentration ratios of EGFR-NPs/isotype-NPs on A431 and 3T3 cells; (c) Concentration ratios of HER2-NPs/isotype-NPs on SkBr3 and 3T3 cells; concentration ratios of EGFR-NPs/isotype-NPs and HER2-NPs/isotype-NPs on (d) A431, (e) SkBr3 and (f) 3T3 cell monolayers.
For A431 cell monolayers, EGFR-NPs and isotype-NPs were used for initial 2-flavor staining experiments. As shown in Fig. 3b, after three rinses the concentration ratio (+NPs/−NPs) increases to 2.4 for A431 monolayers but remains near unity for 3T3 monolayers. For SkBr3 cell monolayers (Fig. 3c), the concentration ratio of HER2-NPs/isotype-NPs increases to 2.6 after three rinses, while remaining near unity for 3T3 cell monolayers. After three rinsing steps, the approximate concentration of the isotype-NPs decreases to 4–6 pM. Since this is near the limit of detection (LOD) of our spectral-detection probe (see Supplementary Fig. 6), data with further rinsing steps are not shown.
In vitro experiments with three multiplexed NP flavors (two positive and one negative control) were carried out to demonstrate the ability to simultaneously assess the expression of multiple cell-surface biomarkers (EGFR and HER2). Here, a mixture of EGFR-NPs, HER2-NPs and isotype-NPs (150 pM per flavor) were applied to stain cell monolayers, followed by 3 rounds of rinsing. Figure 3d–f show the concentration ratios between positive and negative NPs (both the EGFR/isotype ratio and the HER2/isotype ratio) on the different cell monolayers (n = 18 as previously described). These results are consistent with the two-particle staining experiments and demonstrate the utility of ratiometric analyses of multiplexed NPs to rapidly identify the expression of one or more cell-surface proteins.
Ratiometric analysis of multiplexed NPs on tumor explants (ex vivo)
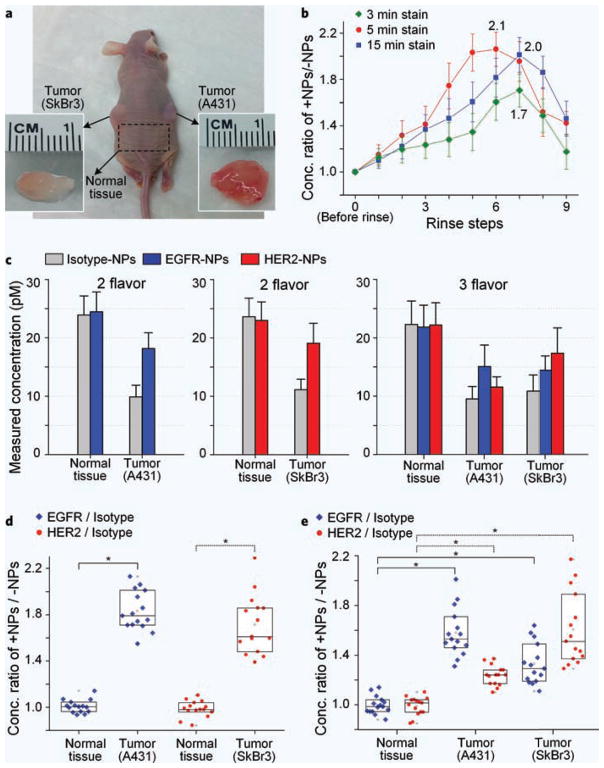
Ex vivo experiments were performed to demonstrate the ability to detect biomarker expression in fresh tissues with topically applied SERS NPs and rapid detection with a miniature probe (see Fig. 2d, Methods, and design optimization in Supplementary Fig. 1). A431 and SkBr3 tumor xenografts (5–10 mm diameter) as well as normal tissues (Fig. 4a) were resected and cut into similarly sized pieces (30–60 mm3) for the measurement of NP concentrations and ratios.
Figure 4.
Ex vivo experiments with xenograft tumors. (a) Nude mouse with xenograft tumors 14 d after subcutaneous injection of 1 × 106 A431 cells and 5 × 106 SkBr3 cells (the lower insets show the explanted tissues). (b) Experiment to optimize the staining of tissues with SERS NPs showing the concentration ratio of a targeted vs. nontargeted NP (EGFR-NP/isotype-NP) after each rinse step (3 s per rinse) following either a 3-, 5-, or 15-min staining duration. (c) Measured absolute concentrations of NPs on tumor and normal tissues after 18 s of rinsing (equivalent to 6 rinse steps) for 2-flavor and 3-flavor multiplexed experiments, showing elevated nonspecific accumulation in normal tissues vs. tumors. However, plotting the concentration ratio with (d) 2-flavor and (e) 3-flavor multiplexing (n = 15) allows for the unambiguous quantification of specific vs. nonspecific binding. *P-value < 0.001.
The staining and rinsing protocols were first optimized to maximize the concentration ratio of +NPs/−NP. For optimization experiments, A431 tumor specimens were immersed into a 50-μL equimolar mixture of EGFR-NPs and isotype-NPs (200 pM per flavor), and allowed to incubate without agitation for 3, 5 or 15 min (3 pieces per staining condition). The tissues were then washed 9 times by submerging them into 20 mL of PBS (with gentle agitation) for 3 s per wash. SERS spectra were collected with the contact probe (integration time: 0.25 s) after each rinse step. Note that before staining, a 4-μL droplet of an equimolar NP mixture was placed on the contact surface of the probe to calibrate subsequent concentration and ratiometric measurements (see SI). Figure 4b plots the calibrated NP concentration ratio after each rinse (n = 6). A 5-min staining duration resulted in the highest NP concentration ratio with the least number of rinse steps and was therefore used for subsequent ex vivo and in vivo experiments.
Results from ex vivo experiments with two multiplexed NP flavors (1 targeted NP and 1 nontargeted control NP) or three multiplexed NP flavors (2 targeted NPs and 1 nontargeted control NP) are shown in Fig. 4c–e. Under identical staining (5 min) and washing conditions (18 s), stronger Raman signals were detected from normal tissues than from tumor tissues (Fig. 4c), indicating that there is enhanced nonspecific retention of topically applied NPs in normal tissues vs. the tumor xenografts. However, Fig. 4d,e show that the concentration ratio between positive and negative NPs (EGFR/isotype and HER2/isotype) on tumors was significantly elevated, indicating preferential binding of targeted NPs to overexpressed cell-surface biomarkers. In normal tissues, the NP ratios remained near unity due to the negligible expression of EGFR and HER2 in these tissues.
Rapid ratiometric analysis of multiplexed NPs on tumor implants (in vivo)
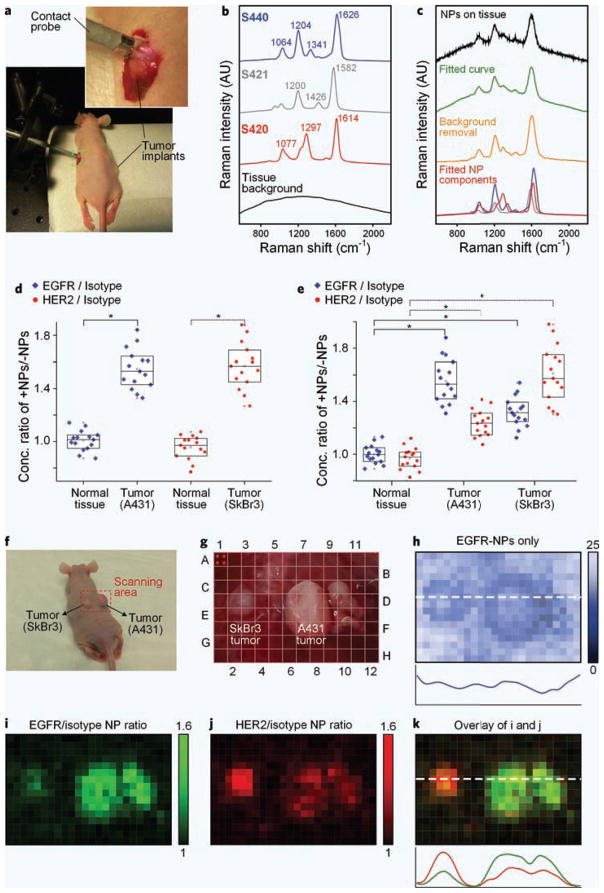
In vivo experiments were performed with a 0.1-s detector integration time (see Supplementary Fig. 10 for a noise analysis) using a flexible version of the spectral-detection device (Fig. 5a) to demonstrate rapid ratiometric quantification of NPs in live mice. Figure 5b shows the reference spectra of the SERS NPs and tissue background (no NPs) that were acquired for these in vivo experiments. Figure 5c provides example spectra to illustrate the DCLS demultiplexing algorithm, including a raw spectrum from a 3-flavor mixture of NPs on tumor tissues acquired with a 0.1-s integration time, a best-fit curve using the DCLS algorithm, the combined spectrum of the NPs after removing the tissue background component, and the demultiplexed spectra from individual NP flavors (blue: EGFR-S440, 17.9 pM; red: HER2-S420, 13.1 pM; gray: isotype-S421, 10.4 pM). Figure 5d,e show the concentration ratio between positive and negative NPs (EGFR/isotype and HER2/isotype) on the tumors and normal tissues of 5 mice (n = 15). The results are similar to the ex vivo results: the ratios on normal tissues remain near unity, but are significantly elevated on tumors. Interestingly, the measurement uncertainty for the in vivo results is less than for the ex vivo results, possibly due to reduced mechanical perturbation and measurement artifact when probing in vivo tissues compared with surgically excised tissues. In order to demonstrate that our demultiplexing algorithm and results are insensitive to any specific choice of NP flavor, we altered the combination of NP flavors that were conjugated to each of the three mAbs (isotype control, EGFR-targeted, and HER2-targeted). In vivo experiments were repeated with two mice (see Supplementary Fig. 11) and yielded results that are consistent with what is presented in Fig. 5e.
Figure 5.
In vivo ratiometric analysis of multiplexed NPs on tumor implants. (a) Mouse with surgically exposed tumors; the inset provides a magnified view of the 2.5-mm diameter flexible Raman probe. (b) Reference Raman spectra of pure SERS NPs (red: S420, gray: S421 and blue: S440) and tissue background with no NPs (black). (c) Raw spectra of NPs applied on tissue acquired with a 0.1-s integration time (black), best-fit curve using a DCLS algorithm (green), spectra of NPs on tissue after tissue-background removal using a DCLS algorithm (orange), and the DCLS-demultiplexed NP spectra (blue: EGFR-S440, red: HER2-S420, gray: isotype-S421). The concentration ratio of targeted and nontargeted NPs topically applied on exposed tumors and normal tissues is plotted for (d) 2-flavor and (e) 3-flavor multiplexed NPs (n = 15). *P-value < 0.001. (f–k) Image-grid experiment. (f) Mouse with two adjacent tumor xenografts. (g) Photograph of stained tissue with a virtual grid of detection locations. Four positions were probed within each grid coordinate (e.g. refer to the red dots in coordinate A1) such that 24 × 16 positions were detected in total; (h) Map of the absolute concentration (pM) of EGFR-NPs; (i) Map of the concentration ratio of EGFR/isotype NPs. (j) Map of the concentration ratio of HER2/isotype NPs. (k) Overlay of EGFR/isotype and HER2/isotype maps.
To further demonstrate the utility of the ratiometric analysis of multiplexed SERS NPs, we scanned a grid of positions over the two tumors (A431 and SkBr3) and surrounding normal tissues on one mouse and mapped the concentration ratios of targeted/nontargeted NPs (Fig. 5f–k). For this experiment, a mouse with A431 and SkBr3 tumors implanted adjacent to each other was utilized (Fig. 5f). Before detection, a mixture of EGFR-NPs, HER2-NPs and isotype-NPs (200 pM per flavor) was topically applied to stain a 4.2 × 2.8 cm2 tissue area for 5 min, followed by a PBS rinse step (20 s). A total of 24 × 16 positions were detected with a 0.1-s integration time at each point (see Fig. 5g). In Fig. 5h, the absolute concentration of EGFR-NPs is mapped to simulate a non-ratiometric imaging strategy using a single tumor-targeted contrast agent. Consistent with our ex vivo results (Fig. 4c), the normal tissues in Fig. 5h show higher absolute intensities compared with the tumors due to enhanced nonspecific accumulation of the NPs, which is potentially misleading because of the apparent inverse contrast. Figure 5i,j are maps of the concentration ratio of EGFR-NP/isotype-NP and HER2-NP/isotype-NP, respectively. Both maps demonstrate improved contrast between tumor (color) and normal tissues (black) and provide quantification of specific binding to molecular targets. These results are consistent with the previous flow cytometry, in vitro monolayer and ex vivo tissue measurements, in which A431 tumors express high levels of EGFR and moderate levels of HER2, whereas SkBr3 tumors express high levels of HER2 and moderate levels of EGFR.
DISCUSSION
Interest in SERS-coded NPs has been accelerating in recent years due to their potential for highly multiplexed detection with negligible photobleaching when illuminated with a single low-power laser source. The relatively large size of these NPs (~120 nm) allows them to be used for the detection of protein biomarkers expressed at the surface of epithelia and surgically exposed tissue with minimal systemic uptake and toxicity39,53. However, to date, there has not been a demonstration of the ability to rapidly quantify the specific binding of multiplexed SERS NPs ex vivo and in vivo following a brief topical application and rinse-removal protocol. Hence, we used a miniature spectral probe and a calibrated ratiometric analysis method to investigate the performance of SERS NPs under clinically relevant conditions, demonstrating the ability to accurately quantify molecularly specific vs. nonspecific NP accumulation on tumor and normal tissues with a spectral integration time of 0.1 s after a 5-min topical application and 20 s of rinsing.
We present in vitro, ex vivo and in vivo data to illustrate the progressively more challenging nature of these experimental conditions. For example, cell suspensions are easily stained and rinsed, yielding specific-to-nonspecific binding ratios of nearly two orders of magnitude in flow cytometry experiments (Fig. 1). These results show an unprecedented ability of the large NPs to bind to cell-surface receptors and were achieved with a short staining duration of only 15 min. Monolayers of adherent cells are less efficient to stain and rinse, and yield specific-to-nonspecific binding ratios of 2- to 3-fold when measured with a miniature probe (Fig. 3). Finally, ex vivo and in vivo tissue xenografts are the most challenging to stain with exogenous molecular contrast agents (Fig. 4,5), as they exhibit nonspecific permeability and retention effects as well as a host of nonspecific chemical targets that reduce detection contrast. For example, Figs. 4c and 5h demonstrate how measuring the absolute concentration of targeted NPs can be misleading due to a larger uptake and retention in normal tissues vs. tumors (i.e. inverse contrast). Additional sources of misleading contrast include variations in probe working distance and illumination power, as well as uneven NP application and rinse removal at different tissue locations.
In light of the potentially misleading nonspecific sources of contrast mentioned in the previous paragraph, calibrated ratiometric detection with multiplexed positive- and negative-control contrast agents is a valuable strategy for accurately quantifying specific vs. nonspecific signals43–45. SERS NPs are particularly well-suited for ratiometric detection, not only because of their high multiplexing capability but also due to the ability of multiple SERS NP flavors to be excited at a single illumination wavelength, ensuring that all NP flavors in a single measurement are interrogated identically in terms of illumination intensity, spot size and effective excitation depth (see SI). In addition, while it is possible under idealized experimental conditions to perform calibrated measurements of absolute NP concentrations (Fig. 4c), these measurements are potentially not as accurate as measurements of concentration ratios. This could be due, for example, to slight differences in sample conditions and measurement geometries between the calibration measurements and the actual experiments. Ratiometric measurements are much less sensitive to these differences, and therefore more accurate, due to the fact that the effects of experimental variations tend to cancel out in the ratios (see SI for additional discussions).
A surprising and significant result of this study is the demonstration that accurate in vivo molecular detection is optimally achieved with a topical application of only 5 min in duration. We hypothesize that the nonspecific transport and binding of these NPs within tissues at longer staining durations impedes the ability to easily remove unbound NPs via rinsing. While brief staining durations are ideal for the translation of these technologies into clinical settings, the binding affinity of the targeted NPs must be very high in order to achieve significant levels of specific binding at such short time scales. Fortunately, the molecularly targeted NPs developed in this study exhibit sufficiently high binding affinities under brief in vitro staining conditions (Fig. 1) that they may also be effectively utilized for the rapid ex vivo and in vivo detection of biomarkers in fresh tissues.
For rapid clinical diagnostics, additional work is necessary to further optimize the binding affinity of targeted SERS NPs, as well as to minimize nonspecific binding. In addition, while the brightness of the SERS NPs used here allows for the detection of low-pM concentrations of NPs at 0.1-s integration times, brighter NPs are needed to apply these contrast agents in a greater variety of biomedical detection and imaging applications. Furthermore, while studies have shown the feasibility of detecting and demultiplexing large panels (5 – 10) of nontargeted SERS NPs in animals and ex vivo human tissues, further work is needed to demonstrate the ability to quantify a large panel of protein biomarkers with targeted NPs21,22,32–36. While there are many challenges that must be resolved to improve SERS NPs for molecular diagnostics, this study optimizes across multiple disciplines — including a bioconjugation protocol, portable-measurement device, quantitative-detection assay, and a topical-staining protocol — to demonstrate the feasibility of rapidly and quantitatively detecting a panel of cancer biomarkers on ex vivo and in vivo tissues with multiplexed SERS NPs under time-constrained conditions. These tools have the potential to greatly improve our ability to investigate the mechanisms of disease progression as well as for early detection, surgical guidance, and for guiding/monitoring personalized therapies in the clinic.
METHODS
Fluorophore and antibody conjugation to SERS NPs
The NPs (Cabot Security Materials Inc, Mountain View, CA) utilize a gold core and are encapsulated in silica. The gold-core diameters are ~60 nm and the overall dimension is ~120 nm. Additional details may be found in the literature9. The silica surface of the NPs are functionalized with thiols to allow conjugation to a variety of targeting molecules (Fig. 6a). Three “flavors” of NPs were used, identified as S420, S421 and S440, each of which emits a characteristic Raman spectrum (Fig. 5b).
Figure 6.
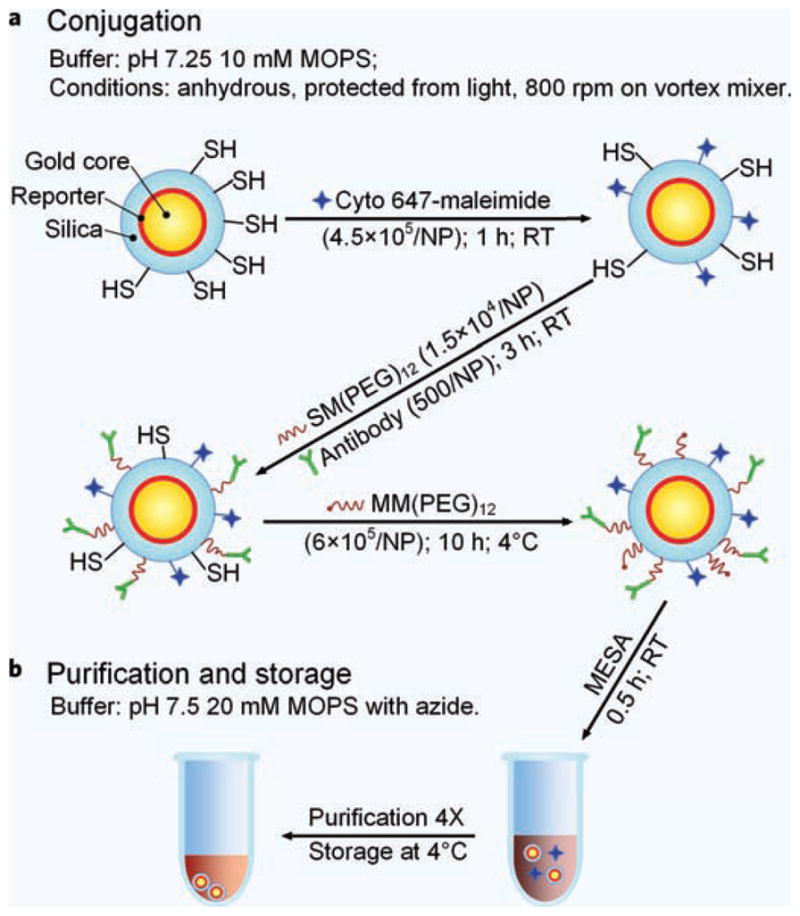
Schematic illustration of the preparation of SERS NPs conjugated to monoclonal antibodies and a fluorophore for flow cytometry. (a) Conjugation and (b) purification.
Stock concentrations of SERS NPs (800 pM in water) were diluted in 10 mM MOPS (Sigma-Aldrich, part No. M1254) buffer, pH 7.25, at a volume ratio of 1:1 (e.g. 200 μL buffer to 200 μL NPs in water), and then reacted with Cyto 647-maleimide (Cytodiagnostics Inc, part No. NF647-3-01), at 4.5 × 105 molar equivalents per NP, at room temperature for 1 h to conjugate Cyto 647 fluorophores to the NP surface (Fig. 6a). Three different monoclonal antibodies (mAb) were used for conjugation: anti-EGFR “panitumumab” (Vectibix, NDC 55513-954-01), anti-HER2 (Thermo Scientific, MS-229-PABX), or an isotype control mAb (Thermo Scientific, MA110407). These mAbs were first eluted over a desalting spin column (Thermo Scientific, 89882) and buffer-exchanged into the MOPS buffer to remove preservatives such as sodium azide. All mAbs were purchased free of protein stabilizers such as BSA or gelatin. Antibodies were added to fluorescent NPs at 500 molar equivalents per NP, along with the heterobifunctional PEG cross linker SM(PEG)12 (Thermo Scientific, 22112), at 1.5 × 104 molar equivalents per NP, and incubated at room temperature for 3 h. Following the primary conjugation reaction, MM(PEG)12 (Thermo Scientific, 22711), at 6 × 105 molar equivalents per NP, was then added to the NPs and reacted over night at 4°C to block residual thiols on the NPs. Note that all reagents were degassed with dry argon gas prior to use and all reactions were conducted under anhydrous conditions in light-protected amber vials (Fisher Scientific, 03-391-36&03-391-17) on a vortex mixer (Fisher Scientific™ Microplate Vortex Mixers, 02-216-101) set at 800 rpm. Finally, the NPs were reacted at room temperature with 2-mercaptoethanesulfonate (Sigma-Aldrich, M1511) for 30 min, at 9 × 105 molar equivalents per NP, and the reaction mixture was then diluted at a volume ratio of 1:1 with storage buffer (20 mM MOPS at pH 7.5 with 0.1% BSA (Jackson Immuno, 001-000-162) and 0.05% sodium azide (Sigma-Aldrich, S2002)). The diluted reaction mixture was purified four times via centrifugation (1000 g for 10 min), in which the supernatant was removed and replaced with storage buffer after each round of centrifugation. The conjugated NPs were stored at 4°C and protected from light. UV-VIS spectrophotometry was used to measure the final concentration of the conjugated NPs (Supplementary Fig. 2).
Customized spectral-imaging device
A miniature spectral-imaging device was developed to quantify the concentration and concentration ratios of SERS NPs applied on cells and tissues. The detection probe is cylindrical with an outer diameter of 2.5 mm. A singlemode fiber at the center of the probe is used to illuminate the sample, and 36 multimode fibers (200-micron core) surrounding the singlemode fiber are used to collect optical signals including Raman-scattered light (additional details about the detection system have been described previously23). A low power 785-nm diode laser (10 mW, well below ANSI safety limits) is used to illuminate the tissue with a spot size of ~200 μm. In order to minimize specular reflections, the probe is angled at 20 to 45 deg with respect to the direction normal to the sample surface (Fig. 2 and Supplementary Fig. 9). A custom spectrometer (Bayspec Inc.) is used to disperse the collected signal onto a cooled deep-depletion spectroscopic CCD (Andor Newton, DU920P-BR-DD). Detector integration times ranging from 0.1 to 2 s were used. The linearity of our device for measurements of 3-flavor NP mixtures (of various ratios) was assessed and is shown in Supplementary Fig. 6.
For monolayer measurements in multiwell plates, a probe holder was used to maintain a constant working distance (~2 mm) and probe angle (20–25 deg) (Supplementary Fig. 9). For ex vivo and in vivo tissue measurements, a glass prism (Tower Optical Inc., 4531-0006) was adhered to the probe tip to create a fixed-angle and fixed-working-distance contact surface with the tissue (refer to Fig. 2d and design optimization in Supplementary Fig. 1).
Cell culture, flow cytometry, and cell monolayers
A431 and 3T3 cells were cultured in DMEM (Lonza, 12-604F) and SkBr3 cells were cultured in Mccoy’s 5A (Lonza, 12-688F), both of which were supplemented with 10% fetal bovine serum (FBS, Thermo Scientific, SH3008803) and 1% penicillin-streptomycin (Lonza, 17-602E). Cells were cultured at 37 °C at a CO2 level of 5.0%. Trypsin EDTA 1X (Mediatech, MT25051CI) was used to detach adherent cells for flow cytometry.
For flow cytometry, 50-uL cell suspensions (0.2 million cells) were reacted with 50 μL of 300-pM antibody-conjugated NPs and incubated for 15 min at room temperature protected from light under gentle agitation with a vortex mixer (Fisher Scientific™ Microplate Vortex Mixers, 02-216-101) at 300 rpm. After staining, the cells were purified three times via centrifugation (400 g for 5 min) and supernatant-replacement (500 μL per rinse) with FACS buffer (20% FBS in PBS), and then stored in 500 μL of FACS buffer. Competitive binding experiments were also performed to verify the specificity of the targeted SERS NPs. For these experiments, A431 and SkBr3 cells (0.2 million in 50 μL) were incubated 4 h prior to NP staining with 50 μL of 10 mg·mL−1 panitumumab and anti-HER2 mAb to competitively inhibit the cell-surface receptor targets (EGFR or HER2).
For in vitro experiments, a 96-well plate with a 200 μm-thick glass bottom was used to culture cells into monolayers. For each cell line, cells were counted and seeded into four wells (1 × 105 cells in 50 μL of media per well for A431 and SkBr3, and 0.5 × 105 cells in 50 μL of media per well for 3T3). After incubation for 24 hours, a similar confluent cell monolayer was observed in all wells (Supplementary Fig. 7). For Raman measurements, 1 of the 4 wells for each cell line was used to acquire a background measurement in the absence of NPs (Supplementary Fig. 8). For the three remaining wells of each cell line, an equimolar mixture of targeted (anti-EGFR or anti-HER2) and nontargeted (isotype control) NPs was added for ratiometric analysis of specific vs. nonspecific binding of the NPs. Both 2-flavor and 3-flavor mixtures were used at a staining concentration of 150 pM per flavor and a total staining volume of 100 μL. Staining was performed for 15 min, followed by up to three rounds of rinsing with PBS. Spectral measurements and ratiometric analyses were performed before and after each rinse step.
Xenograft tumor model
All animal work was approved by the Institutional Animal Care and Use Committee (IACUC) at Stony Brook University. Five male nude mice at 7–9 weeks of age (Taconic Farms Inc, model NCRNU-F) were subcutaneously implanted with tumor cells in their flank under isoflurane inhalation anesthesia (Baxter, 1001936060). 1 × 106 A431 cells suspended in 50% matrigel (BD biosciences, 354234) (200 μL mixture) were injected into one flank and 5 × 106 SkBr3 cells in 50% matrigel (300 μL mixture) were injected into the contralateral side. After 1–4 weeks, the injected cells developed into tumors of approximately 5 to 10 mm in diameter.
For ex vivo imaging, the mice were anesthetized and euthanized by cervical dislocation, followed by the surgical removal of tumors and normal tissues of a similar size. The explanted tumors and normal tissues were cut into small pieces (30–60 mm3) and hydrated in FACS buffer for background acquisition and NP staining. For ex vivo staining, pieces of fresh tumor and normal tissue were submerged for 5 minutes in an equimolar mixture of 2 or 3 flavors of conjugated NPs (200 pM per flavor). After incubation, the pieces were washed up to 7 times by submerging them for 3 s per wash, with gentle agitation, in a large volume of PBS (20–50 mL).
For in vivo imaging, the mice were anesthetized via intraperitoneal injection of ketamine and xylazine and then the tumors were surgically exposed. An equimolar mixture of 2 or 3 flavors of conjugated NPs (200 pM per flavor) was topically applied to tumor and normal tissues for 5 min. After rinsing off unbound NPs with PBS, the tumor and normal tissues were scanned using the contact probe (Fig. 2d). After imaging, the mice were euthanized.
Statistics
For ex vivo and in vivo experiments, a two-sample t-test was performed to determine the significance of the difference in the positive/negative NP ratio (EGFR/isotype or HER2/isotype) between tumors and normal tissues. A p-value < 0.001 was considered significant. Data points represent average values with error bars denoting one standard deviation from the mean. For the box plots in Figs. 4d,e and 5d,e, the bottom and top of the box represent the 1st and 3rd quartiles of the dataset, respectively, and the band inside the box represents the median (2nd quartile) of the data.
Supplementary Material
Acknowledgments
The authors acknowledge support from the NIH/NIBIB – R21 EB015016 (Liu), the Department of Biomedical Engineering, and the Office of the Vice President of Research at Stony Brook University (SUNY).
Footnotes
SUPPLEMENTARY INFORMATION AVAILABLE
Demultiplexing SERS spectra using direct classical least squares; ratiometric quantification of specific vs. nonspecific binding; optimization of a miniature hand-held probe for contact detection of SERS NPs on tissues (Supplementary Fig. 1); calculating the concentration of NP conjugates via UV-VIS spectrophotometry (Supplementary Fig. 2); optimization of conjugation and staining parameters (Supplementary Table 1); flow cytometry comparison of different control NPs with cultured cells (Supplementary Fig. 3); observation of NP distribution on cells via confocal microscopy (Supplementary Fig. 4); confocal microscopic images of different cells stained with conjugated NPs (Supplementary Fig. 5); limit-of-detection (linearity) test for a 3-flavor mixture (Supplementary Fig. 6); microscopic images of confluent cell monolayers of 3T3, A431 and SkBr3 in one well of a 96-well plate (Supplementary Fig. 7); schematic of cell-monolayer experiments on a 96-well plate (Supplementary Fig. 8); experimental platform for the spectral detection of cell monolayers on a well plate (Supplementary Fig. 9); coefficient of variation of the measured concentration of NPs on tissues under different integration times (Supplementary Fig. 10); effect of NP flavor on ratiometric analysis (Supplementary Fig. 11); Raman signal of NPs in different buffers (Supplementary Fig. 12). This material is available free of charge.
AUTHOR CONTRIBUTIONS
Y.W. designed experiments, performed nanoparticle conjugations and experiments, analyzed data, and wrote the manuscript. A.K. set up the Raman hardware, completed the demultiplexing algorithm and Labview software. M.S., D.W., Y.C., S.Y.L., and D.M. provided assistance and training for various experiments. P.Z.M. and B.C.W. provided training and support for nanoparticle conjugations. J.T.C.L. initiated the project, designed experiments, provided training and support for various experiments, analyzed data, supervised and coordinated all investigators on the project, and wrote the manuscript.
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Estimating the world cancer burden: Globocan 2000. Int J Cancer. 2001;94:153–156. doi: 10.1002/ijc.1440. [DOI] [PubMed] [Google Scholar]
- 2.Levin B, et al. Screening and surveillance for the early detection of colorectal cancer and adenomatous polyps, 2008: A joint guideline from the American Cancer Society, the US Multi-Society Task Force on Colorectal Cancer, and the American College of Radiology. CA Cancer J Clin. 2008;58:130–160. doi: 10.3322/CA.2007.0018. [DOI] [PubMed] [Google Scholar]
- 3.Bergholt MS, et al. Characterizing variability in in vivo Raman spectra of different anatomical locations in the upper gastrointestinal tract toward cancer detection. J Biomed Optics. 2011;16:037003–037003-10. doi: 10.1117/1.3556723. [DOI] [PubMed] [Google Scholar]
- 4.Jo JA, et al. In vivo simultaneous morphological and biochemical optical imaging of oral epithelial cancer. IEEE Trans Biomed Eng. 2010;57:2596–2599. doi: 10.1109/TBME.2010.2060485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Dam GM, et al. Intraoperative tumor-specific fluorescence imaging in ovarian cancer by folate receptor-[alpha] targeting: First in-human results. Nat Med. 2011;17:1315–1319. doi: 10.1038/nm.2472. [DOI] [PubMed] [Google Scholar]
- 6.Bird-Lieberman EL, et al. Molecular imaging using fluorescent lectins permits rapid endoscopic identification of dysplasia in Barrett’s esophagus. Nat Med. 2012;18:315–321. doi: 10.1038/nm.2616. [DOI] [PubMed] [Google Scholar]
- 7.Le Q, et al. Spectral imaging with scattered light: From early cancer detection to cell biology. IEEE J Sel Top Quant Electron. 2012;18:1073–1083. doi: 10.1109/JSTQE.2011.2161575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bergholt MS, et al. In vivo diagnosis of gastric cancer using Raman endoscopy and ant colony optimization techniques. Int J Cancer. 2011;128:2673–2680. doi: 10.1002/ijc.25618. [DOI] [PubMed] [Google Scholar]
- 9.Jokerst JV, Miao Z, Zavaleta C, Cheng Z, Gambhir SS. Affi body-functionalized gold–silica nanoparticles for Raman molecular imaging of the epidermal growth factor receptor. Small. 2011;7:625–633. doi: 10.1002/smll.201002291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kircher MF, et al. A brain tumor molecular imaging strategy using a new triple-modality MRI-photoacoustic-Raman nanoparticle. Nat Med. 2012;18:829–834. doi: 10.1038/nm.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y, Seebald JL, Szeto DP, Irudayaraj J. Biocompatibility and biodistribution of surface-enhanced Raman scattering nanoprobes in zebrafish embryos: In vivo and multiplex imaging. ACS Nano. 2010;4:4039–4053. doi: 10.1021/nn100351h. [DOI] [PubMed] [Google Scholar]
- 12.Samoylova T, Morrison N, Globa L, Cox N. Peptide phage display: Opportunities for development of personalized anti-cancer strategies. Anticancer Agents Med Chem. 2006;6:9–17. doi: 10.2174/187152006774755492. [DOI] [PubMed] [Google Scholar]
- 13.Weissleder R, Ross BD, Rehemtulla A, Gambhir SS. Molecular Imaging: Principles and Practice. People’s Medical Publishing House; 2010. [Google Scholar]
- 14.Hellebust A, Richards-Kortum R. Advances in molecular imaging: Targeted optical contrast agents for cancer diagnostics. Nanomedicine Lond. 2012;7:429–445. doi: 10.2217/nnm.12.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nguyen QT, Tsien RY. Fluorescence-guided surgery with live molecular navigation — A new cutting edge. Nat Rev Cancer. 2013;13:653–662. doi: 10.1038/nrc3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weng KC, et al. Targeted tumor cell internalization and imaging of multifunctional quantum dot-conjugated immunoliposomes in vitro and in vivo. Nano Lett. 2008;8:2851–2857. doi: 10.1021/nl801488u. [DOI] [PubMed] [Google Scholar]
- 17.Choi HS, et al. Renal clearance of quantum dots. Nat Biotechnol. 2007;25:1165–1170. doi: 10.1038/nbt1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang YQ, Yan B, Chen LX. SERS tags: Novel optical nanoprobes for bioanalysis. Chem Rev. 2013;113:1391–1428. doi: 10.1021/cr300120g. [DOI] [PubMed] [Google Scholar]
- 19.Mulvaney SP, Musick MD, Keating CD, Natan MJ. Glass-coated, analyte-tagged nanoparticles: A new tagging system based on detection with surface-enhanced Raman scattering. Langmuir. 2003;19:4784–4790. [Google Scholar]
- 20.Sha MY, Xu H, Natan MJ, Cromer R. Surface-enhanced Raman scattering tags for rapid and homogeneous detection of circulating tumor cells in the presence of human whole blood. J Am Chem Soc. 2008;130:17214–17215. doi: 10.1021/ja804494m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zavaleta CL, et al. Multiplexed imaging of surface enhanced Raman scattering nanotags in living mice using noninvasive Raman spectroscopy. Proc Natl Acad Sci. 2009;106:13511–13516. doi: 10.1073/pnas.0813327106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zavaleta CL, et al. A Raman-based endoscopic strategy for multiplexed molecular imaging. Proc Natl Acad Sci USA. 2013;110:E2288–E2297. doi: 10.1073/pnas.1211309110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leigh SY, Som M, Liu JTC. Method for assessing the reliability of molecular diagnostics based on multiplexed SERS-coded nanoparticles. PLoS ONE. 2013;8:e62084. doi: 10.1371/journal.pone.0062084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lutz BR, et al. Spectral analysis of multiplex Raman probe signatures. ACS Nano. 2008;2:2306–2314. doi: 10.1021/nn800243g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van de Sompel D, Garai E, Zavaleta C, Gambhir SS. A hybrid least squares and principal component analysis algorithm for Raman spectroscopy. Conf Proc IEEE Eng Med Biol Soc. 2011;2011:6971–6974. doi: 10.1109/IEMBS.2011.6091762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qian X, et al. In vivo tumor targeting and spectroscopic detection with surface-enhanced Raman nanoparticle tags. Nat Biotechnol. 2007;26:83–90. doi: 10.1038/nbt1377. [DOI] [PubMed] [Google Scholar]
- 27.Wang X, et al. Detection of circulating tumor cells in human peripheral blood using surface-enhanced Raman scattering nanoparticles. Cancer Res. 2011;71:1526–1532. doi: 10.1158/0008-5472.CAN-10-3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee S, et al. Fabrication of SERS-fluorescence dual modal nanoprobes and application to multiplex cancer cell imaging. Nanoscale. 2012;4:124–129. doi: 10.1039/c1nr11243k. [DOI] [PubMed] [Google Scholar]
- 29.Talley CE, et al. Surface-enhanced Raman scattering from individual au nanoparticles and nanoparticle dimer substrates. Nano Lett. 2005;5:1569–1574. doi: 10.1021/nl050928v. [DOI] [PubMed] [Google Scholar]
- 30.Su X, et al. Composite organic-inorganic nanoparticles (coins) with chemically encoded optical signatures. Nano Lett. 2005;5:49–54. doi: 10.1021/nl0484088. [DOI] [PubMed] [Google Scholar]
- 31.Sun L, et al. Composite organic-inorganic nanoparticles as Raman labels for tissue analysis. Nano Lett. 2007;7:351–356. doi: 10.1021/nl062453t. [DOI] [PubMed] [Google Scholar]
- 32.Bohndiek SE, et al. A small animal Raman instrument for rapid, wide-area, spectroscopic imaging. Proc Natl Acad Sci USA. 2013;110:12408–12413. doi: 10.1073/pnas.1301379110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mohs AM, et al. Hand-held spectroscopic device for in vivo and intraoperative tumor detection: Contrast enhancement, detection sensitivity, and tissue penetration. Anal Chem. 2010;82:9058–9065. doi: 10.1021/ac102058k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McVeigh PZ, Mallia RJ, Veilleux I, Wilson BC. Widefield quantitative multiplex surface enhanced Raman scattering imaging in vivo. J Biomed Optics. 2013;18:046011–046011. doi: 10.1117/1.JBO.18.4.046011. [DOI] [PubMed] [Google Scholar]
- 35.Garai E, et al. High-sensitivity, real-time, ratiometric imaging of surface-enhanced Raman scattering nanoparticles with a clinically translatable Raman endoscope device. J Biomed Optics. 2013;18:096008-1–106008-13. doi: 10.1117/1.JBO.18.9.096008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mallia RJ, McVeigh PZ, Veilleux I, Wilson BC. Filter-based method for background removal in high-sensitivity wide-field-surface-enhanced Raman scattering imaging in vivo. J Biomed Optics. 2012;17:076017. doi: 10.1117/1.JBO.17.7.076017. [DOI] [PubMed] [Google Scholar]
- 37.Maiti KK, et al. Multiplex targeted in vivo cancer detection using sensitive near-infrared SERS nanotags. Nano Today. 2012;7:85–93. [Google Scholar]
- 38.Zavaleta CL, et al. Preclinical evaluation of Raman nanoparticle biodistribution for their potential use in clinical endoscopy imaging. Small. 2011;7:2232–2240. doi: 10.1002/smll.201002317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thakor AS, et al. The fate and toxicity of Raman-active silica-gold nanoparticles in mice. Sci Transl Med. 2011;3:79ra33. doi: 10.1126/scitranslmed.3001963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reibel J. Prognosis of oral pre-malignant lesions: Significance of clinical, histopathological, and molecular biological characteristics. Crit Rev Oral Biol Med. 2003;14:47–62. doi: 10.1177/154411130301400105. [DOI] [PubMed] [Google Scholar]
- 41.Jacobs L. Positive margins: The challenge continues for breast surgeons. Ann Surg Oncol. 2008;15:1271–1272. doi: 10.1245/s10434-007-9766-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jeevan R, et al. Reoperation rates after breast conserving surgery for breast cancer among women in England: Retrospective study of hospital episode statistics. BMJ. 2012;345:e4505. doi: 10.1136/bmj.e4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tichauer KM, et al. Improved tumor contrast achieved by single time point dual-reporter fluorescence imaging. J Biomed Optics. 2012;17:066001. doi: 10.1117/1.JBO.17.6.066001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tichauer KM, et al. In vivo quantification of tumor receptor binding potential with dual-reporter molecular imaging. Mol Imag Biol. 2012;14:584–592. doi: 10.1007/s11307-011-0534-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu JTC, et al. Quantifying cell-surface biomarker expression in thick tissues with ratiometric three-dimensional microscopy. Biophys J. 2009;96:2405–2414. doi: 10.1016/j.bpj.2008.12.3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang TD, et al. Functional imaging of colonic mucosa with a fibered confocal microscope for real-time in vivo pathology. Clin Gastroenterol Hepatol. 2007;5:1300–1305. doi: 10.1016/j.cgh.2007.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmidt M, Hynes NE, Groner B, Wels W. A bivalent single-chain antibody-toxin specific for ERBB-2 and the EGF receptor. Int J Cancer. 1996;65:538–546. doi: 10.1002/(SICI)1097-0215(19960208)65:4<538::AID-IJC24>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 48.Wels W, et al. EGF receptor and p185ERBB-2-specific single-chain antibody toxins differ in their cell-killing activity on tumor cells expressing both receptor proteins. Int J Cancer. 1995;60:137–144. doi: 10.1002/ijc.2910600120. [DOI] [PubMed] [Google Scholar]
- 49.Gaborit N, et al. Time-resolved fluorescence resonance energy transfer (TR-FRET) to analyze the disruption of EGFR/HER2 dimers: A new method to evaluate the efficiency of targeted therapy using monoclonal antibodies. J Biol Chem. 2011;286:11337–11345. doi: 10.1074/jbc.M111.223503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Habib AA, Chun SJ, Neel BG, Vartanian T. Increased expression of epidermal growth factor receptor induces sequestration of extracellular signal-related kinases and selective attenuation of specific epidermal growth factor-mediated signal transduction pathways. Mol Cancer Res. 2003;1:219–233. [PubMed] [Google Scholar]
- 51.Moasser MM, Basso A, Averbuch SD, Rosen N. The tyrosine kinase inhibitor ZD1839 (“iressa”) inhibits HER2-driven signaling and suppresses the growth of HER2-overexpressing tumor cells. Cancer Res. 2001;61:7184–7188. [PubMed] [Google Scholar]
- 52.Van De Sompel D, Garai E, Zavaleta C, Gambhir SS. A hybrid least squares and principal component analysis algorithm for Raman spectroscopy. PLoS ONE. 2012;7:e38850. doi: 10.1371/journal.pone.0038850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chithrani BD, Ghazani AA, Chan WCW. Determining the size and shape dependence of gold nanoparticle uptake into mammalian cells. Nano Lett. 2006;6:662–668. doi: 10.1021/nl052396o. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.