Abstract
Mutations in leucine-rich repeat kinase 2 (LRRK2) are the most common cause of familial and idiopathic Parkinson's disease. However, the mechanisms for activating its physiological function are not known, hindering identification of the biological role of endogenous LRRK2. The recent discovery that LRRK2 is highly expressed in cells of the innate immune system and genetic association is a risk factor for autoimmune disorders implies an important role for LRRK2 in pathology outside of the central nervous system. Thus, an examination of endogenous LRRK2 in immune cells could provide insight into the protein's function. Here, we establish that stimulation of specific Toll-like receptors results in a complex biochemical activation of endogenous LRRK2, with early phosphorylation of LRRK2 preceding its dimerization and membrane translocation. Membrane-associated LRRK2 co-localized to autophagosome membranes following either TLR4 stimulation or mTOR inhibition with rapamycin. Silencing of endogenous LRRK2 expression resulted in deficits in the induction of autophagy and clearance of a well-described macroautophagy substrate, demonstrating the critical role of endogenous LRRK2 in regulating autophagy. Inhibition of LRRK2 kinase activity also reduced autophagic degradation and suggested the importance of the kinase domain in the regulation of autophagy. Our results demonstrate a well-orchestrated series of biochemical events involved in the activation of LRRK2 important to its physiological function. With similarities observed across multiple cell types and stimuli, these findings are likely relevant in all cell types that natively express endogenous LRRK2, and provide insights into LRRK2 function and its role in human disease.
INTRODUCTION
Parkinson's disease (PD) is the second most common neurodegenerative disorder, and mutations in leucine-rich repeat kinase 2 (LRRK2) are the leading cause of both familial and sporadic forms of the disease (1). This large ∼280 kDa protein has multiple functional domains including a Ras of complex (Roc) GTPase, a COR (C-terminal of Roc) domain for protein–protein interactions and a MAPKKK-like kinase domain. While primarily a cytosolic monomer (2,3), there is also a smaller dimeric population of LRRK2 with higher kinase activity located at cellular membranes (2,4,5). Low endogenous LRRK2 expression in neurons has often necessitated ectopic overexpression in immortalized cell lines to gain insight into LRRK2 biology. In addition, there is no current consensus on substrates of LRRK2 kinase activity or its overall function in the cell (6–9). The absence of an obvious neurological phenotype in LRRK2 knockout (KO) animals has further complicated efforts to understand the importance of LRRK2 in disease pathogenesis (10,11) and emphasizes the necessity for studying other relevant and endogenous LRRK2-expressing cell types in order to identify a physiologically and pathologically relevant function of LRRK2.
Recent data from multiple groups indicate that LRRK2 dysfunction within the immune system may be a central component in the development of autoimmune diseases. A genome-wide association study (GWAS) revealed a possible involvement of the LRRK2 gene in the autoimmune disorders Crohn's disease and colitis (12). This involvement was further supported by the observation of increased LRRK2 expression in inflamed colonic tissues from patients suffering from Crohn's disease (13). Furthermore, an analysis of experimental colitis in LRRK2 KO animals revealed exacerbated disease severity when compared with normal animals (14). Thus, dysfunction of LRRK2-dependent processes in immune cells could be a foundation for the development of autoimmune diseases, and insights into these processes may prove relevant to the pathological mechanisms of LRRK2 in the PD brain.
In the immune system, monocytic cells such as dendritic cells, macrophages and microglia display high levels of LRRK2 mRNA and protein (13,15), and stimulation of these cells can induce LRRK2 expression and/or its phosphorylation (16,17). Furthermore, results following pharmacological inhibition of LRRK2 kinase activity during monocyte activation suggest an important role for LRRK2 kinase activity in these cells (18). However, we and others have shown that cytokine expression and release from stimulated LRRK2 KO macrophages are no different from wild-type (WT) macrophages (15,17). Other cellular functions of activated monocytes have been ascribed to LRRK2 as well, including reactive oxygen species generation, phagocytosis and cell migration (13,18,19). However, the lack of consensus across these reports suggests a complexity to LRRK2 signaling in monocytes that requires more attention.
The immunologic stimulation of monocytes involves many well-characterized pathways, making these cells potentially ideal for identifying the means and consequences of activating endogenous LRRK2 in the cell. We previously proposed a model of LRRK2 signaling that predicted a cellular stimulus would result in dimerization and membrane recruitment of LRRK2. This would then result in its activation of its kinase activity and participation in a biological function, likely involving membrane dynamics (2). To test this hypothesis, we employed macrophage and microglia cell lines to determine whether monocyte activation would change the biochemical properties of LRRK2 in the specific context of monocyte biology. Here, we demonstrate that immunologic stimulation of two independent monocyte cell lines resulted in increased endogenous LRRK2 phosphorylation and dimerization, and an increase in total LRRK2 at the membrane. This newly recruited pool of LRRK2 was spatially distinct from the membrane-associated LRRK2 at rest, and co-localized with purified autophagosomes. Importantly, these biochemical changes in LRRK2 could be reproduced through direct induction of mTOR-dependent autophagy. Functional analyses exhibited no likely involvement of LRRK2 in phagocytosis, but an assessment of autophagic activity revealed substantial deficits in both LC3-II conversion and autophagic protein turnover following knockdown of endogenous LRRK2 or pharmacological inhibition of LRRK2 kinase activity. Our data demonstrate that distinct alterations in endogenous LRRK2 biochemistry are associated with its functional role regulating autophagy and that these effects are likely to be relevant in all cells that natively express LRRK2.
RESULTS
Activation of macrophages and microglia induces LRRK2 phosphorylation and membrane translocation
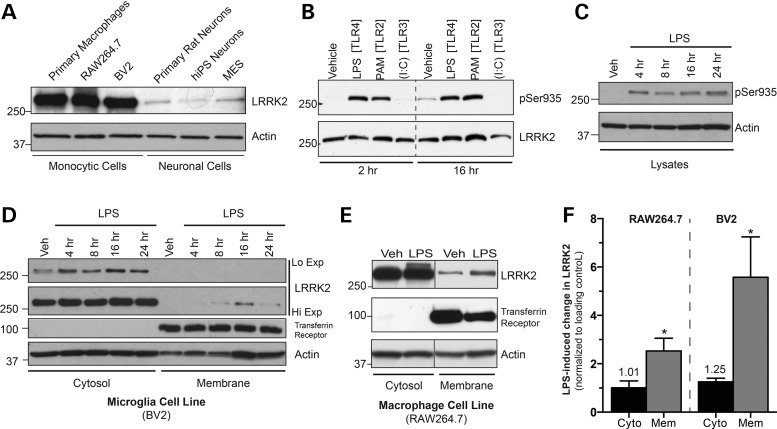
The expression of LRRK2 in various monocytes including primary cultured human macrophages, the murine RAW264.7 macrophage cell line and the murine BV2 microglial cell line was examined and compared with those in neuronal preparations. Expression of LRRK2 was much higher in both monocytic cell lines and primary monocytes compared with neuronal counterparts (Fig. 1A), and notably the two monocyte cell lines (RAW264.7 and BV2) showed comparable endogenous LRRK2 expression to primary cultured human macrophages, validating their suitability as a cellular model of endogenous LRRK2.
Figure 1.
Monocyte activation induces LRRK2 phosphorylation at Ser935 and membrane translocation. (A) Cell lysates from various cell lines and primary cell cultures were immunoblotted for LRRK2 levels. (B) RAW264.7 macrophage cells were treated with TLR agonists LPS (100 ng/ml), PAM3CysSK4 (1 μg/ml) or poly (I:C) (10 μg/ml) for the times listed, followed by western blotting for pSer935 and total LRRK2 levels. (C) BV2 cells were treated with LPS for the shown times, and lysates were immunoblotted for LRRK pSer935 antibody. (D) BV2 cells treated with LPS for different time points and fractionated in the absence of detergent. Cytosolic and membrane fractions were immunoblotted separately to determine LRRK2 location during TLR4 stimulation. (E) Cellular fractionation was performed on RAW264.7 cells following treatment with vehicle or LPS for 16 h. Equal amounts of cytosol and membrane fractions immunoblotted for LRRK2. (F) Quantification of LPS-induced membrane LRRK2 changes compared with changes in cytosolic LRRK2 for RAW264.7 and BV2 cells at 16 h LPS treatment, the time where greatest LRRK2 translocation was observed. Student's t-test from n = 3–5 experiments, *P < 0.05.
Our previous results demonstrated the potential importance of the membrane-associated, dimeric sub-population of LRRK2. Analyses of both exogenous and endogenous LRRK2 suggested that this pool of protein was likely to be the most physiologically relevant form of the protein (2). However, at the time these studies were conducted, there were no means to stimulate endogenous LRRK2 function to formally investigate this prediction. Examining this model in an endogenous setting requires a cell system that can induce biochemical changes in LRRK2 during cellular activation, and we selected two different monocytic cell lines, macrophage (RAW264.7) and microglial (BV2) cells. Activation of monocytes was previously shown to induce expression and phosphorylation of endogenous LRRK2, believed to be an indicator of LRRK2 activation (20–22). Thus, these cells were ideal for testing the hypothesis that membrane recruitment and dimerization of LRRK2 is important for its physiological function.
As expected, stimulation of Toll-like receptors 2 and 4 (TLR2, TLR4) in RAW264.7 cells resulted in rapid and sustained phosphorylation of Ser935, while TLR3 activation had no effect (Fig. 1B). These observations were in agreement with previous results in macrophage cells (17). While LRRK2 activation was observed following stimulation with the TLR2 agonist PAM3CysSK4, the TLR4 agonist lipopolysaccharide (LPS) was used for subsequent experiments due to its robust and reliable effects on LRRK2 phosphorylation across every system we investigated. To test the conservation of LRRK2 phosphorylation in other monocyte cells, we then measured LPS-induced Ser935 phosphorylation in BV2 cells, using a time course of treatment. As with RAW264.7 cells, phosphorylation occurred quickly and reached a peak after 4 h (Fig. 1C), establishing that Ser935 phosphorylation occurred in both of the monocytic lines.
At this time, it remained unclear if LRRK2 membrane recruitment could be induced with TLR4 activation of monocytes. Using our previous LPS time course, we analyzed the levels of LRRK2 in both the cytosol and membrane components of BV2 cells. Modest translocation was observed as early as 8 h, but the effect was most significant at 16 h post-LPS treatment (Fig. 1D). Treating with RAW264.7 cells with LPS for 16 h reproducibly increased LRRK2 at the membrane (Fig. 1E) and validated the use this time point for subsequent experiments. A statistical analysis was performed on both cell types, examining the LPS-induced change in membrane LRRK2 compared with changes in cytosolic LRRK2 across multiple independent experiments. Of note, both cells had a significant increase in membrane LRRK2, with BV2 cells demonstrating a greater effect (Fig. 1F). Thus, immunologic stimulation of monocytes induced LRRK2 phosphorylation prior to an increase in its membrane recruitment.
TLR4 activation stimulates LRRK2 dimerization at the membrane
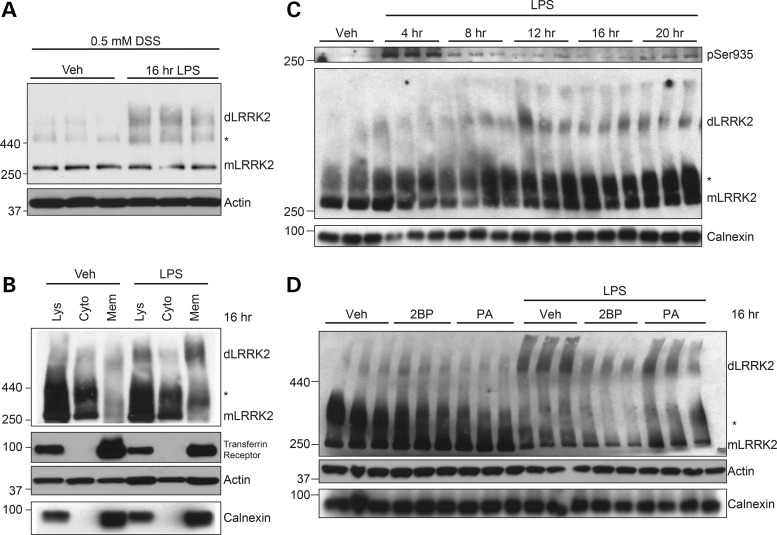
LRRK2 can exist in both a monomeric and dimeric form, with the dimer representing a population that is minor yet more active than its monomeric counterpart (2,5,23). However, conditions that dynamically promote LRRK2 dimerization have not been previously reported. To determine whether monocyte activation results in LRRK2 dimerization, we captured LRRK2 dimers via chemical live-cell crosslinking with disuccinimidyl suberate (DSS). Whenever possible, both actin and calnexin, two proteins not affected by DSS crosslinking, were used as loading controls. Activation of RAW264.7 (data not shown) and BV2 (Fig. 2A) cells with LPS increased the total levels of captured dimeric LRRK2. Subcellular fractionation in resting and LPS-stimulated RAW264.7 cells was conducted following crosslinking to reveal the location of these newly formed dimers. Substantial LPS-induced LRRK2 dimers were found within the cytosol, but a greater fold-change in LPS-induced LRRK2 dimerization was observed at the membrane compared with the cytosol (Fig. 2B). Next, the temporal sequence of these events was determined. In RAW264.7 cells, we observed that peak Ser935 phosphorylation preceded dimerization of LRRK2 by several hours (Fig. 2C), temporally associating dimerization with membrane translocation (Fig. 1). Of note, the RAW264.7 cells showed slight differences in the time course of LRRK2 phosphorylation compared with BV2 cells (Fig. 1C), but the peak phosphorylation observed at 4 h was very similar across the two cell types.
Figure 2.
Membrane-bound LRRK2 dimers form after TLR4 activation. (A) BV2 cells were treated with vehicle or LPS for 16 h prior to live-cell crosslinking with DSS (500 μm) to capture LRRK2 homodimers. Band marked with an asterisk is a high-molecular weight LRRK2 species we have previously reported (2,15). Calnexin and actin were both used as loading controls when possible. (B) RAW264.7 cells were treated with vehicle or LPS for 16 h, prior to live-cell crosslinking with DSS. Cytosol and membrane fractions were isolated and immunoblotted to determine location of TLR4-induced LRRK2 dimers. (C) RAW264.7 cells were treated for times indicated prior to crosslinking with DSS and cell lysis, detecting LRRK2 dimers following immunoblotting with LRRK2 antibody. (D) RAW264.7 cells were treated with vehicle, 2BP (100 μm) or palmitic acid (100 μm), with or without LPS for 16 h. LRRK2 dimers were detected from total cell lysates.
While not fully understood, many proteins can be post-translationally modified via the addition of a palmitoyl group at free cysteine residues, a process thought to increase membrane association (24). The software CSS-PALM 2.0 can predict the likelihood of a palmitoylation sequence in a protein (25), and results compiled from this tool suggested an N-terminal sequence in LRRK2 as a potential target (data not shown). In an attempt to prevent endogenous LRRK2 membrane recruitment and dimer formation, LPS-stimulated RAW264.7 cells were co-treated with 2-bromopalmitate (2BP), an inhibitor of palmitoyl transferase. Palmitic acid, a fatty acid that does not inhibit the enzyme, was used as a negative control. Co-treatment with 2BP inhibited LPS-induced LRRK2 dimerization, while palmitic acid had no effect (Fig. 2D), indicating that palmitoylation of LRRK2 may be required for dimer formation. While successful, 2BP was not used for functional analysis, as other targets of palmitoylation could be affected in addition to LRRK2 and confound data interpretation. Nonetheless, these data indicate that the membrane recruitment and dimerization of LRRK2 may be concomitant during activation of monocytes.
LRRK2 does not regulate phagocytosis in monocytes
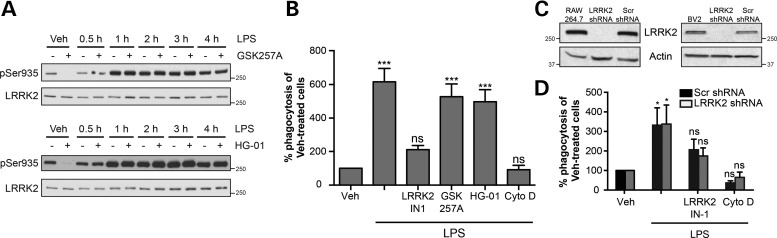
Our results have shown that monocyte activation resulted in the membrane recruitment of LRRK2. To test if this activated population of LRRK2 had a function in monocytes, we examined monocyte phagocytosis as a putatively relevant cellular process that is known to involve dynamic changes in membrane organization. Phagocytosis is a key consequence of monocyte activation, involving the engulfment of extracellular particles at the plasma membrane. Additionally, LRRK2 has been implicated as both a mediator of phagocytosis in microglia and endocytotic mechanisms at synaptic membranes (6,19,26). To examine the role of LRRK2 in TLR4-activated RAW264.7 cells, we tested the effects of two newly-developed LRRK2 inhibitors GSK2578215A and HG-10-102-01 on LPS-induced LRRK2 phosphorylation (27,28). Western blot analysis revealed that the LRRK2 inhibitors prevented phosphorylation at Ser935 in monocytes at rest but were unable to prevent phosphorylation that resulted from TLR4 receptor activation (Fig. 3A), a finding observed in a previous report that employed the well-studied inhibitor LRRK2-IN-1 (17). This was our first indication that kinase inhibition may not be a suitable approach for the prevention of LRRK2 function in TLR4-activated monocytes. We verified this hypothesis by attempting to alter TLR-induced phagocytosis using both GSK2578215A and HG-10-102-01, as well as LRRK2-IN-1, previously shown to prevent the uptake of HIV Tat protein in BV2 cells (19). Co-treatment with LRRK2 inhibitors and LPS for 16 h revealed that LRRK2-IN-1 significantly reduced LPS-induced phagocytosis of FITC-conjugated beads in RAW264.7 cells, in a manner comparable to phagocytosis inhibitor cytochalasin D. However, neither GSK2578215A nor HG-10-102-10 had a significant effect on phagocytosis (Fig. 3B). Similar results were observed in BV2 cells (data not shown).
Figure 3.
LRRK2 is not required for macrophage phagocytosis. (A) RAW264.7 cells were co-treated with LPS and LRRK2 inhibitors GSK2578215A (GSK257A, 1 μm) and HG-10-102-01 (HG-01, 1 μm) for the times shown. Total cell lysates were analyzed for pSer935 levels. (B) Cells were treated with LPS with or without LRRK2 inhibitors (1 μm for each) for 16 h, prior to media change and addition of FITC-conjugated beads. The phagocytosis inhibitor cytochalasin D (Cyto D) was used as a control. Flow cytometry was used to measure degree of phagocytotic activity for each cellular treatment. Data are presented as a percentage of the fluorescent intensity observed in vehicle-treated cells. Data were analyzed by one-way ANOVA, using a Dunnett's post hoc test to compare experimental groups to vehicle treated cells. Graph displays results of three to five independent experiments (***P < 0.001). (C) shRNA lentiviral constructs were used to make stable BV2 and RAW264.7 cells with reduced LRRK2 expression. LRRK2 shRNA was used to lower LRRK2 expression, and a scrambled protein sequence (Scr) was used as a control. (D) RAW264.7 LRRK2 KD cells and RAW264.7 cells with scrambled protein (Scr) were treated with LPS, with and without LRRK2-IN-1, and phagocytosis was measured as previously. One-way ANOVA with Dunnett's post hoc for each cell type was used as in (B; P < 0.05).
The lack of shared effects across multiple LRRK2 inhibitors necessitated the development of additional approaches to assess LRRK2's involvement in monocyte phagocytosis. Therefore, we generated stable LRRK2 knockdown cell lines (LRRK2 shRNA) using commercial shRNA lentiviral constructs. Endogenous LRRK2 expression in our KD cell line was virtually undetectable, while a cell line stably expressing a scramble shRNA sequence (Scr shRNA) had normal protein expression (Fig. 3C). The degree of knockdown was similar in both RAW264.7 and BV2 cells. To further investigate the putative importance of LRRK2 to monocyte phagocytosis, RAW264.7 LRRK2 shRNA or Scr shRNA cells were treated with LPS, with or without LRRK2-IN-1, the only LRRK2 inhibitor that affected phagocytosis in our hands. The dramatically reduced LRRK2 expression in LRRK2 shRNA cells did not change the phagocytotic capacity of these cells, compared with Scr shRNA control. However, LRRK2-IN-1 prevented phagocytosis, even in the LRRK2-deficient knockdown cells (Fig. 3D), indicating that the LRRK2-IN-1 effect on phagocytosis was not LRRK2 specific. These results demonstrating substantial off-target effects of LRRK2-IN1 are in agreement with a recent study observing that this inhibitor had significant off-target effects (29), and that care should be taken when pharmacologically inhibiting LRRK2 kinase activity. More critically, these data show that LRRK2 is not involved in monocyte phagocytosis.
Monocyte stimulation prompts redistribution of LRRK2 to unique membranes
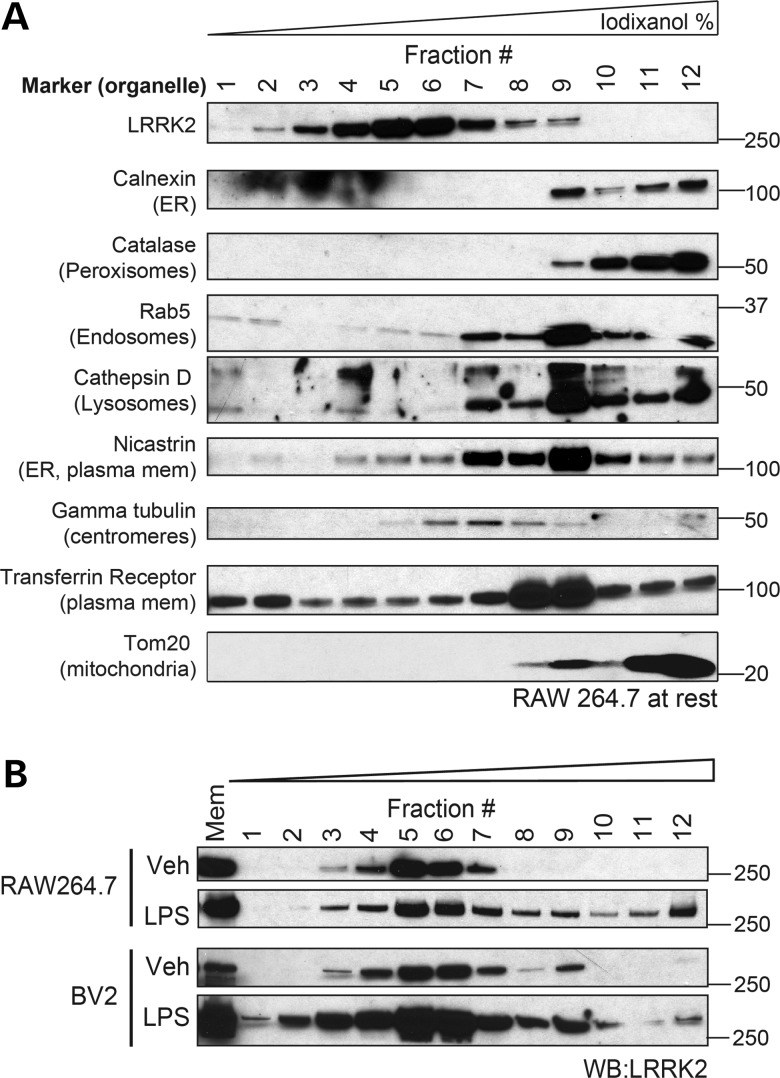
Having failed to identify a role for LRRK2 in monocyte phagocytosis, we turned back to the biochemical changes in LRRK2 we observed following LPS stimulation with the hypothesis that further details of membrane-associated LRRK2 might inform its function. The specific cellular location of membrane-associated LRRK2 has been heavily debated, as LRRK2 has been associated with numerous organelles including endosomes, mitochondria, synaptic vesicles and lipid rafts (30–33). To elucidate the location of membrane LRRK2 in monocytes, native iodixanol ultracentrifuge gradients were used to separate isolated membranous vesicles, prior to immunoblotting with different organelle markers. LRRK2 from unstimulated RAW264.7 cells consistently migrated to the middle of the iodixanol gradient (fractions #5–7), in a region that did not correlate with a specific and isolated subcellular organelle marker (Fig. 4A) and may be consistent with its reported colocalization with numerous subcellular structures in unstimulated cells (32–34). Similar data were obtained from BV2 cells (data not shown). We then analyzed the effects of immune cell activation on LRRK2 membrane location by separating membranes from LPS-activated RAW264.7 and BV2 cells. The stimulation of both BV2 and RAW264.7 cells resulted in the novel appearance of a population of LRRK2 in fractions #9–12, locations where LRRK2 was extremely low or below detection in resting cells (Fig. 4B). These data demonstrate broadly that monocyte stimulation recruits LRRK2 to a membrane compartment distinct from that of basal membrane LRRK2, a location at which it is likely to mediate a specific cellular function.
Figure 4.
Monocyte activation causes recruitment of LRRK2 to a novel membrane compartment. (A) RAW264.7 cells were fractionated in a sucrose buffer to obtain cellular membranes and separated by native iodixanol ultracentrifugation gradients. Equal amounts of each gradient fraction were mixed in sample buffer prior to immunoblot analysis. (B) RAW264.7 cells were treated with vehicle or LPS for 16 h, and membranes were isolated from cell lysates and separated with iodixanol gradients. Immunoblot analysis found LRRK2 in fraction #12 in both RAW264.7 and BV2 cells following LPS treatment.
TLR4 simulation and direct induction of autophagy result in similar LRRK2 membrane translocation
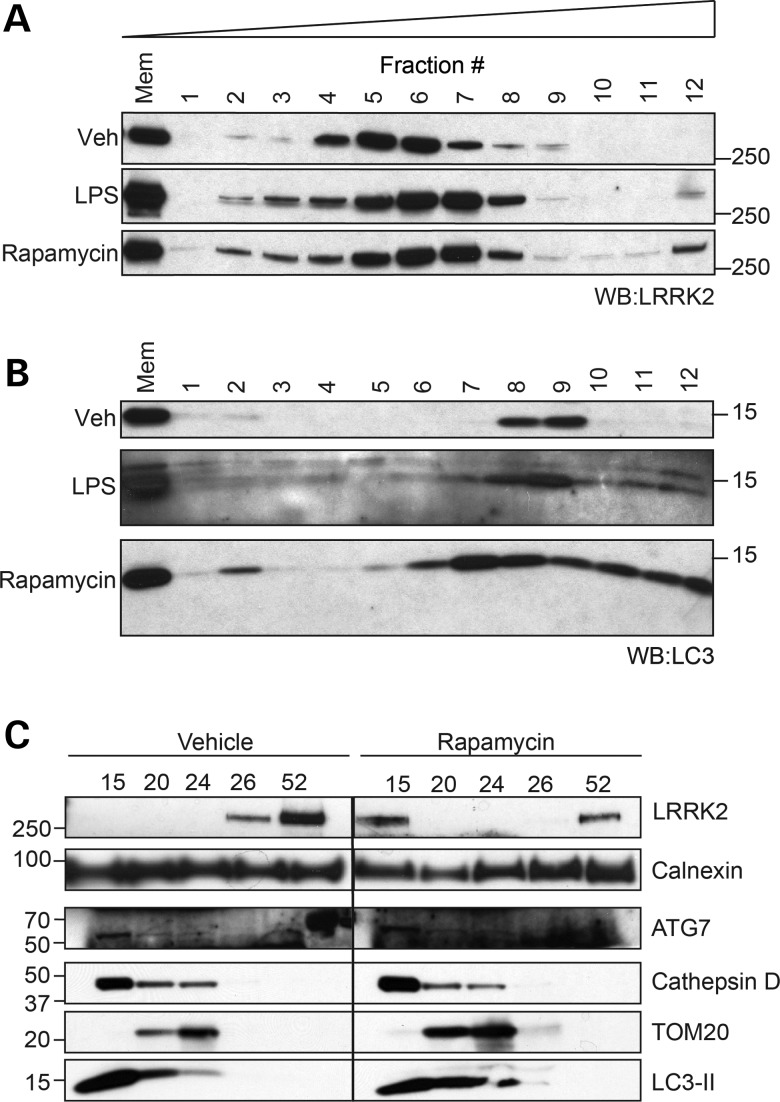
The process of macroautophagy (henceforth referred to as autophagy) involves the formation of double membrane vesicles called autophagosomes, designed for the engulfment and subsequent shuttling of cellular material for lysosomal degradation. This is a major biological function of both macrophages and microglia, called upon to degrade extracellular debris or microorganisms following internalization by phagocytosis in order to maintain immune system or central nervous system (CNS) integrity, respectively (35–39). Upregulation of autophagy is a major consequence of TLR-induced phagocytosis following monocyte activation (40), and migration of LRRK2 to membranes may be required for this process. To examine the biochemical responses of endogenous LRRK2 during monocyte autophagy, RAW264.7 cells were treated with either LPS or the mTOR inhibitor rapamycin, a well-studied and specific inducer of autophagy. Membranes from LPS- and rapamycin-treated RAW264.7 cells were isolated and analyzed by the iodixanol gradient separation technique previously described (41). Direct induction of autophagy with rapamycin caused a similar change in the LRRK2 migration pattern as did TLR4 stimulation (Fig. 5A). A similar response in LRRK2 recruitment with LPS and rapamycin was also observed in BV2 cells (data not shown). In addition, the critical autophagosome component LC3-II migrated to the same fractions as LRRK2 (#9–12) in both LPS and rapamycin-treated cells (Fig. 5B). LC3-II migration during rapamycin treatment suggests that relocalization, as observed in iodixanol gradients, may be a pre-requisite of autophagosome formation and that co-recruitment of LRRK2 may be functionally involved. The fact that direct stimulation of autophagy with rapamycin resulted in greater membrane recruitment of LRRK2 and LC3-II than indirect activation with LPS is also consistent with a primary role for LRRK2 in mTOR-dependent autophagy. These data suggest the potential role for this pool of newly recruited endogenous LRRK2 at cellular membranes to regulate autophagy.
Figure 5.
TLR4 stimulation and mTOR-dependent autophagy alters LRRK2 biochemistry. (A) RAW264.7 cells were treated with LPS or rapamycin (1 μm) for 16 h prior to separation of membrane components by ultracentrifugation, as in Figure 3. (B) RAW264.7 cells were treated and isolated as in (A), immunoblotting for LC3-II. (C) Autophagosomes were isolated from vehicle- and rapamycin-treated BV2 cells using stepwise Histodenz gradients. Equal volumes of membrane fractions concentrating at gradient interfaces were separated by western blot before analyzing with listed markers.
To further establish a physical association between LRRK2 and autophagic membranes, autophagosomes were isolated from the crude membrane fractions of vehicle- and rapamycin-treated BV2 cells using Histodenz ultracentrifugation gradients (42). Western blot analysis of membrane fractions from untreated BV2 cells revealed the co-localization of autophagy and lysosome markers in the interface above the 15% fraction (henceforth referred to as the 15% fraction; Fig. 5C), but no LRRK2 was observed in this location. However, rapamycin treatment caused LRRK2 to appear in the 15% fraction with LC3-II, ATG7 and cathepsin D. Membrane markers calnexin and TOM20, two proteins that were found in the same iodixanol gradient fraction as ‘activated’ LRRK2 (#12), had a different migration pattern than LRRK2, further demonstrating the selective co-enrichment of LRRK2 and autophagosome.
Endogenous LRRK2 regulates autophagic flux in monocytes
The observation that LRRK2 was co-recruited with LC3-II was a strong indicator that LRRK2 plays a functional role in monocyte autophagy. These data are supported by multiple studies in diverse cell types, including EM analysis localizing exogenous LRRK2 to autophagosome membranes (43–48). To address the role of endogenous LRRK2 in monocyte autophagy, we employed stable LRRK2 KD macrophage and microglial cell lines. Rapamycin was used to directly stimulate autophagy in all subsequent experiments.
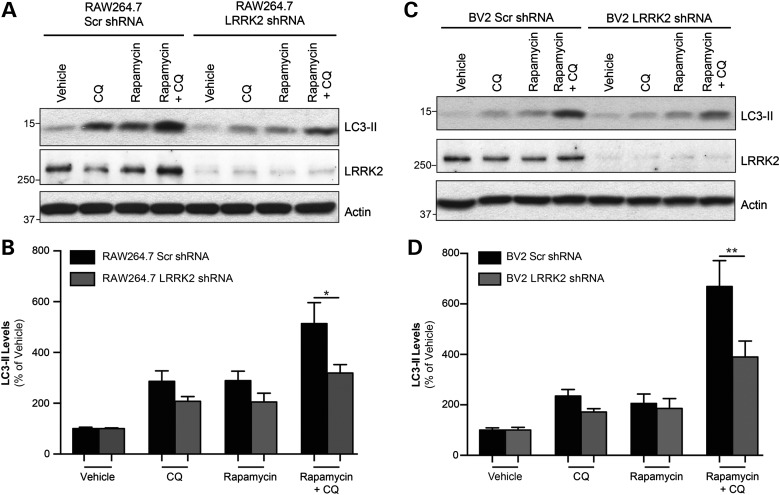
Quantification of LC3-II is a common biochemical measure of autophagy, as it is essential for autophagosome formation (49). However, autophagy is a dynamic process that also results in the rapid degradation of autophagosome proteins including LC3-II, making analysis difficult to interpret. Therefore, inhibition of lysosomal degradation during experimental treatment is often required to prevent LC3-II turnover for an accurate measure of autophagic flux, described as the net increase in autophagic activity within a cell (50). The lysosomal inhibitor chloroquine (CQ) alkalizes lysosomal compartments and thus inhibits protein degradation, and is commonly used to assess autophagic flux (49). For our purposes, autophagic flux was determined by comparing LC3-II levels from rapamycin and CQ co-treated cells with cells treated with CQ alone, giving us the net increase in rapamycin-induced autophagy (49,50) These guidelines were used to examine the contribution of endogenous LRRK2 during induction of autophagy by assessing LRRK2 shRNA and Scr shRNA RAW264.7 and BV2 cells.
Treatment of RAW264.7 Scr shRNA cells with CQ alone increased LC3-II levels by preventing its lysosomal degradation, reflecting the basal autophagic flux in unstimulated control cells (Fig. 6A). The induction of autophagy with rapamycin further increased LC3-II as expected and these levels were greatest with CQ co-treatment. This CQ-captured increase demonstrated the rapamycin-induced autophagic capacity of these control cells. A comparison of LC3-II levels from rapamycin + CQ-treated Scr cells to the same treatment in LRRK2 shRNA cells revealed that LRRK2 knockdown significantly reduced the autophagic capacity of the macrophages (Fig. 6B). This function of endogenous LRRK2 was conserved in BV2 cells, as evidence by the autophagic deficits observed when comparing Scr shRNA and BV2 LRRK2 shRNA cells. As expected, a significant increase in LC3-II levels observed in rapamycin + CQ-treated BV Scr shRNA cells was not matched in BV2 LRRK2 shRNA cells (Fig. 6C and D). These observations demonstrate an influential role for endogenous LRRK2 in the regulation of autophagy and suggest that LRRK2-deficient monocytes would manifest difficulties in removing autophagic substrates. As PD is a disease primarily affecting the CNS, we were especially interested to see if LRRK2-dependent autophagy is important in microglial cells, the resident immune cells of the brain, which play an important role in the clearance of protein aggregates within the brain and are found activated in affected regions of the PD brain (51,52). Thus, the autophagic capacity of LRRK2-deficient BV2 cells warranted further investigation using an independent methodology.
Figure 6.
Knockdown of endogenous LRRK2 decreases LC3-II levels and flux in monocytes. (A) Representative blot of RAW264.7 Scr and LRRK2 KD cells treated with rapamycin for 16 h to activate autophagy, with some cells being treated CQ (20 μm) for the last 6 h to prevent autophagic degradation of LC3-II for analysis of flux. (B) Analysis of LC3-II levels determined by densitometry, expressed as a percentage of vehicle-treated Scr cells. Graph is a compilation of three independent experiments, performed in duplicate (n = 6; *P < 0.05). (C) Representative blot of BV2 Scr and LRRK2 KD cells, treated as in (A). (D) Histogram of LC3-II levels in BV2 cells, derived from three independent experiments, performed in duplicate (n = 6; **P < 0.01).
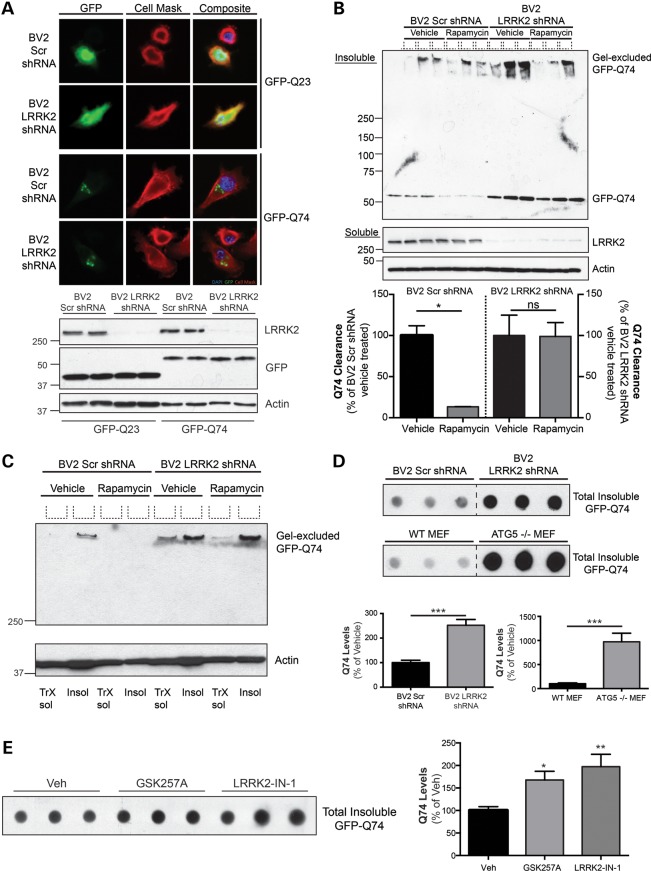
To examine the defects in LRRK2-directed protein degradation in microglia, we assessed whether BV2 LRRK2 shRNA cells had impairments in the ability to remove a well-studied experimental protein aggregate. The GFP-Q74 construct is a research tool derived from a mutation in exon 1 of the gene huntingtin, which expresses a pathogenic polyglutamine expansion that forms protein aggregates in vitro (53). This protein is a target of autophagic degradation and has been successfully employed to quantify rapamycin-induced autophagic flux in cell-based systems (54–56). BV2 Scr and LRRK2-deficient cells were transiently transfected with the GFP-Q74 construct to assess LRRK2-mediated clearance of Q74 aggregates. A GFP-Q23 construct that produces a soluble cytosolic protein derivative of the Q74 protein was used as a transfection control. Microscopic analysis of the GFP-tagged proteins revealed that Q23 expression demonstrated a diffuse cytosolic distribution throughout the cell, while Q74 protein exhibited as distinct puncta indicative of protein aggregation (Fig. 7A, top panel). Western blot analysis revealed that soluble Q23 and Q74 protein expression were equal following transient transfection between stable cell lines, indicating comparable transfection efficiencies (Fig. 7A, bottom panel). A biochemical methodology was employed to specifically examine differences in insoluble GFP-Q74, as defined by the levels of protein resistant to extraction in 1% Triton x-100. Following Q74 transfection, cells were treated with vehicle or rapamycin, and proteins from BV2 Scr shRNA and LRRK2 shRNA cells were sequentially extracted in 1% Triton X-100 followed by solubilization in 2% SDS, to obtain soluble and insoluble proteins, respectively. High levels of GFP-Q74 protein were found in the SDS-extracted pellet of BV2 Scr cells, consistent with its propensity to aggregate (Fig. 7B). Furthermore, treatment of Q74-transfected BV2 Scr shRNA cells with rapamycin significantly reduced this GFP-Q74 pool, indicating that these aggregates could be removed through mTOR-mediated autophagy. However, there was significantly more insoluble Q74 protein in BV2 LRRK2 shRNA cells, and rapamycin had no effect on protein clearance (Fig. 7B). Once again, this demonstrates that LRRK2-deficient microglia have significant impairments in autophagy and protein degradation.
Figure 7.
LRRK2 contributes to autophagic turnover of aggregated protein in microglial cells. (A) Representative images of BV2 Scr and LRRK2 shRNA cells demonstrating the expression pattern of GFP-tagged proteins. GFP-Q23, a native derivate of Q74, was used as a control for protein expression. Western blot of soluble GFP-Q23 and GFP-Q74 demonstrates equality of transfection efficiency between Scr and LRRK2 shRNA cell lines. (B) BV2 Scr and LRRK2 KD cells were transiently transfected with GFP-Q74 construct for 24 h, with wells getting either vehicle or rapamycin (1 μm) for the last 12 h. Cells were sequentially extracted with 1% Triton X-100, to obtain soluble proteins, followed by 2% SDS, to isolate insoluble proteins. Insoluble protein solutions were protein normalized and separated by SDS–PAGE electrophoresis prior to western blotting using a GFP-antibody. Soluble proteins were also separated and immunoblotted for LRRK2 and actin as a loading control. Q74 clearance was quantified by densitometry and analyzed by one-way ANOVA (*P < 0.05). (C) An example of high molecular weight aggregates (‘gel-excluded GFP-Q74’) following 1% Triton X-100 extraction (‘TrX sol’) or 2% SDS extraction (‘insol’). BV2 Scr shRNA and LRRK2 shRNA cells were transiently transfected with Q74 for 24 h and then treated with vehicle or rapamycin (1 μm, last 12 h of 24 h transfection) prior to immunoblotting. (D) BV2 and MEF cell lines were transfected with Q74 constructs for 24 h prior to total protein extraction with 2% SDS. Cell lysates normalized for protein were spotted onto PVDF membrane prior to immunoblotting for insoluble GFP-Q74. Quantification is taken from three separate experiments, performed in triplicate (n = 9, Student's t-test, ***P < 0.0001). (E) BV2 cells were transfected with GFP-Q74 protein for 12 h prior to treatment with vehicle, GSK257A (1 μm) or LRRK2-IN-1 (1 μm). Following treatment, cells were sequentially lysed as previously. Quantification is from three separate experiments, each performed in triplicate (n = 9, one-way ANOVA, Dunnett's post hoc; *P < 0.05; **P < 0.01).
Interestingly, a sub-population of high molecular weight GFP-Q74 aggregate was also detected in both Scr and LRRK2 shRNA cells (Fig. 7B, ‘gel-excluded GFP-Q74’). The detection of this population was variable, likely due to its extremely large size, and caused difficulties in resolution and quantification with traditional SDS–PAGE techniques. There was one instance where the gel-excluded Q74 bands resolved well. Here, the gel-excluded Q74 was found almost exclusively in the insoluble pellet (‘insol’) of BV2 Scr shRNA cells, but was found in both the soluble and insoluble fraction from the LRRK2 KD cells (Fig. 7C). In addition, rapamycin treatment of the control cells resulted in clearance of this protein aggregate but little to no effect in the LRRK2 KD cells (Fig. 7C). While these data further confirmed that reductions in LRRK2 expression confer a deficit in the execution of autophagy, the detection of these two Q74 populations, and the variations in the degree of their separation by SDS–PAGE, required a different approach. Therefore, we employed semi-quantitative dot-blotting of the insoluble fractions, as this method does not suffer from unpredictable electrophoresis of insoluble aggregates of variable size. Dot blot analysis of the insoluble GFP-Q74 from control and LRRK2 knockdown BV2 cells confirmed a significant accumulation of GFP-Q74 in the LRRK2 shRNA cells, compared with control (Fig. 7D, top panel). To confirm that accumulated GFP-Q74 was an indication of autophagy deficits, we analyzed WT and ATG5 KO MEFs, cells totally deficient in autophagy (57). Results showed that insoluble GFP-Q74 was dramatically accumulated in the ATG5 KO cells, compared with its respective control (Fig. 7D, bottom panel). Despite these being different cell types, we observed comparable transfection efficiencies across the BV2 and MEF lines (data not shown). As expected, the increase in insoluble Q74 levels were greater in ATG5-deficient cells than in LRRK2 KD cells, which suggests that unlike ATG5, endogenous LRRK2 is not required for but is an important positive regulator of autophagy.
Reduced LRRK2 expression resulted in autophagic deficits, but it was important to further address the impact of LRRK2 kinase activity on the degradation of insoluble. The LRRK2 inhibitors GSK2578215A or LRRK2-IN-1 were added to BV2 cells transfected with GFP-Q74 constructs (Fig. 7E). Consistent with the effects of silencing LRRK2 expression, both LRRK2 kinase inhibitors demonstrated a similar effect, significantly increasing insoluble Q74 levels compared with vehicle-treated cells (Fig. 7E). These data confirm that LRRK2 kinase activity, at least in part, plays an important role in the LRRK2-dependent regulation of autophagy and the clearance of aggregated proteins.
DISCUSSION
Kinases are typically quiescent proteins that require a specific stimulus to initiate their activity and subsequent role in the cell. Functional analysis of the PD-linked leucine-rich repeat kinase 2 (LRRK2) has been hampered by a poor understanding of the physiological mechanisms that promote its activity. The discoveries that LRRK2 is a risk factor for autoimmune diseases, and that immune cells have significant expression and activity of LRRK2, indicate an unbeknownst role warranting further investigation (12,13,15,18). This study demonstrates, for the first time, that monocyte activation induces the dimerization of endogenous LRRK2, its recruitment to autophagosome-rich membranes, and that these biochemical events are likely important in the LRRK2-dependent regulation of autophagic flux and protein degradation.
At rest, membrane-associated LRRK2 may be found evenly distributed across many organelles and subcellular compartments (33,34,58,59). However, our data reveal that the activation of endogenous LRRK2 resulted in a modest, but significant, increase in its accumulation at cellular membranes (Fig. 1). Further examination suggested that this newly recruited pool of LRRK2 coincided with the novel appearance of LRRK2 physically associated with autophagosomes (Fig. 5C). These data are consistent with those obtained following investigation of exogenous LRRK2 expression (32) and have meaningful implications for both the magnitude of LRRK2 recruitment and the temporal nature of these effects. Autophagosomes comprise <2% of a cell's organic mass (60,61), and thus represent only a fraction of total cellular membrane. Therefore, if the nearly 2- to 5-fold increase in membrane-associated LRRK2 is occurring exclusively at these structures, as demonstrated via two distinct biochemical separation techniques, this would represent a massive increase in LRRK2 levels at these organelles. Therefore, while this newly recruited pool of membrane-associated LRRK2 represents only a small fraction of total LRRK2 in the cell, these biochemical data support the profound functional impact of LRRK2 on autophagy observed in LRRK2-deficient cells and cells treated with LRRK2 kinase inhibitors.
Phosphorylation of the requisite autophagy cofactor, Beclin-1, is a precursor to its association with early autophagosome membranes (62,63). Furthermore, despite the rapid phosphorylation (<1 h) of Beclin-1, induction of autophagic flux can continue to increase for more than 24 h later (49,50). Both the early phosphorylation of Beclin-1 and the latent induction of autophagy parallel our observations regarding LRRK2 phosphorylation and its subsequent association with autophagosome membranes. Unlike Beclin-1, however, our data and others may suggest a physiological role for dimerization of LRRK2 (2,23). LRRK2 is a unique protein in that it contains two functional elements, both a GTPase and a kinase domain. These features, along with its membrane recruitment during activation, may make the behavior of LRRK2 somewhat similar fusion of the GTPase Ras and kinase Raf, where Raf kinase signaling requires the membrane recruitment and dimerization of its cofactor, Ras (64). Future work may elucidate the mechanistic and temporal relationships between LRRK2 dimerization and its membrane association.
Despite being members of the monocyte family and having common functions in their respective organs, macrophages and microglia have noticeable differences in their protein profile and cellular phenotype (65). While largely parallel, our data did reveal subtle differences in LRRK2-related activation between the two cell lines used here. A quick rise and fall (∼4 h) in LRRK2 phosphorylation was often observed in RAW264.7 cells (Fig. 2C), whereas BV2 cells typically showed a more sustained LRRK2 phosphorylation in response to LPS (Fig. 1C). Additionally, we observed a trend for increased LRRK2 expression in stimulated BV2s that was usually absent in the RAW264.7 cell line (Fig. 1B and C). These two cell lines also differed in their capacity to upregulate autophagic flux, with RAW264.7 cells having an ∼5-fold increase in rapamycin-induced autophagy and BV2s having an ∼7-fold increase (Fig. 6B and D). However, the biochemical events and functional role of LRRK2 were similar, with both lines demonstrating a significant increase in membrane-associated LRRK2 and increased dimerization after stimulation. Furthermore, autophagic deficits upon LRRK2 knockdown were seen in both cell types, suggesting that the regulation of autophagy by LRRK2 is a canonical function of the protein.
A role for LRRK2 in regulating autophagy has been suggested previously. However, prior work was based predominantly on the overexpression of WT or PD-linked pathogenic mutant LRRK2 constructs in cell systems with little to no endogenous LRRK2 expression. In most cases, exogenous LRRK2 expression resulted in an increase in autophagy, as determined by LC3-II levels (32,43,46), consistent with the positive regulation of endogenous LRRK2 observed here. Evaluation of endogenous LRRK2 in fibroblasts and astrocytes also suggests a similar role, but contradictory results in some of these studies have made interpretation difficult (44,47,48). Our data confirm the importance of LRRK2 expression in autophagy and represent the first demonstration of endogenous LRRK2-regulated autophagy in both macrophage and microglial cells. In addition, our data linking LRRK2 kinase activity to the positive regulation of autophagy closely match the autophagic deficits in the kidney of LRRK2 KO mice (10), and increased autophagic flux in cells expressing the LRRK2 G2019S mutation, which is known to possess greater kinase activity than WT LRRK2 (2,43,44). Together, these data suggest the organism-wide importance of endogenous LRRK2 in the regulation of autophagy.
The tight regulation of autophagy is critical for maintaining normal cellular homeostasis and either excessive or deficient autophagy can result in tissue pathology and disease. For example, reduced autophagy is thought to be a deleterious consequence of aging and potential contributor to PD pathogenesis (66), while excessive autophagy can lead to cardiomyopathy and heart failure (67). Similarly, both excessive and diminished LRRK2 activity can result in dysregulation of autophagy that could contribute to pathology across multiple organ systems. The G2019S LRRK2 mutation, which increases LRRK2 kinase activity, reduces neurite outgrowth in both cell culture and transgenic animal models (68–70), and this effect is thought to partly involve dysregulation of autophagy (43). Consistent with a broad role for LRRK2 in the regulation of autophagy in multiple organs, LRRK2 KO mice display a severe age-dependent kidney pathology that has been linked to reduced autophagy (10,71). LRRK2 KO mice rats also show pathology associated with decreased surfactant secretion in the lung (72), a process involving a specialized lysosome structure called the lamellar body, which may be related to decreased autophagolysosomal degradation of Q74 observed here. These examples demonstrate that a loss or gain of LRRK2 function likely results in a pathological dysregulation of autophagy with significant consequences in tissues that natively express the LRRK2 protein. For example, deficits in monocyte autophagy have been pathogenically linked to Crohn's disease and colitis (73,74). Relevant to the present data, GWAS studies have linked LRRK2 to these disorders (12) and LRRK2 KO animals demonstrate exacerbated colonic inflammation in an experimental model of colitis (14). The common thread connecting the impaired neurite outgrowth in neurons, age-related kidney dysfunction and colitis-induced colonic pathology in these drastically different tissue systems is the dysregulation of autophagic flux via genetic alterations of LRRK2, and our work and others (43,44,48) directly implicate the kinase activity of LRRK2 in this process.
While these data and others strongly argue that the dominant physiological function of LRRK2 is the regulation of autophagic flux, how this relates to the etiology of LRRK2-dependent PD remains unclear. The intraneuronal accumulation of insoluble α-synuclein into Lewy bodies and Lewy neurites is the pathological hallmark of idiopathic PD (75), and is also characteristic of LRRK2 cases (76) However, multiple lines of experimental evidence suggest that LRRK2 mutations (e.g. G2019S) would likely increase neuronal autophagy and aggregate clearance, but this speculation has not yet been adequately addressed. The observation that LRRK2 expression levels are higher in monocyte populations than neurons, for example, may indicate a non-autonomous relationship between LRRK2 mutations and PD pathogenesis, as recently suggested (77). This hypothesis is further supported by the potent upregulation of microglial LRRK2 during neuroinflammation (18) and the fact that microgliosis is a well-recognized feature of PD (51). A model for LRRK2-induced glial/neuronal co-pathology in PD is also consistent with contemporary views on amyotrophic lateral sclerosis, where disease-linked genes are expressed in both neurons and glia, and dysfunction in all cell types is thought to ultimately contribute to neurodegeneration (78–80). It is likely that the biological interactions between the pathogenic mutations in LRRK2 and α-synuclein homeostasis are likely more complex than we currently appreciate and the present data may provide insight into how to approach this critical next step.
MATERIALS AND METHODS
Cell culture and cell treatments
RAW264.7 cells (ATCC) were cultured in DMEM media supplemented with 10% FBS, 2 mm glutamine and penicillin/streptomycin. BV2 cells (a generous gift from D. Selkoe) were cultured in RPMI media supplemented with FBS, glutamine and pen/strep, as above. WT and ATG5−/− MEFs (a kind gift from N. Mizushima) were maintained in DMEM media containing 10% FBS, minimum essential amino acids, β-mercaptoethanol (0.0008%), glutamine and pen/strep.
Materials and antibodies
TLR agonists (LPS, PAM3CSK4, poly (I:C)), rapamycin, CQ and protease inhibitors were obtained from Sigma. LRRK2 inhibitor GSK2578215A was purchased from Tocris, while inhibitor HG-10-102-01 was a generous gift of N.S. Gray from the Dana-Farber Cancer Institute. Monoclonal LRRK2 antibody clone N241A was purchased from Neuromab, and LRRK2 pSer935 was purchased from Epitomics. LC3 antibody was obtained from MBL International. Antibodies for organelle markers and GFP were obtained from Santa Cruz Biotechnology.
Cell fractionation
Membrane and cytosol fractions were obtained as previously described (2). Briefly, cells were mechanically homogenized in detergent-free lysis buffer (50 mm HEPES, 150 mm NaCl, 1 mm EDTA, 1 mm EGTA, protease inhibitors) for western blot analysis or sucrose buffer (10 mm HEPES pH 7.4, 1 mm EDTA, 0.25 m sucrose, protease inhibitors) for iodixanol gradients. Cell suspensions were centrifuged at 100 000g for 1 h at 4°C. Supernatant was kept as the cytosolic fraction, and the pellet was resuspended in buffer with 1% Triton X-100 to solubilize membrane proteins for subsequent immunoblotting analysis.
Protein crosslinking
Live-cell crosslinking was performed as previously described (2). Briefly, cells were washed twice in PBS with 1 mm Ca2+/1 mm Mg2+ following experimental treatment, and incubated in 500 μm DSS for 30 min at room temperature on an orbital shaker. Cells were then washed with PBS before quenching in 50 mm Tris–HCl, pH 7.4 for 15 min, followed by two more washes prior to cell lysis and western blot analysis.
Phagocytosis assay
Cells were treated for 16 h with LPS and/or LRRK2 inhibitors prior to the assay. For phagocytosis inhibition, cytochalasin D (1 μm) was added for the last 30 min of the time course, prior to addition for beads. Following treatment, FluoSpheres carboxylate, yellow–green (1 μm size, 100 beads/cell) were added and incubated for 2 h. Cells were washed in PBS twice, scraped in PBS-10 mm EDTA and collected in 5 ml tubes. A flow cytometer (FACSCalibur, BD Biosciences) was used to count 30 000 events and levels of fluorescence in the FL1 channel were used as a measure of phagocytotic activity.
Stable LRRK2 knockdown in cell lines
Several lentiviral constructs containing shLRRK2 were tested (data not shown), and the construct with best knockdown of endogenous LRRK2 was used (Sigma # TRCN0000322191). A lentiviral construct containing a scrambled shRNA sequence (Sigma #SHC002) was used as a control (Scr). For virus production, shRNA constructs were mixed with ViraPower Packaging Mix (Invitrogen) and transfected into to HEK293FT cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instruction. Media containing virus was collected 48 h after transfection. Lentiviral constructs were added to RAW264.7 and BV2 cells, along with polybrene (8 μg/ml) to aid in transduction, and incubated for 24 h at 37°C. Media containing virus was removed and fresh DMEM or RPMI media was added, with cells incubated for another 48 h. Cells containing shRNA were selected for with puromycin (4–5 μg/ml) for a minimum of 2 weeks and then kept in maintenance media (DMEM or RPMI/FBS/2–2.5 μg/ml puromycin). Cells were switched to DMEM/RPMI + FBS for experiments.
Membrane separation by iodixanol gradients
Iodixanol gradients were composed as previously shown (41). Briefly, discontinuous gradients of various concentrations of iodixanol (60% Optiprep) were poured as follows: 1 2.5, 2 5, 2 7.5, 2 10, 0.5 12.5, 2 15, 0.5 17.5, 0.5 20 and 0.3 ml 30%. Resuspended membrane vesicles were applied to the top of the gradient and spun at 270 000g for 2.5 h. An 18-gauge needle was used to puncture the bottom of the tube, and 1 ml fractions were collected from the bottom in a drop-wise manner. A portion of each fraction was mixed with 2× Laemmli sample buffer and analyzed by western blot.
Autophagic vacuole purification
The autophagic vacuole purification was performed as previously reported (42), with slight modification. BV2 cells were treated with 1 μm rapamycin or vehicle for 16 h, followed by treatment with 25 μm nocodozole for 2 h to accumulate autophagosomes. Cells were lysed by Dounce homogenization in detergent-free hypotonic lysis buffer (20 mm Hepes pH 7.3, 0.3 m sucrose). The membrane fraction was concentrated via centrifugation at 100 000g for 8 min, and membranes were resuspended in 2 ml of Histodenz resuspension buffer (20 mm Hepes 7.3, 0.3 m sucrose, 52% Histodenz). Resuspended membrane vesicles were applied to the bottom of a Histodenz gradient composed of 3.3 26, 1.6 24, 1.6 20 and 1.6 ml 15%. Gradients were centrifuged at 100 000g for 4 h. The interfaces between gradient fractions (containing different membrane vesicle compartments) were collected, diluted in hypotonic lysis buffer and concentrated via ultracentrifugation at 100 000g for 30 min. The interface membranes were solubilized in hypotonic lysis buffer containing 1% Triton X-100, and a portion of the fraction was mixed with 2× Laemmli sample buffer for western Blot analysis.
Cell transfection
Q23 and Q74 constructs were a generous gift of D.C. Rubinsztein at Cambridge, UK. LRRK2 KD and Scr cells in six-well dishes were exposed to GFP-Q23 or GFP-Q74 constructs:lipofectamine at a ratio of 1:3 (2 μg, 6 μl), in Optimem serum-free media. After 4 h of transfection, media was replaced with RPMI + FBS. Following 12 h of transfection, cells were treated with vehicle or rapamycin (1 μm) for 12 h prior to biochemical analyses. Cells were fixed in 3% paraformaldehyde for 10 min, washed twice and an incubated with the nuclear stain DAPI for 10 min. Mounting media and a cover slip were added, and images were taken using a 40× objective on a LSM710 confocal microscopy.
Q74 biochemistry
Following transfection and treatment with vehicle or rapamycin treatment, cells were solubilized in lysis buffer (see above) containing 1% Triton X-100 for 30 min on ice. Lysates were centrifuged at 10 000g for 10 min, and supernatant was kept as soluble proteins. The remaining pellet was resuspended in lysis buffer containing 2% SDS for 1 h at room temperature. After protein normalization, lysates were mixed with SDS sample buffer (60 mm Tris/HCl pH 6.8, 10% glycerol, 2% SDS final) and immunoblotted in 4–20% Tris–HCl SDS–PAGE gels (Criterion; Bio-Rad). For dot blot analysis, samples were boiled for 10 min and put through an insulin syringe (30 gauge) 5× to fully suspend the mixture, containing insoluble proteins. Samples were assayed for protein levels using a BCA protein assay (Pierce), and equal amounts of each sample were spotted on PVDF membrane encased in a Minifold II system (Schleicher Schuel), attached to a vacuum line. After a 15 min incubation, a vacuum was lightly applied to the manifold for 30 min to bind the protein mixture to the spot and dry. PVDF membranes were immunoblotted as usual with anti-GFP antibody for subsequent analysis.
Statistics
All experiments were executed at least three independent times. Image J was used for densitometry analysis of western blots. When appropriate, statistical analysis was performed using GraphPad Prism software (V6.0), using a one-way ANOVA with Tukey's post hoc test or Student's t-test for immunoblots. For phagocytosis assays and Q74 dot blot analysis, a one-way ANOVA with a Dunnett's post hoc test was performed to compare experimental groups to vehicle-treated controls.
FUNDING
This work was supported by funds from the Michael J. Fox Foundation (M.J.L.); and the National Institutes of Health (NS072604 to M.J.L., AG000222 to J.D.N.).
ACKNOWLEDGEMENTS
We are grateful to N. Gray for the generous contribution of LRRK2 inhibitor compounds, and to C. Muratore and M. Jin for neuronal lysate samples. We would like to thank C. Serhan for enlightening discussions on the project, and R. Charan for critical reading of the manuscript. We would also like to thank J. Schaeffer, A. Gengatharan and M. Berdnt for their technical assistance and data contribution to the project.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Healy D.G., Falchi M., O'Sullivan S.S., Bonifati V., Durr A., Bressman S., Brice A., Aasly J., Zabetian C.P., Goldwurm S., et al. Phenotype, genotype, and worldwide genetic penetrance of LRRK2-associated Parkinson's disease: a case-control study. Lancet Neurol. 2008;7:583–590. doi: 10.1016/S1474-4422(08)70117-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berger Z., Smith K.A., LaVoie M.J. Membrane localization of LRRK2 is associated with increased formation of the highly active LRRK2 dimer and changes in its phosphorylation. Biochemistry. 2010;49:5511–5523. doi: 10.1021/bi100157u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ito G., Iwatsubo T. Re-examination of the dimerization state of leucine-rich repeat kinase 2: predominance of the monomeric form. Biochem. J. 2012;441:987–994. doi: 10.1042/BJ20111215. [DOI] [PubMed] [Google Scholar]
- 4.Webber P.J., Smith A.D., Sen S., Renfrow M.B., Mobley J.A., West A.B. Autophosphorylation in the Leucine-Rich Repeat Kinase 2 (LRRK2) GTPase domain modifies kinase and GTP-binding activities. J. Mol. Biol. 2011;412:94–100. doi: 10.1016/j.jmb.2011.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greggio E., Zambrano I., Kaganovich A., Beilina A., Taymans J.-M., Daniëls V., Lewis P., Jain S., Ding J., Syed A., et al. The Parkinson disease-associated leucine-rich repeat kinase 2 (LRRK2) is a dimer that undergoes intramolecular autophosphorylation. J. Biol. Chem. 2008;283:16906–16914. doi: 10.1074/jbc.M708718200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matta S., Van Kolen K., da Cunha R., van den Bogaart G., Mandemakers W., Miskiewicz K., De Bock P.-J., Morais V.A., Vilain S., Haddad D., et al. LRRK2 controls an EndoA phosphorylation cycle in synaptic endocytosis. Neuron. 2012;75:1008–1021. doi: 10.1016/j.neuron.2012.08.022. [DOI] [PubMed] [Google Scholar]
- 7.Parisiadou L., Xie C., Cho H.J., Lin X., Gu X.-L., Long C.-X., Lobbestael E., Baekelandt V., Taymans J.-M., Sun L., et al. Phosphorylation of ezrin/radixin/moesin proteins by LRRK2 promotes the rearrangement of actin cytoskeleton in neuronal morphogenesis. J. Neurosci. 2009;29:13971–13980. doi: 10.1523/JNEUROSCI.3799-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gillardon F. Leucine-rich repeat kinase 2 phosphorylates brain tubulin-beta isoforms and modulates microtubule stability—a point of convergence in parkinsonian neurodegeneration? J. Neurochem. 2009;110:1514–1522. doi: 10.1111/j.1471-4159.2009.06235.x. [DOI] [PubMed] [Google Scholar]
- 9.Imai Y., Gehrke S., Wang H.-Q., Takahashi R., Hasegawa K., Oota E., Lu B. Phosphorylation of 4E-BP by LRRK2 affects the maintenance of dopaminergic neurons in Drosophila. EMBO J. 2008;27:2432–2443. doi: 10.1038/emboj.2008.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tong Y., Yamaguchi H., Giaime E., Boyle S., Kopan R., Kelleher R.J., Shen J. Loss of leucine-rich repeat kinase 2 causes impairment of protein degradation pathways, accumulation of alpha-synuclein, and apoptotic cell death in aged mice. Proc. Natl Acad. Sci. USA. 2010;107:9879–9884. doi: 10.1073/pnas.1004676107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andres-Mateos E., Mejias R., Sasaki M., Li X., Lin B.M., Biskup S., Zhang L., Banerjee R., Thomas B., Yang L., et al. Unexpected lack of hypersensitivity in LRRK2 knock-out mice to MPTP (1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine) J. Neurosci. 2009;29:15846–15850. doi: 10.1523/JNEUROSCI.4357-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Umeno J., Asano K., Matsushita T., Matsumoto T., Kiyohara Y., Iida M., Nakamura Y., Kamatani N., Kubo M. Meta-analysis of published studies identified eight additional common susceptibility loci for Crohn's disease and ulcerative colitis. Inflamm. Bowel Dis. 2011;17:2407–2415. doi: 10.1002/ibd.21651. [DOI] [PubMed] [Google Scholar]
- 13.Gardet A., Benita Y., Li C., Sands B.E., Ballester I., Stevens C., Korzenik J.R., Rioux J.D., Daly M.J., Xavier R.J., et al. LRRK2 is involved in the IFN-gamma response and host response to pathogens. J. Immunol. 2010;185:5577–5585. doi: 10.4049/jimmunol.1000548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Z., Lee J., Krummey S., Lu W., Cai H., Lenardo M.J. The kinase LRRK2 is a regulator of the transcription factor NFAT that modulates the severity of inflammatory bowel disease. Nat. Immunol. 2011;12:1063–1070. doi: 10.1038/ni.2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hakimi M., Selvanantham T., Swinton E., Padmore R.F., Tong Y., Kabbach G., Venderova K., Girardin S.E., Bulman D.E., Scherzer C.R., et al. Parkinson's disease-linked LRRK2 is expressed in circulating and tissue immune cells and upregulated following recognition of microbial structures. J. Neural. Transm. 2011;118:795–808. doi: 10.1007/s00702-011-0653-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thévenet J., Pescini Gobert R., Hooft van Huijsduijnen R., Wiessner C., Sagot Y.J. Regulation of LRRK2 expression points to a functional role in human monocyte maturation. PLoS ONE. 2011;6:e21519. doi: 10.1371/journal.pone.0021519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dzamko N., Inesta-Vaquera F., Zhang J., Xie C., Cai H., Arthur S., Tan L., Choi H., Gray N., Cohen P., et al. The IkappaB kinase family phosphorylates the Parkinson's disease kinase LRRK2 at Ser935 and Ser910 during toll-like receptor signaling. PLoS ONE. 2012;7:e39132. doi: 10.1371/journal.pone.0039132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moehle M.S., Webber P.J., Tse T., Sukar N., Standaert D.G., Desilva T.M., Cowell R.M., West A.B. LRRK2 inhibition attenuates microglial inflammatory responses. J. Neurosci. 2012;32:1602–1611. doi: 10.1523/JNEUROSCI.5601-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marker D.F., Puccini J.M., Mockus T.E., Barbieri J., Lu S.-M., Gelbard H.A. LRRK2 kinase inhibition prevents pathological microglial phagocytosis in response to HIV-1 Tat protein. J. Neuroinflammat. 2012;9:261. doi: 10.1186/1742-2094-9-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nichols R.J., Dzamko N., Morrice N.A., Campbell D.G., Deak M., Ordureau A., Macartney T., Tong Y., Shen J., Prescott A.R., et al. 14-3-3 binding to LRRK2 is disrupted by multiple Parkinson's disease-associated mutations and regulates cytoplasmic localization. Biochem. J. 2010;430:393–404. doi: 10.1042/BJ20100483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dzamko N., Deak M., Hentati F., Reith A.D., Prescott A.R., Alessi D.R., Nichols R.J. Inhibition of LRRK2 kinase activity leads to dephosphorylation of Ser(910)/Ser(935), disruption of 14–3–3 binding and altered cytoplasmic localization. Biochem. J. 2010;430:405–413. doi: 10.1042/BJ20100784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sheng Z., Zhang S., Bustos D., Kleinheinz T., Le Pichon C.E., Dominguez S.L., Solanoy H.O., Drummond J., Zhang X., Ding X., et al. Ser1292 autophosphorylation is an indicator of LRRK2 kinase activity and contributes to the cellular effects of PD mutations. Sci. Trans. Med. 2012;4:164 ra161–1164ra161. doi: 10.1126/scitranslmed.3004485. [DOI] [PubMed] [Google Scholar]
- 23.Sen S., Webber P.J., West A.B. Dependence of leucine-rich repeat kinase 2 (LRRK2) kinase activity on dimerization. J. Biol. Chem. 2009;284:36346–36356. doi: 10.1074/jbc.M109.025437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salaun C., Greaves J., Chamberlain L.H. The intracellular dynamic of protein palmitoylation. J. Cell Biol. 2010;191:1229–1238. doi: 10.1083/jcb.201008160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ren J., Wen L., Gao X., Jin C., Xue Y., Yao X. CSS-Palm 2.0: an updated software for palmitoylation sites prediction. Protein Eng. Des. Sel. 2008;21:639–644. doi: 10.1093/protein/gzn039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shin N., Jeong H., Kwon J., Heo H.Y., Kwon J.J., Yun H.J., Kim C.-H., Han B.S., Tong Y., Shen J., et al. LRRK2 regulates synaptic vesicle endocytosis. Exp. Cell Res. 2008;314:2055–2065. doi: 10.1016/j.yexcr.2008.02.015. [DOI] [PubMed] [Google Scholar]
- 27.Reith A.D., Bamborough P., Jandu K., Andreotti D., Mensah L., Dossang P., Choi H.G., Deng X., Zhang J., Alessi D.R., et al. GSK2578215A; a potent and highly selective 2-arylmethyloxy-5-substitutent-N-arylbenzamide LRRK2 kinase inhibitor. Bioorg. Med. Chem. Lett. 2012;22:5625–5629. doi: 10.1016/j.bmcl.2012.06.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choi H.G., Zhang J., Deng X., Hatcher J.M., Patricelli M.P., Zhao Z., Alessi D.R., Gray N.S. Brain penetrant LRRK2 inhibitor. ACS Med. Chem. Lett. 2012;3:658–662. doi: 10.1021/ml300123a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luerman G.C., Nguyen C., Samaroo H., Loos P., Xi H., Hurtado-Lorenzo A., Needle E., Stephen Noell G., Galatsis P., Dunlop J., et al. Phosphoproteomic evaluation of pharmacological inhibition of leucine-rich repeat kinase 2 reveals significant off-target effects of LRRK-2-IN-1. J. Neurochem. 2014;128:561–576. doi: 10.1111/jnc.12483. [DOI] [PubMed] [Google Scholar]
- 30.Biskup S., Moore D.J., Rea A., Lorenz-Deperieux B., Coombes C.E., Dawson V.L., Dawson T.M., West A.B. Dynamic and redundant regulation of LRRK2 and LRRK1 expression. BMC Neurosci. 2007;8:102. doi: 10.1186/1471-2202-8-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Higashi S., Moore D.J., Yamamoto R., Minegishi M., Sato K., Togo T., Katsuse O., Uchikado H., Furukawa Y., Hino H., et al. Abnormal localization of leucine-rich repeat kinase 2 to the endosomal-lysosomal compartment in lewy body disease. J. Neuropathol. Exp. Neurol. 2009;68:994–1005. doi: 10.1097/NEN.0b013e3181b44ed8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alegre-Abarrategui J., Christian H., Lufino M.M.P., Mutihac R., Venda L.L., Ansorge O., Wade-Martins R. LRRK2 regulates autophagic activity and localizes to specific membrane microdomains in a novel human genomic reporter cellular model. Hum. Mol. Genet. 2009;18:4022–4034. doi: 10.1093/hmg/ddp346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hatano T., Kubo S.-I., Imai S., Maeda M., Ishikawa K., Mizuno Y., Hattori N. Leucine-rich repeat kinase 2 associates with lipid rafts. Hum. Mol. Genet. 2007;16:678–690. doi: 10.1093/hmg/ddm013. [DOI] [PubMed] [Google Scholar]
- 34.Biskup S., Moore D.J., Celsi F., Higashi S., West A.B., Andrabi S.A., Kurkinen K., Yu S.-W., Savitt J.M., Waldvogel H.J., et al. Localization of LRRK2 to membranous and vesicular structures in mammalian brain. Ann. Neurol. 2006;60:557–569. doi: 10.1002/ana.21019. [DOI] [PubMed] [Google Scholar]
- 35.Deretic V. Autophagy in immunity and cell-autonomous defense against intracellular microbes. Immunol. Rev. 2011;240:92–104. doi: 10.1111/j.1600-065X.2010.00995.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lucin K.M., O'Brien C.E., Bieri G., Czirr E., Mosher K.I., Abbey R.J., Mastroeni D.F., Rogers J., Spencer B., Masliah E., et al. Microglial beclin 1 regulates retromer trafficking and phagocytosis and is impaired in Alzheimer's disease. Neuron. 2013;79:873–886. doi: 10.1016/j.neuron.2013.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaneko Y.S., Nakashima A., Mori K., Nagatsu T., Nagatsu I., Ota A. Microglial activation in neuroinflammation: implications for the etiology of neurodegeneration. Neurodegener. Dis. 2012;10:100–103. doi: 10.1159/000332936. [DOI] [PubMed] [Google Scholar]
- 38.Majumder S., Richardson A., Strong R., Oddo S. Inducing autophagy by rapamycin before, but not after, the formation of plaques and tangles ameliorates cognitive deficits. PLoS ONE. 2011;6:e25416. doi: 10.1371/journal.pone.0025416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takenouchi T., Nakai M., Iwamaru Y., Sugama S., Tsukimoto M., Fujita M., Wei J., Sekigawa A., Sato M., Kojima S., et al. The activation of P2X7 receptor impairs lysosomal functions and stimulates the release of autophagolysosomes in microglial cells. J. Immunol. 2009;182:2051–2062. doi: 10.4049/jimmunol.0802577. [DOI] [PubMed] [Google Scholar]
- 40.Galluzzi L., Kepp O., Kroemer G. Autophagy and innate immunity ally against bacterial invasion. EMBO J. 2011;30:3213–3214. doi: 10.1038/emboj.2011.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xia W., Zhang J., Ostaszewski B.L., Kimberly W.T., Seubert P., Koo E.H., Shen J., Selkoe D.J. Presenilin 1 regulates the processing of β-amyloid precursor protein C-terminal fragments and the generation of amyloid β-protein in endoplasmic reticulum and Golgi. Biochemistry. 1998;37:16465–16471. doi: 10.1021/bi9816195. [DOI] [PubMed] [Google Scholar]
- 42.Marzella L., Ahlberg J., Glaumann H. Isolation of autophagic vacuoles from rat liver: morphological and biochemical characterization. J. Cell Biol. 1982;93:144–154. doi: 10.1083/jcb.93.1.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Plowey E.D., Cherra S.J., Liu Y.-J., Chu C.T. Role of autophagy in G2019S-LRRK2-associated neurite shortening in differentiated SH-SY5Y cells. J. Neurochem. 2008;105:1048–1056. doi: 10.1111/j.1471-4159.2008.05217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bravo-San Pedro J.M., Niso-Santano M., Gómez-Sánchez R., Pizarro-Estrella E., Aiastui-Pujana A., Gorostidi A., Climent V., López de Maturana R., Sanchez-Pernaute R., López de Munain A., et al. The LRRK2 G2019S mutant exacerbates basal autophagy through activation of the MEK/ERK pathway. Cell. Mol. Life Sci. 2013;70:121–136. doi: 10.1007/s00018-012-1061-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Orenstein S.J., Kuo S.-H., Tasset I., Arias E., Koga H., Fernandez-Carasa I., Cortes E., Honig L.S., Dauer W., Consiglio A., et al. Interplay of LRRK2 with chaperone-mediated autophagy. Nat. Neurosci. 2013;16:394–406. doi: 10.1038/nn.3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gómez-Suaga P., Luzón-Toro B., Churamani D., Zhang L., Bloor-Young D., Patel S., Woodman P.G., Churchill G.C., Hilfiker S. Leucine-rich repeat kinase 2 regulates autophagy through a calcium-dependent pathway involving NAADP. Hum. Mol. Genet. 2012;21:511–525. doi: 10.1093/hmg/ddr481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Manzoni C., Mamais A., Dihanich S., Abeti R., Soutar M.P.M., Plun-Favreau H., Giunti P., Tooze S.A., Bandopadhyay R., Lewis P.A. Inhibition of LRRK2 kinase activity stimulates macroautophagy. Biochim. Biophys. Acta. 2013;1833:2900–2910. doi: 10.1016/j.bbamcr.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sánchez-Danés A., Richaud-Patin Y., Carballo-Carbajal I., Jiménez-Delgado S., Caig C., Mora S., Di Guglielmo C., Ezquerra M., Patel B., Giralt A., et al. Disease-specific phenotypes in dopamine neurons from human iPS-based models of genetic and sporadic Parkinson's disease. EMBO Mol. Med. 2012;4:380–395. doi: 10.1002/emmm.201200215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Klionsky D.J., Abdalla F.C., Abeliovich H., Abraham R.T., Acevedo-Arozena A., Adeli K., Agholme L., Agnello M., Agostinis P., Aguirre-Ghiso J.A., et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mizushima N., Yoshimori T., Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tansey M.G., Goldberg M.S. Neuroinflammation in Parkinson's disease: its role in neuronal death and implications for therapeutic intervention. Neurobiol. Dis. 2010;37:510–518. doi: 10.1016/j.nbd.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McGeer P.L., McGeer E.G. Inflammation and neurodegeneration in Parkinson's disease. Parkinsonism Relat. Disord. 2004;10:S3–S7. doi: 10.1016/j.parkreldis.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 53.Ravikumar B., Duden R., Rubinsztein D.C. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum. Mol. Genet. 2002;11:1107–1117. doi: 10.1093/hmg/11.9.1107. [DOI] [PubMed] [Google Scholar]
- 54.Berger Z., Ravikumar B., Menzies F.M., Oroz L.G., Underwood B.R., Pangalos M.N., Schmitt I., Wullner U., Evert B.O., O'Kane C.J., et al. Rapamycin alleviates toxicity of different aggregate-prone proteins. Hum. Mol. Genet. 2006;15:433–442. doi: 10.1093/hmg/ddi458. [DOI] [PubMed] [Google Scholar]
- 55.Ravikumar B., Vacher C., Berger Z., Davies J.E., Luo S., Oroz L.G., Scaravilli F., Easton D.F., Duden R., O'Kane C.J., et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat. Genet. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 56.Wang H. Suppression of polyglutamine-induced toxicity in cell and animal models of Huntington's disease by ubiquilin. Hum. Mol. Genet. 2006;15:1025–1041. doi: 10.1093/hmg/ddl017. [DOI] [PubMed] [Google Scholar]
- 57.Mizushima N., Yamamoto A., Hatano M., Kobayashi Y., Kabeya Y., Suzuki K., Tokuhisa T., Ohsumi Y., Yoshimori T. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J. Cell Biol. 2001;152:657–668. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vitte J., Traver S., Maués De Paula A., Lesage S., Rovelli G., Corti O., Duyckaerts C., Brice A. Leucine-rich repeat kinase 2 is associated with the endoplasmic reticulum in dopaminergic neurons and accumulates in the core of Lewy bodies in Parkinson disease. J. Neuropathol. Exp. Neurol. 2010;69:959–972. doi: 10.1097/NEN.0b013e3181efc01c. [DOI] [PubMed] [Google Scholar]
- 59.Dodson M.W., Zhang T., Jiang C., Chen S., Guo M. Roles of the Drosophila LRRK2 homolog in Rab7-dependent lysosomal positioning. Hum. Mol. Genet. 2012;21:1350–1363. doi: 10.1093/hmg/ddr573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kovács A.L., Reith A., Seglen P.O. Accumulation of autophagosomes after inhibition of hepatocytic protein degradation by vinblastine, leupeptin or a lysosomotropic amine. Exp. Cell Res. 1982;137:191–201. doi: 10.1016/0014-4827(82)90020-9. [DOI] [PubMed] [Google Scholar]
- 61.Schworer C.M., Mortimore G.E. Glucagon-induced autophagy and proteolysis in rat liver: mediation by selective deprivation of intracellular amino acids. Proc. Natl Acad. Sci. USA. 1979;76:3169–3173. doi: 10.1073/pnas.76.7.3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Russell R.C., Tian Y., Yuan H., Park H.W., Chang Y.-Y., Kim J., Kim H., Neufeld T.P., Dillin A., Guan K.-L. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat. Cell Biol. 2013;15:741–750. doi: 10.1038/ncb2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McEwan D.G., Dikic I. The three musketeers of autophagy: phosphorylation, ubiquitylation and acetylation. Trends Cell Biol. 2011;21:195–201. doi: 10.1016/j.tcb.2010.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kolch W. Meaningful relationships: the regulation of the Ras/Raf/MEK/ERK pathway by protein interactions. Biochem. J. 2000;351(Pt 2):289–305. [PMC free article] [PubMed] [Google Scholar]
- 65.Ulvestad E., Williams K., Mørk S., Antel J., Nyland H. Phenotypic differences between human monocytes/macrophages and microglial cells studied in situ and in vitro. J. Neuropathol. Exp. Neurol. 1994;53:492–501. doi: 10.1097/00005072-199409000-00008. [DOI] [PubMed] [Google Scholar]
- 66.Ravikumar B., Sarkar S., Davies J.E., Futter M., Garcia-Arencibia M., Green-Thompson Z.W., Jimenez-Sanchez M., Korolchuk V.I., Lichtenberg M., Luo S., et al. Regulation of mammalian autophagy in physiology and pathophysiology. Physiol. Rev. 2010;90:1383–1435. doi: 10.1152/physrev.00030.2009. [DOI] [PubMed] [Google Scholar]
- 67.De Meyer G.R.Y., De Keulenaer G.W., Martinet W. Role of autophagy in heart failure associated with aging. Heart Fail. Rev. 2010;15:423–430. doi: 10.1007/s10741-010-9166-6. [DOI] [PubMed] [Google Scholar]
- 68.MacLeod D., Dowman J., Hammond R., Leete T., Inoue K., Abeliovich A. The familial Parkinsonism gene LRRK2 regulates neurite process morphology. Neuron. 2006;52:587–593. doi: 10.1016/j.neuron.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 69.Chan D., Citro A., Cordy J.M., Shen G.C., Wolozin B. Rac1 protein rescues neurite retraction caused by G2019S leucine-rich repeat kinase 2 (LRRK2) J. Biol. Chem. 2011;286:16140–16149. doi: 10.1074/jbc.M111.234005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lin C.-H., Tsai P.-I., Wu R.-M., Chien C.-T. LRRK2 G2019S mutation induces dendrite degeneration through mislocalization and phosphorylation of tau by recruiting autoactivated GSK3ß. J. Neurosci. 2010;30:13138–13149. doi: 10.1523/JNEUROSCI.1737-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tong Y., Giaime E., Yamaguchi H., Ichimura T., Liu Y., Si H., Cai H., Bonventre J.V., Shen J. Loss of leucine-rich repeat kinase 2 causes age-dependent bi-phasic alterations of the autophagy pathway. Mol. Neurodegener. 2012;7:2. doi: 10.1186/1750-1326-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Miklavc P., Ehinger K., Thompson K.E., Hobi N., Shimshek D.R., Frick M. Surfactant secretion in LRRK2 knock-out rats: changes in lamellar body morphology and rate of exocytosis. PLoS ONE. 2014;9:e84926. doi: 10.1371/journal.pone.0084926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stappenbeck T.S., Rioux J.D., Mizoguchi A., Saitoh T., Huett A., Darfeuille-Michaud A., Wileman T., Mizushima N., Carding S., Akira S., et al. Crohn disease: a current perspective on genetics, autophagy and immunity. Autophagy. 2011;7:355–374. doi: 10.4161/auto.7.4.13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Strisciuglio C., Duijvestein M., Verhaar A.P., Vos A.C.W., van den Brink G.R., Hommes D.W., Wildenberg M.E. Impaired autophagy leads to abnormal dendritic cell-epithelial cell interactions. J. Crohns Colitis. 2013;7:534–541. doi: 10.1016/j.crohns.2012.08.009. [DOI] [PubMed] [Google Scholar]
- 75.Lee V.M.-Y., Trojanowski J.Q. Mechanisms of Parkinson's disease linked to pathological alpha-synuclein: new targets for drug discovery. Neuron. 2006;52:33–38. doi: 10.1016/j.neuron.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 76.Simón-Sánchez J., Schulte C., Bras J.M., Sharma M., Gibbs J.R., Berg D., Paisán-Ruíz C., Lichtner P., Scholz S.W., Hernandez D.G., et al. Genome-wide association study reveals genetic risk underlying Parkinson's disease. Nat. Genet. 2009;41:1308–1312. doi: 10.1038/ng.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hyun C.H., Yoon C.Y., Lee H.-J., Lee S.-J. LRRK2 as a potential genetic modifier of synucleinopathies: interlacing the two major genetic factors of Parkinson's disease. Exp. Neurobiol. 2013;22:249–257. doi: 10.5607/en.2013.22.4.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nagai M., Re D.B., Nagata T., Chalazonitis A., Jessell T.M., Wichterle H., Przedborski S. Astrocytes expressing ALS-linked mutated SOD1 release factors selectively toxic to motor neurons. Nat. Neurosci. 2007;10:615–622. doi: 10.1038/nn1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Di Giorgio F.P., Carrasco M.A., Siao M.C., Maniatis T., Eggan K. Non-cell autonomous effect of glia on motor neurons in an embryonic stem cell-based ALS model. Nat. Neurosci. 2007;10:608–614. doi: 10.1038/nn1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yamanaka K., Chun S.J., Boillee S., Fujimori-Tonou N., Yamashita H., Gutmann D.H., Takahashi R., Misawa H., Cleveland D.W. Astrocytes as determinants of disease progression in inherited amyotrophic lateral sclerosis. Nat. Neurosci. 2008;11:251–253. doi: 10.1038/nn2047. [DOI] [PMC free article] [PubMed] [Google Scholar]