Abstract
The genetic contribution to the variation in human lifespan is ∼25%. Despite the large number of identified disease-susceptibility loci, it is not known which loci influence population mortality. We performed a genome-wide association meta-analysis of 7729 long-lived individuals of European descent (≥85 years) and 16 121 younger controls (<65 years) followed by replication in an additional set of 13 060 long-lived individuals and 61 156 controls. In addition, we performed a subset analysis in cases aged ≥90 years. We observed genome-wide significant association with longevity, as reflected by survival to ages beyond 90 years, at a novel locus, rs2149954, on chromosome 5q33.3 (OR = 1.10, P = 1.74 × 10−8). We also confirmed association of rs4420638 on chromosome 19q13.32 (OR = 0.72, P = 3.40 × 10−36), representing the TOMM40/APOE/APOC1 locus. In a prospective meta-analysis (n = 34 103), the minor allele of rs2149954 (T) on chromosome 5q33.3 associates with increased survival (HR = 0.95, P = 0.003). This allele has previously been reported to associate with low blood pressure in middle age. Interestingly, the minor allele (T) associates with decreased cardiovascular mortality risk, independent of blood pressure. We report on the first GWAS-identified longevity locus on chromosome 5q33.3 influencing survival in the general European population. The minor allele of this locus associates with low blood pressure in middle age, although the contribution of this allele to survival may be less dependent on blood pressure. Hence, the pleiotropic mechanisms by which this intragenic variation contributes to lifespan regulation have to be elucidated.
INTRODUCTION
Worldwide, human life expectancy has increased remarkably over the last two centuries (1), although the healthy life expectancy lags behind. Citizens of the European Union, for example, spend only 75–80% of their lifespan in good health (2). Families in which longevity clusters form an exception in this sense, by showing beneficial or ‘youthful’ profiles for many metabolic and immune-related parameters (3–7) and a low prevalence of common diseases from middle age onwards (5,8,9). Therefore, the genome of long-lived individuals is investigated to identify variants that promote healthy aging and protect against age-related disease. This is a major challenge because the genetic component of lifespan variation in the population at large has been estimated to be only ∼25% (10,11) and is assumed to be determined by many, still uncharacterized, genes (12,13). Genetic influences on human longevity are expected to reflect longevity assurance mechanisms acting across species (14), as well as more heterogeneous population-specific effects. Although numerous genome-wide association studies (GWAS) have successfully identified loci involved in common, age-related diseases (15), the corresponding susceptibility loci do not explain the genetic component of human longevity (16). GWAS for human longevity have thus far failed to identify genome-wide significant loci, besides the well-known TOMM40/APOE/APOC1 locus (17–19).
In this paper, we conducted a large genome-wide association meta-analysis of human longevity in 14 studies with long-lived cases (≥85 years) and younger controls (<65 years) from European descent. In addition, we performed a subset analysis in cases aged ≥90 years. The novel longevity locus we identified was tested for association with prospective (cause-specific) mortality in a meta-analysis of 11 European cohorts and examined for association with various metabolic traits that may explain the mechanism by which the locus contributes to survival to high ages.
RESULTS
Genome-wide association analysis
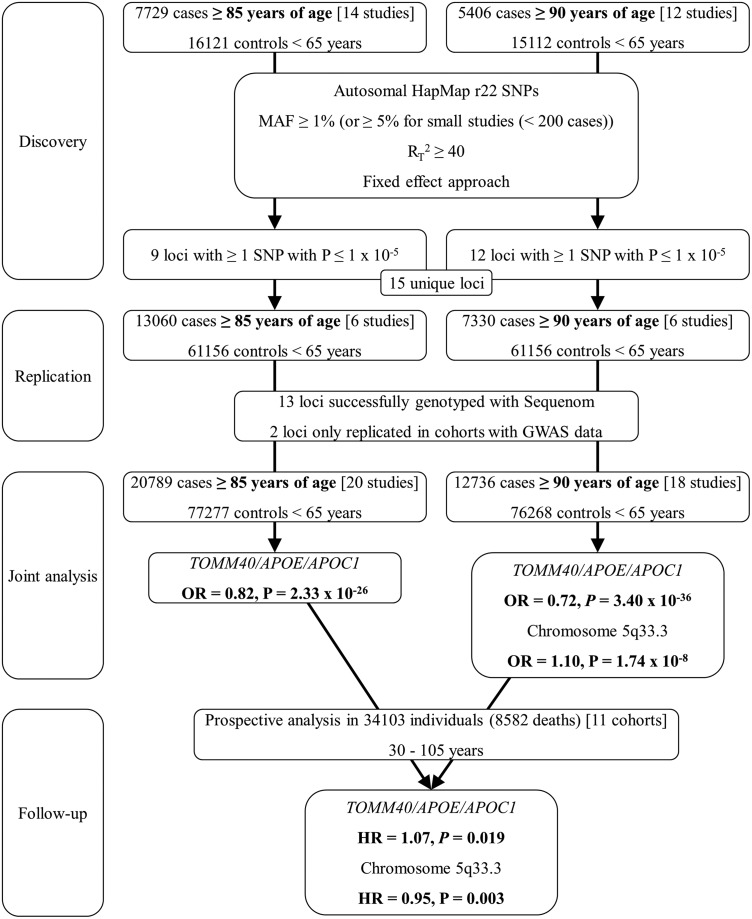
In order to identify novel loci involved in lifespan regulation, we conducted a meta-analysis on GWAS data of 7729 long-lived cases (≥85 years) and 16 121 younger controls (<65 years) from 14 studies originating from 7 European countries (Supplementary Material, Table S1). For each study, cases and controls originated from the same country. Given the higher heritability of longevity at older ages (11,20), we performed a subset analysis in which we compared cases aged ≥90 years (n = 5406) with 15 112 controls (<65 years) from the corresponding control cohorts. Replication was performed in 13 060 cases aged ≥85 years (of which 7330 were ≥90 years) and 61 156 controls from 6 additional studies, of which 3 originated from European countries not represented in the discovery phase meta-analysis (Supplementary Material, Table S1). Analysis of each study was performed using a logistic regression-based method, and results were adjusted for study-specific genomic inflation factors (λ) (Supplementary Material, Table S2). Meta-analysis was performed on 2 480 356 (≥85 years) and 2 470 825 (≥90 years) imputed SNPs using a fixed-effect approach, and results were further adjusted for the overall genomic inflation factor (λ = 1.019) (Supplementary Material, Fig. S1). A flow chart of the consecutive analysis steps is depicted in Figure 1.
Figure 1.
Flow chart of experimental work. The analysis in the cases aged ≥90 years is a subset analysis of the analysis in the cases aged ≥85 years. Twelve out of 14 studies used for the discovery phase analysis of cases aged ≥85 years contained at least 100 cases over 90 years of age and were thus analyzed in the subset analysis of cases aged ≥90 years.
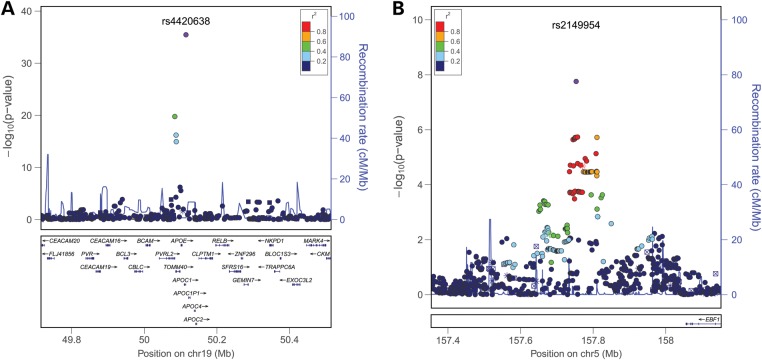
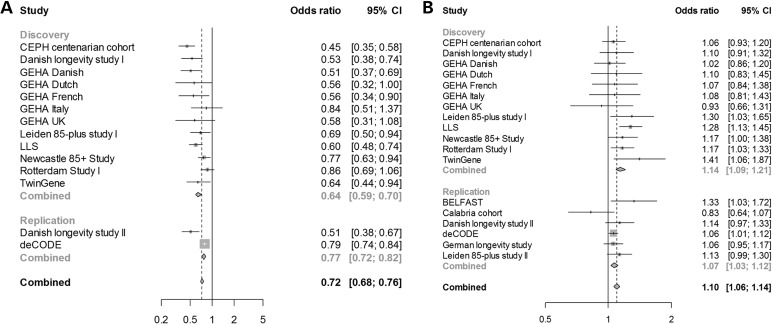
The discovery phase meta-analyses of the cases aged ≥85 years (n = 7729) showed genome-wide significant association with survival into old age at one locus, the previously identified TOMM40/APOE/APOC1 locus (17,21) (rs4420638 (G); odds ratio (OR) = 0.71, P = 6.14 × 10−19; Table 1). No gender-dependent effects were observed in the sex-stratified analysis of the cases aged ≥85 years (Supplementary Material, Table S4). The discovery-phase meta-analysis of the cases aged ≥90 years (n = 5406) showed a similar result, i.e. the TOMM40/APOE/APOC1 locus was the only genome-wide significant locus (OR = 0.64, P = 4.09 × 10−21; Fig. 2 and Table 2). The regional association plot and forest plot for the TOMM40/APOE/APOC1 locus are depicted in Figures 3 and 4, respectively. Although several SNPs on chromosome 19q13.32, which are in moderate linkage disequilibrium (LD) with rs4420638, show additional association with survival into old age, meta-analysis conditional on rs4420638 showed no independent associations among these SNPs (Supplementary Material, Fig S2 and Table S3).
Table 1.
Results of the discovery phase, replication phase and joint analysis of cases aged ≥85 years
| Locus | Lead SNP | Chromosome | Position | Candidate/closest gene | EA | Analysis |
n |
EAF |
OR | 95% CI | P | I2 (%) | Phet | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | Cases | Controls | ||||||||||||
| 1q43 | rs1625040 | 1 | 235 213 002 | MTR, RYR2 | A | Discovery | 7729 | 16 121 | 0.170 | 0.150 | 1.16 | 1.09–1.23 | 3.36 × 10−6 | ||
| Replication | 13 027 | 60 914 | 0.178 | 0.182 | 1.02 | 0.98–1.07 | 0.216 | ||||||||
| Joint | 20 756 | 77 035 | 1.07 | 1.03–1.10 | 3.50 × 10−4 | 31.0 | 0.093 | ||||||||
| 2q24.3 | rs6432832 | 2 | 166 079 072 | CSRNP3 | A | Discovery | 7729 | 16 121 | 0.344 | 0.321 | 1.12 | 1.07–1.17 | 2.79 × 10−6 | ||
| Replication | 13 019 | 60 824 | 0.346 | 0.339 | 1.03 | 1.00–1.07 | 0.029 | ||||||||
| Joint | 20 748 | 76 945 | 1.06 | 1.03–1.09 | 8.73 × 10−6 | 0.0 | 0.467 | ||||||||
| 4q27 | rs13114426 | 4 | 120 942 533 | PDE5A, MAD2L1 | T | Discovery | 7729 | 16 121 | 0.387 | 0.405 | 0.90 | 0.87–0.95 | 2.20 × 10−5 | ||
| Replication | 13 024 | 60 932 | 0.364 | 0.351 | 1.00 | 0.97–1.04 | 0.711 | ||||||||
| Joint | 20 753 | 77 053 | 0.97 | 0.94–0.99 | 0.033 | 46.5 | 0.012 | ||||||||
| 5q33.3 | rs2149954 | 5 | 157 753 180 | EBF1 | T | Discovery | 7729 | 16 121 | 0.388 | 0.360 | 1.12 | 1.07–1.17 | 5.98 × 10−6 | ||
| Replication | 12 973 | 60 262 | 0.365 | 0.352 | 1.04 | 1.01–1.07 | 0.013 | ||||||||
| Joint | 20 702 | 76 383 | 1.07 | 1.04–1.09 | 4.34 × 10−6 | 28.2 | 0.118 | ||||||||
| 8q13.3 | rs10957550a | 8 | 72 457 142 | EYA1 | A | Discovery | 7727 | 16 093 | 0.268 | 0.285 | 0.88 | 0.84–0.93 | 3.61 × 10−6 | ||
| Replication | 10 056 | 56 262 | 0.236 | 0.244 | 0.95 | 0.92–0.99 | 0.012 | ||||||||
| Joint | 17 783 | 72 355 | 0.92 | 0.90–0.95 | 1.41 × 10−6 | 29.4 | 0.130 | ||||||||
| 10q23.33 | rs4466755 | 10 | 96 622 243 | CYP2C19, CYP2C9 | T | Discovery | 7729 | 16 121 | 0.454 | 0.443 | 1.12 | 1.07–1.16 | 2.72 × 10−6 | ||
| Replication | 13 051 | 61 105 | 0.488 | 0.508 | 0.98 | 0.95–1.01 | 0.129 | ||||||||
| Joint | 20 780 | 77 226 | 1.03 | 1.00–1.05 | 0.161 | 65.6 | 2.15 × 10−5 | ||||||||
| 17q23.3 | rs17760362 | 17 | 58 772 399 | TANC2 | A | Discovery | 7729 | 16 121 | 0.252 | 0.233 | 1.13 | 1.07–1.19 | 5.38 × 10−6 | ||
| Replication | 13 007 | 60 679 | 0.252 | 0.249 | 1.04 | 1.00–1.07 | 0.033 | ||||||||
| Joint | 20 736 | 76 800 | 1.07 | 1.04–1.10 | 1.56 × 10−5 | 0.0 | 0.473 | ||||||||
| 19q13.32 | rs4420638a | 19 | 50 114 786 | APOE | G | Discovery | 7728 | 16 111 | 0.157 | 0.195 | 0.71 | 0.67–0.77 | 6.14 × 10−19 | ||
| Replication | 10 165 | 57 126 | 0.180 | 0.202 | 0.87 | 0.83–0.91 | 2.12 × 10−12 | ||||||||
| Joint | 17 893 | 73 237 | 0.82 | 0.79–0.85 | 2.33 × 10−26 | 80.2 | 4.35 × 10−10 | ||||||||
| 20q13.2 | rs8126377 | 20 | 51 590 254 | TSHZ2, ZNF217 | G | Discovery | 7532 | 15 902 | 0.059 | 0.069 | 0.79 | 0.71–0.87 | 1.35 × 10−5 | ||
| Replication | 12 974 | 60 647 | 0.058 | 0.054 | 1.01 | 0.94–1.08 | 0.901 | ||||||||
| Joint | 20 506 | 76 549 | 0.93 | 0.88–0.99 | 0.020 | 51.1 | 0.006 | ||||||||
EA, effect allele; EAF, effect allele frequency after pooling the data of all analyzed individuals; OR, odds ratio for the effect allele; 95% CI, 95% confidence interval; I2, heterogeneity statistic; Phet, P-value for heterogeneity.
aGenotyping of these SNPs with the Sequenom MassARRAY system for the replication phase was unsuccessful. The SNPs in bold overlap with Table 2.
Figure 2.
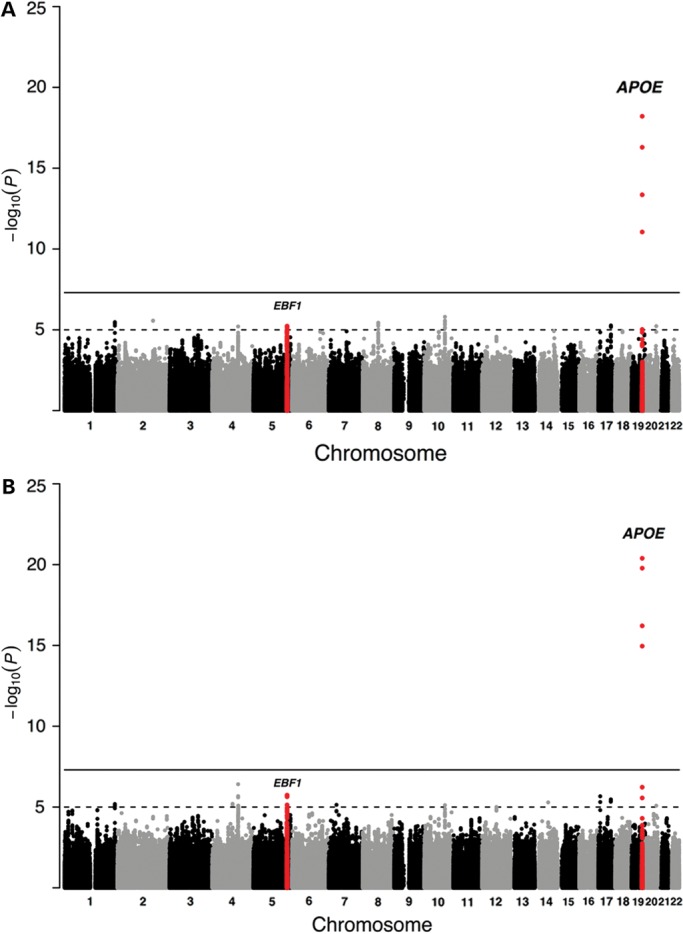
Results of the discovery phase analysis. Manhattan plot presenting the −log10 P-values from the discovery phase analysis of cases aged ≥85 years (A) and ≥90 years (B). The loci that showed a genome-wide significant association after the joint analysis of the discovery and replication phase (chromosome 19q13.32 and 5q33.3) are shown in red.
Table 2.
Results of the discovery phase, replication phase and joint analysis of cases aged ≥90 years
| Locus | Lead SNP | Chromosome | Position | Candidate/closest gene | EA | Analysis |
n |
EAF |
OR | 95% CI | P | I2 (%) | Phet | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | Cases | Controls | ||||||||||||
| 1q43 | rs1625040 | 1 | 235 213 002 | MTR, RYR2 | A | Discovery | 5406 | 15 112 | 0.176 | 0.150 | 1.18 | 1.10–1.26 | 6.53 × 10−6 | ||
| Replication | 7310 | 60 914 | 0.175 | 0.182 | 1.05 | 0.99–1.10 | 0.065 | ||||||||
| Joint | 12 716 | 76 026 | 1.10 | 1.05–1.14 | 2.60 × 10−5 | 9.3 | 0.343 | ||||||||
| 4q22.2 | rs4693331 | 4 | 94 760 609 | GRID2 | C | Discovery | 5406 | 15 112 | 0.416 | 0.444 | 0.89 | 0.84–0.93 | 6.63 × 10−6 | ||
| Replication | 7267 | 60 324 | 0.449 | 0.440 | 1.03 | 0.99–1.07 | 0.095 | ||||||||
| Joint | 12 673 | 75 436 | 0.97 | 0.94–1.00 | 0.139 | 61.3 | 3.51 × 10−4 | ||||||||
| 4q27 | rs13114426 | 4 | 120 942 533 | PDE5A, MAD2L1 | T | Discovery | 5406 | 15 112 | 0.381 | 0.405 | 0.88 | 0.84–0.92 | 2.11 × 10−6 | ||
| Replication | 7305 | 60 932 | 0.369 | 0.351 | 0.98 | 0.94–1.02 | 0.336 | ||||||||
| Joint | 12 711 | 76 044 | 0.94 | 0.91–0.97 | 1.95 × 10−4 | 32.5 | 0.090 | ||||||||
| 5q33.3 | rs2149954 | 5 | 157 753 180 | EBF1 | T | Discovery | 5406 | 15 112 | 0.396 | 0.360 | 1.14 | 1.09–1.21 | 1.85 × 10−6 | ||
| Replication | 7298 | 60 262 | 0.374 | 0.352 | 1.07 | 1.03–1.12 | 5.98 × 10−4 | ||||||||
| Joint | 12 704 | 75 374 | 1.10 | 1.06–1.14 | 1.74 × 10−8 | 28.5 | 0.125 | ||||||||
| 7p14.2 | rs11977641 | 7 | 36 761 949 | AOAH, ELMO1 | C | Discovery | 5406 | 15 112 | 0.062 | 0.076 | 0.78 | 0.70–0.87 | 7.31 × 10−6 | ||
| Replication | 3049 | 4805 | 0.071 | 0.073 | 0.93 | 0.82–1.06 | 0.226 | ||||||||
| Joint | 8455 | 19 917 | 0.84 | 0.77–0.91 | 1.57 × 10−5 | 50.2 | 0.010 | ||||||||
| 10q23.33 | rs4466755 | 10 | 96 622 243 | CYP2C19, CYP2C9 | T | Discovery | 5406 | 15 112 | 0.455 | 0.445 | 1.13 | 1.07–1.18 | 1.30 × 10−5 | ||
| Replication | 7326 | 61 105 | 0.477 | 0.508 | 0.98 | 0.94–1.02 | 0.208 | ||||||||
| Joint | 12 732 | 76 217 | 1.03 | 1.00–1.07 | 0.087 | 55.4 | 0.002 | ||||||||
| 12q15 | rs11834614 | 12 | 67 197 344 | MDM1, RAP1B | C | Discovery | 5406 | 15 112 | 0.138 | 0.155 | 0.85 | 0.79–0.91 | 9.94 × 10−6 | ||
| Replication | 7272 | 60 210 | 0.165 | 0.173 | 1.01 | 0.96–1.07 | 0.603 | ||||||||
| Joint | 12 678 | 75 322 | 0.95 | 0.91–0.99 | 0.023 | 43.9 | 0.024 | ||||||||
| 14q23.2 | rs2784505 | 14 | 61 501 766 | SYT16 | G | Discovery | 5406 | 15 112 | 0.080 | 0.067 | 1.23 | 1.11–1.35 | 8.87 × 10−5 | ||
| Replication | 7323 | 60 979 | 0.070 | 0.066 | 1.10 | 1.02–1.19 | 0.012 | ||||||||
| Joint | 12 729 | 76 091 | 1.15 | 1.08–1.22 | 9.47 × 10−6 | 28.3 | 0.127 | ||||||||
| 17p13.1 | rs940850 | 17 | 8 870 805 | NTN1 | T | Discovery | 5405 | 15 112 | 0.072 | 0.093 | 0.78 | 0.70–0.87 | 4.93 × 10−6 | ||
| Replication | 7276 | 60 146 | 0.109 | 0.118 | 1.03 | 0.97–1.10 | 0.318 | ||||||||
| Joint | 12 681 | 75 258 | 0.95 | 0.90–1.01 | 0.111 | 63.7 | 1.32 × 10−4 | ||||||||
| 17q23.2 | rs2109265 | 17 | 58 307 001 | MARCH10, TANC2 | A | Discovery | 5406 | 15 112 | 0.443 | 0.420 | 1.13 | 1.08–1.19 | 3.34 × 10−6 | ||
| Replication | 7307 | 60 672 | 0.453 | 0.465 | 1.01 | 0.97–1.05 | 0.671 | ||||||||
| Joint | 12 713 | 75 784 | 1.06 | 1.02–1.09 | 0.001 | 34.7 | 0.074 | ||||||||
| 19q13.32 | rs4420638a | 19 | 50 114 786 | APOE | G | Discovery | 5405 | 15 102 | 0.145 | 0.195 | 0.64 | 0.59–0.70 | 4.09 × 10−21 | ||
| Replication | 4861 | 57 126 | 0.165 | 0.202 | 0.77 | 0.72–0.82 | 2.95 × 10−18 | ||||||||
| Joint | 10 266 | 72 228 | 0.72 | 0.68–0.76 | 3.40 × 10−36 | 70.1 | 3.69 × 10−5 | ||||||||
| 20q13.2 | rs8126377 | 20 | 51 590 254 | TSHZ2, ZNF217 | G | Discovery | 5209 | 14 893 | 0.057 | 0.068 | 0.75 | 0.66–0.85 | 3.38 × 10−5 | ||
| Replication | 7278 | 60 647 | 0.063 | 0.054 | 1.04 | 0.95–1.13 | 0.309 | ||||||||
| Joint | 12 487 | 75 540 | 0.94 | 0.87–1.00 | 0.117 | 58.1 | 0.001 | ||||||||
EA, effect allele; EAF, effect allele frequency after pooling the data of all analyzed individuals; OR, odds ratio for the effect allele; 95% CI, 95% confidence interval; I2, heterogeneity statistic; Phet, P-value for heterogeneity.
aGenotyping of this SNP with the Sequenom MassARRAY system for the replication phase was unsuccessful. The SNPs in bold overlap with Table 1.
Figure 3.
Regional association plots for the chromosome 19q13.32 and 5q33.3 loci. Results of the discovery-phase analysis of chromosome 19q13.32 (A) and 5q33.3 (B) in cases aged ≥90 years, generated using LocusZoom (http://csg.sph.umich.edu/locuszoom/) (22). For the two SNPs taken forward to the replication phase (rs4420638 and rs2149954), the results of the joint analysis are plotted. The color of the SNPs is based on the LD with the lead SNP (shown in purple). The blue peaks represent the recombination rates based on HapMap Phase I+II CEU release 22 (hg18/build36), and the RefSeq genes in the region are shown in the lower panel.
Figure 4.
Forest plots for rs4420638 and rs2149954. Forest plots representing the odds ratios with 95% CI of rs4420638 (A) and rs2149954 (B) for the cohorts analyzed in the discovery and replication phase (≥90 years). The size of the boxes represents the sample size of the cohort.
Replication
In addition to the TOMM40/APOE/APOC1 locus, we found eight loci that showed suggestive evidence for association in the discovery-phase meta-analysis of cases aged ≥85 years (P ≤ 1 × 10−5; Table 1), whereas six additional SNPs met this criterion in the meta-analysis of cases aged ≥90 years (Table 2). The most or (when not successfully measured) second most significant SNPs from these 14 loci and the TOMM40/APOE/APOC1 locus were taken forward for replication in 13 060 cases aged ≥85 years (of which 7330 were also ≥90 years) and 61 156 controls from 6 additional studies. In the joint analysis of the discovery and replication phase of the cases aged ≥85 years (9 loci), the TOMM40/APOE/APOC1 locus remained the only genome-wide significant locus (Table 1). The joint analysis of the discovery and replication phase of the cases aged ≥90 years (12 loci), however, showed an additional genome-wide significant locus, rs2149954 (T), on chromosome 5q33.3 (OR = 1.10, P = 1.74 × 10−8; Table 2). Although the association of this SNP with survival up to 85 years is not genome-wide significant (OR = 1.07, P = 4.34 × 10−6; Table 1), the locus likely affects survival from middle age onwards. The regional association plot (based on the discovery phase only) and forest plot of this locus are depicted in Figures 3 and 4, respectively. Conditional analysis of rs4420638 in the discovery phase studies showed that the association of rs2149954 (T) with survival is independent of the TOMM40/APOE/APOC1 locus (P = 7.20 × 10−6 instead of P = 5.98 × 10−6 in the analysis of survival up to 85 years).
Prospective analysis
To determine the association of rs4420638 (TOMM40/APOE/APOC1 locus) and rs2149954 (chromosome 5q33.3 locus) with longitudinal survival, we performed a prospective meta-analysis of the 2 SNPs in 34 103 individuals aged 30–105 years from 11 different cohorts, of which 8582 had died after a mean follow-up time ranging from 2.2 to 17.4 years (Supplementary Material, Table S5). Carriers of the minor allele of rs4420638 (G) showed significantly higher all-cause mortality (hazard ratio (HR) = 1.07, P = 0.019), whereas carriers of the minor allele of rs2149954 (T) demonstrated significantly lower all-cause mortality (HR = 0.95, P = 0.003; Supplementary Material, Table S6).
Association with cardiovascular disease and blood pressure
To gain insight into the mechanism by which the chromosome 5q33.3 locus might promote human longevity, we analyzed the cause-specific mortality of rs2149954. Carriers of the minor allele of rs2149954 have a lower mortality risk for cardiovascular disease (CVD) (HR = 0.86, P = 0.004), which mainly appeared to be caused by protection from stroke (HR = 0.60, P = 2.27 × 10−7). In addition, we observed an effect of this SNP on non-CVD mortality (HR = 0.86, P = 0.002) (Supplementary Material, Table S7). We also examined the Coronary ARtery DIsease Genome-Wide Replication And Meta-Analysis (CARDIoGRAM) GWAS (23), which showed a significant association of rs2149954 with a decreased risk for coronary artery disease (CAD) (OR = 0.96, P = 0.011) (Supplementary Material, Table S8). In addition, two SNPs on chromosome 5q33.3 in high LD with rs2149954, rs9313772 (r2 = 0.928) and rs11953630 (r2 = 0.854) have previously been reported to associate with blood pressure and hypertension (24,25). As expected, examining rs2149954 in the International Consortium for Blood Pressure GWAS (24) showed a significant association of the minor allele with lower diastolic (P = 3.46 × 10−5) and systolic (P = 6.55 × 10−6) blood pressure (Supplementary Material, Table S9). Despite the highly interesting association of the minor allele of rs2149954 with low blood pressure and a decreased risk for CAD, stroke and mortality, its association with decreased all-cause mortality was not influenced by blood pressure in two studies of participants aged ≥75 years (PROSPER and Leiden 85-plus study Cohort II; Supplementary Material, Table S10). This may indicate that at higher ages, this locus influences longevity via pathways additional to those involved in blood pressure regulation.
Phenotypic characterization and pathway analysis
In an attempt to identify the underlying mechanism by which this novel longevity locus at chromosome 5q33.3 could influence human longevity, we examined rs2149954 in the published data of several large GWAS consortia for association with metabolic traits in generally middle-aged individuals. None of the investigated traits, i.e. 2 h glucose (OGTT), Hb1Ac, fasting glucose, fasting insulin, insulin resistance (HOMA-IR), β-cell activity (HOMA-B), total/HDL/LDL cholesterol, triglycerides and type 2 diabetes (26–32), demonstrated evidence for association (all P > 0.05) with rs2149954 (Supplementary Material, Tables S8 and S9).
Gene set enrichment analysis (GSEA) of the meta-analysis results of the discovery-phase analysis of survival aged ≥90 years using Meta-Analysis Gene-set Enrichment of variaNT Associations (MAGENTA) (33), as well as examination of interconnectivity of implicated genes using Gene Relationships Across Implicated Loci (GRAIL) (34) (Supplementary Material, Fig. S3 and Table S11), provided no firm clues for potential pathways involved in human longevity.
Fine mapping and functional characterization
The newly identified longevity locus on chromosome 5q33.3 is located in an intergenic region on chromosome 5q33.3, 302 kb downstream of the EBF1 gene. To determine the functional impact of this locus, we first identified the SNPs in LD with rs2149954 (r2 ≥ 0.8) using the 1000 Genomes CEU Phase 1 data implemented in HaploReg v2 (http://www.broadinstitute.org/mammals/haploreg/haploreg.php) (35). In total, we identified 25 SNPs, spanning a region of ∼22.3 kb (Supplementary Material, Table S12). Subsequently, we examined the potential effects of these SNPs on gene expression using several eQTL databases. None of the SNPs showed an association with gene expression in the various examined tissues, so it is still unclear in which tissue(s) the locus exert its longevity-promoting effect. We did, however, find some promising functional implication of this locus, i.e. the presence of multiple DNase I hypersensitivity sites, transcription factor binding sites and enhancer histone marks, by exploring ENCODE data using HaploReg v2 (35) and RegulomeDB (http://www.regulomedb.org/) (36) (Supplementary Material, Table S12). Very recently, a large intergenic non-coding RNA (lincRNA), RP11-524N5.1, has been annotated right on top of our locus. The poly(A) features of this lincRNA are supported by PolyA-seq reads from liver, muscle and testis. PhastCons 44-way alignment supports conservation of the transcription start site (TSS), 3′ UTR and the third, fifth and last exon of the lincRNA transcript (Fig. 5). The transcript does not align to the mouse genome, but orthologous transcripts are found in other primate genome sequences, suggesting that this is a primate-specific lincRNA.
Figure 5.
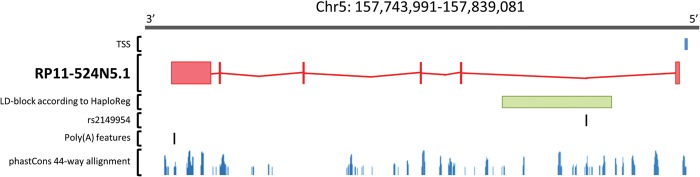
Chromosomal region around rs2149954. The region contains a lincRNA (RP11-524N5.1) for which the poly(A) features are supported by PolyA-seq reads from liver, muscle and testis. RP11-524N5.1 is transcribed from the negative strand, and the phastCons 44-way alignment supports conservation of the TSS, 3′ UTR and the third, fifth and last exon of the transcript. Rs2149954 and the 25 SNPs in high LD (r2 ≥ 0.8, according to HaploReg v2 (35)) are located in the first intron of RP11-524N5.1.
DISCUSSION
We have performed the largest genome-wide association meta-analysis for human longevity, in which a novel locus on chromosome 5q33.3 associating with survival beyond 90 years was identified.
The minor allele of rs2149954 (T) promotes human longevity by reducing the risk of mortality owing to stroke and non-cardiovascular causes. In addition, this allele has previously been associated with low blood pressure, which may explain the protection from CVD mortality risk in middle age. At ages above 80 years, however, low SBP associates with increased mortality (37,38). Hence, the observed blood pressure-independent association of the minor allele with mortality ≥75 years may be due to pleiotropic effects on other mortality-related clinical parameters. Examination of publically available data of several large GWAS consortia for association of the locus with parameters related to glucose and fat metabolism provided as yet no clues for other potentially involved mechanisms.
Rs2149954 is located in an intergenic region on chromosome 5q33.3 between CLINT1 and EBF1. The presence of several regulatory elements in this region implies that transcription factor binding and/or expression of (nearby) genes could be influenced. The currently available eQTL databases did not provide evidence for such effects, which might be due to the limited tissue diversity of the databases. The effects of the chromosome 5q33.3 locus on human longevity might be exerted through the lincRNA, which has recently been annotated right on top of our locus (RP11-524N5.1) and shows evidence for expression in liver, muscle and testis. LincRNAs are involved in chromatin modification and transcriptional regulation (39) and seem to play a role in human disease (40). However, the newly annotated lincRNA is not yet available in the large eQTL databases, and the effect of SNPs in the chromosome 5q33.3 locus on expression of this transcript still needs to be determined. Hence, further functional studies are required to illuminate the mechanism by which this locus influences human longevity.
GWAS has thus far not been a successful approach to identify genome-wide significant hits for human longevity or mortality besides the well-known TOMM40/APOE/APOC1 locus (17–19). The FOXO3A locus, for which the longevity effect is most prominent in individuals aged ≥100 years (41), showed only moderate evidence for association with survival ≥90 years in the discovery phase of our GWAS (lowest P = 1.35 × 10−4 (rs1268161)). Sebastiani and colleagues suggested that human longevity might be explained by a signature consisting of 281 SNPs (42). However, none of the SNPs (except the already known SNP rs2075650 in TOMM40) was significant after adjustment for multiple testing (P < 1.78 × 10−4 (0.05/281)). In addition, we did not observe an enrichment of significant SNPs from their signature in our data (λ = 1.004, Supplementary Material, Fig. S4). Because the association of SNPs other than the TOMM40/APOE/APOC1 locus could not be replicated in this, much larger, GWAS, we have doubts that these signature SNPs are indeed candidate SNPs influencing human longevity. Although we detected merely one novel genome-wide significant locus, the current GWAS had sufficient power, based on our results, to detect lifespan-regulating loci with relatively small effects (OR <0.9 and >1.1).
The genetic component of human longevity is small (∼25%) (10,11) and is assumed to be determined by many genes (12,13). Furthermore, the genetic heterogeneity in ageing and lifespan regulation is expected to be high, because individual genes may contribute by a diversity of late acting deleterious stochastic (germline) variation resulting in a genetic component that is hard to disentangle (13). GWAS of complex late-onset diseases, such as osteoarthritis and Alzheimer's disease, with sample sizes comparable to our current study (43–45), have identified more loci compared with GWAS of longevity. This most likely reflects the greater inherent complexity of the longevity trait, with its diverse spectrum of biological pathways subject to intrinsic and extrinsic (environmental) interactions. Hence, even larger GWAS (>50 000 long-lived individuals) may be required to identify additional longevity loci, preferably in the most stringent phenotype, i.e. the oldest old.
As survival to ages ≥85 or 90 years is relatively common in Western populations, the human longevity trait suffers from etiological heterogeneity. Lifespan extension in the past generations owing to non-genetic factors likely created phenocopies diluting the genetic component of survival to ages ≥85 years. The genetic contribution to survival to ages ≥100 years is higher but will render smaller sample sizes for GWAS. This may explain why the novel locus on chromosome 5q33.3 was only genome-wide significant in the subset analysis of cases aged ≥90 years. For the same reason, a large number of individuals from the control groups (up to 50%, depending on the gender and year of birth of the individuals and demography of the cohort) will live to ages ≥85 years. In 2011, the mean life expectancy at age 65 in Europe was 21.3 years for women and 17.8 years for men (http://epp.eurostat.ec.europa.eu/portal/page/portal/product_details/dataset?P_product_code=TSDDE210), which makes selection of proper controls a challenging issue. The most ideal controls would be individuals from the same birth cohort as the long-lived cases that survived to the mean age of death of that birth cohort. However, for most of these individuals there is no DNA available. Alternatively, we selected controls that have not yet reached the age of 65 years at inclusion to represent the frequency of variants in the general population and minimize selection owing to mortality. Hence, the low contrast between cases and controls likely has reduced our probability of identifying longevity loci.
In addition, there will be differences between case and control cohorts that may have had an impact on our results. An example of a potential confounder is smoking behavior, which was not adequately measured in most elderly cohorts. However, none of the SNPs that were previously associated with smoking behavior in cohorts from European descent (according to the NHGRI GWAS Catalog (http://www.genome.gov/gwastudies/)), namely rs1051730, rs1329650 and rs4105144, show differences between cases (≥85 years) and controls in the joint analysis of the discovery and replication phase (all P > 0.05). We have to note that these SNPs only explain a small proportion of the variance observed in smoking behavior. However, as the frequency of these proxy SNPs for smoking behavior is similar between cases and controls, we expect no obvious differences in smoking behavior between the groups.
In conclusion, besides the previously implicated TOMM40/APOE/APOC1 locus, we identified a novel locus on chromosome 5q33.3 that associates with survival beyond 90 years. Although rs2149954 is associated with survival beyond 90 years at a genome-wide significant level in our study, replication in additional cohorts from European as well as non-European descent is warranted. The minor allele of the lead SNP at this locus, rs2149954, promotes human longevity in a prospective meta-analysis by lowering the risk of mortality owing to stroke and non-cardiovascular causes. The locus harbors a lincRNA and is implicated in blood pressure regulation, but the mechanism by which it influences longevity likely also involves other traits.
MATERIALS AND METHODS
Study populations
The discovery analysis was performed in 7729 cases that survived to ages ≥85 years (of which 5406 also survived to ages ≥90 years) and 16 121 controls below 65 years at baseline, from 14 studies. Replication was performed in 13 060 cases that survived to ages ≥85 years (of which 7330 also survived to ages ≥90 years) and 61 156 controls below 65 years at baseline, from 6 additional studies. All individuals were of European descent. The details of the discovery and replication studies can be found in Supplementary Material, Tables S1 and S2. Some cohorts only provided controls (GOYA, NTR, SU.VI.MAX, TwinsUK and WTCCC2) or only cases (BELFAST, CEPH centenarian cohort, Danish longevity study I/II, Leiden 85-plus Study I/II and Newcastle 85+ Study), whereas others contained both (Calabria cohort, deCODE, EGCUT, GEHA Study, German longevity study, Leiden Longevity Study, Rotterdam Study I/II and TwinGene). The names of the studies in the tables and figures are based on the names of the cohorts containing the cases. The cases and controls used for each study originated from the same country (Supplementary Material, Table S1). The only exception is BELFAST (Northern Ireland), for which we used controls from the NTR (Netherlands). A check in the PROSPER study, which includes individuals from Northern Ireland and the Netherlands, showed that the allele frequencies in control individuals from both countries are similar for our SNPs (data not shown). All participants provided written informed consent, and the study was approved by the relevant institutional review boards.
Genotyping, imputation and genome-wide association analysis
All discovery studies were genotyped using Illumina genotyping arrays, and pre-imputation quality control was performed for each study separately. Imputation was performed using IMPUTE or MACH with reference HapMap Phase I+II CEU release 22 (hg18/build36). Further details about the genotyping, quality control and imputation of each study are summarized in Supplementary Material, Table S2.
Two replication studies (deCODE and the Danish longevity study II) were also genotyped using Illumina genotyping arrays and imputed using IMPUTE with reference HapMap Phase I+II CEU release 22 (hg18/build36) (Danish longevity study II) or deCODE software (deCODE). The other replication studies were genotyped with the Sequenom MassARRAY system using iPLEX Gold genotyping assays (Sequenom, San Diego, CA, USA). More information about the studies used in the replication phase can be found in Supplementary Material, Tables S1 and S2. Of the 15 SNPs measured with the Sequenom MassARRAY system, 13 were successfully genotyped in at least 95% of the samples and the average genotyping call rate was 99.80%. We also checked the concordance between the SNPs measured with the Sequenom MassARRAY system and (imputed) GWAS data of the Leiden 85-plus study I cases, and the average concordance rate was 99.07%. The two SNPs that were not successfully genotyped with the Sequenom MassARRAY system (rs10957550 and rs4420368) were only analyzed in the replication studies, which had imputed GWAS data available (deCODE and the Danish longevity study II).
All studies were analyzed separately using CC-assoc (https://www.msbi.nl/dnn/Research/Genetics/Software/TestsforGWASinrelatedindividuals(cc_assoc).aspx), which is based on a modified version of the score test that takes into account imputation uncertainty and familial relatedness (46). SNPs with a low imputation quality and a MAF of ≤1 or ≤5% (if ncases < 200) were excluded from analysis in the discovery phase. Adjustment for population stratification of the discovery studies was performed by multiplying the -adjusted variances of the score statistic with the genomic inflation factor (λrange = 0.97 – 1.08, Supplementary Material, Table S2) of the study.
Meta-analyses
For the meta-analyses, a fixed-effect approach was used. Scores and variances of the studies were combined to obtain a single meta-statistic, which was adjusted using the genomic inflation factor (λ = 1.019, discovery phase only) (Supplementary Material, Fig. S1). For each analysis, we only used studies with at least 100 cases (Supplementary Material, Table S1). P-values <5 × 10−8 were considered genome-wide significant (47). To determine heterogeneity across the studies, the between-study variance was calculated.
Conditional analysis
To ascertain independent signals at the chromosome 19q13.32 locus, we performed a meta-analysis conditional on rs4420638 in all studies used for the discovery phase analysis in cases aged ≥85 years. The results are depicted in Supplementary Material, Figure S2 and Table S3.
Sex-stratified analysis
Sex-stratified analysis of the cases aged ≥85 years (nwomen = 5400 and nmen = 1865) was performed to investigate the presence of gender-dependent associations. In addition, the 15 loci that showed (suggestive) evidence for association with survival ≥85 and/or ≥90 years were tested for differences between sexes using the formula: The results of this analysis are depicted in Supplementary Material, Table S4.
Prospective analysis
Prospective analysis of rs2149954 and rs4420638 was performed using a Cox proportional hazards model adjusted for age at baseline, sex and study-specific covariates. The details about each of the analyzed cohorts are summarized in Supplementary Material, Table S5.
Pathway analysis
For the pathway analysis, we used GSEA implemented in MAGENTA (http://www.broadinstitute.org/mpg/magenta/) (33). In short, each SNP is mapped to a gene considering a window of 110 kb upstream and 40 kb downstream around the genes. Subsequently, each gene is assigned a gene association score based on the SNP with the lowest P-value, which is mapped to that gene and this score is adjusted for confounding factors like gene size and the amount of SNPs per kb. Genes within the HLA region were removed from analysis owing to high LD and high gene density in that region. The GSEA algorithm tests for over-representation of adjusted gene scores in a given pathway using a pre-defined score rank cutoff (in our case, the 95th and 75th percentile). The generated statistic is then compared with 10 000–1 000 000 gene sets of identical size randomly sampled from the genome to generate an empirical P-value for each pathway. In total, 3216 pathways from Gene Ontology, PANTHER, Ingenuity, KEGG, REACTOME and BIOCARTA were tested. Pathways were considered significant if the FDR-adjusted P-value (the 95th or 75th percentile) was ≤0.05.
To determine the relationship between loci associated with survival ≥90 years, we used GRAIL (http://www.broadinstitute.org/mpg/grail/) (34). In short, this program maps SNPs to genes and subsequently uses a text-mining algorithm on PubMed abstracts to determine connections between these genes. Genes from independent loci, which share informative words, receive a high GRAIL similarity score and are more likely to be functionally related. As we only had a limited number of loci with at least one SNP with a P-value ≤1 × 10−5 (n = 12, Table 2), we decided to perform GRAIL analysis on all loci with at least one SNP with a P-value ≤1 × 10−4 (n = 65).
eQTL analysis
To determine whether rs2149954 or SNPs in LD (r2 ≥ 0.8 based on 1000 Genomes CEU Phase 1 data) influenced gene expression, we searched several eQTL databases, namely (1) the Gutenberg Heart Study database (GHS_Express) (48), which is based on expression data of monocytes; (2) the Genotype-Tissue Expression (GTEx) eQTL database (http://www.ncbi.nlm.nih.gov/gtex/GTEX2/gtex.cgi), which is based on expression data of brain (cerebellum, frontal cortex, temporal cortex and pons), liver and lymphoblastoid cell lines; (3) the GENe Expression VARiation (Genevar) database (http://www.sanger.ac.uk/resources/software/genevar/), which is based on expression data of adipose tissue, fibroblasts, T cells, skin and lymphoblastoid cell lines (49) and (4) the Blood eQTL browser (http://genenetwork.nl/bloodeqtlbrowser/) (50).
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the Augustinus Foundation; Avera Institute for Human Genetics (AIHG); AXA Research Fund; Belfast City Hospital Trust Fund, Research and Education into Ageing-0153; Biobanking and Biomolecular Resources Research Infrastructure (BBMRI –NL, NWO 184.021.007); Biotechnology and Biological Sciences Research Council (BBSRC); Bristol-Myers Squibb; Center for Inherited Disease Research (CIDR); Centre for Medical Systems Biology (CMSB); CERA Foundation; Commissariat à L'Energie Atomique (CEA)-Centre National de Génotypage (CNG); Danish Agency for Science, Technology and Innovation (DASTI)/The Danish Council for Independent Research (DCIR, grant 11-107308); Danish National Research Foundation (DNRF); Department of Health and Social Services (Northern Ireland); DFG-Cluster of Excellence ‘Inflammation at Interfaces’; Dunhill Medical Trust (grant R124/0509); Egmont Foundation; Estonian Science Foundation (grant 7859); Estonian Government (grant SF0180142s08); European Research Council (ERC, advanced grant 230374); European Science Foundation (ESF, EU/QLRT-2001-01254); European Union's Fifth/Sixth/Seventh Framework Programmes (FP5-QLK6-CY-2001-00128, FP6-LIFESCIHEALTH-36894, FP6-LSHM-CT-2004-503270, FP7-HEALTH-2007-B-223004, FP7-HEALTH-F4-2007-201413, FP7-HEALTH-F4-2008-202047, FP7-HEALTH-2009-single-stage-242244 and FP7-HEALTH-2010-two-stage-259679); Fondation Caisse d'Epargne Rhône-Alpes Lyon CERAL (2004–2007); Genetic Association Information Network (GAIN) of the Foundation for the US National Institutes of Health (NIMH, grant MH081802); GenomEUtwin (EU/QLRT-2001-01254; QLG2-CT-2002-01254); Guy's & St Thomas' NHS Foundation Trust; Health Foundation; Heart and Lung foundation (grant 20070481); Innovation-Oriented Research Program on Genomics (SenterNovem, grant IGE05007); Institute for Ageing and Health; Institut National de la Recherche Agronomique (INRA); Institut National de la Santé et de la Recherche Médicale (INSERM); INTERREG 4A programme Syddanmark-Schleswig-K.E.R.N (with EU funds from the European Regional Development Fund); King's College London; Medical Research Council (MRC, grant G0500997 and G0601333); Ministère de l'Enseignement supérieur et de la Recherche (MESR); National Institutes of Health (NIH)/National Institute of Aging (NIA, P01AG08761, R01D0042157-01A and U01DK066134); National Institute for Health Research (NIHR) Newcastle Biomedical Research Centre; NBIC BioAssist (NWO-NBIC/BioAssist/RK/2008.024); Netherlands Consortium for Healthy Ageing (NCHA, grant 050-060-810); Netherlands Genomics Initiative (NGI); Netherlands Heart Foundation (NHF, grant 2001 D 032); Netherlands Organization for Scientific Research (NWO, MagW/ZonMW grant 904-61-090, 904-61-193, 480-04-004, 400-05-717, Spinozapremie 56-464-14192, 175.010.2005.011, 911-03-012, 985-10-002, Addiction-31160008 and Middelgroot-911-09-032); Netspar – Living longer for a good health; NHS North of Tyne (Newcastle Primary Care Trust); Pharmacy Foundation; Regione Autonoma della Sardegna; Rutgers University Cell and DNA Repository (NIMH U24 MH068457-06); Swedish Research Council (grant M-2005-1112); The Competitive Research Funding of the Tampere University Hospital and Academy of Finland; The Danish Interdisciplinary Research Council; The Health Foundation (Helsefonden); The Ministry for Higher Education; The National Program for Research Infrastructure 2007 (grant 09-063256); The March of Dimes Birth Defects Foundation; The Swedish Foundation for Strategic Research (SSF); Unilever Discover Colworth; Université Paris 13; University of Calabria; University of Tartu (grant SP1GVARENG); Velux Foundation; VU University's Institute for Health and Care Research (EMGO+) and Neuroscience Campus Amsterdam (NCA); Wellcome Trust (grant 084762, 085475 and 087436). Funding to pay the Open Access publication charges for this article was provided by IDEAL (FP7-HEALTH-2010-two-stage-259679).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Laurens Wilming from the Wellcome Trust Sanger Institute for the annotation of the lincRNA RP11-524N5.1. This study was undertaken within the framework of European Union's Seventh Framework Programme (FP7/2007-2011) under grant agreement no 259679 (IDEAL). A full list of acknowledgments, including support for each study, is provided in Supplementary Material.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Oeppen J., Vaupel J.W. Demography: broken limits to life expectancy. Science. 2002;296:1029–1031. doi: 10.1126/science.1069675. [DOI] [PubMed] [Google Scholar]
- 2.Jagger C., Gillies C., Moscone F., Cambois E., Van O.H., Nusselder W., Robine J.M. Inequalities in healthy life years in the 25 countries of the European Union in 2005: a cross-national meta-regression analysis. Lancet. 2008;372:2124–2131. doi: 10.1016/S0140-6736(08)61594-9. [DOI] [PubMed] [Google Scholar]
- 3.Barzilai N., Atzmon G., Schechter C., Schaefer E.J., Cupples A.L., Lipton R., Cheng S., Shuldiner A.R. Unique lipoprotein phenotype and genotype associated with exceptional longevity. JAMA. 2003;290:2030–2040. doi: 10.1001/jama.290.15.2030. [DOI] [PubMed] [Google Scholar]
- 4.Derhovanessian E., Maier A.B., Beck R., Jahn G., Hahnel K., Slagboom P.E., de Craen A.J., Westendorp R.G., Pawelec G. Hallmark features of immunosenescence are absent in familial longevity. J. Immunol. 2010;185:4618–4624. doi: 10.4049/jimmunol.1001629. [DOI] [PubMed] [Google Scholar]
- 5.Newman A.B., Glynn N.W., Taylor C.A., Sebastiani P., Perls T.T., Mayeux R., Christensen K., Zmuda J.M., Barral S., Lee J.H., et al. Health and function of participants in the Long Life Family Study: a comparison with other cohorts. Aging. 2011;3:63–76. doi: 10.18632/aging.100242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Slagboom P.E., Beekman M., Passtoors W.M., Deelen J., Vaarhorst A.A., Boer J.M., van den Akker E.B., van H.D., de Craen A.J., Maier A.B., et al. Genomics of human longevity. Philos. Trans. R. Soc. Lond B Biol. Sci. 2011;366:35–42. doi: 10.1098/rstb.2010.0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wijsman C.A., Rozing M.P., Streefland T.C., Le C.S., Mooijaart S.P., Slagboom P.E., Westendorp R.G., Pijl H., van H.D. Familial longevity is marked by enhanced insulin sensitivity. Aging Cell. 2011;10:114–121. doi: 10.1111/j.1474-9726.2010.00650.x. [DOI] [PubMed] [Google Scholar]
- 8.Atzmon G., Schechter C., Greiner W., Davidson D., Rennert G., Barzilai N. Clinical phenotype of families with longevity. J. Am. Geriatr. Soc. 2004;52:274–277. doi: 10.1111/j.1532-5415.2004.52068.x. [DOI] [PubMed] [Google Scholar]
- 9.Westendorp R.G., van Heemst D., Rozing M.P., Frolich M., Mooijaart S.P., Blauw G.J., Beekman M., Heijmans B.T., de Craen A.J., Slagboom P.E. Nonagenarian siblings and their offspring display lower risk of mortality and morbidity than sporadic nonagenarians: The Leiden Longevity Study. J. Am. Geriatr. Soc. 2009;57:1634–1637. doi: 10.1111/j.1532-5415.2009.02381.x. [DOI] [PubMed] [Google Scholar]
- 10.Herskind A.M., McGue M., Holm N.V., Sorensen T.I., Harvald B., Vaupel J.W. The heritability of human longevity: a population-based study of 2872 Danish twin pairs born 1870–1900. Hum. Genet. 1996;97:319–323. doi: 10.1007/BF02185763. [DOI] [PubMed] [Google Scholar]
- 11.Hjelmborg J.V., Iachine I., Skytthe A., Vaupel J.W., McGue M., Koskenvuo M., Kaprio J., Pedersen N.L., Christensen K. Genetic influence on human lifespan and longevity. Hum. Genet. 2006;119:312–321. doi: 10.1007/s00439-006-0144-y. [DOI] [PubMed] [Google Scholar]
- 12.Finch C.E., Tanzi R.E. Genetics of aging. Science. 1997;278:407–411. doi: 10.1126/science.278.5337.407. [DOI] [PubMed] [Google Scholar]
- 13.Kirkwood T.B., Cordell H.J., Finch C.E. Speed-bumps ahead for the genetics of later-life diseases. Trends Genet. 2011;27:387–388. doi: 10.1016/j.tig.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 14.Schachter F., Cohen D., Kirkwood T. Prospects for the genetics of human longevity. Hum. Genet. 1993;91:519–526. doi: 10.1007/BF00205074. [DOI] [PubMed] [Google Scholar]
- 15.Ganna A., Rivadeneira F., Hofman A., Uitterlinden A.G., Magnusson P.K., Pedersen N.L., Ingelsson E., Tiemeier H. Genetic determinants of mortality. Can findings from genome-wide association studies explain variation in human mortality? Hum. Genet. 2013;132:553–561. doi: 10.1007/s00439-013-1267-6. [DOI] [PubMed] [Google Scholar]
- 16.Beekman M., Nederstigt C., Suchiman H.E., Kremer D., van der Breggen R., Lakenberg N., Alemayehu W.G., de Craen A.J., Westendorp R.G., Boomsma D.I., et al. Genome-wide association study (GWAS)-identified disease risk alleles do not compromise human longevity. Proc. Natl. Acad. Sci. USA. 2010;107:18046–18049. doi: 10.1073/pnas.1003540107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deelen J., Beekman M., Uh H.W., Helmer Q., Kuningas M., Christiansen L., Kremer D., van der B.R., Suchiman H.E., Lakenberg N., et al. Genome-wide association study identifies a single major locus contributing to survival into old age; the APOE locus revisited. Aging Cell. 2011;10:686–698. doi: 10.1111/j.1474-9726.2011.00705.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Newman A.B., Walter S., Lunetta K.L., Garcia M.E., Slagboom P.E., Christensen K., Arnold A.M., Aspelund T., Aulchenko Y.S., Benjamin E.J., et al. A meta-analysis of four genome-wide association studies of survival to age 90 years or older: the Cohorts for Heart and Aging Research in Genomic Epidemiology Consortium. J. Gerontol. A Biol. Sci. Med. Sci. 2010;65:478–487. doi: 10.1093/gerona/glq028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deelen J., Beekman M., Capri M., Franceschi C., Slagboom P.E. Identifying the genomic determinants of aging and longevity in human population studies: progress and challenges. Bioessays. 2013;35:386–396. doi: 10.1002/bies.201200148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gavrilova N.S., Gavrilov L.A., Evdokushkina G.N., Semyonova V.G., Gavrilova A.L., Evdokushkina N.N., Kushnareva Y.E., Kroutko V.N., Andreyev A.Y. Evolution, mutations, and human longevity: European royal and noble families. Hum. Biol. 1998;70:799–804. [PubMed] [Google Scholar]
- 21.Nebel A., Kleindorp R., Caliebe A., Nothnagel M., Blanche H., Junge O., Wittig M., Ellinghaus D., Flachsbart F., Wichmann H.E., et al. A genome-wide association study confirms APOE as the major gene influencing survival in long-lived individuals. Mech. Ageing Dev. 2011;132:324–330. doi: 10.1016/j.mad.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 22.Pruim R.J., Welch R.P., Sanna S., Teslovich T.M., Chines P.S., Gliedt T.P., Boehnke M., Abecasis G.R., Willer C.J. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schunkert H., Konig I.R., Kathiresan S., Reilly M.P., Assimes T.L., Holm H., Preuss M., Stewart A.F., Barbalic M., Gieger C., et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat. Genet. 2011;43:333–338. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ehret G.B., Munroe P.B., Rice K.M., Bochud M., Johnson A.D., Chasman D.I., Smith A.V., Tobin M.D., Verwoert G.C., Hwang S.J., et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–109. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wain L.V., Verwoert G.C., O'Reilly P.F., Shi G., Johnson T., Johnson A.D., Bochud M., Rice K.M., Henneman P., Smith A.V., et al. Genome-wide association study identifies six new loci influencing pulse pressure and mean arterial pressure. Nat. Genet. 2011;43:1005–1011. doi: 10.1038/ng.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dupuis J., Langenberg C., Prokopenko I., Saxena R., Soranzo N., Jackson A.U., Wheeler E., Glazer N.L., Bouatia-Naji N., Gloyn A.L., et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat. Genet. 2010;42:105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manning A.K., Hivert M.F., Scott R.A., Grimsby J.L., Bouatia-Naji N., Chen H., Rybin D., Liu C.T., Bielak L.F., Prokopenko I., et al. A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat. Genet. 2012;44:659–669. doi: 10.1038/ng.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morris A.P., Voight B.F., Teslovich T.M., Ferreira T., Segre A.V., Steinthorsdottir V., Strawbridge R.J., Khan H., Grallert H., Mahajan A., et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat. Genet. 2012;44:981–990. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saxena R., Hivert M.F., Langenberg C., Tanaka T., Pankow J.S., Vollenweider P., Lyssenko V., Bouatia-Naji N., Dupuis J., Jackson A.U., et al. Genetic variation in GIPR influences the glucose and insulin responses to an oral glucose challenge. Nat. Genet. 2010;42:142–148. doi: 10.1038/ng.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soranzo N., Sanna S., Wheeler E., Gieger C., Radke D., Dupuis J., Bouatia-Naji N., Langenberg C., Prokopenko I., Stolerman E., et al. Common variants at 10 genomic loci influence hemoglobin A(1)(C) levels via glycemic and nonglycemic pathways. Diabetes. 2010;59:3229–3239. doi: 10.2337/db10-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Strawbridge R.J., Dupuis J., Prokopenko I., Barker A., Ahlqvist E., Rybin D., Petrie J.R., Travers M.E., Bouatia-Naji N., Dimas A.S., et al. Genome-wide association identifies nine common variants associated with fasting proinsulin levels and provides new insights into the pathophysiology of type 2 diabetes. Diabetes. 2011;60:2624–2634. doi: 10.2337/db11-0415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Teslovich T.M., Musunuru K., Smith A.V., Edmondson A.C., Stylianou I.M., Koseki M., Pirruccello J.P., Ripatti S., Chasman D.I., Willer C.J., et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Segre A.V., Groop L., Mootha V.K., Daly M.J., Altshuler D. Common inherited variation in mitochondrial genes is not enriched for associations with type 2 diabetes or related glycemic traits. PLoS Genet. 2010;6:e1001058. doi: 10.1371/journal.pgen.1001058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raychaudhuri S., Plenge R.M., Rossin E.J., Ng A.C., Purcell S.M., Sklar P., Scolnick E.M., Xavier R.J., Altshuler D., Daly M.J. Identifying relationships among genomic disease regions: predicting genes at pathogenic SNP associations and rare deletions. PLoS Genet. 2009;5:e1000534. doi: 10.1371/journal.pgen.1000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ward L.D., Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucl. Acids Res. 2012;40:D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boyle A.P., Hong E.L., Hariharan M., Cheng Y., Schaub M.A., Kasowski M., Karczewski K.J., Park J., Hitz B.C., Weng S., et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Molander L., Lovheim H., Norman T., Nordstrom P., Gustafson Y. Lower systolic blood pressure is associated with greater mortality in people aged 85 and older. J. Am. Geriatr. Soc. 2008;56:1853–1859. doi: 10.1111/j.1532-5415.2008.01948.x. [DOI] [PubMed] [Google Scholar]
- 38.Oates D.J., Berlowitz D.R., Glickman M.E., Silliman R.A., Borzecki A.M. Blood pressure and survival in the oldest old. J. Am. Geriatr. Soc. 2007;55:383–388. doi: 10.1111/j.1532-5415.2007.01069.x. [DOI] [PubMed] [Google Scholar]
- 39.Mercer T.R., Dinger M.E., Mattick J.S. Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 40.Esteller M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 41.Flachsbart F., Caliebe A., Kleindorp R., Blanche H., von Eller-Eberstein H., Nikolaus S., Schreiber S., Nebel A. Association of FOXO3A variation with human longevity confirmed in German centenarians. Proc. Natl. Acad. Sci. USA. 2009;106:2700–2705. doi: 10.1073/pnas.0809594106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sebastiani P., Solovieff N., Dewan A.T., Walsh K.M., Puca A., Hartley S.W., Melista E., Andersen S., Dworkis D.A., Wilk J.B., et al. Genetic signatures of exceptional longevity in humans. PLoS One. 2012;7:e29848. doi: 10.1371/journal.pone.0029848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zeggini E., Panoutsopoulou K., Southam L., Rayner N.W., Day-Williams A.G., Lopes M.C., Boraska V., Esko T., Evangelou E., Hoffman A., et al. Identification of new susceptibility loci for osteoarthritis (arcOGEN): a genome-wide association study. Lancet. 2012;380:815–823. doi: 10.1016/S0140-6736(12)60681-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hollingworth P., Harold D., Sims R., Gerrish A., Lambert J.C., Carrasquillo M.M., Abraham R., Hamshere M.L., Pahwa J.S., Moskvina V., et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer's disease. Nat. Genet. 2011;43:429–435. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Naj A.C., Jun G., Beecham G.W., Wang L.S., Vardarajan B.N., Buros J., Gallins P.J., Buxbaum J.D., Jarvik G.P., Crane P.K., et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer's disease. Nat. Genet. 2011;43:436–441. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Uh H.W., Deelen J., Beekman M., Helmer Q., Rivadeneira F., Hottenga J.J., Boomsma D.I., Hofman A., Uitterlinden A.G., Slagboom P.E., et al. How to deal with the early GWAS data when imputing and combining different arrays is necessary. Eur. J. Hum. Genet. 2012;20:572–576. doi: 10.1038/ejhg.2011.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pe'er I., Yelensky R., Altshuler D., Daly M.J. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet. Epidemiol. 2008;32:381–385. doi: 10.1002/gepi.20303. [DOI] [PubMed] [Google Scholar]
- 48.Zeller T., Wild P., Szymczak S., Rotival M., Schillert A., Castagne R., Maouche S., Germain M., Lackner K., Rossmann H., et al. Genetics and beyond--the transcriptome of human monocytes and disease susceptibility. PLoS One. 2010;5:e10693. doi: 10.1371/journal.pone.0010693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang T.P., Beazley C., Montgomery S.B., Dimas A.S., Gutierrez-Arcelus M., Stranger B.E., Deloukas P., Dermitzakis E.T. Genevar: a database and Java application for the analysis and visualization of SNP-gene associations in eQTL studies. Bioinformatics. 2010;26:2474–2476. doi: 10.1093/bioinformatics/btq452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Westra H.J., Peters M.J., Esko T., Yaghootkar H., Schurmann C., Kettunen J., Christiansen M.W., Fairfax B.P., Schramm K., Powell J.E., et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat. Genet. 2013;45:1238–1243. doi: 10.1038/ng.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.