Abstract
The Maf-family leucine zipper transcription factor NRL is essential for rod photoreceptor development and functional maintenance in the mammalian retina. Mutations in NRL are associated with human retinopathies, and loss of Nrl in mice leads to a cone-only retina with the complete absence of rods. Among the highly down-regulated genes in the Nrl−/− retina, we identified receptor expression enhancing protein 6 (Reep6), which encodes a member of a family of proteins involved in shaping of membrane tubules and transport of G-protein coupled receptors. Here, we demonstrate the expression of a novel Reep6 isoform (termed Reep6.1) in the retina by exon-specific Taqman assay and rapid analysis of complementary deoxyribonucleic acid (cDNA) ends (5′-RACE). The REEP6.1 protein includes 27 additional amino acids encoded by exon 5 and is specifically expressed in rod photoreceptors of developing and mature retina. Chromatin immunoprecipitation assay identified NRL binding within the Reep6 intron 1. Reporter assays in cultured cells and transfections in retinal explants mapped an intronic enhancer sequence that mediated NRL-directed Reep6.1 expression. We also demonstrate that knockdown of Reep6 in mouse and zebrafish resulted in death of retinal cells. Our studies implicate REEP6.1 as a key functional target of NRL-centered transcriptional regulatory network in rod photoreceptors.
INTRODUCTION
The mammalian retina offers a convenient paradigm to investigate complexities of neuronal differentiation and degeneration (1). Six major types of neurons in the mammalian retina are organized in three distinct cell layers to facilitate the capture, integration and processing of visual information. All retinal neurons and Müller glia originate from common pools of multipotent progenitor cells in a conserved order of birth under the stringent control of an intrinsic genetic program and signals from the microenvironment (2–4). Over 70% of the retinal neurons are photoreceptors that are highly specialized to capture photons and initiate the visual process. Rod photoreceptors dominate the retina of most mammals, including humans, and mediate dim light vision, whereas cone photoreceptors are responsible for day light and color vision. In humans, dysfunction and death of photoreceptors are the primary cause of retinal degenerative diseases (5). Rod degeneration is an early indicator of disease even in age-related macular degeneration (6).
Development of rod photoreceptors and their functional maintenance are critically dependent upon the Maf family basic motif leucine zipper transcription factor NRL (3). Abrogation of NRL expression in mice (Nrl−/−) leads to a retina with the complete absence of rods but with dramatically enhanced S-cone photoreceptors (7), whereas ectopic expression of NRL results in a rod-only mouse retina (8). In developing mouse retina, NRL and thyroid hormone receptor β2 (Trβ2) determine the specification of three photoreceptor subtypes (rod, M-cone and S-cone) from postmitotic precursors (9). Other key transcription factors that regulate photoreceptor development include OTX2 (10–12), retinoid-related orphan nuclear receptor RORβ (13) and cone-rod homeobox CRX (14–16). NRL interacts with a number of transcriptional regulatory proteins including CRX, orphan nuclear receptor NR2E3 and NonO/p54nrb to control the expression of rhodopsin and other rod-specific genes (17–19).
Gene profiling analysis of Nrl−/− retina and chromatin immunoprecipitation followed by next generation sequencing (ChIP-Seq) have identified a large number of NRL target genes that include transcription factors (such as NR2E3, MEF2C and ESRRB) as well as rod and cone expressed genes with diverse functions (20–26). These studies have yielded new insights into rod development and provided candidate genes for retinal degenerative diseases (22). Among the top-ranked down-regulated genes in the Nrl−/− retina (20,26) is Reep6, encoding one of the six mammalian REEP family proteins, which are orthologs of DP1/Yop1p family implicated in shaping tubular organelle structures and intracellular trafficking of receptors to membranes (27,28). REEP1 is implicated in enhancing cell surface expression of odorant receptors (29,30), probably by facilitating vesicular transport (31). Mutations in REEP1 cause hereditary spastic paraplegia by disrupting tubular endoplasmic reticulum (ER) shaping and microtubule interactions (32). REEP3, a candidate gene for autism (33), was recently shown to be involved in clearance of ER from metaphase chromatin during cell cycle progression (34). REEP5 (DP1) was identified by its interaction with reticulon, and its yeast homolog Yop1p has been implicated in ER shaping (27). REEP6 (also called Dp1l1) was initially identified in retinal ganglion cells (35), and REEP6 polymorphisms have been associated with colon cancer and inflammatory bowel disease (36). However, little is known about REEP6 function and its role, if any, in human disease.
We recently demonstrated that Reep6 is highly expressed in rod photoreceptors of mouse retina (37). As rod outer segment development and maintenance require high membrane turnover in conjunction with transport of rhodopsin, and REEP proteins are good candidates to perform such roles through their ER–microtubule interaction, we wanted to further explore the regulation and importance of REEP6 in the retina. Here, we report the identification of a novel retina-specific isoform of Reep6. We have identified an intronic enhancer element that binds to NRL and is required for Reep6 transcription in both in vitro and in vivo assays. Furthermore, knockdown experiments demonstrate the requirement of Reep6 in mouse and zebrafish retina. Our studies reveal REEP6 as a key target of NRL for maintaining rod photoreceptor function and homeostasis.
RESULTS
The long isoform of Reep6 is highly expressed in the retina
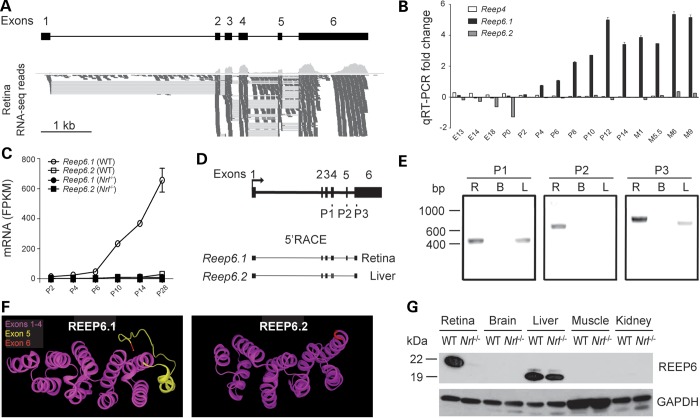
The Reep6 gene encodes several splice variants (ENSMUSG00000035504, Ensembl); of these, two major variants, listed in the NCBI database, differ by an alternate exon 5. To identify the isoform(s) expressed in the mouse retina, we checked the RNA-Seq data generated in our laboratory (38,39). We identified RNA sequence reads corresponding to the longer isoform of Reep6 with exon 5 (RefSeq. Isoform 1; NM_139292.2), termed Reep6.1, in postnatal (P) day-10 mouse retina (Fig. 1A). Although the shorter Reep6 isoform (Reep6.2; NM_001204931.1) lacking exon 5 is detectable in RNA-Seq from early developmental stages, Reep6.1 is the predominant form at P0 and the only form detected at P21 retina (data not shown).
Figure 1.
Reep6 structure, expression profile and identification of a retina-specific isoform. (A) Expression of Reep6 long transcript (Reep6.1), with exon 5, in the retina. The transcript analysis was done by RNA-Seq using WT mouse retina at postnatal (P) Day 10. (B) Developmental expression profile of Reep6.1 (with exon 5), Reep6.2 (without exon 5) and Reep4 transcripts. These transcripts from WT mouse retina, at indicated ages, were quantified by RT-qPCR using exon-specific Taqman probes. (C) Expression levels of Reep6.1 and Reep6.2 transcripts in developing and mature photoreceptors purified by flow-sorting from WT and Nrl−/− retina, based on RNA-Seq quantification. x-axis shows developmental time, and y-axis represents fragments per kilobase of exon per million fragments mapped (FPKM) ± standard error of mean. (D) Reep6 gene structure showing the primers used for 5′-RACE. Exons are shown as black boxes and the connecting lines represent spliced regions. P1, P2 and P3 are reverse primers. P2 primer is specific for exon 5. (E) Agarose gel showing 5′-RACE products developed from retina (R), brain (B) and liver (L) using reverse primers residing in exon 4 (P1), exon 5 (P2) and exon 6 (P3). Exon 5 was recognized with P2 and Reep6.1 transcript was present only in the retina. P3 generated a longer product from retina compared with the product from liver. (F) Protein structure modeling of full length (REEP6.1) and REEP6.2, without exon 5. The amino acid residues coded by exon 5 (shown in yellow) confer additional structural features to the remaining region. (G) REEP6.1 expression in the retina. Immunoblot analysis of various tissues using anti-REEP6 antibody demonstrates retina-specific expression of REEP6.1, which is not detected in the Nrl−/− retina. REEP6.2 is expressed in the liver. No REEP6 protein is observed in brain, muscle and kidney. The analysis was done using tissues from 1-month old WT and Nrl−/− mice. GAPDH was used as a loading control.
We then examined the transcript levels of Reep6.1 (with exon 5) and Reep6.2 (without exon 5) in developing and mature retina using splice-junction specific Taqman probes in quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) assays (Fig. 1B). Reep6.1 transcript is evident at P4, reaching its peak level at P12, whereas Reep6.2 expression was barely detectable (Fig. 1B). The expression of Reep4, another member of the REEP family, is not detected in the mouse retina (Fig. 1B). The increase in Reep6.1 transcript follows the course of rod maturation during mouse retinal development. To test this hypothesis, we performed transcript-level quantification using RNA-Seq data from rod photoreceptors purified from Nrl-green fluorescent protein (GFP) transgenic mouse (26). Consistent with the whole retina data, developmental expression profiling revealed a sharp increase in the expression levels of Reep6.1 after P6 and during rod photoreceptor maturation (Fig. 1C). Moreover, Reep6.1 transcripts were barely detectable in purified photoreceptors from cone-only Nrl−/− retina (Fig. 1C).
To further examine the tissue specificity of Reep6 isoform expression, we performed 5′-RACE using ribonucleic acid (RNA) from retina and liver, two tissues reported to exhibit high levels of Reep6 transcripts (35). As retina is part of the central nervous system, we also tested brain RNA. We used reverse primers P1 (for exon 4) and P3 (for exon 6) to target common exons in Reep6.1 and Reep6.2 isoforms, and P2 (for exon 5) to target exon 5 specifically (Fig. 1D). P1 primer produced products of similar size in 5′-RACE using retina and liver RNA, whereas P2 primer generated a product only when retinal RNA was used (Fig. 1E). As predicted, the P3 primer produced a longer PCR product (with exon 5) from retinal RNA compared with liver RNA (without exon 5) (Fig. 1E). Brain RNA did not generate any product with any of the primers (Fig. 1E). Sequencing of 15 clones generated from Reep6 5′-RACE products showed that transcripts from the retina and liver share the same transcription start site (TSS) located at the beginning of exon 1, and that the retinal transcript, but not the liver transcript, included Reep6 exon 5 (data not shown). These results further confirm that Reep6.1 is expressed specifically in the retina.
The presence of an additional exon in Reep6.1 prompted us to investigate its possible consequence for protein structure. In silico analysis of REEP6.1 and REEP6.2 using I-TASSER (40–42) predicted an additional coil and a loop structure that are displaced from the core as a result of 27 additional amino acids (corresponding to exon 5) in REEP6.1 protein (Fig. 1F, shaded in yellow).
Using anti-REEP6 antibody (37) for immunoblotting, we detected high expression of REEP6 protein in the retina and liver, but not in the brain, muscle and kidney (Fig. 1G). Furthermore, REEP6 protein in the retina showed higher molecular mass compared with that from the liver, consistent with 5′-RACE data and the presence of additional 27 residues in REEP6.1 (see Fig. 1D and E). Concordant with the photoreceptor gene profiling data (see Fig. 1C), REEP6.1 protein is not detectable in the Nrl−/− retina, but REEP6.2 expression is unaffected in the Nrl−/− liver (Fig. 1G).
REEP6.1 is specifically expressed in rod photoreceptors
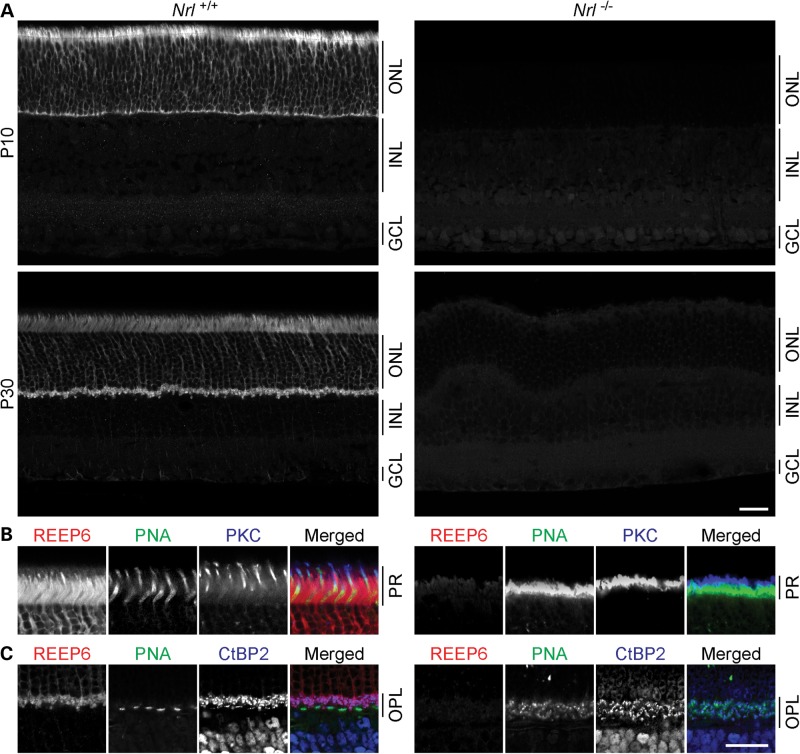
The preceding studies provide strong evidence in favor of specific expression of REEP6.1 in rod photoreceptors and the control of its expression by NRL. To test rod photoreceptor-specific expression of REEP6.1, we performed immunofluorescence analysis of Nrl+/+ [wild-type (WT)] and Nrl−/− retinal sections using REEP6 antibody. In WT retina, REEP6 immunolabeling was present throughout the outer nuclear layer (ONL) as well as in the outer plexiform layer (OPL) both during the development (P10) and in the mature (P30) retina (Fig. 2A). REEP6.1 protein was not detected in Nrl−/− retina (Fig. 2A).
Figure 2.
Reep6 expression in rods requires NRL. (A) Mouse retinas at P10 and P30 labeled with anti-REEP6 antibody. At both ages, REEP6 immunolabeling in the WT retina is restricted to the photoreceptor layer (PR), the ONL and OPL, but is entirely absent in the Nrl−/− retina. (B) Higher magnification micrographs through the photoreceptor OS of WT retina show that REEP6 intensely labels the majority of photoreceptors, yet sparing the cone photoreceptors that are labeled with PNA or antibodies to PKC. In the Nrl−/− retina, the entire PR is labeled for both PNA and PKC and lacks REEP6 labeling. (C) Higher magnification double-labeled images from the OPL confirm that REEP6 labeling is restricted to the rod spherules, evidenced by its absence in the CtBP2 and PNA double-labeled cone pedicles. The OPL of the Nrl−/− retina contains no REEP6 labeling, and the distribution of CtBP2-positive synaptic terminals overlaps that of the PNA-positive (respecified) cone terminals. PR, photoreceptor layer; ONL, outer nuclear layer; OPL, outer plexiform layer; INL, inner nuclear layer; GCL, ganglion cell layer. Scale bar = 25 μm.
Notably, the occasional sporadic absence of REEP6.1 immunolabeling was observed within the outer segments (OS), particularly evident in the mature retina (see Fig. 2A). Double-labeling with cone photoreceptor-specific markers, such as peanut agglutinin [PNA; (43)] and anti-protein kinase C (PKC) antibody (44), revealed that the REEP6.1-negative regions correspond to cone OS embedded among the more densely packed REEP6-positive rod OS (Fig. 2B). Consistent with the exclusion of REEP6.1 staining from cone photoreceptors, no REEP6.1 labeling was present within OS in the Nrl−/− retina, in which all photoreceptors are functional cones (7,45,46).
Next, we examined REEP6.1 expression within the OPL. REEP6.1 labeling was also found in the photoreceptor terminals, evidenced by double-labeling with antibodies to C-terminal binding protein 2 (CtBP2; also called Ribeye), a protein associated with the synaptic ribbons of all photoreceptor cells [(47); Fig. 2C]. REEP6.1 immunolabeling, however, was excluded from CtBP2-labeled photoreceptor terminals that were also PNA-positive, indicating that REEP6.1 is restricted to the terminal endings (the spherules) of the rod photoreceptors only. In the cone-only Nrl−/− retina, no REEP6 labeling was present in the OPL, where the entire population of CtBP2-positive photoreceptor terminals was also PNA-positive (Fig. 2C). In contrast to a previous report (35), no REEP6 immunostaining was observed in ganglion cells both in WT and Nrl−/− retina. Thus, REEP6.1 is specifically expressed in rod photoreceptors of the mouse retina.
NRL binds to an intronic enhancer in Reep6 and increases its promoter activity in an in vitro reporter assay
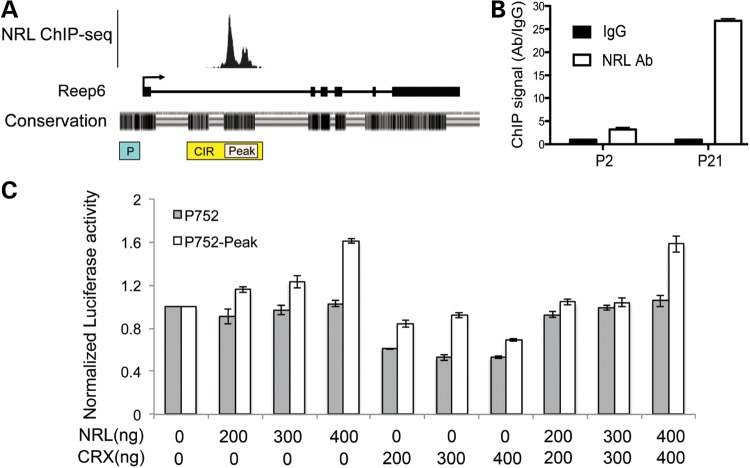
To test whether Reep6 is a transcriptional target of NRL, we examined our published NRL ChIP-Seq data (22). NRL binding was detected within the first intron of the mouse Reep6 gene (Fig. 3A), suggesting that the intronic region could act as an enhancer for NRL-mediated transactivation and produce Reep6.1 transcript in rod photoreceptors. We therefore performed ChIP-qPCR assays using mature (P21) and developing (P2, time of the peak of rod birth) retina. High NRL binding was evident in the intronic region of Reep6 (Fig. 3B), concordant with previous ChIP-Seq results and higher expression of Reep6.1 transcript in mature rod photoreceptors (see Fig. 1C).
Figure 3.
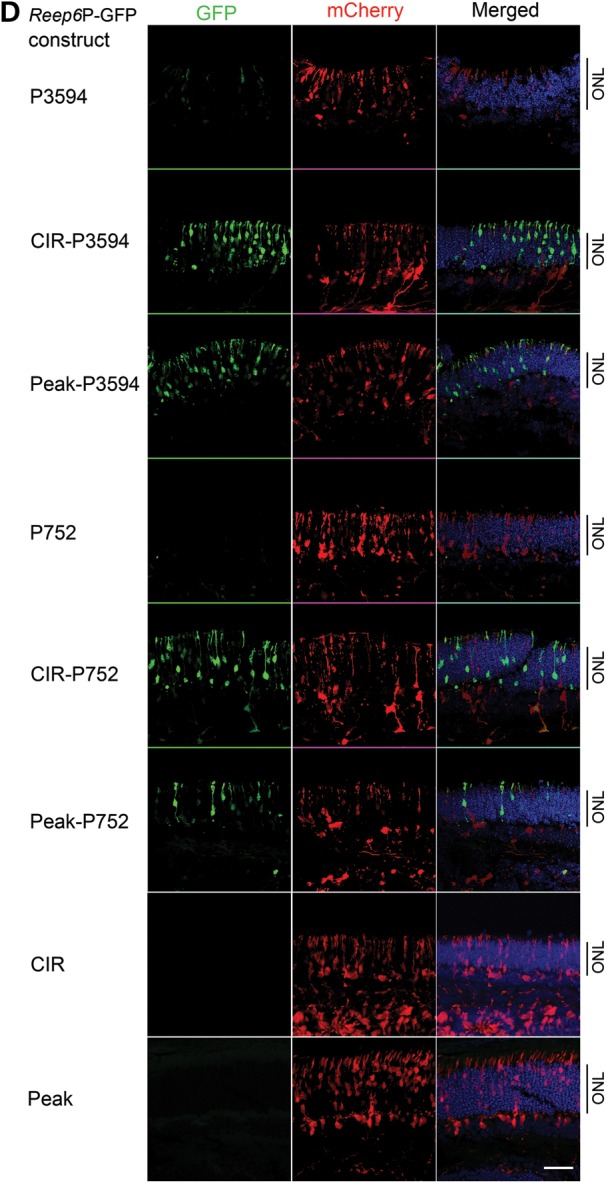
An intronic enhancer in Reep6 is required for NRL binding and its expression in the retina. (A) Mouse Reep6 gene structure with exon:intron boundary is shown. Black boxes are exons and connecting lines indicate introns. A directional arrow indicates TSS. ChIP-seq signal peaks demonstrate NRL binding within Reep6 intron 1 at P28. The sequence conservation between mouse and human is represented with black strips (density is proportional to conservation) below the gene structure. The bottom part shows Reep6 genomic fragments cloned in green fluorescent protein (GFP) constructs. P: promoter, CIR: conserved intronic region (conservation between mouse and human) that encompasses the NRL peak region, Peak: NRL ChIP-Seq peak region. (B) NRL binding to Reep6 intronic region measured by ChIP-qPCR at P2 and P21. The x- and y-axes are age and ChIP signal, respectively. (C) Luciferase assays suggesting the requirement of conserved ‘peak’ sequence for NRL-mediated transcription. HEK293 cells were co-transfected with the luciferase-reporter construct driven by Reep6 basal promoter of 752 bp (P752) with or without ‘peak’ and expression plasmids for human NRL and/or human CRX. Presence of CRX did not affect NRL's action at high concentration. The data were obtained at 48 h after transfection, and represented as mean ± standard deviation. The x-axis shows quantity of NRL and/or CRX used. The y-axis shows normalized signal from luciferase reporter. (D) Representative confocal images of P7 CD1 mouse retina, electroporated in vitro at P0 with indicated constructs. CAG-mCherry (red) was used as the electroporation control. Reep6 promoter (P3594) without CIR generated very weak green signal (top panel), whereas adding CIR or peak significantly increased GFP expression (green signal). Instead of P3594, using a shorter promoter of 752 bp (P752) produced similar results though signal intensity was lesser than with P3594. P752 or CIR or Peak alone did not generate any reporter signal. Nuclei were visualized by 4′,6-diamidino-2-phenylindole (DAPI) (blue). ONL, outer nuclear layer. Scale bar = 10 μm.
To test whether NRL binding enhances Reep6 promoter activity, we cloned the NRL-binding intronic region at the 3′-end of a luciferase reporter gene that is driven by the Reep6 basal promoter (752 bp). Co-transfection of the reporter construct with NRL expression plasmid in HEK293 cells increased luciferase expression in the presence of the enhancer in a dose-dependent manner, whereas NRL co-transfection had no effect on Reep6 basal promoter in the absence of this sequence (Fig. 3C). Co-transfection with CRX, which controls the expression of many rod genes synergistically with NRL (18,22), did not alter Reep6 promoter activity with or without the enhancer and showed no effect on NRL-mediated transactivation of Reep6 enhancer activity (Fig. 3C).
NRL activates Reep6 promoter in the retina through an intronic enhancer element
To investigate the mechanism of Reep6.1 regulation in the retina, we generated GFP reporter constructs under the control of Reep6 promoter and performed in vitro transfection of retina using electroporation (48,49) (Fig. 3D). We first tested genomic regions upstream of TSS for promoter activity and genomic fragments up to (−) 3594 bp (P3594), which did not show significant promoter activity (Fig. 3D). We then cloned a 617 bp intronic region that includes the NRL ChIP-Seq peak (termed ‘Peak’) and a 1368 bp intronic region (CIR) that contains two additional highly conserved sequence clusters (see Fig. 3A), one of which overlaps with the peak region, in the Reep6 promoter-GFP constructs. The CIR significantly increased the activity of a longer Reep6 promoter (P3594) (shown as construct CIR-P3594 in Fig. 3D) and a short promoter (P752) in construct CIR-P752 (Fig. 3D). The ‘Peak’ sequence also increased Reep6 promoter activity though at a lower level (Fig. 3D). Our results suggest that the CIR acts as an enhancer to mediate transactivation of Reep6 by NRL. CIR or ‘Peak’ region alone did not show detectable promoter activity in electroporation assays (Fig. 3D).
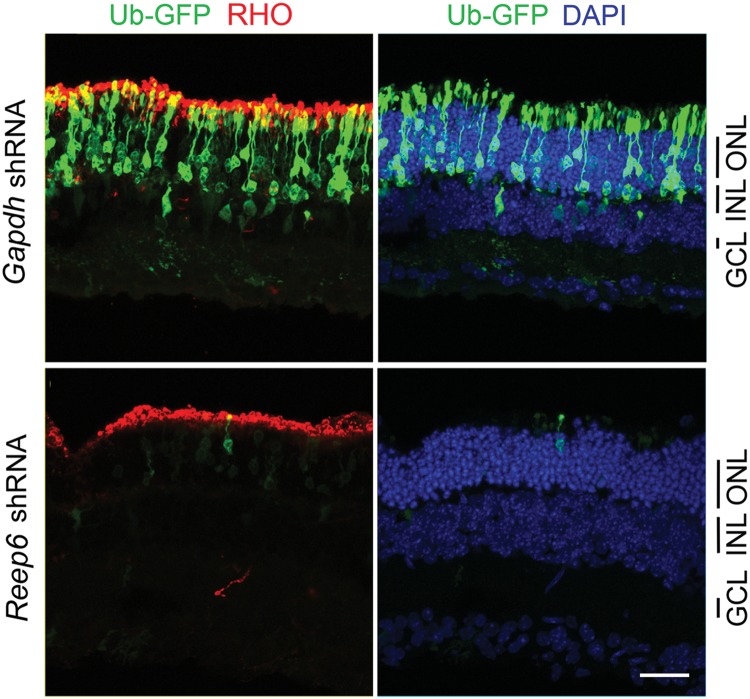
To further validate NRL as a key positive regulator of Reep6 transcription, we used shRNA to knockdown Nrl expression in the mouse retina by in vivo electroporation and monitored CIR-dependent Reep6 promoter activity. Nrl shRNA abolished the expression of GFP while Gapdh shRNA had no effect, demonstrating that NRL is the primary activator of Reep6 transcription (Fig. 4). These results are in concordance with the dramatic reduction of REEP6 protein in Nrl−/− mouse retina (see Figs 1G and 2).
Figure 4.
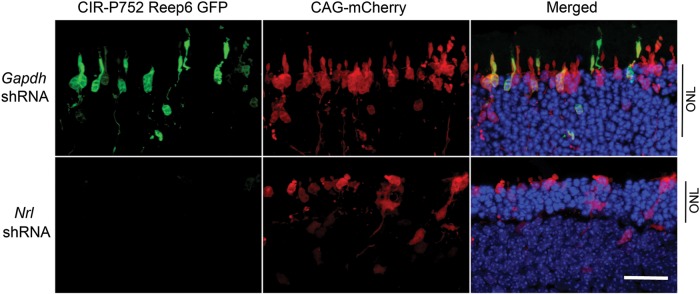
NRL knockdown decreased Reep6 promoter activity in adult mouse retina. Representative confocal images of P21 CD1 mouse retinas, electroporated in vivo at P0 with the Reep6-driven GFP reporter construct (CIR-P752-GFP). mCherry construct driven by CAG promoter (CAG-mCherry) was used as electroporation control. Retina co-transfected with Gapdh shRNA control and GFP reporter showed green signal (top), whereas Nrl-shRNA transfection greatly eliminated Reep6 driven green signal (bottom). GFP, mCherry and DAPI are shown in green, red and blue, respectively. ONL, outer nuclear layer. Scale bar = 10 μm.
Reep6 is required for retinal development in zebrafish and mouse
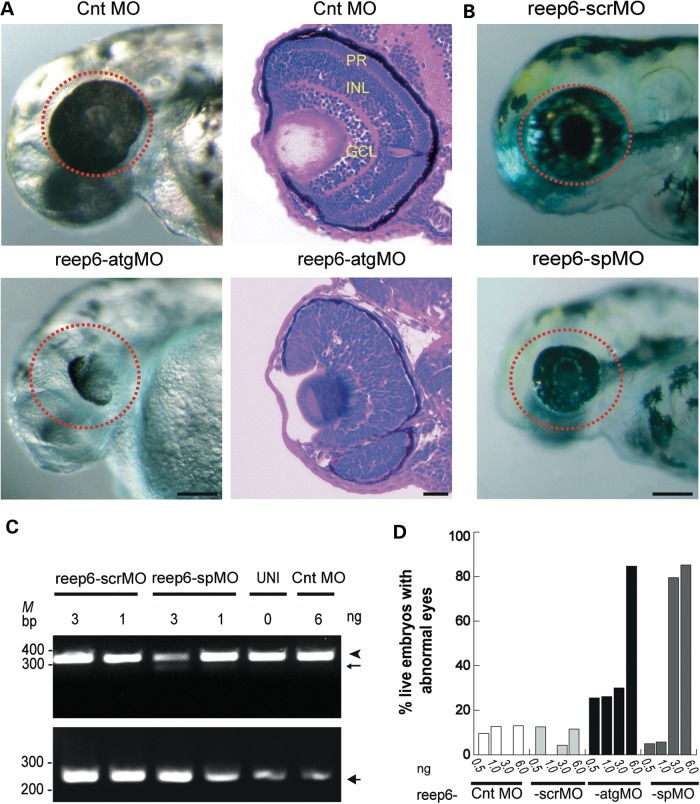
To examine the role of Reep6 in retinal development, we knocked down reep6 in zebrafish using antisense morpholinos (MOs). Knockdown of reep6 by translation blocking MO (reep6-atgMO) led to severely compromised eye development in the morphants compared with the embryos injected with standard negative control morpholino (Fig. 5A). Histological analysis of retina at 72 h postfertilization showed aberrant retinal lamination in reep6 morphants compared with control morpholino injected embryos (Fig. 5A). The specificity of the phenotype observed with reep6-atgMO was confirmed by using a splice-blocking morpholino, termed reep6-spMO, with a scrambled version (reep6-scrMO) as a negative control. Consistent with the partial knockdown of reep6 expression observed by RT-PCR a milder defect was observed in their eye development compared with the reep6-atgMO injected embryos (Fig. 5B and C). The phenotype was observed in a dose-dependent manner for both morpholinos (Fig. 5D).
Figure 5.
reep6 knockdown in zebrafish leads to abnormal eye development. (A) The eyes of morphants injected with 6 ng of control (Cnt MO) or translation blocking (reep6-atgMO) morpholinos observed at 72 hpf (left panels). Red circles denote the compromised eye development in the morphant (reep6-atgMO) compared with the Cnt MO injected embryos. H&E stained sections of retina from the morphants injected with Cnt MO or reep6-atgMO morpholinos (3 ng), at 72 hpf (right panels). The morphants injected with Cnt MO had normally laminated retina with discriminable PR, INL and GCL, whereas reep6-atgMO-injected retina had no proper lamination. Scale bar = 100 μm. (B) The eyes of morphants (denoted by red circles) injected with scrambled (scrMO) or splice blocking (reep6-spMO) morpholino, examined at 72 hpf. The scrMO did not elicit a visible effect on eye development whereas spMO compromised the eye development. Scale bar = 100 μm. (C) The photograph of RT-PCR products showing exon-skipping by reep6-spMO. Total RNA from individual embryos was used for RT-PCR. Splice blocking resulted in an additional smaller band (<300 bp, thin arrow) along with the WT band (343 bp, arrowhead). The bottom panel shows beta-actin (263 bp, thick arrow) control for respective samples. M indicates size maker. (D) Quantification of abnormal eyes injected with Cnt MO, scrMO, atgMO and spMO morpholinos. The x- and y- axes indicate injected morpholino and percentage of live embryos with abnormal eyes, respectively. UNI and Cnt MO indicate uninjected and control MO injected samples, respectively. MO concentration in nanograms is indicated.
We then used shRNA to knockdown Reep6 by in vivo electroporation in P0 mouse retina (Fig. 6). Gapdh shRNA was used as a control. To mark the retinal cells transfected with Reep6 shRNA in retinal cells, Ub-GFP plasmid carrying a GFP reporter under the control of ubiquitin promoter was co-transfected with Reep6 or Gapdh shRNA. Knockdown of Reep6 in mouse retina led to a significant decrease in the number of GFP-expressing cells, which may reflect the death of transfected photoreceptor cells as suggested by the thinner ONL in the electroporated area. Taken together, these results show that REEP6 is required for photoreceptor development and/or survival in zebrafish and mouse retina.
Figure 6.
Reep6 knockdown caused photoreceptor cell death in adult mouse retina. Representative confocal images of P21 mouse retina. Retina of CD1 mice were transfected at P0 with Ub-GFP (electroporation control), Gapdh shRNA (control shRNA) or Reep6 shRNA. The transfection was tracked by GFP (green). Rods are highlighted by rhodopsin immunoreactivity (red), and nuclei were visualized by DAPI (blue). Scale bar = 10 μm.
DISCUSSION
To increase the functional diversity of a limited number of genes, their products are differentially spliced in spatial and/or temporal contexts. Temporal or quantitative control of tissue-specific gene splicing is largely achieved through integrated action of regulatory proteins with multiple cis-regulatory elements, including enhancers (50–52). In this study, we demonstrate that rod photoreceptors specifically express a long isoform of Reep6, Reep6.1, whereas a shorter isoform Reep6.2 is expressed in the liver. Reep6.1 expression is regulated by rod differentiation factor NRL through an intronic enhancer, as evidenced by: (1) complete loss of expression of Reep6 in Nrl−/− mouse retina, (2) increased Reep6 promoter activity by inclusion of CIR or ‘Peak’ region, to which NRL binds, (3) correlation of Reep6 expression with NRL binding to the Reep6 enhancer region, (4) NRL-mediated activation of Reep6 promoter through the intronic enhancer in HEK293 cells, and (5) decreased Reep6 expression following in vivo Nrl knockdown. Furthermore, our gene knockdown experiments in zebrafish and mouse demonstrate that Reep6 is required for retinal development.
Extensive alternative splicing and promoter usage in the mammalian genome generate tremendous transcriptional diversity (53,54), which is particularly evident during the development of complex neuronal functions (55) and in aging and disease (56). We previously reported an alternate transcript of Mef2c, the gene associated with cortical development and myocyte differentiation (57,58), in rod photoreceptors and its regulation by NRL from an alternative promoter (25). Dynamic usage of alternative splicing has also been reported during retinal development (59). Identification of a novel Reep6.1 isoform in rod photoreceptors further highlights the importance of alternate splicing in producing functional diversity.
Splicing regulatory sequences have been identified in both intronic and exonic gene regions (60–62), and differential recruitment of splicing factors can influence the choice of splice sites (63,64). Mutations in specific splicing factors have been associated with retinal and photoreceptor degeneration (5,65,66). The binding of NRL to an enhancer within Reep6 intron 1 leads to rod-specific expression of Reep6.1 isoform with an alternative exon 5, suggesting a possible role of NRL in recruiting gene-specific splicing factors. In concordance, we recently demonstrated functional interaction of NRL with transcription-splicing associated protein NonO and suggested that NonO and its interacting proteins might fine-tune rod-specific gene expression (17). Interestingly, ChIP-Seq data show NonO binding in the Reep6 intron 1 coinciding the binding region for NRL (Supplementary Material, Fig. S1). We thus hypothesize that the binding of NRL together with NonO in intron 1 leads to rod-specific Reep6.1 transcript with the inclusion of a novel exon 5. Further investigations are necessary to test whether NRL-NonO interaction facilitates the recruitment of specific splicing complexes for producing rod photoreceptor-specific transcripts.
REEP family proteins are implicated in trafficking of transmembrane proteins, including G-protein coupled receptors (GPCRs) (28,30). While the relevance of REEP1 to microtubule dynamics and its association with hereditary spastic paraplegias has been elegantly demonstrated (32), the function of other REEP proteins, particularly REEP6, is poorly understood. The specific expression of REEP6.1 in rod photoreceptors ((37), and this report), and not in retinal ganglion cells (35), suggests its role in trafficking of membrane proteins (such as GPCRs) to distinct cellular compartments unique to photoreceptors. Daily renewal of outer segment membrane disks, which have a high concentration of rhodopsin (67), requires an active and efficient transport system that is critical for photoreceptor function (68–70). Not surprisingly, a substantial fraction of retinal degenerative diseases are related to transport defects in rod photoreceptors (5). Subcellular localization of REEP6 in rods (see Fig. 2) supports its likely function in transport of proteins to different membrane compartments, including the pre-synaptic region. Our results on Reep6 knockdown in zebrafish and mouse retina (see Figs 5 and 6) are also consistent with the postulated function of REEP6 in rod photoreceptors. The inclusion of exon 5 in Reep6.1 introduces 27 additional amino acids that are predicted to generate a very short coil and a long loop domain in the REEP6 protein. This new domain may provide an interaction surface for rod photoreceptor-specific modulation of REEP6. Thus, a better understanding of REEP6.1 function, including its protein interactions, should yield new insights into rod homeostasis and disease.
MATERIALS AND METHODS
Animal care and use
All animal work was conducted according to national and international guidelines. Animal Care and Use Committee of the National Eye Institute and National Human Genome Research Institute approved mouse and zebrafish protocols, respectively. C57BL/6J and CD1 mice were purchased from the Jackson Laboratory (Bar Harbor, ME) and from Charles River Laboratory (Wilmington, MA), respectively. Nrl-GFP and Nrl-GFP;Nrl−/− mice have been described earlier (26).
Immunoblot analysis
Freshly dissected mouse tissues were lysed by homogenization and sonication in radioimmunoprecipitation buffer supplemented with 20 mm N-ethylmaleimide and protease inhibitor mixture (Roche Applied Science). After centrifugation, the protein concentration of the supernatant was measured by the bicinchoninic acid assay (Thermo Scientific, Waltham, MA). Equal amounts of lysates were boiled in 2× sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer (Invitrogen), resolved by SDS-PAGE and transferred to nitrocellulose membrane (Invitrogen). The membrane was probed sequentially with anti-REEP6 and anti-GAPDH antibodies and visualized by enhanced chemiluminescence (Thermo Scientific).
5′ Rapid analysis of cDNA ends derived from full-length RNA (RACE)
Total RNA was isolated from mouse tissues (retina, brain and liver) using TRIzol reagent (Invitrogen). 5′-RACE analysis of the full-length RNA was conducted using the GeneRacer™ kit (Invitrogen) as described by the vendor. Briefly, full-length RNA was treated with tobacco acid pyrophosphatase to remove end caps and then ligated to the GeneRacer RNA oligos. Reverse transcription was performed using hexamers. To amplify fragments containing the 5′ end of the transcripts, the GeneRacer™ 5′ primer was paired with one of the Reep6 specific reverse primers (P1: 5′-GGCGCACTTGCC CGCGTAGTAGAAAG-3′, P2: 5′-CCCAGCTGCTAGGTCCAATGCTCTTC-3′, P3: 5′-AGCGGCTGGGGGTTCCGATGTTGATGCT-3′). The RACE PCR products were resolved on 1.5% agarose gel, purified and cloned for sequencing using the TOPO TA cloning kit (Invitrogen). Minimum of 15 clones were sequenced for each RACE PCR reaction.
Cloning of the mouse Reep6 promoter and enhancer constructs
Mouse Reep6 promoters (3594 bp and 752 bp), an intronic region of 1368 bp (intron 1368 bp) and an intron region of 617 bp where NRL ChIP-Seq peak resides (peak) were amplified from C57BL/6J mouse genomic DNA using Accuprime High Fidelity Taq (Invitrogen) and cloned into a GFP vector and a luciferase vector (Rep4-Luc), respectively. Please see Supplementary Material, Table 1 for primers and restriction sites used for cloning.
Chromatin immunoprecipitation (ChIP)-quantitative PCR (qPCR)
ChIP was performed using retinas from CL57BL/6J mice at postnatal Day 2 (P2) or P28 as previously described (22). ChIP DNA was quantified as a percentage of input DNA signal in triplicate using SYBR Green (Bio-Rad) and graphed as a ratio of ChIP signal from NRL antibody to the signal from normal IgG (negative control).
Gene expression assays
Total RNA was isolated from C57BL/6J mouse retina using TRIzol reagent (Invitrogen), and cDNA was synthesized using the AffinityScript QPCR cDNA Synthesis Kit (Agilent Technologies, La Jolla, CA). Gene expression was detected by qPCR using Taqman probes (IDs: Mm01170350_g1, Mm01173489_m1, Mm01315649_m1) and 2×-Gene Expression Master mix (Applied Biosystems, Foster City, CA).
RNA-Seq analysis
Total RNA (15–50 ng) from whole retina and from flow sorted rod photoreceptors was used to construct RNA-Seq libraries and for next generation sequencing on an Illumina GAIIx (Illumina, Inc., San Diego, CA), as previously described. Transcript quantitation was performed with eXpress v1.3.1 by streaming Bowtie2 v2.1.0 (71) aligned pass filter reads to GRCm38.p2 Ensembl release 73 annotation. This study utilized the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health (http://biowulf.nih.gov).
Cell culture, transient transfection and dual luciferase assay
HEK293 cells (American Type Culture Collection) were cultured in minimum essential medium-alpha medium supplemented with 10% fetal bovine serum (FBS) and 100 units/ml penicillin/streptomycin at 37°C under 5% CO2. The cells were seeded into 12-well plates (350 000 cells/well) at 24 h before transfection. The cells were transfected with Fugene 6 (Promega) according to the manufacturer's instruction. Triplicate wells were co-transfected with the Firefly-luciferase reporter constructs (200 ng), the transfection control plasmid: Renilla-luciferase reporter pRL-CMV (10 ng), and expression plasmid for mouse NRL and/or expression plasmid for mouse CRX as indicated in figure legend. The empty vector pcDNA3.1/HisC was used to adjust the total amount of transfected DNA. Cells were incubated for 48 h and then harvested for analysis of Firefly- and Renilla-luciferase activities using the Dual-Reporter Luminescence Reagent (Promega) and a Turner dual-injector luminometer system (Promega). Renilla-luciferase activity served as an internal control to normalize transfection efficiency. All experiments were repeated at least three times.
In vitro electroporation and retina explant culture
Retinas of P0 CD1 pups were dissected and electroporated in a DNA mixture containing Reep6 promoter-GFP constructs (1 μg/μl) and the electroporation control CAG-mCherry (1 μg/μl). The retina explants were cultured in Dulbecco's modified Eagle's medium/F12 medium (Invitrogen) supplemented with 10% FBS and 100 units/ml penicillin/streptomycin at 37°C under 5% CO2 for 7 days. The retina explant was fixed in 4% paraformadehyde, cryoprotected in 30% sucrose. Cryosections (12 µm) were imaged with an Olympus FluoView FV1000 confocal laser scanner.
In vivo electroporation
Transfection of retina was achieved by injecting DNA into the sub-retinal space of CD1 mouse pups at P0 and followed by electroporation, as previously described (22). Briefly, equal amount of plasmids (Reep-P712-Intron-GFP and CAG-mCherry) were mixed with either Nrl shRNA or Gapdh shRNA control and injected into the sub-retinal space of CD1 pups at P0. The Nrl shRNA plasmid was used to knockdown Nrl expression, and Gapdh shRNA (Open Biosystems) was used as a negative control in knockdown experiments. The DNA concentration was 400 nm, and the total injection volume was 0.2 µl. Square electric pulses (80 V, 1 Hz, 5 pulses) were applied across the heads of pups using an ECM830 square wave electroporator and 10 mm diameter BTX Tweezertrode electrodes (Holliston, MA). Retina was dissected at P21, fixed in 4% paraformadehyde, cryoprotected in 30% sucrose. Cryosections (12 µm) were imaged with an Olympus FluoView FV1000 confocal laser scanner. Knockdown of Reep6 by in vivo shRNA electroporation was performed at P0 and examined at P20.
Immunofluoresence and confocal microscopy
Eyes from P10 and P30 Nrl−/− mice (7), which had been previously backcrossed onto a C57BL/6J background for over 10 generations, and WT control mice were immersed in 4% paraformaldehyde for 30 min. The retinas were dissected and sectioned at 150 µm on a Vibratome. Immunofluorescent staining was performed as described (37), using the following primary antibodies: a custom rabbit polyclonal antibody to REEP6 [1:1000, (37)], a mouse monoclonal antibody to CtBP2 (1:500; cat# 612044, BD Transduction), and a mouse monoclonal antibody to PKC (1:50; cat# RPN 536, Amersham). Sections were also labeled with the lectin peanut agglutinin preconjugated to an AlexaFluor fluorophore (PNA; 1:1000; cat# L32460; Invitrogen). Secondary antibodies raised in donkey to either rabbit or mouse IgG and conjugated to AlexaFluor dyes (1:200; Life Technologies) were used to detect the primary antibodies. Retinal sections were imaged using an Olympus FluoView FV1000 confocal laser scanner and processed in Adobe Photoshop CS3 to adjust contrast.
Morpholino knockdown in zebrafish embryos
Fluorescein-tagged morpholinos (MOs) were procured from Gene Tools Inc. (OR, USA). A standard negative control (Cnt-MO) and custom-designed translation blocking (reep6-atgMO-5′-ACATGGTGACTTAAAATAAACTCGT- 3′), splice blocking (reep6-spMO - 5′- TCTGGCATTGCTGAACTTACCTTGC- 3′) and scrambled control reep6-scrMO (5′-TGCTCAAATAAAATTCAGTGGTACA-3′) against zebrafish reep6 were obtained in lyophilized form, re-suspended in distilled water, and quantified spectrophotometrically (NanoDrop Tech Inc., DE, USA).
Zebrafish (Danio rerio) were maintained under an approved animal use protocol. Staged WT embryos of EK strain between 2–8 cells were microinjected with 0.4–1.2 nl of morpholinos into the yolk sac using pneumatic pico pump (WPI, FL, USA). Morpholino studies were performed as described earlier (72).
Hematoxylin and eosin staining
At 72 hpf, zebrafish larvae were fixed with 4% glutaraldehyde for 30 min at RT, then fixed with 4% paraformaldehyde overnight at 4°C. Subsequently, they were washed with PBS and embedded in OCT compound Tissue-Tek (SakuraFinetek USA, Inc., CA, USA) and 10 μm sections were cut. The sections were stained with standard H&E staining protocol.
SUPPLEMENTARY MATERIAL
FUNDING
These studies were supported by intramural research programs of the National Eye Institute and National Human Genome Research Institute, National Institutes of Health, and by NIH extramural support (EY019968).
Supplementary Material
ACKNOWLEDGEMENTS
We thank N-NRL members, especially Matthew Brooks, for constructive discussions and technical support. We acknowledge Robert N. Fariss and Chun Gao for confocal microscopy training.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Dowling J.E. The Retina: an Approachable Part of the Brain. Cambridge, MA: Belknap Press of Harvard University Press; 1987. [Google Scholar]
- 2.Livesey F.J., Cepko C.L. Vertebrate neural cell-fate determination: lessons from the retina. Nat. Rev. Neurosci. 2001;2:109–118. doi: 10.1038/35053522. [DOI] [PubMed] [Google Scholar]
- 3.Swaroop A., Kim D., Forrest D. Transcriptional regulation of photoreceptor development and homeostasis in the mammalian retina. Nat. Rev. Neurosci. 2010;11:563–576. doi: 10.1038/nrn2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Agathocleous M., Harris W.A. From progenitors to differentiated cells in the vertebrate retina. Annu. Rev. Cell Dev. Biol. 2009;25:45–69. doi: 10.1146/annurev.cellbio.042308.113259. [DOI] [PubMed] [Google Scholar]
- 5.Wright A.F., Chakarova C.F., Abd El-Aziz M.M., Bhattacharya S.S. Photoreceptor degeneration: genetic and mechanistic dissection of a complex trait. Nat. Rev. Genet. 2010;11:273–284. doi: 10.1038/nrg2717. [DOI] [PubMed] [Google Scholar]
- 6.Jackson G.R., Owsley C., Curcio C.A. Photoreceptor degeneration and dysfunction in aging and age-related maculopathy. Ageing Res. Rev. 2002;1:381–396. doi: 10.1016/s1568-1637(02)00007-7. [DOI] [PubMed] [Google Scholar]
- 7.Mears A.J., Kondo M., Swain P.K., Takada Y., Bush R.A., Saunders T.L., Sieving P.A., Swaroop A. Nrl is required for rod photoreceptor development. Nat. Genet. 2001;29:447–452. doi: 10.1038/ng774. [DOI] [PubMed] [Google Scholar]
- 8.Oh E.C., Khan N., Novelli E., Khanna H., Strettoi E., Swaroop A. Transformation of cone precursors to functional rod photoreceptors by bZIP transcription factor NRL. Proc. Natl. Acad. Sci. U. S. A. 2007;104:1679–1684. doi: 10.1073/pnas.0605934104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ng L., Lu A., Swaroop A., Sharlin D.S., Swaroop A., Forrest D. Two transcription factors can direct three photoreceptor outcomes from rod precursor cells in mouse retinal development. J. Neurosci. 2011;31:11118–11125. doi: 10.1523/JNEUROSCI.1709-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nishida A., Furukawa A., Koike C., Tano Y., Aizawa S., Matsuo I., Furukawa T. Otx2 homeobox gene controls retinal photoreceptor cell fate and pineal gland development. Nat. Neurosci. 2003;6:1255–1263. doi: 10.1038/nn1155. [DOI] [PubMed] [Google Scholar]
- 11.Omori Y., Katoh K., Sato S., Muranishi Y., Chaya T., Onishi A., Minami T., Fujikado T., Furukawa T. Analysis of transcriptional regulatory pathways of photoreceptor genes by expression profiling of the Otx2-deficient retina. PLoS One. 2011;6:e19685. doi: 10.1371/journal.pone.0019685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roger J.E., Hiriyanna A., Gotoh N., Hao H., Cheng D.F., Ratnapriya R., Kautzmann M.A., Chang B., Swaroop A. OTX2 loss causes rod differentiation defect in CRX-associated congenital blindness. J. Clin. Invest. 2014;124:631–643. doi: 10.1172/JCI72722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jia L., Oh E.C., Ng L., Srinivas M., Brooks M., Swaroop A., Forrest D. Retinoid-related orphan nuclear receptor RORbeta is an early-acting factor in rod photoreceptor development. Proc. Natl. Acad. Sci. U. S. A. 2009;106:17534–17539. doi: 10.1073/pnas.0902425106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corbo J.C., Lawrence K.A., Karlstetter M., Myers C.A., Abdelaziz M., Dirkes W., Weigelt K., Seifert M., Benes V., Fritsche L.G., et al. CRX ChIP-seq reveals the cis-regulatory architecture of mouse photoreceptors. Genome Res. 2010;20:1512–1525. doi: 10.1101/gr.109405.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hennig A.K., Peng G.H., Chen S. Regulation of photoreceptor gene expression by Crx-associated transcription factor network. Brain Res. 2008;1192:114–133. doi: 10.1016/j.brainres.2007.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Furukawa T., Morrow E.M., Li T., Davis F.C., Cepko C.L. Retinopathy and attenuated circadian entrainment in Crx-deficient mice. Nat. Genet. 1999;23:466–470. doi: 10.1038/70591. [DOI] [PubMed] [Google Scholar]
- 17.Yadav S.P., Hao H., Yang H.J., Kautzmann M.A., Brooks M., Nellissery J., Klocke B., Seifert M., Swaroop A. The transcription-splicing protein NonO/p54nrb and three NonO-interacting proteins bind to distal enhancer region and augment rhodopsin expression. Hum. Mol. Genet. 2014;23:2132–2144. doi: 10.1093/hmg/ddt609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mitton K.P., Swain P.K., Chen S., Xu S., Zack D.J., Swaroop A. The leucine zipper of NRL interacts with the CRX homeodomain. A possible mechanism of transcriptional synergy in rhodopsin regulation. J. Biol. Chem. 2000;275:29794–29799. doi: 10.1074/jbc.M003658200. [DOI] [PubMed] [Google Scholar]
- 19.Cheng H., Aleman T.S., Cideciyan A.V., Khanna R., Jacobson S.G., Swaroop A. In vivo function of the orphan nuclear receptor NR2E3 in establishing photoreceptor identity during mammalian retinal development. Hum. Mol. Genet. 2006;15:2588–2602. doi: 10.1093/hmg/ddl185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoshida S., Mears A.J., Friedman J.S., Carter T., He S., Oh E., Jing Y., Farjo R., Fleury G., Barlow C., et al. Expression profiling of the developing and mature Nrl−/− mouse retina: identification of retinal disease candidates and transcriptional regulatory targets of Nrl. Hum. Mol. Genet. 2004;13:1487–1503. doi: 10.1093/hmg/ddh160. [DOI] [PubMed] [Google Scholar]
- 21.Yu J., He S., Friedman J.S., Akimoto M., Ghosh D., Mears A.J., Hicks D., Swaroop A. Altered expression of genes of the Bmp/Smad and Wnt/calcium signaling pathways in the cone-only Nrl−/− mouse retina, revealed by gene profiling using custom cDNA microarrays. J. Biol. Chem. 2004;279:42211–42220. doi: 10.1074/jbc.M408223200. [DOI] [PubMed] [Google Scholar]
- 22.Hao H., Kim D.S., Klocke B., Johnson K.R., Cui K., Gotoh N., Zang C., Gregorski J., Gieser L., Peng W., et al. Transcriptional regulation of rod photoreceptor homeostasis revealed by in vivo NRL targetome analysis. PLoS Genet. 2012;8:e1002649. doi: 10.1371/journal.pgen.1002649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Onishi A., Peng G.H., Poth E.M., Lee D.A., Chen J., Alexis U., de Melo J., Chen S., Blackshaw S. The orphan nuclear hormone receptor ERRbeta controls rod photoreceptor survival. Proc. Natl. Acad. Sci. U. S. A. 2010;107:11579–11584. doi: 10.1073/pnas.1000102107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oh E.C., Cheng H., Hao H., Jia L., Khan N.W., Swaroop A. Rod differentiation factor NRL activates the expression of nuclear receptor NR2E3 to suppress the development of cone photoreceptors. Brain Res. 2008;1236:16–29. doi: 10.1016/j.brainres.2008.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hao H., Tummala P., Guzman E., Mali R.S., Gregorski J., Swaroop A., Mitton K.P. The transcription factor neural retina leucine zipper (NRL) controls photoreceptor-specific expression of myocyte enhancer factor Mef2c from an alternative promoter. J. Biol. Chem. 2011;286:34893–34902. doi: 10.1074/jbc.M111.271072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Akimoto M., Cheng H., Zhu D., Brzezinski J.A., Khanna R., Filippova E., Oh E.C., Jing Y., Linares J.L., Brooks M., et al. Targeting of GFP to newborn rods by Nrl promoter and temporal expression profiling of flow-sorted photoreceptors. Proc. Natl. Acad. Sci. U. S. A. 2006;103:3890–3895. doi: 10.1073/pnas.0508214103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shibata Y., Hu J., Kozlov M.M., Rapoport T.A. Mechanisms shaping the membranes of cellular organelles. Annu. Rev. Cell Dev. Biol. 2009;25:329–354. doi: 10.1146/annurev.cellbio.042308.113324. [DOI] [PubMed] [Google Scholar]
- 28.Mainland J., Matsunami H. RAMP like proteins: RTP and REEP family of proteins. Adv. Exp. Med. Biol. 2012;744:75–86. doi: 10.1007/978-1-4614-2364-5_7. [DOI] [PubMed] [Google Scholar]
- 29.Zhuang H., Matsunami H. Synergism of accessory factors in functional expression of mammalian odorant receptors. J. Biol. Chem. 2007;282:15284–15293. doi: 10.1074/jbc.M700386200. [DOI] [PubMed] [Google Scholar]
- 30.Saito H., Kubota M., Roberts R.W., Chi Q., Matsunami H. RTP family members induce functional expression of mammalian odorant receptors. Cell. 2004;119:679–691. doi: 10.1016/j.cell.2004.11.021. [DOI] [PubMed] [Google Scholar]
- 31.Calero M., Whittaker G.R., Collins R.N. Yop1p, the yeast homolog of the polyposis locus protein 1, interacts with Yip1p and negatively regulates cell growth. J. Biol. Chem. 2001;276:12100–12112. doi: 10.1074/jbc.M008439200. [DOI] [PubMed] [Google Scholar]
- 32.Park S.H., Zhu P.P., Parker R.L., Blackstone C. Hereditary spastic paraplegia proteins REEP1, spastin, and atlastin-1 coordinate microtubule interactions with the tubular ER network. J. Clin. Invest. 2010;120:1097–1110. doi: 10.1172/JCI40979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Castermans D., Vermeesch J.R., Fryns J.P., Steyaert J.G., Van de Ven W.J., Creemers J.W., Devriendt K. Identification and characterization of the TRIP8 and REEP3 genes on chromosome 10q21.3 as novel candidate genes for autism. Eur. J. Hum. Genet. 2007;15:422–431. doi: 10.1038/sj.ejhg.5201785. [DOI] [PubMed] [Google Scholar]
- 34.Schlaitz A.L., Thompson J., Wong C.C., Yates J.R., 3rd, Heald R. REEP3/4 ensure endoplasmic reticulum clearance from metaphase chromatin and proper nuclear envelope architecture. Dev. Cell. 2013;26:315–323. doi: 10.1016/j.devcel.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sato H., Tomita H., Nakazawa T., Wakana S., Tamai M. Deleted in polyposis 1-like 1 gene (Dp1l1): a novel gene richly expressed in retinal ganglion cells. Invest. Ophthalmol. Vis. Sci. 2005;46:791–796. doi: 10.1167/iovs.04-0867. [DOI] [PubMed] [Google Scholar]
- 36.Wellmann A., Fogt F., Hollerbach S., Hahne J., Koenig-Hoffmann K., Smeets D., Brinkmann U. Polymorphisms of the apoptosis-associated gene DP1L1 (deleted in polyposis 1-like 1) in colon cancer and inflammatory bowel disease. J Cancer Res. Clin. Oncol. 2010;136:795–802. doi: 10.1007/s00432-009-0719-9. [DOI] [PubMed] [Google Scholar]
- 37.Keeley P.W., Luna G., Fariss R.N., Skyles K.A., Madsen N.R., Raven M.A., Poche R.A., Swindell E.C., Jamrich M., Oh E.C., et al. Development and plasticity of outer retinal circuitry following genetic removal of horizontal cells. J. Neurosci. 2013;33:17847–17862. doi: 10.1523/JNEUROSCI.1373-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brooks M.J., Rajasimha H.K., Swaroop A. Retinal transcriptome profiling by directional next-generation sequencing using 100 ng of total RNA. Methods Mol. Biol. 2012;884:319–334. doi: 10.1007/978-1-61779-848-1_23. [DOI] [PubMed] [Google Scholar]
- 39.Brooks M.J., Rajasimha H.K., Roger J.E., Swaroop A. Next-generation sequencing facilitates quantitative analysis of wild-type and Nrl(-/-) retinal transcriptomes. Mol. Vis. 2011;17:3034–3054. [PMC free article] [PubMed] [Google Scholar]
- 40.Roy A., Kucukural A., Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Proto. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roy A., Yang J., Zhang Y. COFACTOR: an accurate comparative algorithm for structure-based protein function annotation. Nucl. Acids Res. 2012;40:W471–W477. doi: 10.1093/nar/gks372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blanks J.C., Johnson L.V. Selective lectin binding of the developing mouse retina. J. Comp. Neurol. 1983;221:31–41. doi: 10.1002/cne.902210103. [DOI] [PubMed] [Google Scholar]
- 44.Wikler K.C., Stull D.L., Reese B.E., Johnson P.T., Bogenmann E. Localization of protein kinase C to UV-sensitive photoreceptors in the mouse retina. Vis. Neurosci. 1998;15:87–95. doi: 10.1017/s0952523898151155. [DOI] [PubMed] [Google Scholar]
- 45.Daniele L.L., Lillo C., Lyubarsky A.L., Nikonov S.S., Philp N., Mears A.J., Swaroop A., Williams D.S., Pugh E.N., Jr Cone-like morphological, molecular, and electrophysiological features of the photoreceptors of the Nrl knockout mouse. Invest. Ophthalmol. Vis. Sci. 2005;46:2156–2167. doi: 10.1167/iovs.04-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roger J.E., Ranganath K., Zhao L., Cojocaru R.I., Brooks M., Gotoh N., Veleri S., Hiriyanna A., Rachel R.A., Campos M.M., et al. Preservation of cone photoreceptors after a rapid yet transient degeneration and remodeling in cone-only Nrl-/- mouse retina. J. Neurosci. 2012;32:528–541. doi: 10.1523/JNEUROSCI.3591-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmitz F., Konigstorfer A., Sudhof T.C. RIBEYE, a component of synaptic ribbons: a protein’s journey through evolution provides insight into synaptic ribbon function. Neuron. 2000;28:857–872. doi: 10.1016/s0896-6273(00)00159-8. [DOI] [PubMed] [Google Scholar]
- 48.Matsuda T., Cepko C.L. Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proc. Natl. Acad. Sci. U. S. A. 2004;101:16–22. doi: 10.1073/pnas.2235688100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kautzmann M.A., Kim D.S., Felder-Schmittbuhl M.P., Swaroop A. Combinatorial regulation of photoreceptor differentiation factor, neural retina leucine zipper gene NRL, revealed by in vivo promoter analysis. J. Biol. Chem. 2011;286:28247–28255. doi: 10.1074/jbc.M111.257246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Heintzman N.D., Ren B. The gateway to transcription: identifying, characterizing and understanding promoters in the eukaryotic genome. Cell. Mol. Life Sci. 2007;64:386–400. doi: 10.1007/s00018-006-6295-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Black D.L. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 52.Irimia M., Rukov J.L., Penny D., Roy S.W. Functional and evolutionary analysis of alternatively spliced genes is consistent with an early eukaryotic origin of alternative splicing. BMC Evol. Biol. 2007;7:188. doi: 10.1186/1471-2148-7-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Blekhman R., Marioni J.C., Zumbo P., Stephens M., Gilad Y. Sex-specific and lineage-specific alternative splicing in primates. Genome Res. 2010;20:180–189. doi: 10.1101/gr.099226.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marinov G.K., Williams B.A., McCue K., Schroth G.P., Gertz J., Myers R.M., Wold B.J. From single-cell to cell-pool transcriptomes: stochasticity in gene expression and RNA splicing. Genome Res. 2014;24:496–510. doi: 10.1101/gr.161034.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pal S., Gupta R., Kim H., Wickramasinghe P., Baubet V., Showe L.C., Dahmane N., Davuluri R.V. Alternative transcription exceeds alternative splicing in generating the transcriptome diversity of cerebellar development. Genome Res. 2011;21:1260–1272. doi: 10.1101/gr.120535.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tollervey J.R., Wang Z., Hortobagyi T., Witten J.T., Zarnack K., Kayikci M., Clark T.A., Schweitzer A.C., Rot G., Curk T., et al. Analysis of alternative splicing associated with aging and neurodegeneration in the human brain. Genome Res. 2011;21:1572–1582. doi: 10.1101/gr.122226.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Leifer D., Krainc D., Yu Y.T., McDermott J., Breitbart R.E., Heng J., Neve R.L., Kosofsky B., Nadal-Ginard B., Lipton S.A. MEF2C, a MADS/MEF2-family transcription factor expressed in a laminar distribution in cerebral cortex. Proc. Natl. Acad. Sci. U. S. A. 1993;90:1546–1550. doi: 10.1073/pnas.90.4.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin Q., Schwarz J., Bucana C., Olson E.N. Control of mouse cardiac morphogenesis and myogenesis by transcription factor MEF2C. Science. 1997;276:1404–1407. doi: 10.1126/science.276.5317.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wan J., Masuda T., Hackler L., Jr, Torres K.M., Merbs S.L., Zack D.J., Qian J. Dynamic usage of alternative splicing exons during mouse retina development. Nucl. Acids Res. 2011;39:7920–7930. doi: 10.1093/nar/gkr545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hastings M.L., Wilson C.M., Munroe S.H. A purine-rich intronic element enhances alternative splicing of thyroid hormone receptor mRNA. RNA. 2001;7:859–874. doi: 10.1017/s1355838201002084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gallego M.E., Gattoni R., Stevenin J., Marie J., Expert-Bezancon A. The SR splicing factors ASF/SF2 and SC35 have antagonistic effects on intronic enhancer-dependent splicing of the beta-tropomyosin alternative exon 6A. EMBO J. 1997;16:1772–1784. doi: 10.1093/emboj/16.7.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kornblihtt A.R. Coupling transcription and alternative splicing. Adv. Exp. Med. Biol. 2007;623:175–189. doi: 10.1007/978-0-387-77374-2_11. [DOI] [PubMed] [Google Scholar]
- 63.Cramer P., Caceres J.F., Cazalla D., Kadener S., Muro A.F., Baralle F.E., Kornblihtt A.R. Coupling of transcription with alternative splicing: RNA pol II promoters modulate SF2/ASF and 9G8 effects on an exonic splicing enhancer. Mol. Cell. 1999;4:251–258. doi: 10.1016/s1097-2765(00)80372-x. [DOI] [PubMed] [Google Scholar]
- 64.Simard M.J., Chabot B. SRp30c is a repressor of 3′ splice site utilization. Mol. Cell. Biol. 2002;22:4001–4010. doi: 10.1128/MCB.22.12.4001-4010.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Graziotto J.J., Farkas M.H., Bujakowska K., Deramaudt B.M., Zhang Q., Nandrot E.F., Inglehearn C.F., Bhattacharya S.S., Pierce E.A. Three gene-targeted mouse models of RNA splicing factor RP show late-onset RPE and retinal degeneration. Invest. Ophthalmol. Vis. Sci. 2011;52:190–198. doi: 10.1167/iovs.10-5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mordes D., Luo X., Kar A., Kuo D., Xu L., Fushimi K., Yu G., Sternberg P., Jr, Wu J.Y. Pre-mRNA splicing and retinitis pigmentosa. Mol. Vis. 2006;12:1259–1271. [PMC free article] [PubMed] [Google Scholar]
- 67.Wright W.E., Brown P.K., Wald G. Orientation of intermediates in the bleaching of shear-oriented rhodopsin. J. Gen. Physiol. 1973;62:509–522. doi: 10.1085/jgp.62.5.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Deretic D., Wang J. Molecular assemblies that control rhodopsin transport to the cilia. Vis. Res. 2012;75:5–10. doi: 10.1016/j.visres.2012.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Najafi M., Calvert P.D. Transport and localization of signaling proteins in ciliated cells. Vis. Res. 2012;75:11–18. doi: 10.1016/j.visres.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lamb T.D. Evolution of phototransduction, vertebrate photoreceptors and retina. Prog. Retin. Eye Res. 2013;36:52–119. doi: 10.1016/j.preteyeres.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 71.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Veleri S., Bishop K., Dalle Nogare D.E., English M.A., Foskett T.J., Chitnis A., Sood R., Liu P., Swaroop A. Knockdown of Bardet–Biedl syndrome gene BBS9/PTHB1 leads to cilia defects. PLoS One. 2012;7:e34389. doi: 10.1371/journal.pone.0034389. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.