Abstract
Genetic factors are thought to be an important determinant of thyroid function and autoimmunity. However, there are limited data on genetic variants in Asians. In this study, we performed a genome-wide association study on plasma thyroid-stimulating hormone (TSH) and free thyroxine (fT4) concentration and anti-thyroid peroxidase (anti-TPO) antibody positivity in 4238 Korean subjects. In the Stage 1 genome scan, 3396 participants from the Ansung cohort were investigated using 1.42 million genotyped or imputed markers. In the Stage 2 follow-up, 10 markers were genotyped in 842 participants from the Korean Longitudinal Study on Health and Aging cohort. An intronic variant in VAV3, rs12126655, which has been reported in Europeans, was significantly associated with plasma TSH concentration in the joint Stages 1 and 2 analyses (P = 2.2 × 10−8). We observed that a novel variant, rs2071403, located 75 bp proximal to the translational start site of TPO was significantly associated with plasma anti-TPO antibody positivity in the joint Stages 1 and 2 analyses (P = 1.3 × 10−10). This variant had a marginal sex-specific effect, and its association was more significant in females. Subjects possessing the rs2071403A allele, associated with an absence of the anti-TPO antibody, had decreased TPO mRNA expression in their thyroid tissue. Another intronic variant of HLA-DPB2, rs733208, had a suggestive association with anti-TPO antibody positivity (P = 4.2 × 10−7). In conclusion, we have identified genetic variants that are strongly associated with TSH level and anti-TPO antibody positivity in Koreans. Further replications and meta-analysis are required to confirm these findings.
INTRODUCTION
Thyroid hormone has diverse physiologic functions, including fetal development, oxygen consumption, thermogenesis and glucose and lipid metabolism (1,2). Within an individual, thyroid hormone and thyroid-stimulating hormone (TSH) levels are maintained within a relatively constant range over a long period (low intra-individual variation) (3). However, the variation between different individuals is larger (high inter-individual variation) (3). Inter-individual variability is partially explained by genetic factors. In a recent study, the heritability of free thyroxine (fT4) and TSH was 89 and 49%, respectively (3). The advent of genome-wide association studies (GWAS) have enabled us to identify the genetic variants associated with TSH and fT4 concentrations (4–10). A large-scale meta-analysis of GWAS in Europeans confirmed 20 genetic variants associated with TSH and six variants associated with fT4 concentrations. However, the genetic association of TSH and fT4 in Asians is not fully understood.
The dysregulation of thyroid hormone, resulting in hypo- or hyperthyroidism, is a relatively common condition. The most common clinical problem is subclinical hypothyroidism (defined as an increased TSH level with normal fT4 concentration), which occurs in 4.3% of the US population (11) and 11.7% of the Korean population (12). A high concentration of anti-thyroid peroxidase (TPO) antibody is a well-known risk factor associated with the development of thyroid dysfunction (13). Anti-TPO antibody is observed in nearly all patients with Hashimoto's thyroiditis (also known as chronic lymphocytic thyroiditis). The presence of this antibody suggests the pathogenic role of autoimmunity in the development of Hashimoto's thyroiditis. Several studies have investigated genetic variants associated with Hashimoto's thyroiditis in Europeans (10,12). The genetic association of anti-TPO antibody positivity has not been investigated on a genome-wide scale.
The aim of this study was to investigate the genetic variants associated with plasma TSH and fT4 concentrations and anti-TPO antibody positivity in Koreans. We performed a two-staged GWAS in 4238 Korean participants recruited from two independent community-based cohorts.
RESULTS
Stage 1 genome scan
The participants in the Stage 1 genome scan were from the Ansung cohort comprising the Korean Genome Epidemiology Study (KoGES). The clinical characteristics of the participants are shown in Table 1. To test for an association between the genetic variants and TSH and fT4 concentrations, we performed a linear regression analysis adjusting for age and sex in 2789 participants. For anti-TPO antibody positivity, we used a logistic regression adjusting for age and sex in a case-controlled manner of 3396 participants. A total of 351 669 single-nucleotide polymorphism (SNP) variants were actually genotyped and passed our stringent quality control filters. After the imputation, we were able to use 1 418709 SNPs for analyses. The quantile–quantile plots and Manhattan plots from the association tests are shown in the Supplementary Material, Figure S1. Ten independent variants were selected for suggestive associations according to our predefined threshold of P < 1.0 × 10−5, except for rs17111090 (P = 2.4 × 10−3) in TRHDE (thyrotropin-releasing hormone degrading enzyme) which was selected based on its biological plausibility (Table 2). The full list of variants that showed associations with P < 1.0 × 10−5 are listed in the Supplementary Material, Tables S1–S3. To eliminate hidden population stratification and cryptic relatedness, a variance component approach using EMMAX (http://www.sph.umich.edu/csg/kang/emmax/) was used to test the associations (Supplementary Material, Tables S1–S3) (14). The EMMAX association results were similar to the original analyses.
Table 1.
Clinical characteristics of study participants
| Stage 1 genome scan (Ansung cohort) |
Stage 2 follow-up (KLoSHA cohort) |
|||||
|---|---|---|---|---|---|---|
| Men | Women | P | Men | Women | P | |
| N (%) | 1527 (45.0%) | 1869 (55.0%) | 362 (43.0%) | 480 (57.0%) | ||
| Age (years)a | 56.1 ± 8.7 | 55.8 ± 8.9 | 0.100 | 76.5 ± 8.9 | 76.7 ± 9.1 | 0.715 |
| TSH (μIU/L)b | 1.62 (1.08–2.34) | 2.36 (1.53–3.51) | <0.001 | 2.56 (1.77–3.67) | 2.66 (1.70–3.74) | 0.815 |
| fT4 (ng/dl)b | 0.99 (0.91–1.07) | 0.97 (0.90–1.05) | 0.001 | 1.24 (1.06–1.45) | 1.15 (0.94–1.36) | <0.001 |
| Positive anti-TPO antibody, N (%)b | 108 (7.1%) | 269 (14.4%) | <0.001 | 32 (9.0%) | 76 (15.9%) | 0.003 |
aData are shown as mean ± SD and t-test was used to compare means between men and women.
bData are shown as median (interquartile range) and Mann–Whitney U test was used to compare means between men and women.
Table 2.
Association between genetic variants and TSH, fT4 concentration and anti-TPO antibody positivity
| Variant | Chr | Position (bp) | A1 | Nearby gene | Stage 1 genome scan |
Stage 2 follow-up | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | MAF |
β | SE | P | n | MAF |
β | SE | P | |||||||
| TSH concentration | ||||||||||||||||
| rs12126655 | 1 | 108 158 343 | G | VAV3 | 2665 | 0.35 | 0.135 | 0.028 | 1.7 × 10−6 | 834 | 0.36 | 0.147 | 0.051 | 3.7 × 10−3 | ||
| rs17042140 | 1 | 212 741 548 | T | PTPN14 | 2750 | 0.46 | 0.129 | 0.026 | 6.6 × 10−7 | 833 | 0.43 | 0.019 | 0.049 | 7.0 × 10−1 | ||
| rs11026407 | 11 | 22 061 606 | C | ANO5 | 2640 | 0.26 | −0.149 | 0.030 | 6.1 × 10−7 | 832 | 0.28 | −0.038 | 0.057 | 5.0 × 10−1 | ||
| rs1203867 | 20 | 22 479 141 | T | FOXA2 | 2607 | 0.13 | −0.175 | 0.039 | 9.0 × 10−6 | 830 | 0.17 | −0.023 | 0.065 | 7.2 × 10−1 | ||
| FT4 concentration | ||||||||||||||||
| rs6834538 | 4 | 113 763 601 | T | C4orf21 | 2754 | 0.03 | −0.351 | 0.074 | 2.4 × 10−6 | 826 | 0.04 | −0.117 | 0.133 | 3.8 × 10−1 | ||
| rs7785730 | 7 | 28 062 724 | G | JAZF1 | 2787 | 0.48 | −0.122 | 0.027 | 4.6 × 10−6 | 823 | 0.49 | −0.050 | 0.049 | 3.1 × 10−1 | ||
| rs7951105 | 11 | 38 988 369 | T | – | 2771 | 0.15 | −0.181 | 0.036 | 6.8 × 10−7 | 827 | 0.17 | −0.092 | 0.068 | 1.8 × 10−1 | ||
| n | MAF | OR | SE | P | N | MAF | OR | SE | P | |||||||
| Cases | Controls | Cases | Controls | |||||||||||||
| Anti-TPO antibody positivity | ||||||||||||||||
| rs2071403 | 2 | 1 396 251 | A | TPO | 3391 | 0.27 | 0.38 | 0.626 | 0.087 | 8.6 × 10−8 | 827 | 0.27 | 0.40 | 0.541 | 0.169 | 2.7 × 10−4 |
| rs733208 | 6 | 33 190 286 | A | HLA-DPB2 | 3300 | 0.39 | 0.30 | 1.450 | 0.081 | 4.6 × 10−6 | 825 | 0.38 | 0.31 | 1.385 | 0.151 | 3.0 × 10−2 |
| rs17111090 | 12 | 71 035 409 | A | TRHDE | 3378 | 0.03 | 0.01 | 2.144 | 0.251 | 2.4 × 10−3 | 829 | 0.00 | 0.01 | 0.754 | 1.062 | 7.9 × 10−1 |
| Variant | Chr | Position (bp) | A1 | Nearby gene | Joint stage 1 and 2 | |||||||||||
| n | β | SE | P | I2 | Het P | |||||||||||
| TSH concentration | ||||||||||||||||
| rs12126655 | 1 | 108 158 343 | G | VAV3 | 3499 | 0.138 | 0.025 | 2.2 × 10−8 | 0 | 0.837 | ||||||
| rs17042140 | 1 | 212 741 548 | T | PTPN14 | 3583 | 0.105 | 0.023 | 4.4 × 10−6 | 75 | 0.048 | ||||||
| rs11026407 | 11 | 22 061 606 | C | ANO5 | 3472 | −0.125 | 0.026 | 2.1 × 10−8 | 66 | 0.085 | ||||||
| rs1203867 | 20 | 22 479 141 | T | FOXA2 | 3437 | −0.134 | 0.034 | 6.7 × 10−5 | 75 | 0.045 | ||||||
| FT4 concentration | ||||||||||||||||
| rs6834538 | 4 | 113 763 601 | T | C4orf21 | 3580 | −0.296 | 0.065 | 5.2 × 10−6 | 58 | 0.124 | ||||||
| rs7785730 | 7 | 28 062 724 | G | JAZF1 | 3610 | 0.106 | 0.023 | 6.1 × 10−6 | 41 | 0.195 | ||||||
| rs7951105 | 11 | 38 988 369 | T | – | 3598 | −0.161 | 0.032 | 4.9 × 10−7 | 25 | 0.250 | ||||||
| n | OR | SE | P | I2 | Het P | |||||||||||
| Anti-TPO antibody positivity | ||||||||||||||||
| rs2071403 | 2 | 1 396 251 | A | TPO | 4218 | 0.607 | 0.078 | 1.3 × 10−10 | 42 | 0.190 | ||||||
| rs733208 | 6 | 33 190 286 | A | HLA-DPB2 | 4125 | 1.435 | 0.071 | 4.2 × 10−7 | 0 | 0.530 | ||||||
| rs17111090 | 12 | 71 035 409 | A | TRHDE | 4207 | 2.029 | 0.244 | 3.8 × 10−3 | 65 | 0.092 | ||||||
Associations were tested with minor alleles by linear regression for TSH and fT4 concentration and by logistic regression for anti-TPO antibody positivity. For TSH and fT4 concentration, the effect size is shown as β and for anti-TPO antibody positivity as odds ratio (OR). The minor allele and its physical position are indexed to the positive strand of the National Center for Biotechnology Information (NCBI) build 36. The nearby gene is defined as the gene nearest to the variant within the boundary of the 50-kb distance. Chr, chromosome number; A1, minor allele; N, number of subjects; MAF, minor allele frequency; β, effect size of linear regression; SE, standard error; I2, heterogeneity estimate; Het P, P-value for heterogeneity.
Stage 2 follow-up and joint Stages 1 and 2 analyses
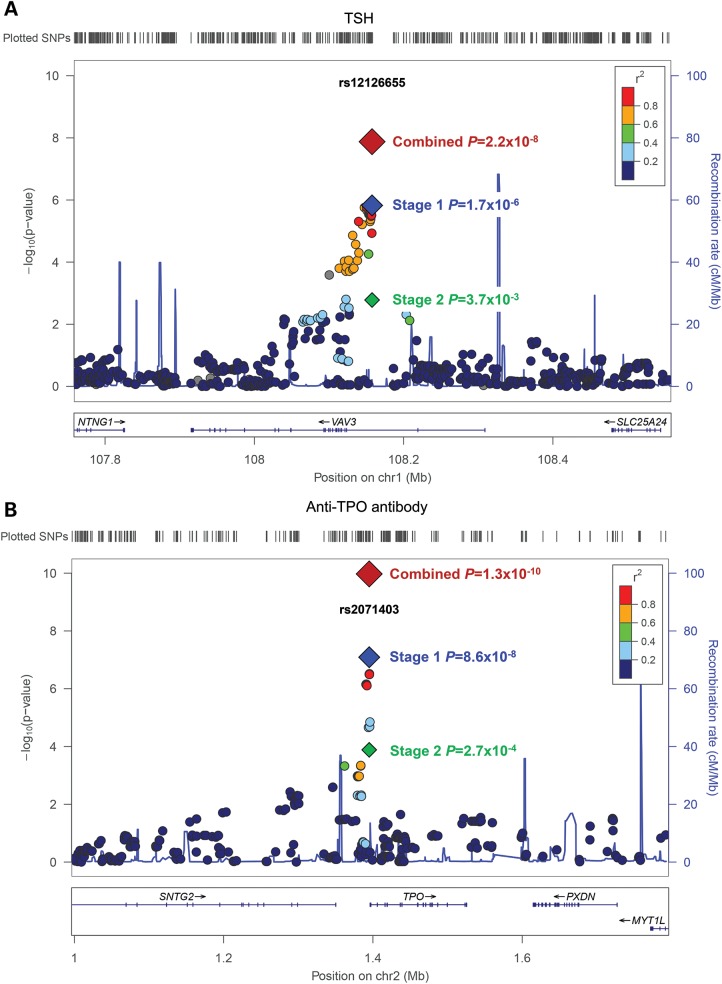
The 10 variants were further evaluated in the Stage 2 analysis. The 829 participants for the Stage 2 follow-up were from the Korean Longitudinal Study on Health and Aging (KLoSHA) cohort, and their clinical characteristics are shown in Table 1. Among the 10 variants, three showed significant associations (P < 0.05) (Table 2). The rs12126655 variant in the intron of VAV3 (vav 3 guanine nucleotide exchange factor) was associated with TSH concentration (P = 3.7 × 10−3). The rs2071403 variant in the 5′ untranslated region (UTR) of TPO (thyroid peroxidase) and rs733208 in the intron of HLA-DPB2 (major histocompatibility complex, class II, DP beta 2 (pseudogene)) were associated with anti-TPO antibody positivity (P = 2.7 × 10−4 and 3.0 × 10−2). In the joint Stages 1 and 2 meta-analyses, we observed a genome-wide significance for an association between the rs12126655 variant in VAV3 and TSH concentration (P = 2.2 × 10−8) and the rs2071403 variant in TPO and anti-TPO antibody positivity (P = 1.3 × 10−10). The rs12126655 variant was in strong linkage disequilibrium (LD) [r2 = 0.84 in HapMap Han Chinese in Beijing, China (CHB) and Japanese in Tokyo, Japan (JPT)] with rs17020124, which was recently reported to be associated with TSH concentration in Europeans (8). Regional association plots of the variants near rs12126655 and rs2071403 are shown in Figure 1. Two additional variants showed a suggestive genome-wide significant association with fT4 (rs7951195 in 11p12, P = 4.9 × 10−7) and anti-TPO antibody positivity (rs733208 in HLA-DPB2, P = 4.2 × 10−7). We also limited our subjects to those who had normal TSH, and normal fT4 concentrations, and were absent of anti-TPO antibody (N = 2794) and observed a similar association trend (Supplementary Material, Table S4).
Figure 1.
A regional association plot showing associations with TSH concentration (A) and anti-TPO antibody (B). The hash marks above the panel show the location of each variant. The P-values from the association testing are shown in the negative log10 scale on the y-axis. The blue diamond indicates the variant with the most significant association in the Stage 1 genome scan. The green and red diamonds represent the association results in Stage 2 and joint Stages 1 and 2 analyses, respectively. The variants in LD with the most significant SNPs are color-coded to represent their LD strength in HapMap CHB/JPT. The locations of genes, exons and their transcription direction are shown in the lower panel (derived from the Human Genome hg18 build).
Subgroup analysis according to sex or subclinical hypothyroidism
We investigated whether there was a sex-specific effect of the two variants that reached genome-wide significance (rs12126655 and rs2071403) (Table 3). Although the effect size of the rs12126655 variant was larger in females compared with males, the difference in effect size was not significant between the two groups. Regarding the rs2071403 variant, the effect size was also larger in females compared with males, and there was a marginal significance in difference of effect size (P = 0.063). We further performed a case–control analysis by comparing subgroups of euthyroid subjects (N = 3056) and those who had subclinical hypothyroidism (N = 480) (Supplementary Material, Table S5). The rs12126655 variant in VAV3 was associated with subclinical hypothyroidism with a significance of P < 0.05 in both Stages 1 and 2 analyses, thus resulting in a joint Stages 1 and 2 significance of P = 1.1 × 10−3. The rs2071403 variant in TPO was not associated subclinical hypothyroidism (P = 0.404). The phenotypic variances explained by genome-wide SNPs in the Stage 1 participants were analysed with a polygenic mixed linear model using Genome-Wide Complex Trait Analysis (GCTA) (15). We estimated that the variance explained by common variants across the genome was 17.1% for TSH, 27.6% for fT4, and 15.1% for anti-TPO antibody positivity.
Table 3.
Sex-specific effect of rs1212665 and rs2071403 on TSH concentration and anti-TPO antibody positivity
| Male |
Female |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Variant | Chr | Position (bp) | A1 | Nearby gene | N | β/OR | SE | P | N | β/OR | SE | P |
| rs12126655 | 1 | 108 158 343 | G | VAV3 | 1535 | 0.108 | 0.036 | 2.7 × 10−3 | 1964 | 0.157 | 0.034 | 2.7 × 10−6 |
| rs2071403 | 2 | 1 396 251 | A | TPO | 1881 | 0.749 | 0.135 | 3.2 × 10−2 | 2337 | 0.551 | 0.095 | 4.1 × 10−10 |
| Joint male and female | ||||||||||||
| N | β/OR | SE | P | I2 | Het P | |||||||
| rs12126655 | 3499 | 0.134 | 0.025 | 6.1 × 10−8 | 0 | 0.322 | ||||||
| rs2071403 | 4218 | 0.610 | 0.078 | 2.3 × 10−10 | 71 | 0.063 | ||||||
Associations were tested with minor alleles by linear regression for TSH concentration and by logistic regression for anti-TPO antibody positivity. For TSH concentration, the effect size is shown as β and for anti-TPO antibody positivity as odds ratio (OR). The minor allele and its physical position are indexed to the positive strand of the NCBI build 36. The nearby gene is defined as the gene nearest to the variant within the boundary of the 50-kb distance. Chr, chromosome number; A1, minor allele; N, number of subjects; MAF, minor allele frequency; β, effect size of linear regression; SE, standard error; I2, heterogeneity estimate; Het P, P-value for heterogeneity.
Pathway analysis
We performed pathway analysis using Database for Annotation, Visualization and Integrated Discovery (DAVID) with genes annotated by top listed variants (16,17). Using the functional annotation tool we found that ‘Antigen processing and presentation of peptide antigen’ was the most significantly enriched annotation term in Gene Onotology (GO) biological process database (P = 1.2 × 10−5) (18) and ‘Autoimmune thyroid disease’ was the most significantly enriched term in Kyoto Encyclopedia of Genes and Genomes (KEGG) database (P = 1.8 × 10−6) (19) regarding anti-TPO antibody (Table 4). However, we were not able to identify a significantly enriched GO term or KEGG pathway using the genes annotated from the TSH or fT4 association results.
Table 4.
Pathway analysis of top genes associated with anti-TPO antibody positivity (FDR by Benjamini–Hochberg <0.5)
| Database code | Annotation term | No. of genes | % | P | Genes |
|---|---|---|---|---|---|
| Gene Ontology Biological Process Level 3 | |||||
| GO:0048002 | Antigen processing and presentation of peptide antigen | 4 | 12.5 | 1.21 × 10−5 | MICA, HLA-B, HLA-G, HLA-F |
| GO:0006952 | Defense response | 4 | 12.5 | 7.67 × 10−2 | MICA, TACR1, HLA-B, HLA-G |
| GO:0009408 | Response to heat | 2 | 6.2 | 8.77 × 10−2 | MICA, TACR1 |
| KEGG Pathway | |||||
| hsa05320 | Autoimmune thyroid disease | 5 | 15.6 | 1.80 × 10−6 | TPO, HLA-DPB1, HLA-B, HLA-G, HLA-F |
| hsa05330 | Allograft rejection | 4 | 12.5 | 3.78 × 10−5 | HLA-DPB1, HLA-B, HLA-G, HLA-F |
| hsa05332 | Graft-versus-host disease | 4 | 12.5 | 4.82 × 10−5 | HLA-DPB1, HLA-B, HLA-G, HLA-F |
| hsa04940 | Type I diabetes mellitus | 4 | 12.5 | 6.04 × 10−5 | HLA-DPB1, HLA-B, HLA-G, HLA-F |
| hsa05416 | Viral myocarditis | 4 | 12.5 | 2.92 × 10−4 | HLA-DPB1, HLA-B, HLA-G, HLA-F |
| hsa04612 | Antigen processing and presentation | 4 | 12.5 | 4.63 × 10−4 | HLA-DPB1, HLA-B, HLA-G, HLA-F |
| hsa04514 | Cell adhesion molecules | 4 | 12.5 | 1.80 × 10−3 | HLA-DPB1, HLA-B, HLA-G, HLA-F |
| hsa04650 | Natural killer cell mediated cytotoxicity | 3 | 9.4 | 2.66 × 10−2 | MICA, HLA-B, HLA-G |
| hsa04144 | Endocytosis | 3 | 9.4 | 4.84 × 10−2 | HLA-B, HLA-G, HLA-F |
DAVID analysis for functional annotation was performed with GO, and KEGG database. The P-value is from the modified Fisher exact test provided from the DAVID system. The genes submitted as input were: ASTN2, BCL2L11, C11orf41, C4orf35, C6orf10, CAPRIN2, COL11A2, CTNND2, HEATR1, HLA-B, HLA-DPB1, HLA-DPB2, HLA-F, HLA-G, IPO8, KCNIP1, LARGE, MICA, NELL2, PDCD10, PIGN, PKNOX2, POLE4, RGS9, RPS6KA2, RSPO2, SDK1, TACR1, TPO, TSH, UBD, WDR49 and ZNF8.
The expression of TPO in human thyroid tissue
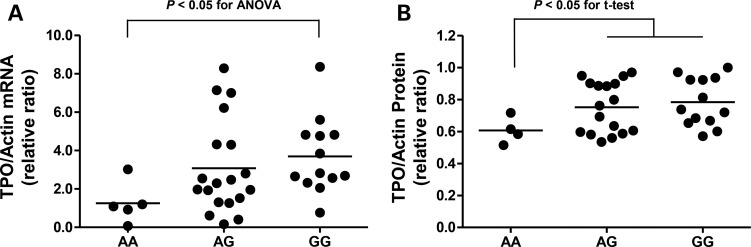
Among the two variants that passed the threshold for the genome-wide significance, rs12126655 variant was located in intron of VAV3 and it was not feasible to perform a functional analysis. Regarding the rs2071403 in the TPO gene, we studied the putative functional role of this variant by comparing the expression level of TPO in human thyroid tissues according to the genotypes. Thyroid tissues were used from 37 participants who had thyroid tumours (benign or malignant) and underwent thyroidectomy. The participants who had the rs2071403A variant, which was associated with lower anti-TPO antibody positivity, had a significantly decreased TPO mRNA expression level in their thyroid tissue (Fig. 2A). However, the protein level was only decreased in participants with the AA genotype compared to those with the AG or GG genotype (Fig. 2B).
Figure 2.
The relative expression of TPO mRNA (A) and protein (B) in human thyroid tissues according to the rs2071403 genotypes. There was a significant difference in TPO mRNA expression according to the three genotype groups [P < 0.05 for the analysis of variance (ANOVA)]. TPO protein expression was decreased in the AA group compared to the AG and GG groups (P < 0.05 for Student's t-test).
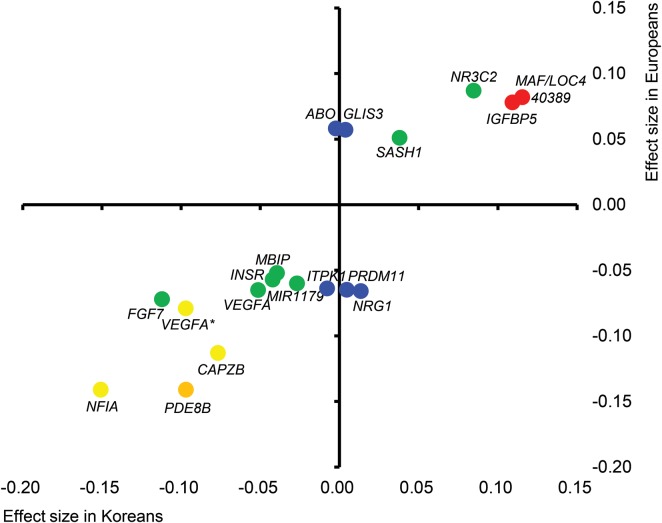
A comparison with the European GWAS results
The effect sizes (β) reported in the recent meta-analysis of the GWAS regarding TSH in Europeans (7) were compared with those from our Stage 1 genome scan (Fig. 3 and Supplementary Material, Table S6). For TSH concentration, 20 variants reported in Europeans were available for analyses in our study, and six variants showed significant associations with P < 0.05 in the identical direction of association. There was a significant positive correlation between effect sizes in the two populations (Pearson's correlation coefficient 0.864 and P = 4.0 × 10−6). We also investigated the variants that were confirmed to be associated with autoimmune thyroid disease (including Graves' disease and Hashimoto's thyroiditis) (Supplementary Material, Table S7) (11–13). Among the 15 variants investigated, three were nominally (P < 0.05) associated with plasma TSH concentration and three other variants were associated with anti-TPO antibody positivity (P < 0.05).
Figure 3.
A comparison of variant effect size associated with TSH between Koreans and Europeans. For the variants associated with TSH concentrations in a recent European meta-GWAS (9), the effect sizes (β) for Koreans (x-axis) and Europeans (y-axis) are plotted with the corresponding Korean P-values. P < 0.0001, red; 0.0001 ≤ P < 0.01, orange; 0.01 ≤ P < 0.10, yellow; 0.10 ≤ P < 0.50, green; and 0.50 ≤ P, blue. The two VEGFA variants are distinguished by VEGFA* for rs9472138 and VEGFA for rs11755845.
DISCUSSION
Using two-stage genome-wide association analyses, we replicated and confirmed that the rs12126655 variant in VAV3 was associated with TSH concentration and observed a novel genome-wide significant association between the rs2071403 variant at the 5′ UTR of TPO and anti-TPO antibody positivity in a Korean population. We also identified two variants that had suggestive genome-wide significant associations (P < 5.0 × 10−7) with fT4 concentration (rs7951105 in 11p12) and anti-TPO antibody positivity (rs733208 in HLA-DPB2). To our best knowledge, this is the first study in Asians to investigate genetic variants affecting TSH and fT4 concentrations as a continuous trait at a genome-wide scale. In addition, this is the first study to identify a presumptive functional variant affecting anti-TPO antibody positivity with unequivocal significance.
The VAV3 encodes for vav 3 guanine nucleotide exchange factor, which activates Rho and Rac GTPase. VAV3 is involved in immune function, sympathetic hyperactivity and oncogenesis (20–22). The functional role of VAV3 in TSH regulation is not fully understood. VAV3 is widely expressed in several tissues, including peripheral lymphocytes, the spleen, brain and thyroid gland (23). In 27 758 Icelanders, the rs17020124 variant, which is in strong LD (r2 = 0.84 in HapMap CHB/JPT) with rs12126655, was associated with TSH concentration with genome-wide significance (P = 4.3 × 10−17) (8). In another recent GWAS involving 39 248 Europeans, the rs4915077 variant in VAV3 was associated with self-reported hypothyroidism (P = 7.5 × 10−10) (24). This variant was also in strong LD with rs12126655 (r2 = 0.86 in HapMap CHB/JPT). The rs12126655 was associated with subclinical hypothyroidism in both our Stages 1 and 2 participants with nominal significance. These results indicate that variants in VAV3 are important determinants of TSH concentration and affect thyroid dysfunction across different ethnicities.
A notable result of this study was the association between the rs2071403 variant at the 5′ UTR of TPO gene and anti-TPO antibody positivity. The TPO gene encodes for TPO and is highly expressed in human thyroid tissue. This gene plays a critical role in thyroid hormone synthesis by iodinating tyrosine residues in thyroglobulin and phenoxy-ether formation between iodinated tyrosine residues (25,26). Inactivating mutations in the TPO gene is known to cause an iodide organification defect that results in congenital hypothyroidism (27,28). Because the rs2071403 variant was located at the 5′ UTR of the TPO gene 75 bp proximal to the translation start site, we thought that it could influence the expression level of the TPO gene. We investigated the mRNA and protein expression levels in human thyroid tissue and observed that the expression levels are different according to the rs2071403 genotype. However, the G to A variation of rs2071403 does not appear to alter transcription factor-binding site (29), and a detailed molecular mechanism of how this variant affects expression levels should be further investigated. Currently, it is unclear whether the rs2071403 variant is merely a marker that is in LD with a nearby functional variant. It could be postulated that the rs2071403 variant is associated with a decreased expression of the TPO gene in thyroid tissues and thus results in a lower rate of anti-TPO antibody positivity. The variant was also marginally associated with decreased fT4 concentration in our joint Stages 1 and 2 analyses (β = −0.044, P = 0.071). The decrease in fT4 concentration might also be attributed to decreased TPO gene expression and altered hormonal synthesis in the thyroid tissue.
When we compared the TSH-associated variants reported in Europeans with our Stage 1 analysis, we observed a significant positive linear correlation of effect size between the two populations. Among the variants reported in Europeans, rs3813582 near MAF/LOC440389 and rs13015993 near IGFBP5 showed strong associations in our Stage 1 analysis, albeit not passing the significance threshold of the Stage 1 analysis (P = 4.4 × 10−5 and 2.9 × 10−5). The most significantly associated genetic variant for TSH in Europeans was rs6885099, which is located in the intron of PDE8B (phosphodiesterase 8B). This variant also had a nominal association (P = 5.9 × 10−3) with TSH in our Stage 1 subjects. These results imply that the genetic variants affecting thyroid function may be similar between the two populations. It is not clear whether the difference in the prevalence of subclinical hypothyroidism between the US and the Korean population could be explained by genetic predisposition. Excessive iodine intake is associated with higher risk of subclinical hypothyroidism (30). As iodine intake in Korean population is amongst the highest except for the Japanese, the difference might be more likely associated with difference in iodine intake (31).
A major limitation of our study regards the relatively modest sample size for genome-wide association analyses. For the quantitative trait analyses, we had 87% power to detect a genome-wide significant association for variants with a minor allele frequency of 0.35 and an effect size of 0.15 in 3500 subjects. However, the power decreased as the effect size reduced to 0.14 and 0.13 by 75 and 60%, respectively. For the case–control analyses, we only had sufficient power to detect association results for variants with odds ratios of 1.6 or more, assuming a minor allele frequency of 0.35 and an anti-TPO antibody positive rate of 0.11 in 4200 subjects. Therefore, we might have missed a significant portion of true positive associations. Further replications and meta-analysis would be required to confirm our findings and to discover additional genetic variants regarding thyroid function and anti-TPO antibody positivity.
In conclusion, we performed a genome-wide association analysis on TSH and fT4 concentrations and anti-TPO antibody positivity in Koreans. We confirmed that an intronic variant of VAV3, rs12126655, was associated with TSH concentration with genome-wide significance. We also identified a novel association between a variant at the 5′ UTR of the TPO gene and anti-TPO antibody positivity. This variant showed marginal sex-specific effects and was associated with TPO mRNA expression in human thyroid tissues. We hope these results can expand our knowledge of thyroid hormone synthesis, secretion, and autoimmunity and eventually provide a basis for personalized medicine in autoimmune thyroid disease.
MATERIALS AND METHODS
Study participants for the Stage 1 genome scan
Participants for the Stage 1 genome scan were recruited from the Ansung cohort based on Anusung-Si, GyeongGi-Do, Korea, as a component of the KoGES (32,33). The initial investigation began in 2001 with 5018 participants aged 40–69. Participants with a previous history of thyroid disease or on medications (thyroid hormone, anti-thyroid medications, steroid, estrogen and contraceptives) that could affect thyroid hormone concentration were excluded. The total number of subjects with plasma TSH and fT4 concentration was 2789, and the number of subjects with anti-TPO antibody measurement was 3396. The baseline characteristics of the study subjects are shown in Table 1.
Study participants for the Stage 2 follow-up
Participants for the Stage 2 follow-up were from the KLoSHA cohort (34). The cohort was initiated in 2005 for 953 participants 65 years old or older who lived in SeongNam-Si, GyeongGi-Do, Korea. Participants with a previous history of thyroid disease or taking the above medications were also excluded. A total of 829 subjects had plasma TSH, fT4 and anti-TPO antibody measurements. The institutional review board of the Biomedical Research Institute at Seoul National University Hospital approved the study protocol (H-1309-075-521), and written informed consent was obtained from each subject. All clinical investigations were conducted according to the principles expressed in the Declaration of Helsinki.
Measurements of TSH, fT4 and anti-TPO antibody
For both Stages 1 and 2 participants, the plasma TSH concentrations were measured using a radioimmunoassay (RIA) kit from CIS Bio International S.A., Gif-sur-Yvette, France (normal range 0.4–4.1 μIU/l), and the plasma fT4 concentrations were measured using an RIA kit from DiaSorin S.p.A., Saluggia, Italy (normal range 0.7–1.8 ng/dl). Plasma anti-TPO antibody concentrations were measured with an RIA kit from RSR Ltd, Pentwyn, UK (normal range <0.3 μIU/l) in Stage 1 participants and with an RIA kit from BRAHMS, Hennigsdorf, UK (normal range <25 μIU/l) in Stage 2 subjects. Participants with an anti-TPO antibody concentration above the normal range were classified as anti-TPO antibody positive cases and the remaining participants were controls. Because the variables showed a significant deviation from a Gaussian distribution, each of the TSH and fT4 concentrations was inverse normal transformed before the analyses.
Genotyping and imputation
We extracted genomic DNA from the participants' peripheral leukocytes. The Stage 1 genome scan was performed using an Affymetrix Genome-Wide Human SNP Array 5.0. Only unrelated subjects with genotype missingness <5% were included in the analysis. Markers with significant deviations from the Hardy–Weinberg equilibrium (P < 1.0 × 10−6), a genotype call rate <0.95 and minor allele frequency <0.01 were excluded. A total of 351 669 SNPs were directly genotyped and were available for analyses. For the Stage 2 follow-up genotyping, we used the TaqMan assay from Applied Biosystems, Carlsbad, CA, USA. Overall, the TaqMan genotyping success rate was 99.5%, and the concordance rate based on 15 blind duplicate comparisons was 100% in the Stage 2 study. Imputation was performed using IMPUTE software (https://mathgen.stats.ox.ac.uk/impute) (35). The HapMap phased genotype information of CHB and JPT (build 36 release 22) was used as a reference (36). Imputed SNPs with high genotype information content (info >0.5) were used. The identical quality control criteria were applied for the imputed SNPs as in the genotyped SNPs. After filtering, a total of 1 418 709 SNPs were available for the analyses.
Statistical analyses
Most of the association testing was performed using PLINK version 1.07 (http://pngu.mgh.harvard.edu/purcell/plink/) (37). For plasma TSH and fT4 concentrations, a linear regression assuming an additive genotypic model was used. For the plasma anti-TPO antibody concentration, a logistic regression (anti-TPO antibody positive versus negative subjects) was used. Age and sex were used as covariates. The full result of the Stage 1 genome scan can be downloaded in our webpage (http://bri.snuh.org/bench/_/notice/5250/view.do). Variants that passed the significance threshold (P < 1.0 × 10−5) in the Stage 1 genome scan were further genotyped in Stage 2. The significance threshold for the Stage 2 follow-up was P < 0.05. A joint analysis of Stages 1 and 2 was performed with METAL (http://www.sph.umich.edu/csg/abecasis/Metal/) using an inverse-variance meta-analysis method assuming fixed effects (38). Genome-wide significance was considered as P < 5.0 × 10−8. To account for possible cryptic relatedness and population stratification in the Stage 1 genome scan, we performed additional analyses using EMMAX (http://www.sph.umich.edu/csg/kang/emmax/) (14). To identify sex-specific effects, the analyses were performed separately for each sex, and we tested heterogeneity between effect sizes (P < 0.05 to be significant). The estimated proportion of phenotypic variance explained by the genome-wide SNPs was calculated using GCTA software (http://www.complextraitgenomics.com/software/gcta) (37). The effect sizes of variants associated with TSH in the recent European meta-GWAS (27) and our Stage 1 genome scan were compared. For the pathway analysis, we used functional annotation tool in the DAVID system (http://david.abcc.ncifcrf.gov/) (17,18). We searched the GO (18) and KEGG database (19). The genes that are annotated by the variants that had false discovery rate (FDR) of <0.5 by Benjamini–Hochberg procedure in our Stage 1 genome scan were selected for input. The corresponding cut-off P-values for the association with TSH, fT4 and anti-TPO antibody were 1.3 × 10−5, 1.7 × 10−4 and 1.2 × 10−4, respectively. The P–value of <0.01 for the modified Fisher exact test provided from the DAVID system was considered to be significant.
Thyroid tissue preparation
Thyroid tissue was prepared from 37 participants who had thyroid tumours and underwent thyroidectomy from February 2012 to March 2013. The normal thyroid tissue in a 1 × 1 × 1 cm3 volume was dissected from the contralateral lobe without a tumour and was immediately stored in a liquid nitrogen tank. Thyroid tissues were collected for the biobank program of the Department of Surgery, Seoul National University Hospital. The biobank study protocol was approved by the institutional review board of the Biomedical Research Institute at Seoul National University Hospital (H-0809-097-258). Each participant provided written informed consent for the study.
RNA extraction and real-time PCR analysis
Total RNA was isolated from normal thyroid tissues using TRizol reagent (Ambion, Foster City, CA, USA), and M-MLV reverse transcriptase (Invitrogen, Carlsbad, CA, USA) was used for cDNA synthesis. Real-time PCR was performed using SYBR-master mix (Takara, Otsu, Shiga, Japan) and an ABI 7500 real-time PCR system (Applied Biosystems, Forster City, CA, USA). Each sample was analysed in duplicate. The following primer sequences were used for the real-time PCR: for TPO (189 bp), forward primer 5′-CGGGTCATCTGTGACAACAC-3′, reverse primer 5′-GTGCACAAAGTCCCCATTCT-3′; and for beta-actin (205 bp), forward primer 5′-TGACGTGGACATCCGC AAAG-3′, reverse primer 5′-CTGGAAGGTGGACAGCGAGG-3′.
Western blot analysis
Normal thyroid tissues were lysated with 20 mm Tris (pH 7.4), 5 mm EDTA and 1% NP-40, supplemented with a protease inhibitor mixture in 1 : 100 dilution (Sigma–Aldrich, St Louis, MO, USA), separated by SDS–PAGE and transferred to a Hybond ECL nitrocellulose membrane (Amersham, Piscataway, NJ, USA). To assess TPO expression, the nitrocellulose membranes were incubated with an anti-TPO antibody (Abcam, Cambridge, MA, USA) at a 1 : 3000 dilution overnight at 4°C, and then subsequently incubated with horseradish peroxidase-conjugated goat anti-rabbit IgG (Cell Signaling, Boston, MA, USA) at a 1 : 5000 dilution for 1 h at room temperature. The signals were detected with the ECL Western Blotting Analysis System (Thermo Fisher Scientific, Waltham, MA, USA). For a loading control, β-actin immunoreactivity was detected with a monoclonal anti-β-actin antibody (Sigma–Aldrich, St Louis, MO, USA) at a 1 : 5000 dilution.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by a grant from the National Project for Personalized Genomic Medicine, Ministry for Health and Welfare (grant no. A111218-GM09); the National Genome Research Institute, Korean Center for Disease Control and Prevention (contract #2001-347-6111-221, 2002-347-6111-221, 2003-347-6111-221, 2004-E71001-00, 2005-E71001-00, 2006-E71005-00, 2006-E71006-00, 2007-E71001-00, 2007-E71003-00, 2008-E71001-00, 2008-E71005-00, 2009-E71002-00, 2009-E71007-00, 2010-E71001-00, 2010-E71004-00, 2011-E71004-00, 2011-E71008-00, 2012-E71008-00, 2012-E71005-00, and grant no. 4845-301); and an intramural grant from the Korea National Institute of Health (grant no. 2012-N73002-00), Republic of Korea.
Supplementary Material
ACKNOWLEDGEMENT
We thank Dr Jung Hun Ohn for his advice and comments on performing the pathway analysis. This study was supported by a grant from the Korean Health Technology R&D Project, Ministry for Health, Welfare and Family Affairs, Republic of Korea (grant no. A092077).
Conflict of Interest statement. None declared.
REFERENCES
- 1.Bianco A.C., Kim B.W. Deiodinases: implications of the local control of thyroid hormone action. J. Clin. Invest. 2006;116:2571–2579. doi: 10.1172/JCI29812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Visser W.E., Friesema E.C., Visser T.J. Minireview: thyroid hormone transporters: the knowns and the unknowns. Mol. Endocrinol. 2011;25:1–14. doi: 10.1210/me.2010-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andersen S., Pedersen K.M., Bruun N.H., Laurberg P. Narrow individual variations in serum T(4) and T(3) in normal subjects: a clue to the understanding of subclinical thyroid disease. J. Clin. Endocrinol. Metab. 2002;87:1068–1072. doi: 10.1210/jcem.87.3.8165. [DOI] [PubMed] [Google Scholar]
- 4.Dayan C.M., Panicker V. Novel insights into thyroid hormones from the study of common genetic variation. Nat. Rev. Endocrinol. 2009;5:211–218. doi: 10.1038/nrendo.2009.19. [DOI] [PubMed] [Google Scholar]
- 5.Arnaud-Lopez L., Usala G., Ceresini G., Mitchell B.D., Pilia M.G., Piras M.G., Sestu N., Maschio A., Busonero F., Albai G., et al. Phosphodiesterase 8B gene variants are associated with serum TSH levels and thyroid function. Am. J. Hum. Genet. 2008;82:1270–1280. doi: 10.1016/j.ajhg.2008.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Panicker V., Wilson S.G., Walsh J.P., Richards J.B., Brown S.J., Beilby J.P., Bremner A.P., Surdulescu G.L., Qweitin E., Gillham-Nasenya I., et al. A locus on chromosome 1p36 is associated with thyrotropin and thyroid function as identified by genome-wide association study. Am. J. Hum. Genet. 2010;87:430–435. doi: 10.1016/j.ajhg.2010.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lowe J.K., Maller J.B., Pe'er I., Neale B.M., Salit J., Kenny E.E., Shea J.L., Burkhardt R., Smith J.G., Ji W., et al. Genome-wide association studies in an isolated founder population from the Pacific Island of Kosrae. PLoS Genet. 2009;5:e1000365. doi: 10.1371/journal.pgen.1000365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gudmundsson J., Sulem P., Gudbjartsson D.F., Jonasson J.G., Masson G., He H., Jonasdottir A., Sigurdsson A., Stacey S.N., Johannsdottir H., et al. Discovery of common variants associated with low TSH levels and thyroid cancer risk. Nat. Genet. 2012;44:319–322. doi: 10.1038/ng.1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Porcu E., Medici M., Pistis G., Volpato C.B., Wilson S.G., Cappola A.R., Bos S.D., Deelen J., den Heijer M., Freathy R.M., et al. A meta-analysis of thyroid-related traits reveals novel loci and gender-specific differences in the regulation of thyroid function. PLoS Genet. 2013;9:e1003266. doi: 10.1371/journal.pgen.1003266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simmonds M.J. GWAS in autoimmune thyroid disease: redefining our understanding of pathogenesis. Nat. Rev. Endocrinol. 2013;9:277–287. doi: 10.1038/nrendo.2013.56. [DOI] [PubMed] [Google Scholar]
- 11.Chu X., Pan C.M., Zhao S.X., Liang J., Gao G.Q., Zhang X.M., Yuan G.Y., Li C.G., Xue L.Q., Shen M., et al. A genome-wide association study identifies two new risk loci for Graves’ disease. Nat. Genet. 2011;43:897–901. doi: 10.1038/ng.898. [DOI] [PubMed] [Google Scholar]
- 12.Cooper J.D., Simmonds M.J., Walker N.M., Burren O., Brand O.J., Guo H., Wallace C., Stevens H., Coleman G., Franklyn J.A., et al. Seven newly identified loci for autoimmune thyroid disease. Hum. Mol. Genet. 2012;21:5202–5208. doi: 10.1093/hmg/dds357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao S.X., Xue L.Q., Liu W., Gu Z.H., Pan C.M., Yang S.Y., Zhan M., Wang H.N., Liang J., Gao G.Q., et al. Robust evidence for five new Graves’ disease risk loci from a staged genome-wide association analysis. Hum. Mol. Genet. 2013;22:3347–3362. doi: 10.1093/hmg/ddt183. [DOI] [PubMed] [Google Scholar]
- 14.Kang H.M., Sul J.H., Service S.K., Zaitlen N.A., Kong S.Y., Freimer N.B., Sabatti C., Eskin E. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 2010;42:348–354. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang J., Lee S.H., Goddard M.E., Visscher P.M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 17.Huang da W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanehisa M., Goto S., Kawashima S., Okuno Y., Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dumont C., Corsoni-Tadrzak A., Ruf S., de Boer J., Williams A., Turner M., Kioussis D., Tybulewicz V.L. Rac GTPases play critical roles in early T-cell development. Blood. 2009;113:3990–3998. doi: 10.1182/blood-2008-09-181180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sauzeau V., Sevilla M.A., Rivas-Elena J.V., de Alava E., Montero M.J., Lopez-Novoa J.M., Bustelo X.R. Vav3 proto-oncogene deficiency leads to sympathetic hyperactivity and cardiovascular dysfunction. Nat. Med. 2006;12:841–845. doi: 10.1038/nm1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang K.H., Sanchez-Aguilera A., Shen S., Sengupta A., Madhu M.N., Ficker A.M., Dunn S.K., Kuenzi A.M., Arnett J.L., Santho R.A., et al. Vav3 collaborates with p190-BCR-ABL in lymphoid progenitor leukemogenesis, proliferation, and survival. Blood. 2012;120:800–811. doi: 10.1182/blood-2011-06-361709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Movilla N., Bustelo X.R. Biological and regulatory properties of Vav-3, a new member of the Vav family of oncoproteins. Mol. Cell. Biol. 1999;19:7870–7885. doi: 10.1128/mcb.19.11.7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eriksson N., Tung J.Y., Kiefer A.K., Hinds D.A., Francke U., Mountain J.L., Do C.B. Novel associations for hypothyroidism include known autoimmune risk loci. PloS ONE. 2012;7:e34442. doi: 10.1371/journal.pone.0034442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taurog A., Dorris M.L., Doerge D.R. Mechanism of simultaneous iodination and coupling catalyzed by thyroid peroxidase. Arch. Biochem. Biophys. 1996;330:24–32. doi: 10.1006/abbi.1996.0222. [DOI] [PubMed] [Google Scholar]
- 26.Nunez J. Thyroid hormones: mechanism of phenoxy ether formation. Methods Enzymol. 1984;107:476–488. doi: 10.1016/0076-6879(84)07032-4. [DOI] [PubMed] [Google Scholar]
- 27.Bikker H., Vulsma T., Baas F., de Vijlder J.J. Identification of five novel inactivating mutations in the human thyroid peroxidase gene by denaturing gradient gel electrophoresis. Hum. Mutat. 1995;6:9–16. doi: 10.1002/humu.1380060104. [DOI] [PubMed] [Google Scholar]
- 28.Park S.M., Chatterjee V.K. Genetics of congenital hypothyroidism. J. Med. Genet. 2005;42:379–389. doi: 10.1136/jmg.2004.024158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heinemeyer T., Wingender E., Reuter I., Hermjakob H., Kel A.E., Kel O.V., Ignatieva E.V., Ananko E.A., Podkolodnaya O.A., Kolpakov F.A., et al. Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic Acids Res. 1998;26:362–367. doi: 10.1093/nar/26.1.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Teng W., Shan Z., Teng X., Guan H., Li Y., Teng D., Jin Y., Yu X., Fan C., Chong W., et al. Effect of iodine intake on thyroid diseases in China. N. Engl. J. Med. 2006;354:2783–2793. doi: 10.1056/NEJMoa054022. [DOI] [PubMed] [Google Scholar]
- 31.Kim J.Y., Moon S.J., Kim K.R., Sohn C.Y., Oh J.J. Dietary iodine intake and urinary iodine excretion in normal Korean adults. Yonsei Med. J. 1998;39:355–362. doi: 10.3349/ymj.1998.39.4.355. [DOI] [PubMed] [Google Scholar]
- 32.Cho Y.S., Go M.J., Kim Y.J., Heo J.Y., Oh J.H., Ban H.J., Yoon D., Lee M.H., Kim D.J., Park M., et al. A large-scale genome-wide association study of Asian populations uncovers genetic factors influencing eight quantitative traits. Nat. Genet. 2009;41:527–534. doi: 10.1038/ng.357. [DOI] [PubMed] [Google Scholar]
- 33.Hong K.W., Kim S.S., Kim Y. Genome-wide association study of orthostatic hypotension and supine-standing blood pressure changes in two Korean populations. Genomics Inform. 2013;11:129–134. doi: 10.5808/GI.2013.11.3.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moon J.H., Park Y.J., Kim T.H., Han J.W., Choi S.H., Lim S., Park D.J., Kim K.W., Jang H.C. Lower-but-normal serum TSH level is associated with the development or progression of cognitive impairment including mild cognitive impairment and dementia in elderly: a result from Korean Longitudinal Study on Health and Aging (KLoSHA) J. Clin. Endocrinol. Metab. 2013;99:424–432. doi: 10.1210/jc.2013-3385. [DOI] [PubMed] [Google Scholar]
- 35.Marchini J., Howie B., Myers S., McVean G., Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 36.The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 37.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Willer C.J., Li Y., Abecasis G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.