Abstract
RASopathies, a family of disorders characterized by cardiac defects, defective growth, facial dysmorphism, variable cognitive deficits and predisposition to certain malignancies, are caused by constitutional dysregulation of RAS signalling predominantly through the RAF/MEK/ERK (MAPK) cascade. We report on two germline mutations (p.Gly39dup and p.Val55Met) in RRAS, a gene encoding a small monomeric GTPase controlling cell adhesion, spreading and migration, underlying a rare (2 subjects among 504 individuals analysed) and variable phenotype with features partially overlapping Noonan syndrome, the most common RASopathy. We also identified somatic RRAS mutations (p.Gly39dup and p.Gln87Leu) in 2 of 110 cases of non-syndromic juvenile myelomonocytic leukaemia, a childhood myeloproliferative/myelodysplastic disease caused by upregulated RAS signalling, defining an atypical form of this haematological disorder rapidly progressing to acute myeloid leukaemia. Two of the three identified mutations affected known oncogenic hotspots of RAS genes and conferred variably enhanced RRAS function and stimulus-dependent MAPK activation. Expression of an RRAS mutant homolog in Caenorhabditis elegans enhanced RAS signalling and engendered protruding vulva, a phenotype previously linked to the RASopathy-causing SHOC2S2G mutant. Overall, these findings provide evidence of a functional link between RRAS and MAPK signalling and reveal an unpredicted role of enhanced RRAS function in human disease.
INTRODUCTION
Signalling elicited by activated cell surface receptors and transduced through RAS proteins to the RAF/MEK/ERK and PI3K/AKT cascades is central to cell proliferation, survival, differentiation and metabolism (1,2). Owing to this nodal role, enhanced traffic through RAS proteins and their downstream effectors has been established to have a major impact on oncogenesis (3,4). This signalling network also controls early and late developmental processes (e.g. organogenesis, morphology determination, synaptic plasticity and growth), and germline mutations in a number of genes encoding transducers and modulatory proteins participating in the RAS/MAPK signalling pathway have been causally linked to Noonan syndrome (NS) (5), one of the most common diseases affecting development and growth, and a group of clinically related syndromes, the so-called RASopathies (6–8). In this family of disorders, constitutional dysregulation of RAS signalling can be caused by enhanced activation of HRAS, KRAS and NRAS (RAS proteins hereafter), aberrant function of upstream signal transducers or effectors (PTPN11/SHP2, SOS1, SHOC2, RAF1, BRAF, MAP2K1/MEK1 and MAP2K2/MEK2) or inefficient down modulation by feedback mechanisms (CBL, NF1 and SPRED1). More recently, RIT1, encoding a monomeric GTPase structurally linked to RAS proteins, was identified as disease gene implicated in NS (9), extending the concept of ‘RASopathy gene’ to a transducer that contributes to signal propagation through RAS effector pathways but does not belong to the RAS/MAPK signalling backbone.
Clinical manifestations of RASopathies include postnatal reduced growth, a wide spectrum of cardiac defects, facial dysmorphism, ectodermal and skeletal anomalies and variable cognitive deficits (5,8,10). Consistent with the key role of most RASopathy genes in oncogenesis, these disorders are also characterized by variably increased risk for certain haematologic malignancies and other paediatric cancers (6,7,11,12). Most of these conditions are genetically heterogeneous, and the underlying disease gene has not been identified yet for a still significant fraction of cases. Based on the strict mechanistic link between the molecular events controlling development and contributing to oncogenesis, these ‘missing’ genes represent excellent candidate oncogenes/tumour suppressors.
Here, we report that constitutional dysregulation of RRAS function is associated with a Mendelian trait within the RASopathy spectrum and that somatically acquired mutations in the same gene occur in an aggressive form of juvenile myelomonocytic leukaemia (JMML), a rare childhood myeloproliferative/myelodysplastic neoplasm representing the archetypal somatic RASopathy (13), rapidly progressing to acute myeloid leukaemia (AML). We also demonstrate that RASopathy-causing RRAS mutations are activating and promote signalling perturbation by enhancing stimulus-dependent MEK, ERK and, at a lower extent, AKT phosphorylation.
RESULTS
Identification of candidate disease genes and RRAS mutation analysis
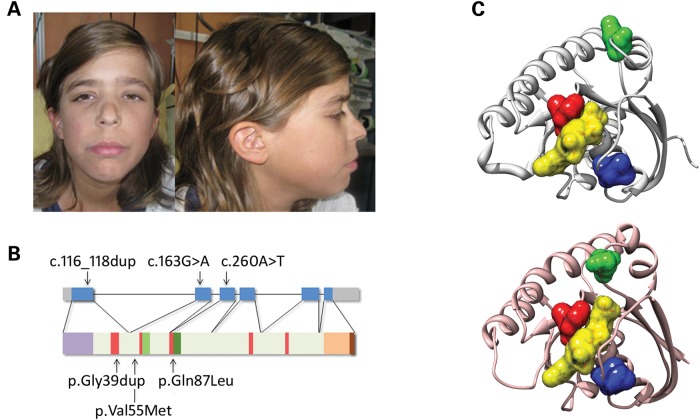
While the core of the machinery implicated in RAS signalling has been characterized comprehensively, signal propagation through this network is likely to include a larger number of proteins playing a modulatory or structural role (14), whose aberrant or defective function is expected to perturb development and contribute to oncogenesis. Based on this assumption, we used a protein interaction/functional association network analysis to select a panel of genes encoding proteins functionally linked to the RAS signalling network as candidates for NS or a related RASopathy (15). Candidate gene selection was based on the use of the previously identified RASopathy genes as ‘seed’ proteins (i.e. proteins used to build the interaction/functional networks), and considering a panel of databases to construct functional subnetworks (Supplementary material, Table S1 and Fig. S1). Sequence scanning of the best candidates in a RASopathy cohort including 96 unrelated subjects negative for mutations in known disease genes allowed the identification of a functionally relevant RRAS change (c.163G>A, p.Val55Met) (Supplementary material, Fig. S2) in an adult subject with clinical features suggestive of NS but lacking sufficient characteristics to allow a definitive diagnosis (Supplementary material, Table S2). Parental DNA was not available for segregation analysis. The mutation was not identified among >400 population-matched unaffected individuals, indicating that it did not represent a common polymorphic nucleotide substitution. This change, rs368625677 (dbSNP 138), had been described in 1/13,006 alleles in the NHLBI Exome Sequencing Project (http://eversusgs.washington.edu/EVS/). Of note, similar frequencies have been reported in the same database for recurrent RASopathy-causing mutations (e.g. c.922A>G in PTPN11, and c.1259G>A in CBL). Mutation analysis was extended to additional 408 patients with NS or a clinically related phenotype tested negative for mutations in the major NS disease genes (see Materials and Methods), allowing to identify one sporadic case heterozygous for a three-nucleotide duplication (c.116_118dup, p.Gly39dup) (Supplementary material, Fig. S2). Parental DNA sequencing of the relevant exon demonstrated the de novo origin of the variant, and STR genotyping confirmed paternity. In this subject, the duplication was documented in DNA obtained from skin fibroblasts, excluding a somatic event restricted to haematopoietic cells. The subject had features reminiscent of NS (Fig. 1A and Supplementary material, Table S2), with onset of AML suspected to represent a blast crisis of JMML (Supplementary material, Table S3 and Fig. S3). In this patient, exome sequencing performed on leukaemic and non-leukaemic DNA failed to disclose any additional relevant germline/somatic change affecting genes known to be mutated in RASopathies and JMML, as well as genes directly linked to the RAS signalling network, further supporting the causal role of the identified RRAS lesion. Based on this association, the occurrence of RRAS mutations was also explored in a panel of genomic DNAs obtained from bone marrow aspirates/circulating leukocytes of 110 subjects with JMML. Heterozygosity for the previously identified Gly39 duplication and the c.260A>T (p.Gln87Leu) change was observed in two patients with JMML rapidly progressing to AML (Supplementary material, Table S3 and Fig. S3). Both lesions were absent in non-leukaemic DNA, indicating their somatic origin (Supplementary material, Fig. S2). These subjects also carried a somatic NRAS mutation, suggesting that the two hits might cooperate with this particularly severe form of disease. Sequencing of isolated JMML myeloid colonies in patient 14385 showed that NRAS and RRAS mutations coexisted in the same progenitors but failed to establish their sequence of appearance during leukaemogenesis, not allowing to discriminate whether the latter was involved in initiation or progression of disease.
Figure 1.
RASopathy-causing and leukaemia-associated RRAS mutations. (A) Facial features of the affected subject (9802) heterozygous for the de novo germline c.116_118dup. (B) RRAS exon–intron arrangement with coding exons as blue boxes. RRAS functional motifs include the GTP/GDP binding domain (G1 to G5, starting from the N-terminus) (red), switch I (light green), switch II (dark green) and hypervariable region (light brown) with the C-terminal CAAX motif (dark brown). The unique N-terminal region is also shown (violet). Location of disease-associated mutations is reported. (C) Position of affected residues on the three-dimensional structure of RRAS in its GDP-bound, inactive state (PDB: 2FN4) (above) and that of non-hydrolysable GTP analogue (GppNHp)-bound, active HRAS (PDB: 5P21) (below). The red surface indicates Gly39 and Val40 (Gly13 and Val14, in HRAS), whereas Val55 (Val29) and Gln87 (Gln61) are shown in blue and green, respectively. GDP is reported as semi-transparent yellow surface.
Structural analyses
RRAS encodes a 23-kD a membrane-bound monomeric GTPase with 55–60% amino acid identity to RAS proteins (16). This highly conserved structure is flanked by a unique 26-amino acid region at the N-terminus (Fig. 1B). Similarly to the other RAS family proteins, RRAS binds to GTP and GDP with high affinity and specificity and functions as a molecular switch by cycling between active, GTP-bound and inactive, GDP-bound states (17). RRAS is activated by guanine nucleotide exchange factors (GEFs) in response to signals elicited by cell surface receptors. In the GTP-bound state, two functionally conserved regions, switch I and switch II (Fig. 1B), undergo a conformational change enabling RRAS to bind to and activate effector proteins. This interaction is terminated by hydrolysis of GTP to GDP, which is promoted by GTPase-activating proteins (GAPs) and results in switching towards the inactive conformation. Disease-associated RRAS mutations affected residues highly conserved among orthologs and paralogs (Supplementary material, Fig. S4) residing in the GTP-binding pocket (Fig. 1C) and were predicted to be damaging with high confidence (Supplementary material, Table S4). Among them, Gln87, homolog of Gln61 in RAS proteins, is directly involved in catalysis (18,19). The p.Gln87Leu substitution had previously been reported as a rare somatic event in lung carcinoma, and mutations affecting Gln61 are among the most recurrent oncogenic lesions in RAS genes (COSMIC database, http://cancer.sanger.ac.uk/cancergenome/projects/cosmic/). Likewise, p.Gly39dup altered the G1 motif participating in GTP/GDP binding and GTPase activity (Fig. 1B). Within this motif, Gly12 and Gly13 (Gly38 and Gly39 in RRAS) represent major mutation hot-spots in human cancer (COSMIC database) and account for the majority of germline HRAS mutations causing Costello syndrome (20). Of note, analogous insertions in RAS proteins have been reported in JMML and other malignancies (21–24). In contrast, no somatic/germline RAS mutation affecting Val29, homolog of Val55 in RRAS, had previously been reported. Val55 side-chain is not directly involved in GTP/GDP binding, GTP hydrolysis or interaction with effectors. However, it has been reported that H-bonds are possible between the backbone of Val29 in HRAS and GDP/GTP (25). Furthermore, it has been suggested that Val29 can play a role in the transition between the GDP- and GTP-bound states (26), as supported by the evidence that the Val29Gly substitution in HRAS accelerates the GDP/GTP exchange in vitro (27).
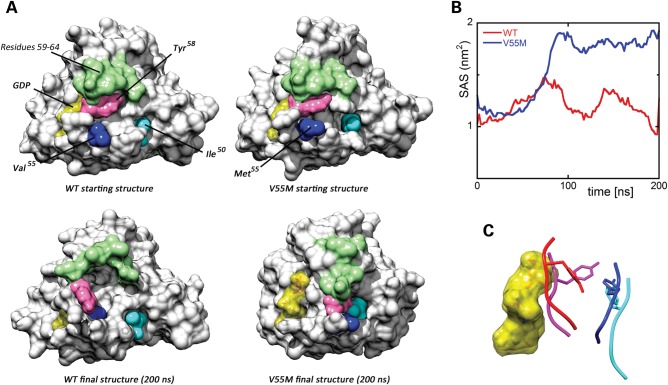
Molecular dynamics (MD) simulations were performed to predict in silico the effects of p.Val55Met on the structure and dynamics of RRAS (Fig. 2). The mutation was introduced in the available crystallographic structure of RRAS in complex with GDP and Mg2+, and the system was simulated in water for 200 ns. For comparison, MD simulations were also performed using the wild-type protein, which maintained a stable structure along the whole simulation, as expected (Fig. 2A, left panel). In contrast, a dramatic local structural transition extending up to the switch I region (residues 58–64), which mediates effector binding, was documented for the RRASV55M mutant, after ∼80 ns (Fig. 2A, right panel). This conformational transition resulted in an increased solvent exposure of Met55, in agreement with the higher hydrophilicity of this residue compared with Val, and was accompanied by the formation of a stable cluster involving residues Ile50, Met55 and Tyr58 (Fig. 2A and Supplementary material, Table S5) permitted by the unbranched and long side-chain of Met55. No further significant conformational changes were observed for the remaining interval of the simulation. The major effect of this structural rearrangement was to increase exposure of GDP to the solvent (Fig. 2B), with an almost doubled solvent accessible surface area of the nucleotide after the conformational transition. This structural rearrangement was accompanied by a perturbation of the intermolecular H-bond network stabilizing GDP binding, with loss of the H-bonds between residues at codons 55 and 56, and GDP (Supplementary material, Table S5). Of note, a possible impact of the described structural transition on RRAS binding to GEF proteins, which bind to this region and mediate GDP release, was also noticed. Specifically, we observed that after the conformational rearrangement, the RRASV55M region implicated in GEF binding populated a structure similar to that assumed in RAS/GEF complexes (Fig. 2C), suggesting a possible enhanced interaction of the disease-associated RRAS mutant with GEFs. In particular, Tyr58 was observed to adopt a side-chain orientation very similar to that of the RAS homolog Tyr32 in the HRAS/SOS1 complex, which has been shown to contribute to the structural rearrangements of switch I and interaction with GEFs (28–30).
Figure 2.
Molecular dynamics (MD) simulations. (A) Structural perturbations promoted by the p.Val55Met substitution as obtained from MD simulations of the RRAS/GDP complex. The wild-type (WT) protein is also shown for comparison. Top panels report the protein structures at the beginning of simulations, whereas the final structures (200 ns) are shown at the bottom. The final structure of RRASV55M is well representative of the last 120 ns of the trajectory. The protein surface of RRAS is shown with GDP (yellow). The mutated residues and those forming a cluster in the simulation of mutated RRAS are coloured as follows: Val55/Met55 (blue), Tyr58 (pink) and Ile50 (cyan). Residues 59–64, which, together with Tyr58, form the switch I region, are coloured in green. (B) Solvent accessible surface of GDP in the MD simulations of wild-type (red) and mutant (blue) RRAS/GDP complexes. (C) Conformation of the loop comprised between Val55/Met55 and Asp59 in wild-type (red) and mutant (blue) RRAS/GDP complexes obtained from MD simulations. GDP is reported as semi-transparent yellow surface. Superimposed conformations of the corresponding loop (residues 29–33) in GDP-bound HRAS (violet) (PDB: 4Q21) and GDP-bound HRAS complexed with SOS1 (cyan) (PDB: 1BKD) are shown for comparison. The side chains of Tyr58 and the corresponding residue in HRAS, Tyr32, are displayed as sticks.
Overall, these data supported an activating role of p.Val55Met through enhanced GDP release as a result of a decreased affinity for the nucleotide and/or enhanced interaction with a GEF.
Biochemical and functional characterization of RRAS mutants
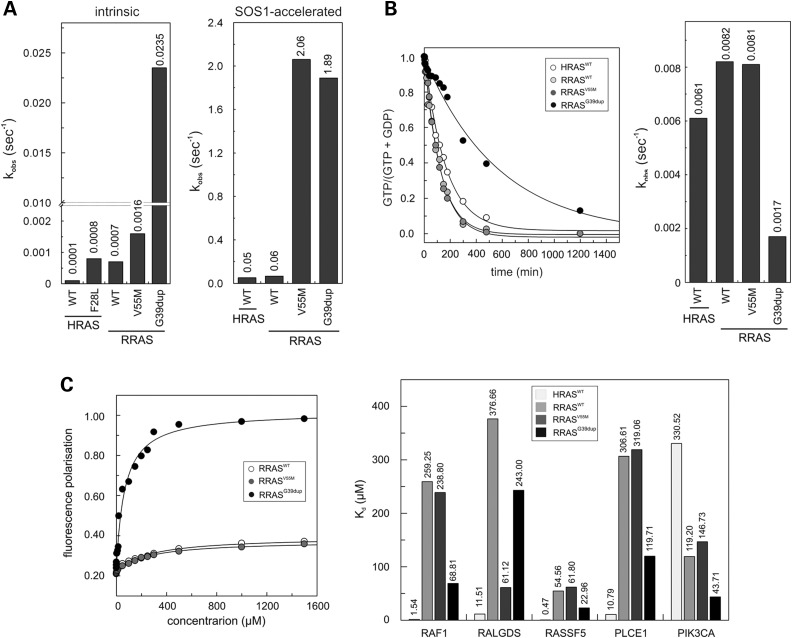
Previous studies documented the gain-of-function role of p.Gln87Leu on RRAS function, and MAPK and PI3K/AKT signalling (31). To characterize the impact of p.Val55Met and p.Gly39dup on protein function, we analysed the intrinsic and GEF-accelerated nucleotide exchange reaction of these mutants. Dissociation kinetics analysis demonstrated a dramatically increased intrinsic (RRASG39dup) and GEF-stimulated (RRASG39dup and RRASV55M) dissociation rate of mantGDP, indicating a facilitated nucleotide release in both mutants (Fig. 3A). RAS proteins exhibit low intrinsic GTPase activity that is enhanced by GAPs (32). Assessment of RRASG39dup and RRASV55M GTPase activity documented a significantly reduced intrinsic and GAP-stimulated GTP hydrolysis in the former (Fig. 3B and Supplementary material, Fig. S5). Finally, the interaction of RRAS proteins with various effectors was analysed by fluorescence polarization (Fig. 3C). While RRASWT was found to bind to RAF1, RALGDS, RASSF5 and PLCE1 less efficiently than HRAS, an increased binding affinity to PIK3CA was observed. Compared with RRASWT, aberrant binding behaviour of the two RRAS mutants was demonstrated, with RRASG39dup exhibiting an increased binding affinity towards PIK3CA, RAF1, PLCE1 and RASSF5, and RRASV55M to RALGDS.
Figure 3.
In vitro biochemical characterization of the RRASG39dup and RRASV55M mutants. (A) Intrinsic (left) and SOS1-accelerated (right) mantGDP nucleotide dissociation measured in the presence of 20-fold excess of non-labelled GDP. The decrease in mant fluorescence was fitted by single exponentials to obtain kobs values for nucleotide dissociation. RRASG39dup exhibited a 35-fold increased intrinsic dissociation of mantGDP, whereas SOS1-accelerated mantGDP dissociation was augmented by ∼30-fold for both mutants, compared with RRASWT and HRASWT. The Kobs values are an average of five to seven independent measurements. (B) Intrinsic GTP hydrolysis kinetics (left) and rate constants (right) of RRASG39dup and RRASV55M proteins, documenting the impaired catalytic activity in the former. The Kobs values are an average of five to seven independent measurements. (C) Binding of RRASWT, RRASV55M and RRASG39dup to the RAS-binding domain of RAF1 measured as variation in fluorescence polarization of each mantGppNHp-bound RRAS protein at increasing concentrations of RAF1-RBD (left), and dissociation constants (Kd) for the interaction of HRASWT and RRAS proteins to the RBDs of RAF1, RALGDS, PLCE1, PIK3CA and RASSF5 (right). Kd values were obtained by quadratic fitting of the concentration-dependent binding curves from fluorescence polarization measurement as exemplified for RAF1-RBD. Of note, RRASWT binds to RAF1, RALGDS, RASSF5 and PLCE1 less efficiently than HRAS, whereas an increased binding affinity to PIK3CA is observed.
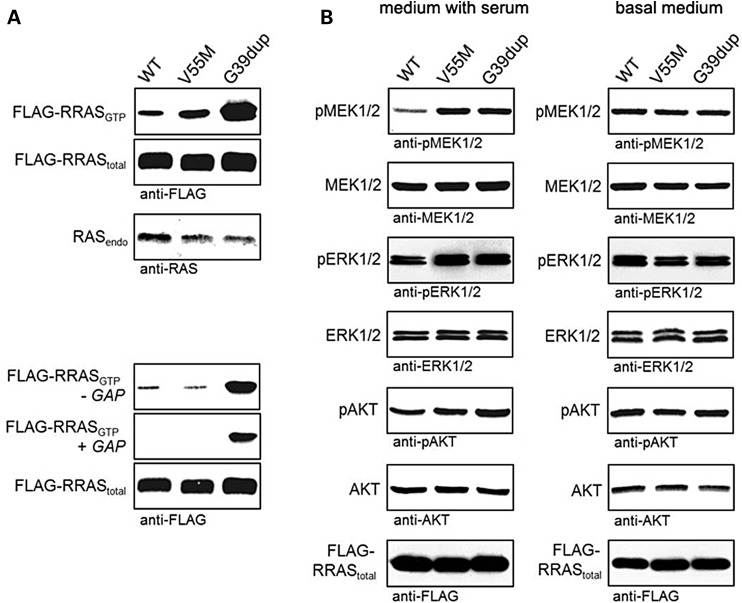
To gain further insights into the impact of disease-causing mutations on RRAS functional dysregulation and explore their effects on RAS signalling, the activation state of RRAS proteins and extent of signalling through the MAPK and PI3K/AKT cascades were evaluated using transient expression in COS-7 cells. Consistent with the above-mentioned findings, pull-down assays revealed a variably higher proportion of active, GTP-bound form for both mutants (Fig. 4A). Moreover, similarly to what observed under cell-free conditions, RRASG39dup was resistant to GAP stimulation. Expression of both mutants promoted enhanced serum-dependent MEK, ERK and AKT phosphorylation (Fig. 4B), which was more evident in cells expressing the RRASG39dup mutant.
Figure 4.
RRASG39dup and RRASV55M signalling activities in cells. (A) Determination of GTP-bound RRAS levels in COS-7 cells transiently expressing wild-type or mutant FLAG-tagged RRAS proteins. Assays were performed in the presence of serum (above), and in serum-free conditions (−GAP) or in the presence of the neurofibromin GAP domain (+GAP) (below). RRASG39dup was predominantly present in the active GTP-bound form and was resistant to GAP stimulation, whereas a slightly increased level of GTP-bound RRASV55M was observed in the presence of serum. Representative blots of three performed experiments are shown. (B) Determination of MEK, ERK and AKT phosphorylation levels (pMEK, pERK and pAKT) in transiently transfected COS-7 cells cultured in medium with serum (left) or basal medium (right). Expression of each RRAS mutant resulted in variably enhanced MEK, ERK and also partially AKT phosphorylation after stimulation. Total MEK, ERK and AKT in cell lysates are shown for equal protein expression and loading. Expression levels of exogenous, FLAG-tagged RRAS in cell lysates are shown for each experiment. Representative blots of three performed experiments are shown.
Caenorhabditis elegans studies
To explore further the functional impact of the RASopathy causative RRAS mutants on RAS signalling in vivo, we used the nematode C. elegans as an experimental model. In C. elegans, the role of ras-1, the RRAS ortholog (33), has not been characterized yet. On the contrary, proper signalling through LET-60, the C. elegans ortholog of RAS proteins, has been established to play a crucial role in vulval development (34). In particular, LET-60/RAS is known to mediate the priming signal (LIN-3/EGF) released by the anchor cell to induce the three nearby vulval precursor cells (VPCs), P5.p, P6.p and P7.p, to generate a normal vulva. Enhanced and decreased signalling through LET-60 and the MAPK cassette results in multiple ectopic pseudovulvae (multivulva phenotype) and a failure in VPC induction (vulvaless phenotype), respectively (34,35).
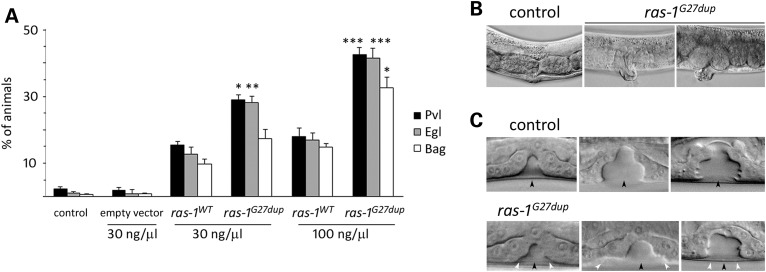
Multiple transgenic lines were generated to conditionally express the wild-type ras-1 cDNA (ras-1WT) or the allele homologous to the disease-associated three-nucleotide duplication (ras-1G27dup), which was identified to occur both as a germline and somatic event. Exogenous RAS-1 expression was induced by heat shock at early L3 larval stage to investigate the effects of the mutant protein on vulval development. Animals expressing ras-1G27dup displayed abnormal vulval morphogenesis resulting in the formation of a protruding vulva (Pvl) (Fig. 5A and B and Supplementary Material, Table S6), a phenotype associated with aberrant traffic through different signalling cascades (36,37). Of note, this phenotype had previously been reported in worms expressing the RASopathy causative SHOC2S2G mutant (38). Like those animals, ras-1G27dup worms showed decreased egg-laying efficiency (Egl phenotype), and accumulation of larvae inside the mother (Bag-of-worms phenotype). A significantly less penetrant phenotype was observed in animals expressing ras-1WT. These findings, together with the observation that animals lacking ras-1 do not exhibit any vulval defect (WormBase, http://www.wormbase.org/, and our personal assessment), supported the gain-of-function role of the mutation on RAS-1 function. At the late L3/early L4 larval stage, vulva morphogenesis normally begins with the descendants of VPC P6.p detaching from the cuticle and forming a symmetric invagination (Fig. 5C) (34). Animals in which the expression of ras-1WT had been induced at early L3 largely maintained this pattern (17/20). In contrast, in larvae expressing ras-1G27dup, descendants of VPCs P5.p and/or P7.p more frequently detached from the cuticle, resulting in larger and more asymmetric invaginations (10/30). This morphogenesis defect was the earliest detectable effect of the ras-1G27dup allele on vulval development, similarly to that previously documented in transgenic lines expressing SHOC2S2G (38).
Figure 5.
Consequences of ras-1G27dup expression on C. elegans vulval development. (A) Heat-shock-driven expression of ras-1WT and ras-1G27dup at early L3 stage results in protruding vulva (Pvl), egg laying defective (Egl) and bag-of-worms (Bag) phenotypes. Isogenic animals that had lost the transgene (control group) and worms expressing the heat shock-inducible vector (empty vector) were subjected to heat shock and scored in parallel for comparison. The dose at which the transgene has been injected is reported at the bottom. Error bars indicate SD of three independent experiments. Asterisks indicate significant differences compared with ras-1WT at the corresponding dose of injection (*P < 0.05; **P < 0.005; ***P < 0.0005; Fisher's Exact Test). (B) A proper vulva develops in heat-shocked control animals (left), whereas a protruding vulva is observed in heat-shocked ras-1G27dup young adults (middle) and adult worms (right). (C) Nomarski images of vulval precursor cells in late L3 (left), early L4 (middle) and mid-late L4 (right) stages from synchronized animals heat-shocked at early L3. In control animals (N = 48), only P6.p descendants invaginate (upper panel), whereas in 10 of 30 analysed ras-1G27dup-expressing worms, P5.p and/or P7.p descendants also detach from the cuticle, generating asymmetric invaginations (lower panel). Black arrowheads point to P6.p descendant invagination, whereas white arrowheads point to P5.p and P7.p descendant invagination. Anterior is to the left and dorsal is up, in all images.
Genetic interaction between the RAS-1/RRAS mutant and LET-60/RAS was also investigated. While expression of the RAS-1G27dup mutant was able to exacerbate the multivulva phenotype associated with a hyperactive let-60 allele (n1046), expression of wild-type RAS-1 failed to do so (Table 1). Similarly, a significant, although partial rescue of the VPC induction defect associated with a let-23/EGFR hypomorphic allele (sy1) was observed in animals expressing the activating RAS-1G27dup mutant, but not in worms expressing the wild-type counterpart (Table 1). Overall, these experiments provided evidence of a positive modulatory role of the RAS-1/RRAS mutant on LET-60/RAS signalling.
Table 1.
Vulva phenotypes in C. elegans mutant strains expressing wild-type RAS-1 or the disease-associated RAS-1G27dup mutant
| Genotype | Transgene | N | Muv (%) | Vul (%) | Pvl (%) | N | P6.p induction (%) |
|---|---|---|---|---|---|---|---|
| wild-type | none | 207 | 0 | 0 | 1.0 | 48 | 100 |
| let-60(n1046) | none | 201 | 77.9 | – | 0.5 | 50 | 100 |
| let-60(n1046) | ras-1wt | 244 | 76.4 | – | 2.8 | 43 | 100 |
| let-60(n1046) | ras-1G27dup | 231 | 87.1a | – | 3.0 | 50 | 100 |
| let-23(sy1) | none | 194 | – | 87.8 | 3.6 | 178 | 13.4 |
| let-23(sy1) | ras-1WT | 169 | – | 84.3 | 4.1 | 156 | 14.0 |
| let-23(sy1) | ras-1G27dup | 282 | – | 83.3 | 10.3b | 128 | 24.2c |
Strains: let-60(n1046) is a gain-of-function allele of let-60 (ortholog of the human HRAS, KRAS and NRAS genes); let-23(sy1) is a hypomorphic allele of let-23 (ortholog of the human EGFR gene). ras-1WT and ras-1G27dup indicate hsp-16.41::ras-1WT- and hsp-16.41::ras-1G27dup-containing constructs injected at 100 ng/μl, respectively. After each cross, isogenic worms that had lost the transgene were cloned separately and used as controls.
Animals were grown at 20°C and heat-shocked in parallel at early L3 stage. N indicates the number of animals scored. Multivulva (Muv), vulvaless (Vul) and protruding vulva (Pvl) phenotypes are expressed as percentage of worms with ectopic pseudovulvae, animals lacking a vulva and adults with a protruding vulva, respectively. Induction of vulval cell fate is expressed as percentage of P6.p that has been induced to invaginate.
In all comparisons, P-values were calculated using two-tailed Fisher's exact test.
aSignificantly different from let-60(n1046) (P < 0.02).
bSignificantly different from let-23(sy1) (P < 0.01) and let-23(sy1);ras-1WT (P < 0.02).
cSignificantly different from let-23(sy1) (P = 0.02) and let-23(sy1);ras-1WT (P < 0.05).
DISCUSSION
Mutations of genes coding for proteins with role in RAS signalling and the RAF/MEK/ERK cascade have been identified as the molecular cause underlying a group of clinically related developmental disorders, the RASopathies. Here, we used a gene candidacy approach based on large-scale protein–protein interaction/functional network analysis to identify RRAS as a novel gene implicated in a condition with features within the RASopathy spectrum. Disease-causing RRAS mutations are activating and act by maintaining the GTPase in its GTP-bound active state. Aberrant RRAS function was demonstrated to perturb variably intracellular signal flow through the RAF/MEK/ERK cascade, and to a certain extent also the PI3K/AKT pathway. Of note, these gain-of-function mutations are likely to define a novel leukaemia-prone condition. Consistent with this view, the same class of RRAS lesions was identified to occur as acquired somatic event in JMML, characterizing a subset of this myeloproliferative/myelodysplastic disorder with rapid progression to AML.
RRAS shares several biochemical properties with HRAS, NRAS and KRAS, as well as some common function, including stimulation of cell proliferation, survival and transformation (19,39). Despite these similarities, however, previous observations have emphasized the role of RRAS in cell adhesion, spreading and migration, and its modulatory function on effectors distinct from those used by ‘classical’ RAS proteins (40,41). While PI3K/AKT has been recognized as a major effector pathway of RRAS, only a minor impact on MAPK signalling had been reported (41,42). The present in vitro findings provide evidence that disease-associated RRAS mutants enhance the activation of the MAPK cascade, at least in response to specific stimuli. On the other hand, the identification of RRAS as a novel disease gene implicated in a RASopathy disorder further emphasizes the relevance of dysregulated signalling controlling cell spreading and migration in certain features of NS (e.g. congenital heart defects and lymphedema) and JMML (leukocyte infiltration in non-haematopoietic tissues) (43–45).
Caenorhabditis elegans studies provided evidence for a genetic interaction between the RAS-1G27dup/RRASG39dup and LET-60/RAS in vivo. Specifically, expression of the RAS-1 mutant protein was able to rescue, in part, the VPC induction defect resulting from a hypomorphic LET-23 mutant and enhanced the multivulva phenotype associated with a LET-60 gain-of-function genetic background. No impact of wild-type RAS-1/RRAS expression was observed in both models. We also observed that worms expressing ras-1G27dup displayed abnormal vulval morphogenesis (protruding vulva), possibly resulting from aberrant morphogenetic movements of the VPC descendant cells. Of note, we observed an equivalent phenotype in transgenic lines expressing SHOC2S2G (38) and a PTPN11/SHP2 gain-of-function mutant (our unpublished data), suggesting functional equivalence of these mutants. Genetic studies support the view that these vulva defects arise, in part, through perturbation of signalling mediated by the RHO-related GTPase, RAC, which plays a critical role in vulval morphogenesis (37). This finding is in line with the established role of RRAS on RAC signalling (40,41) and with preliminary data indicating enhanced migration and chemotactic capabilities in cells stably expressing the disease-associated RRAS mutants (our unpublished data).
The biochemical characterization of disease-associated RRAS mutations provided strong evidence for the existence of distinct structural and mechanistic effects resulting in an overall enhancement of RRAS signalling. Function of RAS family proteins in signal transduction is controlled by two events, the GDP/GTP exchange and GTP hydrolysis. Any perturbation of these processes can affect dramatically the fine-tuned balance of the GTPase interaction with effectors and signal output. The majority of gain-of-function mutations affecting RAS proteins, including those contributing to oncogenesis, trigger the accumulation of these GTPases in the active state by impairing intrinsic GTPase activity, and/or conferring resistance to GAPs (17). This is also the case of two of the three mutations identified in this study, p.Gly39dup and p.Gln87Leu, the latter corresponding to the p.Gln61Leu in RAS proteins (present study and ref. 18,19). The characterization of the biochemical behaviour of RRASG39dup, however, also demonstrated a dramatic increase in both the intrinsic and GEF-catalysed nucleotide exchange as a process contributing to the accumulation of this mutant in its GTP-bound state. Aberrant GEF-accelerated nucleotide exchange dynamics was identified as the event driving functional dysregulation in the RRASV55M mutant, which was documented to be hyper responsive to GEF stimulation, but retained stimulus-dependency. Remarkably, the RRASG39dup and RRASV55M mutants were demonstrated to exhibit a diverse binding behaviour to effectors suggesting a differential impact of mutations on downstream signalling cascades, including PI3K/AKT and RALGDS/RAL, whose biological significance and impact, however, require further studies.
The clinical phenotype of the two subjects with germline RRAS mutations was reminiscent of NS. The individual with the Gly39 duplication displayed pulmonic stenosis, reduced growth, café-au-lait spots, mild motor delay and low-set ears, which recur in NS (5). Facial features, however, were distinctive, and not typical of NS. In contrast, the patient heterozygous for the p.Val55Met substitution exhibited a very mitigated phenotype characterized by suggestive facial characteristics (triangular face, downslanting palpebral fissures and low-set ears), low posterior hairline, broad chest and borderline cognitive abilities, without cardiac involvement or defective growth, indicating that clinical features associated with RRAS mutations might be quite subtle. Of note, the milder phenotype associated with the p.Val55Met change is consistent with the weaker perturbing effect of the RRASV55M mutant on MAPK and PI3K/AKT signalling compared with the RRASG39dup protein. Additional information on the spectrum of germline RRAS mutations, their associated phenotype and their functional impact on signalling, however, is necessary to establish clinically relevant genotype–phenotype correlations.
JMML is a clonal myeloproliferative/myelodysplastic disorder of childhood characterized by overproduction of immature myeloid cells that variably retain the capacity to differentiate. Upregulation of RAS/MAPK signalling owing to germline and somatic mutations in PTPN11, NRAS, KRAS, NF1 and CBL is a major event implicated in this malignancy (13,46,47). Our data document that upregulated RRAS function represents a novel event contributing to JMML pathogenesis and/or disease progression. Notably, somatic RRAS mutations co-occurred with acquired NRAS lesions in atypical JMML characterized by late onset and rapid progression to AML. While JMML is generally an aggressive malignancy, a subset of NRAS/KRAS mutation-positive patients has been reported to exhibit a mild course, with spontaneous remission despite the RAS-mutated clone persisting for years (48–50, our unpublished data). This suggests that in some instances, certain NRAS mutations are not sufficient to support full leukaemogenesis, requiring synergism with a second RAS signalling targeting event. In line with this view, NRAS mutations have been documented to co-exist with defects in other RASopathy genes (e.g. PTPN11) in some cases resulting in a particularly aggressive disease resembling AML with myelodysplasia-related changes (51,52), as that observed in the present cases. Other studies, however, are required to appreciate more precisely the role of enhanced RRAS function in leukaemogenesis as well as its clinical relevance in haematological malignancies.
In conclusion, our findings document that germline activating mutations in RRAS underlie a condition within the RASopathy family that may resemble NS phenotypically. In the examined cohorts, RRAS lesions were found to account for only a small portion of cases, which might be related to their severe consequences on embryonic/foetal development and/or to the biased selection of the subjects included in this study. Based on the present findings, however, RRAS mutations are expected to be more common among subjects with clinical features only partially overlapping NS, and particularly in patients with syndromic JMML/AML not associated with mutations in the PTPN11, NF1, CBL, KRAS and NRAS genes. While further efforts are required to characterize more precisely the clinical impact of germline and somatic mutations affecting RRAS, our findings suggest an unpredicted role of this GTPase in development and haematopoiesis. Consistent with the recent identification of RIT1 as disease gene implicated in a significant proportion of NS (9), our findings further extend the concept of ‘RASopathy gene’ to a transducer whose dysregulated function perturbs signal flow through the MAPK cascade but does not belong to the core RAS/MAPK signalling cassette.
MATERIALS AND METHODS
Selection of RASopathy candidate genes
A web-based tool, Genes2FANs (http://actin.pharm.mssm.edu/genes2FANs), using a large-scale protein–protein interaction network coupled to a panel of functional association networks (FANs) was utilized to build a subnetwork connecting proteins to the known RASopathy genes (i.e. PTPN11, SOS1, NF1, SPRED1, CBL, NRAS, KRAS, HRAS, RAF1, BRAF, SHOC2, MAP2K1 and MAP2K2), as seed proteins. Gene Ontology (biological process tree), mammalian phenotype browser, and Connectivity Map (drug-associated gene expression signatures), ChEA and TRANSFAC (transcription factor networks) databases were selected to construct the functional subnetworks utilized for prioritization of candidates (15). The programme allows to calculate z-scores for intermediate nodes (i.e. candidates), which are ranked based on their connections to the seed proteins.
Subjects and mutation analysis
Three cohorts of patients were considered in the study. A first group including 96 subjects with clinical features within the RASopathy spectrum and without mutation in previously identified RASopathy genes (i.e. CBL, PTPN11, SOS1, KRAS, HRAS, NRAS, RAF1, BRAF, SHOC2, MAP2K1 and MAP2K2) was screened for a selected panel of candidates. A second cohort including 408 subjects with NS or a closely related phenotype previously tested negative for mutations in a heterogeneous subset of RASopathy genes was scanned for RRAS mutations. All subjects included in this cohort had been screened for mutations in PTPN11, SOS1 and RAF1 genes. In both cohorts, the clinical diagnosis was made on the basis of standardized clinical criteria assessed by experienced clinical geneticists and paediatricians. Clinical features for the majority of subjects satisfied diagnostic criteria reported for NS, LEOPARD syndrome and cardiofaciocutaneous syndrome (53–57), but individuals who lacked sufficient features for a definitive diagnosis were also included. RRAS mutation analysis was also carried out on a cohort including 110 subjects with non-syndromic JMML that had prospectively been collected and genotyped (58). Mutation screening was performed on the entire RRAS coding sequence and flanking intronic stretches (NC_000019.10, 49635295.. 49640143, complement; NM_006270.3; NP_006261.1) on genomic DNA extracted from circulating leukocytes (cohorts I, II and III) or bone marrow aspirates (cohort III) by denaturing high-performance liquid chromatography (DHPLC) (3100 or 3500HT WAVE DNA fragment analysis system, Transgenomic, Omaha, NE, USA) and/or direct bidirectional sequencing (ABI Prism 3130, 3730 and 3500 Genetic Analyzers; Applied Biosystems, Foster City, CA, USA). Primer pairs, PCR and DHPLC conditions are available upon request. dbSNP137 (http://www.ncbi.nlm.nih.gov/projects/SNP/snp_summary.cgi), HapMap (rel.27) (http://hapmap.ncbi.nlm.nih.gov/) and 1000 Genomes (http://www.1000genomes.org/) databases were used to annotate the identified sequence variants. SIFT (http://sift.jcvi.org/), PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/) and MutationTaster (http://www.mutationtaster.org/) were used to predict the functional impact of the identified variants. Paternity was confirmed by STR genotyping, using the PowerPlex 16 System (Promega). DNA from leukocytes, hair bulb cells, bone marrow aspirates and skin fibroblasts was extracted using standard protocols. DNA specimens were collected under Institutional Review Board-approved protocols. Informed consent for DNA storage and genetic analyses was obtained from all subjects. Permission was obtained to publish picture of patient 9802, whereas subject NS1166 denied consent for picture publication.
Exome sequencing
Targeted enrichment and massively parallel sequencing were performed on genomic DNA extracted from circulating leukocytes and fibroblasts of patient 9802. Exome capture was carried out using the SureSelect Human All Exon V4+UTRs (Agilent), and sequencing with a HiSeq2000 instrument (Illumina). Image analysis and base calling were performed using the Real Time Analysis (RTA) pipeline v. 1.14 (Illumina). Paired-end reads alignment to the reference human genome (UCSC GRCh37/hg19) and variant calling were carried out using the CASAVA v. 1.8 pipeline (Illumina). Variant annotation, SNP filtering (dbSNP135, 1000 Genomes, HapMap and IntegraGen Exome databases) and patient-matched germline variant filtering were attained using an in-house pipeline by IntegraGen (Evry, France).
Molecular dynamics analyses
Starting coordinates for MD simulations were obtained from the RRAS crystallographic structure in complex with GDP and Mg2+ (PDB: 2FN4; RCSB Protein Data Bank, http://www.rcsb.org/pdb/home/home.do). The N-terminus and C-terminus of RRAS, absent in the crystal, were not considered in simulations. Deep View software was used to add atoms missing in the PDB file, belonging to residues 31, 96, 114, 121 and 192 as well as to substitute residue 55 (simulation of the p.Val55Met mutant) (59). All MD simulations were performed with GROMACS 4.5 package, by using the GROMOS96 43a1 force field parameters for the protein. Parameters for GDP were taken from the GROMACS website (http://www.gromacs.org). Simulations were performed as previously described (60,61), except for some details. Briefly, the proteins were initially placed in a dodecahedral box, solvated with ∼6700 SPC water molecules, and six Na+ ions were added to neutralize the protein charge. Following initial energy minimization and a 100-ps MD run, during which the protein atoms were position restrained, the temperature of solute and solvent was raised from 50 to 300 K in a stepwise manner. 200-ns-long simulations were performed for wild-type RRAS and the RASopathy causative mutant. The particle mesh Ewald method was used to evaluate electrostatic interactions (62), whereas a cut-off scheme was employed for Van der Waals interactions. Temperature and pressure were kept constant at 300 K and 1 bar by using the Berendsen weak-coupling method (63), using separate temperature baths for protein–GDP complex and solvent, with a relaxation time of 0.1 ps for temperature and 1 ps for pressure. A time step of 2 fs was employed. Root mean square deviations were calculated according to standard definitions. UCSF Chimera (http://www.cgl.ucsf.edu/chimera/) was used for molecular graphics and structures superposition, by using the MatchMaker option.
Biochemical and functional characterization of RRAS mutants
The generation of constructs, and preparation and purification of proteins were as previously described (64). The intrinsic activities of the RAS proteins, their modulation by GEFs and GAPs and their interaction with different effector proteins were determined as described earlier (65,66). Dissociation of mantGDP from RAS proteins (0.1 µm) was measured in the presence of 20 µm GDP in 30 mm Tris–HCl, pH 7.5, 10 mm KH2PO4/K2HPO4, 5 mm MgCl2, 3 mm dithioerythritol (DTE), at 25 °C, using a Perkin Elmer fluorimeter at 366 nm (excitation wavelength) and 450 nm (emission wavelength). Observed rate constants (kobs) of dissociation were obtained by single exponential fitting of the data. GEF-accelerated mantGDP dissociation from RAS proteins (0.1 µm) was measured as mentioned earlier, in the presence of the catalytic domain of SOS1, Cdc25 (5 µm), using stopped-flow instrument.
The intrinsic GTPase reaction was performed by mixing 70 µm nucleotide-free RAS proteins (HRAS, RRASWT, RRASV55M and RRASG39dup) with 50 µm GTP in 30 mm Tris–HCl, pH 7.5, 10 mm KH2PO4/K2HPO4, 10 mm MgCl2, 3 mm DTE, at 25°C, using HPLC assay as previously described (67). Samples were taken at different time points and analysed by HPLC for their GDP and GTP contents to determine the relative GTP content [(GTP)/(GDP + GTP)]. Intrinsic GTP hydrolysis kobs of proteins were obtained by single exponential fitting of the data. For determination of GAP (neurofibromin, residues 1–333)-stimulated GTPase activity, GDP bound to HRAS and RRAS proteins was exchanged with excess mantGTP in the presence of alkaline phosphatase. Unbound nucleotides were removed by NAP5 column, and the RAS/mantGTP proteins were snap-frozen in liquid nitrogen (66). GAP-stimulated GTP hydrolysis of RAS proteins (0.2 µm) was measured in 30 mm Tris–HCl, pH 7.5, 10 mm MgCl2, 3 mm DTE at 25°C using a Hightech TgK Scientific stopped-flow instrument. Reactions measured the decrease in fluorescence owing to hydrolysis of mantGTP. This decay was fit by a single exponential.
Effector binding assays were performed in 30 mm Tris–HCl, pH 7.5, 100 mm NaCl, 5 mm MgCl2, 3 mm DTE at 25°C using a Fluoromax 4 fluorimeter in polarization mode. Increasing amounts of GST-tagged RAS-binding domains (RBD) of RAS effectors were titrated to 0.3 µm mantGppNHp-bound RAS proteins resulting in an increase of polarization (64). The dissociation constants (Kd) were calculated by fitting the concentration-dependent binding curve using a quadratic ligand binding equation.
For cell-based assays, COS-7 cells were transiently transfected with FLAG-tagged RRASWT, RRASV55M or RRASG39dup by the DEAE-dextran method. For serum conditions, cells were incubated for 48 h in 10% FCS. In serum-starved conditions, serum was changed to basal medium midway between the transfection and harvesting. Transfected COS-7 cells were harvested and lysed in fishing buffer [50 mm Tris–HCl, pH 7.5, 2 mm MgCl2, 100 mm NaCl, 1% IGEPAL CA-630, 10% glycerol, EDTA-free protease inhibitor cocktail (Roche, 1 tablet/50 ml buffer), 20 mm disodium β-glycerol phosphate and 1 mm Na3VO4]. Cleared cell lysates were incubated with GSH-beads loaded with GST-RAF1-RBD. GTP-bound proteins and total recombinant proteins were analysed by immunoblotting with anti-FLAG antibody. Antibodies against MEK1/2, ERK1/2, AKT, phospho-MEK1/2 (Ser217/221), phospho-ERK1/2 (Thr202/Tyr204) and phospho-AKT (Thr308) were purchased from Cell Signaling Technology (68).
Caenorhabditis elegans studies
Culture and maintenance of animals were as previously described (69). The let-60(n1046) (let-60/RAS gain-of-function allele) and let-23(sy1) (let-23/EGFR hypomorphic allele) strains were provided by the Caenorhabditis Genetics Center (University of Minnesota). The three-nucleotide insertion, c.81_82insGGC (ras-1G27dup), corresponding to c.116_118dup in RRAS, was introduced in the wild-type cDNA (ras-1WT) (C. elegans ORF clone AAB03320, Thermo Scientific) by site-directed mutagenesis (QuikChange Site-Directed Mutagenesis Kit, Stratagene). ras-1 cDNAs were subcloned into the pPD49.83 heat shock-inducible vector (a gift of A. Fire, Stanford University School of Medicine). Germline transformation was performed as described (70). pJM371 plasmid [pelt-2::NLS::RFP] (a gift from J.D. McGhee, University of Calgary), which drives red fluorescent protein (RFP) expression in intestinal cell nuclei, was used as co-injection marker (30 ng/μl). Two different doses of constructs were injected (30 and 100 ng/μl). Animals from at least three independent transgenic lines for each construct and each dose of injection (i.e. six lines expressing ras-1WT and six lines expressing ras-1G27dup) were heat-shocked in parallel and scored blindly at a Leica MZ10F dissecting microscope to check for the presence of protruding vulvae (Pvl phenotype) and multiple ectopic pseudovulvae (Muv phenotype), count the number of eggs retained in the uterus (Egl phenotype) and identify animals that had become bag-of-worms (Bag phenotype). Isogenic worms that had lost the transgene were cloned separately and used as controls. Following heat shock, all the transgenic lines expressing ras-1WT or ras-1G27dup showed a variable degree of these phenotypes. Lines gbEx555a[hsp-16.41::ras-1WT;pelt-2::NLS::RFP] and gbEx557a[hsp-16.41::ras-1G27dup;pelt-2::NLS::RFP] were scored quantitatively in triplicate experiments at the compound microscope and used for further analyses. Genetic crosses were performed according to standard methods (69). The genotype of individual alleles was confirmed by direct sequencing of the appropriate genomic region. After each cross, isogenic worms that had lost the transgene were used as controls.
To investigate VPCs induction and vulva morphogenesis, synchronized hermaphrodites carrying each transgene and the corresponding isogenic controls were heat-shocked in parallel at early L3 stage (33°C, 1 h, followed by 30°C, 1 h). Animals were scored at the compound microscope for vulval induction at late L3 and L4 stages, and for Pvl/Egl/Bag phenotypes at the adult stage. Microscopy observations were performed with a Nikon Eclipse 80i instrument equipped with Nomarski differential interference contrast optics on live animals mounted on 2% agarose pads containing 10 mm sodium azide as anaesthetic.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by grants from the ERA-Net for research programmes on rare diseases 2009 (NSEuroNet to M.Z., H.C., M.R.A. and M.T.), Telethon-Italy (GGP10020 and GGP13107 to M.T.), AIRC (IG 13360 to M.T.), NGFNplus program of the German Ministry of Science and Education (01GS08100 to M.R.A.), German Research Foundation through the Collaborative Research Center 974 (Communication and Systems Relevance during Liver Injury and Regeneration to M.R.A.) and NIH (HL071207 to B.D.G.). F.P. was recipient of a research fellowship from ‘Associazione Italiana Sindromi di Costello e cardiofaciocutanea’. Funding to pay the Open Access publication charges for this article was provided by Telethon-Italy.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to the participating patients and their families. We thank Serenella Venanzi (Istituto Superiore di Sanità, Rome, Italy), Michela Bonaguro (Policlinico S.Orsola-Malpighi, Bologna, Italy), Federica Consoli (Istituto Mendel, Rome, Italy) and Cédric Vignal and Sabrina Pereira (Hôpital Robert Debré, Paris, France) for skilful technical assistance, and the Open Laboratory (IGB-CNR, Naples, Italy) for experimental support. We also thank Paolo Bazzicalupo (IGB-CNR) for critical reading of the manuscript, paediatricians from the Société Française des Cancers de l'Enfant (SFCE) for providing biological material from their patients and CINECA for computational resources. Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center (University of Minnesota, Minneapolis, MN, USA) funded by the NIH Office of Research Infrastructure Programs (P40OD010440).
Conflict of Interest statement. None declared.
REFERENCES
- 1.Mitin N., Rossman K.L., Der C.J. Signaling interplay in Ras superfamily function. Curr. Biol. 2005;15:R563–R574. doi: 10.1016/j.cub.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 2.Mendoza M.C., Er E.E., Blenis J. The Ras-ERK and PI3K-mTOR pathways: cross-talk and compensation. Trends Biochem. Sci. 2011;36:320–328. doi: 10.1016/j.tibs.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harris T.J.R., McCormick F. The molecular pathology of cancer. Nat. Rev. Clin. Oncol. 2010;7:251–265. doi: 10.1038/nrclinonc.2010.41. [DOI] [PubMed] [Google Scholar]
- 4.Pylayeva-Gupta Y., Grabocka E., Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat. Rev. Cancer. 2011;11:761–774. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roberts A.E., Allanson J.E., Tartaglia M., Gelb B.D. Noonan syndrome. Lancet. 2013;381:333–342. doi: 10.1016/S0140-6736(12)61023-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schubbert S., Shannon K., Bollag G. Hyperactive Ras in developmental disorders and cancer. Nat. Rev. Cancer. 2007;7:295–308. doi: 10.1038/nrc2109. [DOI] [PubMed] [Google Scholar]
- 7.Tartaglia M., Gelb B.D. Disorders of dysregulated signal traffic through the RAS-MAPK pathway: phenotypic spectrum and molecular mechanisms. Ann. N. Y. Acad. Sci. 2010;1214:99–121. doi: 10.1111/j.1749-6632.2010.05790.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rauen K.A. The RASopathies. Annu. Rev. Genomics Hum. Genet. 2013;14:355–369. doi: 10.1146/annurev-genom-091212-153523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aoki Y., Niihori T., Banjo T., Okamoto N., Mizuno S., Kurosawa K., Ogata T., Takada F., Yano M., Ando T., et al. Gain-of-function mutations in RIT1 cause Noonan syndrome, a RAS/MAPK pathway syndrome. Am. J. Hum. Genet. 2013;93:173–180. doi: 10.1016/j.ajhg.2013.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tartaglia M., Gelb B.D., Zenker M. Noonan syndrome and clinically related disorders. Best Pract. Res. Clin. Endocrinol. Metab. 2011;25:161–179. doi: 10.1016/j.beem.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kratz C.P., Rapisuwon S., Reed H., Hasle H., Rosenberg P.S. Cancer in Noonan, Costello, cardiofaciocutaneous and LEOPARD syndromes. Am. J. Med. Genet. C Semin. Med. Genet. 2011;157C:83–89. doi: 10.1002/ajmg.c.30300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gripp K.W. Tumor predisposition in Costello syndrome. Am. J. Med. Genet. C Semin. Med. Genet. 2005;137C:72–77. doi: 10.1002/ajmg.c.30065. [DOI] [PubMed] [Google Scholar]
- 13.Loh M.L. Recent advances in the pathogenesis and treatment of juvenile myelomonocytic leukaemia. Br. J. Haematol. 2011;152:677–687. doi: 10.1111/j.1365-2141.2010.08525.x. [DOI] [PubMed] [Google Scholar]
- 14.McKay M.M., Morrison D.K. Integrating signals from RTKs to ERK/MAPK. Oncogene. 2007;26:3113–3121. doi: 10.1038/sj.onc.1210394. [DOI] [PubMed] [Google Scholar]
- 15.Dannenfelser R., Clark N.R., Ma'ayan A. Genes2FANs: connecting genes through functional association networks. BMC Bioinformatics. 2012;13:156. doi: 10.1186/1471-2105-13-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lowe D.G., Capon D.J., Delwart E., Sakaguchi A.Y., Naylor S.L., Goeddel D.V. Structure of the human and murine R-ras genes, novel genes closely related to ras proto-oncogenes. Cell. 1987;48:137–146. doi: 10.1016/0092-8674(87)90364-3. [DOI] [PubMed] [Google Scholar]
- 17.Wennerberg K., Rossman K.L., Der C.J. The Ras superfamily at a glance. J. Cell Sci. 2005;118:843–846. doi: 10.1242/jcs.01660. [DOI] [PubMed] [Google Scholar]
- 18.Krengel U., Schlichting I., Scherer A., Schumann R., Frech M., John J., Kabsch W., Pai E.F., Wittinghofer A. Three-dimensional structures of H-ras p21 mutants: molecular basis for their inability to function as signal switch molecules. Cell. 1990;62:539–548. doi: 10.1016/0092-8674(90)90018-a. [DOI] [PubMed] [Google Scholar]
- 19.Saez R., Chan A.M., Miki T., Aaronson S.A. Oncogenic activation of human R-ras by point mutations analogous to those of prototype H-ras oncogenes. Oncogene. 1994;9:2977–2982. [PubMed] [Google Scholar]
- 20.Aoki Y., Niihori T., Kawame H., Kurosawa K., Ohashi H., Tanaka Y., Filocamo M., Kato K., Suzuki Y., Kure S., et al. Germline mutations in HRAS proto-oncogene cause Costello syndrome. Nat. Genet. 2005;37:1038–1040. doi: 10.1038/ng1641. [DOI] [PubMed] [Google Scholar]
- 21.Bollag G., Adler F., elMasry N., McCabe P.C., Conner E., Jr, Thompson P., McCormick F., Shannon K. Biochemical characterization of a novel KRAS insertion mutation from a human leukemia. J. Biol. Chem. 1996;271:32491–32494. doi: 10.1074/jbc.271.51.32491. [DOI] [PubMed] [Google Scholar]
- 22.Reimann C., Arola M., Bierings M., Karow A., van den Heuvel-Eibrink M.M., Hasle H., Niemeyer C.M., Kratz C.P. A novel somatic K-Ras mutation in juvenile myelomonocytic leukemia. Leukaemia. 2006;20:1637–1638. doi: 10.1038/sj.leu.2404303. [DOI] [PubMed] [Google Scholar]
- 23.Murugan A.K., Hong N.T., Cuc T.T., Hung N.C., Munirajan A.K., Ikeda M.A., Tsuchida N. Detection of two novel mutations and relatively high incidence of H-RAS mutations in Vietnamese oral cancer. Oral. Oncol. 2009;45:e161–e166. doi: 10.1016/j.oraloncology.2009.05.638. [DOI] [PubMed] [Google Scholar]
- 24.Sartori G., Cavazza A., Sgambato A., Marchioni A., Barbieri F., Longo L., Bavieri M., Murer B., Meschiari E., Tamberi S., et al. EGFR and K-ras mutations along the spectrum of pulmonary epithelial tumors of the lung and elaboration of a combined clinicopathologic and molecular scoring system to predict clinical responsiveness to EGFR inhibitors. Am. J. Clin. Pathol. 2009;131:478–489. doi: 10.1309/AJCPH0TRMPXVZW2F. [DOI] [PubMed] [Google Scholar]
- 25.Milburn M.V., Tong L., deVos A.M., Brünger A., Yamaizumi Z., Nishimura S., Kim S.-H. Molecular switch for signal transduction: structural differences between active and inactive forms of protooncogenic ras proteins. Science. 1990;247:939–945. doi: 10.1126/science.2406906. [DOI] [PubMed] [Google Scholar]
- 26.Diaz J.F., Wroblowski B., Schlitter J., Engelborghs Y. Calculation of pathways for the conformational transition between the GTP- and GDP-bound states of the Ha-ras-p21 protein: calculations with explicit solvent simulations and comparison with calculations in vacuum. Proteins. 1997;28:434–451. [PubMed] [Google Scholar]
- 27.Kuppens S., Díaz J.F., Engelborghs Y. Characterization of the hinges of the effector loop in the reaction pathway of the activation of ras-proteins. Kinetics of binding of beryllium trifluoride to V29G and I36G mutants of Ha-ras-p21. Prot. Sci. 1999;8:1860–1866. doi: 10.1110/ps.8.9.1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma J., Karplus M. Molecular switch in signal transduction: reaction paths of the conformational changes in ras p21. Proc. Natl. Acad. Sci. USA. 1997;94:11905–11910. doi: 10.1073/pnas.94.22.11905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gorfe A.A., Grant B.J., McCammon J.A. Mapping the nucleotide and isoform-dependent structural and dynamical features of Ras proteins. Structure. 2008;16:885–896. doi: 10.1016/j.str.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hall B.E., Yang S.S., Boriack-Sjodin P.A., Kuriyan J., Bar-Sagi D. Structure-based mutagenesis reveals distinct functions for Ras switch 1 and switch 2 in Sos-catalyzed guanine nucleotide exchange. J. Biol. Chem. 2001;276:27629–27637. doi: 10.1074/jbc.M101727200. [DOI] [PubMed] [Google Scholar]
- 31.Suzuki J., Kaziro Y., Koide H. An activated mutant of R-Ras inhibits cell death caused by cytokine deprivation in BaF3 cells in the presence of IGF-I. Oncogene. 1997;15:1689–1697. doi: 10.1038/sj.onc.1201333. [DOI] [PubMed] [Google Scholar]
- 32.Scheffzek K., Ahmadian M.R., Kabsch W., Wiesmüller L., Lautwein A., Schmitz F., Wittinghofer A. The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science. 1997;277:333–338. doi: 10.1126/science.277.5324.333. [DOI] [PubMed] [Google Scholar]
- 33.Lundquist E.A. 2006. Small GTPases. In WormBook . The C. elegans Research Community, WormBook (ed.), http://www.wormbook.org . . [DOI]
- 34.Sternberg P.W. 2005. Vulval development. In Wormbook . The C. elegans Research Community (ed.), http://www.wormbook.org. . [DOI]
- 35.Sundaram M.V. 2006. RTK/Ras/MAPK signaling. In Wormbook . The C. elegans Research Community (ed.), http://www.wormbook.org . . [DOI]
- 36.Eisenmann D.M., Kim S.K. Protruding vulva mutants identify novel loci and Wnt signaling factors that function during Caenorhabditis elegans vulva development. Genetics. 2000;156:1097–1116. doi: 10.1093/genetics/156.3.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kishore R.S., Sundaram M.V. ced-10 Rac and mig-2 function redundantly and act with unc-73 trio to control the orientation of vulval cell divisions and migrations in Caenorhabditis elegans. Dev. Biol. 2002;241:339–348. doi: 10.1006/dbio.2001.0513. [DOI] [PubMed] [Google Scholar]
- 38.Cordeddu V., Di Schiavi E., Pennacchio L.A., Ma'ayan A., Sarkozy A., Fodale V., Cecchetti S., Cardinale A., Martin J., Schackwitz W., et al. Mutation of SHOC2 promotes aberrant protein N-myristoylation and causes Noonan-like syndrome with loose anagen hair. Nat. Genet. 2009;41:1022–1026. doi: 10.1038/ng.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cox A.D., Brtva T.R., Lowe D.G., Der C.J. R-Ras induces malignant, but not morphologic, transformation of NIH3T3 cells. Oncogene. 1994;9:3281–3288. [PubMed] [Google Scholar]
- 40.Wozniak M.A., Kwong L., Chodniewicz D., Klemke R.L., Keely P.J. R-Ras controls membrane protrusion and cell migration through the spatial regulation of Rac and Rho. Mol. Biol. Cell. 2005;16:84–96. doi: 10.1091/mbc.E04-04-0277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Osada M., Tolkacheva T., Li W., Chan T.O., Tsichlis P.N., Saez R., Kimmelman A.C., Chan A.M. Differential roles of Akt, Rac, and Ral in R-Ras-mediated cellular transformation, adhesion, and survival. Mol. Cell. Biol. 1999;19:6333–6344. doi: 10.1128/mcb.19.9.6333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marte B.M., Rodriguez-Viciana P., Wennström S., Warne P.H., Downward J. R-Ras can activate the phosphoinositide 3-kinase but not the MAP kinase arm of the Ras effector pathways. Curr. Biol. 1997;7:63–70. doi: 10.1016/s0960-9822(06)00028-5. [DOI] [PubMed] [Google Scholar]
- 43.Jopling C., van Geemen D., den Hertog J. Shp2 knockdown and Noonan/LEOPARD mutant Shp2-induced gastrulation defects. PLoS Genet. 2007;3:e225. doi: 10.1371/journal.pgen.0030225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang S., Yu W.M., Zhang W., McCrae K.R., Neel B.G., Qu C.K. Noonan syndrome/leukemia-associated gain-of-function mutations in SHP-2 phosphatase (PTPN11) enhance cell migration and angiogenesis. J. Biol. Chem. 2009;284:913–920. doi: 10.1074/jbc.M804129200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen P.C., Wakimoto H., Conner D., Araki T., Yuan T., Roberts A., Seidman C., Bronson R., Neel B., Seidman J.G., et al. Activation of multiple signalling pathways causes developmental defects in mice with a Noonan syndrome-associated Sos1 mutation. J. Clin. Invest. 2010;120:4353–4365. doi: 10.1172/JCI43910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Emanuel P.D. Juvenile myelomonocytic leukemia and chronic myelomonocytic leukemia. Leukemia. 2008;22:1335–1342. doi: 10.1038/leu.2008.162. [DOI] [PubMed] [Google Scholar]
- 47.Niemeyer C.M., Kratz C.P. Paediatric myelodysplastic syndromes and juvenile myelomonocytic leukaemia: molecular classification and treatment options. Br. J. Haematol. 2008;140:610–624. doi: 10.1111/j.1365-2141.2007.06958.x. [DOI] [PubMed] [Google Scholar]
- 48.Matsuda K., Shimada A., Yoshida N., Ogawa A., Watanabe A., Yajima S., Iizuka S., Koike K., Yanai F., Kawasaki K., et al. Spontaneous improvement of hematologic abnormalities in patients having juvenile myelomonocytic leukemia with specific RAS mutations. Blood. 2007;109:5477–5480. doi: 10.1182/blood-2006-09-046649. [DOI] [PubMed] [Google Scholar]
- 49.Flotho C., Kratz C.P., Bergsträsser E., Hasle H., Starý J., Trebo M., van den Heuvel-Eibrink M.M., Wójcik D., Zecca M., Locatelli F., et al. Genotype-phenotype correlation in cases of juvenile myelomonocytic leukemia with clonal RAS mutations. Blood. 2008;111:966–967. doi: 10.1182/blood-2007-09-111831. [DOI] [PubMed] [Google Scholar]
- 50.Takagi M., Piao J., Lin L., Kawaguchi H., Imai C., Ogawa A., Watanabe A., Akiyama K., Kobayashi C., Mori M., et al. Autoimmunity and persistent RAS-mutated clones long after the spontaneous regression of JMML. Leukaemia. 2013;27:1926–1928. doi: 10.1038/leu.2013.82. [DOI] [PubMed] [Google Scholar]
- 51.Park H.D., Lee S.H., Sung K.W., Koo H.H., Jung N.G., Cho B., Kim H.K., Park I.A., Lee K.O., Ki C.S., et al. Gene mutations in the Ras pathway and the prognostic implication in Korean patients with juvenile myelomonocytic leukemia. Ann. Hematol. 2012;91:511–517. doi: 10.1007/s00277-011-1326-9. [DOI] [PubMed] [Google Scholar]
- 52.Sakaguchi H., Okuno Y., Muramatsu H., Yoshida K., Shiraishi Y., Takahashi M., Kon A., Sanada M., Chiba K., Tanaka H., et al. Exome sequencing identifies secondary mutations of SETBP1 and JAK3 in juvenile myelomonocytic leukemia. Nat. Genet. 2013;45:937–941. doi: 10.1038/ng.2698. [DOI] [PubMed] [Google Scholar]
- 53.Van der Burgt I., Berends E., Lommen E., van Beersum S., Hamel B., Mariman E. Clinical and molecular studies in a large Dutch family with Noonan syndrome. Am. J. Med. Genet. 1994;53:187–191. doi: 10.1002/ajmg.1320530213. [DOI] [PubMed] [Google Scholar]
- 54.Allanson J.E. Noonan syndrome. J. Med. Genet. 1987;24:9–13. doi: 10.1136/jmg.24.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sarkozy A., Digilio M.C., Dallapiccola B. LEOPARD syndrome. Orphanet J. Rare Dis. 2008;3:13. doi: 10.1186/1750-1172-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Voron D.A., Hatfield H.H., Kalkhoff R.K. Multiple lentigines syndrome: case report and review of the literature. Am. J. Med. 1976;60:447–456. doi: 10.1016/0002-9343(76)90764-6. [DOI] [PubMed] [Google Scholar]
- 57.Roberts A., Allanson J., Jadico S.K., Kavamura M.I., Noonan J., Opitz J.M., Young T., Neri G. The cardiofaciocutaneous syndrome. J. Med. Genet. 2006;43:833–842. doi: 10.1136/jmg.2006.042796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pérez B., Kosmider O., Cassinat B., Renneville A., Lachenaud J., Kaltenbach S., Bertrand Y., Baruchel A., Chomienne C., Fontenay M., et al. Genetic typing of CBL, ASXL1, RUNX1, TET2 and JAK2 in juvenile myelomonocytic leukaemia reveals a genetic profile distinct from chronic myelomonocytic leukaemia. Br. J. Haematol. 2010;151:460–468. doi: 10.1111/j.1365-2141.2010.08393.x. [DOI] [PubMed] [Google Scholar]
- 59.Guex N., Peitsch M.C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 60.Martinelli S., Torreri P., Tinti M., Stella L., Bocchinfuso G., Flex E., Grottesi A., Ceccarini M., Palleschi A., Cesareni G., et al. Diverse driving forces underlie the invariant occurrence of the T42A, E139D, I282V and T468M SHP2 amino acid substitutions causing Noonan and LEOPARD syndromes. Hum. Mol. Genet. 2008;17:2018–2029. doi: 10.1093/hmg/ddn099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bocchinfuso G., Stella L., Martinelli S., Flex E., Carta C., Pantaleoni F., Pispisa B., Venanzi M., Tartaglia M., Palleschi A. Structural and functional effects of disease-causing amino acid substitutions affecting residues Ala72 and Glu76 of the protein tyrosine phosphatase SHP-2. Proteins. 2007;66:963–974. doi: 10.1002/prot.21050. [DOI] [PubMed] [Google Scholar]
- 62.Darden T., York D., Pedersen L. Particle mesh Ewald: an N · log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 63.Berendsen H.J.C., Postma J.P.M., van Gunsteren W.F., Di Nola A., Haak J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 64.Gremer L., Merbitz-Zahradnik T., Dvorsky R., Cirstea I.C., Kratz C.P., Zenker M., Wittinghofer A., Ahmadian M.R. Germline KRAS mutations cause aberrant biochemical and physical properties leading to developmental disorders. Hum. Mutat. 2011;32:33–43. doi: 10.1002/humu.21377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hemsath L., Ahmadian M.R. Fluorescence approaches for monitoring interactions of Rho GTPases with nucleotides, regulators, and effectors. Methods. 2005;37:173–182. doi: 10.1016/j.ymeth.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 66.Jaiswal M., Dubey B.N., Koessmeier K.T., Gremer L., Ahmadian M.R. Biochemical assays to characterise Rho GTPases. Methods Mol. Biol. 2012;827:37–58. doi: 10.1007/978-1-61779-442-1_3. [DOI] [PubMed] [Google Scholar]
- 67.Eberth A., Ahmadian M.R. In vitro GEF and GAP assays. Curr. Protoc. Cell Biol. 2009;43:14.9.1–14.9.25. doi: 10.1002/0471143030.cb1409s43. [DOI] [PubMed] [Google Scholar]
- 68.Cirstea I.C., Gremer L., Dvorsky R., Zhang S.C., Piekorz R.P., Zenker M., Ahmadian M.R. Diverging gain-of-function mechanisms of two novel KRAS mutations associated with Noonan and cardio-facio-cutaneous syndromes. Hum. Mol. Genet. 2013;22:262–270. doi: 10.1093/hmg/dds426. [DOI] [PubMed] [Google Scholar]
- 69.Sulston J.E., Hodgkin J. The Community of C. elegans Researchers. Methods. In: Wood W.B., editor. The Nematode Caenorhabditis Elegans. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. pp. 587–606. [Google Scholar]
- 70.Mello C.C., Kramer J.M., Stinchcomb D., Ambros V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.