Abstract
Cerebral cavernous malformations (CCMs) are vascular lesions affecting the central nervous system. CCM occurs either sporadically or in an inherited, autosomal dominant manner. Constitutional (germline) mutations in any of three genes, KRIT1, CCM2 and PDCD10, can cause the inherited form. Analysis of CCM lesions from inherited cases revealed biallelic somatic mutations, indicating that CCM follows a Knudsonian two-hit mutation mechanism. It is still unknown, however, if the sporadic cases of CCM also follow this genetic mechanism. We extracted DNA from 11 surgically excised lesions from sporadic CCM patients, and sequenced the three CCM genes in each specimen using a next-generation sequencing approach. Four sporadic CCM lesion samples (36%) were found to contain novel somatic mutations. Three of the lesions contained a single somatic mutation, and one lesion contained two biallelic somatic mutations. Herein, we also describe evidence of somatic mosaicism in a patient presenting with over 130 CCM lesions localized to one hemisphere of the brain. Finally, in a lesion regrowth sample, we found that the regrown CCM lesion contained the same somatic mutation as the original lesion. Together, these data bolster the idea that all forms of CCM have a genetic underpinning of the two-hit mutation mechanism in the known CCM genes. Recent studies have found aberrant Rho kinase activation in inherited CCM pathogenesis, and we present evidence that this pathway is activated in sporadic CCM patients. These results suggest that all CCM patients, including those with the more common sporadic form, are potentially amenable to the same therapy.
INTRODUCTION
Cerebral cavernous malformations (CCMs, OMIM #116860, 603284, 603285) are vascular lesions affecting the central nervous system. CCM lesions consist of grossly dilated, capillary-like structures lined by endothelium and lacking mature vascular wall structure. Lesions can be found in ∼0.5% of the general population (1–4) and affected individuals have a lifetime risk of focal neurological deficits and hemorrhagic stroke. Sporadic lesions are typically solitary, whereas familial CCM is characterized by multifocal cerebral lesions in the setting of autosomal dominant mutations in one of three genes, KRIT1 (CCM1), CCM2 and PDCD10 (CCM3) (5–9).
A Knudson two-hit mutation mechanism has been suggested in the familial forms of CCM, where both copies of a particular gene must be mutated in order for disease to occur (10). Lesions in familial cases would result from an inherited, constitutional (germline) mutation and biallelic somatic mutations. This mechanism would also suggest that lesions in sporadic cases may result from two independent, biallelic, somatic mutations in CCM genes occurring in the same cell.
Our laboratory and others have identified biallelic somatic mutations in lesions from familial cases (11–13), with one mutation being the inherited constitutional mutation, and the other occurring somatically in the wild-type (WT) copy of the corresponding gene in the vascular lesional cells. These data demonstrated that the familial forms can be caused by a Knudsonian, two-hit mutation mechanism. However, although these studies focused on the inherited form of CCM, the majority of CCM patients are sporadic cases (14). Based on the two-hit genetic mechanism, we hypothesize that sporadic CCM lesions are caused by two independent, biallelic, somatic mutations in one of the known CCM genes. We investigated this hypothesis through next-generation sequencing (NGS) of the three CCM genes using genomic DNA isolated from the lesions of sporadic CCM patients.
Previous studies on somatic mutations in CCM lesions focused on single nucleotide changes or relatively small indels (11–13). Mutant alleles in these samples were present at frequencies between 5 and 20%. If a single mutant cell became a CCM lesion by clonal expansion (such as that seen in cancerous tumors), then the frequency of mutant alleles within the samples would be expected to approach 50%. The lower than expected values observed for somatic mutations in CCM lesions are in part because of the cellular heterogeneity of the cerebrovascular lesion. However, the somatic mutation frequency remains well below 50% even in laser capture-microdissected lesion endothelial cells (11–13), suggesting that the CCM lesions do not develop from a purely clonal expansion of a mutant cell. The low frequencies of mutations within the lesions also demonstrate the need for using sensitive techniques to identify the somatic mutation.
In examining sporadic CCM lesions for two somatic mutations, both of which are likely to be found at low frequencies, a NGS strategy was used. Multiple sporadic CCM lesions were found to contain somatic mutations, including one lesion with two biallelic, somatic mutations. These findings from sporadic lesion samples, along with a somatic mosaic patient and a case of CCM lesion regrowth, support a somatic mutation mechanism involving the known CCM genes, suggesting a common pathway for CCM lesion pathogenesis.
RESULTS
For our NGS strategy, the coding exons (with >10 bp of flanking intronic sequence) of all three CCM genes were amplified by polymerase chain reaction (PCR). All coding exons were successfully amplified from each sample, ruling out constitutional or high-frequency trans deletions in the samples examined. PCR products were pooled and sequencing on a Roche 454 GS-FLX Titanium platform. Fourteen lesion samples were analyzed in this manner. A total of 379 394 reads aligned with the human reference sequence, with an average of 596 reads per amplicon, though this value ranged from 0 to 8292 reads and a standard deviation of 776 reads. Since the vast majority of constitutional CCM mutations that have been identified severely alter protein function (premature stop codons, splice-site mutations, insertions/deletions leading to frameshifts, etc.), the sequences from these sporadic CCM lesions were analyzed for these same classes of somatic mutations. Thus, we may have missed bona fide somatic missense mutations, but our approach was designed to generate few false-positive mutant calls. The results are summarized in Table 1.
Table 1.
Summary of CCM sample mutations
| Sample no. | Type | Constitutional mutation |
Somatic mutation |
||||||
|---|---|---|---|---|---|---|---|---|---|
| No. of Mut readsa | Total readsa | Age of mut allelesa | No. of Mut readsa | Total readsa | Age of mut allelesa | ||||
| 2049b | Constitutional | KRIT1, c.1363C>T, p.Q455X | 39 | 94 | 41.5% | KRIT1, c.1271_1274delTATAfs*12 | 4 | 94 | 4.3% |
| 4035b | Constitutional | PDCD10, c.474+1G>A, splice | NS | NS | NS | PDCD10, c.205_211insAfs*18 | NS | NS | NS |
| 4362 | Constitutional | KRIT1, c.213_214CG>AT, p.Y71X | 321 | 703 | 45.7% | KRIT1, c.1890G>A, p.W630X | 53 | 2097 | 2.5% |
| 4384 | Constitutional | CCM2, c.472+1G>T, splice | 43 | 82 | 52.4% | None found | |||
| 4258 | Somatic mosaic | None found | NS | NS | NS | KRIT1, LOH | NS | NS | NS |
| 4310 | Sporadic | None found | CCM2, c.355_369del | 6 | 528 | 1.1% | |||
| 4311 | Sporadic | None found | None found | ||||||
| 4358 | Sporadic | None found | CCM2, c.611_622del | 2 | 39 | 5.1% | |||
| 4360 | Sporadic | None found | None found | ||||||
| 4364 | Sporadic | None found | None found | ||||||
| 4382 | Sporadic | None found | None found | ||||||
| 4386 | Sporadic | None found | KRIT1, c.1659_1688delins | 3 | 702 | 0.4% | |||
| 4388 | Sporadic | None found | None found | ||||||
| 4390 | Sporadic | None found | None found | ||||||
| 4392 | Sporadic | None found | KRIT1, c.993T>G, p.Y331X; KRIT1, c.1159C>T, p.Q387X | 44; 35 | 610; 578 | 7.2%; 6.1% | |||
| 4394 | Sporadic | None found | None found | ||||||
del, deletion; delins, insertion/deletion; splice, splice-site mutation; ins, insertion; fs, frameshift; Mut, mutant; NS, not sequenced by NGS; LOH, loss of heterozygosity.
aMeasured by NGS.
bPreviously published (13).
CCM lesions with constitutional mutations
Of the 14 CCM samples analyzed, 13 were from patients exhibiting a single CCM lesion, a common feature of sporadic CCM. The remaining CCM lesion was a sample from an inherited case previously analyzed by our laboratory (13) and served as a positive control for our sequencing strategy. Sample 2049 was resected from a family member harboring the common Hispanic constitutional mutation (13) in KRIT1 (c.1363C>T, p.Q455X). In the original study, we utilized Sanger sequencing of individual cloned exonic amplicons from the lesion DNA to identify the trans somatic mutation in KRIT1 (c.1271_1274delTATAfs*12) in 4.6% of the clones. Using NGS for the same sample, the constitutional mutation was identified in 39 of 94 reads (42% frequency) and the somatic mutation was identified in 4 of 94 reads (4.3% frequency). Since both mutations occur in the same exon, it was evident that the mutations are indeed in trans as no read contained both mutations. This positive control sample served as a proof of principle that NGS was sensitive enough to detect one of our previously reported somatic mutations, even though it can only be found at low frequency in the bulk lesion sample.
In nine of the CCM lesion samples, 10 heterozygous single nucleotide polymorphisms (SNPs) were identified (Supplementary Material, Table S1), each with an associated reference number. For SNPs with coverage of at least 50 reads, the minor alleles ranged in frequency from 45.0 to 58.4%. From these data, we can surmise that heterozygous variants would be found at similar frequencies and somatic variants would be found at lower frequencies.
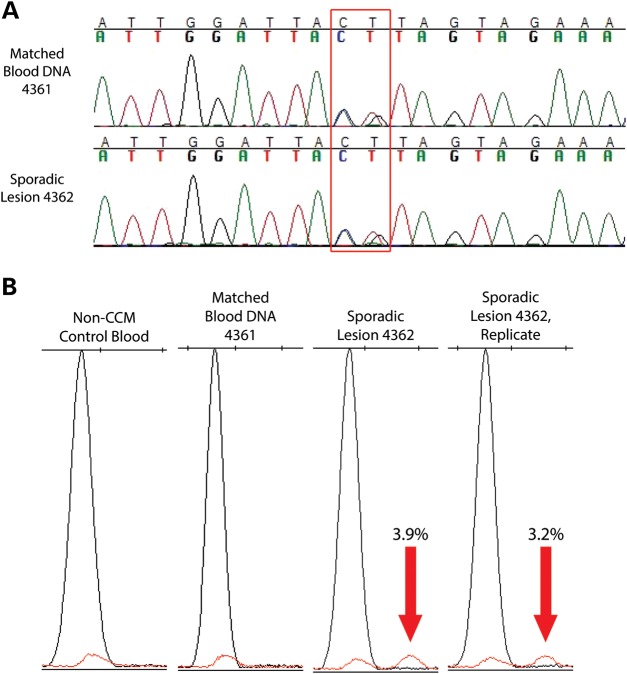
Surprisingly, analysis of the sequencing data revealed that 2 of the 13 CCM lesion samples (15%) from patients harboring a single lesion instead contained high-frequency mutations in one of the CCM genes. In lesion 4362, a mutation was identified in KRIT1 (c.213_214CG>AT, p.Y71X) in 46% of the reads (321 of 703) and confirmed by Sanger sequencing (Fig. 1A). Analysis of a matched blood DNA sample from this patient revealed the presence of the c.213_214CG>AT mutation, confirming that this high-frequency variant is a constitutional, heterozygous mutation. In the lesion sample 4362, a somatic mutation was also identified in KRIT1 (c.1890G>A, p.W630X) at a frequency of 2.5% (53 of 2097 reads).
Figure 1.
CCM lesion sample 4362 contained one constitutional and one somatic mutation. (A) Sanger sequencing chromatogram shows a heterozygous, nonsense mutation in KRIT1 (c.213_214CG>AT, p.Y71X) in both lesion sample 4362 and a matched blood DNA sample from the same patient. (B) SNaPshot assay confirms the presence of a somatic nonsense mutation in KRIT1 (c.1890G>A, p.W630X) in the lesion DNA sample (red peaks indicated by red arrows) but not in unmatched and matched blood DNA control samples. Numbers indicate the proportion of mutant alleles within the sample.
A single-base extension assay (SNaPshot, Applied Biosystems) was used to validate these mutations. First, primers are designed to anneal adjacent to the base of interest, and then an extension reaction adds a single nucleotide to the primer. The resulting fragments are separated by size, and different alleles of the same amplicon appear as adjacent peaks. A diagram depicting SNaPshot results is shown in Supplementary Material, Figure S1, and it should be noted that only the data from the relevant bases are shown in the remaining figures. One of the major benefits of this assay is that it can be used to quantify the frequency of alleles at a particular DNA base.
As shown in Figure 1B and Supplementary Material, Figure S2, the c.1890G>A mutation in KRIT1 was confirmed by SNaPshot in sample 4362. The mutant allele appeared only in DNA from the lesion sample (two replicates, 3.0 and 2.8% frequencies of the mutant allele). Only the WT allele was present in DNA from this patient's blood (sample 4361) and in a non-CCM control DNA sample.
A high-frequency mutation was also found in lesion 4384, a splice-site mutation in CCM2 (c.472+1G>T, splice donor site) occurring in 52% of the reads (43/82). This heterozygous mutation was verified by Sanger sequencing (data not shown). No obvious somatic mutation was identified in this sample.
Although these samples were obtained from CCM patients with no apparent family history and who exhibited a single lesion on magnetic resonance imaging (MRI), both hallmarks of sporadic CCM disease, these patients did harbor high-frequency CCM gene mutations, indicating that they were either cryptic familial cases with constitutional mutations or possibly cases of somatic mosaicism. For patient sample 4362, we were able to confirm the presence of the constitutional mutation in a matched blood sample (Fig. 1A), so this patient is likely to be a cryptic familial case of CCM. Since most of the samples were obtained through an anonymous tissue bank, we were unable to follow up to determine whether MRI analysis of the entire family might identify hidden cases of CCM or whether the individuals instead represent de novo constitutional mutations. Regardless, our sequence analysis identified another case of biallelic somatic and constitutional mutations in an inherited case of CCM.
Sporadic CCM lesion containing two biallelic, somatic mutations
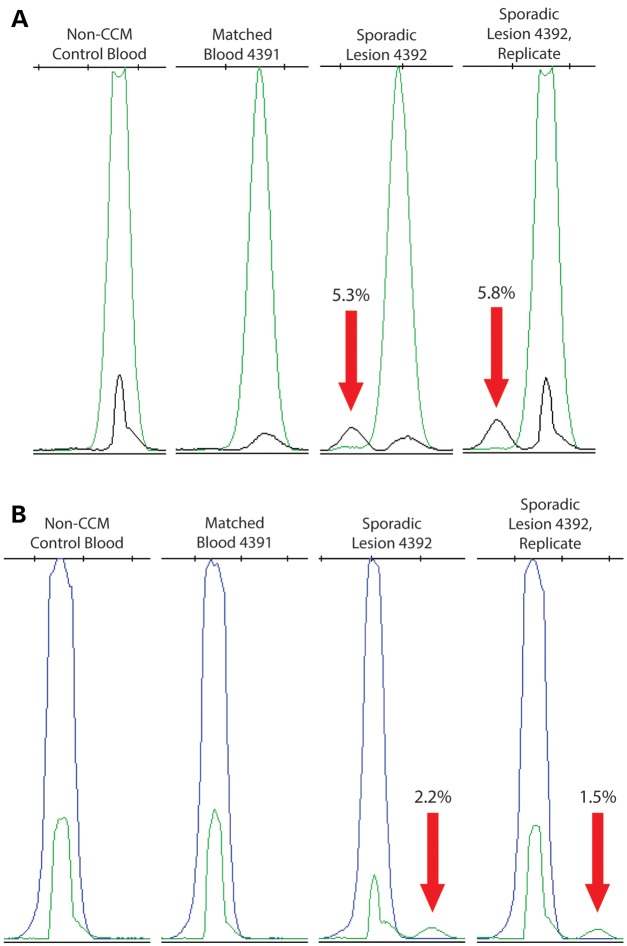
Of the 11 remaining sporadic CCM lesion samples analyzed by NGS, 5 somatic mutations were identified in 4 samples. Two somatic mutations in KRIT1 were identified in sample 4392. The first somatic mutation was a premature stop codon in coding exon 8 (c.993T>G, p.Y331X) occurring in 44/610 reads (7.2% frequency). The second somatic mutation was a different premature stop codon in coding exon 9 (c.1159C>T, p.Q387X) occurring in 35 of 578 reads (6.1% frequency).
By SNaPshot, the exon 8 and exon 9 mutant alleles appeared in the original lesion DNA sample (4392) at frequencies of 5.3 and 2.2%, respectively (Fig. 2; Supplementary Material, Fig. S3). A second, separate PCR amplification also replicated these results with the exon 8 mutation measured at 8.5% frequency and the exon 9 mutation measured at 1.5% frequency. Mutant alleles were not apparent in a matched blood DNA sample (4391) or a control DNA sample (non-CCM case).
Figure 2.
CCM lesion sample 4392 contained two somatic mutations. (A) SNaPshot assay confirms the presence of a somatic nonsense mutation in KRIT1 (c.993T>G, p.Y331X) in the lesion DNA sample (black peaks indicated by red arrows) but not in unmatched and matched blood DNA control samples. Numbers indicate the proportion of mutant alleles within the sample. (B) SNaPshot assay also confirms the presence of a somatic nonsense mutation in KRIT1 (c.1159C>T, p.Q387X) in the lesion DNA sample (green peaks indicated by red arrows) but not in unmatched and matched blood DNA control samples.
To test whether these two somatic mutations were in trans, a 1 kb region surrounding the two mutations was amplified using the high-fidelity (15) PfuTurbo polymerase (Agilent) and sequenced using the Pacific Biosystems RS platform. The PacBio RS NGS system was selected because it can produce longer reads than other platforms. The exon 8 (c.993T>G) and exon 9 (c.1159C>T) mutations were detected by the PacBio RSII system at frequencies of 4.6 and 5.0%, respectively. Of the 7611 reads with the mutant G allele at c.993 that were long enough to span both mutations, 96.2% had the WT C allele at c.1159 (compared with the background base frequencies of 2.2% A, 1.1% T and 0.4% G at that position). Of the 8357 reads with the mutant T allele at c.1159, 98.3% had the WT T allele at c.993 (compared with 1.0% G, 0.5% A and 0.2% C at that position). No sequence read contained both somatic mutations at a frequency that was any different than the background error rate of this sequencing platform, determined by the substitution of other (random) nucleotides at the relevant position. Thus, the two somatic mutations found in sporadic lesion sample 4392 occur on different alleles of KRIT1 and are therefore in trans.
Sporadic CCM lesions with single somatic mutations
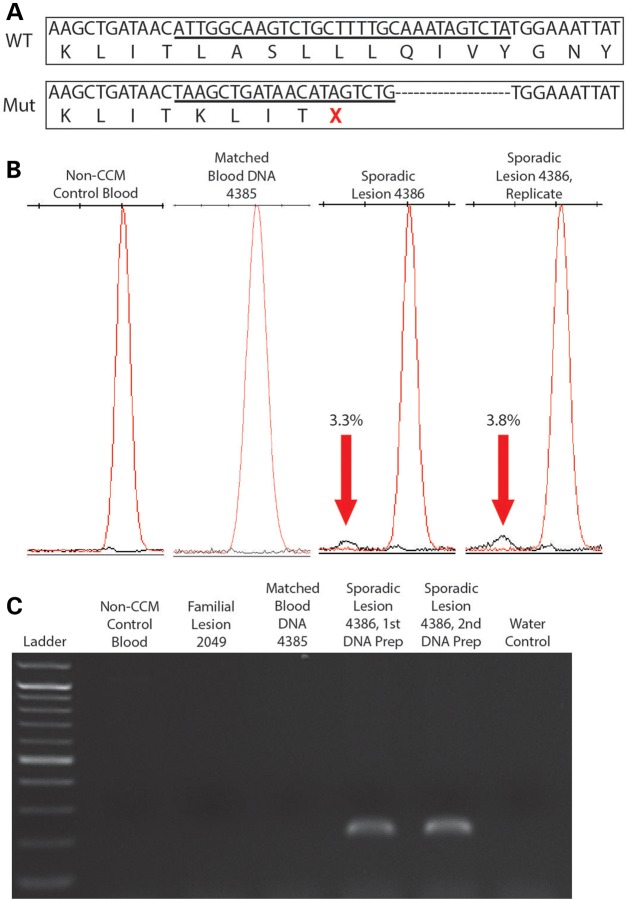
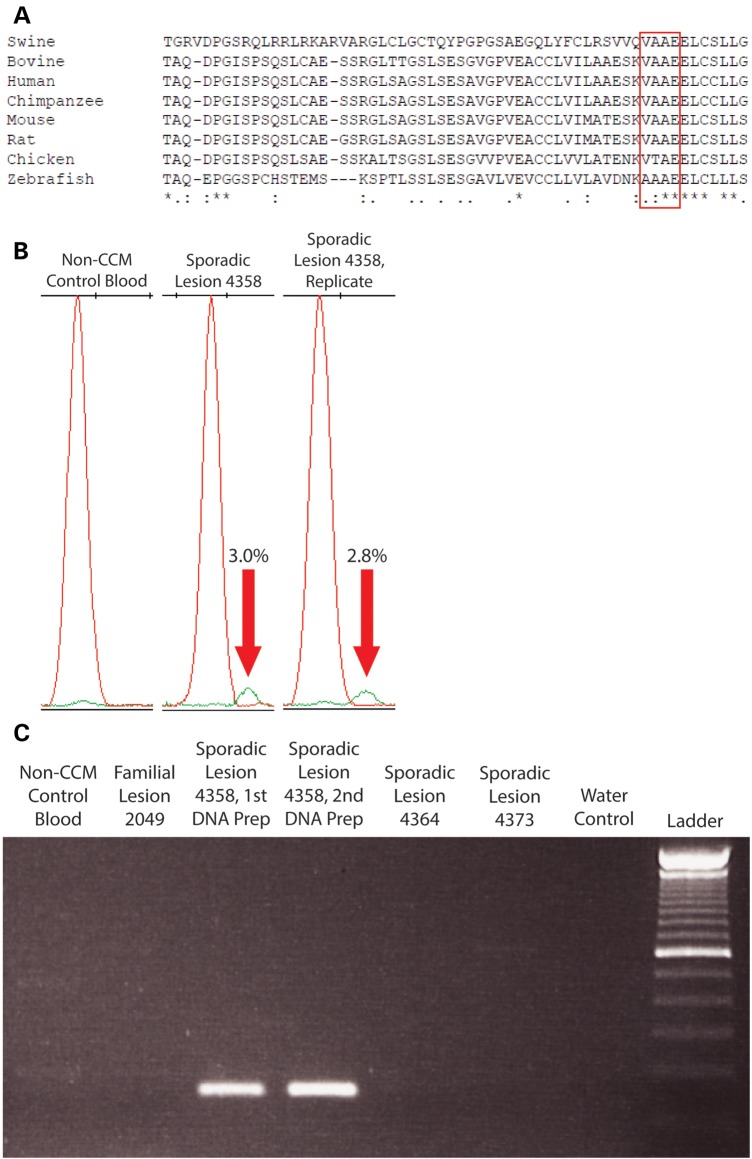
Three sporadic lesion samples (4386, 4310 and 4358) contained one identifiable somatic mutation each. Sample 4386 bore an insertion–deletion mutation in KRIT1 (c.1659_1688delins) in 3/702 reads (0.4%), causing a frameshift and a premature stop codon after four altered amino acid residues (Fig. 3A). Although originally identified at a very low frequency, this mutation was validated by a SNaPshot assay (Fig. 3B; Supplementary Material, Fig. S4), showing that the mutant allele only appears in sample 4386 (3.3% frequency in the original amplification and 3.8% in the replicate) and not in control samples including a matched blood DNA sample from this patient. For further confirmation of this insertion/deletion mutation, PCR primers were designed to specifically amplify the mutant sequence. Based on this PCR strategy, bands on an electrophoresis gel were only apparent for sample 4386 and not for control samples (Fig. 3C). Sanger sequencing was used to confirm that these bands contained the mutated sequence (data not shown).
Figure 3.
CCM lesion sample 4386 contained a single somatic mutation. (A) NGS revealed a mutant allele in sample 4386 in which 30 bp of WT (WT) sequence were deleted (top box, underlined sequence, translated protein sequence appears below) and 20 bp of mutant (Mut) sequence were inserted (bottom box, underlined sequence, translated protein sequence appears below), causing a premature stop codon (red X). (B) SNaPshot assay confirms the presence of the somatic insertion/deletion (indel) mutation in KRIT1 (c.1659_1688delins) in the lesion DNA sample (black peaks indicated by red arrows) but not in unmatched and matched blood DNA control samples. Numbers indicate the proportion of mutant alleles within the sample. (C) A deletion-specific PCR strategy confirms that the somatic indel mutation only appears in sporadic lesion sample 4386 and not in DNA preparations of matched blood from this patient, blood from a non-CCM patient, familial lesion 2049, a different sporadic lesion (4384) or a control reaction without DNA.
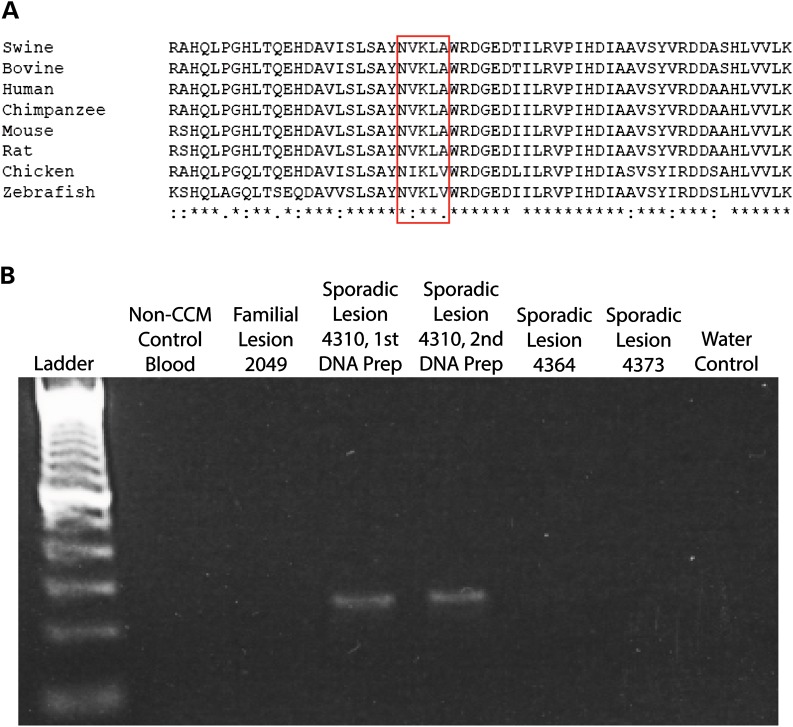
Sample 4310 contained one somatic mutation in CCM2 (c.355_369del) in 6/528 reads (1.1%). Similar to sample 4386, a mutation-specific PCR (Fig. 4B) and Sanger sequencing (data not shown) confirmed that this somatic mutation was present in sample 4310 and not in control samples.
Figure 4.
CCM lesion sample 4310 contained a single somatic mutation in CCM2 (c.355_369del). (A) The in-frame deletion mutation in sample 4310 would remove five highly conserved amino acid residues (red box) from the mature CCM2 protein. (B) A deletion-specific PCR strategy confirms that the somatic deletion mutation only appears in sporadic lesion sample 4310 and not in DNA preparations of non-CCM patient blood, familial lesion 2049, different sporadic lesions (4364 or 4373) or a control reaction without DNA.
Sample 4358 contained a single mutation in CCM2 (c.611_622del) in 2/39 reads (5.1%). This mutation was confirmed to be only in sample 4358 by SNaPshot (Fig. 5B; Supplementary Material, Fig. S5), mutation-specific PCR (Fig. 5C) and Sanger sequencing (data not shown).
Figure 5.
CCM lesion sample 4358 contained a single somatic mutation in CCM2 (c.611_622del). (A) The in-frame deletion mutation in sample 4358 would remove four moderately conserved amino acid residues (red box) from the mature CCM2 protein. (B) SNaPshot assay confirms that sample 4358 contained a somatic deletion mutation at varying frequencies in multiple DNA preparations from the same CCM lesion sample (green peaks indicated by red arrows). The mutation was not found in a non-CCM patient blood control sample. Numbers indicate the proportion of mutant alleles within the sample. (C) A deletion-specific PCR strategy confirms that the somatic deletion mutation only appears in sporadic lesion sample 4310 and not in DNA preparations of non-CCM patient blood, familial lesion 2049, different sporadic lesions (4364 or 4373) or a control reaction without DNA.
Unlike the somatic mutation in 4386, the somatic deletion mutations present in samples 4310 and 4358 are in frame and do not cause frameshifts or premature stop codons. The CCM2 mutation in sample 4310 (c.355_369del) deletes five amino acid residues (119-NVKLA-123) that are moderately to highly conserved in mammals and other vertebrates (Fig. 4A). The CCM2 mutation in sample 4358 (c.611_622del) deletes four moderately conserved amino acid residues (204-VAAE-207, Fig. 5A). The c.611_622del mutation also occurs near the intron–exon border of the sixth coding exon of CCM2, so it is possible that this mutation could alter splicing of the mRNA transcript. Both of these small deletions were found in the phosphotyrosine-binding domain of CCM2, which mediates interactions with other proteins (16) including KRIT1 (17).
Heterogeneous frequency of somatic mutations within CCM lesions
To prepare DNA from the sporadic CCM lesions, the frozen block tissue was bisected and a portion of the sample near the middle was used for DNA extraction. For samples with confirmed somatic mutations by NGS and the SNaPshot assay, additional preparations of DNA were generated and examined (Supplementary Material, Figs S2–S5). The frequency of the somatic mutations within these additional DNA preparations varied considerably. For example, in sample 4386 the somatic mutation appeared at frequencies of 3.3, 11.9, and 10.3% in three different preparations (Supplementary Material, Fig. S4). In sample 4362, the somatic mutation appeared at frequencies of between 3.2 and 5.3% in three separate DNA preparations and could not be seen (0%) in a fourth preparation. Since tissue was first sampled from the middle of the CCM lesion and subsequent samples were removed farther away from the middle, it is likely that this final preparation from 4362 consisted almost entirely of WT draining or feeding vessels resected with the CCM lesion proper. The variation in mutation frequency within a single lesion represents the heterogeneity of the complex, multicavernous vascular structure of the CCM lesion.
CCM patient with a somatic mosaic mutation in KRIT1
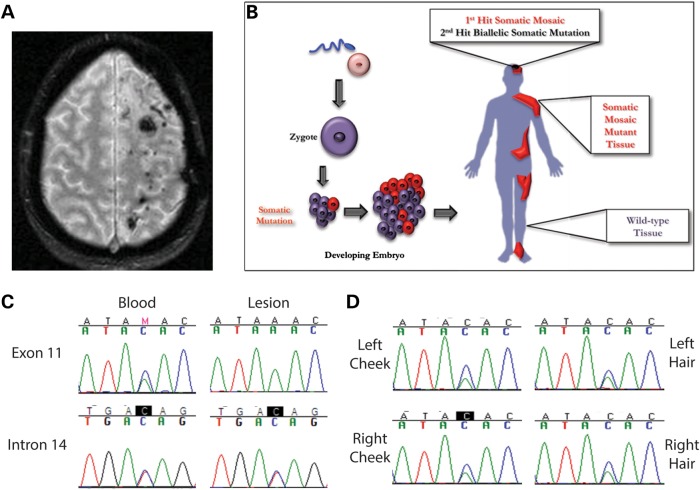
Subsequent to the identification of biallelic somatic mutations in sporadic CCM lesions, we sought to determine whether the two-hit mutation mechanism extended to other non-inherited forms of the disease. A female patient was identified with an unusual presentation of CCM lesions (18). Despite having no family history of the disease (which would normally classify this as a sporadic case of CCM), this patient has over 130 CCM lesions. The patient originally presented with a large intracerebral hemorrhage that had severe mass effect and occurred during the third trimester of a pregnancy. Interestingly, the lesions reside exclusively within the left cerebral hemisphere of the patient's brain and have progressively increased in number since the original presentation (Fig. 6A). Based on these observations, we hypothesize that the first-hit mutation in this case is a somatic mutation which was acquired early in development such that it caused somatic mosaicism (Fig. 6B).
Figure 6.
Somatic mosaicism of a LOH mutation in a CCM patient. (A) Magnetic resonance image of the brain of a patient with over 100 CCM lesions in the left cerebral hemisphere. (B) A schematic of somatic mosaicism. A somatic mutation occurs during development that propagates in subsequent tissues. The result is that some cells have two WT alleles (WT tissue) and some cells are heterozygous (somatic mosaic mutant tissue). (C) A heterozygous missense variant in KRIT1 exon 11 (c.1436A>C, p.K479 T) appeared in blood DNA but not in lesion DNA, indicating a somatic mosaic LOH mutation. A SNP variant in KRIT1 intron 14 (rs34344935) was heterozygous both in blood and lesion DNA, indicating a region where heterozygosity was maintained. (D) Further samples were obtained from this patient and analyzed for evidence of LOH. Left and right cheek swabs and hair samples from the left and right side of the head all displayed heterozygosity for the KRIT1 exon 11 variant (c.1436A>C, p.K479T), implying that the somatic mosaic mutation was isolated to the left cerebral hemisphere.
We obtained a lesion sample from this patient (4285) as well as a blood sample for control DNA. Sanger sequencing of the CCM genes in the blood revealed only a non-synonymous amino acid substitution in coding exon 11 of KRIT1 (c.1436A>C, p.K479T). We examined this mutation for possible effects on mRNA splicing using RT–PCR, but this sequence variation did not appear to affect splicing (data not shown). However, while the c.1436A>C variant appeared heterozygous in blood DNA, lesion DNA only contained the WT A allele (Fig. 6C). Somatic mutations in CCM lesion tissue have been reported at frequencies <21% (11–13), and we have never seen a clear case of complete loss of heterozygosity (LOH) in CCM lesion tissue. We hypothesized that the complete loss of the C allele in lesion DNA represented an LOH event, but since we observed complete LOH only in the CCM lesion that this event represented a somatic mosaic mutation as the first-hit mutation.
To test our somatic mosaicism hypothesis, we sequenced regions of the CCM genes by Sanger sequencing from the blood and lesion samples. A comparison of the coding sequence of KRIT1 revealed regions where heterozygous SNPs in the blood DNA appeared as single alleles in the lesion DNA (Fig. 6C). Heterozygous SNPs in other regions of KRIT1 in the blood DNA, such as intron 14, displayed heterozygosity in the lesion as well. Table 2 summarizes these results identifying a 12–18 kb somatic mosaic deletion that would effectively remove four coding exons from KRIT1. The extensive molecular genetic analyses required to identify this deletion depleted the archived tissue from this CCM lesion, precluding further attempts to fine-map this deletion.
Table 2.
LOH in the somatic mosaic CCM sample 4258
| Chromosomal position (build GRCh37) | Location in KRIT1 | Variant | Heterozygosity status |
|---|---|---|---|
| 91 865 662 | Intron 3 | rs17164451 | MOH |
| 91 853 777 | Intron 9 | rs3793327 | MOH |
| 91 851 343 | Exon 11 | LOH | |
| 91 843 060 | Intron 3 | rs74603220 | LOH |
| 91 842 554 | Exon 14 | rs11542682 | LOH |
| 91 839 202 | Intron 14 | rs11762710 | LOH |
| 91 835 436 | Intron 14 | rs34344935 | MOH |
| 91 829 700 | 3′ UTR | rs1140724 | MOH |
| 91 809 657 | Intergenic | rs2040499 | MOH |
MOH, maintenance of heterozygosity; LOH, loss of heterozygosity.
To determine the anatomic extent of this first-hit somatic mosaic mutation, we sought to investigate the genotype of other tissues from this patient. With the CCM lesions occurring almost exclusively on one side of the brain, we hypothesized that the somatic event may have occurred only in one cerebral hemisphere after establishment of a primitive vascular plexus in the developing brain. We obtained tissue samples through non-invasive methods, such as buccal swabs from the left and right sides of the mouth, and hair samples from each side of the patient's head and eyebrows. DNA from all of these samples was examined for LOH by amplifying coding exon 11 of KRIT1 and sequencing for the c.1436A>C variant. As shown in Figure 6D, all of the tissues sampled displayed heterozygosity at this locus. These data demonstrate that the somatic mosaic mutation is not present in the ectoderm-derived tissues that were available for sampling and thus, we could not conclude when it occurred in relation to development of the cerebrovascular circulation.
We further sought to identify a second-hit somatic mutation from this somatic mosaic CCM lesion using our previously published cloning and Sanger sequencing strategy (13). Sequencing at least 42 clones per coding exon of KRIT1 revealed no identifiable single nucleotide or short-indel somatic mutations.
An aggressive regrowth of a CCM lesion contains the original somatic mutation
Lesion sample 4035 was obtained from an inherited case of CCM. Biallelic constitutional and somatic mutations were identified in PDCD10 (c.474+1G>A and c.205_211insAfs*18), as previously published (13). Recently, this patient required a second surgery to remove what appeared to be a regrowth of this aggressive CCM lesion, and we obtained a sample of the regrowth lesion tissue.
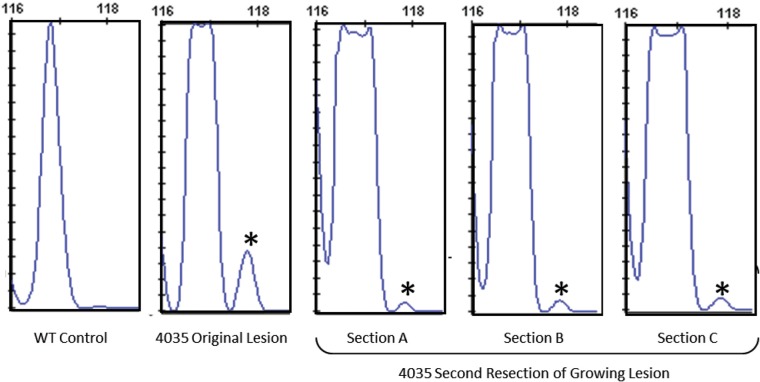
To determine whether this regrowth lesion contained the same somatic mutation as the original lesion sample 4035, we used a fragment analysis assay to detect the presence of the insertion somatic mutation. To account for tissue sample heterogeneity within the lesion, DNA was prepared from three distinct sections of the sample. Following PCR amplification, fragment analysis revealed that the somatic mutation (PDCD10 c.205_211insAfs*18) was present in all three sections of the re-growth lesion sample (Fig. 7). Interestingly, the somatic mutation was present at markedly reduced frequency in the re-growth compared with the original lesion sample 4035. In the original sample, the somatic mutation was detected in 7% of the alleles sampled, but in the regrowth sample it was detected in only 1% of the total alleles. These results suggest that while the somatic mutation may have initiated regrowth of this lesion, other genetic and non-genetic factors may have driven the aggressive growth phenotype. It is possible that another somatic mutation occurred within the endothelium of the lesion and/or hemodynamic factors, inflammation or particular signaling pathways may have contributed to the lesion regrowth.
Figure 7.
Aggressive regrowth of CCM lesion 4035 showed the presence of the original mutation, though at a reduced frequency. Fragment analysis of the original lesion sample 4035 quantified the presence of the somatic mutation (118 bp fragment compared with the WT 117 bp fragment, indicated by asterisks) at 7% of the total alleles. Three independent sections, (A–C), of the regrown lesion show the presence of the somatic mutation allele at 1% frequency.
Increased Rho kinase activity in sporadic lesion endothelial cells
siRNA silencing of any one of the CCM genes causes an increase in the expression and activity of the small GTPase RhoA (16,19,20). The RhoA pathway, including its downstream effector Rho kinase (ROCK), is activated in the lesions seen in mouse models of CCM (21,22) which mimic the genetics of the inherited form of the disease. There is also evidence of increased ROCK activity in human lesions from familial cases of CCM (23). However, it remains uncertain if sporadic human CCM lesions also demonstrate similar involvement of the RhoA pathway. A previous study examined ROCK activity in one inherited and two sporadic CCM lesions (23), and we set out to confirm these results and further profile a larger panel of sporadic and familial CCM patients. We hypothesized that since the sporadic lesions show evidence of somatic mutations or epigenetic silencing in the same CCM genes as in the inherited cases, the RhoA pathway will be activated in the endothelial cells lining the caverns of these lesions from sporadic cases.
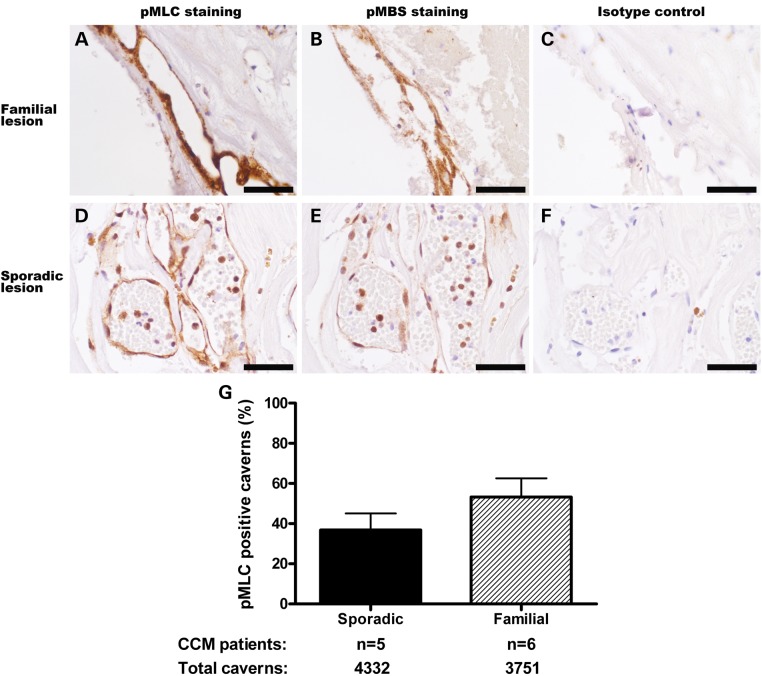
We analyzed CCM lesions from sporadic patients and patients with constitutional mutations in the three CCM genes. Sections of lesions were examined with antibodies against phosphorylation targets of ROCK, phosphorylated myosin light chain (pMLC) and phosphorylated myosin-binding subunit (pMBS) (24,25). Lesions from inherited cases of CCM have increased ROCK activity in the lesion endothelial cells (Fig. 8A and B) compared with isotype control (Fig. 8C). Sporadic lesion endothelial cells also had increased ROCK activity (Fig. 8D and E) compared with isotype control (Fig. 8F). In comparing the prevalence of ROCK-positive caverns within these samples (Fig. 8G), lesions from sporadic patients bore the same amount of ROCK activity (37% of caverns with pMLC staining by immunohistochemistry) as lesions from patients with constitutional mutations in the three CCM genes (53% ROCK-positive caverns, P = 0.23). The RhoA pathway, therefore, is activated in sporadic lesions in a similar manner to lesions from inherited cases of CCM.
Figure 8.
ROCK activity in the endothelial cells of human CCM lesions. (A–F) Immunohistochemical staining of human CCM samples. Strong pMLC (A) and pMBS (B) staining (brown) of lesion endothelial cells in a CCM sample from a familial patient with a constitutional mutation in KRIT1 indicates highly elevated ROCK activity. Staining of a human sporadic CCM sample reveals increased ROCK activity in lesion endothelial cells (D and E). Isotype control antibody of adjacent sections from these samples revealed no staining (C and F). Quantification of the CCM cavern endothelia with ROCK activity (G) demonstrates that all CCM lesions, sporadic or inherited, show evidence of increased ROCK activity. Sporadic CCM lesions show similar levels of ROCK activity to lesions from patients with constitutional mutations in any of the three known CCM genes (P = 0.23).
DISCUSSION
Four CCM lesion samples of the 11 examined (36%) were found to contain a total of 5 somatic mutations. All of the mutations identified are novel and do not appear in the publicly available databases (such as Ensembl and 1000 Genomes). One sample, 4392, had two biallelic somatic mutations in the same gene, as was predicted by the Knudson two-hit mutation hypothesis. Three other samples, 4310, 4358 and 4386, each contained a single somatic mutation. While analyzing additional sporadic CCM lesions for somatic mutations is warranted, it is clear from these four lesion samples that at least some cases of sporadic CCM disease involve (somatic) mutation in the same genes as are mutated in the inherited cases.
Our methods did not find two somatic mutations in each of the samples, but this does not indicate that these lesions contain only one or no somatic mutations. Our NGS approach can only identify single-base substitutions and small insertions/deletions, whereas other types of mutations will remain undetected. For example, constitutional and somatic LOH events such as large deletions of one or more exons would not be found using this method. The major limitation in detecting somatic mutations within CCM lesions is that mutant alleles are found at low frequencies. The highest frequency reported for a somatic mutation in a CCM is 21% (11–13). One group recently identified an LOH event in a glomuvenous malformation sample, a form of inherited vascular malformation, using SNP-chip-based genotyping (26). The copy number data in this sample indicate the LOH mutation was present at 30% frequency within the lesion. Since the frequency of mutations within CCM lesions is much lower, it becomes more difficult to measure LOH in these samples. Our NGS approach, then, is limited to a particular class of mutations and does not exclude the possibility of other types of somatic mutations. However, this approach was sensitive enough to identify multiple low-frequency, somatic mutations from CCM lesions.
While the primary inclusion criterion for lesions in this study was that they were surgically removed from patients with only a single lesion, it is likely that 2 of the 14 CCM lesions analyzed (14.3%) were from inherited cases of the disease. It is also possible that these patients had de novo constitutional mutations or somatic mosaic mutations early in development, but that is not possible to determine without DNA samples from the patients' parents. Most inherited cases of CCM present with multiple CCM lesions, but some inherited cases show lower penetrance (27), owing either to the stochastic nature of this disease or to mutation- or environment-specific factors affecting lesion growth. As such, it would not be unexpected to find inherited cases of CCM among these biobank samples.
In this study, NGS was a reliable method for identifying mutations in both sporadic and inherited cases of CCM. Indeed, NGS identified somatic mutations at frequencies below those previously observed with our cloning and sequencing strategy (13). It should be noted, however, that the frequencies of the somatic mutations reported here varied between different lesions and even within the same lesion. Different somatic mutation frequencies were found in multiple DNA preparations from the same lesion, reflecting the heterogeneous and complex structure of the lesions. Different assays (NGS, SNaPshot) using the same DNA sample also found the somatic mutations at varying frequencies. While it is difficult to determine which of these assays most closely approximates the ‘true’ mutation frequency, NGS was able to qualitatively identify the presence or absence of these somatic mutations.
In addition to the sporadic CCM patients with single lesions and no apparent family history, we also examined the genetic mechanism underlying a patient with over 100 CCM lesions exclusively in one hemisphere of the brain (18). Though no second-hit mutation was identified, it was apparent that the first genetic hit was a somatic mosaic LOH event, deleting 12–18 kb of KRIT1. The presence of a somatic mosaic mutation potentially explains why this patient presented with CCM lesions localized to one side of the brain: endothelial cells in the affected region of the brain would require only one further somatic mutation to cause CCM genesis, whereas the unaffected portion of the brain was still effectively WT, requiring two, independent somatic mutations for lesions to form.
Lesion sample 4035 was previously reported in a study of somatic mutations in familial cases of CCM (13). After publication of these results, this patient underwent further surgeries to remove apparent regrowth of a CCM lesion in the same stereotactic location of the brain. Samples of this regrowth lesion retained the somatic mutation signature of the original resected lesion. We hypothesize that cells harboring the original somatic mutation evaded surgical resection and subsequently initiated the regrowth of the CCM lesion. This is the first report of a CCM lesion demonstrating regrowth potential similar to a tumor. Further investigation of this phenomenon may reveal that more aggressive treatments are necessary in some cases to prevent regrowth and hemorrhage of CCM lesions.
Multiple groups have previously found evidence of a two-hit mutation mechanism in inherited cases of CCM (11–13,28). From these experiments, we have provided evidence that at least some sporadic CCM lesions harbor somatic mutations in the same genes, with one case showing biallelic somatic mutations. Since our approach can only identify certain classes of mutations, we suggest that many sporadic CCMs may also be caused by a two-hit mutational mechanism. Furthermore, we have presented other cases of somatic mosaicism and lesion regrowth that, while less typical, nonetheless reinforce that the distinct presentations of CCM are all caused by a Knudsonian two-hit mechanism in the known CCM genes.
Examination of CCM lesions from humans (23) and CCM mouse models (21–23) has revealed activation of the RhoA pathway through its effector ROCK. Through these data, we have demonstrated that CCM lesions from both sporadic and familial cases of the disease contain endothelial cells with increased levels of ROCK activity. The ROCK activity in sporadic lesions closely resembled that in the lesions from familial patients. Importantly, increased ROCK activity was heterogeneous within the CCM lesions, with ∼40% of the caverns showing phosphorylation of MLC or MBS. The loss of CCM proteins due to biallelic mutations and aberrant ROCK activity, then, appear to be cell-autonomous events in CCM pathogenesis.
Novel therapeutic strategies are currently being tested in animal models of CCM based on the genetics of the inherited form of the disease (22). Previously, it was unknown if sporadic CCM patients, who represent the majority of CCM cases (29), would benefit from the pharmacological inhibition treatments currently under investigation. Since these data demonstrate that sporadic form of CCM also follows an underlying genetic mechanism and shows activation of the RhoA pathway similar to inherited cases of CCM, it is likely that these novel therapies will also be effective for the larger population with sporadic CCM.
MATERIALS AND METHODS
CCM samples
All CCM samples were obtained from the Angioma Alliance Tissue Bank in accordance with Institutional Review Board standards. Lesions were bisected and tissue samples were removed from the center of the lesion. DNA was isolated using the Puregene tissue protocol (Gentra).
Next-generation sequencing
PCR primers (IDT) were designed to amplify the coding exons (along with >10 bp of flanking intronic sequence) of all three CCM genes. During primer design, 5′ overhangs were created in accordance to 454 sequencing protocols (Roche), including the addition of a 4 bp identification sequence used for multiplexing samples (four samples per run). Regions of interest were amplified by PCR and a small aliquot was checked for quality control by gel electrophoresis. Where necessary, PCR products of interest were analyzed by gel electrophoresis and isolated using the GeneClean Turbo kit (MP Biomedicals).
PCR products were pooled and submitted to the Duke Genome Sequencing and Analysis Core for quality control and sequencing on a Roche 454 GS-FLX Titanium platform. The GS Data Analysis Software Package (Roche) was used to split the multiplexed sample into the four independent datasets. The SWAP454 toolkit (30) was then used to generate a coverage map and SNP calls from the amplified sections of the human CCM genes. Briefly, SWAP454 aligned the individual reads to the sequences of the amplified regions recording only the unique alignments. A coverage map was then constructed from the aligned reads after filtering based upon their neighborhood-quality score method (MIN_QUAL = 20; NQ = 15). Individual SNPs were then called that had a coverage of at least two reads, with at least one read mapping to both the forward and reverse strand, and a minimum variant/reference ratio of 0.02. A separate analysis of these data was also performed using Geneious software (Biomatters) (31).
For analysis of phase of somatic mutations in the same sporadic CCM sample, PCR products were first amplified using the high-fidelity polymerase PfuTurbo (Agilent), and then purified by gel extraction (Geneclean Turbo, Qbiogene). Samples were run on the Pacific Biosystems (PacBio) RSII platform. PacBio reads were first trimmed to remove adapter sequences and low-quality (≤0.75) bases from the ends of the reads. Reads that were at least 50 nt in length were aligned to the KRIT1 sequence using the blasr tool from the SMRT Analysis Toolkit (Pacific Biosciences; default parameters). The number of reference and variant calls for each of the two somatic SNP locations was then counted. Reads that had a base call at both locations were then used to identify the monoallelic or biallelic nature of the variants.
SNaPshot analysis
PCR products with potential mutations were analyzed by SNaPshot (Applied Biosystems). Primers were designed adjacent to the base of interest, and SNaPshot was performed according to Applied Biosystems protocols. Results were visualized and quantified using GeneMapper software (Applied Biosystems). Allele frequencies were calculated by dividing the area under the peak of a particular allele by the total area under both allele peaks.
Sanger sequencing
Samples were analyzed by Sanger sequencing using the BigDye reaction kit (Applied Biosystems) on the 3130 Genetic Analyzer (Applied Biosystems). Sequences were examined using Sequencher software (Gene Codes Corporation).
ROCK activity assay
The ROCK activity was assessed by the expression of pMLC and pMBS, which are ROCK substrates. Excised CCM lesions from five sporadic patients (obtained from the University of Chicago) and six familial patients (including two patients with constitutional mutations in KRIT1, two patients with constitutional mutations in CCM2, and two patients with constitutional mutations in PDCD10) were embedded in paraffin and cut into serial 5 µm sections. After antigen retrieval, sections were blocked using PBS supplemented with 0.5% fish skin gelatin (Sigma–Aldrich), 5% goat serum (Invitrogen) and an avidin/biotin blocking kit (Vector Laboratories). Slides were probed with either rabbit polyclonal anti-pMLC (Thr18/Ser19) antibody (Cell Signaling Technology, 1:250) or rabbit polyclonal anti-pMBS (MYPT1, Thr853) antibody (MyBioSource, 1:2000 for sporadic samples and 1:5000 for familial samples) overnight at 4°C, followed by biotinylated goat anti-rabbit antibody (Vector Laboratories, 1:400) for 2 h, as previously described (21–23). Isotype controls were used as negative controls and run simultaneously with all specimens. Quantification of pMLC expression was performed in each cavern of CCM lesions from a total of 11 patients. A negative cavern was defined as complete absence of pMLC staining, whereas a positive cavern was defined as definite but diffuse, intense and confluent staining. Student's t-test was used to compare the prevalence of positive pMLC caverns of CCM lesions between sporadic patient and familial patient samples.
Fragment analysis
The presence of the single-base insertion somatic mutation in sample 4035 was validated by fragment analysis assay. PCR products for exon 6 of PDCD10 were amplified using one fluorescently labeled primer, 5′-6-FAM-CTCACACAAGACATCATTATG, and one unlabeled primer, 5′-CCATACGAAGAAGGGACTCC or 5′-AAACAAGGTTCTTCTGTCCGTTA. The high-fidelity DNA polymerase Phusion (Thermo Scientific) was used for these reactions. The resulting PCR products were resuspended in formamide (Applied Biosystems) and were characterized on an Applied Biosystems 3130 sequencer. Subsequent analysis of fragment seizes was performed using GeneMapper software (Applied Biosystems). Positive and negative controls were colony PCR products from clones with sequence showing either WT (negative control) or somatic mutation (positive control) sequences.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the National Institutes of Health (R01-NS060748 and R01-NS077957 to D.A.M. and I.A.W., F31-NS077702 to D.A.M. and F31-NS061468 to A.L.A.).
Supplementary Material
ACKNOWLEDGEMENTS
We thank St Joseph's Hospital and Medical Center's Human Specimen Procurement Service as well as Angioma Alliance's DNA/Tissue Bank (www.angioma.org/DNA) for providing the CCM lesion samples used in this study.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Rigamonti D., Hadley M.N., Drayer B.P., Johnson P.C., Hoenig-Rigamonti K., Knight J.T., Spetzler R.F. Cerebral cavernous malformations. Incidence and familial occurrence. N. Engl. J. Med. 1988;319:343–347. doi: 10.1056/NEJM198808113190605. [DOI] [PubMed] [Google Scholar]
- 2.Otten P., Pizzolato G.P., Rilliet B., Berney J. 131 cases of cavernous angioma (cavernomas) of the CNS, discovered by retrospective analysis of 24,535 autopsies. Neurochirurgie. 1989;35:82–83. [PubMed] [Google Scholar]
- 3.Del Curling O., Jr, Kelly D.L., Jr, Elster A.D., Craven T.E. An analysis of the natural history of cavernous angiomas. J. Neurosurg. 1991;75:702–708. doi: 10.3171/jns.1991.75.5.0702. [DOI] [PubMed] [Google Scholar]
- 4.Robinson J.R., Awad I.A., Little J.R. Natural history of the cavernous angioma. J. Neurosurg. 1991;75:709–714. doi: 10.3171/jns.1991.75.5.0709. [DOI] [PubMed] [Google Scholar]
- 5.Laberge-le Couteulx S., Jung H.H., Labauge P., Houtteville J.P., Lescoat C., Cecillon M., Marechal E., Joutel A., Bach J.F., Tournier-Lasserve E. Truncating mutations in CCM1, encoding KRIT1, cause hereditary cavernous angiomas. Nat. Genet. 1999;23:189–193. doi: 10.1038/13815. [DOI] [PubMed] [Google Scholar]
- 6.Sahoo T., Johnson E.W., Thomas J.W., Kuehl P.M., Jones T.L., Dokken C.G., Touchman J.W., Gallione C.J., Lee-Lin S.Q., Kosofsky B., et al. Mutations in the gene encoding KRIT1, a Krev-1/rap1a binding protein, cause cerebral cavernous malformations (CCM1) Hum. Mol. Genet. 1999;8:2325–2333. doi: 10.1093/hmg/8.12.2325. [DOI] [PubMed] [Google Scholar]
- 7.Liquori C.L., Berg M.J., Siegel A.M., Huang E., Zawistowski J.S., Stoffer T., Verlaan D., Balogun F., Hughes L., Leedom T.P., et al. Mutations in a gene encoding a novel protein containing a phosphotyrosine-binding domain cause type 2 cerebral cavernous malformations. Am. J. Hum. Genet. 2003;73:1459–1464. doi: 10.1086/380314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Denier C., Goutagny S., Labauge P., Krivosic V., Arnoult M., Cousin A., Benabid A.L., Comoy J., Frerebeau P., Gilbert B., et al. Mutations within the MGC4607 gene cause cerebral cavernous malformations. Am. J. Hum. Genet. 2004;74:326–337. doi: 10.1086/381718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bergametti F., Denier C., Labauge P., Arnoult M., Boetto S., Clanet M., Coubes P., Echenne B., Ibrahim R., Irthum B., et al. Mutations within the programmed cell death 10 gene cause cerebral cavernous malformations. Am. J. Hum. Genet. 2005;76:42–51. doi: 10.1086/426952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Knudson A.G., Jr Mutation and cancer: statistical study of retinoblastoma. Proc. Natl. Acad. Sci. USA. 1971;68:820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gault J., Awad I.A., Recksiek P., Shenkar R., Breeze R., Handler M., Kleinschmidt-DeMasters B.K. Cerebral cavernous malformations: somatic mutations in vascular endothelial cells. Neurosurgery. 2009;65:138–144. doi: 10.1227/01.NEU.0000348049.81121.C1. (discussion 144–135) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gault J., Shenkar R., Recksiek P., Awad I.A. Biallelic somatic and germ line CCM1 truncating mutations in a cerebral cavernous malformation lesion. Stroke. 2005;36:872–874. doi: 10.1161/01.STR.0000157586.20479.fd. [DOI] [PubMed] [Google Scholar]
- 13.Akers A.L., Johnson E., Steinberg G.K., Zabramski J.M., Marchuk D.A. Biallelic somatic and germline mutations in cerebral cavernous malformations (CCMs): evidence for a two-hit mechanism of CCM pathogenesis. Hum. Mol. Genet. 2009;18:919–930. doi: 10.1093/hmg/ddn430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pozzati E., Acciarri N., Tognetti F., Marliani F., Giangaspero F. Growth, subsequent bleeding, and de novo appearance of cerebral cavernous angiomas. Neurosurgery. 1996;38:662–669. (discussion 669–670) [PubMed] [Google Scholar]
- 15.Shafikhani S. Factors affecting PCR-mediated recombination. Environ. Microbiol. 2002;4:482–486. doi: 10.1046/j.1462-2920.2002.00326.x. [DOI] [PubMed] [Google Scholar]
- 16.Crose L.E., Hilder T.L., Sciaky N., Johnson G.L. Cerebral cavernous malformation 2 protein promotes smad ubiquitin regulatory factor 1-mediated RhoA degradation in endothelial cells. J. Biol. Chem. 2009;284:13301–13305. doi: 10.1074/jbc.C900009200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zawistowski J.S., Stalheim L., Uhlik M.T., Abell A.N., Ancrile B.B., Johnson G.L., Marchuk D.A. CCM1 and CCM2 protein interactions in cell signaling: implications for cerebral cavernous malformations pathogenesis. Hum. Mol. Genet. 2005;14:2521–2531. doi: 10.1093/hmg/ddi256. [DOI] [PubMed] [Google Scholar]
- 18.Reid P.J., Campbell S.S., Vates G.E., Allende R. Extreme de novo appearance of cerebral cavernous malformations: case report. Neurosurgery. 2008;62:E969–E970. doi: 10.1227/01.neu.0000318184.25783.b9. (discussion E970) [DOI] [PubMed] [Google Scholar]
- 19.Borikova A.L., Dibble C.F., Sciaky N., Welch C.M., Abell A.N., Bencharit S., Johnson G.L. Rho kinase inhibition rescues the endothelial cell cerebral cavernous malformation phenotype. J. Biol. Chem. 2010;285:11760–11764. doi: 10.1074/jbc.C109.097220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Whitehead K.J., Chan A.C., Navankasattusas S., Koh W., London N.R., Ling J., Mayo A.H., Drakos S.G., Marchuk D.A., Davis G.E., et al. The cerebral cavernous malformation signaling pathway promotes vascular integrity via Rho GTPases. Nat. Med. 2009;15:177–184. doi: 10.1038/nm.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McDonald D.A., Shenkar R., Shi C., Stockton R.A., Akers A.L., Kucherlapati M.H., Kucherlapati R., Brainer J., Ginsberg M.H., Awad I.A., et al. A novel mouse model of cerebral cavernous malformations based on the two-hit mutation hypothesis recapitulates the human disease. Hum. Mol. Genet. 2011;20:211–222. doi: 10.1093/hmg/ddq433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McDonald D.A., Shi C., Shenkar R., Stockton R.A., Liu F., Ginsberg M.H., Marchuk D.A., Awad I.A. Fasudil decreases lesion burden in a murine model of cerebral cavernous malformation disease. Stroke. 2012;43:571–574. doi: 10.1161/STROKEAHA.111.625467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stockton R.A., Shenkar R., Awad I.A., Ginsberg M.H. Cerebral cavernous malformations proteins inhibit Rho kinase to stabilize vascular integrity. J. Exp. Med. 2010;207:881–896. doi: 10.1084/jem.20091258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grassie M.E., Moffat L.D., Walsh M.P., MacDonald J.A. The myosin phosphatase targeting protein (MYPT) family: a regulated mechanism for achieving substrate specificity of the catalytic subunit of protein phosphatase type 1delta. Arch. Biochem. Biophys. 2011;510:147–159. doi: 10.1016/j.abb.2011.01.018. [DOI] [PubMed] [Google Scholar]
- 25.Hudson C.A., Heesom K.J., Lopez Bernal A. Phasic contractions of isolated human myometrium are associated with Rho-kinase (ROCK)-dependent phosphorylation of myosin phosphatase-targeting subunit (MYPT1) Mol. Hum. Reprod. 2012;18:265–279. doi: 10.1093/molehr/gar078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amyere M., Aerts V., Brouillard P., McIntyre B.A., Duhoux F.P., Wassef M., Enjolras O., Mulliken J.B., Devuyst O., Antoine-Poirel H., et al. Somatic uniparental isodisomy explains multifocality of glomuvenous malformations. Am. J. Hum. Genet. 2013;92:188–196. doi: 10.1016/j.ajhg.2012.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Denier C., Labauge P., Bergametti F., Marchelli F., Riant F., Arnoult M., Maciazek J., Vicaut E., Brunereau L., Tournier-Lasserve E. Genotype–phenotype correlations in cerebral cavernous malformations patients. Ann. Neurol. 2006;60:550–556. doi: 10.1002/ana.20947. [DOI] [PubMed] [Google Scholar]
- 28.Pagenstecher A., Stahl S., Sure U., Felbor U. A two-hit mechanism causes cerebral cavernous malformations: complete inactivation of CCM1, CCM2 or CCM3 in affected endothelial cells. Hum. Mol. Genet. 2009;18:911–918. doi: 10.1093/hmg/ddn420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aboian M.S., Daniels D.J., Rammos S.K., Pozzati E., Lanzino G. The putative role of the venous system in the genesis of vascular malformations. Neurosurg. Focus. 2009;27:E9. doi: 10.3171/2009.8.FOCUS09161. [DOI] [PubMed] [Google Scholar]
- 30.Brockman W., Alvarez P., Young S., Garber M., Giannoukos G., Lee W.L., Russ C., Lander E.S., Nusbaum C., Jaffe D.B. Quality scores and SNP detection in sequencing-by-synthesis systems. Genome Res. 2008;18:763–770. doi: 10.1101/gr.070227.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Drummond A.J.A.B., Buxton S., Cheung M., Cooper A., Heled J., Kearse M., Moir R., Stones-Havas S., Sturrock S., Thierer T., Wilson A. Geneious v5.3. 2010 Available from http://www.geneious.com. (date last accessed, 9 April 2014) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.