Abstract
A number of genetic variants have been linked to increased risk of breast cancer. Little is, however, known about the prognostic significance of hereditary factors. Here, we investigated the frequency and prognostic significance of two ERBB4 promoter region variants, −782G>T (rs62626348) and −815A>T (rs62626347), in a cohort of 1010 breast cancer patients. The frequency of nine previously described somatic ERBB4 kinase domain mutations was also analyzed. Clinical material used in the study consisted of samples from the phase III, adjuvant, FinHer breast cancer trial involving 1010 women. Tumor DNA samples were genotyped for ERBB4 variants and somatic mutations using matrix-assisted laser desorption ionization/time of flight mass spectrometry. Paraffin-embedded tumor sections from all patients were immunohistochemically stained for ErbB4 expression. Association of ERBB4 genotype to distant disease-free survival (DDFS) was assessed using Kaplan-Meier and Cox regression analyses. Genotyping was successful for 91–93% of the 1010 samples. Frequencies observed for the ERBB4 variants were 2.5% and 1.3% for −782G>T and −815A>T, respectively. Variant −815A>T was significantly associated with poor survival (HR = 2.86 [95% CI 1.15–6.67], P = 0.017). In contrast, variant −782G>T was associated with well-differentiated cancer (P = 0.019). Two (0.2%) ERBB4 kinase domain mutations were found, both of which have previously been shown to be functional and promote cancer cell growth in vitro. These data present the germ-line ERBB4 variant −815A>T as a novel prognostic marker in high-risk early breast cancer and indicate the presence of rare but potentially oncogenic somatic ERBB4 mutations in breast cancer.
Introduction
Breast cancer is the most frequently diagnosed cancer and the leading cause of death due to a malignancy among women [1]. Hereditary genetic factors are thought to account for approximately 5 to 10% of breast cancers due to germ-line variants in genes that increase the risk for breast cancer, such as BRCA1 and BRCA2 [2]. Although a number of germ-line variants have been linked to increased risk of breast cancer [2], less is known about the prognostic significance of hereditary variants.
ErbB4 is a member of the EGF receptor (EGFR) subfamily of receptor tyrosine kinases including EGFR (ErbB1), ErbB2 (HER2, neu), ErbB3 (HER3), and ErbB4 (HER4) [3]. Despite of active research on ErbB4 biology in normal mammary tissue and breast cancer, significance of ErbB4 for breast carcinogenesis is still poorly understood. ErbB4 expression is typically associated with ER- and PR-positivity, ErbB2 receptor-negativity, well-differentiated phenotype and favorable outcome [4]–[7]. On the other hand, ErbB4 overexpression has been associated with shorter relapse-free survival in early, node-negative tumors [8] and with decreased survival in patients with node-positive tumors [9]. Treatment with ErbB4-targeted monoclonal antibody suppresses the growth of breast cancer cells [10], suggesting a possible oncogenic role of ErbB4 in breast cancer.
The role of ERBB4 gene variation in breast cancer has been less extensively studied. We have previously shown that a single nucleotide polymorphism (SNP) −782 G>T in the promoter region of ERBB4 gene is a novel risk variant for breast cancer in German population [11]. Two recent studies have also discovered variants of ERBB4 gene that are associated with increased risk for breast cancer. ERBB4 SNP rs13393577 was implicated as a new risk variant in a genome-wide association study (GWAS) conducted in Korean population [12], and three ERBB4 risk variants (rs905883, rs7564590, and rs7558615) were identified in a family-based GWAS in patients of the Framingham heart study [13]. However, no studies have addressed the possible prognostic or predictive value of ERBB4 variants.
According to the Catalogue Of Somatic Mutations In Cancer (COSMIC), somatic ERBB4 mutations in breast cancer are rare, as only 1.4% of breast cancers harbor ERBB4 missense mutations (17 out of 1200 patients) [14]. Although the functional consequences of ERBB4 breast cancer mutations have not been studied, one ERBB4 kinase domain mutation (E872K) initially found in breast cancer [15] has later been shown to be functionally active in metastatic melanoma [16].
Here, we analyzed the frequencies and prognostic value of two ERBB4 promoter variants, −782G>T and −815A>T in a large phase III clinical trial data set of high-risk early breast cancer patients (n = 1010). Frequency of nine specific ERBB4 kinase domain mutations [15] was also analyzed. The results indicate that the ERBB4 variant −815A>T was significantly associated with poor distant disease-free survival, indicating for the first time a possible prognostic significance for a genetic variant of ERBB4 in cancer. The frequency of the analyzed ERBB4 kinase domain mutations was low (0.2%). However, both somatic mutations had previously been shown to be functional and promote cancer cell growth in vitro [16], [17], suggesting a presence of rare but oncogenic ERBB4 mutations in breast cancer.
Materials and Methods
Patient DNA and tumor tissue samples
Study material consisted of DNA and formalin-fixed, paraffin-embedded tissue samples from primary tumors of 1010 women with high-risk early breast cancer who participated in the adjuvant phase III FinHer trial (International Standard Ran- domised Controlled Trial number, ISRCTN76560285) [18]. The key inclusion criteria in the FinHer trial were histologically confirmed invasive breast cancer, age 65 or less, macroscopically complete surgery for breast cancer, presence of at least one positive axillary lymph node or a node-negative breast cancer with tumor diameter at least 20 mm and a negative immunostaining for progesterone steroid hormone receptors. Patients with distant metastases at the time of randomization were excluded. Most (89%) of the study patients had axillary lymph node-positive cancer [18]. All patients were randomly assigned to receive three cycles of vinorelbine or docetaxel together with fluorouracil, epirubicin and cyclophosphamide. Patients with ERBB2-positive tumors (n = 232) were also assigned to receive or not to receive adjuvant trastuzumab. The patients signed an informed consent for use of breast tumor tissue samples for research purposes prior to entry to the clinical trial. The protocol of the present study was approved by an Ethics Committee of the Helsinki University Central Hospital.
Analysis of ERBB4 variants and somatic mutations
Two ERBB4 germline single nucleotide variants, −782G>T and −815A>T [11], and nine previously described somatic ERBB4 mutations [15] were genotyped to establish allele and genotype frequencies in the FinHer cohort. Genotyping was carried out with matrix-assisted laser desorption ionization/time of flight mass spectrometry using SpectroCHIP microarray and Bruker Autoflex (Sequenom) as well as MTP Anchor Chip 400/384 TF and Bruker Ultraflex (Bruker Daltonics) [11]. The ERBB4 variant analyses were conducted using tumor DNA, as no DNA from non-neoplastic tissue was available. However, when the variants were initially identified from tumor DNA of colorectal cancer patients, the variants were confirmed to be germ-line in all cases [11].
ErbB4 immunohistochemistry
Paraffin-embedded tumor sections were stained for ErbB4 using HFR-1 monoclonal antibody (2 µg/ml; Abcam), anti-mouse Envision+ System HRP secondary antibody (code K4001; Dako Cytomation), and DAB+ (code K3468; Dako Cytomation) peroxidase substrate. All incubations were carried out in room temperature, and all steps were followed by a rinsing step in 50 mM Tris-HCl pH 7.6 containing 0.05% Tween-20. Sections were counterstained with hematoxylin. ErbB4-positive breast cancer control sections were used as positive controls for each staining series.
Statistical analyses
Frequency tables were analyzed using the chi-squared test or Fisher's exact test. Survival analyses were carried out with Kaplan-Meier statistics, and survival between groups was compared with the log-rank test. The hazard ratio was computed using a univariable Cox model. Distant disease-free survival was calculated from the date of randomization to the date of detection of distant recurrence of breast cancer or to the date of death whenever death preceded distant recurrence, censoring patients who were alive without distant recurrence on the date of last follow-up [19]. All P-values are 2-tailed.
Results
Frequencies of ERBB4 promoter region variants −782G>T and −815A>T
To investigate the prevalence of two ERBB4 promoter region SNPs, −782G>T and −815A>T, tumor DNA samples from 1010 women with high-risk early breast cancer were analyzed. Successful genotype was obtained from 936 (93%) and 932 (92%) patient samples for ERBB4 promoter positions −782 and −815, respectively. From these patients, 23 (2.5%) were genotyped to harbor the ERBB4 −782G>T variant whereas 12 patients (1.3%) harbored the −815A>T variant. All genotypes were heterozygous, with the exception of one homozygous −782TT genotype that was not included in the subsequent statistical analyses.
Associations of ERBB4 variants with clinicopathological features and ErbB4 protein expression
When the ERBB4 promoter region SNP status was compared with clinicopathological characteristics, the −782G>T variant was associated with well-differentiated cancer (P = 0.018; Table 1). Neither of the SNPs was significantly associated with primary tumor diameter, axilliary nodal status, histology, tumor grade, ER or PR expression, or ERBB2 amplification (Table 1). Sixteen (69.6%) out of the 23 cancers with the −782G>T variant were ER-positive and ERBB2-negative in immunohistochemical stainings (an approximation for the luminal A biological subtype) as compared with 548 (62.8%) out of the 872 cancers that did not harbor this variant (P = 0.510; one of the seven remaining −782G>T cases was ER+/ERBB2+, three ER−/ERBB2+, and three ER−/ERBB2−). Nine (75.0%) of the 12 cancers with the −815A>T variant were ER+ and ERBB2− (the remaining three were ER–/ERBB2–) as compared with 553 (63.2%) out of the 874 cancers that did not harbor −815A>T (P = 0.551). All 8 tumors with the −815A>T variant stained by immunohistochemistry for the p53 protein expression stained negative, whereas 4 (20.0%) out of the 20 cases with the −782G>T variant stained positively for p53 [20].
Table 1. Clinicopathological features of patients harboring ERBB4 variants −782G>T or −815A>T.
| −782 G>T | −815 A>T | |||||
| GG | GT | AA | AT | |||
| n (%) | n (%) | P-value | n (%) | n (%) | P-value | |
| Frequency | 872 (98) | 23 (2) | 879 (99) | 12 (1) | ||
| pT | ||||||
| ≤20 mm | 364 (98) | 9 (2) | 366 (99) | 5 (1) | ||
| >20 mm | 508 (98) | 13 (2) | 0.938 | 512 (99) | 7 (1) | 0.999 |
| pN | ||||||
| negative | 91 (97) | 3 (3) | 95 (99) | 1 (1) | ||
| positive | 781 (98) | 20 (2) | 0.726 | 784 (99) | 11 (1) | >0.999 |
| Histology | ||||||
| ductal | 682 (97) | 19 (3) | 689 (99) | 9 (1) | ||
| lobular/other | 190 (98) | 4 (2) | 0.799 | 190 (98) | 3 (2) | 0.729 |
| Grade | ||||||
| 1 | 121 (94) | 8 (6) | 0.018 | 125 (98) | 3 (2) | 0.408 |
| 2 | 352 (98) | 7 (2) | 356 (99) | 3 (1) | ||
| 3 | 362 (98) | 7 (2) | 361 (98) | 6 (2) | ||
| ER | ||||||
| - | 242 (97) | 7 (3) | 243 (99) | 3 (1) | ||
| + | 630 (98) | 16 (2) | 0.777 | 636 (99) | 9 (1) | >0.999 |
| PR | ||||||
| - | 373 (97) | 10 (3) | 371 (98) | 6 (2) | ||
| + | 498 (97) | 13 (3) | 0.959 | 507 (99) | 6 (1) | 0.590 |
| ERBB2 | ||||||
| - | 675 (97) | 19 (3) | 678 (77) | 12 (2) | ||
| + | 197 (98) | 4 (2) | 0.555 | 201 (23) | 0 (0) | 0.079 |
| ErbB4 IHC | ||||||
| negative | 138 (96) | 6 (4) | 140 (97) | 4 (3) | ||
| positive | 697 (98) | 15 (2) | 0.145 | 701 (99) | 7 (1) | 0.098 |
To address whether the two SNPs regulated ErbB4 expression levels in primary tumors, tumor sections from all the 1010 patients were immunohistochemically stained with a monoclonal antibody recognizing the C-terminus of ErbB4 (HFR-1). However, no associations were found between ERBB4 SNP status and ErbB4 staining intensity (Table 1). Also, no statistically significant associations were found when cytoplasmic and nuclear ErbB4 staining intensities were separately scored and compared with the ERBB4 SNP status (data not shown). ErbB4 protein expression did not correlate with patient survival (P = 0.826, n = 926), but was strongly associated with ER-positivity (P = 0.003) (Supplementary table 1).
Prognostic significance of ERBB4 variants
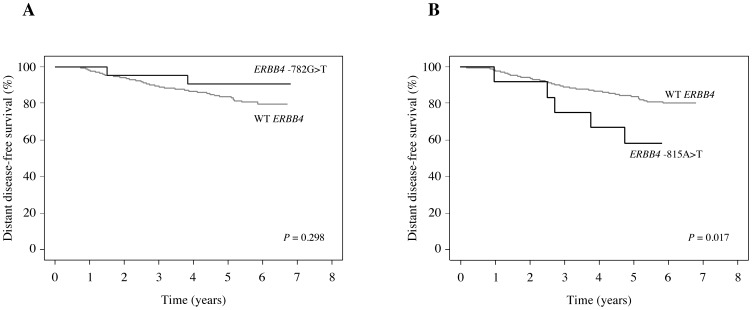
The prognostic significance of the ERBB4 variants was assessed by analyzing the association of ERBB4 SNP status with distant disease-free survival (DDFS). No association between ERBB4 −782G>T and DDFS was found (Figure 1A). In contrast, the ERBB4 variant −815A>T was significantly associated with poor prognosis (HR = 2.86 [95% CI 1.15-6-67], P = 0.017; Figure 1B).
Figure 1. Prognostic associations of the ERBB4 variants.
Kaplan-Meier plots of distant disease-free survival of patients harboring wild-type ERBB4 or ERBB4 variants −782G>T (A) or −815A>T (B).
Frequency of ERBB4 kinase domain mutations
The frequency of nine previously reported [15] somatic ERBB4 kinase domain mutations V721I, A773S, R782Q, G802dup, E810K, P854Q, D861Y, E872K, and T926M, including a mutation previously found in breast cancer (E872K), was also analyzed from the tumor DNA samples. The different point mutations were successfully analyzed from 91–93% of the 1010 samples. Two tumors out of all the genotyped tumors were found to harbor ERBB4 kinase domain mutations G802dup or E872K.
Discussion
Clinical studies on association of ErbB4 expression with breast cancer patient survival are contradictory [4]–[9], [21], despite in vitro as well as in vivo mouse xenograft data suggesting an oncogenic role for ErbB4 in breast cancer [10], [22], [23]. However, the prognostic or predictive role of germ-line or somatic ERBB4 mutations in breast cancer has not been addressed. Here we analyzed the frequencies and prognostic significance of two ERBB4 genetic variants, −782G>T and −815A>T [11] in a cohort of 1010 patients with high-risk early breast cancer. The frequencies of nine specific ERBB4 kinase domain mutations [15] was also investigated.
The frequencies of the two ERBB4 variants were 2.5% (23 out of 936 patients) and 1.3% (12 out of 932 patients) for 782G>T and −815A>T, respectively. In our previous study using samples from German GENICA breast cancer colletion, the frequencies were 5.3% and 1% for 782G>T and −815A>T, respectively [11]. The variant −815A>T was significantly associated with poor prognosis. Interestingly, the variant −782G>T, which was implicated as a risk factor for breast cancer in our previous study [11], was not associated with distant disease-free survival, but with well-differentiated cancer. These data suggest that the heterozygous genotype ERBB4 −815A/T could be a prognostic marker in high-risk early breast cancer. This is the first indication of prognostic significance for a genetic variant of ERBB4 in cancer. However, these findings should be confirmed in an independent large patient cohort. The association between the ERBB4 −815A/T polymorphism and clinical outcome serves the hypothesis that ErbB4-targeted therapy could be beneficial for a subgroup of breast cancer patients in the adjuvant setting.
Immunohistochemical analysis of ErbB4 protein expression levels in the tumors demonstrated that neither of the ERBB4 variants induced significant changes in ErbB4 expression or subcellular localization in the primary tumors. Total ErbB4 expression also did not associate with DDFS of the patients, but correlated with ER-positivity. This is in accordance with reports associating ErbB4 protein expression with markers of favorable prognosis [4]–[7].
Our analysis of specific ERBB4 kinase domain mutations revealed two patients harboring somatic ErbB4 mutations G802dup and E872K, respectively. Interestingly, E872K is a mutation initially found in breast cancer [15] that was later also detected in melanoma [16]. The other mutation, G802dup, has previously been reported in non-small cell lung cancer [15]. Both mutations have been shown to be functional and promote cancer cell/tumor growth in vitro [16], [17], suggesting the presence of rare but potentially oncogenic ERBB4 mutations in breast cancer. Although the observed ERBB4 mutation frequency (0.2%) is low, it corresponds to the frequency of ERBB2 kinase domain mutations (0.5%) in the same patient cohort [24]. Rare ERBB2 kinase domain mutations have recently been suggested to serve as predictive markers for ErbB2-targeted therapy in the absence of ERBB2 amplification [25], indicating that rare mutations may have clinical relevance in high-incidence cancers such as breast cancer.
Taken together, this study presents a genetic ERBB4 variant as a novel prognostic marker in high-risk early breast cancer and indicates the presence of rare but potentially oncogenic ERBB4 mutations in breast cancer.
Supporting Information
Associations of immunohistochemical ErbB4 staining intensity with clinicopathological parameters.
(PDF)
Acknowledgments
We thank Maria Tuominen and Minna Santanen for excellent technical assistance.
Data Availability
The authors confirm that, for approved reasons, there are access restrictions on the data underlying the findings. Relevant data are within the paper and its Supporting Information files. However, there is no permission from the relevant ethical committee to make the patient data of FinHer trial freely available. This data is available upon request pending the decisions of the relevant ethical committee and requests may be sent to the corresponding author.
Funding Statement
This work was supported by the following sources of funding: The Academy of Finland (ref. 137845; http://www.aka.fi; KE); Finnish Cancer Organizations (http://www.cancer.fi; KE); Sigrid Juselius Foundation (http://www.sigridjuselius.fi/foundation; KE); Turku University Hospital (ref. 13785; http://www.vsshp.fi/fi/evo-rahoitus; KE); Finnish Cultural Foundation (http://www.skr.fi; KK); Ida Montin Foundation (http://www.idamontininsaatio.fi; KK); and the Jenny and Antti Wihuri Foundation (http://www.wihurinrahasto.fi; KK). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Jemal A, Bray F, Ferlay J (2011) Global Cancer Statistics. CA Cancer J Clin 61: 69–90. [DOI] [PubMed] [Google Scholar]
- 2. Lalloo F, Evans DG (2012) Familial breast cancer. Clin Genet 82: 105–114. [DOI] [PubMed] [Google Scholar]
- 3. Hynes NE, MacDonald G (2009) ErbB receptors and signaling pathways in cancer. Curr Opin Cell Biol 21: 177–184. [DOI] [PubMed] [Google Scholar]
- 4. Bacus SS, Chin D, Yarden Y, Zelnick CR, Stern DF (1996) Type 1 receptor tyrosine kinases are differentially phosphorylated in mammary carcinoma and differentially associated with steroid receptors. Am J Pathol 148: 549–558. [PMC free article] [PubMed] [Google Scholar]
- 5. Kew TY, Bell JA, Pinder SE, Denley H, Srinivasan R, et al. (2000) c-erbB-4 protein expression in human breast cancer. Br J Cancer 82: 1163–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sassen A, Rochon J, Wild P, Hartmann A, Hofstaedter F, et al. (2008) Cytogenetic analysis of HER1/EGFR, HER2, HER3 and HER4 in 278 breast cancer patients. Breast Cancer Res 10: R2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koutras AK, Kalogeras KT, Dimopoulos M, Wirtz RM, Dafni U, et al.. (2008) Evaluation of the prognostic and predictive value of HER family mRNA expression in high-risk early breast cancer: A Hellenic Cooperative Oncology Group (HeCOG) study: 1775–1785. [DOI] [PMC free article] [PubMed]
- 8. Bièche I, Onody P, Tozlu S, Driouch K, Vidaud M, et al. (2003) Prognostic value of ERBB family mRNA expression in breast carcinomas. Int J Cancer 106: 758–765. [DOI] [PubMed] [Google Scholar]
- 9. Lodge AJ, Anderson JJ, Gullick WJ, Haugk B, Leonard RCF, et al. (2003) Type 1 growth factor receptor expression in node positive breast cancer: adverse prognostic significance of c-erbB-4. J Clin Pathol 56: 300–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hollmén M, Määttä JA, Bald L, Sliwkowski MX, Elenius K (2009) Suppression of breast cancer cell growth by a monoclonal antibody targeting cleavable ErbB4 isoforms. Oncogene 28: 1309–1319. [DOI] [PubMed] [Google Scholar]
- 11. Rokavec M, Justenhoven C, Schroth W, Istrate MA, Haas S, et al. (2007) A novel polymorphism in the promoter region of ERBB4 is associated with breast and colorectal cancer risk. Clin Cancer Res 13: 7506–7514. [DOI] [PubMed] [Google Scholar]
- 12. Kim H, Lee J-Y, Sung H, Choi J-Y, Park SK, et al. (2012) A genome-wide association study identifies a breast cancer risk variant in ERBB4 at 2q34: results from the Seoul Breast Cancer Study. Breast Cancer Res 14: R56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Murabito JM, Rosenberg CL, Finger D, Kreger BE, Levy D, et al. (2007) A genome-wide association study of breast and prostate cancer in the NHLBI's Framingham Heart Study. BMC Med Genet 8 Suppl 1 S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Catalogue of Somatic Mutations In Cancer (COSMIC) website. Available: http://cancer.sanger.ac.uk. Accessed 2014 Jun 23.
- 15. Soung YH, Lee JW, Kim SY, Wang YP, Jo KH, et al. (2006) Somatic mutations of the ERBB4 kinase domain in human cancers. Int J Cancer 118: 1426–1429. [DOI] [PubMed] [Google Scholar]
- 16. Prickett TD, Agrawal NS, Wei X, Yates KE, Lin JC, et al. (2009) Analysis of the tyrosine kinome in melanoma reveals recurrent mutations in ERBB4. Nat Genet 41: 1127–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tvorogov D, Sundvall M, Kurppa K, Hollmén M, Repo S, et al. (2009) Somatic mutations of ErbB4: selective loss-of-function phenotype affecting signal transduction pathways in cancer. J Biol Chem 284: 5582–5591. [DOI] [PubMed] [Google Scholar]
- 18. Joensuu H, Kellokumpu-Lehtinen P-L, Bono P, Alanko T, Kataja V, et al. (2006) Adjuvant docetaxel or vinorelbine with or without trastuzumab for breast cancer. N Engl J Med 354: 809–820. [DOI] [PubMed] [Google Scholar]
- 19. Joensuu H, Bono P, Kataja V, Alanko T, Kokko R, et al. (2009) Fluorouracil, epirubicin, and cyclophosphamide with either docetaxel or vinorelbine, with or without trastuzumab, as adjuvant treatments of breast cancer: final results of the FinHer Trial. J Clin Oncol 27: 5685–5692. [DOI] [PubMed] [Google Scholar]
- 20. Joensuu H, Isola J, Lundin M, Salminen T, Holli K, et al. (2003) Amplification of erbB2 and erbB2 Expression Are Superior to Estrogen Receptor Status As Risk Factors for Distant Recurrence in pT1N0M0 Breast Cancer: A Nationwide Population-based Study. Clin Cancer Res 9: 923–930. [PubMed] [Google Scholar]
- 21. Sundvall M, Iljin K, Kilpinen S, Sara H, Kallioniemi O-P, et al. (2008) Role of ErbB4 in breast cancer. J Mammary Gland Biol Neoplasia 13: 259–268. [DOI] [PubMed] [Google Scholar]
- 22.Liu P, Kurppa K, Wildiers H, Reinvall I, Vandorpe T, et al.. (2012) Proteolytic Processing of ErbB4 in Breast Cancer. 7. [DOI] [PMC free article] [PubMed]
- 23. Muraoka-Cook RS, Sandahl MA, Strunk KE, Miraglia LC, Husted C, et al. (2009) ErbB4 splice variants Cyt1 and Cyt2 differ by 16 amino acids and exert opposing effects on the mammary epithelium in vivo. Mol Cell Biol 29: 4935–4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Loi S, Michiels S, Lambrechts D, Fumagalli D, Claes B, et al. (2013) Somatic mutation profiling and associations with prognosis and trastuzumab benefit in early breast cancer. J Natl Cancer Inst 105: 960–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bose R, Kavuri SM, Searleman AC, Shen W, Shen D, et al. (2013) Activating HER2 mutations in HER2 gene amplification negative breast cancer. Cancer Discov 3: 224–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Associations of immunohistochemical ErbB4 staining intensity with clinicopathological parameters.
(PDF)
Data Availability Statement
The authors confirm that, for approved reasons, there are access restrictions on the data underlying the findings. Relevant data are within the paper and its Supporting Information files. However, there is no permission from the relevant ethical committee to make the patient data of FinHer trial freely available. This data is available upon request pending the decisions of the relevant ethical committee and requests may be sent to the corresponding author.